Abstract
Several intracellular pathogens, including a key etiological agent of chronic periodontitis, Porphyromonas gingivalis, infect blood myeloid dendritic cells (mDCs). This infection results in pathogen dissemination to distant inflammatory sites (i.e., pathogen trafficking). The alteration in chemokine-chemokine receptor expression that contributes to this pathogen trafficking function, particularly toward sites of neovascularization in humans, is unclear. To investigate this, we utilized human monocyte-derived DCs (MoDCs) and primary endothelial cells in vitro, combined with ex vivo-isolated blood mDCs and serum from chronic periodontitis subjects and healthy controls. Our results, using conditional fimbria mutants of P. gingivalis, show that P. gingivalis infection of MoDCs induces an angiogenic migratory profile. This profile is enhanced by expression of DC-SIGN on MoDCs and minor mfa-1 fimbriae on P. gingivalis and is evidenced by robust upregulation of CXCR4, but not secondary lymphoid organ (SLO)-homing CCR7. This disruption of SLO-homing capacity in response to respective chemokines closely matches surface expression of CXCR4 and CCR7 and is consistent with directed MoDC migration through an endothelial monolayer. Ex vivo-isolated mDCs from the blood of chronic periodontitis subjects, but not healthy controls, expressed a similar migratory profile; moreover, sera from chronic periodontitis subjects expressed elevated levels of CXCL12. Overall, we conclude that P. gingivalis actively “commandeers” DCs by reprogramming the chemokine receptor profile, thus disrupting SLO homing, while driving migration toward inflammatory vascular sites.
INTRODUCTION
The “keystone” (1) anaerobic mucosal pathogen Porphyromonas gingivalis is a main etiological agent of chronic periodontitis, a disease that affects more than one-half of the adult U.S. population (2). Chronic periodontitis is characterized by formation of a biofilm that triggers host-mediated tissue and bone destruction and increases the overall risk for several inflammatory diseases (3). Treatment involves the debridement and removal of the pathogenic biofilm that continually builds over time (4). P. gingivalis is not eliminated from these infections, but rather induces reoccurring bouts of low-grade inflammation, which further increases tissue destruction (5). It is not presently clear how this pathogen evades elimination by the adaptive immune system.
Dendritic cells (DCs) are a unique group of leukocytes that are critical for antigen clearance and for mounting efficient adaptive immune responses (6). Circulating blood DCs generally are divided into two subpopulations, the plasmacytoid (pDC) and myeloid (mDC) lineages. Specifically, mDCs are believed to be important in both the clearance and processing of bacterial pathogens and apoptotic cells, along with the associated antigen display (7, 8). Blood mDCs navigate in and out of tissues to survey the environment and control homeostatic and immune responses (9). Proper homing of DCs is dependent on their maturation state and the resulting profile of chemokine receptors expressed (10, 11). DC migration is highly regulated and crucial for host immune maintenance (12). Dysregulation of immune cell migration impairs immune response and contributes to human inflammatory diseases (13, 14), including asthma (15), periodontitis (16, 17), and coronary artery disease (18, 19).
DCs are highly migratory but lack robust bactericidal capabilities (20, 21). Our group has previously reported that P. gingivalis takes advantage of these DC characteristics by infecting DCs and being routed into a protective niche that shields P. gingivalis from more-efficient extracellular killing (22). Optimal infection of DCs by P. gingivalis is dependent on its glycosylated minor (mfa-1) fimbrial adhesin, which targets the c-type lectin dendritic cell specific ICAM-3 grabbing nonintegrin (DC-SIGN) for entry (23). Previously, this has been shown to mediate suboptimal DC maturation and modulate the effector response from a proinflammatory Th1-biased to a Th2-biased response (24). Another aspect of maturation is the shift in chemokine receptor expression that drives directed migration of DCs toward secondary lymphoid organs (SLO). This is particularly pertinent in view of evidence that P. gingivalis disseminates from its oral mucosal niche to distant sites, such as the respiratory tract (25), atherosclerotic plaques (26, 27), arthritic lesions (28), and the gut (29). We have shown that blood mDCs disseminate P. gingivalis to atherosclerotic plaques (22), but the mechanisms are not clear.
A major control point for DC migration is their activation and maturation status. This is initiated upon migration of immature DCs into peripheral tissues, in a process controlled by chemokine receptor-ligand pairs CCR2-CCL2, CCR5-CCL5 (RANTES), and CCR6-CCL20 (MIP3α). As DCs migrate and acquire antigen, CCR2, CCR5, and CCR6 are downregulated (30), triggering efflux from peripheral tissues. Further maturation upregulates CCR7 to facilitate SLO homing (31) toward CCL19/21 (MIP3β, 6Ckine), which guide DCs through afferent lymphatics into T cell-rich zones of draining lymph nodes. CCR7 facilitates DC interaction with naive or antigen-specific T cells (32). DCs can then present antigen to T lymphocytes (33) and dictate the magnitude and specificity of immune responses. The lack of CCR7 expression disrupts SLO homing of DCs (34). CXCR4, in contrast, is involved in proangiogenic mobilization (35) into peripheral blood (36). It is broadly expressed on hematopoietic stem cells and vascular endothelial cells (37) and is expressed on early-stage DCs (38). CXCR4 is highly expressed on metastasizing cancer cells (39). In animal models, specific blocking of CXCR4 prevents tumor neovascularization (40). A major characteristic of chronic periodontitis is extensive vascular remodelling and angiogenesis (41). Both CXCR4 (42) and CXCL12 (43, 44) have been found to be elevated locally in the tissues during chronic periodontitis.
We show here, using wild-type or conditional knockout P. gingivalis strains that are cultured in media containing selective antibiotics to prevent expression of DC adhesive fimbriae, that P. gingivalis infection of monocyte-derived DCs (MoDCs) is fimbria dependent and disrupts functional maturation of DCs in vitro, as evidenced by robust upregulation of CXCR4, but not CCR7. This phenotype is consistent with migratory function of infected MoDCs through a membrane or through internal thoracic aortic endothelial cells (ITECs) in response to the respective chemokine. Blood mDCs isolated from patients with chronic periodontitis express a similar migratory phenotype. Moreover, sera from chronic periodontitis subjects express elevated levels of CXCR4 ligand CXCL12 compared to those of healthy controls. These data, combined with our previously published evidence for P. gingivalis dissemination within mDCs to coronary artery plaques (22), provide a mechanistic framework for how this intracellular pathogen directs its dissemination in DCs in vivo.
MATERIALS AND METHODS
Patient study population.
Analysis of patient peripheral blood mDCs was conducted with approval from the Office of Human Research Protection (OHRP) at Georgia Regents University. The cohort of chronic periodontitis subjects consisted of 8 subjects with moderate to severe disease as determined by exhibition of the following: probing depth of >4 mm, attachment loss of >3 mm, bleeding on probing, and alveolar bone crest >3 mm from the cementoenamel junction. Protocols for healthy mDC and sera were approved by the Committee on Research Involving Human Subjects (CORIHS) at Stony Brook University. The study population and demographics have been previously outlined (22).
Bacterial culture growth conditions, bacterial labeling, and MoDC infection.
Wild-type (WT) Pg381, which expresses both minor (min) and major (maj) fimbriae (Pg min+/maj+), isogenic major fimbria-deficient mutant DPG3, which expresses only the minor fimbriae (Pg min+/maj−), and the double fimbria mutant MFB (Pg min−/maj−) were maintained anaerobically (10% H2, 10% CO2, and 80% N2) at 37°C (45, 46) in Acumedia Wilkins-Chalgren anaerobe broth. Erythromycin (5 μg/ml) and tetracycline (2 μg/ml) were added according to the selection requirements of the strains (47). Bacteria were washed once in phosphate-buffered saline (PBS) and stained with carboxyfluorescein succinimidyl ester (CFSE) (eBioscience) as described for flow cytometry (48). Briefly, bacteria in PBS were stained with CFSE at a final concentration of 5 μM for 30 min at 37°C and protected from light. The bacterial suspension was washed three times in PBS and resuspended to an optical density at 660 nm (OD660) of 0.11, which had previously been determined to be equal to 5 × 107 CFU (49). MoDCs were pulsed with P. gingivalis strains at a multiplicity of infection (MOI) of 1 for 24 h. Low MOI values were used to mimic a natural infection as well as to avoid overwhelming the host response (22). Mock infections did not utilize any microbial compound and consisted of PBS alone. For positive controls, cells were treated with 100 nM Escherichia coli lipopolysaccharide (LPS) (Sigma L4391) and 10 nM tumor necrosis factor alpha (TNF-α) (Sigma T6674) or whole, inactivated E. coli (Life Technologies product number P35361).
Generation of immature MoDCs.
The Committee of Research Involving Human Subjects (CORIHS) at Stony Brook University approved all studies involving human blood samples from healthy volunteers. Written informed consent was obtained from all volunteers, and all research was conducted in accordance with the guidelines provided by the Declaration of Helsinki. MoDCs were generated as we have described previously (50, 51). Briefly, monocytes were isolated from mononuclear fractions of the peripheral blood of healthy donors and seeded in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) (100 ng/ml) and interleukin-4 (IL-4) (25 ng/ml) at a concentration of 1 × 105 to 2 × 105 cells/ml in RPMI 1640 (Cellgro) containing 10% HI-FBS (Lonza) and antibiotic/antimycotic (HyClone) for 5 to 7 days, after which flow cytometry was performed to confirm the immature DC phenotype (CD14loCD83−CD1c+DC-SIGN+). Cell surface markers of DCs were evaluated by four-color immunofluorescence staining with the following monoclonal antibodies (MAbs): eBiosciences CD83 (product number 11-0839-42), CD14 (17-0149-42), and CD209 (45-2099042) and MACS CD1c (130-090-507). After 5 to 7 days of culture, monocytes displayed 95 to 99% differentiation into MoDCs.
Flow cytometry gating strategies.
All flow samples were analyzed using an Accuri C6 flow analyzer and with CFlow Plus analysis software. Gates were chosen using suspension cell forward scatter and side scatter characteristics. Nonadherent MoDCs typically fall within a forward scatter height (fsc-h) of 300 to 600 and a side scatter height (ssc-h) of 100 to 200 (× 1,000). Antibody staining was determined by running an unstained control and single-stain controls to determine necessary compensation. Undifferentiated monocytes, lymphocyte carryover, and debris were excluded from analysis. All cells were blocked in 5% bovine serum albumin (BSA) containing Human FcR blocker (MACS 130-059-901), and single-stained antibodies were used for compensation.
Analysis of surface migration receptors.
Cells were prepared for analysis by flow cytometry as previously described. Levels of CCR2, CCR5, CCR6, CXCR4, and CCR7 expression were determined using the following MAbs: R&D Systems CCR2 (FAB151C), BD CCR5 (555992), and eBioscience CXCR4 (12-9999-42), CCR6 (12-1969-42), and CCR7 (17-1979-42). DCs were gated based on size and scatter characteristics and analyzed for receptor expression. For the percent positive DC populations, negative and positive populations were determined using unstained and single-stained controls, and cells falling into the positive populations were represented as the percentage of total cells. For evaluation of mean fluorescence intensity (MFI), the MFI of each DC population was reported after applying compensation from single stains.
Chemotaxis and endothelial migration assays.
For endothelial migration assays, 6.5-mm transwell inserts with 5-μm pore size (Costar product number 3421) were used in 24-well plates. The bottom reservoir of these plates contained 600 μl of RPMI medium (Costar) with either 100 ng of CCL19 or CXCL12 or 1 μg of CCL19 for forced matured MoDCs (PeproTech). Before ITEC seeding, transwell inserts were coated with 5 μg/ml fibronectin (Sigma) for 45 min and allowed to dry. ITECs were then seeded in the inserts at a concentration of 1 × 105 ITECs/100 μl serum-free medium (Cell Applications catalog no. 113PR) and grown as a monolayer for 24 h. To confirm confluence of ITECs, inserts were monitored by inverted light microscopy (Evos XL Core). After 24 h, medium was removed, and P. gingivalis-infected MoDCs were loaded into the upper chambers at a concentration of 1 × 105 MoDCs in 100 μl of medium and allowed to migrate for 3 h at 37°C and 5% CO2. After 3 h, bottoms of the inserts were washed with ice-cold PBS to remove migrated but attached cells, and the reservoir medium was collected. The migrating cells were counted by flow cytometry for a fixed volume of 100 μl at a constant rate. The number of cells migrating was determined using the following equation: (number of treated cells migrating) − (number of untreated cells migrating). DC migration distance was determined using a 48-well microchemotaxis chamber (P48; NeuroProbe) (52). Migration distance (μm) was determined with leading-edge analysis after hematoxylin staining by using a 40× objective lens and subtracting migration distance without chemokine from migration distance with chemokine. MoDCs were infected as in the flow experiments described above. Cells were washed and resuspended in 0.5% BSA at a concentration of 1 × 105 cells/50 μl. Cells were loaded into upper compartments and allowed to migrate through nitrocellulose filters (SCB8; NeuroProbe) for 3 h. Chemoattractants CXCL12 and MIP3β were also resuspended in 0.5% BSA and loaded into the upper chamber with MoDCs or in the bottom compartment as indicated. The synthetic compound AMD3100 (Tocris) was used at a final concentration of 25 μM to block CXCR4 on MoDCs (53). Filters were stained using Harris modified hematoxylin (SH30-500D; Fisher), and leading-edge migration depths were determined under a 40× objective lens (54). All samples were loaded and assays performed in triplicate.
CXCL12 gene expression.
ITECs were plated at 1.0 × 105 in 6-well plates and were infected with Pg381 at MOIs of 0.1, 1, 10, and 100 for 24 h in serum-free medium (catalog number 113PR; Cell Applications). After 24 h, supernatants were collected and cells were used for RNA isolation and quantitative PCR (qPCR). For qPCR analysis, RNA was isolated from ITECs (catalog number 74134; Qiagen), and mRNA was quantified by One-Step qPCR (product number A10316; Invitrogen) on a StepOne Plus thermocycler (AP Biosystems) with StepOne software v2.2.2. CXCL12 (TaqMan Hs00171022_m1) primers were used to detect respective gene amplification.
CXCL12 ELISA.
Human umbilical vein endothelial cells (HUVEC) were isolated via collagenase perfusion of umbilical vein as previously described (55) and plated at 1 × 105 cells/ml of serum-free EBM-2 medium (CC-3156; Lonza) in 6-well plates. HUVEC were treated with Pg381, DPG-3, MFB, or 100 ng/ml IL-1β for 24 h in serum-free medium before culture supernatants were collected. Supernatants from cell cultures were collected, centrifuged to remove extracellular debris, and measured using a CXCL12 enzyme-linked immunosorbent assay (ELISA) kit (product number 100637; Abcam) according to the manufacturer's instructions. ITECs from the above-described treatments and HUVECs were each measured in triplicate on a microplate reader (Epoch; BioTek) at a wavelength of 450 nm with a correction wavelength of 540 nm. Samples were run in triplicate alongside standards to calculate secreted protein in pg/ml. The means (± standard errors of the means [SEM]) were compared using the Holm-Sidak unpaired Student t test to find significant differences among the various populations.
Statistical analysis.
For statistical analyses of receptor expression, experimental samples were compared with uninfected controls or between P. gingivalis strains of infection using unpaired Student t tests of the means from at least 5 different flow experiments. Statistical significance was calculated using GraphPad Prism 6 software, and only results with a P value of <0.05 were reported as significant (α, P < 0.05; *, P < 0.01). Holm-Sidak unpaired t tests were used for the comparison of multiple populations of treatments for in vitro experiments. All in vitro experiments are from at least 3 individual experiments. A multiple-comparison two-way analysis of variance (ANOVA) test was performed for ex vivo experiments comparing chronic periodontitis to healthy control patient groups.
RESULTS
P. gingivalis-infected MoDCs do not express mature chemokine receptors.
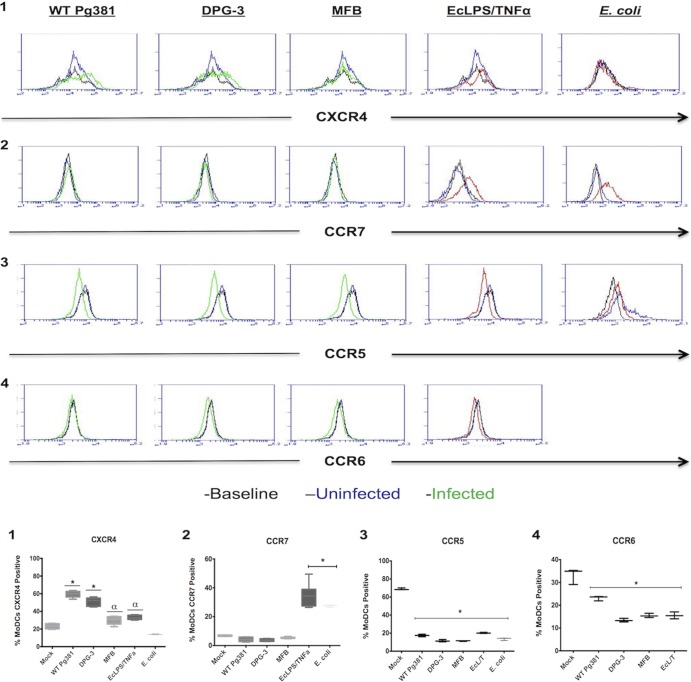
Surface protein expression of CXCR4, CCR7, CCR5, and CCR6 were analyzed on human MoDCs after treatment with P. gingivalis strains. The strains (green trace) included the following: DPG-3, which only expresses the DC-SIGN-targeting mfa-1 fimbrial adhesin; Pg381, a wild-type strain that expresses both fimA and mfa-1; and MFB, a double fimbria mutant. Fimbria-mediated P. gingivalis invasion of MoDCs has been shown to be highly efficient, while nonfimbriated uptake, as with strain MFB, is close to zero (56). For positive controls (red trace), we utilized a cocktail of E. coli LPS/TNF-α or whole, inactivated E. coli. Other intracellular pathogens that target DC-SIGN (57–59) disrupt DC maturation by dampening accessory molecule upregulation (60, 61), but the effects on DC migration have not been reported. A multiplicity of infection of 1 was used for these experiments to reproduce the natural infection rate of human blood mDCs in patients with chronic periodontitis in vivo (22). Summary data are provided as the percentages of MoDCs expressing the indicated chemokine receptor, as we find that the populations of MoDCs highly upregulate CXCR4 but do not have enhanced expression on a cell-to-cell basis. Both Pg381 and DPG-3 induce a more robust upregulation of CXCR4 than the nonfimbriated MFB strain (Fig. 1, panel 1), without significant differences observed between Pg381 and DPG-3. None of the P. gingivalis strains induced CCR7 upregulation on MoDCs; however, MoDCs are capable of CCR7 upregulation in response to either E. coli LPS/TNF-α or whole E. coli (Fig. 1, panel 2). All treatments resulted in decreased CCR5 and CCR6 expression (Fig. 1, panels 3 and 4), albeit slightly lower CCR6 downregulation was observed in the Pg381-treated MoDCs.
FIG 1.
P. gingivalis-infected MoDCs do not express mature chemokine receptors. MoDCs were infected for 24 h with WT Pg381, fimbria-deficient DPG-3 (mfa-1+), nonfimbriated MFB, or fixed E. coli at an MOI of 1 or stimulated with a cocktail of E. coli LPS and TNF-α. Representative histogram plots are shown for each of the analyzed chemokine receptors, with percentage-based cumulative data shown in the lower part of the figure. Baseline expression of MoDCs at the time of infection is indicated with a black trace, uninfected MoDCs are indicated with a blue trace, and infected MoDCs are indicated with a green trace. Our controls of whole E. coli or DC maturation stimuli E. coli LPS and TNF-α are indicated by a red trace (panel 1). Mfa-1+ strains (WT, DPG-3) caused a more significant upregulation of CXCR4 than other treatments. E. coli LPS/TNF-α and E. coli treatment did not cause CXCR4 upregulation (panel 2). P. gingivalis did not cause significant upregulation of CCR7 as in controls (panels 3 and 4). Treatments resulted in a significant downregulation of CCR5 and CCR6. α, P < 0.05; *, P < 0.01.
CFSE-stained intracellular P. gingivalis in MoDCs correlates with increased CXCR4 but not CCR7.
To determine if surface CXCR4 expression in MoDCs correlates with bacterial burden, strains were fluorescently labeled with CFSE before infections (Fig. 2). As previously reported, fimbriated strains of P. gingivalis show much higher uptake in MoDCs than the nonfimbriated strain. MoDCs with visible P. gingivalis uptake (green dots) had high expression of CXCR4 (red dots) relative to basal levels (Fig. 2A and B). The nonfimbriated MFB P. gingivalis or the inflammatory cocktail did cause some CXCR4 upregulation over the control level, but this was significantly less than the fimbriated P. gingivalis infections (Fig. 2B). In Fig. 2C, the same CXCR4+ and CFSE-Pg+ MoDC populations were analyzed for CCR7 upregulation. We observed that CXCR4-expressing, P. gingivalis-infected cells (green and red dots) have a slight increase in CCR7 (Fig. 2C). Only control MoDCs treated with the inflammatory cocktail, which were not infected with P. gingivalis, showed a significant upregulation of CCR7 (Fig. 2D). Taken together, these results showed that P. gingivalis infection drives CXCR4 expression in MoDCs, but not CCR7 expression.
FIG 2.
CXCR4 and CCR7 MoDC surface expression in CFSE-positive P. gingivalis infections. MoDCs were infected for 24 h with CFSE-labeled WT Pg381, fimbria-deficient DPG-3 (mfa-1+), and nonfimbriated MFB at an MOI of 1 or stimulated with E. coli LPS and TNF-α. (A) Representative scatter graphs show that bacterial uptake is highest in live, fimbriated P. gingivalis (green and red), and this correlates with CXCR4 upregulation (red). Nonfimbriated MFB is not efficiently taken up by MoDCs, and the subsequent CXCR4 levels are only slightly increased from control levels. (B) Cumulative data from panel A are shown, indicating CXCR4 expression of infected cells. The control E. coli LPS- and TNF-α-treated MoDCs are shown but are not infected with Pg. WT Pg381 and DPG-3 P. gingivalis infection leads to CXCR4 upregulation that is increased significantly over controls. (C) Representative scatter graphs show that P. gingivalis-infected MoDCs (green and red) do not upregulate CCR7 simultaneously with CXCR4, as cells appear only in the upper left quadrant of the positive control. (D) Cumulative data from panel C are shown, indicating CCR7 expression of infected cells. Significant CCR7 upregulation is observed in our positive control. α, P < 0.05; *, P < 0.01; ns, nonsignificant.
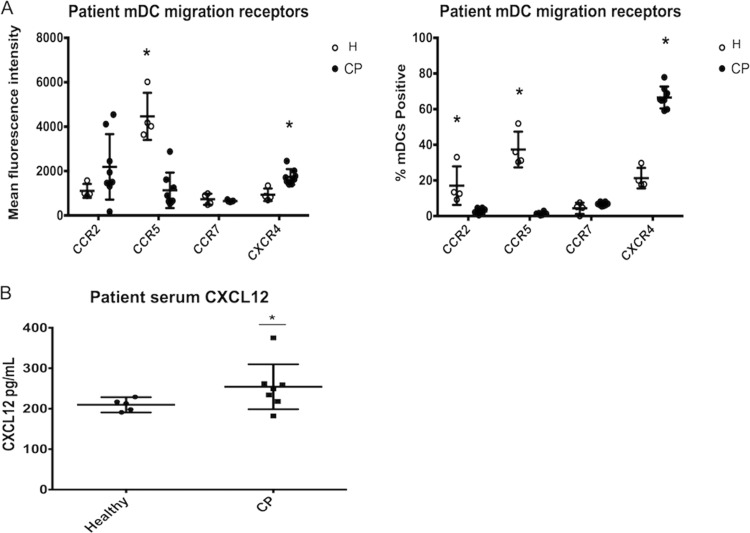
Chronic periodontitis patient blood mDCs express an angiogenic migratory phenotype, and chronic periodontitis sera have increased levels of CXCL12.
To determine the in vivo relevance of this chemokine receptor profile, we analyzed peripheral blood mDCs from patients with chronic periodontitis. At the time of patient mDC isolation, antibody to CCR2 was available, so we analyzed CCR2 expression, as opposed to CCR6, as a similar mediator of immature mDC recruitment into inflamed tissues toward chemokine gradient (55, 62). Relative to healthy controls, the chronic periodontitis patients' mDCs expressed significantly lower levels of the immature receptors CCR2 and CCR5, did not have significant changes in the levels of CCR7 expression, and displayed significantly elevated CXCR4 expression (Fig. 3A). Interestingly, CCR2 is expressed on a lower number of mDCs, but at higher intensity, in the chronic periodontitis group. As CCR2 is expressed slightly earlier in sequence than CCR6, this could suggest that chronic inflammation or bacteremia may lead to slight, but attenuated, maturation of circulating DCs. We next analyzed patient sera and found that CXCR4 ligand CXCL12 was elevated in chronic periodontitis sera compared to healthy controls (Fig. 3B).
FIG 3.
Chronic periodontitis mDCs express an angiogenic migratory profile, and chronic periodontitis sera have elevated CXCL12. Patient peripheral blood samples were collected, and blood mDCs were purified by automated separation. The mean fluorescence intensity of chronic periodontitis patient mDCs (CP, solid circle) is compared to healthy controls (H, open circle). The immature receptor CCR5 is significantly elevated in H versus CP, whereas CXCR4 is significantly elevated in CP versus H. The data are also presented as percentages in the right panel. Immature receptors CCR2 and CCR5 are significantly elevated in H versus CP, while CXCR4 is significantly upregulated in CP versus H. (C) Patient serum samples were collected using serum separator clot activator tubes and analyzed by ELISA. The chronic periodontitis cohort had a significant elevation of CXCL12 compared to the healthy controls. Error bars are included for each sample in panels A, B, and C but are not always clear in the figure due to close clustering of certain data points. *, P < 0.01.
MoDCs infected with P. gingivalis are responsive to the CXCR4 ligand CXCL12 but not to the CCR7 ligand CCL19.
Next, we assessed the functionality of this chemokine receptor phenotype by analysis of the directed migration of P. gingivalis-infected MoDCs toward CXCL12 and CCL19. This was conducted using both a 48-well microchemotaxis Boyden chamber (Fig. 4A to C) and 24-well chemotaxis transwell inserts (Fig. 4D). This allowed us to quantitate the relative distances traveled (Fig. 4A to C) and analyze the total number of MoDCs migrating (Fig. 4D). P. gingivalis infection of MoDCs did not cause significant chemotaxis toward SLO-homing chemokine CCL19 with any of our strains (Fig. 4A). Only the inflammatory cocktail drove significant CCL19-directed migration of MoDCs (Fig. 4A). MoDC migration toward CXCL12 upon infection with fimbriated P. gingivalis was significantly increased over that of untreated MoDCs (Fig. 4B). Controls show a basal level of CXCL12-directed migration due to basal CXCR4 expression, but the CXCL12-driven migration was significantly higher in WT Pg381-infected MoDCs at increasing concentrations of CXCL12 (Fig. 4B). To further confirm that this migration was CXCR4 specific, P. gingivalis-infected MoDCs were pretreated with the CXCR4-specific blocking compound AMD3100. Treatment with AMD3100 completely abrogated migration of MoDCs in response to CXCL12 (Fig. 4C).
FIG 4.
Efficient chemotactic and endothelial migration of P. gingivalis-infected MoDCs toward CXCL12 but not CCL19. After 24 h of treatment as indicated, MoDCs were migrated through a filter using a Boyden chamber to determine relative MoDC migration distance (A, B, C) or through transwell inserts to determine the number of MoDCs undergoing transendothelial migration (D) in response to either the CXCR4 ligand CXCL12 or the CCR7 ligand CCL19. (A to C) MoDCs (1 × 105/well) were loaded in triplicate into the top chamber of a microchemotaxis chamber and migrated through a nitrocellulose membrane toward a CCL19 or CXCL12 chemokine gradient. (A) P. gingivalis-infected MoDCs do not actively migrate to the 100-ng/ml CCL19 gradient. Only the MoDCs treated with the inflammatory cocktail migrated toward the CCL19 significantly compared to untreated MoDCs. (B) MoDCs infected with live WT Pg381 show a significant increase in migration to CXCL12 over migration of untreated MoDCs at a low concentration. With an increased concentration of CXCL12, MoDCs infected with heat-killed Pg381 show significant increases in migration toward CXCL12, but migration of MoDCs treated with live Pg381 is still significantly higher. (C) The chemical inhibitor of CXCR4, AMD3100, was incubated with MoDCs for 1 h prior to migration. Treatments with AMD3100 completely abrogated MoDCs' migration in response to CXCL12. (D) Pg381-infected MoDCs (1 × 105/well) were migrated through a monolayer of ITECs in response to 100 ng/ml of either CXCL12 or CCL19 in the bottom compartment of the transwell inserts. Only infected MoDCs responding to CXCL12 were able to significantly migrate through the endothelial barrier. *, P < 0.01.
A transwell insert chemotaxis assay was then implemented to determine the ability of infected MoDCs to migrate through an endothelial barrier (Fig. 4D). Pg381-infected MoDCs were applied to a monolayer of internal thoracic endothelial cells (ITECs), with the addition of chemokines to the bottom compartment, and the relative number of MoDCs migrating through the barrier was calculated. The presence of the endothelial monolayer was ensured using inverted light microscopy and trypan blue staining. When CXCL12 was added to the bottom compartment, MoDCs were highly migratory through the ITEC barrier (Fig. 4D). Yet, when the same concentration of CCL19 was placed in the bottom compartment, MoDCs did not significantly migrate through the ITEC barrier (Fig. 4D).
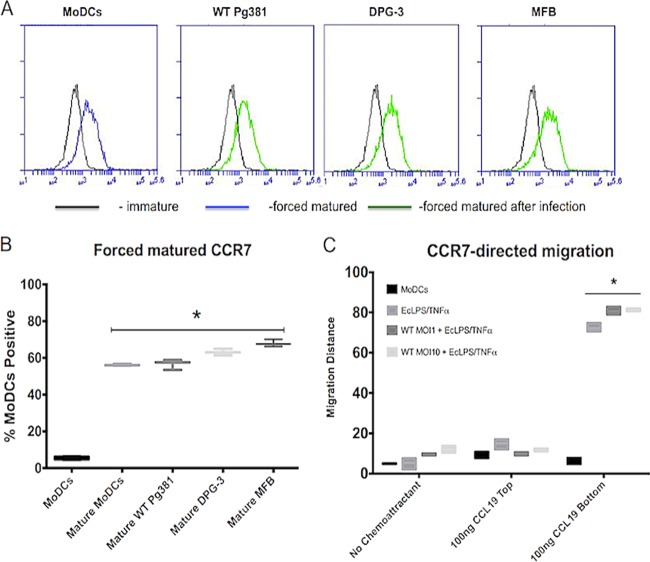
P. gingivalis-infected MoDCs are capable of expressing CCR7 and migrate toward CCL19 when stimulated with inflammatory cocktail treatment.
To determine if this migratory blockade can be overcome by forced maturation, P. gingivalis-infected MoDCs were treated with E. coli LPS and TNF-α for an additional 24 h. This additional treatment resulted in increased CCR7 expression on MoDCs (Fig. 5A and B) and restored CCL19-directed migration of infected MoDCs (Fig. 5C).
FIG 5.
Forced maturation induces CCR7 expression and CCL19-directed migration of P. gingivalis-infected MoDCs. MoDCs that had been infected as indicated for 24 h were treated with E. coli LPS/TNF-α for an additional 24 h to force maturation. (A) Representative histograms are shown for each MoDC infection treatment for CCR7 expression. The inflammatory cocktail leads to significant upregulation of surface CCR7 expression. (B) Summary data for CCR7 expression are shown, representing the additional 24-h treatment with the inflammatory cocktail. The treatment causes significant upregulation of CCR7 in all of our infected MoDCs, which matches with what occurs in typical matured MoDCs. (C) After treatment with inflammatory cocktail, cells were collected and loaded into the upper compartment of a microchemotaxis Boyden chamber (1 × 105/well) and migrated toward a CCL19 chemokine gradient. Migration distance (μm) was determined with leading-edge analysis after hematoxylin staining by using a 40× objective lens and subtracting migration distance without chemokine from migration distance with chemokine. The infected MoDCs show an increased response to CCL19 after the additional treatment compared to untreated controls. *, P < 0.01.
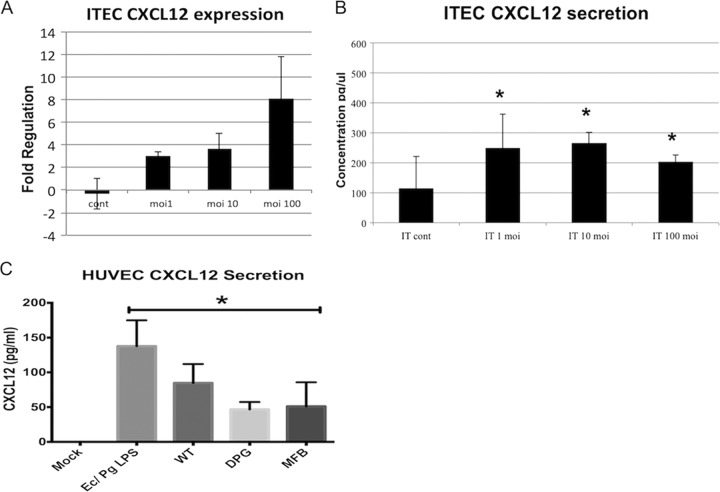
Vascular endothelial cells as a source of CXCL12.
We hypothesized that the vascular endothelium was a significant source of CXCL12 driving DC angiogenic migration. We therefore investigated CXCL12 secretion by human umbilical vein endothelial cells (HUVEC) and ITECs in response to P. gingivalis infection and inflammatory stimulation. CXCL12 was induced by WT Pg381 at both transcript and protein levels in ITECs, in a dose-dependent manner (Fig. 6A and B). P. gingivalis-treated and LPS cocktail-treated HUVECs both had upregulated CXCL12 protein secretion compared to mock, untreated HUVEC (Fig. 6C).
FIG 6.
Vascular endothelial cells produce CXCL12 after stimulation. Internal thoracic endothelial cells (ITECs) or human umbilical vein endothelial cells (HUVEC) were seeded in 6-well plates (1 × 105 cells per well) with serum-free medium and infected with WT Pg381 for 24 h. Serum was collected for ELISA, and ITECs were processed for quantitative PCR. (A) ITECs (IT) upregulate CXCL12 gene expression in a P. gingivalis dose-dependent fashion from uninfected controls. (B) The secreted protein levels in culture supernatants (pg/μl) were subsequently analyzed, and it was found that CXCL12 secretion was also increased in P. gingivalis-infected ITECs (IT). (C) HUVEC stimulated with P. gingivalis strains or with an E. coli LPS and P. gingivalis LPS cocktail displayed upregulated CXCL12 secretion from controls, which fell below the ELISA detectable limits.
DISCUSSION
We have shown that both human MoDCs and peripheral blood mDCs from patients with the oral mucosal infection chronic periodontitis have a unique migratory profile, characterized by high CXCR4 expression but low expression of SLO-homing CCR7, compared to uninfected or healthy controls. This lymphoid-homing “blockade” was observed after infection of MoDCs with live, fimbriated P. gingivalis, which did not drive CCR7 upregulation typical of other infections (63–65). The migration patterns of P. gingivalis-infected MoDCs matched this receptor profile, with directed migration toward the CXCR4 ligand CXCL12, but not toward CCR7 ligand CCL19. Upon forced maturation, however, P. gingivalis-infected DCs can upregulate CCR7 and become responsive to CCL19, and thus this migratory deficiency can be overcome. Additionally, HUVECs and ITECs challenged with P. gingivalis and activating stimuli were observed to activate CXCL12 protein secretion. Patient sera from a chronic periodontitis cohort also displayed increased CXCL12 compared to healthy controls. This migratory blockade should have functional consequences for immune homeostasis, i.e., by preventing P. gingivalis-infected DCs from homing to secondary lymphoid organs, instead directing these DCs to sites of vascularization.
CCR7 is essential for trafficking of antigen-bearing DCs to T cell zones in lymph nodes. While other chemokine receptors can aid in DC migration through lymphatics, only CCR7 can provide the ability for correct compartmental positioning and stimulation of robust CD4 and CD8 T cell responses (66). We show that DC-SIGN ligation by mfa-1+ P. gingivalis may disrupt DC homeostasis by disrupting the migratory ability of DCs toward CCL19 and promote DC migration toward CXCL12. This is evident by the comparable upregulation of CXCR4 driven by wild-type Pg381 and the mfa-1+ DPG-3 strain, but not by the nonfimbriated MFB strain. Moreover, the chemokine expression profiles of Pg381- and DPG-3-treated MoDCs are similar, albeit Pg381 led to dampened CCR6 downregulation. Hence, it appears the mfa-1/DC-SIGN interaction is important for driving CXCR4 upregulation, without stimulating CCR7 expression, but other bacterial factors may also contribute to the loss of immature receptors. Previous work indicates that CXCL12 is elevated during chronic periodontitis (67), and upregulation has already been shown to be crucial to mobilizing stem cells into the blood by following a CXCL12 gradient toward blood circulation (68). CXCL12 recruits endothelial progenitors to sites of neovascularization. Our data provide a plausible mechanism of pathogen-trafficking through the blood within DCs, as we have previously shown in vivo, but until now the mechanism was unclear (22). We propose that low-grade, chronic infections can disrupt DC homeostasis (69), preventing resolution of infection, and thereby prolonging inflammatory diseases (70, 71) and potentially contributing to autoimmune disease (72, 73).
To further examine the chemokine receptor expression on subpopulations of P. gingivalis-infected MoDCs, we labeled our bacteria with CFSE prior to infection. This revealed that mfa1+ P. gingivalis strains had rates of infection in MoDCs at an MOI that mimics natural clinical infection rates. The CFSE-positive MoDCs displayed upregulated surface CXCR4 but did not induce any detectable CCR7 expression over immature controls. Thus, P. gingivalis infection of MoDCs correlates with high CXCR4 expression and low CCR7 expression. This disruption of chemokine receptor switching is evident in both infected MoDCs and mDCs from chronic periodontitis patients, as it appears that infection leads to a downregulation of early chemokine receptors CCR5/CCR6 and CCR2, respectively, but in the absence of CCR7 upregulation. The infected MoDCs were capable of expressing CCR7 and responding to CCL19 after we induced maturation with E. coli LPS and TNF-α. LPS is a strong inducer of NF-κB in activating DCs. This secondary stimulation restores typical antigen-bearing DC migration and demonstrates that mfa-1+ P. gingivalis does not actively suppress CCR7 but fails to induce expression. This potentially demonstrates the importance of the location of the DC and the abundance of inflammatory signals for proper resolution of infection and also demonstrates a potential therapeutic approach to alleviating this dysregulation. We show that this oral pathogen fails to properly upregulate CCR7 after capture, which greatly impedes adaptive immunity and has broad consequences for many chronic infections.
High prevalence rates of chronic periodontitis stress the potential impact of P. gingivalis-laden DCs on human systemic health, most notably the DC migration patterns. During chronic periodontitis, it has been shown that the peripheral blood myeloid DC (mDC) pool expands, commensurate with detectable intracellular P. gingivalis (22). DCs have been shown to provide a protective niche for this anaerobe, whereas other immune cells, such as neutrophils, are very efficient at killing P. gingivalis (22). Due to the constant low-grade bacteremia during chronic periodontitis (74), P. gingivalis translocation may be a periodic and ongoing event. Not surprisingly, DC infiltrates are a common occurrence at inflammatory sites (75), revealing a mechanism that can apply to a broad range of pathogenic organisms (76). Pathogens that can manipulate DCs and cause reentry into the bloodstream generate a mode of dissemination to various distant sites within the body. The events leading to reentry of DCs into the bloodstream remain unclear, but hypoxia, common in diseased gingival pockets, has been shown to increase the reverse transmigration capabilities of DCs and production of CXCL12 during chronic periodontitis (77).
In summary, our data show a mechanism whereby an intracellular pathogen may exploit and redirect the highly migratory function of DCs to sites of neovascularization, such as atherosclerotic plaques (22), all the while preventing immunoelimination by the acquired immune response. Chronic periodontitis is strongly associated with various extraoral systemic inflammatory diseases, and here, we provide a model for the possible contribution of coinfection leading to exacerbation of widespread inflammation. As many chronic infections and inflammatory sites are characterized by increased vascularization, we believe that this can be a broad microbial pathogen strategy to escape localized immunity and sequester into the bloodstream. By disrupting the highly regulated DC migration process, pathogens can direct their own dissemination to distant sites in the body and exacerbate systemic disease.
ACKNOWLEDGMENTS
B.M. and C.W.C. designed and discussed experiments. B.M. and E.S. performed cell surface receptor expression. B.M. and A.E.-A. performed chemotactic analysis. B.M. and I.Z. performed chemokine ELISAs. B.M. obtained clinical data. B.M., J.C., J.C.O., A.R., C.S., J.K.S., and C.W.C. designed and performed the protocols involving human subjects. B.M. and C.W.C. wrote the manuscript.
This work was supported by U.S. Public Health Service Grants from the National Institutes of Health/National Institute of Dental and Craniofacial Research (R01 DE014328 and R21 DE020916 to C.W.C, K23 DE018187 to J.C., and F30 DE021649-01 to E.S.). The funders did not have a role in study design, data collection, manuscript preparation, or publishing.
The authors declare that they have no conflicts of interest.
Footnotes
Published ahead of print 14 October 2013
REFERENCES
- 1.Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10:717–725. 10.1038/nrmicro2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albandar J. M. 2005. Epidemiology and risk factors of periodontal diseases. Dent. Clin. North Am. 49:517–532, v–vi [DOI] [PubMed] [Google Scholar]
- 3.Offenbacher S, Madianos PN, Champagne CME, Southerland JH, Paquette DW, Williams RC, Slade G, Beck JD. 1999. Periodontitis-atherosclerosis syndrome: an expanded model of pathogenesis. J. Periodontal Res. 34:346–352. 10.1111/j.1600-0765.1999.tb02264.x [DOI] [PubMed] [Google Scholar]
- 4.Holt SC, Kesavalu L, Walker S, Genco CA. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168–238. 10.1111/j.1600-0757.1999.tb00162.x [DOI] [PubMed] [Google Scholar]
- 5.Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8:481–490. 10.1038/nrmicro2337 [DOI] [PubMed] [Google Scholar]
- 6.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu Y-J, Pulendran B, Palucka K. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767. 10.1146/annurev.immunol.18.1.767 [DOI] [PubMed] [Google Scholar]
- 7.Shortman K, Liu Y-J. 2002. Mouse and human dendritic cell subtypes. Nat. Rev. Immunol. 2:151–161. 10.1038/nri746 [DOI] [PubMed] [Google Scholar]
- 8.Chen M, Wang Y-H, Wang Y, Huang L, Sandoval H, Liu Y-J, Wang J. 2006. Dendritic cell apoptosis in the maintenance of immune tolerance. Science 311:1160–1164. 10.1126/science.1122545 [DOI] [PubMed] [Google Scholar]
- 9.Ishibashi S, Maric D, Mou Y, Ohtani R, Ruetzler C, Hallenbeck JM. 2009. Mucosal tolerance to E-selectin promotes the survival of newly generated neuroblasts via regulatory T-cell induction after stroke in spontaneously hypertensive rats. J. Cereb. Blood Flow Metab. 29:606–620. 10.1038/jcbfm.2008.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dieu-Nosjean MC, Vicari A, Lebecque S, Caux C. 1999. Regulation of dendritic cell trafficking: a process that involves the participation of selective chemokines. J. Leukoc. Biol. 66:252–262 [DOI] [PubMed] [Google Scholar]
- 11.Sozzani S, Allavena P, Vecchi A, Mantovani A. 1999. The role of chemokines in the regulation of dendritic cell trafficking. J. Leukoc. Biol. 66:1–9 [DOI] [PubMed] [Google Scholar]
- 12.Cutler CW, Jotwani R, Palucka KA, Davoust J, Bell D, Banchereau J. 1999. Evidence and a novel hypothesis for the role of dendritic cells and Porphyromonas gingivalis in adult periodontitis. J. Periodontal Res. 34:406–412. 10.1111/j.1600-0765.1999.tb02274.x [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki A. 2007. Mucosal dendritic cells. Annu. Rev. Immunol. 25:381–418. 10.1146/annurev.immunol.25.022106.141634 [DOI] [PubMed] [Google Scholar]
- 14.Luster AD, Alon R, von Andrian UH. 2005. Immune cell migration in inflammation: present and future therapeutic targets. Nat. Immunol. 6:1182–1190. 10.1038/ni1275 [DOI] [PubMed] [Google Scholar]
- 15.Fainaru O, Shseyov D, Hantisteanu S, Groner Y. 2005. Accelerated chemokine receptor 7-mediated dendritic cell migration in Runx3 knockout mice and the spontaneous development of asthma-like disease. Proc. Natl. Acad. Sci. U. S. A. 102:10598–10603. 10.1073/pnas.0504787102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cutler CW, Jotwani R. 2004. Antigen-presentation and the role of dendritic cells in periodontitis. Periodontol. 2000 35:135–157. 10.1111/j.0906-6713.2004.003560.x [DOI] [PubMed] [Google Scholar]
- 17.Cutler CW, Teng Y-TA. 2007. Oral mucosal dendritic cells and periodontitis: many sides of the same coin with new twists. Periodontol. 2000 45:35–50. 10.1111/j.1600-0757.2007.00222.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niessner A, Weyand CM. 2010. Dendritic cells in atherosclerotic disease. Clin. Immunol. 134:25–32. 10.1016/j.clim.2009.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scannapieco FA, Bush RB, Paju S. 2003. Associations between periodontal disease and risk for atherosclerosis, cardiovascular disease, and stroke. A systematic review. Ann. Periodontol. 8:38–53. 10.1902/annals.2003.8.1.38 [DOI] [PubMed] [Google Scholar]
- 20.Jiao X, Lo-Man R, Guermonprez P, Fiette L, Dériaud E, Burgaud S, Gicquel B, Winter N, Leclerc C. 2002. Dendritic cells are host cells for mycobacteria in vivo that trigger innate and acquired immunity. J. Immunol. 168:1294–1301 [DOI] [PubMed] [Google Scholar]
- 21.Nagl M, Kacani L, Müllauer B, Lemberger E-M, Stoiber H, Sprinzl GM, Schennach H, Dierich MP. 2002. Phagocytosis and killing of bacteria by professional phagocytes and dendritic cells. Clin. Diagn. Lab. Immunol. 9:1165–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrion J, Scisci E, Miles B, Sabino GJ, Zeituni AE, Gu Y, Bear A, Genco CA, Brown DL, Cutler CW. 2012. Microbial carriage state of peripheral blood dendritic cells (DCs) in chronic periodontitis influences DC differentiation, atherogenic potential. J. Immunol. 189:3178–3187. 10.4049/jimmunol.1201053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeituni AE, McCaig W, Scisci E, Thanassi DG, Cutler CW. 2010. The native 67-kilodalton minor fimbria of porphyromonas gingivalis is a novel glycoprotein with DC-SIGN-targeting motifs. J. Bacteriol. 192:4103–4110. 10.1128/JB.00275-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeituni AE, Jotwani R, Carrion J, Cutler CW. 2009. Targeting of DC-SIGN on human dendritic cells by minor fimbriated Porphyromonas gingivalis strains elicits a distinct effector T cell response. J. Immunol. 183:5694–5704. 10.4049/jimmunol.0901030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scannapieco FA. 1999. Role of oral bacteria in respiratory infection. J. Periodontol. 70:793–802. 10.1902/jop.1999.70.7.793 [DOI] [PubMed] [Google Scholar]
- 26.Dorn BR, Dunn WA, Progulske-Fox A. 2001. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect. Immun. 69:5698–5708. 10.1128/IAI.69.9.5698-5708.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozarov EV, Dorn BR, Shelburne CE, Dunn WA, Progulske-Fox A. 2005. Human atherosclerotic plaque contains viable invasive Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Arterioscler. Thromb. Vasc. Biol. 25:e17–e18. 10.1161/01.ATV.0000155018.67835.1a [DOI] [PubMed] [Google Scholar]
- 28.Liao F, Li Z, Wang Y, Shi B, Gong Z, Cheng X. 2009. Porphyromonas gingivalis may play an important role in the pathogenesis of periodontitis-associated rheumatoid arthritis. Med. Hypotheses 72:732–735. 10.1016/j.mehy.2008.12.040 [DOI] [PubMed] [Google Scholar]
- 29.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, Cordum H, Van Brunt A, Kim K, Fulton RS, Fulton LA, Clifton SW, Wilson RK, Knight RD, Gordon JI. 2007. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 5:e156. 10.1371/journal.pbio.0050156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randolph GJ, Ochando J, Partida-Sanchez S. 2008. Migration of dendritic cell subsets and their precursors. Annu. Rev. Immunol. 26:293–316. 10.1146/annurev.immunol.26.021607.090254 [DOI] [PubMed] [Google Scholar]
- 31.Dieu M-C, Vanbervliet B, Vicari A, Bridon J-M, Oldham E, Aït-Yahia S, Brière F, Zlotnik A, Lebecque S, Caux C. 1998. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J. Exp. Med. 188:373–386. 10.1084/jem.188.2.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forster R, Davalos-Misslitz AC, Rot A. 2008. CCR7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol. 8:362–371. 10.1038/nri2297 [DOI] [PubMed] [Google Scholar]
- 33.Pulendran B, Banchereau J, Maraskovsky E, Maliszewski C. 2001. Modulating the immune response with dendritic cells and their growth factors. Trends Immunol. 22:41–47. 10.1016/S1471-4906(00)01794-4 [DOI] [PubMed] [Google Scholar]
- 34.Ohtani T, Mizuashi M, Nakagawa S, Sasaki Y, Fujimura T, Okuyama R, Aiba S. 2009. TGF-beta1 dampens the susceptibility of dendritic cells to environmental stimulation, leading to the requirement for danger signals for activation. Immunology 126:485–499. 10.1111/j.1365-2567.2008.02919.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGrath KE, Koniski AD, Maltby KM, McGann JK, Palis J. 1999. Embryonic expression and function of the chemokine SDF-1 and its receptor, CXCR4. Dev. Biol. 213:442–456. 10.1006/dbio.1999.9405 [DOI] [PubMed] [Google Scholar]
- 36.Ma Q, Jones D, Springer TA. 1999. The chemokine receptor CXCR4 is required for the retention of b lineage and granulocytic precursors within the bone marrow microenvironment. Immunity 10:463–471. 10.1016/S1074-7613(00)80046-1 [DOI] [PubMed] [Google Scholar]
- 37.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. U. S. A. 96:5215–5220. 10.1073/pnas.96.9.5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luster AD. 1998. Chemokines—chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 338:436–445. 10.1056/NEJM199802123380706 [DOI] [PubMed] [Google Scholar]
- 39.Sun X, Cheng G, Hao M, Zheng J, Zhou X, Zhang J, Taichman R, Pienta K, Wang J. 2010. CXCL12 / CXCR4 / CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 29:709–722. 10.1007/s10555-010-9256-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H. 2005. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 65:967–971 [PMC free article] [PubMed] [Google Scholar]
- 41.Chapple CC, Kumar RK, Hunter N. 2000. Vascular remodelling in chronic inflammatory periodontal disease. J. Oral Pathol. Med. 29:500–506. 10.1034/j.1600-0714.2000.291004.x [DOI] [PubMed] [Google Scholar]
- 42.Jotwani R, Muthukuru M, Cutler CW. 2004. Increase in HIV receptors/co-receptors/α-defensins in inflamed human gingiva. J. Dent. Res. 83:371–377. 10.1177/154405910408300504 [DOI] [PubMed] [Google Scholar]
- 43.Havens AM, Chiu E, Taba M, Wang J, Shiozawa Y, Jung Y, Taichman LS, D'Silva NJ, Gopalakrishnan R, Wang C, Giannobile WV, Taichman RS. 2008. Stromal-derived factor-1alpha (CXCL12) levels increase in periodontal disease. J. Periodontol. 79:845–853. 10.1902/jop.2008.070514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hosokawa Y, Hosokawa I, Ozaki K, Nakae H, Murakami K, Miyake Y, Matsuo T. 2005. CXCL12 and CXCR4 expression by human gingival fibroblasts in periodontal disease. Clin. Exp. Immunol. 141:467–474. 10.1111/j.1365-2249.2005.02852.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuboniwa M, Amano A, Hashino E, Yamamoto Y, Inaba H, Hamada N, Nakayama K, Tribble G, Lamont R, Shizukuishi S. 2009. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis. BMC Microbiol. 9:105. 10.1186/1471-2180-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malek R, Fisher JG, Caleca A, Stinson M, van Oss CJ, Lee JY, Cho MI, Genco RJ, Evans RT, Dyer DW. 1994. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol. 176:1052–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Njoroge T, Genco RJ, Sojar HT, Hamada N, Genco CA. 1997. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect. Immun. 65:1980–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuominen-Gustafsson H, Penttinen M, Hytonen J, Viljanen MK. 2006. Use of CFSE staining of borreliae in studies on the interaction between borreliae and human neutrophils. BMC Microbiol. 6:92. 10.1186/1471-2180-6-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cutler CW, Kalmar JR, Arnold RR. 1991. Phagocytosis of virulent Porphyromonas gingivalis by human polymorphonuclear leukocytes requires specific immunoglobulin G. Infect. Immun. 59:2097–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chapuis F, Rosenzwajg M, Yagello M, Ekman M, Biberfeld P, Gluckman JC. 1997. Differentiation of human dendritic cells from monocytes in vitro. Eur. J. Immunol. 27:431–441. 10.1002/eji.1830270213 [DOI] [PubMed] [Google Scholar]
- 51.Jotwani R, Pulendran B, Agrawal S, Cutler CW. 2003. Human dendritic cells respond to Porphyromonas gingivalis LPS by promoting a Th2 effector response in vitro. Eur. J. Immunol. 33:2980–2986. 10.1002/eji.200324392 [DOI] [PubMed] [Google Scholar]
- 52.Vegh Z, Kew RR, Gruber BL, Ghebrehiwet B. 2006. Chemotaxis of human monocyte-derived dendritic cells to complement component C1q is mediated by the receptors gC1qR and cC1qR. Mol. Immunol. 43:1402–1407. 10.1016/j.molimm.2005.07.030 [DOI] [PubMed] [Google Scholar]
- 53.Donzella GA, Schols D, Lin SW, Este JA, Nagashima KA, Maddon PJ, Allaway GP, Sakmar TP, Henson G, De Clercq E, Moore JP. 1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 4:72–77. 10.1038/nm0198-072 [DOI] [PubMed] [Google Scholar]
- 54.Zigmond SH, Hirsch JG. 1973. Leukocyte locomotion and chemotaxis. J. Exp. Med. 137:387–410. 10.1084/jem.137.2.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Osterholzer JJ, Ames T, Polak T, Sonstein J, Moore BB, Chensue SW, Toews GB, Curtis JL. 2005. CCR2 and CCR6, but not endothelial selectins, mediate the accumulation of immature dendritic cells within the lungs of mice in response to particulate antigen. J. Immunol. 175:874–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jotwani R, Cutler CW. 2004. Fimbriated Porphyromonas gingivalis is more efficient than fimbria-deficient P. gingivalis in entering human dendritic cells in vitro and induces an inflammatory Th1 effector response. Infect. Immun. 72:1725–1732. 10.1128/IAI.72.3.1725-1732.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bergman MP, Engering A, Smits HH, van Vliet SJ, van Bodegraven AA, Wirth H-P, Kapsenberg ML, Vandenbroucke-Grauls CMJE, van Kooyk Y, Appelmelk BJ. 2004. Helicobacter pylori modulates the T helper cell 1/T helper cell 2 balance through phase-variable interaction between lipopolysaccharide and DC-SIGN. J. Exp. Med. 200:979–990. 10.1084/jem.20041061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geijtenbeek TBH, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GCF, Middel J, Cornelissen IL, Nottet HS, KewalRamani VN, Littman DR, Figdor CG, van Kooyk Y. 2000. DC-SIGN, a dendritic cell–specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587–597. 10.1016/S0092-8674(00)80694-7 [DOI] [PubMed] [Google Scholar]
- 59.Soilleux EJ, Sarno EN, Hernandez MO, Moseley E, Horsley J, Lopes UG, Goddard MJ, Vowler SL, Coleman N, Shattock RJ, Sampaio EP. 2006. DC-SIGN association with the Th2 environment of lepromatous lesions: cause or effect? J. Pathol. 209:182–189. 10.1002/path.1972 [DOI] [PubMed] [Google Scholar]
- 60.Fantuzzi L, Purificato C, Donato K, Belardelli F, Gessani S. 2004. Human immunodeficiency virus type 1 gp120 induces abnormal maturation and functional alterations of dendritic cells: a novel mechanism for AIDS pathogenesis. J. Virol. 78:9763–9772. 10.1128/JVI.78.18.9763-9772.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hodges A, Sharrocks K, Edelmann M, Baban D, Moris A, Schwartz O, Drakesmith H, Davies K, Kessler B, McMichael A, Simmons A. 2007. Activation of the lectin DC-SIGN induces an immature dendritic cell phenotype triggering Rho-GTPase activity required for HIV-1 replication. Nat. Immunol. 8:569–577. 10.1038/ni1470 [DOI] [PubMed] [Google Scholar]
- 62.Vanbervliet B, Homey B, Durand I, Massacrier C, Aït Yahia S, de Bouteiller O, Vicari A, Caux C. 2002. Sequential involvement of CCR2 and CCR6 ligands for immature dendritic cell recruitment: possible role at inflamed epithelial surfaces. Eur. J. Immunol. 32:231–242. [DOI] [PubMed] [Google Scholar]
- 63.Smed-Sörensen A, Loré K, Vasudevan J, Louder MK, Andersson J, Mascola JR, Spetz A-L, Koup RA. 2005. Differential susceptibility to human immunodeficiency virus type 1 infection of myeloid and plasmacytoid dendritic cells. J. Virol. 79:8861–8869. 10.1128/JVI.79.14.8861-8869.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitchell P, Germain C, Fiori PL, Khamri W, Foster GR, Ghosh S, Lechler RI, Bamford KB, Lombardi G. 2007. Chronic exposure to Helicobacter pylori impairs dendritic cell function and inhibits Th1 development. Infect. Immun. 75:810–819. 10.1128/IAI.00228-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hansson M, Lundgren A, Elgbratt K, Quiding-Järbrink M, Svennerholm A-M, Johansson E-L. 2006. Dendritic cells express CCR7 and migrate in response to CCL19 (MIP-3β) after exposure to Helicobacter pylori. Microbes Infect. 8:841–850. 10.1016/j.micinf.2005.10.007 [DOI] [PubMed] [Google Scholar]
- 66.Martín-Fontecha A, Sebastiani S, Höpken UE, Uguccioni M, Lipp M, Lanzavecchia A, Sallusto F. 2003. Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J. Exp. Med. 198:615–621. 10.1084/jem.20030448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Havens AM, Chiu E, Taba M, Jr, Wang J, Shiozawa Y, Jung Y, Taichman LS, D'Silva NJ, Gopalakrishnan R, Wang C. 2008. Stromal-derived factor-1α (CXCL12) levels increase in periodontal disease. J. Periodontol. 79:845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petit I, Szyper-Kravitz M, Nagler A, Lahav M, Peled A, Habler L, Ponomaryov T, Taichman RS, Arenzana-Seisdedos F, Fujii N, Sandbank J, Zipori D, Lapidot T. 2002. G-CSF induces stem cell mobilization by decreasing bone marrow SDF-1 and up-regulating CXCR4. Nat. Immunol. 3:687–694. 10.1038/ni813 [DOI] [PubMed] [Google Scholar]
- 69.Kornman KS, Page RC, Tonetti MS. 1997. The host response to the microbial challenge in periodontitis: assembling the players. Periodontol. 2000 14:33–53. 10.1111/j.1600-0757.1997.tb00191.x [DOI] [PubMed] [Google Scholar]
- 70.Giacona MB, Papapanou PN, Lamster IB, Rong LL, D'Agati VD, Schmidt AM, Lalla E. 2004. Porphyromonas gingivalis induces its uptake by human macrophages and promotes foam cell formation in vitro. FEMS Microbiol. Lett. 241:95–101 [DOI] [PubMed] [Google Scholar]
- 71.Gibson FC, Yumoto H, Takahashi Y, Chou H-H, Genco CA. 2006. Innate immune signaling and porphyromonas gingivalis-accelerated atherosclerosis. J. Dent. Res. 85:106–121. 10.1177/154405910608500202 [DOI] [PubMed] [Google Scholar]
- 72.Angeli V, Llodrá J, Rong JX, Satoh K, Ishii S, Shimizu T, Fisher EA, Randolph GJ. 2004. Dyslipidemia associated with atherosclerotic disease systemically alters dendritic cell mobilization. Immunity 21:561–574. 10.1016/j.immuni.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 73.Drakesmith H, Chain B, Beverley P. 2000. How can dendritic cells cause autoimmune disease? Immunol. Today 21:214–217. 10.1016/S0167-5699(00)01610-8 [DOI] [PubMed] [Google Scholar]
- 74.Forner L, Larsen T, Kilian M, Holmstrup P. 2006. Incidence of bacteremia after chewing, tooth brushing and scaling in individuals with periodontal inflammation. J. Clin. Periodontol. 33:401–407. 10.1111/j.1600-051X.2006.00924.x [DOI] [PubMed] [Google Scholar]
- 75.Bobryshev YV. 2005. Dendritic cells in atherosclerosis: current status of the problem and clinical relevance. Eur. Heart J. 26:1700–1704. 10.1093/eurheartj/ehi282 [DOI] [PubMed] [Google Scholar]
- 76.Steinman RM. 2000. DC-SIGN: a guide to some mysteries of dendritic cells. Cell 100:491–494. 10.1016/S0092-8674(00)80684-4 [DOI] [PubMed] [Google Scholar]
- 77.Schioppa T, Uranchimeg B, Saccani A, Biswas SK, Doni A, Rapisarda A, Bernasconi S, Saccani S, Nebuloni M, Vago L, Mantovani A, Melillo G, Sica A. 2003. Regulation of the chemokine receptor CXCR4 by hypoxia. J. Exp. Med. 198:1391–1402. 10.1084/jem.20030267 [DOI] [PMC free article] [PubMed] [Google Scholar]