Abstract
Asthma has a strong genetic component. The final disease phenotype results from complex interactions between environment and multiple genes of small-to-modest effects. We investigated whether the polymorphism in genes encoding inflammatory mediators and cytokines is important for solving the onset and progression of asthma. We investigated whether 31 single nucleotide polymorphisms (SNPs) in genes encoding cytokines or monokines (interleukin [IL]-5R, matrix metalloproteinase [MMP] 8, beta2 adrenergic receptor, cytotoxic T-lymphocyte–associated antigen 4, IL-3, C-reactive protein, cytochrome P450 (CYP) 2C9, CYP3A4, a disintegrin and metalloproteinase [ADAM] 33, cysteinyl leukotriene receptor [CysLTR] 1, CysLTR2, eosinophilic cationic protein, glucocorticoid receptor, and leukotriene A 4 hydrolase) are related to asthma development in 206 Japanese bronchial asthma patients and 127 healthy controls. Using multifactor dimensionality reduction (MDR), we identified rs17099451 in MMP8, using a single locus model, with a mean cross-validation of 87.0%. Using a two-locus model, combinations of MMP8 and rs44707 in ADAM33, and MMP8 and rs40401 in IL-3, were identified, with mean cross-validation consistencies reaching 45.0%. Of the SNPs selected by the MDR method, rs17099451 in MMP8 and rs40401 in IL-3 were regarded as the most significant results in a 2 × 2 dominant model analysis. The finding that an MMP8 allele was most strongly related to asthma development indicates that metalloproteinase function is crucial to the airflow limitation process involved in this disease.
Keywords: Asthma, Japanese population, multifactor dimensionality reduction (MDR), rs17099451 in MMP8, rs44707 in ADAM33, rs40401 in IL-3, single nucleotide polymorphism (SNP)
The main pathogenic mechanism of bronchial asthma involves a particular form of chronic inflammation, in which the increase in eosinophils, lymphocytes, and mast cells in the respiratory tract membrane and exfoliation of the respiratory epithelium cause bronchial hyperresponsiveness, followed by airway remodeling.1 These pathological changes result from the interactions between the inflammatory mediators and the various cytokines produced by the infiltrated cells or the structural cells.2
Asthma has a strong genetic component and complex interactions between the environment and multiple genes. It should therefore be possible to specify the risk and possibly prevent the development of the disease by investigating genetic polymorphisms; thus, the investigation of polymorphisms in genes encoding inflammatory mediators and cytokines is important. For example, genes encoding the high-affinity IgE receptor (Fc epsilon RI), an atopy-related gene3; eosinophil cationic protein, an inflammation-related gene4; interleukin (IL)-4 and IL-4 receptor α-chain5; IL-5 receptor α-subunit6 and IL-107 as Th2-related cytokines; and cysteinyl leukotriene receptor (CysLTR) 1–related gene8 have been reported to be involved in asthma. Moreover, an a disintegrin and metalloproteinase (ADAM) 33 mutant has been reported to be related to airway responsiveness and bronchial remodeling.9
In the study, we investigated the role played by 31 single nucleotide polymorphisms (SNPs), located in genes encoding 14 cytokines or monokines, in asthma development and the onset of bronchial asthma in Japanese bronchial asthma patients.
MATERIALS AND METHODS
Subjects
The study protocol was approved by the Ethics Committee of Fukuoka National Hospital; all participants received verbal and written study information before giving their informed consent (approval number 23-14).
We studied 206 Japanese subjects (102 men and 104 women) with newly diagnosed bronchial asthma at the Fukuoka National Hospital and 127 controls (46 men and 81 women) from the Japanese general population (Table 1).
Table 1.
Clinical profile of study subjects

*One sample is not included in the multifactor dimensionality reduction and association analysis because of missing genotype of rs1130864.
All asthma patients were diagnosed according to the Global Initiative for Asthma standard with asthma-related symptoms (recurrent cough, wheezing, or dyspnea), with bronchial hyperresponsiveness (PC20 to acetylcholine was <8 mg/mL), and with airway reversibility (forced expiratory volume at 1 second (FEV1) increased with 12% and 200 mL or more after salbutamol inhalation). None of these individuals were current smokers, had chest infections, had used antibiotic or oral prednisone in the previous 4 weeks, or had heart or lung disease other than asthma. Controls had no current disease or abnormal chest radiograms and were self-reported as healthy.
Selection of Polymorphisms
Using public databases, e.g., PubMed and Online Mendelian Inheritance in Man,10 we selected 14 genes that have been implicated in the onset of asthma or the asthma phenotype (Table 2), viz., IL-5R, matrix metalloproteinase (MMP) 8, β2 adrenergic receptor, cytotoxic T-lymphocyte–associated antigen 4, IL-3, C-reactive protein, cytochrome P450 (CYP)2C9, CYP3A4, ADAM33, CysLTR1, CysLTR2, eosinophil cationic protein, glucocorticoid receptor, and leukotriene A 4 hydrolase.
Table 2.
SNPs included in the MDR analysis
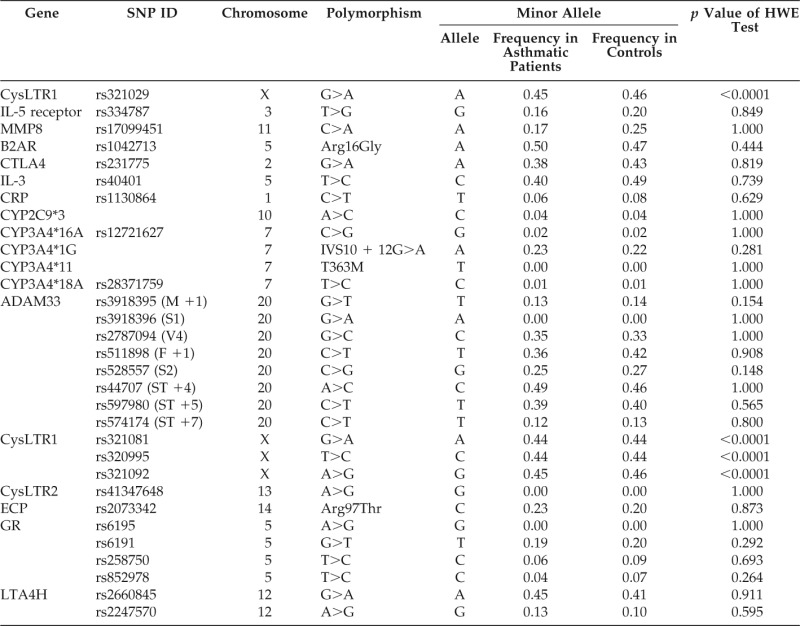
IL = interleukin; MMP8 = matrix metalloproteinase 8; CTLA4 = cytotoxic T-lymphocyte–associated antigen 4; CRP = C-reactive protein; ADAM33 = a disintegrin and metalloproteinase; CysLTR1 = cysteinyl leukotriene receptor 1; CysLTR2 = cysteinyl leukotriene receptor 2; ECP = eosinophil cationic protein; GR = glucocorticoid receptor; LTA4H = leukotriene A4 hydrolase; HWE = Hardy-Weinberg equilibrium; SNP = single nucleotide polymorphism; MDR = multifactor dimensionality reduction; B2AR = β2 adrenergic receptor.
Genotyping
Venous blood (7 mL) was collected from each subject into tubes containing 50 mmol/L of EDTA; from these samples, genomic DNA was isolated with a FlexiGene DNA Kit (Qiagen, Tokyo, Japan). Genotypes at 31 polymorphisms in the 14 genes were determined with a fluorescence or colorimetry-based allele-specific DNA-primer–probe assay system (Toyobo Gene Analysis, Tsuruga, Japan).
Polymorphic regions of each gene were determined using the intercalator-mediated fluorescence resonance energy transfer probe method (Toyobo Gene Analysis). In brief, the reaction was performed in a volume of 25 μL containing 20 ng of genomic DNA, enzyme reaction buffer, 2.5 mmol/L of MgCl2, 0.2 mmol/L of deoxynucleotide triphosphates, and 1.25 U of rTaq, containing anti-Taqplus High (Toyobo Gene Analysis). The amplification protocol was performed in a PE 9700 thermocycler (Applied Biosystems, Tokyo, Japan) and included an initial denaturation at 95°C for 5 minutes; 40 or 45 cycles of denaturation at 95°C for 30 seconds, annealing at 65°C for 30 seconds, and primer extension at 72°C for 30 seconds; and a final extension at 72°C for 2 minutes. The polymerase chain reaction products were then mixed with 5 mL of a solution containing 40 mmol/L of EDTA, 20 pmol of Texas Red-labeled probe, and 1/2000 SYBR Green I (Invitrogen, Carlsbad, CA); these samples were then transferred to an ABI Prism 7900 instrument (Applied Biosystems, Foster City, CA) for measurement of the melting point as temperatures were increased to 80°C over a period of 10 minutes.
To confirm the accuracy of genotyping by this method, we randomly selected 50 DNA samples and subjected them to polymerase chain reaction and restriction fragment length polymorphism analysis or to direct DNA sequencing. In each instance, the genotype determined by the allele-specific DNA primer–probe assay system was identical to that determined by the confirmatory methods.
Statistical Analysis
Hardy-Weinberg equilibrium was tested separately in the patients and controls using χ2-tests. To analyze interactions between SNPs and between SNPs and asthma development risk, the multifactor dimensionality reduction (MDR) approach was used.11–14 MDR was implemented with a cross-validation framework (typically, 10-fold, although any number of intervals, including 1-fold, can be used). In step 1 of the process, the data were divided into a training set (nine-tenths of the data) and a testing set (one-tenth of the data). Next, a set of “n” genetic and/or environmental factors was selected from the set of all possible factors (i.e., in the study n was 1, 2, 3, and 4 for SNPs); each of n individuals in the training set was grouped according to its state at each of the n factors (i.e., for two loci with three possible genotypes, there were 9 possible combinations; for three loci, there were 27, etc.). Then, each genotype combination was classified as high or low risk depending on the ratio of cases to controls with that genotype combination. If the ratio was <1 (i.e., there were less cases than controls), the genotype combination was classified as low risk. If the ratio was >1, the genotype combination was classified as high risk. This collection of high and low risk groups formed the MDR model for that combination of factors, and the fitness of that model was determined by classification error (the number of individuals incorrectly labeled by the model). To evaluate the predictive ability of a model, its prediction error was calculated based on the proportion of individuals in the testing set that were labeled incorrectly by that model. Each combination of variables specified by the user was evaluated in this fashion, and the entire procedure was performed 10 times, using one-tenth of the data as a testing set and nine-tenths as a training set. To evaluate MDR analysis, we evaluated balanced accuracy ([sensitivity + specificity])/2).15 The analysis was repeated 10 times after reordering individuals, and the averages of cross-validation consistency (CVC), training, and test accuracies are presented. To decrease random effects due to the arranging of data sets, the 10-fold cross-validation was repeated 10 times using different random numbers. To avoid the effect of linkage disequilibrium (LD) among SNPs in the same gene, the SNPs of which the pairwise LD coefficient r2 exceeded 0.8 were omitted from the analysis. For the SNPs that had been selected by MDR methods, we compared differences in the allele frequencies and genotype distribution of each polymorphism between the case and control subjects by using a 2 × 2 (allele) or 2 × 3 (encompassing dominant, codominant, and recessive genotype models) contingency χ2-test, with 1 or 2° of freedom. For the aforementioned analyses, the exact p values were calculated using Fisher's exact test. The empiric p values were also calculated by permutation tests (10,000 permutations) for the allelic analyses. All of the analyses other than MDR analyses were performed with PLINK Version 1.0.6 (MIT Broad Institute, Cambridge, MA).16
RESULTS
All SNPs were in Hardy-Weinberg equilibrium (p > 0.05) in both the study groups, except for the CysLTR1 polymorphism, because it was located on the X chromosome (Table 2).
MDR Analysis
Ten repetitions of 10-fold cross-validation resulted in a mean CVC of <30% when a model involving a combination of three or more loci could not detect a significant gene model. An SNP, rs17099451, in MMP8 was selected by the single gene locus model; the cross-validation mean for this model was 87.0%. The two-gene loci model selected two combinations, viz., MMP8 and ADAM33 SNP rs44707, as well as MMP8 and IL-3 SNP rs40401, of which the CVC means reached 45.0%
Using the aforementioned selected SNPs, the single gene locus model was found to be superior to the two-locus model, based on balanced accuracy and sensitivity evaluations, when used to predict bronchial asthma development in the testing data. The balanced accuracy means for these SNPs were 55.7 and 54.5%, and the sensitivity means were 68.7 and 56.2%, respectively. The mean specificity values were 42.7% for the single gene locus model and 52.8% for the two-gene locus model (Table 3).
Table 3.
Results from MDR analysis
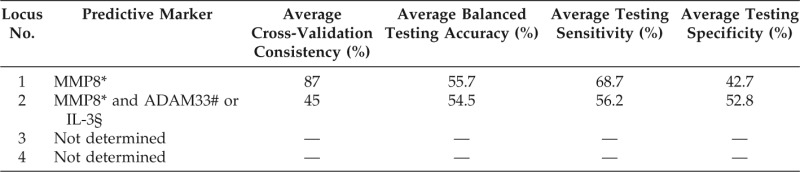
*MMP8 (rs17099451).
#ADAM33 (rs44707).
§IL-3 (rs40401).
MMP8 = matrix metalloproteinase 8; ADAM33 = a disintegrin and metalloproteinase 33; IL = interleukin.
Association Analysis of MMP8 and IL-3 Genotypes with Asthma
For the three SNPs selected by the MDR method, association analysis of the MMP8 rs17099451 and IL-3 rs40401 yielded the most significant result using the 2 × 2 dominant model (Table 4). The minor alleles were A and C in the MMP8 and IL-3 SNPs, respectively (Table 1); the risk alleles were the major alleles.
Table 4.
Association analysis of MMP8 and IL-3 genotypes with asthma
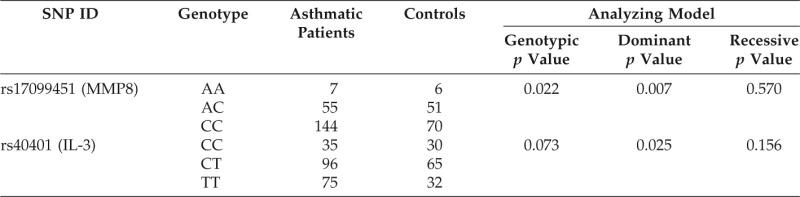
MMP8 = matrix metalloproteinase 8; IL = interleukin; SNP = single nucleotide polymorphism.
Association Analysis of MMP8 and IL-3 Alleles with Asthma
MMP8 rs17099451 was associated with an increased risk of asthma development (odds ratio [OR], 1.64; 95%CI, 1.07–2.51; p = 0.012; Table 5). The empiric p value calculated by permutation testing was 0.014. In the same manner, IL-3 rs40401 was associated with an increased risk of asthma development (OR, 1.44; 95%CI, 1.05–1.97; p = 0.025). The empiric p value for this association was 0.049.
Table 5.
Association analysis of MMP8 and IL-3 alleles with bronchial asthma
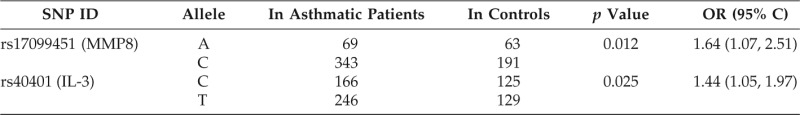
MMP8 = matrix metalloproteinase 8; IL = interleukin; SNP = single nucleotide polymorphism; OR = odds ratio.
Association Analysis of Each MMP8 and IL-3 Haplotype with Asthma
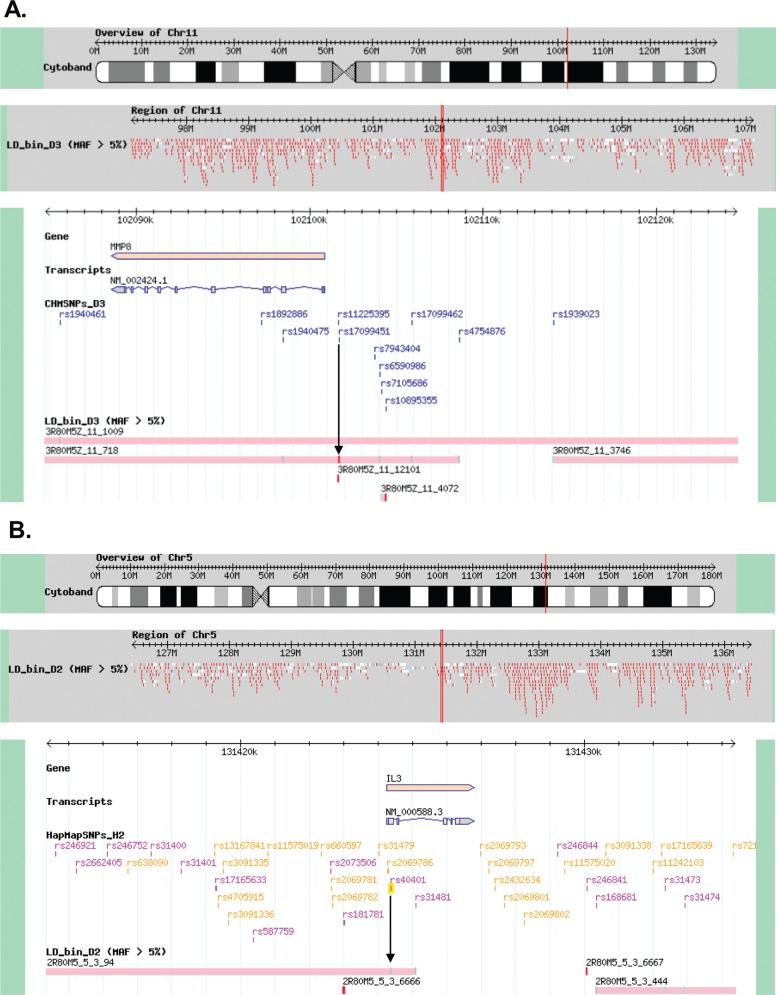
According to the gene database,17 MMP8 rs17099451 was the best tagSNP for the LD bin that included MMP8, and IL-3 rs40401 was located within an LD bin that contained IL-3 (Fig. 1). Each of these SNPs was used to construct a haplotype with neighboring markers with which it was in LD; in particular, IL-3 rs40401 was in complete LD with another IL-3 SNP, rs31480.
Figure 1.
According to the gene database, (A) matrix metalloproteinase (MMP) 8 rs17099451 was the best tag-single nucleotide polymorphism (SNP) for the linkage disequilibrium (LD) bin that included MMP8 (red). (B) Interleukin (IL)-3 rs40401 was located within an LD bin that contained IL-3.
Haplotype analysis revealed that distributions of the MMP8 rs11225395 and rs17099451 GC haplotype and that of the IL-3 TT haplotype were significantly higher in asthma patients in comparison with controls (OR, 1.52 and 1.43; Table 6).
Table 6.
Association analysis of MMP8 and IL-3 haplotypes with asthma

MMP8 = matrix metalloproteinase 8; IL = interleukin; SNP = single nucleotide polymorphism; OR = odds ratio.
DISCUSSION
In this study, we investigated whether any of 31 SNPs in genes encoding cytokines or monokines are associated with asthma development in a Japanese population. In a single locus model, we found that MMP8 rs17099451, and in a two-locus model, the combination of this rs17099451 in MMP8 SNP and rs44707 in ADAM33, as well as this MMP8 SNP and rs40401 IL-3, were selected as significant asthma development risk markers.
The single locus model of the MMP8 was consistently selected, and the balanced accuracy and sensitivity evaluation against the test data were superior in the single locus model compared with those in the two-locus model. In these analyses, MMP8 rs17099451 and IL-3 rs40401 had the most significant effect on asthma development according to the 2 × 2 dominant model. It is known that MMP8 is involved in the pathogenesis of some diseases, such as cancer, chronic obstructive pulmonary disease, and bronchiectasis.18–20 Until recently, MMP8 has been thought to be produced only by neutrophils, but it is now known to be expressed in some other cell types, including articular chondrocytes, fibroblasts, endothelial cells, corneal epithelial cells, smooth muscle cells, and cytotrophoblasts.21–25
Using an MMP8 knockout mouse model, Gueders et al. revealed an important role for MMP8 in the control of neutrophilic and eosinophilic infiltration during allergen-induced lung inflammation; they also showed that the anti-inflammatory effect of MMP8 is partly caused by regulation of inflammatory cell apoptosis.26 In asthma patients, a significant inverse correlation between bronchoalveolar lavage fluid MMP8 levels and FEV1 and bronchoalveolar lavage fluid–activated MMP8 forms and FEV1 was detected.27 Inhaled budesonide normalized the MMP8/tissue inhibitor of MMP-1 ratio in asthmatic children via up-regulation of tissue inhibitor of MMP-1 production and down-regulation of MMP8 production by airway macrophages.28 Overall, these data suggest that MMP8 and its activation have an important role in the airway destruction, healing, remodeling, and treatment response in asthma.
Our results suggested that the minor A allele of rs17099451 in MMP8 may have a suppressing effect on asthma development. Conversely, in a person carrying the major C allele of this SNP, the promoter activity and the ensuing gene expression may be altered, so that suppression of inflammation is lost, resulting in an increase in airway inflammation and remodeling and the development of asthma.
MMP8 forms part of a cluster of MMP genes, MMP1, -3, -7, -10, -12, -13, -20, etc., located on chromosome 11q22.3.29 In this study, we found that rs17099451 in MMP8 was in LD with another MMP8 SNP, rs11225395; it is also possible that this SNP is in LD with other SNPs that may contribute to asthma development.
Furthermore, IL-3 is a cytokine produced by active T cells, mast cells, and eosinophils, which stimulates the proliferation of hematopoietic progenitors.30 Moreover, IL-3 mRNA-expressing cells and IL-3 production are increased in asthma patients; this increase in IL-3 is independent of eosinophil number.31 Park et al.32 reported that one exonic SNP (in exon 1), IL3 + 79T > C (Ser27Pro) showed significant association with the risk of asthma and atopy. The Pro-allele had dominant, protective effects against the development of asthma in nonatopic subjects (p = -.002) and also showed significant association with the risk of atopy in normal control subjects (p = 0.007).32 We also found that rs40401 in IL-3 had the most significant effect in the 2 × 2 dominant model analysis, which suggests that the minor C allele may have a suppressing effect on asthma development.
Van Eerdewegh et al. reported that ADAM33 on chromosome 20p13 was linked to asthma and bronchial hyperresponsiveness, using a genomewide scan of 460 white families.9 The ADAM33 gene group appears to be related to proliferation, differentiation, or migration of fibroblasts, muscle fibroblasts, or smooth muscle cells, factors that are related to bronchial hyperresponsiveness or bronchial remodeling. However, these proteins do not appear to be involved in the respiratory epithelium, which is related to asthmatic immunologic or inflammatory components.9,33 Although a previous genomewide association study in a Japanese population did not implicate ADAM33 among the three new susceptibility loci for adult asthma,34 another study suggested the involvement of ADAM33 in the development of childhood asthma among the Japanese.35
Our finding that the MDR-based two-locus model analysis implicated rs44707 in ADAM33 as a significant factor also supports the relation of this gene to asthma development. Moreover, our finding that the combination of MMP8 and IL-3 polymorphisms and MMP8 and ADAM33 polymorphisms were also selected by two-locus model analysis may suggest that several gene polymorphisms may be involved in the onset of asthma.
CONCLUSIONS
The finding that an MMP8 allele was most strongly associated with asthma indicates that metalloproteinase function is very important for the airflow limitation process of the disease. Gene variation may be important to the differences in protein expression that allow the shedding of cytokines or cytokine receptors in this disease.
ACKNOWLEDGMENTS
The authors thank Miss Kojima and Miss Oda for their technical assistance in doing this study.
Footnotes
The authors have no conflicts of interest to declare pertaining to this article
REFERENCES
- 1. Global Initiative for Asthma (GINA) Global strategy for asthma management and prevention. NHLBI/WHO Report, NIH Publication 02-3659, Updated 2006, National Institutes of Health, National Heart, Lung and Blood Institute, Bethesda, MD: Available online at www.ginasthma.org; accessed September 3, 2013 [Google Scholar]
- 2. Haahtela T, Laitinen A, Laitinen LA. Using biopsies in the monitoring of inflammation in asthmatic patients. Allergy 48:65–69, 1993 [DOI] [PubMed] [Google Scholar]
- 3. Sandford AJ, Shirakawa T, Moffatt MF, et al. Localisation of atopy and beta subunit of high affinity IgE receptor (Fc epsilon RI) on chromosome 11q. Lancet 341:332–334, 1993 [DOI] [PubMed] [Google Scholar]
- 4. Munthe-Kaas MC, Gerritsen J, Carlsen KH, et al. Eosinophil cationic protein (ECP) polymorphisms and association with asthma, s-ECP levels and related phenotypes. Allergy 62:429–436, 2007 [DOI] [PubMed] [Google Scholar]
- 5. Beghé B, Barton S, Rorke S, et al. Polymorphisms in the interleukin-4 and interleukin-4 receptor alpha chain genes confer susceptibility to asthma and atopy in a Caucasian population. Clin Exp Allergy 33:1111–1117, 2003 [DOI] [PubMed] [Google Scholar]
- 6. Cheong HS, Kim LH, Park BL, et al. Association analysis of interleukin 5 receptor alpha subunit (IL5RA) polymorphisms and asthma. J Hum Genet 50:628–634, 2005 [DOI] [PubMed] [Google Scholar]
- 7. Hobbs K, Negri J, Klinnert M, et al. Interleukin-10 and transforming growth factor-beta promoter polymorphisms in allergies and asthma. Am J Respir Crit Care Med 158:1958–1962, 1998 [DOI] [PubMed] [Google Scholar]
- 8. Hao L, Sayers I, Cakebread JA, et al. The cysteinyl-leukotriene type 1 receptor polymorphism 927T/C is associated with atopy severity but not with asthma. Clin Exp Allergy 36:735–741, 2006 [DOI] [PubMed] [Google Scholar]
- 9. Van Eerdewegh P, Little RD, Dupuis J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature 418:426–430, 2002 [DOI] [PubMed] [Google Scholar]
- 10. Online Mendelian Inheritance in Man. Available online at www.ncbi.nlm.nih.gov/Omim/; accessed September 3, 2013
- 11. Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 19:376–382, 2003 [DOI] [PubMed] [Google Scholar]
- 12. Moore JH. Computational analysis of gene-gene interactions using multifactor dimensionality reduction. Expert Rev Mol Diagn 4:795–803, 2004 [DOI] [PubMed] [Google Scholar]
- 13. Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol 24:150–157, 2003 [DOI] [PubMed] [Google Scholar]
- 14. Ritchie MD, Hahn LW, Roodi N, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet 69:138–147, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Velez DR, White BC, Motsinger AA, et al. A balanced accuracy function for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet Epidemiol 31:306–315, 2007 [DOI] [PubMed] [Google Scholar]
- 16. Purcell S, Neale B, Todd-Brown K, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kyushu University Definitive Haplotype Database (D-HaploDB). Available online at www.orca.gem.kyushu-u.ac.jp; accessed September 3, 2013
- 18. Stadlmann S, Pollheimer J, Moser PL, et al. Cytokine-regulated expression of collagenase-2 (MMP8) is involved in the progression of ovarian cancer. Eur J Cancer 39:2499–2505, 2003 [DOI] [PubMed] [Google Scholar]
- 19. Vernooy JH, Lindeman JH, Jacobs JA, et al. Increased activity of matrix metalloproteinase-8 and matrix metalloproteinase-9 in induced sputum from patients with COPD. Chest 126:1802–1810, 2004 [DOI] [PubMed] [Google Scholar]
- 20. Zheng L, Lam WK, Tipoe GL, et al. Overexpression of matrix metalloproteinase-8 and -9 in bronchiectatic airways in vivo. Eur Respir J 20:170–176, 2002 [DOI] [PubMed] [Google Scholar]
- 21. Arner EC, Decicco CP, Cherney R, Tortorella MD. Cleavage of native cartilage aggrecan by neutrophil collagenase (MMP8) is distinct from endogenous cleavage by aggrecanase. J Biol Chem 272:9294–9299, 1997 [DOI] [PubMed] [Google Scholar]
- 22. Cole AA, Chubinskaya S, Schumacher B, et al. Chondrocyte matrix metalloproteinase-8. Human articular chondrocytes express neutrophil collagenase. J Biol Chem 271:11023–11026, 1996 [DOI] [PubMed] [Google Scholar]
- 23. Hanemaaijer R, Sorsa T, Konttinen YT, et al. Matrix metalloprotein-8 is expressed in rheumatoid synovial fibroblasts and endothelial cells. Regulation by tumor necrosis factor-alpha and doxycycline. J Biol Chem 272:31504–31509, 1997 [DOI] [PubMed] [Google Scholar]
- 24. Li DQ, Shang TY, Kim HS, et al. Regulated expression of collagenases MMP-1, -8 and -13 and stromelysins MMP-3, -10, and -11 by human corneal epithelial cells. Invest Ophthalmol Vis Sci 44:2928–2936, 2003 [DOI] [PubMed] [Google Scholar]
- 25. Arechavaleta-Velasco F, Marciano D, Diaz-Cueto L, Parry S. Matrix metalloproteinase-8 is expressed in human chorion during labor. Am J Obstet Gynecol 190:843–850, 2004 [DOI] [PubMed] [Google Scholar]
- 26. Gueders MM, Balbin M, Rocks N, et al. Matrix metalloproteinase-8 deficiency promotes granulocytic allergen-induced airway inflammation. J Immunol 175:2589–2597, 2005 [DOI] [PubMed] [Google Scholar]
- 27. Prikk K, Maisi P, Pirilä E, et al. Airway obstruction correlates with collagenase-2 (MMP8) expression and activation in bronchial asthma. Lab Invest 82:1535–1545, 2002 [DOI] [PubMed] [Google Scholar]
- 28. Obase Y, Rytilä P, Metso T, et al. Effects of inhaled corticosteroids on metalloproteinase-8 and tissue inhibitor of metalloproteinase-1 in the airways of asthmatic children. Int Arch Allergy Immunol 151:247–254, 2010 [DOI] [PubMed] [Google Scholar]
- 29. MMP8 matrix metallopeptidase 8 (neutrophil collagenase) [Homo sapiens], Gene ID 4317, updated on July 26, 2012. Available online at www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4317; accessed September 3, 2013
- 30. Arai KI, Lee F, Miyajima A, et al. Cytokines: Coordinators of immune and inflammatory responses. Annu Rev Biochem 59:783–836, 1990 [DOI] [PubMed] [Google Scholar]
- 31. Du T, Martin JG, Xu LJ, et al. IL-3 does not affect the allergic airway responses and leukotriene production after allergen challenge in rats. Eur Respir J 13:970–975, 1999 [DOI] [PubMed] [Google Scholar]
- 32. Park BL, Kim LH, Choi YH, et al. Interleukin 3 (IL-3) polymorphisms associated with decreased risk of asthma and atopy. J Hum Genet 49:517–527, 2004 [DOI] [PubMed] [Google Scholar]
- 33. Umland SP, Garlisi CG, Shah H, et al. Human ADAM33 messenger RNA expression profile and post-transcriptional regulation. Am J Respir Cell Mol Biol 29:571–582, 2003 [DOI] [PubMed] [Google Scholar]
- 34. Hirota T, Takahashi A, Kubo M, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet 31:43:893–896, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noguchi E, Ohtsuki Y, Tokunaga K, et al. ADAM33 polymorphisms are associated with asthma susceptibility in a Japanese population. Clin Exp Allergy 36:602–608, 2006 [DOI] [PubMed] [Google Scholar]