Abstract
Extraintestinal Escherichia coli (ExPEC) organisms are the leading cause of Gram-negative bacterial bloodstream infections. These bacteria adapt to survival in the bloodstream through expression of factors involved in scavenging of nutrients and resisting the killing activity of serum. In this study, the transcriptional response of a prototypic ExPEC strain (CFT073) to human serum was investigated. Resistance of CFT073 to the bactericidal properties of serum involved increased expression of envelope stress regulators, including CpxR, σE, and RcsB. Many of the upregulated genes induced by active serum were regulated by the Rcs two-component system. This system is triggered by envelope stress such as changes to cell wall integrity. RcsB-mediated serum resistance was conferred through induction of the exopolysaccharide colanic acid. Production of this exopolysaccharide may be protective while cell wall damage caused by serum components is repaired.
INTRODUCTION
Bloodstream infections are predominantly caused by extraintestinal pathogenic Escherichia coli (ExPEC) and are associated with high morbidity and mortality (1). Indeed, E. coli is the most frequently isolated Gram-negative bacterial pathogen from blood cultures of both community-associated and nosocomial cases (2, 3). The majority of bloodstream infections caused by ExPEC originate in the urinary tract. Upon ascension to the kidneys, the infecting bacteria can breach the parenchyma, allowing entry into the bloodstream (4). Although ExPEC can be found among the commensals of the gut, these strains possess a wide variety of specialized virulence factors that confer pathogenesis outside the gastrointestinal tract (5). These virulence factors are required for adhesion and invasion within the urinary tract and evasion of the host defenses.
The presence of E. coli in the bloodstream may lead to powerful host inflammatory responses resulting in sepsis (6). Bacteria present in the bloodstream must be able to resist defense mechanisms of the host, including phagocytosis, the complement system, and antimicrobial peptides. The complement system constitutes one of the major innate defenses in the body (7). Complement activation results in a cascade of enzymatic cleavage and protein-protein interactions of complement components that are deposited on the bacterial surface to form the membrane attack complex. Insertion of the membrane attack complex into the cell membranes of susceptible bacteria creates transmembrane pores and causes osmotic lysis of cells (8). Serum lysozyme has been suggested to enhance lysis of susceptible E. coli cells in the presence of complement. Lysozymes target the bacterial cell wall, hydrolyzing β-(1-4) glycosidic bonds between N-acetylmuramic acid and N-acetylglucosamine (9).
Certain virulence factors antagonize the bactericidal activity of serum complement. A major mechanism of defense against complement-mediated killing is production of group 2 capsular polysaccharide K antigen. Capsular polysaccharides are major components of the bacterial cell envelope that are firmly attached to the cell surface. They play an important role in interactions between bacteria and their immediate environment by providing a protective steric barrier (4, 10). ExPEC can also produce group 1 extracellular polysaccharides, including enterobacterial common antigen and colanic acid (11, 12). These play an important role in survival outside the host during biofilm formation and also provide resistance against desiccation (11). Other virulence factors that have been reported to protect ExPEC against complement-mediated killing include outer membrane protein A, the O78 serogroup lipopolysaccharide, and the TraT and Iss serum resistance outer membrane proteins (13–17).
E. coli CFT073 is a prototypical urosepsis isolate and has been shown to resist the bactericidal effects of serum. Resistance to serum is mediated, in part, by the production of a K2 capsule (4). However, much is unknown about how ExPEC such as E. coli CFT073 adapt to survive in human blood. To answer these questions, we defined the transcriptome of this isolate in response to bactericidal as opposed to nonbactericidal sera. These experiments revealed a role for the RcsB-regulated colanic acid exopolysaccharide in protecting ExPEC from serum bactericidal activity. This appears to be an additional mechanism that can augment the protective properties of the K2 capsule.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
E. coli strains and plasmids used in this study are described in Table 1. The prototypic uropathogenic E. coli (UPEC) strain CFT073 was originally isolated from the blood of a patient with acute pyelonephritis (18). Bacteria were routinely cultured at 37°C in Lennox broth (LB) or agar supplemented with appropriate antibiotics. Antibiotics were used at the following concentrations: carbenicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; and kanamycin, 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| E. coli strain or plasmid | Descriptiona | Source |
|---|---|---|
| Strains | ||
| CFT073 | Urosepsis isolate, serotype O6:K2:H1 | 15 |
| HB101 | K-12 laboratory strain | 58 |
| CFT073 ΔkpsC | CFT073 ΔkpsC, unmarked | This study |
| CFT073 ΔrcsB | CFT073 ΔrcsB::kan | This study |
| CFT073 ΔrcsB* | CFT073 ΔrcsB, unmarked | This study |
| CFT073 ΔrpoE | CFT073 ΔrpoE::kan | This study |
| CFT073 ΔcpxR | CFT073 ΔcpxR::kan | This study |
| CFT073 Δwca | CFT073 ΔwcaDE::kan | This study |
| CFT073 Δwca* | CFT073 ΔwcaDE, unmarked | This study |
| CFT073 ΔyjbE | CFT073 ΔyjbE::kan | This study |
| CFT073 ΔydhA | CFT073 ΔydhA::kan | This study |
| CFT073 ΔrcsA | CFT073 ΔrcsA::kan | This study |
| CFT073 ΔrcsC | CFT073 ΔrcsC::kan | This study |
| CFT073 ΔkpsC rcsB | CFT073 ΔkpsC, ΔrcsB::kan | This study |
| CFT073 ΔkpsC Δwca | CFT073 ΔkpsC, ΔwcaDE::kan | This study |
| CFT073 ΔkpsC ΔyjbE | CFT073 ΔkpsC, ΔyjbE::kan | This study |
| RS218 | Neonatal meningitis isolate, serotype O18:K1:H7 | 59 |
| RS218 Δwca | RS218 ΔwcaDE::kan | This study |
| Plasmids | ||
| pKD4 | FLP recombination target-flanked kanamycin resistance cassette | 22 |
| pKD46 | Phage λ red recombinase expression, temp sensitive | 22 |
| pCP20 | Thermal induction of FLP recombinase expression, Ampr | 60 |
| pCA24N | IPTG-inducible vector, Cmr | 23 |
| pCA24N rcsB | pCA24N containing rcsB gene, Cmr | 23 |
| pTRC99a | Cloning vector with IPTG-inducible Ptrc promoter, Ampr | 61 |
| pTRC99a wca | pTRC99a containing wcaDE, Ampr | This study |
Ampr, ampicillin resistant; Cmr, chloramphenicol resistant.
Preparation of serum.
Blood was collected from 6 healthy volunteers who had not received antibiotic treatment in the prior 3 months into serum clot activator Vacuette tubes (Greiner Bio-One). Ethical approval for the use of human bloods was sought and granted from the St. James Hospital Ethics committee. Cells were separated by centrifugation at 1,000 × g for 10 min, and the serum fractions from the 6 samples were collected, pooled, and stored in aliquots at −80°C. To disrupt the classical and alternative pathways of the complement cascade, serum was heat inactivated (HIS) at 55°C for 30 min.
Adsorption of complement from serum by zymosan treatment.
Complement was removed from serum samples by treatment with the yeast polysaccharide zymosan (Sigma). Zymosan was added to serum samples at 100 mg/ml, and the samples were incubated at 37°C for 30 min with stirring. The serum-zymosan mixture was centrifuged at 12,000 × g for 1 min to pellet the complement-coated zymosan particles (19). The supernatant was collected and used in subsequent assays.
RNA extraction.
Logarithmic-phase cultures were collected by centrifugation and adjusted to 3 × 108 CFU/ml in either LB supplemented with 50% normal human serum (NHS) or 50% HIS. Fifty percent serum was used, since the quality of RNA decreased when CFT073 was exposed to higher concentrations of serum. The samples were incubated at 37°C for 45 min, after which RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. The RNA obtained was treated with DNase I (Ambion).
Microarray.
RNA samples were prepared for microarray analysis using the FairPlay III Labeling kit (Agilent) according to the manufacturer's instructions. Briefly, first-strand cDNA was generated from 5 μg of RNA by reverse transcription incorporating amino allyl dUTP. Cy5 or Cy3 monoreactive dyes (GE Healthcare) were coupled to the reactive amino allyl groups, and the samples were purified of unincorporated dye using Illustra Microspin G-50 columns (GE Healthcare). Dye swaps were performed on sample replicates to eliminate any dye bias. Labeled cDNA (300 ng) was hybridized to the Agilent E. coli gene expression microarray, which includes probes specific for the CFT073 genome (Agilent Technologies).
Scanning and feature extraction was performed using a GenePix Personal 4100A scanner (Axon Laboratories), and data were extracted using GenePix Pro, version 6.1 (Axon Laboratories). Features with fluorescent intensities within 2 standard deviations of background levels were excluded from analysis. Raw data were exported to GeneSpring GX11, and signals for each replicate spot were background corrected and normalized using Loess normalization. Log2 fluorescence ratios were generated for each replicate spot and averaged. Average fold changes in gene expression of 4 biological replicate experiments were determined. Significance of these expression values was determined using a one-sample t test, and genes with P values of <0.05 were selected for analysis. The expression fold change of a given gene was calculated as the ratio of the signal intensity of the test sample (CFT073 exposed to NHS) to the signal intensity of the control sample (CFT073 exposed to HIS). The transcripts of predominantly upregulated or downregulated genes were identified using a ±2-fold expression cutoff.
qRT-PCR.
Primers used are listed in Table S1 in the supplemental material. RNA was reversed transcribed into cDNA using the Superscript VILO cDNA synthesis kit (Invitrogen). Ten nanograms of the resulting cDNA was used in quantitative real-time PCR (qRT-PCR) assays using the SensiMix Syber Low-ROX kit (Bioline) and a 7500 Real Time PCR system (Applied Biosystems). Melt curve analysis was performed to verify product homogeneity. All reactions included four replicates. Data were normalized by the 2−ΔΔCT method (20) using the rplT housekeeping gene of E. coli as a reference (21).
Construction of CFT073 deletion mutants.
Deletion mutants of CFT073 were constructed using one-step chromosomal gene inactivation (22). Briefly, primers were designed to amplify the kanamycin cassette of plasmid pKD4 with up to 50 bp of overhanging flanking sequence specific to the target gene (see Table S1 in the supplemental material). PCR products were electroporated into CFT073 carrying the plasmid pKD46, which encodes lambda Red recombinase, to facilitate homologous recombination. Transformants were selected at 37°C overnight on L agar supplemented with kanamycin. Successful gene deletion was confirmed by PCR and sequencing. The kanamycin resistance marker was eliminated using a helper plasmid expressing the FLP recombinase, pCP20, as previously described (22).
Complementation of CFT073 deletion mutants.
Plasmids pCA24N and pCA24N rcsB were obtained from the ASKA collection and introduced into strain CFT073 ΔrcsB* by electroporation (23). Strains carrying the pCA24N plasmids were grown with chloramphenicol and induced with 10 μM IPTG (isopropyl-β-d-thiogalactopyranoside). To restore expression of the wcaDE mutant, the corresponding region was amplified from CFT073 genomic DNA using primers FpTRCwca and RpTRCwca and cloned into plasmid pTRC99a. Strains carrying pTRC99a and pTRC99a wca were grown with carbenicillin and induced with 1 mM IPTG.
Bactericidal serum assay.
The ability of all strains and deletion mutants to survive in NHS was assessed using a bactericidal serum assay (24). Briefly, bacteria from logarithmic-phase cultures were collected by centrifugation and adjusted to 3 × 108 CFU/ml in LB supplemented with 50% NHS (vol/vol) or 50% HIS (vol/vol). Samples were incubated at 37°C for 45 min or longer. Tenfold serial dilutions of each sample were prepared, and populations were quantified (CFU/ml) by the Miles and Misra method.
Statistical analysis.
The data presented in this study represent the means of three experiments ± standard errors of the means (SEM) unless otherwise stated. The unpaired t test was used to determine the significance of differences in serum survival between strains, with significance defined as P values of <0.05.
Microarray data accession number.
Microarray data are available at http://www.ebi.ac.uk/arrayexpress under accession number E-MTAB-1985.
RESULTS
E. coli CFT073 is resistant to 50% human serum.
It has been previously shown that strain CFT073 can resist the bactericidal effects of 10% serum and that removal of the K2 capsule results in decreased viability of CFT073 in serum (4). In order to establish if CFT073 could resist higher serum concentrations, serum killing assays were carried out with 50% serum. CFT073 displayed no drop in viability when exposed to 50% serum; in fact, cell numbers slowly increased over 180 min. In contrast, the viability of the K2 capsule mutant, CFT073 ΔkpsC, was reduced by at least 4 log when exposed to NHS for 180 min (Fig. 1). Survival of CFT073 ΔkpsC in nonbactericidal HIS was comparable to that of the wild type (WT). These results indicate that CFT073 is resistant to 50% serum in a manner largely dependent of capsule production.
FIG 1.
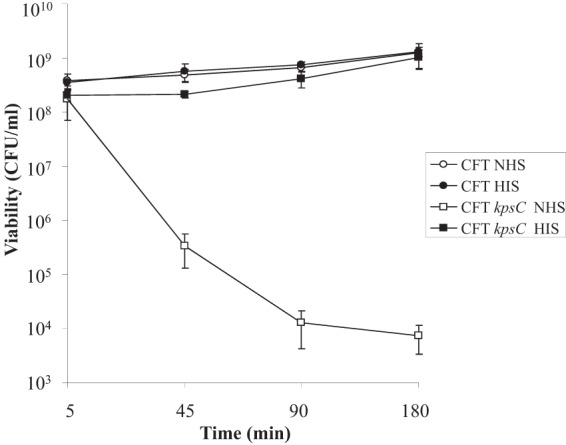
Serum survival of CFT073 and a K2 capsule mutant. CFT073 and a deletion mutant lacking K2 capsule (CFT073 Δkps) were adjusted to the same concentration in LB and incubated with 50% NHS or HIS at 37°C for 180 min. Samples were taken at the indicated time points and incubated overnight at 37°C. Viability was assessed by the Miles and Misra method and is presented in CFU/ml.
E. coli K-12 laboratory isolates such as HB101, which do not produce capsule, are completely sensitive to serum, resulting in no surviving cells in serum killing assays (25; data not shown). Unlike that of HB101, the viability of CFT073 ΔkpsC was not reduced to zero by serum exposure, suggesting that mechanisms of serum resistance other than capsule production are present. We therefore sought to define the transcriptome of CFT073 in response to 50% serum and, more specifically, to the bactericidal effects of serum.
Transcriptional response of CFT073 to the bactericidal activity of serum.
Microarray analysis was carried out to determine how ExPEC strain CFT073 responds to the bactericidal action of serum. The transcriptional response of E. coli CFT073 was compared in bacteria exposed to LB supplemented with either 50% NHS or HIS (which has no bactericidal activity [Fig. 1]) for 45 min. This time point was chosen because the viability of a serum-sensitive strain drops most rapidly in the first 45 min upon exposure to serum (Fig. 1). Exposure of logarithmic-phase cells to bactericidal serum resulted in upregulation of 84 genes and downregulation of 23 genes ≥2-fold.
A definable characteristic of the genes upregulated by bactericidal serum was that regulators of cell envelope integrity (and their regulons) were activated. These included the CpxR and σE regulators that are involved in the cell envelope stress response (Table 2). Upregulation of cpxR and rpoE in NHS compared to HIS was confirmed by qRT-PCR (Fig. 2A; P < 0.0001). In active serum, the CpxR, CpxP, and DegP components of the Cpx two-component system were upregulated. Both the alternate sigma factor σE and anti-sigma factor RseA were upregulated in response to NHS (Table 2).
TABLE 2.
CFT073 envelope stress response regulators and Rcs-regulated genes induced in response to the bactericidal properties of serum
| Gene | Function | Fold induction |
|---|---|---|
| Regulators | ||
| rpoE | RNA polymerase sigma factor E | 3 |
| rseA | Anti-RNA polymerase sigma factor E | 3 |
| cpxR | Envelope stress response regulator | 3 |
| cpxP | Periplasmic stress response protein | 5 |
| Rcs regulated | ||
| osmB | Lipoprotein, multistress response | 19 |
| bdm | Biofilm-dependent modulation protein | 13 |
| yjbE | Exopolysaccharide production | 12 |
| yjbF | Exopolysaccharide production | 4 |
| wcaD | Colanic acid biosynthesis | 7 |
| wcaE | Colanic acid biosynthesis | 4 |
| gmd | Colanic acid biosynthesis | 3 |
| wcaF | Colanic acid biosynthesis | 3 |
| ugd | Colanic acid biosynthesis | 4 |
| ydhA and mliC | Inhibitor of c-type lysozyme | 6 |
| ykfE and ivy | Inhibitor of vertebrate c-type lysozyme | 5 |
FIG 2.
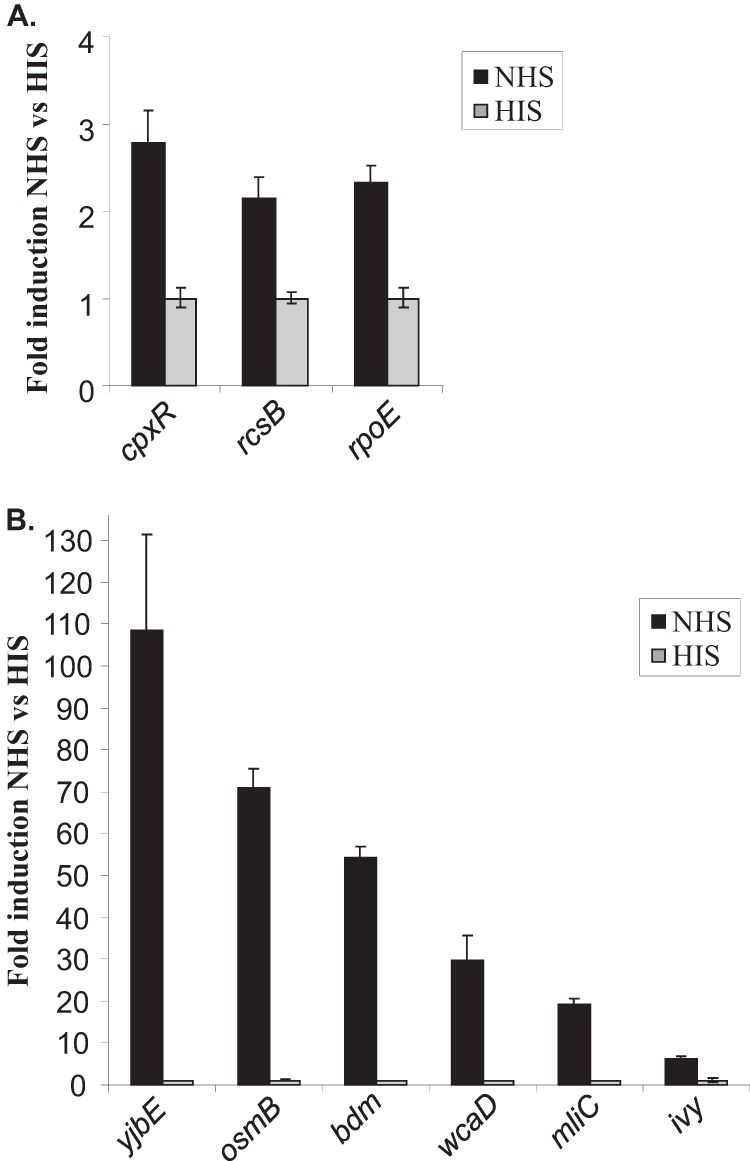
Validation of microarray data by qRT-PCR. qRT-PCR analysis confirmed upregulation of envelope stress response regulators (A) and RcsB-regulated genes (B) of CFT073 in response to NHS as a ratio to HIS.
Twenty-three percent of the genes whose expression was induced by active serum are known to be regulated by the two-component Rcs phosphorelay. Although the Rcs phosphorelay components were not detected by the microarray, qRT-PCR demonstrated that the response regulator RcsB was 2-fold upregulated by active serum (Fig. 2A; P < 0.0001).
A number of RcsB-regulated genes that contribute to the production of an extracellular polysaccharide capsule were found upregulated by CFT073 exposed to active serum (Table 2). The RcsB-regulated genes of the wca operon are involved in the biosynthesis of colanic acid. Colanic acid is the main extracellular polysaccharide produced by E. coli in response to environmental signals and is composed of glucose, galactose, fucose, and glucuronic acid, together with acetate and pyruvate groups. It has been shown to be protective against desiccation and other adverse environmental conditions such as osmotic and oxidative stress. Five genes were upregulated in the colanic acid biosynthesis operon, and upregulation of wcaD was confirmed by qRT-PCR (Table 2 and Fig. 2B; P < 0.0001).
The yjbE and yjbF genes were also upregulated by active serum. This result was confirmed for yjbE by qRT-PCR (Fig. 2B; P < 0.0001). YjbE and YjbFG are reported to be induced by osmotic stress and transcribed as a single operon regulated by RcsB (11, 26). These genes are involved in production of an extracellular polysaccharide distinct from colanic acid (11).
Two Rcs-regulated lysozyme inhibitors were upregulated by active serum, ydhA (also known as mliC) and ivy (inhibitor of vertebrate lysozyme). Expression of these genes was confirmed to be induced by NHS when measured by qRT-PCR (Fig. 2B; P < 0.0001). Ivy has been shown to be protective in human saliva and egg whites, while recent studies have shown that MliC is required for serum resistance of avian-pathogenic E. coli (27).
Downregulated genes.
Only 23 genes were downregulated 2-fold or more in response to bactericidal serum. A number of genes for outer membrane porin proteins were decreased ∼2- to 3-fold in expression, including ompC, ompF, ompN, and nmpC. The cadB gene, which encodes a cadaverine-lysine antiporter, was the most strongly downregulated gene, with a 4.5-fold decrease in expression.
Contribution of envelope stress response regulators to serum survival.
In order to determine whether the envelope stress regulators identified in the microarray study contribute to serum survival, strains lacking rcsB, rpoE, and cpxR were constructed. These were compared to the WT CFT073 strain and the serum-sensitive K2 capsule mutant, CFT073 ΔkpsC. Of the envelope stress response regulators, rpoE had the greatest effect on survival in active serum, with a 41-fold decrease compared to the WT (P = 0.0007; Fig. 3). Although CFT073 ΔrpoE appeared to be decreased in survival in HIS, compared to the WT under the same conditions this difference was not statistically significant (P = 0.07). Survival of CFT073 ΔcpxR was 2.6-fold (P = 0.02) decreased in NHS and could be restored to WT levels in HIS.
FIG 3.
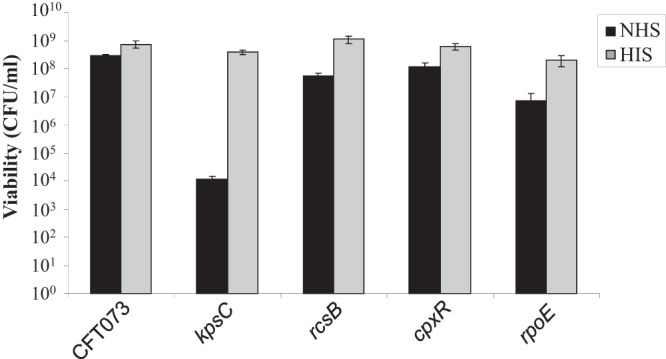
Response regulators contribute to serum resistance. CFT073 and deletion mutants (CFT073 ΔkpsC, CFT073 ΔrcsB, CFT073ΔcpxR, and CFT073ΔrpoE) were adjusted to the same concentration in LB and incubated with 50% NHS or HIS at 37°C for 45 min. Samples were plated and incubated overnight at 37°C. Viability was assessed by the Miles and Misra method and is presented in CFU/ml.
Strain CFT073 ΔrcsB had a 5.3-fold (P = 0.002; Fig. 3) reduction in serum survival, while survival in HIS was similar to that of WT. Deletions of two other genes of the Rcs phosphorelay rcsA and rcsC had no effect on serum survival (see Fig. S1 in the supplemental material).
Contribution of RcsB-regulated genes to serum survival.
Since 23% of genes induced by active serum belong to the Rcs regulon and RcsB was found to be protective in NHS, the effect of RcsB-regulated genes on serum survival was investigated (Fig. 3). Strains with deletions in wcaDE (colanic acid), yjbE (extracellular polysaccharide), and ydhA (lysozyme inhibitor) were constructed. Among the RcsB-regulated genes, deletion of wcaDE of the colanic acid operon had the greatest effect on serum survival (Fig. 4A). A deletion of the same region was previously shown to eliminate colanic acid production in E. coli O157:H7 (28). CFT073 deficient in colanic acid exhibited a 32-fold drop in serum survival (P = 0.002). Eliminating yjbE and mliC did not have a significant effect on survival compared to the WT. In HIS, survival of the mutants was comparable to that of the WT. These results indicate that colanic acid production regulated by RcsB contributes to survival of CFT073 in human serum.
FIG 4.
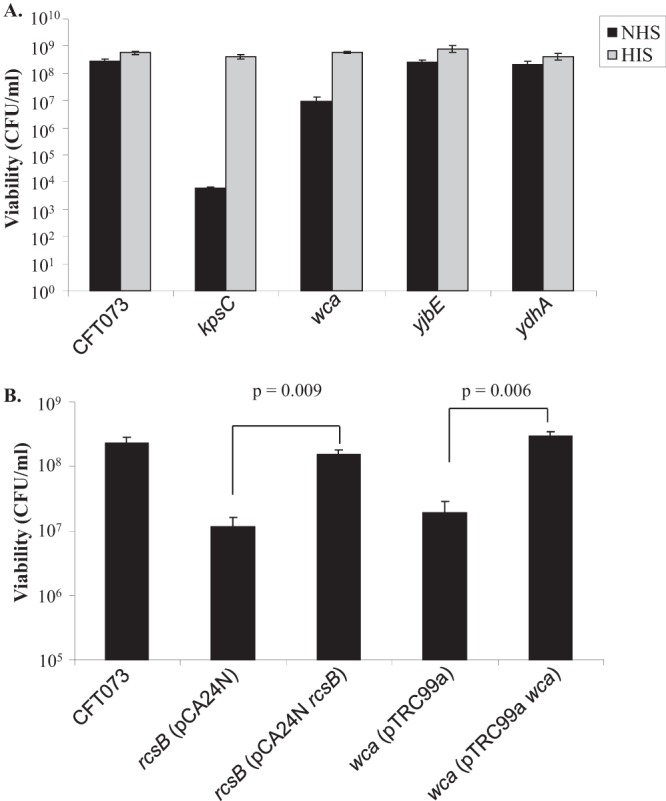
The role of RcsB-regulated genes in serum survival. (A) CFT073 with deletions in exopolysaccharide (wca and yjbE) and lysozyme (ydhA) production genes were adjusted to ∼3 × 108 CFU/ml in LB. (B) CFT073 and deletion mutants (CFT073 ΔrcsB* and CFT073 Δwca*) carrying either empty or recombinant vector were adjusted to ∼3 × 108 CFU/ml in LB. Bacterial cells were incubated with 50% NHS or HIS at 37°C. Viability of bacteria is plotted in CFU/ml and tested by the paired Student t test.
The two most highly upregulated genes identified by the microarray analysis were osmB and bdm (Table 2). Induction of osmB and bdm in NHS was confirmed by qRT-PCR (Fig. 2B; P < 0.0001). These genes are also Rcs regulated; however, they appeared to have no role in serum resistance as determined by testing deletion mutants in a serum killing assay (data not shown).
Complementation of rcsB and wca mutations.
Plasmids encoding rcsB and wcaDE were introduced into strains CFT073 ΔrcsB* and CFT073 Δwca* in order to restore expression of the RcsB regulator and colanic acid. Expression of rcsB from the pCA24N plasmid and wcaDE from pTRC99a restored WT levels of serum survival in CFT073 ΔrcsB* and CFT073 Δwca*, respectively (Fig. 4B). Survival was increased 13-fold (P = 0.009) when CFT073 ΔrcsB* (pCA24N rcsB) was compared to the same strain carrying the empty vector control. Similarly, a 15-fold increase in serum survival was observed for CFT073 Δwca* (pTRC99a wca) compared to CFT073 Δwca* (pTRC99a) (Fig. 4B).
Contribution of exopolysaccharide to serum survival in a K2 capsule mutant.
It is possible that the K2 capsule and the exopolysaccharide capsule regulated by RcsB can act synergistically to protect CFT073 from the bactericidal properties of serum. In order to test this hypothesis, the kpsC mutation was combined with mutations in either rcsB or wca and yjbE. Eliminating rcsB or wcaDE in combination with kpsC resulted in 50-fold and 84-fold reductions in survival, respectively, compared to the sensitive CFT073 ΔkpsC strain (Fig. 5). Deleting yjbE in combination with kpsC did not have a statistically significant effect on survival (P = 0.09). These results confirm that in combination with the K2 capsule, production of the extracellular polysaccharide colanic acid is protective against active serum in ExPEC strain CFT073.
FIG 5.
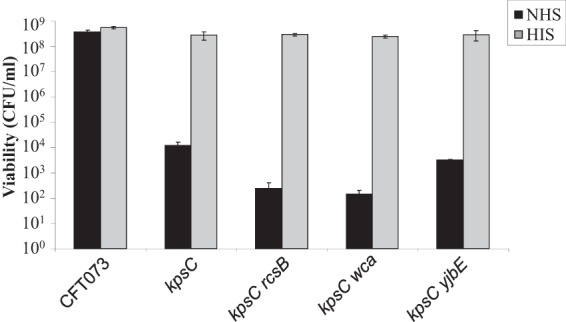
The K2 capsule and colanic acid contribute to serum resistance. CFT073 and mutants (CFT073 ΔkpsC, CFT073 ΔkpsC ΔrcsB, CFT073 ΔkpsC Δwca, and CFT073 ΔkpsC ΔyjbE) were adjusted to the same concentration in LB and incubated with 50% NHS or HIS at 37°C for 45 min. Bacteria were enumerated by the method of Miles and Misra after incubation with sera.
Serum factors necessary for induction of colanic acid expression.
Heating serum inactivates components of the complement system. To demonstrate that the presence of the complement system induces expression of colanic acid, complement was depleted from serum by treatment with zymosan. Zymosan is a yeast-derived polysaccharide isolated from Saccharomyces cerevisiae that activates complement by the alternative pathway (19). Accumulation of complement components on the polysaccharide surface results in a depletion of complement proteins from serum. qRT-PCR was utilized to determine levels of wcaD transcript from cDNA prepared from bacteria incubated in NHS, HIS, and zymosan-treated serum. Levels of wcaD transcript were similar in both zymosan-treated serum and HIS (Fig. 6). When incubated with NHS, levels of wcaD expression from CFT073 were 88-fold higher than in HIS and 95-fold higher than in zymosan-treated serum. These results indicate that an active complement system is required to induce colanic acid expression.
FIG 6.
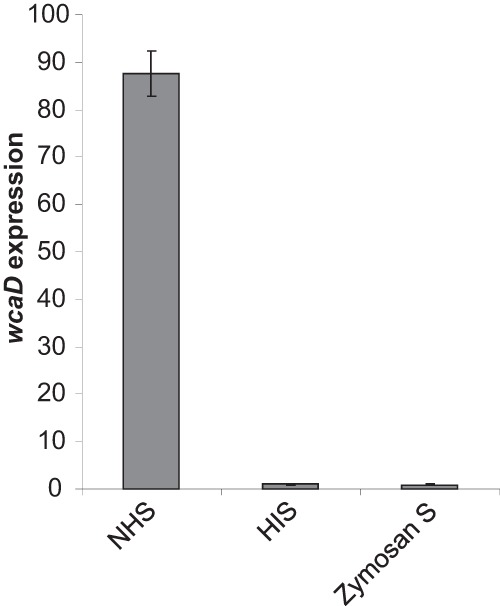
Expression of wcaD in complement-depleted serum. Analysis of wcaD expression was carried out by qRT-PCR with cDNA prepared from bacteria incubated in NHS, HIS, and zymosan-treated serum.
DISCUSSION
To grow and survive in serum, E. coli must modify expression of protective factors that promote survival in a hostile environment. In this study, two major contributors to serum survival were identified upon exposure of ExPEC to human serum. These included the extracytoplasmic stress response pathways and the exopolysaccharide colanic acid.
Several extracytoplasmic stress response pathways have been identified in E. coli, many of which play crucial roles in maintaining membrane integrity and function. Three of these (Rcs, Cpx, and σE) were induced by bactericidal serum (Fig. 3). Previous studies have reported that multiple stress pathway responses can be activated by the same stimuli. There is some overlap in the genes regulated by these three pathways, although each is involved in specific aspects of cell envelope function.
The Cpx two-component system consists of the sensor histidine kinase CpxA and the cytoplasmic response regulator, CpxR (29). CpxP, an inhibitor of the system, is inactivated by the DegP protease. Bactericidal serum induced expression of CpxR, CpxP, and DegP. Also upregulated was the spheroplast protein y (Spy; 8-fold induced), which shares 29% sequence identity with CpxP. Spy is a periplasmic chaperone protein that is induced under envelope stress conditions and has been reported to be regulated by the Cpx and Rcs two-component systems (30, 31). It has been proposed that the main role of Cpx may be as a modulator of the other pathways, and in this study a cpxR mutation had the least effect on serum survival of the three envelope stress regulators tested (31).
The σE regulon plays an important role in maintaining the structure of the outer membrane of Gram-negative bacteria by regulating genes involved in assembly and maintenance of lipopolysaccharide and outer membrane porins (32). Although CpxAR is known to inhibit the rpoE-rseABC operon, expression of both σE and RseA was induced in active serum (29). σE has been shown to be involved in the virulence of many bacterial pathogens (33). In E. coli, it has been implicated in extraintestinal pathogenesis in mouse models of urinary tract infection and peritonitis (34). In this study, σE was found to be protective in human serum, implying a possible role in bacteremia.
The Rcs two-component system controls expression of over 150 genes involved in cell envelope functions (35). Changes to the bacterial cell surface trigger activation of this system and are detected by RcsF on the outer membrane. Detection of activating signals results in autophosphorylation of membrane-spanning RcsC. The phosphoryl group is transferred from RcsC to RscD, which activates the response regulator, RcsB (36). In active serum, the response regulator of the Rcs two-component system rcsB was upregulated and was shown to increase survival of CFT073 in NHS (Fig. 3).
In Salmonella enterica, there is evidence to support a role for the Rcs phosphorelay in virulence, particularly in the transition from localized to systemic infection. Studies have shown that disruption of the Rcs phosphorelay in S. enterica decreased virulence in mice and that this effect was partially dependent on colanic acid production (37, 38).
In this study, RcsB-mediated serum survival seemed to be conferred through induction of the exopolysaccharide colanic acid (Fig. 4A and 5). Other RcsB-regulated genes encoding production of another distinct exopolysaccharide or a lysozyme inhibitor did not make a significant contribution to serum survival. The main role of colanic acid has been proposed to be in biofilm maturation and survival outside the host, since the expression of this exopolysaccharide is usually restricted at 37°C. This restriction is due to the instability of the RcsA protein at 37°C. RcsA forms heterodimers with activated RscB and enhances expression of colanic acid (35). In CFT073, deletion of rcsA had no effect on serum survival at 37°C (see Fig. S1 in the supplemental material). Thus, production of colanic acid does not appear to be dependent on RcsA under the conditions used in this study. RcsB is known to activate expression of other genes such as bdm, osmC, ftsZ, and matA independently of RcsA (39–41). It has also been proposed that increasing the copy number of RcsB can bypass the requirement for RcsA in colanic acid production (42). The increased expression of RcsB in active serum seen in this study may help to explain the lack of requirement for RcsA.
Serum survival was also unaffected by elimination of the sensor kinase of the Rcs phosphorelay, RcsC (see Fig. S1 in the supplemental material). RcsB can be phosphorylated in a RcsC-independent manner by alternate donors such as acetyl phosphate (43). It has also been shown that RcsB can function independently from the phosphorelay components RcsD and RcsC and acetyl phosphate to activate expression of mat fimbriae (42). This indicates that some other phosphate donor may exist or that RscB can activate mat transcription in an unphosphorylated state. In this study, active serum at physiological temperatures induced expression of RcsB-regulated genes, including those of the colanic acid operon. The induction of colanic acid at high temperatures, which did not require RcsA and RcsC, suggests an atypical induction of RcsB, possibly via acetyl phosphate or an alternate mechanism.
Earlier studies examining the role of colanic acid in pathogenesis found no significant effect on serum survival in E. coli strains CP9 and DH1 (44, 45). However, two recent studies have found a role for colanic acid in pathogenesis. In a signature-tagged mutagenesis experiment, colanic acid contributed to septicemia caused by an avian-pathogenic strain of E. coli (46). Colanic acid was also implicated in human serum survival in a recent study investigating the serum resistome of E. coli ST131 (47). To confirm that the protective effect of colanic acid on serum survival was not limited to strain CFT073, the wcaDE genes were deleted in E. coli RS218, a prototype strain causing neonatal meningitis. Colanic acid was also protective against serum killing in this ExPEC strain (data not shown). Confounding results from earlier studies may be a result of strain selection and differences in colanic acid regulation.
The transcriptional response to bactericidal serum observed in this study was strikingly similar to that reported previously in E. coli treated with β-lactam antibiotics and colicin M (48, 49). These have a common target in synthesis of peptidoglycan (PG). Agents that target PG such as β-lactam antibiotics and lysozyme are known inducers of the Rcs phosphorelay (48, 50). Cell wall structure maintenance, including recovery from cell wall damage by de novo synthesis of PG, involves this two-component system. E. coli spheroplasts depleted of PG and lacking rcsB are unable to resynthesize cell wall and require colanic acid for survival (51, 52).
The evidence presented by this study suggests that permeabilization of the cell envelope by complement allows a serum component to access peptidoglycan and trigger the Rcs response. A likely candidate is lysozyme, which is known to target the cell wall and induce the Rcs two-component system. The presence of lysozyme activity in the serum used in this study was confirmed by measuring the lytic activity of HIS on Micrococcus lysodeikticus (data not shown). Alternatively, antimicrobial peptides that target the cell wall may be responsible. Since the strain used in this study is serum resistant, mechanisms to repair the damage caused by lysozyme or other serum factors must exist. Upregulation of genes involved in peptidoglycan synthesis indicates that cell wall recovery may be triggered by bactericidal serum. Expression of penicillin binding protein 6b encoded by dacD and the ybjG gene, which has been implicated in recycling of undecaprenyl pyrophosphase, was induced (53, 54). The production of colanic acid may be protective to E. coli while cell wall damage is being repaired.
A number of outer membrane porins were downregulated in response to bactericidal serum. This may serve as a means of evading complement since loss of OmpC has been shown to protect E. coli against the bactericidal effects of serum (55). Decreased expression of OmpF and OmpC may explain the downregulation of the lysine-cadaverine antiporter encoded by cadB. E. coli strains with mutations in both ompF and ompC have reduced expression of cadB (56). Other studies have found that CadC, a transcriptional activator that regulates the cadAB locus, also regulates ompC and ompF (57).
The protective factors identified in this study are not uniquely induced by exposure to serum. Other stressful environments are also known to induce the envelope stress regulators that promote membrane integrity. Similarly, although colanic acid was shown to be protective against serum killing in this study, it is traditionally associated with survival outside the host. Uropathogenic E. coli strains such as CFT073 are opportunistic pathogens of the bloodstream. Utilization of factors that allow survival in other harsh environments may also allow these pathogens to grow and survive in the blood.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Health Research Board of Ireland grant (grant number HRA.POR.2010.33).
We thank the C. Dorman and J. Hinton laboratories for assistance with the microarray protocol. We are also grateful to Fionan Magee and Aimi Mohamad for technical assistance. The assistance of NBRP (Japan) for supplying plasmids is gratefully acknowledged.
Footnotes
Published ahead of print 28 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00800-13.
REFERENCES
- 1.Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5:449–456. 10.1016/S1286-4579(03)00049-2 [DOI] [PubMed] [Google Scholar]
- 2.Fluit AC, Jones ME, Schmitz FJ, Acar J, Gupta R, Verhoef J. 2000. Antimicrobial susceptibility and frequency of occurrence of clinical blood isolates in Europe from the SENTRY antimicrobial surveillance program, 1997 and 1998. Clin. Infect. Dis. 30:454–460. 10.1086/313710 [DOI] [PubMed] [Google Scholar]
- 3.Stelling JM, Travers K, Jones RN, Turner PJ, O'Brien TF, Levy SB. 2005. Integrating Escherichia coli antimicrobial susceptibility data from multiple surveillance programs. Emerg. Infect. Dis. 11:873–882. 10.3201/eid1106.041160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckles EL, Wang X, Lane MC, Lockatell CV, Johnson DE, Rasko DA, Mobley HL, Donnenberg MS. 2009. Role of the K2 capsule in Escherichia coli urinary tract infection and serum resistance. J. Infect. Dis. 199:1689–1697. 10.1086/598524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiles TJ, Kulesus RR, Mulvey MA. 2008. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 85:11–19. 10.1016/j.yexmp.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. 2000. Bad bugs and beleaguered bladders: interplay between uropathogenic Escherichia coli and innate host defenses. Proc. Natl. Acad. Sci. U. S. A. 97:8829–8835. 10.1073/pnas.97.16.8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor RP, Burge J, Horgan CD, Shasby M. 1983. The complement-mediated binding of soluble antibody/dsDNA immune complexes to human neutrophils. J. Immunol. 130:2656–2662 [PubMed] [Google Scholar]
- 8.Cooper NR. 1999. Biology of the complement system, p 281–315 In Gallin JJ, Snyderman R. (ed), Inflammation: basic principles and clinical correlates. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 9.Callewaert L, Michiels CW. 2010. Lysozymes in the animal kingdom. J. Biosci. 35:127–160. 10.1007/s12038-010-0015-5 [DOI] [PubMed] [Google Scholar]
- 10.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39–68. 10.1146/annurev.biochem.75.103004.142545 [DOI] [PubMed] [Google Scholar]
- 11.Ferrieres L, Aslam SN, Cooper RM, Clarke DJ. 2007. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology 153:1070–1080. 10.1099/mic.0.2006/002907-0 [DOI] [PubMed] [Google Scholar]
- 12.Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasadarao NV, Blom AM, Villoutreix BO, Linsangan LC. 2002. A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J. Immunol. 169:6352–6360 http://www.jimmunol.org/content/169/11/6352 [DOI] [PubMed] [Google Scholar]
- 14.Mellata M, Dho-Moulin M, Dozois CM, Curtiss R, Lehoux B, Fairbrother JM. 2003. Role of avian pathogenic Escherichia coli virulence factors in bacterial interaction with chicken heterophils and macrophages. Infect. Immun. 71:494–503. 10.1128/IAI.71.1.494-503.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JR. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waters VL, Crosa JH. 1991. Colicin V virulence plasmids. Microbiol. Rev. 55:437–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambrozic J, Ostroversnik A, Starcic M, Kuhar I, Grabnar M, Zgur-Bertok D. 1998. Escherichia coli CoIV plasmid pRK100: genetic organization, stability and conjugal transfer. Microbiology 144(Part 2):343–352. 10.1099/00221287-144-2-343 [DOI] [PubMed] [Google Scholar]
- 18.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford I, Douglas CW, Heath J, Rees C, Preston FE. 1996. Evidence for the involvement of complement proteins in platelet aggregation by Streptococcus sanguis NCTC 7863. Br. J. Haematol. 94:729–739. 10.1046/j.1365-2141.1996.d01-1857.x [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 21.Mangan MW, Lucchini S, Danino V, Croinin TO, Hinton JC, Dorman CJ. 2006. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59:1831–1847. 10.1111/j.1365-2958.2006.05062.x [DOI] [PubMed] [Google Scholar]
- 22.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291–299. 10.1093/dnares/dsi012 [DOI] [PubMed] [Google Scholar]
- 24.Leying H, Suerbaum S, Kroll HP, Stahl D, Opferkuch W. 1990. The capsular polysaccharide is a major determinant of serum resistance in K-1-positive blood culture isolates of Escherichia coli. Infect. Immun. 58:222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bliska JB, Falkow S. 1992. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc. Natl. Acad. Sci. U. S. A. 89:3561–3565. 10.1073/pnas.89.8.3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ionescu M, Belkin S. 2009. Overproduction of exopolysaccharides by an Escherichia coli K-12 rpoS mutant in response to osmotic stress. Appl. Environ. Microbiol. 75:483–492. 10.1128/AEM.01616-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanderkelen L, Ons E, Van Herreweghe JM, Callewaert L, Goddeeris BM, Michiels CW. 2012. Role of lysozyme inhibitors in the virulence of avian pathogenic Escherichia coli. PLoS One 7:e45954. 10.1371/journal.pone.0045954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao Y, Doyle MP, Chen J. 2001. Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:3811–3815. 10.1128/JB.183.12.3811-3815.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogt SL, Raivio TL. 2012. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol. Lett. 326:2–11. 10.1111/j.1574-6968.2011.02406.x [DOI] [PubMed] [Google Scholar]
- 30.Quan S, Koldewey P, Tapley T, Kirsch N, Ruane KM, Pfizenmaier J, Shi R, Hofmann S, Foit L, Ren G, Jakob U, Xu Z, Cygler M, Bardwell JC. 2011. Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat. Struct. Mol. Biol. 18:262–269. 10.1038/nsmb.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bury-Mone S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, Jacq A, Bouloc P. 2009. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 5:e1000651. 10.1371/pgen.1000651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. 2006. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 4:e2. 10.1371/pbio.0040002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowley G, Spector M, Kormanec J, Roberts M. 2006. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 4:383–394. 10.1038/nrmicro1394 [DOI] [PubMed] [Google Scholar]
- 34.Redford P, Roesch PL, Welch RA. 2003. DegS is necessary for virulence and is among extraintestinal Escherichia coli genes induced in murine peritonitis. Infect. Immun. 71:3088–3096. 10.1128/IAI.71.6.3088-3096.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clarke DJ. 2010. The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol. 5:1173–1184. 10.2217/fmb.10.83 [DOI] [PubMed] [Google Scholar]
- 36.Hagiwara D, Sugiura M, Oshima T, Mori H, Aiba H, Yamashino T, Mizuno T. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735–5746. 10.1128/JB.185.19.5735-5746.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Calderon CB, Garcia-Quintanilla M, Casadesus J, Ramos-Morales F. 2005. Virulence attenuation in Salmonella enterica rcsC mutants with constitutive activation of the Rcs system. Microbiology 151:579–588. 10.1099/mic.0.27520-0 [DOI] [PubMed] [Google Scholar]
- 38.Mouslim C, Delgado M, Groisman EA. 2004. Activation of the RcsC/YojN/RcsB phosphorelay system attenuates Salmonella virulence. Mol. Microbiol. 54:386–395. 10.1111/j.1365-2958.2004.04293.x [DOI] [PubMed] [Google Scholar]
- 39.Francez-Charlot A, Castanie-Cornet MP, Gutierrez C, Cam K. 2005. Osmotic regulation of the Escherichia coli bdm (biofilm-dependent modulation) gene by the RcsCDB His-Asp phosphorelay. J. Bacteriol. 187:3873–3877. 10.1128/JB.187.11.3873-3877.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379–405. 10.1146/annurev.micro.59.050405.101230 [DOI] [PubMed] [Google Scholar]
- 41.Lehti TA, Heikkinen J, Korhonen TK, Westerlund-Wikstrom B. 2012. The response regulator RcsB activates expression of Mat fimbriae in meningitic Escherichia coli. J. Bacteriol. 194:3475–3485. 10.1128/JB.06596-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brill JA, Quinlan-Walshe C, Gottesman S. 1988. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 170:2599–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fredericks CE, Shibata S, Aizawa S, Reimann SA, Wolfe AJ. 2006. Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol. Microbiol. 61:734–747. 10.1111/j.1365-2958.2006.05260.x [DOI] [PubMed] [Google Scholar]
- 44.Russo TA, Sharma G, Weiss J, Brown C. 1995. The construction and characterization of colanic acid deficient mutants in an extraintestinal isolate of Escherichia coli (O4/K54/H5). Microb. Pathog. 18:269–278 [DOI] [PubMed] [Google Scholar]
- 45.Allen PM, Fisher D, Saunders JR, Hart CA. 1987. The role of capsular polysaccharide K21b of Klebsiella and of the structurally related colanic-acid polysaccharide of Escherichia coli in resistance to phagocytosis and serum killing. J. Med. Microbiol. 24:363–370 [DOI] [PubMed] [Google Scholar]
- 46.Li G, Laturnus C, Ewers C, Wieler LH. 2005. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect. Immun. 73:2818–2827. 10.1128/IAI.73.5.2818-2827.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phan MD, Peters KM, Sarkar S, Lukowski SW, Allsopp LP, Moriel DG, Achard ME, Totsika M, Marshall VM, Upton M, Beatson SA, Schembri MA. 2013. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet. 9:e1003834. 10.1371/journal.pgen.1003834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 190:2065–2074. 10.1128/JB.01740-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamensek S, Zgur-Bertok D. 2013. Global transcriptional responses to the bacteriocin colicin M in Escherichia coli. BMC Microbiol. 13:42. 10.1186/1471-2180-13-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callewaert L, Vanoirbeek KG, Lurquin I, Michiels CW, Aertsen A. 2009. The Rcs two-component system regulates expression of lysozyme inhibitors and is induced by exposure to lysozyme. J. Bacteriol. 191:1979–1981. 10.1128/JB.01549-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glover WA, Yang Y, Zhang Y. 2009. Insights into the molecular basis of L-form formation and survival in Escherichia coli. PLoS One 4:e7316. 10.1371/journal.pone.0007316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ranjit DK, Young KD. 2013. The Rcs stress response and accessory envelope proteins are required for de novo generation of cell shape in Escherichia coli. J. Bacteriol. 195:2452–2462. 10.1128/JB.00160-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baquero MR, Bouzon M, Quintela JC, Ayala JA, Moreno F. 1996. dacD, an Escherichia coli gene encoding a novel penicillin-binding protein (PBP6b) with DD-carboxypeptidase activity. J. Bacteriol. 178:7106–7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatar LD, Marolda CL, Polischuk AN, van Leeuwen D, Valvano MA. 2007. An Escherichia coli undecaprenyl-pyrophosphate phosphatase implicated in undecaprenyl phosphate recycling. Microbiology 153:2518–2529. 10.1099/mic.0.2007/006312-0 [DOI] [PubMed] [Google Scholar]
- 55.Liu YF, Yan JJ, Lei HY, Teng MH, Wang C, Tseng CC, Wu JJ. 2012. Loss of outer membrane protein C in Escherichia coli contributes to both antibiotic resistance and escaping antibody-dependent bactericidal activity. Infect. Immun. 80:1815–1822. 10.1128/IAI.06395-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bekhit A, Fukamachi T, Saito H, Kobayashi H. 2011. The role of OmpC and OmpF in acidic resistance in Escherichia coli. Biol. Pharm. Bull. 34:330–334. 10.1248/bpb.34.330 [DOI] [PubMed] [Google Scholar]
- 57.Samartzidou H, Delcour AH. 1999. Excretion of endogenous cadaverine leads to a decrease in porin-mediated outer membrane permeability. J. Bacteriol. 181:791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boyer HW, Roulland-Dussoix D. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459–472 [DOI] [PubMed] [Google Scholar]
- 59.Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver RP. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect. Immun. 39:315–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 61.Amann E, Ochs B, Abel KJ. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301–315 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


