Abstract
A bacterium's ability to acquire nutrients from its host during infection is an essential component of pathogenesis. For the Gram-positive pathogen Streptococcus pyogenes, catabolism of the amino acid arginine via the arginine deiminase (ADI) pathway supplements energy production and provides protection against acid stress in vitro. Its expression is enhanced in murine models of infection, suggesting an important role in vivo. To gain insight into the function of the ADI pathway in pathogenesis, the virulence of mutants defective in each of its enzymes was examined. Mutants unable to use arginine (ΔArcA) or citrulline (ΔArcB) were attenuated for carriage in a murine model of asymptomatic mucosal colonization. However, in a murine model of inflammatory infection of cutaneous tissue, the ΔArcA mutant was attenuated but the ΔArcB mutant was hyperattenuated, revealing an unexpected tissue-specific role for citrulline metabolism in pathogenesis. When mice defective for the arginine-dependent production of nitric oxide (iNOS−/−) were infected with the ΔArcA mutant, cutaneous virulence was rescued, demonstrating that the ability of S. pyogenes to utilize arginine was dispensable in the absence of nitric oxide-mediated innate immunity. This work demonstrates the importance of arginine and citrulline catabolism and suggests a novel mechanism of virulence by which S. pyogenes uses its metabolism to modulate innate immunity through depletion of an essential host nutrient.
INTRODUCTION
Nutrient acquisition by a pathogen is necessary for the colonization of a host and represents an important aspect of bacterial pathogenesis. Identification of bacterial metabolic pathways utilized during an infection can reveal unique targets for novel therapeutics, as well as provide insight into the environmental cues being sensed by the pathogen. Additionally, the metabolism of a microbe can play an intricate role in pathogenesis by shuttling key metabolites away from or toward the host. This complex interaction can have an important influence on the host immune response and subsequent disease outcome. The challenge, then, becomes identifying those metabolic pathways active during an infection and deciphering their specific contributions to pathogenesis.
The group A streptococcus (Streptococcus pyogenes) is a common Gram-positive pathogen responsible for a large number of diseases that range in severity and invasiveness (1). Noninvasive soft tissue infections include the common bacterial pharyngitis and impetigo; less common, but invasive and often life-threatening, soft tissue infections include necrotizing fasciitis (1). Gene expression studies conducted during soft tissue infection have suggested that S. pyogenes encounters an environment that is likely deficient in glucose but with an abundance of amino acids and peptides (2–4). Adaptation to this nitrogen-rich environment includes the enhanced expression of amino acid and peptide transporters, as well as catabolic pathways that can process these substrates (3, 4). One metabolic operon that is prominently upregulated during growth in a mouse model of soft tissue infection and in human blood is the arginine deiminase (ADI) pathway (3, 4).
Studies conducted in vitro have shown that the ADI pathway of S. pyogenes, encoded by the Arc operon, contributes to metabolism through the production of cellular energy and protection against acid stress (5, 6). The pathway is composed of the three key enzymes: ArcA, ArcB, and ArcC, corresponding to arginine deiminase, ornithine carbamoyl transferase, and carbamate kinase, respectively (7–9) (Fig. 1). The overall reaction converts arginine to ornithine, CO2, ammonia, and one molecule of ATP (Fig. 1). The ArcD antiporter transports ornithine out and concomitantly brings arginine into the cell (10) (Fig. 1). In addition to generating ATP, the production of two ammonia molecules can be utilized to buffer against acid stress (5, 11, 12), which can be endogenously generated by the fermentative metabolism of S. pyogenes and the accumulation of its lactic acid end product (13). ArcB's production of carbamoyl phosphate, a precursor in pyrimidine biosynthesis, may also contribute to synthesis of pyrimidines. These functions suggest that arginine metabolism makes an important contribution to S. pyogenes pathogenesis.
FIG 1.
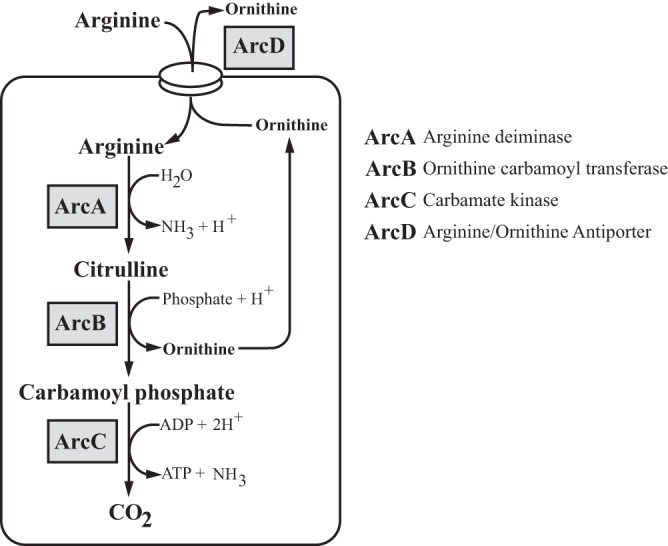
Arginine and citrulline catabolism in S. pyogenes. Through the multienzyme pathway, arginine is transported into the cell via the antiporter ArcD and catabolized by ArcA, ArcB, and ArcC to produce one molecule of carbon dioxide, one molecule of ATP, and two molecules of ammonia.
Arginine metabolism is also a key component of host innate immunity, serving as the substrate for the enzymatic generation of the antimicrobial molecule nitric oxide (NO·), a potent radical that can elicit damage through a broad range of reactions that result in the modification of a number of cellular targets, including heme, protein thiols, tyrosine residues, lipids, and DNA (14–17). Production of NO· for antimicrobial purposes is typically via the inducible nitric oxide synthase (iNOS) (18). Commonly expressed by macrophages, iNOS utilizes arginine to produce NO· and citrulline. Given its requirement for arginine, the expression and activity of iNOS are regulated by the extracellular availability of this substrate (19, 20). To control iNOS activity, the host regulates the amount of available arginine by expression of arginase, an alternative arginine-consuming enzyme that competes with iNOS for substrates (21, 22). Many pathogens exploit this host regulatory interaction by manipulating host arginase expression and limiting arginine availability in order to block production of NO· (21, 22).
The recent identification of macrophages as an important immune mediator for controlling S. pyogenes in soft tissue (23, 24) suggests a role for iNOS during infection and, by extension, host arginine metabolism. To investigate the possible intersection of host and pathogen arginine metabolism in S. pyogenes pathogenesis, we constructed mutants defective for each of the individual genes of the ADI pathway and analyzed their behavior in murine models of asymptomatic mucosal colonization and symptomatic infection of subcutaneous tissue. These studies revealed that arginine metabolism contributes to both S. pyogenes colonization and disease, demonstrating a tissue-specific role for citrulline catabolism in pathogenesis and a novel mechanism for modulating innate immunity using the ADI pathway to deplete a critical host nutrient.
MATERIALS AND METHODS
E. coli strains, media, and growth conditions.
Routine molecular cloning and plasmid propagation utilized E. coli DH5α, which was cultured in Luria-Bertani medium at 37°C. When appropriate, erythromycin was added to the medium at 750 μg/ml.
Streptococcus pyogenes strains, media, and growth conditions.
Unless otherwise indicated, experiments utilized Streptococcus pyogenes HSC5 (25) and mutant derivatives of this strain (Table 1). Routine culture employed Todd-Hewitt medium (Difco) supplemented with 0.2% yeast extract (Difco) (THY medium). When indicated, strains were cultured in C medium (0.5% protease peptone 3 [Difco], 1.5% yeast extract [Difco], 10 mM K2HPO4, 0.4 mM MgSO4, 17 mM NaCl). Growth for infection of macrophages is detailed below. For experiments involving amino acid supplementation, filter-sterilized 0.5 M stock solutions were used to add l-arginine or l-citrulline (Sigma) to a final concentration of 10 mM in media that had been sterilized in an autoclave. All growth experiments, unless otherwise stated, utilized sealed culture tubes at 37°C under static conditions. Solid medium was prepared by the addition of 1.4% Bacto agar (Difco) and was cultured anaerobically in sealed jars in the presence of commercially available gas-generating packets (GasPak catalog no. 70304; BBL). When appropriate, antibiotics were added at the following concentrations: erythromycin, 1 μg/ml, and streptomycin, 1,000 μg/ml.
TABLE 1.
S. pyogenes strains used in this studya
| Strain | Relevant genotype | Description | Reference |
|---|---|---|---|
| HSC5 | Wild type | Wild type | 25 |
| ZC156 | HSC5 arcAΔ10–409 | In-frame deletion of arcA | This work |
| ZC512 | HSC5 arcBΔ13–326 | In-frame deletion of arcB | This work |
| ZC498 | HSC5 arcCΔ9–312 | In-frame deletion of arcC | This work |
| ZC500 | HSC5 arcDΔ7–490 | In-frame deletion of arcD | This work |
| ZC604 | HSC5 arcAΔ10–409 arcBΔ13–326 | In-frame deletion of arcA and arcB | This work |
| HSC12 | Wild type | Spontaneous streptomycin-resistant derivative of HSC5 | 34 |
| MEW124 | HSC5 arcAΔ10–409 | Spontaneous streptomycin-resistant derivative of ZC156 | This work |
| MEW126 | HSC5 arcBΔ13–326 | Spontaneous streptomycin-resistant derivative of ZC512 | This work |
Gene names are based on current annotation of the genome of S. pyogenes SF370 in the NCBI database (http://www.ncbi.nlm.gov).
Construction of deletion mutants.
All references to genomic loci are based upon the genome of S. pyogenes SF370 (26). In-frame deletion mutations in genes encoding ArcA (SPy_1547), ArcB (SPy_1544), ArcC (SPy_1541), and ArcD (SPy_1543) were generated by allelic replacement using the allelic-replacement vector pJRS233 (27) as described previously (28) and listed in Table 1. Each deletion allele was generated by PCR using 5′ phosphorylated primers for three-way ligations or through a process of overlap extension PCR (29) with the primers listed in Table 2. The correct chromosomal structure in all mutants was verified using PCR.
TABLE 2.
Primers used in this study
| Name | Purpose | Sequencea | Template | Plasmid |
|---|---|---|---|---|
| ZC78 | In-frame deletion of arcA | TTTTGTCGACTTTGATTAATTATTTTTTGTGAGAT | HSC5 | pZC32 |
| ZC66 | 5PHOS-GAAGATATTTAATAAGCTAT | |||
| ZC67 | In-frame deletion of arcA | 5PHOS-GGTGTTTGAGCAGTCAT | HSC5 | pZC32 |
| ZC65 | TTTCTGCAGCTCGAGTTAC | |||
| ZC317 | In-frame deletion of arcB | CCCCCTCGAGGTCGACGGTAAAGGTGGTTGTAGGTCAG | HSC5 | pZC141 |
| ZC318 | GGAATAAAGAGGTTACCAAGGGTTGCGCTAGGAAGCTACGTCCTTGAAATACTTGTGTCAT | |||
| ZC319 | In-frame deletion of arcB | GCAACCCTTGGTAACCTCTTTATTCCAAAAGTGTAATAAGGACTGGTACTCCTTAACTCATTTC | HSC5 | pZC141 |
| ZC320 | GATCCCCCGGGCTGCAGTGTTTTTACGAACCACTGACGCAATCCC | |||
| ZC279 | In-frame deletion of arcC | AGGGTCGACCTTGCCAAGGCTTTAGTGGTCCTTG | HSC5 | pZC137 |
| ZC280 | 5PHOS-ACTACGATTTTTTGTTTCGTCATAATTACTCCTC | |||
| ZC281 | In-frame deletion of arcC | 5PHOS-GGAACACAAATTATCGCAGGGTAATCAAGAGG | HSC5 | pZC137 |
| ZC282 | TTTTCTGCAGCAAGACATTGACAGATACTGGATTTTC | |||
| ZC324 | In-frame deletion of arcD | CCCCCTCGAGGTCGACTATCCTTATCACAGAAGATGCTGACGAAGC | HSC5 | pZC15 |
| ZC325 | CTATCTGTTAAAAGAATGTTGCAACTACAAGTCCGTTTTTTTTCTTCTGTCATAATGATTACCTC | |||
| ZC326 | In-frame deletion of arcD | ACTTGTAGTTGCAACATTCTTTTAACAGATAGGAGGTTCCTATGGAATCCTATATCACACCTAAAC | HSC5 | pZC53 |
| ZC327 | GATCCCCCGGGCTGCAGGCAAACCCAAAGGTAGCCTGTTCTTCTAC | |||
| iNOS Forward | RT-PCR primers for iNOS | GCAAACATCACATTCAGATCC | ||
| iNOS Reverse | TCAGCCTCATGGTAAACACG | |||
| GAPDH Forward | RT-PCR primers for GAPDH | CCACCCAGAAGACTCTGGAT | ||
| GAPDH Reverse | CACATTGGGGGTAGGAACAC |
Engineered restriction sites are underlined. 5PHOS represents a phosphate moiety attached to the 5′ end of the oligonucleotide.
Growth curve and determination of maximum doubling time.
Indicated bacterial strains were back-diluted 1:50 into 1 ml of fresh C medium. Cultures were mixed briefly by vortexing, and 200 μl was placed in triplicate into a 96-well plate (catalog no. 655-180; Greiner); plates were sealed using transparent adhesive film (catalog no. 60941-078, VWR). Monitoring of growth at 37°C was performed in a Tecan Infinite M200 Pro plate reader. During growth the plate was shaken every 10 min for 60 s, followed by a 30-s wait period and measurement of the optical density at 600 nm (OD600). To determine the maximum doubling time, the normalized data were input into Prism Graphpad. Next, the initial period of exponential growth was fit to a growth equation (Y = Y0ekt, where Y is OD600, Y0 is OD600 at time zero, k is exponential rate constant, t is time, and e is the constant, Euler's number) using nonlinear regression curve analysis to determine the exponential rate constant, which was then used to calculate the doubling time. The average doubling time was calculated from each replicate from at least three independent experiments.
Preparation of S. pyogenes for macrophage infection.
Following overnight growth in THY medium, the culture was back-diluted to an OD600 of 0.05 in 50 ml of fresh THY medium and the incubation continued for approximately 3 to 4 h until the culture reached an OD600 of 0.2. Bacteria were collected by centrifugation, washed once with 10 ml of a sterile 0.9% endotoxin-free saline solution (Teknova), and then resuspended in 1 ml of the saline solution. The suspension was placed on ice and then subjected to brief sonication to disrupt the streptococcal chains to individual cells as described previously (30); these were enumerated by microscopy using a hemocytometer (catalog no. 3900; Hausser Scientific). The suspension concentration was adjusted to 1 × 109 cells/ml and diluted in the saline solution to vary the multiplicity of infection (MOI). Bacterial CFU in the suspension was determined by serial dilution in phosphate-buffered saline (PBS) and plating on THY medium, with enumeration of CFU following overnight growth.
Infection of macrophages.
Macrophage infection was performed as previously described (31, 32). Briefly, approximately 2.5 × 105 cells of the RAW 264.7 murine macrophage line were plated in each well of a six-well tissue culture plate (catalog no. CC7682-7506; Techno Plastic Products) and maintained in a culture medium consisting of RPMI 1640 (Life Technologies) with 10% fetal bovine serum (FBS) (Lenova). Cultures were incubated in a humidified 5% CO2 atmosphere for 2 days prior to infection. Twenty-four hours prior to infection, macrophages were activated with 100 ng/ml of murine gamma interferon (IFN-γ; Sigma). Prior to infection, macrophages were then washed once with Dulbecco's phosphate-buffered saline (DPBS; Life Technologies), and fresh culture medium was added. Bacterial suspensions were prepared as described above, and aliquots were added to each macrophage-containing well to obtain the MOIs described below. For comparison, lipopolysaccharide (LPS) from E. coli (catalog no. L4391; Sigma) was added to an uninfected well to a final concentration of 1 μg/ml. Incubation was then continued for 60 min at 37°C in 5% CO2. All wells were then washed with 2 ml of DPBS to remove bacteria, followed by addition of fresh culture medium containing penicillin (100 U/ml) and streptomycin (100 μg/ml), and the incubation was continued for an additional 3 h. Macrophages were then harvested in 1 ml of DPBS using a cell scraper, and the number of recovered macrophages was determined by microscopy using a hemocytometer. The macrophages were collected by centrifugation and were placed at −80°C for storage. For measurement of NO·, macrophages were prepared as described above and plated in wells of a 12-well tissue culture plate (catalog no. 92412; TPP). Following a 60-min infection with S. pyogenes or incubation with LPS (1 μg/ml), the wells were washed with DPBS, and fresh medium supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml) was added. The incubation was continued for an additional 24 h, followed by determination of NO· concentrations as described below.
Analysis of conditioned medium.
The influence of S. pyogenes arginine metabolism on the production of NO· by cultured macrophages was determined as follows. The wild type and various Arc mutants were cultured overnight in C medium and then back-diluted 1/100 with RPMI 1640 supplemented with 10% FBS and 0.2% yeast extract in a 15-ml conical tube. Following growth at 37°C for 18 h, the streptococcal cells were removed by centrifugation, the pH was adjusted by the addition of 1 M HEPES (pH 7.5) to a final concentration of 100 mM, and the entire solution was sterilized by filtration using a 0.2-μm filter. This conditioned medium was mixed with cell culture medium (RPMI 1640 plus 10% FBS) at the various concentrations indicated below and used to replace the media of tissue culture plates containing approximately 106 RAW 264.7 macrophages per well. These cultures were then stimulated by the addition of 1 μg/ml of LPS. Following an additional 24 h of incubation, the production of NO· was determined as described below.
Isolation of RNA and real-time RT-PCR.
Total RNA was isolated from macrophages using a Qiagen QIAshredder and RNeasy minikit per the manufacturer's protocol. RNA isolated from mouse lesions were first subjected to mechanical disruption before being further processed using the Qiagen QIAshredder and RNeasy minikit. RNA was subjected to reverse transcription (RT) using iScript (Bio-Rad) per the manufacturer's instructions. RT-PCR analysis of cDNA samples was performed using iQ SYBR green Supermix (Bio-Rad) and the primers listed in Table 2. Relative transcript levels were determined using the threshold cycle (ΔΔCT) method using a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript as a standard and are presented in comparison to those in uninfected cells. The data shown are the means and the standard deviations derived from triplicate determinations of two separate biological samples prepared from at least two independent experiments.
Measurement of NO· concentration.
The concentration of by-products formed by the oxidation of NO·, NO3−, and NO2− were measured using the Griess reagent system (Promega) per the manufacturer's instructions. Absolute concentrations were determined by comparison to a standard curve prepared in cell culture medium.
Infection of mice.
Inflammatory infection of murine subcutaneous tissue was conducted as described in detail previously (30, 33) using 6- to 8-week old female SKH1 mice obtained from Charles River Laboratories and C57BL/6 (catalog no. 000664) or B6.129P2-Nos2tm1Lau/J (iNOS−/−; catalog no. 002609) mice obtained from the Jackson Laboratory. Mice received a subcutaneous injection of 107 bacterial CFU of the bacterial strains indicated below. Following infection, the resulting ulcers were imaged over the course of several days as indicated and the areas of the irregular lesions calculated as described in detail elsewhere (30). Where noted, the number of CFU recovered in tissues was measured as described previously (30). The ability of strains to establish asymptomatic colonization of the murine vaginal mucosa was evaluated in C57BL/6 mice, as described in detail elsewhere (34). Colonization was assessed at selected time points over the course of 34 days by monitoring the number of CFU recoverable in a 50-μl vaginal wash. In all of these cases, data presented are pooled from at least 2 independent experiments with a total of 10 mice per experimental group.
Ethics statement.
This study was carried out in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals and AAALAC accreditation guidelines. The protocols were approved by Washington University in St. Louis' Animal Studies Committee (Animal Welfare Assurance number A-3381-01, protocol 20100186, and protocol 20080079).
Statistical analyses.
Unless otherwise indicated, differences between experimental groups were tested for significance using the Mann-Whitney U test. Difference between groups for recovery of CFU in vaginal washes over time was tested for significance using a repeated-measures analysis of variance (34). For all tests, the null hypothesis was rejected for P values of >0.05. Computation of test statistics utilized the resources available in GraphPad Prism software (GraphPad Software Inc.) or the VassarStats Site for Statistical Computation by Richard Lowry (http://faculty.vassar.edu/lowry/VassarStats.html).
RESULTS
The ADI pathway contributes to growth.
Arginine catabolism can contribute to bacterial metabolism and pathogenesis in a number of different ways, including the production of ATP, ammonia, and carbamoyl phosphate and, potentially, the inhibition of iNOS (12, 32). To investigate the role of arginine catabolism in the pathogenesis of S. pyogenes, individual in-frame deletion mutations were constructed in arcA (SPy_1547), arcB (SPy_1544), arcC (SPy_1541), and arcD (SPy_1543), which encodes the arginine-ornithine antiporter. By comparing each mutant to the ΔArcA mutant, it should be possible to dissect the specific contributions of arginine catabolism to pathogenesis. First, growth phenotypes were analyzed during in vitro culture. For wild-type S. pyogenes, the addition of 10 mM arginine to C medium resulted in a significant increase in growth yield (Fig. 2A). In comparison, the growth yields from all four mutant strains (ΔArcA, ΔArcB, ΔArcC, ΔArcD mutants) did not benefit from additional arginine, with yields that were nearly identical to those obtained in the absence of added arginine (Fig. 2A). Despite the reduction in the final growth yield, all four Arc mutants grew similarly to the wild type (see Fig. S1 in the supplemental material) and retained a maximum doubling time comparable to that of the wild-type strain (Fig. 2A), suggesting that any measurable growth defect is the result of a specific disruption of the ADI pathway. Furthermore, complementation of each Arc mutant restored the ability to grow to yields comparable to those of the wild type in both the presence and absence of arginine (see Fig. S2 in the supplemental material). Previous research has demonstrated that added citrulline can be utilized by S. pyogenes for protection against acid stress (12). To determine if it could also be utilized for growth, we next measured citrulline's ability to increase the growth yield of the wild type in C medium. Addition of 10 mM citrulline resulted in no measurable increase in growth yield as measured by OD600 for the wild type or any Arc mutant (Fig. 2B). These results demonstrate that arginine but not citrulline can be utilized for growth via the ADI pathway and that this growth requires all four proteins.
FIG 2.
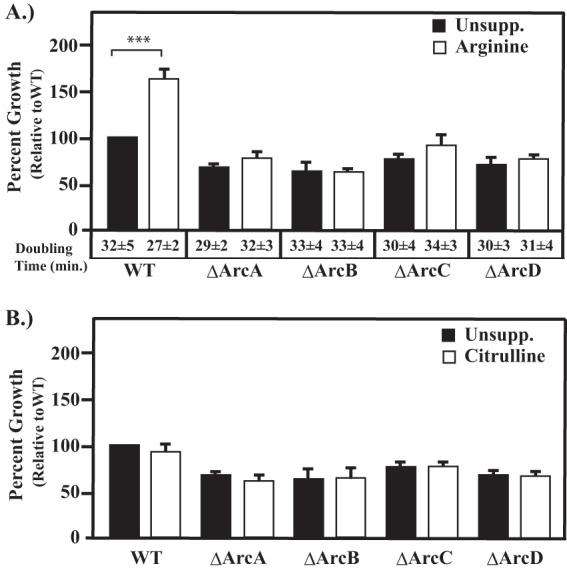
Disruption of arginine catabolism in Arc mutants. Wild-type strain HSC5 (WT) and the indicated mutant strains were cultured in unsupplemented C medium (Unsupp.) or C medium supplemented with 10 mM arginine (A) or 10 mM citrulline (B). Following 16 h of incubation, growth yields were measured by OD600. For details of each strain, consult Table 1. Data are presented as percent growth relative to that of the wild type grown in unsupplemented C medium and are the means and standard deviations from at least three independent experiments. Differences in growth yield between each supplemented culture versus unsupplemented culture were tested for significance using the Mann-Whitney U test (***, P < 0.001). Doubling times were calculated as described in Materials and Methods.
ΔArcA and ΔArcB mutants are attenuated in a murine model of mucosal colonization.
Despite its inability to supplement growth, it has been reported that citrulline can protect S. pyogenes against acid stress as effectively as arginine (12). Given that ArcA− mutants retain the ability to utilize citrulline (12), comparison of the wild type and ΔArcA and ΔArcB mutants should allow a dissection of the relative contribution of arginine versus citrulline catabolism to mucosal colonization. Using a recently established model of asymptomatic carriage in the murine vaginal mucosa (34), each of these strains was vaginally inoculated into pre-estrogenized C57BL/6J mice and colonization monitored by determining numbers of viable CFU from vaginal washes over the course of 34 days. In this model, the wild-type strain maintains a high level of colonization for more than 20 days in the absence of obvious symptomatology before CFU rapidly decreases (34) (Fig. 3). Comparison of CFU revealed a significant decrease in carriage density for both the ΔArcA and ΔArcB mutants compared to the wild type (Fig. 3). Since the ΔArcA mutant retains the ability to use citrulline, while the ΔArcB mutant cannot utilize either citrulline or arginine, it would be expected that a requirement for host-derived citrulline would result in a higher level of attenuation for the ΔArcB mutant. However, the two mutants were attenuated to similar degrees (Fig. 3). Thus, this observation indicates that the ability to acquire arginine, but not citrulline, from host tissue is important for colonization and carriage in an environment that mimics the mucosal surface more typically encountered by S. pyogenes.
FIG 3.
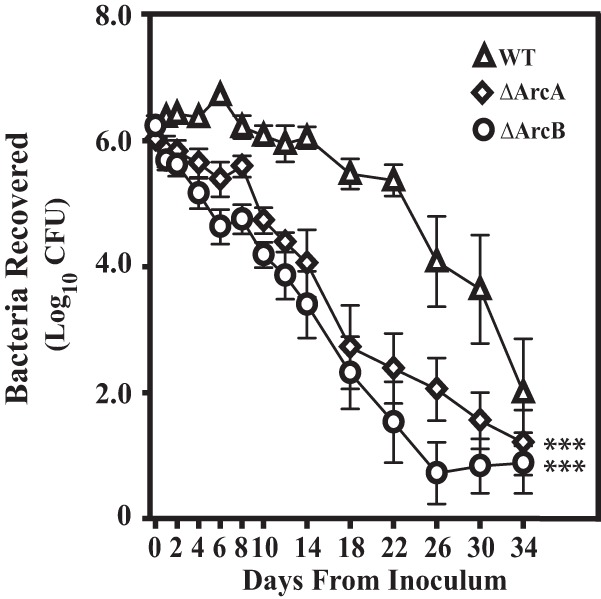
Arc mutants are attenuated in a model of mucosal colonization. Estrogenized C57BL/6J mice were vaginally inoculated on day 0 with 1 × 106 CFU of streptomycin-resistant wild-type S. pyogenes HSC12 (WT) or the ΔArcA or ΔArcB mutant. Vaginal washes were collected at the time points indicated and processed for determination of CFU. Each symbol represents the mean and standard error of the mean derived from 10 mice per group. A repeated-measures analysis indicated that recoveries of both the ΔArcA and ΔArcB mutants from vaginal washes were significantly attenuated compared to that of HSC12 (***, P < 0.0001).
The ΔArcA mutant is attenuated in a model of inflammatory soft tissue infection.
Given the influence of arginine in supporting asymptomatic colonization, we next wished to examine whether utilization of this amino acid contributes to symptomatic infection. Following inoculation into the subcutaneous tissue on the flank of a mouse, the HSC5 wild-type strain typically forms a localized lesion characterized by ulceration and formation of an eschar within 24 h. The lesion expands in size and reaches a peak area by day 3 postinoculation (30). Monitoring the lesion area formed by infection with both the wild type and the ΔArcA mutant revealed that for the 5-day period examined, the mutant formed significantly smaller lesions on all days monitored (Fig. 4A).
FIG 4.
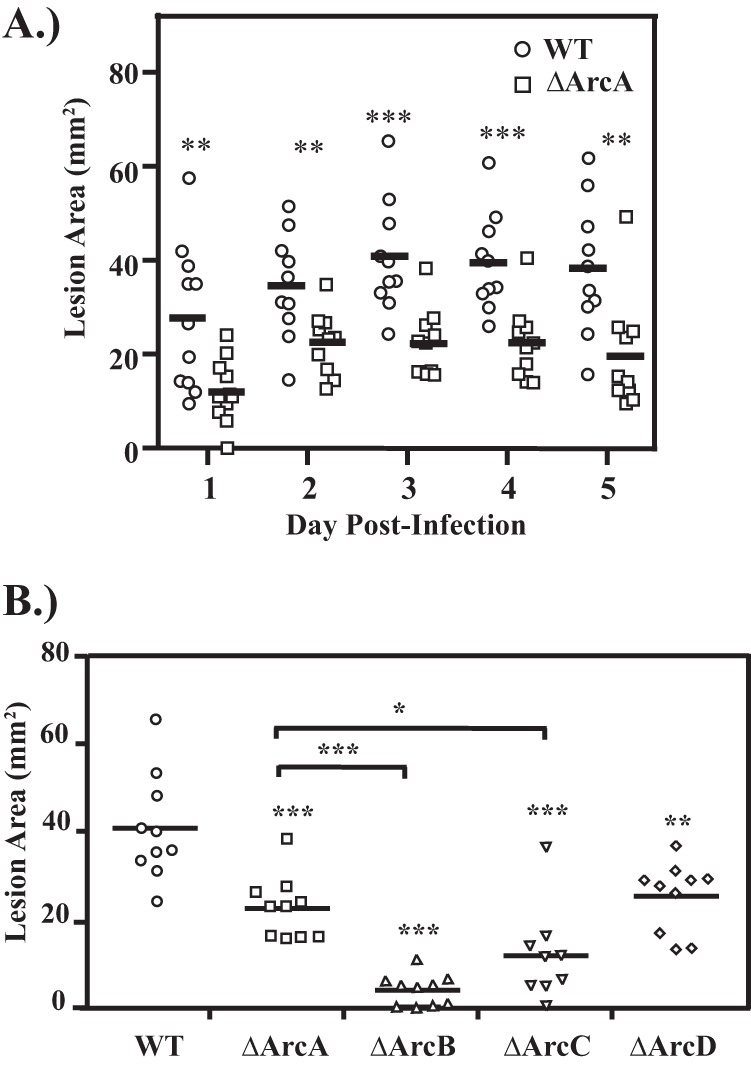
(A) Deletion of arcA disrupts virulence in S. pyogenes. Hairless SKH1 mice were infected subcutaneously with 107 CFU of wild-type (WT) or ArcA mutant (ΔArcA) S. pyogenes. Areas of the resulting ulcers were determined at the various time points indicated. Data shown are pooled from at least 2 independent experiments, with the mean indicated by a bar. (B) Deletion of individual arc genes influences virulence to various degrees. The virulence of strains with individual in-frame deletions in various genes of the Arc operon were compared to that of wild-type strain HSC5 (WT) using the murine subcutaneous infection model. Hairless SKH1 mice were inoculated with 107 CFU of each strain and the area of the resulting ulcer determined 3 days later. Each symbol plotted represents the value derived from an individual animal. Data shown are pooled from a minimum of 2 independent experiments, with the mean indicated by a bar. The asterisks indicate significance as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001).
The ΔArcB and ΔArcC mutants produce significantly smaller lesions than the ΔArcA mutant.
Given that these enzymes function in a discrete metabolic pathway, it was expected that deletion of genes both upstream and downstream of arcA would phenocopy the ΔArcA mutant, as seen in the murine model of mucosal colonization. This was observed for infection by the mutant lacking the arginine antiporter, which produced lesions equivalent to those produced by the ΔArcA mutant (Fig. 4B, ΔArcD). However, the ΔArcB and ΔArcC mutants were both hyperattenuated, forming significantly smaller lesions than the already attenuated lesions formed by the ΔArcA mutant (Fig. 4B). One explanation for this phenotype is that the loss of ArcB or ArcC results in the accumulation of intermediates detrimental to growth. In this case, deletion of an upstream gene should prevent the formation of any detrimental intermediates generated by a gene downstream. However, infection by a ΔArcA ΔArcB double mutant resulted in ΔArcB-like hyperattenuated lesions (Fig. 5A). Differences in virulence correlated to bacterial burden in tissue, with the number of CFU recovered from the ΔArcA mutant significantly less than that of the wild type and those from ΔArcB and the double mutant measurably less than that of the ΔArcA mutant (Fig. 5B). Thus, hyperattenuation was due to the lack of a functional ArcB and not due to the accumulation of intermediates. Taken together, these results suggest that attenuation likely resulted from the loss of an ability to utilize arginine while still retaining an ability to use citrulline, while hyperattenuation resulted from the loss of the ability to utilize both substrates.
FIG 5.
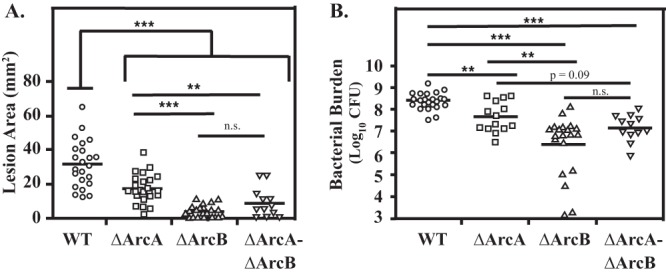
ΔArcA ΔArcB phenocopies ΔArcB. The virulence of mutants with an in-frame deletion in arcA (ΔArcA) or arcB (ΔArcB) was compared with those of the wild type (WT) and a mutant with a deletion of both genes (ΔArcA ΔArcB) using the murine subcutaneous infection model. Hairless SKH1 mice were inoculated with 107 CFU of each strain, and on day 3 the area of the resulting lesion was recorded (A) and the number of recoverable CFU determined (B). Data are pooled from a minimum of three independent experiments, with the bar indicating the mean for lesion area or geometric mean for bacterial burden. Each symbol plotted represents the value derived from an individual animal. Differences between each mutant compared to the wild type were tested for significance using the Mann-Whitney U test (**, P < 0.01; ***, P < 0.001).
iNOS is upregulated during an S. pyogenes infection.
Macrophages are often recruited to a site of active infection and have been demonstrated to be essential for controlling dissemination of S. pyogenes (23, 24). Macrophages target bacteria through a number of different bactericidal effectors, including NO·. However, if macrophages produce NO· in response to S. pyogenes, then both pathogen and host cells would be in competition for arginine. To examine the macrophage response to Streptococcus pyogenes, RAW 264.7 cells were challenged with S. pyogenes in vitro. Following infection with the wild-type strain at various MOIs, expression of iNOS was quantified using real-time RT-PCR. Analysis of transcript abundance revealed that iNOS expression increased in a dose-dependent manner, with an MOI of 1 eliciting expression comparable to that obtained with LPS, a known iNOS agonist (Fig. 6A). These results were confirmed by direct measurement of NO· following infection with S. pyogenes, which was produced to levels similar to those of LPS (Fig. 6A, inset), thus demonstrating that IFN-γ-stimulated macrophages can respond to S. pyogenes by producing NO·. To further validate the upregulation of iNOS in response to S. pyogenes, we next examined the relative expression of iNOS in the tissue surrounding the site of a subcutaneous infection. Comparison of iNOS transcripts from mice infected with S. pyogenes to those from mock-infected mice revealed an increase in transcript to levels comparable to those in in vitro infection of RAW 264.7 macrophages (Fig. 6A), demonstrating the upregulation of iNOS in the presence of S. pyogenes during a subcutaneous infection.
FIG 6.
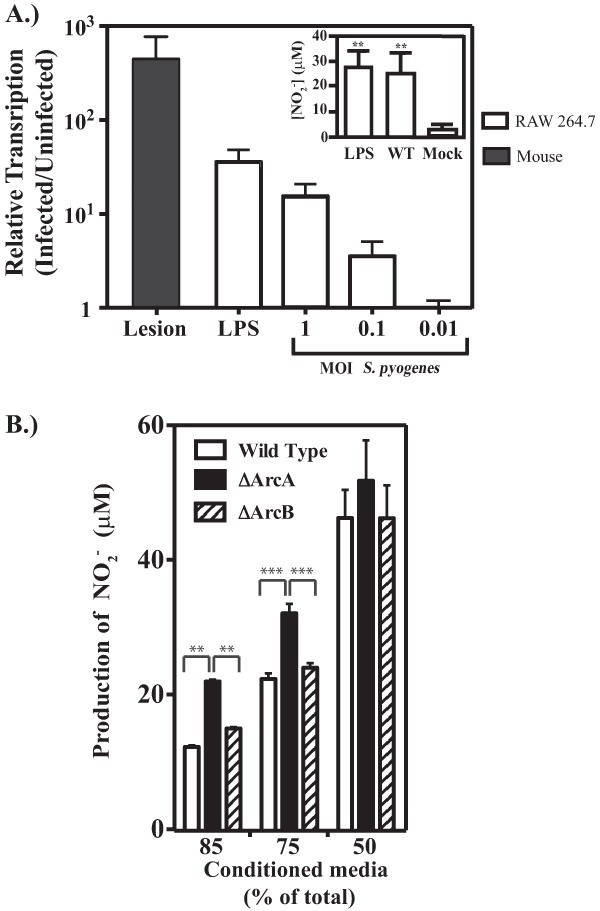
Activation of iNOS in response to S. pyogenes. (A) Cultured RAW 264.7 cells were stimulated with IFN-γ and then infected with wild-type strain HSC5 at the several MOIs indicated for comparison to LPS (1 μg/ml). Extracellular bacteria were removed after 60 min as described in Materials and Methods, and the abundance of the iNOS transcript was determined by real-time RT-PCR following an additional 3 h of incubation. Transcript abundance is reported relative to that detected in uninfected macrophages. For the black bar, SKH1 mice were inoculated with either wild-type HSC5 or 0.9% sodium chloride solution. At day 1 postinfection, samples were processed and analyzed. Transcript abundance is reported relative to that in mock-infected mice. Data represent the means and standard deviations of at least two independent experiments with samples analyzed in triplicate. The generation of NO· following infection (MOI = 1) was also determined in culture supernatant using the Griess reagent and compared to a standard curve (inset). (B) Influence of arginine catabolism on NO production. Cell-free-conditioned medium generated as described in Materials and Methods was added to wells with RAW 264.7 cells in combination with fresh cell culture medium. The cells were then stimulated with 1 μg/ml of LPS. Following 24 h of stimulation, the production of nitric oxide was measured. Data are presented as the means and standard deviations from at least two independent infections, each measured in triplicate. Asterisks indicate significance as follows: **, P < 0.01, and ***, P < 0.001. n.s., not significant.
NO· production by RAW 264.7 macrophages is influenced by S. pyogenes arginine catabolism.
Since S. pyogenes stimulated macrophage NO· production, it was of interest to examine how this activity may be influenced by S. pyogenes arginine metabolism. To look more specifically at arginine metabolism in the absence of macrophage death induced by unrestricted extracellular streptococcal growth and its associated acidification of media, NO· production in response to LPS stimulation was examined in pH-adjusted cell culture medium that had been preconditioned by growth of wild-type S. pyogenes or various Arc mutants. Given that other essential nutrients would also be depleted, conditioned medium was mixed with fresh medium at several different ratios. For all strains, a decrease in the concentration of conditioned medium resulted in a stepwise increase in macrophage NO· production, reaching a maximum at 50% conditioned medium (Fig. 6B). However, conditioned medium from the ΔArcA mutant supported significantly more NO· production than that derived from either the wild type or the ΔArcB mutant (Fig. 6B). Since they do not differ in growth rates, the only difference between the ΔArcA and ΔArcB mutants is that the latter still possesses the arginine-consuming enzyme arginine diminase. Thus, these data indicate that ArcA-mediated consumption of arginine by S. pyogenes can modulate macrophage NO· production.
ΔArcA mutant virulence is restored in iNOS−/− mice.
To further investigate the role of NO· in streptococcal disease, infection of iNOS−/− mice was conducted to determine if arginine utilization by S. pyogenes in vivo actively inhibited production of NO· via substrate depletion. Both wild-type and iNOS−/− C57BL/6 mice were infected subcutaneously with wild-type, ΔArcA, or ΔArcB S. pyogenes, and skin lesions were compared. No significant differences were observed between infections by wild-type S. pyogenes in wild-type or iNOS-deficient mice as assessed by lesion area or recoverable CFU (Fig. 7). Similarly, infection with ΔArcB S. pyogenes produced lesions that were equally attenuated in wild-type or mutant mice compared to those obtained with wild-type S. pyogenes. However, the ΔArcA mutant was significantly more virulent for iNOS−/− mice than for wild-type mice based on both lesion size (Fig. 7) and CFU recovered (Fig. 7). Thus, in the absence of iNOS, ArcA is not required for virulence, suggesting that its principal role is to inhibit production of NO· via depletion of its arginine substrate. In wild-type mice, the ΔArcA mutant cannot deplete arginine, resulting in its attenuation. Furthermore, since the virulence of the ΔArcB mutant is unaltered in iNOS−/− mice, its attenuation is due not to an increase in NO· production but rather to the disruption of citrulline catabolism. Taken together, these data support a role for citrulline catabolism in pathogenesis and demonstrate that arginine catabolism plays an important role in the manipulation of the innate immune response against S. pyogenes.
FIG 7.
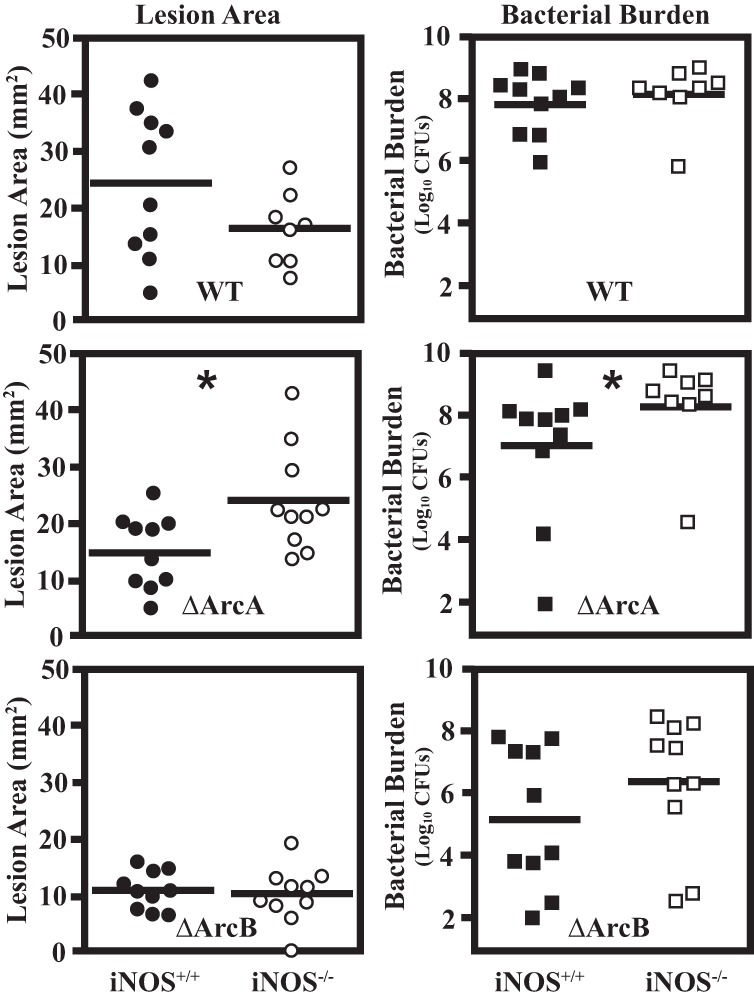
The ΔArcA mutant forms wild-type lesions in iNOS−/− mice. The murine subcutaneous infection model was used to evaluate the influence of iNOS on the course of infection in response to the wild type (WT) andΔArcA and ΔArcB mutants. Each S. pyogenes strain was used to infect both parental (iNOS+/+) and knockout (iNOS−/−) C57BL/6 mice. Lesion area was measured at day 3 postinfection, and tissue was excised for determination of recoverable CFU (bacterial burden). Data are pooled from at least two independent experiments. Each symbol plotted represents the value derived from an individual animal. The bar indicates the mean lesion area or geometric mean of bacterial CFU. For each S. pyogenes strain, differences in virulence observed between the parental and iNOS−/− mice were tested for significance using the Mann-Whitney U test (*, P < 0.05).
DISCUSSION
The ADI pathway has an interesting history in S. pyogenes. For example, ArcA was originally isolated as a protein with antitumor activity from cellular extracts of S. pyogenes and was known as streptococcal acid glycoprotein (35, 36). Studies that have characterized ArcA− mutants as having an altered ability to invade into host epithelial cells have implicated the pathway in pathogenesis. However, these analyses have reported both reduced (12) and enhanced (37) invasion, leaving its role unclear. In the present study, we examined a panel of mutants with mutations in the ADI pathway to show that arginine metabolism contributes to the ability of S. pyogenes to colonize the mucosa, that catabolism of both arginine and citrulline contributes to pathogenesis in soft tissue, and that an ability to consume arginine manipulates innate immunity.
Utilization of arginine and citrulline can aid in both the production of ATP, as well as through protection against acid stress (38). However, given that the abilities to use these two substrates both reside within the ADI pathway, these phenotypes are difficult to separate. In this regard, it is interesting that while the ΔArcA mutant was attenuated for mucosal colonization and in soft tissue infection, the ΔArcB mutant was hyperattenuated in cutaneous tissue. One possible explanation for this hyperattenutation could be the disruption of a secondary function of ArcB that resides outside arginine catabolism. Recently it has been demonstrated that deletion of arcD in S. pneumoniae has a negative impact on pathogenesis independent of its canonical function within the ADI pathway (39). However, given that deletion of arcC in S. pyogenes downstream of arcB was also hyperattenuated in the cutaneous model, the data presented are more consistent with a tissue-specific requirement for citrulline metabolism. Since utilization of citrulline in vitro does not increase growth yield but can contribute to acid stress resistance, the latter property is most likely to be of importance for growth in tissue. Consistent with this, we have utilized the panel of mutants constructed in this study to dissect the ADI pathway's contribution to acid stress resistance and found that the ΔArcB and ΔArcC mutants are unable to use citrulline for protection against acid stress and that while the ADI pathway's ability to produce ammonia contributes to resistance, its production of ATP is critical (Z. Cusumano and M. Caparon, submitted for publication). Histological analysis of murine cutaneous lesions has revealed that S. pyogenes grows confined within a large abscess-like cavity (40). The accumulation of a high density of bacteria undergoing fermentation would be expected to result in production of significant quantities of organic acids and a marked reduction in local pH. This is supported by analyses of the S. pyogenes transcriptome during murine soft tissue infection, which have revealed a signature characteristic of adaptation to an acidic environment (2). Taken together, these observations suggest that the ability to utilize citrulline is necessary to endure the acidic conditions experienced during a soft tissue infection.
Citrulline has long been recognized as an important metabolic intermediate in the urea cycle (41). However, since it is a nonessential amino acid that is not incorporated into proteins, it has received relatively little attention in pathogenesis. The observation that citrulline has a role in fine-tuning arginine homeostasis in response to diet has resulted in an appreciation that citrulline is a ubiquitous amino acid present in all tissues (42), and more recent studies have begun to explore how its ready availability may be exploited by pathogens. For example, in Francisella tularensis, catabolism of citrulline via the enzyme citrulline ureidase (43) produces ammonia that contributes to the inhibition of phagosomal maturation (44) and has also been implicated in inhibiting production of NO· through depletion of citrulline (31), which can be recycled to arginine by the host enzymes arginosuccinate synthase and arginosuccinate lyase (42) to produce a steady supply of arginine for iNOS (45).
Substrate depletion has emerged as a common mechanism by which pathogens inhibit the activity of iNOS. This can occur by pathogen upregulation of host arginase (46) or production of arginase by the pathogen itself (32). In the case of Helicobacter pylori, arginase also works in concert with bacterial urease to create ammonia to protect against acid stress (47). Similar to work with H. pylori, the work described here involves a dual purpose for arginine catabolism for protection against both acid stress and production of NO·. However, an important difference is that rather than using an arginase, S. pyogenes has adapted a catabolic pathway with additional roles in production of energy. In addition, we found that while arginine consumption was dispensable for virulence in iNOS−/− mice, citrulline utilization was still required. Furthermore, despite the fact that it lacks a functional ADI pathway, the ArcB mutant retained the ability to inhibit macrophage NO· production. Since this mutant retains a functional ArcA enzyme, it is likely that the ΔArcB mutant can still deplete arginine. Recently ArcA has been found to locate to the surface of S. pyogenes cells (48), suggesting a mechanism by which ArcA may gain access to extracellular arginine independent of a functional ADI pathway.
Pathogen manipulation of host arginine metabolism can have additional influences on pathogenesis. For example, pathogen-induced upregulation of host arginase also results in an increase in downstream metabolites, including ornithine, which is converted into polyamines such as spermidine and spermine (49). These metabolites can inhibit proinflammatory gene expression to enhance survival of some pathogens (50) and are also detrimental to certain pathogens, such as Staphylococcus aureus, that are sensitive to polycations (51). Interestingly, strains of S. aureus frequently associated with skin infections have a high association of carriage of the arginine catabolic mobile element (ACME), which, in addition to bearing genes necessary for arginine catabolism, also bears a polyamine resistance gene, speG, which has recently been shown to aid in a mouse model of staphylococcal skin infections (52). It will be of interest to determine how polycations may influence S. pyogenes's metabolism and how S. pyogenes's arginine catabolism may also influence host polycation production.
Arginine metabolism also had tissue-specific effects on S. pyogenes pathogenesis, as shown by comparing asymptomatic mucosal carriage versus inflammatory soft tissue infection. Tissue-specific effects on activation of host iNOS expression have also been reported for several other animal models of S. pyogenes infection. During intraperitoneal infection of mice, iNOS expression is not induced in macrophages at time points as late as 16 h postinfection (53). In contrast, an in vitro infection of bone marrow-derived macrophages demonstrated a Toll-like receptor 9 (TLR9)-dependent increase in NO· and reactive oxygen species in the presence of S. pyogenes (54). Additionally, serum NO· levels have also been shown to increase during an intravenous infection of mice (55). Examination of gene expression in the murine soft tissue infection model has also revealed both time-resolved alterations in the S. pyogenes transcriptome (56, 57) and host proinflammatory cytokine expression, with levels of some cytokines not reaching their peak until 48 h postinfection (24). Finally, in addition to the route of infection, iNOS activity has also been found to vary between mouse strains, suggesting an additional host genetic component in murine models (55). Together these experiments demonstrate that the host interaction with S. pyogenes is complex and likely multifactorial in terms of determining the ultimate final host response. In the current study, iNOS was found to be highly upregulated in murine tissue on day 1 following S. pyogenes infection, suggesting that during a cutaneous infection the host may attempt to upregulate iNOS to impede S. pyogenes outgrowth, as seen with other Gram-positive organisms (52, 58). However, depletion of arginine by S. pyogenes via the ADI pathway inhibits the production of NO·, rendering iNOS ineffective. This ineffectiveness is highlighted by the lack of a significant increase in virulence in an iNOS-deficient mouse compared to that in a wild-type mouse. Interestingly, examination of an arcA mutant revealed that inactivation of iNOS resulted in an increase in both lesion size and CFU compared to those in the wild-type mouse strain, further supporting our model in which the ability of iNOS to produce NO· is inhibited by wild-type S. pyogenes through substrate depletion. This unique phenotype reveals an ongoing discussion between the host and pathogen in terms of sharing available metabolites and reveals a unique outcome determined by the pathogen's metabolism. Further investigation into the host innate immune response during a soft tissue infection, and examination of the effect of metabolites on this response, will be essential in understanding the influence of the metabolism of S. pyogenes on host-pathogen interactions and pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gary Port for discussions and scrutiny of the manuscript.
This study was supported by Public Health Service grants NIH 5R01 AI064721 and 5R01 AI070759 from the National Institutes of Health.
Footnotes
Published ahead of print 21 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00916-13.
REFERENCES
- 1.Cunningham MW. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470–511. 10.1128/CMR.13.3.470-511.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loughman JA, Caparon M. 2006. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J. Bacteriol. 188:399–408. 10.1128/JB.188.2.399-408.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham MR, Virtaneva K, Porcella SF, Barry WT, Gowen BB, Johnson CR, Wright FA, Musser JM. 2005. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am. J. Pathol. 166:455–465. 10.1016/S0002-9440(10)62268-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham MR, Virtaneva K, Porcella SF, Gardner DJ, Long RD, Welty DM, Barry WT, Johnson CA, Parkins LD, Wright FA, Musser JM. 2006. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am. J. Pathol. 169:927–942. 10.2353/ajpath.2006.060112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter PD, Hill C. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429–453. 10.1128/MMBR.67.3.429-453.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdelal AT. 1979. Arginine catabolism by microorganisms. Annu. Rev. Microbiol. 33:139–168. 10.1146/annurev.mi.33.100179.001035 [DOI] [PubMed] [Google Scholar]
- 7.Arena ME, Manca de Nadra MC, Munoz R. 2002. The arginine deiminase pathway in the wine lactic acid bacterium Lactobacillus hilgardii X1B: structural and functional study of the arcABC genes. Gene 301:61–66. 10.1016/S0378-1119(02)01083-1 [DOI] [PubMed] [Google Scholar]
- 8.Zúñiga M, Champomier-Verges M, Zagorec M, Perez-Martinez G. 1998. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J. Bacteriol. 180:4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lüthi E, Baur H, Gamper M, Brunner F, Villeval D, Mercenier A, Haas D. 1990. The arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa contains an additional gene, arcD, encoding a membrane protein. Gene 87:37–43. 10.1016/0378-1119(90)90493-B [DOI] [PubMed] [Google Scholar]
- 10.Driessen AJ, Poolman B, Kiewiet R, Konings W. 1987. Arginine transport in Streptococcus lactis is catalyzed by a cationic exchanger. Proc. Natl. Acad. Sci. U. S. A. 84:6093–6097. 10.1073/pnas.84.17.6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsui R, Cvitkovitch D. 2010. Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol. 5:403–417. 10.2217/fmb.09.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Degnan BA, Fontaine MC, Doebereiner AH, Lee JJ, Mastroeni P, Dougan G, Goodacre JA, Kehoe MA. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68:2441–2448. 10.1128/IAI.68.5.2441-2448.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vrancken G, Rimaux T, De Vuyst L, Leroy F. 2008. Kinetic analysis of growth and sugar consumption by Lactobacillus fermentum IMDO 130101 reveals adaptation to the acidic sourdough ecosystem. Int. J. Food Microbiol. 128:58–66. 10.1016/j.ijfoodmicro.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 14.Wink DA, Kasprzak KS, Maragos CM, Elespuru RK, Misra M, Dunams TM, Cebula TA, Koch WH, Andrews AW, Allen JS. 1991. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science 254:1001–1003. 10.1126/science.1948068 [DOI] [PubMed] [Google Scholar]
- 15.Butler AR, Megson IL. 2002. Non-heme iron nitrosyls in biology. Chem. Rev. 102:1155–1166. 10.1021/cr000076d [DOI] [PubMed] [Google Scholar]
- 16.Radi R, Beckman JS, Bush KM, Freeman BA. 1991. Peroxynitrite-induced membrane lipid peroxidation: the cytotoxic potential of superoxide and nitric oxide. Arch. Biochem. Biophys. 288:481–487. 10.1016/0003-9861(91)90224-7 [DOI] [PubMed] [Google Scholar]
- 17.Radi R, Beckman JS, Bush KM, Freeman BA. 1991. Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J. Biol. Chem. 266:4244–4250 [PubMed] [Google Scholar]
- 18.Nathan C, Shiloh MU. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U. S. A. 97:8841–8848. 10.1073/pnas.97.16.8841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori M. 2007. Regulation of nitric oxide synthesis and apoptosis by arginase and arginine recycling. J. Nutr. 137:1616S–1620S [DOI] [PubMed] [Google Scholar]
- 20.Pautz A, Art J, Hahn S, Nowag S, Voss C, Kleinert H. 2010. Regulation of the expression of inducible nitric oxide synthase. Nitric Oxide 23:75–93. 10.1016/j.niox.2010.04.007 [DOI] [PubMed] [Google Scholar]
- 21.Bronte V, Zanovello P. 2005. Regulation of immune responses by L-arginine metabolism. Nat. Rev. Immunol. 5:641–654. 10.1038/nri1668 [DOI] [PubMed] [Google Scholar]
- 22.Das P, Lahiri A, Chakravortty D. 2010. Modulation of the arginase pathway in the context of microbial pathogenesis: a metabolic enzyme moonlighting as an immune modulator. PLoS Pathog. 6:e1000899. 10.1371/journal.ppat.1000899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldmann O, Rohde M, Chhatwal GS, Medina E. 2004. Role of macrophages in host resistance to group A streptococci. Infect. Immun. 72:2956–2963. 10.1128/IAI.72.5.2956-2963.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishalian I, Ordan M, Peled A, Maly A, Eichenbaum MB, Ravins M, Aychek T, Jung S, Hanski E. 2011. Recruited macrophages control dissemination of group A Streptococcus from infected soft tissues. J. Immunol. 187:6022–6031. 10.4049/jimmunol.1101385 [DOI] [PubMed] [Google Scholar]
- 25.Hanski E, Horwitz PA, Caparon MG. 1992. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect. Immun. 60:5119–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, Lai HS, Lin SP, Qian Y, Jia HG, Najar FZ, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton SW, Roe BA, McLaughlin R. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658–4663. 10.1073/pnas.071559398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez-Casal J, Price JA, Maguin E, Scott JR. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809–819. 10.1111/j.1365-2958.1993.tb01628.x [DOI] [PubMed] [Google Scholar]
- 28.Ruiz N, Wang B, Pentland A, Caparon M. 1998. Streptolysin O and adherence synergistically modulate proinflammatory responses of keratinocytes to group A streptococci. Mol. Microbiol. 27:337–346. 10.1046/j.1365-2958.1998.00681.x [DOI] [PubMed] [Google Scholar]
- 29.Horton RM, Cai ZL, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535 [PubMed] [Google Scholar]
- 30.Brenot A, King KY, Janowiak B, Griffith O, Caparon MG. 2004. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect. Immun. 72:408–413. 10.1128/IAI.72.1.408-413.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahawar M, Kirimanjeswara GS, Metzger DW, Bakshi CS. 2009. Contribution of citrulline ureidase to Francisella tularensis strain Schu S4 pathogenesis. J. Bacteriol. 191:4798–4806. 10.1128/JB.00212-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, Cheng Y, Mobley HL, Wilson KT. 2001. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. U. S. A. 98:13844–13849. 10.1073/pnas.241443798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bunce C, Wheeler L, Reed G, Musser J, Barg N. 1992. Murine model of cutaneous infection with gram-positive cocci. Infect. Immun. 60:2636–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson ME, Jr, Nielsen HV, Hultgren SJ, Caparon MG. 2013. A murine vaginal colonization model for investigating asymptomatic mucosal carriage of Streptococcus pyogenes. Infect. Immun. 81:1606–1617. 10.1128/IAI.00021-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higuchi Y, Shoin S. 1988. Purification and characterization of a novel cytotoxic substance from cell-free extract of Streptococcus pyogenes. Biochim. Biophys. Acta 966:239–247. 10.1016/0304-4165(88)90117-1 [DOI] [PubMed] [Google Scholar]
- 36.Yoshida J, Takamura S, Nishio M. 1998. Characterization of a streptococcal antitumor glycoprotein (SAGP). Life Sci. 62:1043–1053. 10.1016/S0024-3205(97)01142-9 [DOI] [PubMed] [Google Scholar]
- 37.Marouni MJ, Ziomek E, Sela S. 2003. Influence of group A streptococcal acid glycoprotein on expression of major virulence factors and internalization by epithelial cells. Microb. Pathog. 35:63–72. 10.1016/S0882-4010(03)00094-9 [DOI] [PubMed] [Google Scholar]
- 38.Cotter PD, Hill C. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429–453. 10.1128/MMBR.67.3.429-453.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta R, Yang J, Dong Y, Swiatlo E, Zhang JR, Metzger DW, Bai G. 2013. Deletion of arcD in Streptococcus pneumoniae D39 impairs its capsule and attenuates virulence. Infect. Immun. 81:3903–3911. 10.1128/IAI.00778-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang SO, Caparon MG, Cho KH. 2010. Virulence gene regulation by CvfA, a putative RNase: the CvfA-enolase complex in Streptococcus pyogenes links nutritional stress, growth-phase control, and virulence gene expression. Infect. Immun. 78:2754–2767. 10.1128/IAI.01370-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romero MJ, Platt DH, Caldwell RB, Caldwell RW. 2006. Therapeutic use of citrulline in cardiovascular disease. Cardiovasc. Drug Rev. 24:275–290. 10.1111/j.1527-3466.2006.00275.x [DOI] [PubMed] [Google Scholar]
- 42.Curis E, Nicolis I, Moinard C, Osowska S, Zerrouk N, Benazeth S, Cynober L. 2005. Almost all about citrulline in mammals. Amino Acids 29:177–205. 10.1007/s00726-005-0235-4 [DOI] [PubMed] [Google Scholar]
- 43.Fleming DE, Foshay L. 1955. Studies on the physiology of virulence of Pasteurella tularensis. I. Citrulline ureidase and deamidase activity. J. Bacteriol. 70:345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, Garcia E, Halltorp G, Johansson D, Isherwood KE, Karp PD, Larsson E, Liu Y, Michell S, Prior J, Prior R, Malfatti S, Sjostedt A, Svensson K, Thompson N, Vergez L, Wagg JK, Wren BW, Lindler LE, Andersson SG, Forsman M, Titball RW. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153–159. 10.1038/ng1499 [DOI] [PubMed] [Google Scholar]
- 45.Bryk J, Ochoa JB, Correia MI, Munera-Seeley V, Popovic PJ. 2008. Effect of citrulline and glutamine on nitric oxide production in RAW 264.7 cells in an arginine-depleted environment. J. Parenter. Enteral Nutr. 32:377–383. 10.1177/0148607108319807 [DOI] [PubMed] [Google Scholar]
- 46.Lahiri A, Das P, Chakravortty D. 2008. Arginase modulates Salmonella induced nitric oxide production in RAW264.7 macrophages and is required for Salmonella pathogenesis in mice model of infection. Microbes Infect. 10:1166–1174. 10.1016/j.micinf.2008.06.008 [DOI] [PubMed] [Google Scholar]
- 47.McGee DJ, Radcliff FJ, Mendz GL, Ferrero RL, Mobley HL. 1999. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J. Bacteriol. 181:7314–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henningham A, Chiarot E, Gillen CM, Cole JN, Rohde M, Fulde M, Ramachandran V, Cork AJ, Hartas J, Magor G, Djordjevic SP, Cordwell SJ, Kobe B, Sriprakash KS, Nizet V, Chhatwal GS, Margarit IY, Batzloff MR, Walker MJ. 2012. Conserved anchorless surface proteins as group A streptococcal vaccine candidates. J. Mol. Med. 90:1197–1207 [DOI] [PubMed] [Google Scholar]
- 49.Kepka-Lenhart D, Mistry SK, Wu G, Morris SM., Jr 2000. Arginase I: a limiting factor for nitric oxide and polyamine synthesis by activated macrophages? Am. J. Physiol. Regul. Integr. Comp. Physiol. 279:R2237–R2242 [DOI] [PubMed] [Google Scholar]
- 50.Bussière FI, Chaturvedi R, Cheng Y, Gobert AP, Asim M, Blumberg DR, Xu H, Kim PY, Hacker A, Casero RA, Jr, Wilson KT. 2005. Spermine causes loss of innate immune response to Helicobacter pylori by inhibition of inducible nitric-oxide synthase translation. J. Biol. Chem. 280:2409–2412 [DOI] [PubMed] [Google Scholar]
- 51.Joshi SG, Paff M, Friedman G, Fridman G, Fridman A, Brooks AD. 2010. Control of methicillin-resistant Staphylococcus aureus in planktonic form and biofilms: a biocidal efficacy study of nonthermal dielectric-barrier discharge plasma. Am. J. Infect. Control 38:293–301 [DOI] [PubMed] [Google Scholar]
- 52.Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107. 10.1016/j.chom.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldmann O, von Kockritz-Blickwede M, Holtje C, Chhatwal GS, Geffers R, Medina E. 2007. Transcriptome analysis of murine macrophages in response to infection with Streptococcus pyogenes reveals an unusual activation program. Infect. Immun. 75:4148–4157. 10.1128/IAI.00181-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zinkernagel AS, Hruz P, Uchiyama S, von Kockritz-Blickwede M, Schuepbach RA, Hayashi T, Carson DA, Nizet V. 2012. Importance of Toll-like receptor 9 in host defense against M1T1 group A Streptococcus infections. J. Innate Immun. 4:213–218. 10.1159/000329550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldmann O, Chhatwal GS, Medina E. 2003. Immune mechanisms underlying host susceptibility to infection with group A streptococci. J. Infect. Dis. 187:854–861. 10.1086/368390 [DOI] [PubMed] [Google Scholar]
- 56.Kietzman CC, Caparon MG. 2010. CcpA and LacD. 1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect. Immun. 78:241–252. 10.1128/IAI.00746-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kietzman CC, Caparon MG. 2011. Distinct time-resolved roles for two catabolite-sensing pathways during Streptococcus pyogenes infection. Infect. Immun. 79:812–821. 10.1128/IAI.01026-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gantwerker EA, Hom DB. 2011. Skin: histology and physiology of wound healing. Facial Plast. Surg. Clin. North Am. 19:441–453. 10.1016/j.fsc.2011.06.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


