SUMMARY
Residues located outside of the active site of cytochromes P450 2B have exhibited importance in ligand binding, structural stability, and drug metabolism. However, contributions of non-active site residues to the plasticity of these enzymes are not known. Thus, a systematic investigation was undertaken of unique residue-residue interactions found in crystal structures of P450 2B4 in complex with 4-(4-chlorophenyl)imidazole (4-CPI), a closed conformation, or in complex with bifonazole, an expanded conformation. Nineteen mutants distributed over eleven sites were constructed, expressed in E. coli, and purified. Most mutants showed significantly decreased expression, especially in the case of interactions found in the 4-CPI structure. Six mutants (H172A, H172F, H172Q, L437A, E474D, and E474Q) were chosen for detailed functional analysis. Among these, the Ks of H172F for bifonazole was ~20-times higher than wild type 2B4, and the Ks of L437A for 4-CPI was ~50-times higher than wild type, leading to significantly altered inhibitor selectivity. Enzyme function was tested with the substrates 7-ethoxy-4-(trifluoromethyl)coumarin (7-EFC), 7-methoxy-4-(trifluoromethyl)coumarin (7-MFC), and 7-benzyloxyresorufin (7-BR). H172F was inactive with all three substrates, and L437A did not turn over 7-BR. Furthermore, H172A, H172Q, E474D and E474Q showed large changes in kcat/KM for each of the three substrates, in some cases up to 50-fold. Concurrent molecular dynamics simulations yield distances between some of the residues in these putative interaction pairs that are not consistent with contact. The results indicate that small changes in the protein scaffold lead to large differences in solution behavior and enzyme function.
Keywords: cytochrome P450, CYP2B, polymorphism, site-directed mutagenesis
INTRODUCTION
Cytochrome P450 (P450)-dependent monooxygenases (EC 1.14.14.1) are members of a ubiquitous superfamily of heme containing proteins and are responsible for oxidation of a broad range of substrates in the biogenesis of sterols and hormones and metabolism of xenobiotic compounds [1, 2]. Some mammalian P450s can accept a wide variety of hydrophobic substrates of differing shapes and sizes and render them more hydrophilic for excretion or subsequent conjugation. Additionally, P450s mediate the conversion of prodrugs to the respective bioactive compounds [3-5].
While P450s accept a broad range of substrates, the protein fold of P450s, consisting of a large, primarily alpha helical single domain is well conserved across families [6, 7], and a high degree of flexibility in these enzymes has previously been observed [7-9]. P450s form compact structures around small ligands or ligand-free active sites, as seen in P450 2B4 complexed with 4-(4-chlorophenyl)imidazole (4-CPI) [10], the P450 2A subfamily [11, 12], and P450 2C5 and P450 2C9 [13, 14]. Some P450s also appear to be able to accommodate ligands of greater volume, as demonstrated with P450 2B4 with bifonazole [15] or P450 3A4 with erythromycin, ketoconazole or ritonavir [16].
Much of this conformational plasticity is exhibited in the P450 2B subfamily of enzymes. These proteins show low catalytic conservation across species, providing a good model system to examine structure-function relationships in P450s [8]. Studies of the P450 2B subfamily have yielded considerable biochemical and biophysical insight into substrate binding, protein-protein interactions, and the catalytic mechanisms of microsomal monooxygenases. Crystal structures of an engineered P450 2B4 show a remarkable amount of plasticity is possible while retaining enzyme function [8, 17]. Experiments utilizing hydrogen/deuterium exchange coupled to mass spectrometry (DXMS) reinforced the view concerning plasticity of P450 2B4 [18].
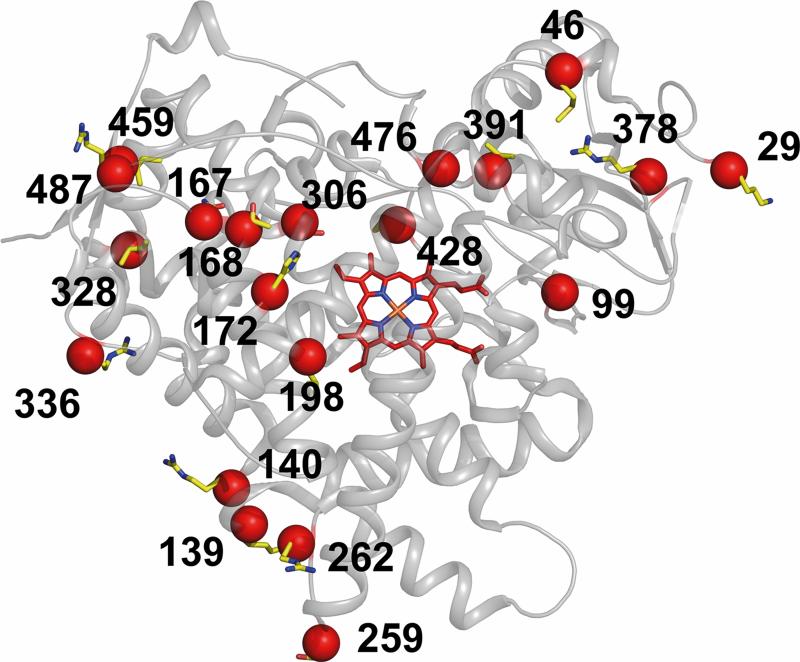
One member of this subfamily, P450 2B6, plays an important role in human drug metabolism [19, 20] and is highly polymorphic [5, 19, 21, 22]. Furthermore, some of these single nucleotide polymorphisms (SNPs) (Fig. 1) have been linked to differential metabolism by P450 2B6 of a variety of drugs [23-25]. Most of the amino acid changes are far from the active site of the enzyme [23]. Previous studies of subfamily 2B enzymes demonstrated that mutations located outside of the active site of P450 2B enzymes significantly alter enzyme function [17, 26-29].
Figure 1.
Ribbon diagram showing the location of known SNPs (yellow sphere and sticks) in 2B6. The heme is shown as red sticks. All the known coding sequence variants of 2B6 are located outside of the active site.
Given the demonstrated importance of non-active site residues in P450 enzymes, an investigation of the functional and structural role of non-active site residues possibly involved in plasticity through residue-residue interactions was undertaken. Residues of interest were selected from interactions unique to either the structure of 2B4 complexed with 4-CPI or with bifonazole [30]. Site-directed mutagenesis was used to interrupt four interactions from the 4-CPI structure and five interactions from the bifonazole complex. One of the interactions in the bifonazole complex involved a site known to be polymorphic in P450 2B6, H172, so another site known to have large effects on ligand binding and catalysis was also investigated, R262. Mutants were characterized by expression level, thermal stability, thermal inactivation, H2O2-mediated heme depletion, binding affinity for 4-CPI and bifonazole, steady-state kinetics with a variety of substrates, enzyme inhibition in the presence of 4-CPI or bifonazole, and molecular dynamics simulation.
RESULTS
Identification of interactions of interest
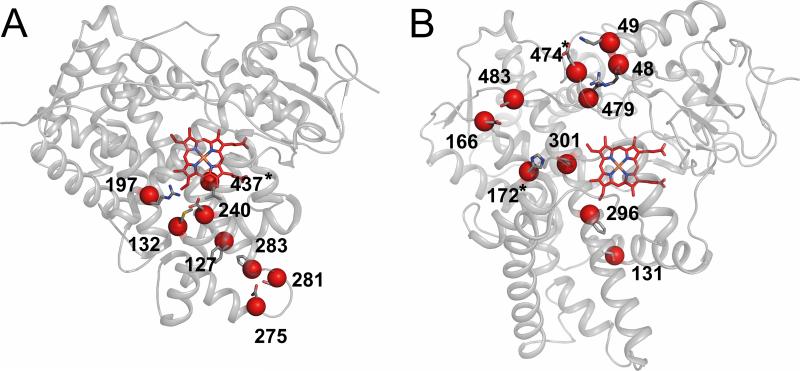
Residue-residue interactions involving two or more amino acid side chains within 4 Å of each other were analyzed in structures of P450 2B4 in the open ligand-free conformation (PDB ID: 1PO5), in complex with 4-CPI (PDB ID: 1SUO), and in complex with bifonazole (PDB ID: 2BDM). Interactions were grouped as conserved (found in all three structures) and alternate (unique to the 4-CPI-bound or bifonazole-bound structure) [30]. Eight alternate interactions are unique to the 4-CPI complex, and seven are unique to the bifonazole complex (Table 1). To simplify interpretation of the effects of mutants, interactions consisting of two amino acid residues were chosen as initial candidates for interruption (Fig. 2). Interestingly, one of the observed interactions (H172-E301) occurs at a SNP (Q172H) in the closely related P450 2B6. Another SNP in P450 2B6 (K262R) involves a residue hypothesized to participate in a hydrogen bonding network in the complex of this enzyme with the small molecule 4-CPI [31]. P450 2B6 Q172H shows marked differences in metabolism of cancer and HIV chemotherapeutics, and P450 2B6 K262R shows altered in vitro binding of clopidogrel, itraconazole, raloxifine, and sertraline [19, 25]. The corresponding mutations (H172Q and R262K) were therefore introduced into P450 2B4.
Table 1.
Unique residue-residue interactions in crystal structures of 2B4.
| 4-CPI complex1 | Bifonazole complex2 | ||
|---|---|---|---|
| Interaction3 | Mutants | Interaction3 | Mutants |
| F115-F108-L290 | None | *R48-N479 | N479Q, N479R |
| R120-E282-N287 | None | *K49-E474 | E474D, E474Q |
| *F127-F283 | F127A, F283A | *T131-F296 | T131A, T131S |
| *M132-L437 | M132A, L437A | *D166-S483 | S483A, S483T |
| F184-F296-F202-L241 | None | *H172-E301 | H127A, H172F |
| *R197-E240 | E240A, E240D | Y268-I289-Q286 | None |
| Q204-S205-Q233-N237 | None | E403-T404-N409-H412 | None |
| *D275-S281 | S281A, S281T | ||
Interactions selected for mutational analysis
Crystal structure of 2B4 in complex with 4-CPI (1SUO)
Crystal structure of 2B4 in complex with bifonazole (2BDM)
Residues within 4 Å of each other in the crystal structure.
Figure 2.
Location of interactions interrupted by mutation. A) Interactions in the 2B4-4-CPI structure (1SUO). B) Interactions in the 2B4-bifonazole structure (2BDM). Residues in each interaction are shown as sticks with a red sphere for the alpha carbon. Positions for focused analysis from SNP sites (H172A/F/Q), the 2B4-4-CPI structure (L437A), and the 2B4-bifonazole structure (E474D/Q) are marked with an asterisk (*).
Expression and purification of 2B4 mutants
Mutants were first expressed and purified as previously described [17]. Each mutation resulted in lower expression levels than wild type enzyme (Table 2). Seven mutants (T131S, E240Q, S281T, F283A, L437A, E474Q, and S483A) showed greater than 50% of wild type protein expression levels, and seven more mutants (T131A, H172A, H172F, H172Q, E474D, N479Q, and S483T) expressed at 20-50% of wild type. E240D, S281A, and N479R showed expression levels less than 20% of wild type. F127A and M132A did not show measurable expression levels as measured by CO difference spectra. For those mutants that expressed, P420 content was measured as described in the Materials and Methods section, and most had levels between 5% less than and 10% more than wild type P420 values, which were ~9%. However, R262K expressed as 100% P420.
Table 2.
Expression levels of 2B4 mutants
| 4-CPI complex | Bifonazole complex | ||
|---|---|---|---|
| Mutant | Expression Level (nmoles P450/L) | Mutant | Expression Level (nmoles P450/L) |
| Wild type | 530 ± 20 | Wild type | 522 ± 39 |
| F127A | N.D. | T131A | 164 ± 15 |
| M132A | N.D. | T131S | 277 ± 112 |
| E240D | 95 ± 15 | #H172A | 156 ± 51 |
| E240Q | 391 ± 65 | #H172F | 220 ± 39 |
| S281A | 15 ± 5 | #H172Q | 154 ± 20 |
| S281T | 351 ± 12 | E474D | 184 ± 35 |
| F283A | 420 ± 22 | E474Q | 324 ± 70 |
| L437A | 264 ± 23 | N479Q | 165 ± 70 |
| N479R | 107 ± 20 | ||
| S483A | 275 ± 15 | ||
| #R262K | N.D. | S483T | 236 ± 30 |
Location of genetic polymorphism in 2B6
Results are the mean ± standard deviation (n=3).
N.D. – not detectable
Enzyme stability
Mutations in P450s resulting in lower expression levels generally indicate less stable enzymes. Therefore, the thermal stability of the 14 mutants that expressed at greater than 20% of wild type levels was assessed with the exception of S483A, which was 100% P420 after purification and not studied further. The TM for each of the other mutants except S483T was within ± 3 °C of P450 2B4 wild type (Fig. 3A). In addition, thermal inactivation showed greater deviation from wild type values, but no clear trends in effects of mutation at a specific residue arose (Fig. 3B).
Figure 3.
Stability of 2B4 wild type and mutant proteins. A) Thermal melting temperature. B) Thermal inactivation rate constant. C) Hydrogen peroxide mediated heme depletion rate constant. Black bars represent P450 inactivation; gray bars represent P420 inactivation.
In the interests of focusing efforts, further characterization of mutants was limited to: H172A/F/Q, representative of a SNP in CYP 2B6; L437A, representing the 4-CPI structure mutants; and E474D/Q, representing the bifonazole structure mutants. Hydrogen peroxide mediated heme destruction was monitored as a measure of water access to the active site pocket. Each mutant showed at least a 2-fold decrease in the P450 kinact, and most mutants at least a small decrease in P420 kinact (Fig. 3C). Interestingly, substituting phenylalanine at residue 172 increased the P420 kinact by 50%.
Ligand interaction
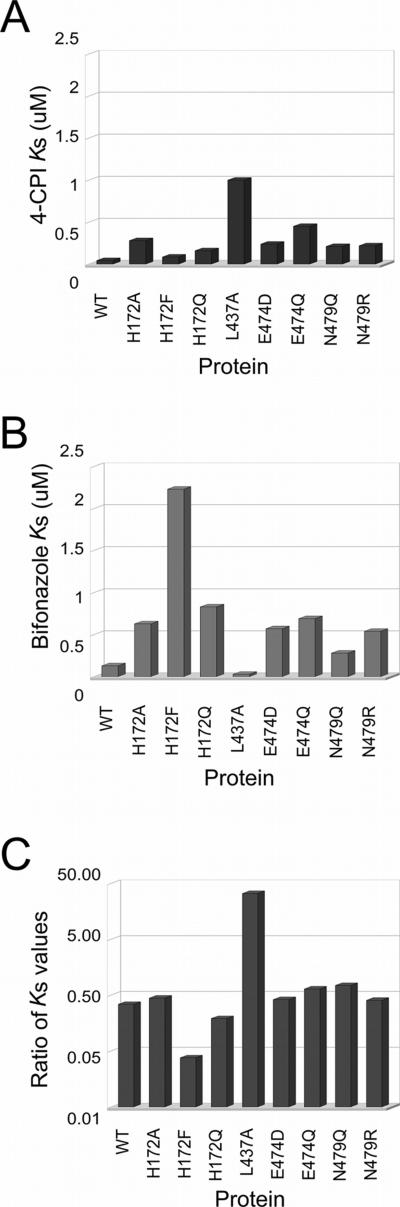
To investigate effects of these mutations on ligand binding, catalysis, and inhibition, we used 7-EFC, 7-MFC, and 7-BR as substrates and 4-CPI and bifonazole as inhibitors. Since mutants were created to interrupt interactions found either in the 2B4-4-CPI complex or the 2B4-bifonazole complex, spectral binding titrations were performed using these two small molecules. The spectral dissociation constant (Ks) values for both imidazole inhibitors generally increase with the introduction of a mutation (Fig. 4A-B). Comparing the ratio of Ks for 4-CPI to Ks for bifonazole for each mutant shows a large change for H172F and L437A (Fig. 4C). Furthermore, a difference in relative specificity for these two imidazole inhibitors was created relative to wild type 2B4, with H172F showing increased selectivity for 4-CPI and L437A increased selectivity for bifonazole (Fig. 4C).
Figure 4.
Spectral binding of imidazole ligands to 2B4 wild type and mutant proteins. A) Ks for 4-CPI. B) Ks for bifonazole. C) Ratio of constants for 4-CPI to bifonazole.
While most of the mutations caused less than 3-fold changes in Ks for the two imidazole molecules, there were greater effects on enzyme function. The substrates 7-EFC, 7-MFC and 7-BR were turned over by many of the mutants (Table 3). However, H172F was inactive with all three compounds, and L437A showed no activity with 7-BR. For 7-EFC, changes in kcat/KM for H172A and L437A appear to be due to a 2- to 3-fold lower kcat. H172Q and E474Q showed a ~5- to ~7-fold lower kcat, and E474D showed a >20-fold decrease in kcat. These three mutants also showed a 2- to 3-fold increase in KM. As with 7-EFC as a substrate, changes in kcat/KM for these mutants using 7-MFC as a substrate seem to be mainly due to changes in kcat. With the exception of the inactive H172F, each mutant showed <2-fold change in KM for 7-MFC. H172A and H172Q showed ~2-fold lower kcat and L437A, E474D and E474Q a 4- to 5-fold decrease. Turnover of 7-BR is affected more by these mutations, as the kcat/S50 values change 6- to 40-fold, mainly due to changes in kcat. The mutants that turned over 7-BR exhibited a 2- to 3-fold increase in S50. E474D and E474Q displayed ~2.5- and ~4.5-fold changes in kcat, while H172A and H172Q showed larger effects on kcat, ~19- and ~15-fold changes, respectively.
Table 3.
Steady-state kinetics of 2B4 mutants
| Protein | 7-EFC | 7-MFC | 7-BR | ||||||
|---|---|---|---|---|---|---|---|---|---|
| K M 1 | k cat 2 | kcat/KM | K M 1 | k cat 2 | kcat/KM | S 50 1 | k cat 2 | kcat/S50 | |
| WT | 25 ± 3 | 9.6 ± 0.4 | 0.36 | 132 ± 6 | 11.3 ± 0.2 | 0.08 | 2.3 ± 0.5 | 7.6 ± 0.3 | 3.3 |
| H172A | 26 ± 4 | 4.2 ± 0.3 | 0.15 | 126 ± 11 | 5.2 ± 0.6 | 0.04 | 5.0 ± 0.8 | 0.4 ± 0.1 | 0.08 |
| H172F | N.D. | 0 | 0 | N.D. | 0 | 0 | N.D. | 0 | 0 |
| H172Q | 48 ± 6 | 1.7 ± 0.3 | 0.04 | 115 ± 15 | 7.4 ± 0.5 | 0.06 | 8.2 ± 1.0 | 0.5 ± 0.1 | 0.06 |
| L437A | 25 ± 4 | 2.7 ± 0.2 | 0.12 | 85 ± 8 | 1.8 ± 0.2 | 0.02 | N.D. | 0 | 0 |
| E474D | 77 ± 10 | 0.4 ± 0.1 | 0.01 | 185 ± 14 | 3.8 ± 0.2 | 0.02 | 6.2 ± 0.7 | 3.1 ± 0.2 | 0.5 |
| E474Q | 64 ± 8 | 1.2 ± 0.2 | 0.02 | 178 ± 17 | 2.4 ± 0.3 | 0.01 | 4.8 ± 0.5 | 1.7 ± 0.3 | 0.35 |
Standard errors for fit to the respective equations are shown. Results are representative of at least two independent determinations.
Km and S50 are measured in μM.
kcat is measured in min−1.
N.D., not detectable
Inhibition of 7-EFC O-deethylation was also affected by mutation of non-active site residues (Table 4). Alteration of the non-active site residues generally increases the IC50 for 4-CPI and causes small changes in the IC50 of bifonazole. The ratio of the two values also increases, since the 4-CPI IC50 generally increases more than the bifonazole IC50 changes. L437A is unique in that this ratio changes drastically and with a significant decrease in the bifonazole IC50. This single non-active site mutant thus shows a 100-fold decrease in selectivity of 4-CPI:bifonazole.
Table 4.
Inhibition of non-active site mutants by imidazole inhibitors.
| Protein | 4-CPI IC50 (μM) | Bifonazole IC50 (μM) | Ratio 4-CPI IC50/Bifonazole IC50 |
|---|---|---|---|
| WT | 0.04 | 0.90 | 0.04 |
| H172A | 0.31 | 0.58 | 0.53 |
| H172Q | 0.22 | 0.76 | 0.29 |
| L437A | 0.97 | 0.05 | 3.88 |
| E474D | 0.24 | 0.62 | 0.39 |
| E474Q | 0.50 | 0.81 | 0.62 |
All experiments were completed using 50 μM 7-EFC as substrate.
Results are the mean of at least two independent determinations.
Molecular dynamics simulation
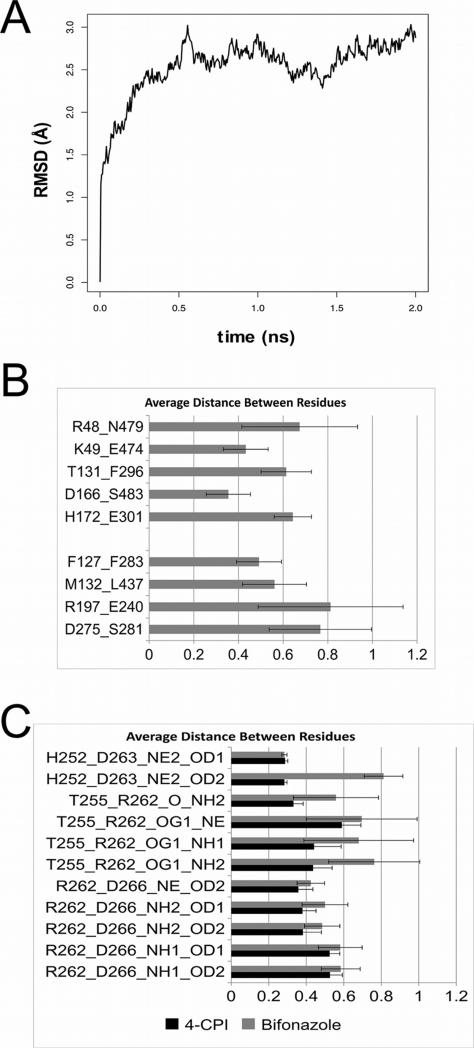
With these findings, a molecular dynamics simulation was employed to explore whether the interactions taken from crystal structures actually occur in a dynamic setting. Using the 4-CPI-bound 2B4 structure (PDB ID: 1PO5) or the bifonazole bound 2B4 structure (PDB ID: 2BDM) as a starting structure for a 2 ns simulation using GROMACS, the backbone of 2B4 appears to breathe in a similar manner to previous simulations (Fig. 5A) [18]. In the interest of examining interaction partners, the closest heteroatoms in each pair from the respective crystal structures were used to measure the distance between residues during the course of the simulation (Fig. 5B and Supplementary Fig. S1). With a 0.6 nm cutoff, four interactions are close enough to persist for the entirety of the simulation (K49-E474, D166-S483 from the 2BDM and F127-F283, M132-L437 from 1SUO), and the remaining interactions are transient to varying degrees (R48-N479, T131-F296, H172-E301 in 2BDM and R197-E240, D275-S281 in 1SUO). Most interaction pairs are within a fairly close range of distances from each other; however R48-N479, R197-E240, and D275-S281 have wide swings in distance between the interaction partners.
Figure 5.
Molecular dynamics simulation of 2B4 wild type. A) Representative change in backbone RMSD over 2 ns simulation for wild type protein using the 4-CPI structure (1PO5) as the starting structure; this change is representative of changes found in simulations of mutant proteins or in the simulation using the bifonazole structure (2BDM) as the starting structure. B) Distance between closest atoms for each interaction found in the crystal structures of 2B4 complexed with bifonazole or 4-CPI for mutants interrupting interactions from the respective structure. The five interactions from the bifonazole structure are listed at the top, and the four interactions from the 4-CPI structure are on the bottom. C) Distance between residues found in the proposed hydrogen bonding network in cytochromes 2B from Gay et al. [31].
The interaction of residues in the proposed hydrogen bonding network around R262 were also examined. The inactive R262K mutant does not exhibit a change in the distance between H252 and D263 (Fig. 5C). However, the T255-R262 and R262-D266 interactions show increased distance between the interaction partner and residue 262 when the latter is Lys (not shown). This likely leads to a disruption of any stabilizing benefit of the hydrogen bonding network provided by R262.
X-ray crystal structure
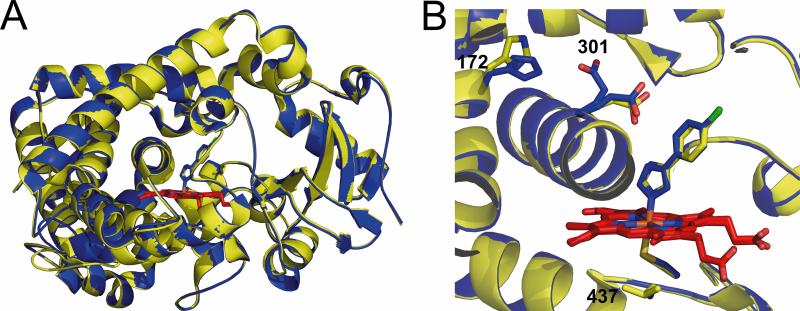
Because of the marked changes in sensitivity to inhibition by 4-CPI vs. bifonazole, we solved an X-ray crystal structure of the 2B4 L437A mutant in complex with 4-CPI (PDB ID: 3TK3) (Table 5). Comparison with the X-ray crystal structure of wild type 2B4 in complex with 4-CPI (PDB ID: 1SUO) yields an RMSD of 0.26 Å, showing the structures are virtually superimposable (Fig. 6A). The orientation of active site residues is maintained in the mutant; however, the density for E301 indicates split occupancy, where one orientation points into the active site to hydrogen-bond with the azole nitrogen of 4-CPI, while the other orientation flips out to interact with H172 (Fig. 6B). The Cβ carbons of residue 437 for the wild type and L437A proteins in complex with 4-CPI are in the same orientation.
Table 5.
X-ray Data Collection & Refinement Statistics
| Crystal Data | |
| Space Group | P3 |
| Unit Cell | |
| a, b, c (Å) | 232.9, 232.9, 57.0 |
| α, β, γ (°) | 90, 90, 120 |
| Molecules per AU | 4 |
| Data Collection | |
| X-ray source | SSRL BL 11-1 |
| Wavelength (Å) | 0.98 |
| Resolution Range (Å) | 76.2-2.80 |
| Total Observations | 354,184 |
| Unique Observations (F>0) | 81,306 |
| Completeness (%)a | 99.4 (99.4) |
| Redundancya | 4.4 (4.2) |
| I/σa | 5.1 (1.6) |
| Rmerge (%)a,b | 20.1 (85.7) |
| Refinement Statistics | |
| R-factor (%)c | 19.30 |
| Rfree (%)c | 24.08 |
| RMS bond lengths (Å) | 0.016 |
| RMS bond angles (°) | 1.356 |
| Number of Atoms | |
| Proteind | 14,727 (52.6) |
| Hemed | 172 (41.4) |
| 4-CPI | 48 (50.4) |
| Waterd | 290 (41.9) |
Values for the highest resolution shell are in parentheses.
Rmerge = [ΣhΣi|Ih-Ihi|/ΣhΣiIhi] where Ih is the mean of Ihi observations of reflection h.
R-factor & Rfree = Σ∥Fobs|-|Fcalc∥/Σ|Fobs| × 100 for 95% of the recorded data (R-factor) and 5% of data (Rfree).
Average B-factors (Å2) are in parentheses.
Figure 6.
Overlay of wild type and mutant 2B4 in complex with 4-CPI. A) Overlay of Cα backbone of wild type 2B4 (yellow, PDB ID: 1SUO) and the L437A mutant (blue, PDB ID: 3TK3). B) The active site and heme support of wild type 2B4 and the L437A mutant.
DISCUSSION
Adapting mammalian P450s for biomedical and biotechnological applications via improved activity and/or stability has been of considerable recent interest [32-34]. Previous studies focused on active site and substrate access channel residues [8, 35-37]. However, the highly plastic nature of P450s suggests that non-active site residues stabilize different conformations through residue-residue contacts. Recent studies of the P450 2B enzymes have demonstrated the importance of non-active site residues in enzyme inhibition, catalysis, stability, and expression [17, 26-29, 38]. Furthermore, most of the known SNPs in human 2B6 are distal from the active site [23]. In light of this information, this study focused on the role of non-active site residues in protein-ligand interactions involving P450 2B4. Introduction of mutants outside of the enzyme active site led in general to small changes in binding affinity but much larger affects on substrate catalysis and enzyme inhibition.
Comparison of interactions of two residues from each of three crystal structures of 2B4 identified four unique interactions in the 4-CPI-bound structure (1SUO) and five in the bifonazole-bound structure (2BDM). Not surprisingly, interrupting these interactions by site directed mutagenesis changed expression levels of the protein to varying degrees. The mutants that did express did not have significantly altered thermal stability or protection from H2O2 heme degradation. In general, interrupting interactions found in the bifonazole-bound structure produced more mutants that yielded significant levels of P450 than interruptions of interactions found in the 4-CPI-bound structure. These results suggest that some of the interactions seen exclusively in the 4-CPI-bound complex also exist in solution in the absence of ligand and may be important for enzyme stability.
Conversely, changes in protein-ligand interactions were more pronounced. Small perturbations in binding affinities of the 4-CPI and bifonazole were seen in most of the six mutants tested. Interestingly, two mutants (H172F and L437A) had large changes in binding affinity for one or both inhibitors. H172F showed increased affinity for 4-CPI, while L437A showed increased affinity for bifonazole. Both mutants had greater than 9-fold changes in the ratio of 4-CPI Ks to bifonazole Ks. While the remaining mutants showed less than 3-fold changes in this ratio, larger changes were seen in steady-state kinetic assays using 7-EFC, 7-MFC, and 7-BR. The two mutants with greatest change in imidazole binding were inactive with one (L437A) or all (H172F) substrates tested. Moreover, the changes in imidazole binding affinity of E474D and E474Q (~5-fold) are accompanied by changes in enzyme activity (>30-fold). While small changes in binding affinity for either 4-CPI or bifonazole were seen for most of the mutants (<2-fold), much greater changes were seen in IC50 values for each of the compounds.
While little structural rearrangement is seen in the crystal structure, replacement of Leu 437 with Ala may create a cavity allowing for water access to or change in redox potential of the heme leading to a decreased kcat; this cavity could also allow for greater porphyrin mobility and resultant affects on kinetics of imidazole binding. Interestingly, profound effects on ligand interaction were recently observed upon mutation of the P450 BM3 residue just C-terminal from the heme cysteine [39], which corresponds to residue 437 in 2B4. Specifically, the BM3 I401P mutant is highly active towards non-natural substrates, and the crystal structure of the ligand-free mutant resembles substrate-bound forms of the wild type enzyme.
Interaction behavior of the residue pairs was accessed using MD simulations. A small number of interaction pairs remained within 6 Å of each other for the entire simulation: F127-F283, D166-S483, and K49-E474. Except for K49-E474, these interactions involve residues in the conserved regions of the protein, which does not show large conformational changes between crystal structures (residues 58-100, 141-176, 189-202, 299-473, and 481-491) [8], the I-helix or very close to the heme cysteine. Portions of the protein showing large conformational changes among crystal structures were previously termed plastic regions [8]. Interestingly, the K49-E474 interaction from the bifonazole structure involves residues in plastic region 1 (residues 39-57) and plastic region 5 (residues 474-480). Another subset of residue pairs (T131-F296, M132-L437, and H172-E301) was transiently within 6 Å of each other in a narrow range of distances over the course of the simulation. These residues are in the conserved region, plastic region 2 (residues 101-140) and plastic region 4 (residues 203-298). The remaining interactions, R48-N479, R197-E240, and D275-S281, have average distances greater than 6 Å and have a wide variance, and they involve residues in the conserved region, plastic region 1, plastic region 4, and plastic region 5. Interestingly, the interactions in the 4-CPI bound structure involving residues in plastic region 4 are highly transient (R197-E240 and D275-S281). R197-E240 involves residues in the F-G cassette of 2B4, which provides variable volume to the active site through coordinated movement around the E-F and G-H loops [40]. D275-S281 is between residues in the H-I loop or transiently in the I-helix. These regions and plastic region 2, consisting of the B`-C loop and the C-helix, show the greatest amount of movement between various crystal structures of the enzyme [8, 40] and have the greatest difference in solvent protection in DXMS experiments. Furthermore, the S281 and F283 are in the H-I loop or the N-terminal end of the I-helix; previous work with 2B1 showed that mutating residues in these regions has detrimental consequences for expression, stability, and kinetic fidelity [41].
In conclusion, mutations to certain non-active site residues changed the relative selectivity of 4-CPI and bifonazole and, in some cases, caused profound functional changes. Interestingly, changes to active site residues produced much smaller changes, generally ~2-fold or less, in previous work [42]. A likely mechanism is disruption of residue-residue interactions that stabilize an active conformation of the enzyme. A similar explanation may account for the altered function of several genetic variants of human P450 2B6 [31, 43]. Moreover, future studies involving mutations based on crystal structures of cytochromes P450 should utilize molecular dynamics simulations to assess the likelihood in solution of residue-residue interactions inferred from the structure.
MATERIALS AND METHODS
Materials
7-hydroxy-4-(trifluoromethyl)coumarin (7-HFC), 7-methoxy-4-(trifluoromethyl)coumarin (7-MFC), and 7-ethoxy-4-(trifluoromethyl)coumarin (7-EFC) were purchased from Invitrogen (Carlsbad, CA). Sodium hydrosulfite, β-mercaptoethanol (βME), phenylmethylsulphonyl fluoride (PMFS), and NADPH were obtained from Sigma (St. Louis, MO). Recombinant NADPH-cytochrome P450 reductase (CPR) and cytochrome b5 (b5) from rat liver were prepared as described previously [44]. Oligonucleotide primers for PCR were obtained from Sigma (St. Louis, MO). Phusion High-Fidelity DNA Polymerase was purchased from New England Biolabs (Ipswich, MA). Nickel-nitrilotriacetic acid (Ni2+-NTA) affinity resin was purchased from Qiagen (Valencia, CA). All other chemicals were of the highest grade available and were used without further purification.
Cytochrome P450 and P420 quantification
During protein purification, the P450 concentration of the supernatant was measured using the reduced CO difference spectrum and a molar extinction coefficient of 91 mM−1 cm−1 [45, 46]. For crystallization trials, the P450 concentration was measured using the oxidized P450 spectrum and a molar extinction coefficient of 106 mM−1 cm−1. For P420 content, thermal stability studies, and H2O2-supported P450 heme depletion, total concentration of the P450 was gauged by nonlinear least-squares approximation of the spectra [47] using a linear combination of spectral standards of P450 2B4 low-spin P450, high-spin P450, and P420 states [48, 49] using the previously described SPECTRALAB software package [50] or Igor Pro version 6.1 (Wavemetrics, Inc., Lake Oswego, OR).
Site directed mutagenesis, protein expression, and purification
Single mutants in 2B4 were created using the 2B4dH plasmid as the template and appropriate forward and reverse primers (Supplementary Table S1). Constructs were sequenced at Retrogen, Inc. (San Diego, CA). Mutants were generated by polymerase chain reaction (PCR) using Phusion High-Fidelity DNA Polymerase and a standard site directed mutagenesis protocol.
For biophysical assays, 2B4 was expressed in TOPP3 cells as described previously [51] and purified by the same protocol. The pellet was resuspended in 10% of the original culture volume in buffer containing 20 mM potassium phosphate (pH 7.4 at 4 °C), 20% (v/v) glycerol, 10 mM βME, and 0.5 mM PMSF. The resuspended cells were further treated with lysozyme (0.3 mg/ml) and stirred for 30 min, followed by a brief centrifugation for 30 min at 7000 rpm in a JA-14 rotor using a Beckman Coulter Avanti J-26 XPI centrifuge. After decanting of the supernatant, spheroplasts were resuspended in 5% of the original culture volume in buffer containing 500 mM potassium phosphate (pH 7.4 at 4 °C), 20% (v/v) glycerol, 10 mM βME, and 0.5 mM PMSF and were sonicated for three times for 45 s on ice. The membrane pellet was separated by centrifugation for 10 min at 7000 rpm, and sodium cholate was added to the supernatant at a final concentration of 0.5% (w/v). This was allowed to stir for 30 min at 4 °C prior to ultracentrifugation for 45 min at 41,000 rpm using a fixed-angle Ti 50.2 rotor in a Beckman Coulter Optima L-80 XP ultracentrifuge.
The supernatant was applied to a nickel-nitrilotriacetic acid column. The column was washed with lysis buffer containing 100 mM potassium phosphate (pH 7.4 at 4 °C), 100 mM NaCl, 20% (v/v) glycerol, 10 mM βME, 0.5 mM PMSF, 0.5% (w/v) sodium cholate, and 5 mM histidine, and the protein was eluted using buffer containing 10 mM potassium phosphate (pH 7.4 at 4 °C), 100 mM NaCl, 20% (v/v) glycerol, 10 mM βME, 0.5 mM PMSF, 0.5% (w/v) sodium cholate, and 60 mM histidine. The P450-containing fractions were pooled and dialyzed against buffer containing 10 mM potassium phosphate (pH 7.4 at 4 °C), 10% (v/v) glycerol, and 1 mM ethylenediaminetetraacetic acid (EDTA) with two changes.
For crystallization trials, protein was expressed, harvested, and resuspended as described above. After lysosome treatment, centrifugation, and sonication, 3-[(3-cholaminopropyl)dimethylammonio]-1-propane sulfonate (CHAPS) was added to the solution at a concentration of 0.8% (w/v). This was allowed to stir for 90 min at 4 °C prior to ultracentrifugation for 45 min at 45,000 rpm using a fixed-angle Ti 50.2 rotor in a Beckman Coulter Optima L-80 XP ultracentrifuge.
The supernatant was applied to a nickel-nitrilotriacetic acid column. The column was washed with lysis buffer containing 100 mM potassium phosphate (pH 7.4 at 4 °C), 100 mM NaCl, 20% (v/v) glycerol, 10 mM βME, 0.5 mM PMSF, 0.5% (w/v) sodium cholate, and 5 mM histidine, and the protein was eluted using buffer containing 10 mM potassium phosphate (pH 7.4 at 4 °C), 100 mM NaCl, 20% (v/v) glycerol, 10 mM βME, 0.5 mM PMSF, 0.5% (w/v) sodium cholate, and 60 mM histidine. The P450-containing fractions were pooled and diluted 10-fold in buffer with 5 mM potassium phosphate (pH 7.4 at 4°C), 20% (v/v) glycerol, 1 mM EDTA, 0.2 mM DTT, 0.5 mM PMSF, and 0.5% (w/v) CHAPS before loading onto a Macroprep CM cation exchange column. The column was washed using 5 mM potassium phosphate (pH 7.4 at 4°C), 20 mM NaCl, 20% (v/v) glycerol, 1 mM EDTA, and 0.2 mM DTT and the protein was eluted with high-salt buffer containing 50 mM potassium phosphate (pH 7.4 at 4°C), 500 mM NaCl, 20% (v/v) glycerol, 1 mM EDTA, and 0.2 mM DTT. Protein fractions with the highest A417/A280 ratios were pooled.
Spectral studies of ligand binding
The absorbance spectra were measured with an MC2000-2 multi-channel CCD rapid scanning spectrometer (Ocean Optics, Dunedin, FL, USA) equipped with one absorbance and one fluorescence channel, a pulsed Xe-lamp PX-2 light source, and a home-made thermostated cell chamber with a magnetic stirrer. A semi-micro quartz cell with a stirring compartment (10 × 4 mm light path) from Hellma GmbH (Müllheim, Germany) was used in the titration experiments. All titration experiments were carried out at 25 °C with continuous stirring in buffer containing 50 mM potassium phosphate (pH 7.4 at 4 °C), 500 mM NaCl, 1 mM EDTA, and 0.2 mM DTT. A baseline was recorded between 350 and 700 nm using this buffer. A spectrum was recorded after the addition of protein to the buffer. Spectra were recorded after the addition of a series of 5 μl aliquots of inhibitor (100 μM) to the sample cuvette. The spectral dissociation constants (Ks) were obtained by fitting the data to the equation for “tight binding” 2ΔA = (ΔAmax / [E0]) ((KD + [I0] + [E0]) + ((KD + [I0] + [E0]))2 - 4[E0][I0])1/2) for high affinity ligands Igor Pro version 6.1.
Enzyme assay
The standard NADPH-dependent assay for 7-MFC, 7-EFC, or 7-BR O-dealkylation by 2B4 was carried out as described previously [26, 52]. The reconstituted system contained the following recombinant proteins at a molar ratio of 1:4:2: 2B4, rat NADPH-cytochrome P450 reductase [44], and rat cytochrome b5 [53]. Reactions were carried out using the reconstituted system in a 100 μL final volume and substrate at varying concentrations (0–150 μM for 7-MFC/7-EFC; 0-10 μM for 7-BR). Assays were performed in buffer containing 50 mM HEPES (pH 7.4) and 15 mM MgCl2, initiated by addition of NADPH (1 mM) and incubated for 10 min at 37 °C. Aqueous 20 % (v/v) trichloroacetic acid was added to quench the reaction. An aliquot of the reaction was then transferred to a glass tube containing 0.1 M Tris (pH 9.0), and fluorescence was determined with excitation at 410 nm and emission at 510 nm for 7-EFC and 7-MFC and with excitation at 530 nm and emission at 585 nm for 7-BR using a Cary Eclipse Fluorimeter (Agilent, Santa Clara, CA). A blank was run for each sample, and the final activity was calculated by comparison to a standard curve for the respective substrate. Steady state kinetic parameters were determined by regression analysis using Igor Pro version 6.1. The kcat and KM values were determined using the Michaelis-Menten equation, and the S50 and n values were determined using the Hill equation. Kinetic experiments included wild type and mutant enzymes for more accurate comparison of the data.
Thermal stability studies
Inactivation of P450 was monitored as described earlier [27]. The reaction mixture contained 1 μM protein in 100 mM NaOH-HEPES buffer (pH 7.4). Absorbance spectra were measured using a Shimadzu-2600 spectrophotometer (Shimadzu, Kyoto, Japan). Thermal inactivation was carried out by measuring a series of absorbance spectra in the 340- to 700-nm range as a function of temperature between 25 and 70 °C with 2.5–5 °C intervals and a 2 min equilibration at each temperature. For inactivation kinetics, the samples were treated at 45 °C, and the spectra (340–700 nm) were recorded at different time intervals. All data treatment and fitting of the titration curves were performed with Igor Pro version 6.1. Fitting of the temperature profile and time-dependent inactivation curves was performed by regression analysis using Igor Pro version 6.1. The inactivation profiles were fit to a two-state model to obtain the mid-point of the thermal transition temperature (Tm); a simple pseudo–first-order equation was used to determine the kinact values [27].
H2O2-supported P450 heme depletion
Determination of the kinetics of P450 2B4 heme depletion in the presence of H2O2 was conducted under conditions similar to those described previously for P450 2B1dH [27]. The reaction was carried out at 25°C in 100 mM NaOH-HEPES buffer (pH 7.4) in a 1 mL semi-micro spectrophotometric cell with constant stirring using a MC2000-2 multi-channel CCD rapid scanning spectrometer from Ocean Optics. The reaction mixture contained 1 μM protein and 60 mM H2O2. A series of absorbance spectra in the 340- to 700-nm range over 5 minutes were recorded following addition of H2O2 to the hemoprotein buffer mixture. Determination of the total concentration of the heme protein and data fitting to determine the rate constants were performed as described under “Cytochrome P450 and P420 quantification” and “Thermal stability studies”, respectively.
Molecular dynamics simulations
Molecular dynamics (MD) simulations were performed with starting coordinates of the 4-CPI-bound 2B4 structure (PDB ID: 1PO5) or the bifonazole bound 2B4 structure (PDB ID: 2BDM) using the molecular dynamics software package GROningen MAchine for Chemical Simulation (GROMACS) Version 4.07 [54]. Residues not found in the 4-CPI complex or the bifonazole complex (residues 20-27 and 493-495) were added using the homology modeling program Modeller and the complete amino acid sequence of the protein [55]. The topology files used in the energy minimization and MD simulation were modified to reflect the cysteinyl ligation to heme [56, 57]. The 2B4 structure was immersed in a simulated water box with 120 Å sides and containing ~70,000-80,000 waters, corresponding to twice the length of the longest diagonal of the protein (~65 Å). The structure was energy minimized by the method of steepest descent to remove Van der Waals contact between overlapping waters and the amino acids of the protein. Simulations were run with Berendsen temperature and pressure coupling (a.k.a. “bath”) [58] at a simulated temperature of 300 K using the GROMOS 53a6 force field [59] and periodic boundary conditions in all directions. Electrostatics of the system were measured using the particle-mesh Ewald method [60]. Then, simulations were conducted using a Linux server at the University of California, San Diego. During the first 250 ps of the MD simulation, the protein was position restrained to allow the waters to fill in the cavities. After that, the MD simulation was continued without restraints for another 2000 ps.
Crystallization and data collection
Pooled L437A protein was diluted to 18 μM in 50 mM potassium phosphate (pH 7.4 at 4 °C), 500 mM NaCl, 500 mM sucrose, 1 mM EDTA, and 0.2 mM DTT. 4-CPI was added to this solution at a concentration of 180 μM and allowed to bind overnight at 4 °C. The protein-ligand complex was then concentrated to 550 μM and supplemented with an additional 1.0 mM of 4-CPI. Cymal-5 and 225-chol were added to this solution to final concentrations of 4.8 mM and 0.063% (w/v), respectively. The protein and detergents were allowed to equilibrate for approximately 20 min before mixing with crystallization reagents. Screening for crystallization conditions was performed by sitting drop vapor diffusion using the Wizard I high throughput kit from Emerald Biosystems (Bainbridge Island, WA) by mixing equal volumes of protein solution and well solution. Drops were equilibrated against the well solutions at 18 °C. Rod shaped crystals suitable for X-ray diffraction appeared over the course of approximately 2 weeks in drops containing 0.1 M MES (pH 6.0), 20% (w/v) PEG 8000, and 0.2 M calcium acetate. Crystals were briefly transferred to a solution of mother liquor supplemented with 335 mM sucrose before flash freezing in liquid nitrogen. 140° of data were collected using 1° oscillations and 20 s exposures at 100 K on a Quantum CCD detector (Area Detector Systems Corp., Poway, CA) at BL 11-1 of the Stanford Synchrotron Radiation Lightsource (SSRL) (Stanford, CA). The data were processed to 2.80 Å using iMOSFLM [61] and SCALA [62].
Structure determination and refinement
Phases were obtained by molecular replacement using the previously determined wild type 2B4−4CPI complex (PDB ID: 1SUO) (with the inhibitor molecular removed from the coordinates) in Phaser [63]. The structure solution was found in space group P3 containing 70.2% solvent, assuming four molecules per asymmetric unit. The initial model was first subjected to a simulated annealing step followed by restrained refinement in PHENIX [64] using non-crystallographic symmetry restraints to remove model bias. Model building was performed in Coot [65] using both 2Fo-Fc and Fo-Fc electron density maps contoured to 1-σ and 3-σ, respectively. The model was modified to fit the electron density and refined in an iterative manner until a final R-factor of 19.3% and an Rfree of 24.1% were reached. Non-crystallographic symmetry restraints were slowly released during the refinement process. Structure refinement statistics are summarized in Table 5.
Protein figures
All protein figures were generated using PyMol [66].
Supplementary Material
Acknowledgements
This work was supported, in whole or in part, by National Institutes of Health Grant ES003619 (to J.R.H.). P.R.W. is supported by the Training Grant in Heme and Blood Proteins (T32-DK07233). We also thank the staff at the Stanford Synchrotron Radiation Lightsource, operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences for assistance with data collection. The Stanford Synchrotron Radiation Lightsource is supported by the National Institute of Health, the National Center for Research Resources, the Biomedical Technology Program, and the United States Department of Energy of Biological and Environmental Research. We thank Dr. Santosh Kumar, University of Missouri, Kansas City, for helpful discussions during the early stages of the project.
Abbreviations and textual footnotes
- P450
cytochrome P450
- 4-CPI
4-(4-chlorophenyl)imidazole
- BIF
bifonazole
- 2B4dH
an N-terminal truncated and modified and C-terminal 4-His-Tagged form of cytochrome P450 2B4
- pdb
protein data bank
- 7-BR
7-benzyloxyresorufin
- SNP
single nucleotide polymorphism
- DXMS
hydrogen/deuterium exchange coupled to mass spectrometry
- 7-HFC
7-hydroxy-4-(trifluoromethyl)coumarin
- 7-MFC
7-methoxy-4-(trifluoromethyl)coumarin
- 7-EFC
7-ethoxy-4-(trifluoromethyl)coumarin
- Ni2+-NTA
nickel-nitrilotriacetic acid
- ITC
isothermal titration calorimetry
- βME
β-mercaptoethanol
- PMSF
phenylmethylsulphonyl fluoride
- NADPH
nicotinamide adenine dinucleotide phosphate
- CPR
recombinant NADPH-cytochrome P450 reductase
- b5
cytochrome b5
- PCR
polymerase chain reaction
- EDTA
ethylenediaminetetraacetic acid
- CO
carbon monoxide
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- CHAPS
3-[(3-cholaminopropyl)dimethylammonio]-1-propane sulfonate
Footnotes
Enzymes: rabbit (Oryctolagus cuniculus) cytochrome P450-dependent monooxygenase 2B4, EC 1.14.14.1, UniProt #: P00178
Database: The atomic coordinates and structure factors have been deposited into the RCSB Protein Data Bank under accession number 3TK3.
REFERENCES
- 1.Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, Ball SE. Drug-drug interactions for UDP-glucuronosyltransferase substrates: A pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab Dispos. 2004;32:1201–1208. doi: 10.1124/dmd.104.000794. [DOI] [PubMed] [Google Scholar]
- 2.Johnson EF, Stout CD. Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem Biophys Res Commun. 2005;338:331–336. doi: 10.1016/j.bbrc.2005.08.190. [DOI] [PubMed] [Google Scholar]
- 3.Al Omari A, Murry DJ. Pharmacogenetics of the cytochrome P450 enzyme system: Review of current knowledge and clinical significance. J Pharm Pract. 2007;20:206–218. [Google Scholar]
- 4.Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 5.Ingelman-Sundberg M. Human drug metabolising cytochrome P450 enzymes: Properties and polymorphisms. N-S Arch Pharmacol. 2004;369:89–104. doi: 10.1007/s00210-003-0819-z. [DOI] [PubMed] [Google Scholar]
- 6.Anzenbacher P, Anzenbacherova E, Lange R, Skopalik J, Otyepka M. Active sites of cytochromes P450: What are they like? Acta Chim Slov. 2008;55:63–66. [Google Scholar]
- 7.Poulos TL. Structural and functional diversity in heme monooxygenases. Drug Metab Dispos. 2005;33:10–18. doi: 10.1124/dmd.104.002071. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y, Halpert JR. Structure-function analysis of cytochromes P450 2B. BBA-Gen Subjects. 2007;1770:402–412. doi: 10.1016/j.bbagen.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Poulos TL. Structural biology of heme monooxygenases. Biochem Biophys Res Commun. 2005;338:337–345. doi: 10.1016/j.bbrc.2005.07.204. [DOI] [PubMed] [Google Scholar]
- 10.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-Å resolution: Insight into the range of P450 conformations and the coordination of redox partner binding. J Biol Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 11.Yano JK, Hsu M-H, Griffin KJ, Stout CD, Johnson EF. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- 12.Smith BD, Sanders JL, Porubsky PR, Lushington GH, Stout CD, Scott EE. Structure of the human lung cytochrome P450 2A13. J Biol Chem. 2007;282:17306–17313. doi: 10.1074/jbc.M702361200. [DOI] [PubMed] [Google Scholar]
- 13.Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, Johnson EF. The structure of human cytochrome P4502C9 complexed with flurbiprofen at 2.0-angstrom resolution. J Biol Chem. 2004;279:35630–35637. doi: 10.1074/jbc.M405427200. [DOI] [PubMed] [Google Scholar]
- 14.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Microsomal cytochrome P450 2C5: Comparison to microbial P450s and unique features. J Inorg Biochem. 2000;81:183–190. doi: 10.1016/s0162-0134(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: Insight into P450 conformational plasticity and membrane interaction. J Biol Chem. 2006;281:5973–5981. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- 16.Ekroos M, Sjögren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. P Natl Acad Sci USA. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S, Zhao Y, Sun L, Negi SS, Halpert JR, Muralidhara BK. Rational engineering of human cytochrome P450 2B6 for enhanced expression and stability: Importance of a Leu264->Phe substitution. Mol Pharmacol. 2007;72:1191–1199. doi: 10.1124/mol.107.039693. [DOI] [PubMed] [Google Scholar]
- 18.Wilderman PR, Shah MB, Liu T, Li S, Hsu S, Roberts AG, Goodlett DR, Zhang Q, Woods VL, Jr., Stout CD, et al. Plasticity of cytochrome P450 2B4 as investigated by hydrogen-deuterium exchange mass spectrometry and X-Ray crystallography. J Biol Chem. 2010;285:38602–38611. doi: 10.1074/jbc.M110.180646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mo SL, Liu YH, Duan W, Wei MQ, Kanwar JR, Zhou SF. Substrate specificity, regulation, and polymorphism of human cytochrome P450 2B6. Curr Drug Metab. 2009;10:730–753. doi: 10.2174/138920009789895534. [DOI] [PubMed] [Google Scholar]
- 20.Wang HB, Tompkins LM. CYP2B6: New insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab. 2008;9:598–610. doi: 10.2174/138920008785821710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daly AK. Pharmacogenetics of the major polymorphic metabolizing enzymes. Fund Clin Pharmacol. 2003;17:27–41. doi: 10.1046/j.1472-8206.2003.00119.x. [DOI] [PubMed] [Google Scholar]
- 22.Ekhart C, Doodeman VD, Rodenhuis S, Smits PHM, Beijnen JH, Huitema ADR. Influence of polymorphisms of drug metabolizing enzymes (CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5, GSTA1, GSTP1, ALDH1A1 and ALDH3A1) on the pharmacokinetics of cyclophosphamide and 4-hydroxycyclophosphamide. Pharmacogenet Genom. 2008;18:515–523. doi: 10.1097/FPC.0b013e3282fc9766. [DOI] [PubMed] [Google Scholar]
- 23.Zanger UM, Klein K, Saussele T, Blievernicht J, H Hofmann M, Schwab M. Polymorphic CYP2B6: Molecular mechanisms and emerging clinical significance. Pharmacogenomics. 2007;8:743–759. doi: 10.2217/14622416.8.7.743. [DOI] [PubMed] [Google Scholar]
- 24.Restrepo JG, Garcia-Martin E, Martinez C, Agundez JAG. Polymorphic drug metabolism in anaesthesia. Curr Drug Metab. 2009;10:236–246. doi: 10.2174/138920009787846305. [DOI] [PubMed] [Google Scholar]
- 25.Talakad JC, Kumar S, Halpert JR. Decreased susceptibility of the cytochrome P450 2B6 variant K262R to inhibition by several clinically important drugs. Drug Metab Dispos. 2009;37:644–650. doi: 10.1124/dmd.108.023655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Chen CS, Waxman DJ, Halpert JR. Directed evolution of mammalian cytochrome P450 2B1: Mutations outside of the active site enhance the metabolism of several substrates, including the anticancer prodrugs cyclophosphamide and ifosfamide. J Biol Chem. 2005;280:19569–19575. doi: 10.1074/jbc.M500158200. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Sun L, Liu H, Muralidhara BK, Halpert JR. Engineering mammalian cytochrome P450 2B1 by directed evolution for enhanced catalytic tolerance to temperature and dimethyl sulfoxide. Protein Eng Des Sel. 2006;19:547–554. doi: 10.1093/protein/gzl042. [DOI] [PubMed] [Google Scholar]
- 28.Sun L, Chen CS, Waxman DJ, Liu H, Halpert JR, Kumar S. Re-engineering cytochrome P450 2B11dH for enhanced metabolism of several substrates including the anti-cancer prodrugs cyclophosphamide and ifosfamide. Arch Biochem Biophys. 2007;458:167–174. doi: 10.1016/j.abb.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talakad JC, Wilderman PR, Davydov DR, Kumar S, Halpert JR. Rational engineering of cytochromes P450 2B6 and 2B11 for enhanced stability: Insights into structural importance of residue 334. Arch Biochem Biophys. 2009;494:151–158. doi: 10.1016/j.abb.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Halpert JR. 16th International Symposium on Microsomes and Drug Oxidations. Budapest Hungary; 2006. Conserved and alternative residue-residue interactions and their role in structure function of CYP2B enzymes. p. 86. [Google Scholar]
- 31.Gay SC, Shah MB, Talakad JC, Maekawa K, Roberts AG, Wilderman PR, Sun L, Yang JY, Huelga SC, Hong W-X, et al. Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-Å resolution. Mol Pharmacol. 2010;77:529–538. doi: 10.1124/mol.109.062570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillam EM. Exploring the potential of xenobiotic-metabolising enzymes as biocatalysts: Evolving designer catalysts from polyfunctional cytochrome P450 enzymes. Clin Exp Pharmacol Physiol. 2005;32:147–152. doi: 10.1111/j.1440-1681.2005.04165.x. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Halpert JR. Use of directed evolution of mammalian cytochromes P450 for investigating the molecular basis of enzyme function and generating novel biocatalysts. Biochem Biophys Res Commun. 2005;338:456–464. doi: 10.1016/j.bbrc.2005.08.080. [DOI] [PubMed] [Google Scholar]
- 34.Gillam EM. Extending the capabilities of nature's most versatile catalysts: Directed evolution of mammalian xenobiotic-metabolizing P450s. Arch Biochem Biophys. 2007;464:176–186. doi: 10.1016/j.abb.2007.04.033. [DOI] [PubMed] [Google Scholar]
- 35.Domanski TL, Halpert JR. Analysis of mammalian cytochrome P450 structure and function by site-directed mutagenesis. Curr Drug Metab. 2001;2:117–137. doi: 10.2174/1389200013338612. [DOI] [PubMed] [Google Scholar]
- 36.Kumar S, Scott EE, Liu H, Halpert JR. A rational approach to re-engineer cytochrome P450 2B1 regioselectivity based on the crystal structure of cytochrome P450 2C5. J Biol Chem. 2003;278:17178–17184. doi: 10.1074/jbc.M212515200. [DOI] [PubMed] [Google Scholar]
- 37.Halpert JR, He YA. Engineering of cytochrome P450 2B1 specificity. Conversion of an androgen 16 beta-hydroxylase to a 15 alpha-hydroxylase. J Biol Chem. 1993;268:4453–4457. [PubMed] [Google Scholar]
- 38.Harlow GR, He YA, Halpert JR. Functional interaction between amino-acid residues 242 and 290 in cytochromes P-450 2B1 and 2B11. Biochim Biophys Acta. 1997;1338:259–266. doi: 10.1016/s0167-4838(96)00209-9. [DOI] [PubMed] [Google Scholar]
- 39.Whitehouse CJC, Yang W, Yorke JA, Rowlatt BC, Strong AJF, Blanford CF, Bell SG, Bartlam M, Wong L-L, Rao Z. Structural Basis for the Properties of Two Single-Site Proline Mutants of CYP102A1 (P450BM3). ChemBioChem. 2010;11:2549–2556. doi: 10.1002/cbic.201000421. [DOI] [PubMed] [Google Scholar]
- 40.Gay SC, Sun L, Maekawa K, Halpert JR, Stout CD. Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: Ligand-induced structural response through α-helical repositioning. Biochemistry. 2009;48:4762–4771. doi: 10.1021/bi9003765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott EE, Liu H, Qun He Y, Li W, Halpert JR. Mutagenesis and molecular dynamics suggest structural and functional roles for residues in the N-terminal portion of the cytochrome P450 2B1 I helix. Arch Biochem Biophys. 2004;423:266–276. doi: 10.1016/j.abb.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 42.Hernandez CE, Kumar S, Liu H, Halpert JR. Investigation of the role of cytochrome P450 2B4 active site residues in substrate metabolism based on crystal structures of the ligand-bound enzyme. Arch Biochem Biophys. 2006;455:61–67. doi: 10.1016/j.abb.2006.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah MB, Pascual J, Zhang Q, Stout CD, Halpert JR. Structures of Cytochrome P450 2B6 Bound to 4-Benzylpyridine and 4-(4-Nitrobenzyl)pyridine: Insight into Inhibitor Binding and Rearrangement of Active Site Side Chains. Mol Pharmacol. 2011 doi: 10.1124/mol.111.074427. DOI:10.1124/mol.111.074427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harlow GR, Halpert JR. Alanine-scanning mutagenesis of a putative substrate recognition site in human cytochrome P450 3A4. Role of residues 210 and 211 in flavonoid activation and substrate specificity. J Biol Chem. 1997;272:5396–5402. doi: 10.1074/jbc.272.9.5396. [DOI] [PubMed] [Google Scholar]
- 45.Omura T, Sato R. Carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 46.Omura T, Sato R. Carbon monoxide-binding pigment of liver microsomes. 2. Solubilization purification + properties. J Biol Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- 47.Arthurs TD. Algorithm 176: least squares surface fit. Commun ACM. 1963;6:313. [Google Scholar]
- 48.Davydov DR, Botchkareva AE, Davydova NE, Halpert JR. Resolution of two substrate-binding sites in an engineered cytochrome P450eryF bearing a fluorescent probe. Biophys J. 2005;89:418–432. doi: 10.1529/biophysj.104.058479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davydov DR, Halpert JR, Renaud JP, Hui Bon Hoa G. Conformational heterogeneity of cytochrome P450 3A4 revealed by high pressure spectroscopy. Biochem Biophys Res Commun. 2003;312:121–130. doi: 10.1016/j.bbrc.2003.09.247. [DOI] [PubMed] [Google Scholar]
- 50.Davydov DR, Deprez E, Hui Bon Hoa G, Knyushko TV, Kuznetsova GP, Koen YM, Archakov AI. High-pressure-induced transitions in microsomal cytochrome P450 2B4 in solution: Evidence for conformational inhomogeneity in the oligomers. Arch Biochem Biophys. 1995;320:330–344. doi: 10.1016/0003-9861(95)90017-9. [DOI] [PubMed] [Google Scholar]
- 51.Scott EE, Spatzenegger M, Halpert JR. A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch Biochem Biophys. 2001;395:57–68. doi: 10.1006/abbi.2001.2574. [DOI] [PubMed] [Google Scholar]
- 52.He YQ, He YA, Halpert JR. Escherichia coli expression of site-directed mutants of cytochrome-P450 2B1 from 6 substrate recognition sites - Substrate-specificity and inhibitor selectivity studies. Chem Res Toxicol. 1995;8:574–579. doi: 10.1021/tx00046a011. [DOI] [PubMed] [Google Scholar]
- 53.Holmans PL, Shet MS, Martin-Wixtrom CA, Fisher CW, Estabrook RW. The high-level expression in Escherichia coli of the membrane-bound form of human and rat cytochrome b5 and studies on their mechanism of function. Arch Biochem Biophys. 1994;312:554–565. doi: 10.1006/abbi.1994.1345. [DOI] [PubMed] [Google Scholar]
- 54.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 55.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 56.Oda A, Yamaotsu N, Hirono S. New AMBER force field parameters of heme iron for cytochrome P450s determined by quantum chemical calculations of simplified models. J Comput Chem. 2005;26:818–826. doi: 10.1002/jcc.20221. [DOI] [PubMed] [Google Scholar]
- 57.Froelich JW, Hearshen DO, Halpert RD, Patel S. Nuclear magnetic resonance: current and future clinical applications. Henry Ford Hosp Med J. 1985;33:122–127. [PubMed] [Google Scholar]
- 58.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 59.Oostenbrink C, Soares TA, van der Vegt NFA, van Gunsteren WF. Validation of the 53A6 GROMOS force field. Eur Biophys J Biophy. 2005;34:273–284. doi: 10.1007/s00249-004-0448-6. [DOI] [PubMed] [Google Scholar]
- 60.Darden T, York D, Pedersen L. Particle Mesh Ewald - An N.log(N) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 61.Leslie AGW. Integration of macromolecular diffraction data. Acta Crystallogr Sect D-Biol Crystallogr. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 62.Bailey S. The CCP4 Suite - Programs for protein crystallography. Acta Crystallogr Sect D-Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 63.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 65.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MacPyMOL, editor. The PyMOL molecular graphics system. 1.3 Schrodinger, LLC; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.