Abstract
Pseudomonas aeruginosa exoenzyme S (ExoS) ADP-ribosylates multiple eukaryotic targets to promote cytopathology and bacterial colonization. ADP-ribosylation of the small GTPase Rab5 has previously been shown to block fluid-phase endocytosis and trafficking of plasma membrane receptors to the early endosomes as well as inhibit phagocytosis of the bacterium. In this study, ExoS is shown to be capable of ADP-ribosylating 6 candidate arginine residues that are located in the effector binding region or in the C terminus of Rab5. Two Rab5 derivatives were engineered, which contained Arg→Ala mutations at four Arg residues within the effector binding region (EF) or two Arg residues within the C-terminal tail (TL). Expression of Rab5(TL) does not affect the ability of ExoS to modify intracellular trafficking, while expression of Rab5(EF) rescued the ability of ExoS to inhibit intracellular trafficking. ADP-ribosylation of effector arginines likely uncouples Rab5 signaling to downstream effectors. This is a different mechanism for inhibition than observed for the ADP-ribosylation of Ras by ExoS, where ADP-ribosylated Ras loses the ability to bind guanine nucleotide exchange factor (GEF). Other experiments showed that expression of dominant negative Rab5(Ser34Asn) does not inhibit ExoS trafficking to the perinuclear region of intoxicated cells. This study provides insight into a mechanism for how ExoS ADP-ribosylation of Rab5 inhibits Rab5 function.
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen, which causes disease primarily among the immunocompromised, patients with cystic fibrosis, or burn patients (1). P. aeruginosa uses a type III secretion system (T3SS) to deliver four effector proteins, which promote an intracellular environment favorable for bacterial survival and replication within the infected organism (2, 3). Exoenzyme S (ExoS), ExoT, ExoU, and ExoY are injected into the target cells, traffic to their respective cellular locations, and associate with eukaryotic protein cofactors to elicit cytopathic effects (4). ExoU is a potent A2 phospholipase (5) and uses ubiquitin as a host cofactor (6), while ExoY is an adenylate cyclase with an unknown host cofactor (7). ExoS and ExoT are bifunctional enzymes, with a Rho GTPase-activating protein domain (RhoGAP) (8–12) and an arginine-specific ADP-ribosyltransferase (ADPr) domain (13–16). The ADPr domains of ExoS and ExoT are activated by 14-3-3 protein factor activating ExoS (FAS) (17, 18). Although sharing 76% sequence identity, ExoS and ExoT ADP-ribosylate different targets (19). ExoT ADP-ribosylates CrkI and CrkII proteins, which uncouples integrin signaling and results in cytoskeleton rearrangement (20, 21). ExoS has broad substrate specificity, with targets including the ezrin, radixin, and moesin (ERM) proteins as well as the small GTPases Ras, Rac, Rab5, Rab7, Rab8, and Rab11. ExoS ADPr activity induces cytotoxicity, cytoskeletal rearrangement, and disruption of vesicle trafficking (22–28).
Rab proteins are members of the Ras superfamily of small GTPases and are involved in intracellular trafficking. Like other GTPases, Rabs cycle between inactive GDP-bound and active GTP-bound forms (29). Rab proteins interact with downstream effector proteins to promote trafficking, tethering, and vesicle fusion mediated by the state of the bound nucleotide, as structural rearrangements in the switch I or switch II region allow interactions with downstream effectors when in the GTP-bound form but not when in the GDP-bound form (30). Different Rab isoforms target cellular cargoes to specific membranes and cellular compartments, often through differing prenylation sequences at their carboxy termini or differing residues in the switch I/II regions (31). Rab5 regulates trafficking of endocytosed cargo from the plasma membrane, transporting clathrin-coated vesicles (CCVs) to the early endosomal (EE) compartment, promoting vesicle tethering and fusion through interaction with the early endosome antigen 1 (EEA1) (32–35).
T3SS delivery of ExoS is clathrin, caveolin, and dynamin independent. Postinjection, ExoS traffics from the plasma membrane to a perinuclear localization to ADP-ribosylate target substrates, utilizing a microtubule-dependent mechanism and associating with Rab-positive vesicles (36–38). In vitro, ExoS-mediated ADP-ribosylation of Rab5 inhibits fluid-phase uptake and blocks the Rab5-EEA1 interaction (28). Intoxication of eukaryotic cells with P. aeruginosa expressing a RhoGAP-deficient mutant of ExoS [ExoS(G−A+)] also inhibits fluid-phase uptake, indicating a role for ExoS ADP-ribosyltransferase activity in inhibition of endocytosis (39). Additionally, ExoS ADPr activity blocks receptor-mediated trafficking by trapping plasma membrane-associated protein receptors in a clathrin-coated vesicle, as shown previously for epithelial growth factor (EGF) receptor (EGFR) (39). EGF binding results in receptor endocytosis through a clathrin-dependent mechanism and utilization of a Rab5- and Rab7-dependent trafficking pathway through the early and late endosomes, resulting in final degradation of the complex in the lysosome (40–42). Recent data suggest that inhibition of Rab5 activity also inhibits macrophage phagocytosis of Pseudomonas aeruginosa and that this inhibition is dependent on ExoS ADPr activity (43).
In this study, the residues of Rab5 ADP-ribosylated by ExoS have been identified, which allowed assessment of the mechanism by which ExoS ADPr activity uncouples endocytic trafficking and blocks phagocytosis of P. aeruginosa. This study also presents data suggesting that ExoS does not require functional Rab5 activity to traffic to the perinuclear region of the cell and exert cytotoxic effects.
MATERIALS AND METHODS
Plasmid vectors and mutagenesis.
Rab5, ExoS, and 14-3-3 protein-coding sequences were cloned into the Escherichia coli pET15B expression vector for protein purification. The wild-type Rab5 construct and mutated Rab5 derivatives were subcloned into pEGFP-C1 (Clontech) for mammalian cell expression studies. The full-length ExoS gene was cloned into pEGFP-N1 (Clontech), with deletions of the enhanced green fluorescent protein (EGFP) gene such that EGFP is not expressed in pExoS. Site-directed mutagenesis of the indicated arginine residues was performed by using Quikchange site-directed mutagenesis (Agilent Technologies). Mutagenesis primers are as follows: 5′-CTGCCAGAGGAGCAGGAGTAGACCTTAC-3′ (positive sense) and 5′-GTAAGGTCTACTCCTGCTCCTCTGGCAG-3′ (negative sense) for R197A, 5′-GGGATACAGCTGGTCAAGAAGCATACCATAGCCTAGC-3′ (positive sense) and 5′-GCTAGGCTATGGTATGCTTCTTGACCAGCTGTATCCC-3′ (negative sense) for R81A, 5′-GCACCAATGTACTACGCAGGAGCACAAGCAGCC-3′ (positive sense) and 5′-GGCTGCTTGTGCTCCTGCGTAGTACATTGGTGC-3′ (negative sense) for R91A, 5′-GAGGAGTCCTTTGCAGCAGCAAAAAATTGGG-3′ (positive sense) and 5′-CCCAATTTTTTGCTGCTGCAAAGGACTCCTC-3′ (negative sense) for R110A, 5′-ATTGGGTTAAAGAACTTCAGGCGCAAGCAAGTCCTAACATTG-3′ (positive sense) and 5′-CAATGTTAGGACTTGCTTGCGCCTGAAGTTCTTTAACCCAAT-3′ (negative sense) for R120A, 5′-GGAGCAAATTCTGCCGCAGGAGCAGGAGTAGACCTTACCG-3′ (positive sense) and 5′-CGGTAAGGTCTACTCCTGCTCCTGCGGCAGAATTTGCTCC-3′ (negative sense) for R195A, 5′-GGGATACAGCTGGTCAAGAAAAATACCATAGCCTAGC-3′ (positive sense) and 5′-GCTAGGCTATGGTATTTTTCTTGACCAGCTGTATCCC-3′ (negative sense) for R81K, and 5′-GCACCAATGTACTACAAAGGAGCACAAGCAGCC-3′ (positive sense) and 5′-GGCTGCTTGTGCTCCTTTGTAGTACATTGGTGC-3′ (negative sense) for R91K.
Expression and purification of recombinant proteins.
His6–14-3-3, His6-ExoS(70–453), and His6-Rab5ΔCAAX proteins in pET15B were expressed in E. coli BL21(DE3) and purified as described previously (44). Briefly, bacteria were plated overnight at 37°C with ampicillin (250 μg/ml). Cells were added to 400 ml L broth plus ampicillin and cultured for 2 h at 30°C, when 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and the cells were cultured for an additional 2 h at 30°C. Cells were harvested; suspended in binding buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, and 5 mM imidazole) with bacterial protease inhibitor cocktail (Sigma), RNase (20 μg/ml), and DNase (20 μg/ml); and broken in a French pressure cell. The lysate was centrifuged (30,000 × g for 20 min), and the soluble fraction was passed through a 0.45-μm filter. The filtrate was subjected to Ni2+ affinity resin chromatography (Qiagen). The His6 fusion proteins were eluted in binding buffer with 250 mM imidazole, and the eluate was subjected to size-exclusion fractionation by Sephacryl S200 (Sigma) in a solution containing 10 mM Tris-HCl (pH 7.6) plus 20 mM NaCl. Peak fractions were dialyzed into a solution containing 10 mM Tris-HCl, 20 mM NaCl, and 40% glycerol and stored at −80°C.
In vitro ADP-ribosylation assay.
Reaction mixtures contained 10 mM Tris-HCl (pH 7.6) and 20 mM NaCl with 2 μg His6-Rab5ΔCAAX, 20 μM ExoS, and 40 μM NAD+ with or without 150 nM FAS. Reaction mixtures were incubated at room temperature for the indicated times, after which each reaction mixture was boiled for 5 min, followed by SDS-PAGE and Coomassie staining. The amount of ADP-ribosylated Rab5 was quantified by densitometry using AlphaView SA software (Cell Biosciences Inc.). Briefly, the band comprising the no-FAS or 0-min time point was boxed, indicative of the electrophoretic mobility of unmodified (non-ADP-ribosylated) Rab5. A box with an identical size was placed in the same location for each additional condition or time point. The percentage of Rab5 which was no longer present due to a differing electrophoretic mobility, as a result of ADP-ribosylation, was then compared to the total Rab5 population. Calculations and statistical analyses were performed with GraphPad Prism 5 software (GraphPad Software Inc.).
Cell culture.
HeLa cells (ATCC CCL-2) were cultured in minimal essential medium (MEM) (Life Technologies) supplemented with 10% fetal calf serum, nonessential amino acids, sodium pyruvate, sodium bicarbonate, and penicillin-streptomycin and maintained humidified at 37°C in 5% CO2. HeLa cells (24-well plates at 70 to 80% confluence) were transiently transfected with Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol.
P. aeruginosa maintenance and HeLa cell infection.
PA103(ΔexoU exoT::Tc)(pUCP-ExoS) was maintained and cultured as described previously (37). pUCP-ExoS(G−A+)-HA contained a hemagglutinin (HA) epitope, and pUCP-ExoS(G−A−)-FLAG contained a 3× FLAG epitope. Intoxication experiments were performed at a multiplicity of infection of 16:1 (bacteria to cells). Plates were centrifuged at 450 × g for 10 min at room temperature to synchronize the infection and then incubated at 37°C for the indicated times of intoxication.
EGFR trafficking experiments.
HeLa cells were mock transfected or transfected with the indicated EGFP-Rab5 construct for 20 h. Cells were then intoxicated by PA103(ΔexoU exoT::Tc)(pUCP-ExoS) for 3 h, washed twice with phosphate-buffered saline (PBS), and then stimulated with 100 ng/ml of AlexFluor555-labeled epithelial growth factor (EGF555) (Life Technologies) at 37°C. EGF555 binds to and stimulates the entry of the epithelial growth factor receptor (EGFR), allowing visualization of EGF-EGFR endocytosis and trafficking, a model for ligand-mediated endocytosis (39, 41, 45). After 30 min, cells were fixed and processed for immunofluorescence microscopy as described below.
ExoS trafficking experiments.
HeLa cells were mock transfected or transfected with pEGFP-ExoS(G−A+)-HA or pEGFP-Rab5(Ser34Asn) (dominant negative Rab5) for 20 h and then infected with PA103(ΔexoU exoT::Tc)[pUCP-ExoS(G−A−)-FLAG] for 60 min. Cells were then processed for immunofluorescence as described below.
Immunofluorescence microscopy.
HeLa cells were washed twice in PBS (plus MgCl and KCl) and then fixed with 4% paraformaldehyde in PBS for 15 min at room temperature, permeabilized with 4% formaldehyde plus 0.1% Triton X-100 for 15 min, and incubated with 150 mM glycine for 15 min. Wells were blocked with PBS containing 2.5% bovine serum albumin (BSA), 10% fetal bovine serum (FBS), 0.05% Tween 20, and 0.1% Triton X-100 for 1 h at room temperature, followed by primary antibody for 1 h at room temperature.
Immunofluorescence.
For immunofluorescence, mouse anti-EEA1 IgG (dilution, 1:1,000; BD Transduction Laboratories) followed by goat anti-mouse IgG647, rabbit anti-Rab5 (1:1,000; Abcam) followed by goat anti-rabbit IgG488, mouse anti-3× FLAG (1:10,000; Sigma) followed by goat anti-mouse IgG568, and rat anti-HA (1:500; Roche) followed by goat anti-rat IgG488 were used. Antibodies were diluted in PBS containing 1% BSA, 5% FBS, 0.05% Tween 20, and 0.1% Triton X-100. Alexa-labeled secondary antibodies were obtained from Life Technologies and used at a 1:500 final dilution. Stained cells were washed 3 times with PBS and mounted with ProLong Gold antifade reagent (Life Technologies). Cells were viewed with a 60× oil immersion objective, and images were taken with a CoolSnap HQ charge-coupled-device (CCD) camera (Photometrics) using Metamorph software (Molecular Devices) and processed with ImageJ software (NIH).
(ii) Colocalization between Rab5 and EEA1.
ImageJ plug-in intensity correlation analysis (ICA) was used (National Institutes of Health, Bethesda, MD), according to the ImageJ instruction manual. Briefly, intensity correlation quotient (ICQ) values of a user-defined region (transfected cells) of the analyzed figures were calculated by ICA. Typical values are as follows: random or independent staining, ICQ = 0; dependent staining, 0 < ICQ < 0.5; segregated staining, 0 > ICQ > −0.5. At least 20 cells were scored under each condition in three independent experiments.
Mass spectroscopy.
Recombinant Rab5 was ADP-ribosylated to saturation and resolved by SDS-PAGE. The protein band comprising ADP-ribosylated-Rab5 was excised and subjected to in-gel trypsinization, followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a linear ion trap-Orbitrap hybrid mass spectrometer (University of Massachusetts—Worcester Proteomic Center). Scaffold 3 (Proteome Software) was used to assign peptides.
RESULTS
ExoS ADP-ribosylates Rab5 at unique sites different from Ras.
Initial experiments attempted to identify the sites of ADP-ribosylation on Rab5 based on previous studies which showed ExoS to ADP-ribosylate Ras at multiple arginine residues (44). The primary ADP-ribosylation site on Ras was shown to be Arg41, the ADP-ribosylation of which uncoupled guanine nucleotide exchange factor (GEF) binding, with Arg128 as a secondary site of ADP-ribosylation (46). While ClustalW amino acid sequence alignment showed that Rab5 contained homologous arginines at positions 39 and 120, these residues did not structurally align with Arg41 and Arg128 in Ras. GTP-bound H-Ras (PDB accession number 5P21) and GTP-bound Rab5A (PDB accession number 1N6H) were used in the structural alignment.
To assess ExoS ADP-ribosylation of Rab5, an in vitro ADP-ribosylation assay was performed, as previously utilized for Ras (46). His6-Rab5 was expressed in E. coli and purified via immobilized metal ion chromatography. ExoS(70–453), which lacks the T3SS sequence and a membrane localization domain that traffics ExoS within cells but retains RhoGAP and ADP-ribosyltransferase activity, was used in this in vitro analysis (47). As ExoS is an arginine-specific ADP-ribosyltransferase (16), alanine mutations engineered into Rab5 at Arg39 or Arg120, as suggested by amino acid sequence alignment, confirmed that neither Arg was a primary site of ADP-ribosylation, as no alteration in the electrophoretic mobility of the mutated proteins was observed (data not shown).
ExoS ADP-ribosylates multiple sites on Rab5.
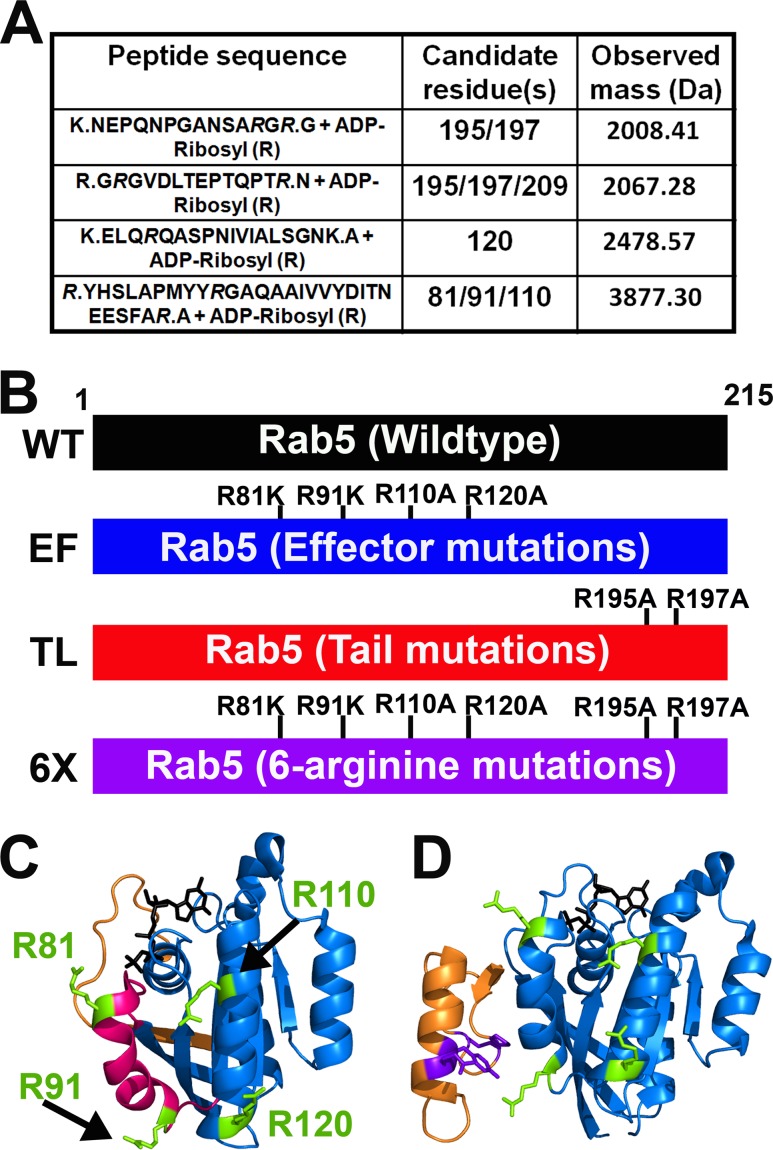
To locate sites of ADP-ribosylation, Rab5 was ADP-ribosylated to saturation and subjected to mass spectrometry analysis. The detected ADP-ribosylated peptides and the candidate arginine residues are shown in Fig. 1A. Engineering of individual alanine mutations at each of the indicated residues through site-directed mutagenesis showed little change in the electrophoretic mobility of ADP-ribosylated Rab5, indicating that ExoS ADP-ribosylated Rab5 at multiple arginines. Structural alignment identified two structural groupings of the arginine residues that were identified as being ADP-ribosylated by mass spectrometry data. One group contained Arg195 and Arg197 and was termed the “tail” (TL) mutations (Arg195Ala and Arg197Ala), since these residues are located on the flexible C terminus near the CAAX box prenylation site of Rab5 (Fig. 1B). The second group was termed the “effector” (EF) mutations due to their location proximal to the switch regions of Rab5 (Arg81Lys, Arg91Lys, Arg110Ala, and Arg120Ala) (Fig. 1B). While distanced in sequence location, these residues cluster in close structural proximity on two helices within or near the switch II region of Rab5 (Fig. 1C), which is known to directly interact with nucleotide exchange factors, such as Rabex5, or downstream effector proteins, such as EEA1 (Fig. 1D).
FIG 1.
Engineered Rab5 constructs. (A) Rab5 was ADP-ribosylated by ExoS to saturation in vitro. A band comprising ADP-ribose–Rab5 was excised and subjected to in-gel trypsinization and LC-MS/MS analysis. Peptides with a mass increase corresponding to ADP-ribose–Arg are shown with the candidate Rab5 arginine residues and observed mass values (Da). (B) Single-amino-acid point mutations were engineered into the wild-type (WT) Rab5 sequence based on mass spectrometry data. Arginine residues were mutated to alanine or lysine to remove ADP-ribosylation sites. Note that Arg81Lys and Arg91Lys mutations were used due to cellular mislocalization of Arg81Ala- and Arg91Ala-transfected constructs. The mutations were grouped into two locations: a group of four in the effector-interacting region (switch II), with a second group of two at the C-terminal tail. These constructs were designated effector mutant (EF) and tail mutant (TL), respectively. A third construct with all six residues mutated was also engineered and named 6-Arg (6×) mutant. These constructs were engineered into pET-15b for recombinant protein production or into pEGFP-C1 for mammalian cell expression. Mutations were confirmed by DNA sequencing. (C) Crystal structure of Rab5a showing Rab5 (blue), the switch I (orange) and switch II (pink) regions, and the effector mutation residues (lime). GTP is shown in black. Note that the tail mutation residues are not labeled, as the flexible C-terminal tail of Rab5 is not amenable to crystallization (adapted from the structure reported under PDB accession number 1N6H). (D) Cocrystal structure of the Rab5-EEA1 interaction showing Rab5 (blue), EEA1 (orange), the effector mutation residues (lime), and EEA1 residues required for interaction with Rab5 (purple). GTP is shown in black. ADPr of EF arginines likely inhibits EEA1-Rab5 contacts necessary for proper vesicle tethering and fusion (adapted from the structure reported under PDB accession number 3MJH).
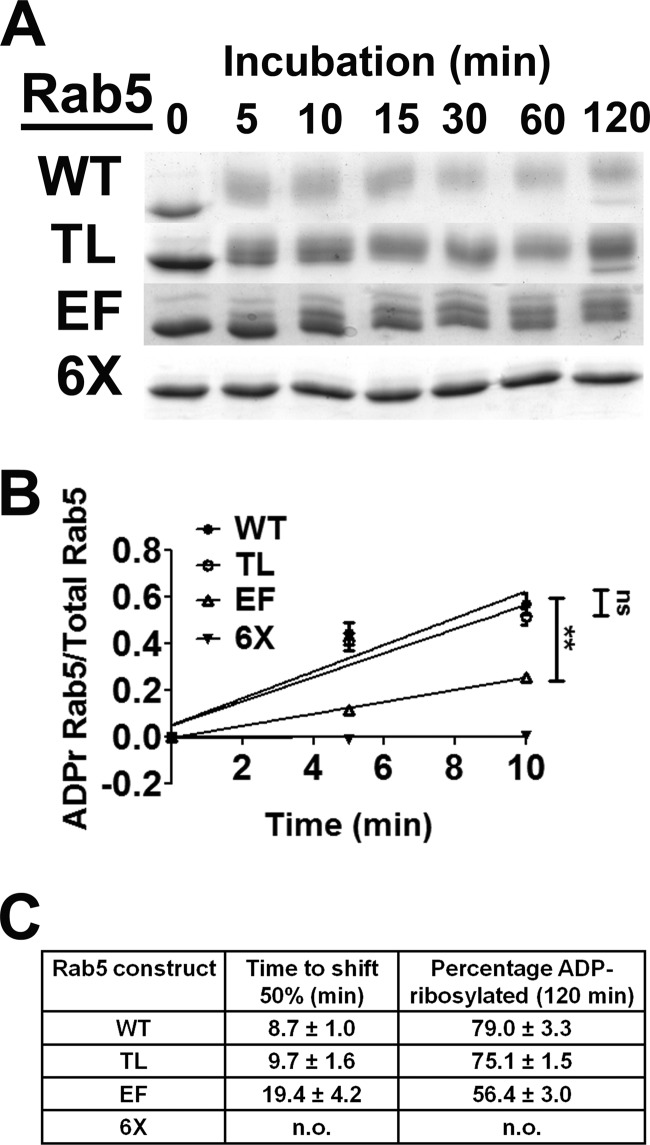
Initial experiments showed that the ADP-ribosylation of Rab5 required FAS (Fig. 2A). Addition of ADP-ribose imparts both an increase in molecular mass (544 Da) and an additional −2 charge, which reduces the electrophoretic mobility of ADP-ribosylated Rab5 relative to that of native Rab5 (Fig. 2A). Mutation of both residues in the TL or the four EF residues resulted in distinct changes in electrophoretic mobility upon ADP-ribosylation by ExoS relative to the ADP-ribosylation of wild-type Rab5 but did not completely block Rab5 ADP-ribosylation (Fig. 2A). Mutation of all six arginine residues comprising the TL and EM (termed 6×) completely blocked ExoS ADP-ribosylation of Rab5 (Fig. 2A). Linear velocity reactions were conducted to compare the relative rates of ADP-ribosylation of the wild type, TL, EF, and 6× proteins to assess the preferred sites of ExoS ADP-ribosylation. The electrophoretic shift of ADP-ribosylated Rab5 was used to standardize the assay, as unmodified Rab5 derivatives display the same electrophoretic mobility. Approximately 80% of wild-type Rab5 was ADP-ribosylated by ExoS in 60 min, while Rab5(TL) showed a rate of ADP-ribosylation similar to that of wild-type Rab5 (Fig. 2B). In contrast, the rate of Rab5(EF) was significantly reduced relative to that of wild-type Rab5, with ∼50% of Rab5(EF) being ADP-ribosylated in the 60-min assay (Fig. 2B). Rab5(6×) was not ADP-ribosylated by ExoS, indicating that these 6 arginines within the EF and TL regions of Rab5 were the sites of ADP-ribosylation by ExoS.
FIG 2.
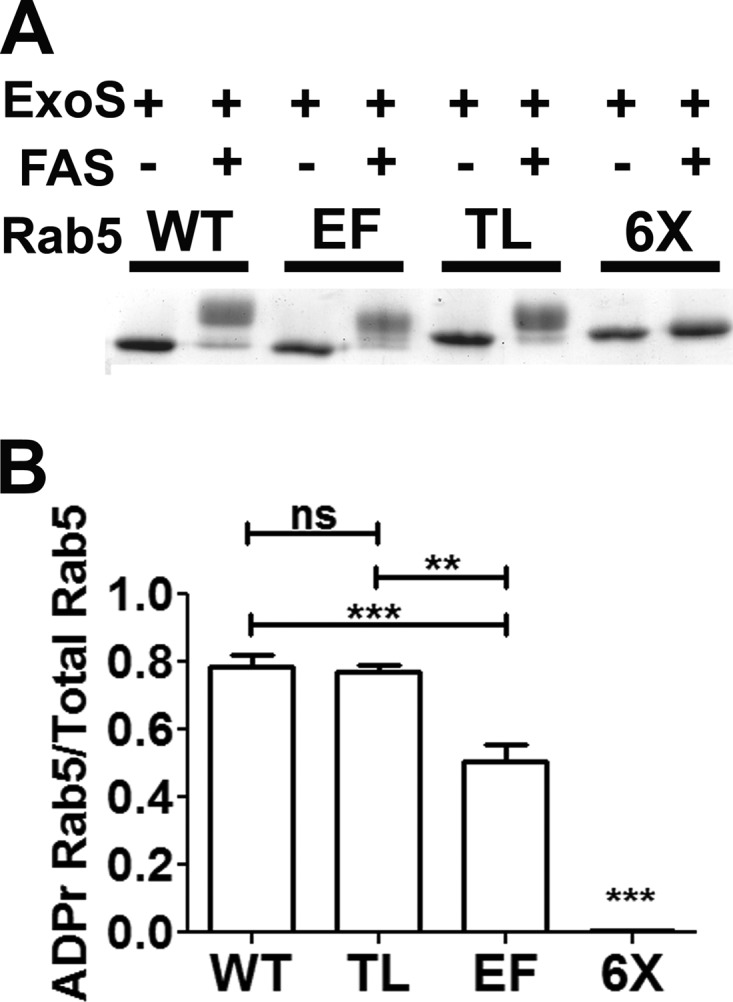
ExoS ADP-ribosylation of Rab5 and Rab5 derivatives. (A) ADP-ribosylation of Rab5 and mutant derivatives. The indicated Rab5 proteins were ADP-ribosylated by ExoS in the presence or absence of 14-3-3 (FAS). The reactions were stopped by addition of Laemmli buffer and boiling for 5 min. Samples were resolved by SDS-PAGE and visualized by Coomassie staining. (B) The amount of ADP-ribosylated Rab5 was quantified as a percentage of the total Rab5 protein by densitometry (see Materials and Methods for a detailed description of the analysis) from each of four separate experiments. The tail mutant shows levels of ADP-ribosylation similar to those of wild-type Rab5, while the effector mutant displays reduced ADP-ribosylation. The 6× arginine mutant showed no detectable ADP-ribosylation. All columns were compared by one-way analysis of variance (∗∗, P < 0.01; ∗∗∗, P < 0.001; ns, not significant).
ExoS ADP-ribosylates Rab5(EF) at a lower rate than wild-type Rab5 or Rab5(TL).
The rates of ADP-ribosylation of Rab5 and Rab5 derivatives were compared over 120 min to further assess the affinity of ExoS for the EF and TL arginines (Fig. 3A). Rab5(TL) was ADP-ribosylated at a rate similar to that of wild-type Rab5, with ∼50% and ∼60% of the respective proteins being ADP-ribosylated in the first 10 min (Fig. 3B). In contrast, Rab5(EF) showed a reduced rate of ADP-ribosylation, with ∼25% of the respective proteins being ADP-ribosylated in the first 10 min. Rab5(6×) did not show any detectable ADP-ribosylation. Over the time course, wild-type Rab5 and Rab5(TL) showed similar reaction rates, which were higher than those of Rab5(EF) (Fig. 3C). This indicated that arginines within the EF region were the primary sites of ADP-ribosylation by ExoS.
FIG 3.
Arginine residues in the Rab5 effector region are the preferred sites of ADP-ribosylation by ExoS. (A) In vitro time course of Rab5 ADP-ribosylation by ExoS. Rab5 and mutant derivatives were incubated with ExoS, 14-3-3, and NAD+. Aliquots were removed at the indicated time points, and enzyme activity was inhibited by addition of Laemmli buffer and boiling. The amount of modified Rab5 at each time point was quantified by densitometry as the fraction of ADP-ribosylated Rab5/total Rab5. (B) Quantification of the kinetics of Rab5 ADP-ribosylation. The fraction of ADP-ribosylated Rab5 as a proportion of total Rab5 was determined at the indicated time points to 10 min. Best-fit lines and statistical significance of the data points were calculated by using GraphPad software (∗∗, P > 0.01). (C) Percentage of total Rab5 ADP-ribosylated at 120 min and time required to ADP-ribosylate 50% of total Rab5 were calculated by using the data in panel A, analyzed by GraphPad software, and displayed in tabular format with the calculated standard errors of the means. n.o., not observed.
Rab5(EF) rescues blockade of EGF trafficking induced by ExoS(G−A+).
To examine the cellular effects of ADP-ribosylation at the EF and TL regions, the effects of transient transfections of EGFP-Rab5 fusion proteins containing the TL, EF, and 6× mutations were determined. Assay conditions were established such that the expression levels of EGFP-Rab5 and mutated Rab5 derivatives were each ∼2-fold higher than the endogenous Rab5 expression levels (not shown), to limit off-target effects of overexpression. To ensure the mutations did not affect Rab5 cellular location, colocalization between endogenous Rab5, EGFP-Rab5, or EGFP-Rab5 derivatives with the Rab5 effector and early endosome marker EEA1 was assessed. Endogenous Rab5 and each of the EGFP-Rab5 fusion proteins showed similar colocalization with EEA1 (data not shown). The Rab5-EEA1 ICQ colocalization values were approximately 0.1 in each transfected cell, indicating that the engineered mutations in the EGFP-Rab5 derivatives did not disrupt Rab5 localization.
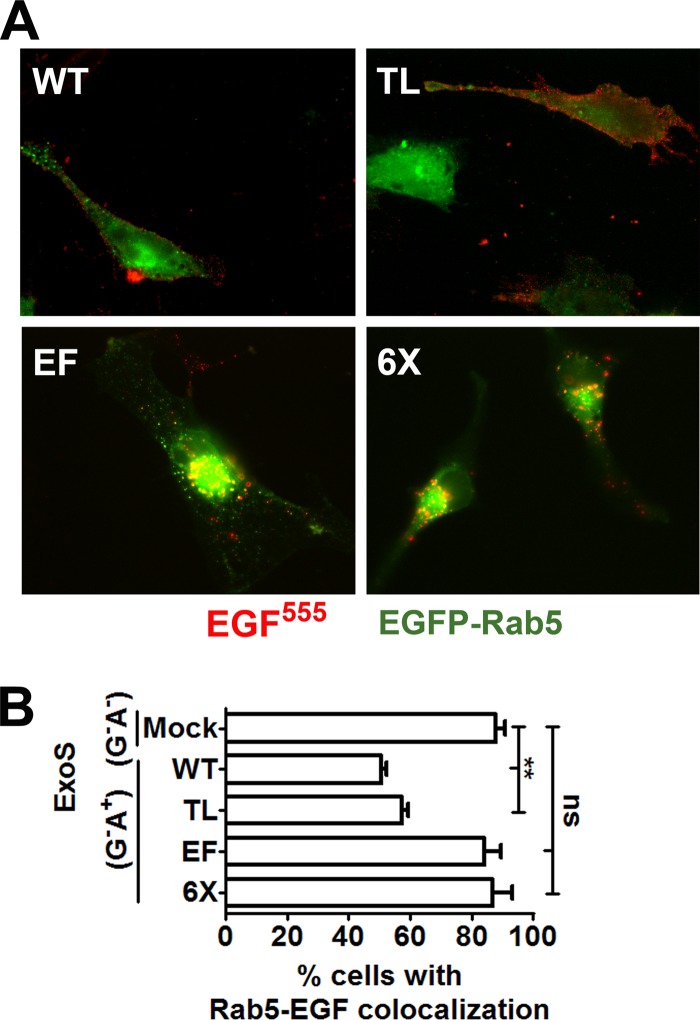
The effect of Rab5 expression on ExoS inhibition of EGFR intracellular trafficking was examined by utilizing intracellular expression of ExoS(G−A+), which is RhoGAP deficient and ADP-ribosyltransferase competent. In HeLa cells transfected with wild-type Rab5 or Rab5(TL), intracellular expression of ExoS(G−A+) blocked trafficking of EGFR activated by EGF555, trapping the EGFR at the plasma membrane (Fig. 4A). However, in cells expressing Rab5(EF) or Rab5(6×) and ExoS(G−A+), EGF colocalized with Rab5-positive vesicles (Fig. 4A), indicating that expression of Rab5 derivatives that cannot be ADP-ribosylated in the effector region rescued ExoS blockade of EGFR trafficking. Since Rab5(EF) alone rescues EGFR trafficking, this suggests the TL mutations do not play a role in ExoS-mediated trafficking inhibition. Quantification of the colocalization of EGF and Rab5 showed that expression of Rab5(EF) or Rab5(6×) resulted in ∼90% of transfected cells with EGF colocalized with Rab5, similar to mock-transfected cells (88%), while in cells expressing wild-type Rab5 or Rab5(TL), the number of cells with colocalized EGF and Rab5 was significantly reduced to ∼55% of transfected cells (Fig. 4B). These experiments show that expression of Rab5 that is not ADP-ribosylated in the effector region rescues the ability of ExoS to block EGFR intracellular trafficking.
FIG 4.
Rab5(EF) and Rab5(6×) rescue EGF trafficking in HeLa cells expressing ExoS(G−A+). (A) EGF trafficking in HeLa cells expressing EGFP-Rab5 or derivatives and pExoS. HeLa cells were cultured to 70% confluence and transfected with the indicated pEGFP-Rab5 construct. After 20 h, cells were transfected with pExoS. After 6 h, EGF555 was added, and the cells were incubated for 30 min, when they were processed for immunofluorescence. For merged images, green indicates Rab5, and red indicates EGF555. (B) Quantification of the number of transfected cells displaying EGF and Rab5 colocalization. Approximately 50 EGFP fluorescent cells were scored for EGFR traffic for each construct in three independent experiments. Columns were compared by one-way analysis of variance (∗∗, P < 0.01).
Rab5(EF) rescues blockade of EGF trafficking induced by T3SS-delivered ExoS.
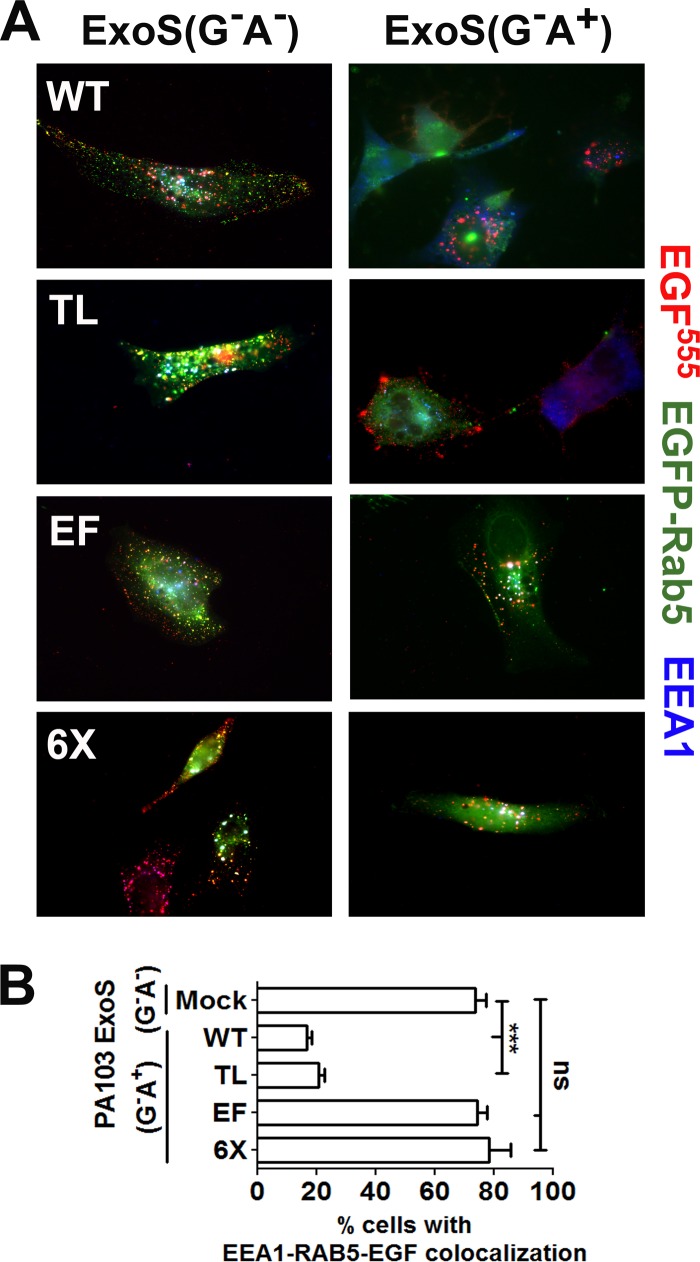
Next, the effect of Rab5 expression on the inhibition of EGFR trafficking by T3SS-delivered ExoS was tested. HeLa cells transfected with wild-type Rab5 or Rab5 derivatives were intoxicated by PA103(ΔexoU exoT::Tc) carrying ExoS(G−A+) or ExoS(G−A−) as an ADPr control. Following the addition of EGF for 30 min, ∼70% of HeLa cells mock transfected and then intoxicated with ExoS(G−A−) displayed EGF-EEA1-Rab5 triple-positive vesicles (Fig. 5A). In comparison, in HeLa cells expressing wild-type Rab5 or Rab5(TL), T3SS-delivered ExoS(G−A+) reduced EGFR trafficking to EEA1-Rab5-containing vesicles significantly, with <20% of cells possessing EGFR-EEA1-Rab5 triple-positive vesicles. As before, expression of Rab5(EF) or Rab5(6×) rescued the ability of HeLa cells intoxicated with ExoS(G−A+) to traffic EGFR to EEA1-Rab5-containing vesicles, with ∼75% of cells showing EGF-EEA1-Rab5 triple-positive vesicles (Fig. 5B). Thus, Rab5 that is not ADP-ribosylated in the effector region rescues the inhibition of EGFR intracellular trafficking induced by T3SS-delivered ExoS ADPr activity, while the TL residues play no appreciable role in ExoS trafficking inhibition.
FIG 5.
Rab5(EF) and Rab5(6×) rescue EGFR trafficking in cells intoxicated with T3SS-delivered ExoS. (A) HeLa cells transfected with the indicated pEGFP-Rab5 construct were intoxicated with PA103(ΔexoU exoT::Tc)[pUCP-ExoS(G−A+)]. After 180 min, EGF555 was added for 30 min, when cells were fixed for immunofluorescence analysis as described in Materials and Methods. Triple colocalization of EGF, Rab5, and EEA1 appears as white puncta. (B) Percentage of transfected cells with triple colocalization of EGF-EEA1-Rab5. Approximately 50 EGFP fluorescent cells from three independent experiments were scored for each construct. Results were compared by one-way analysis of variance (∗∗∗, P < 0.001).
Intracellular trafficking of ExoS is independent of Rab5.
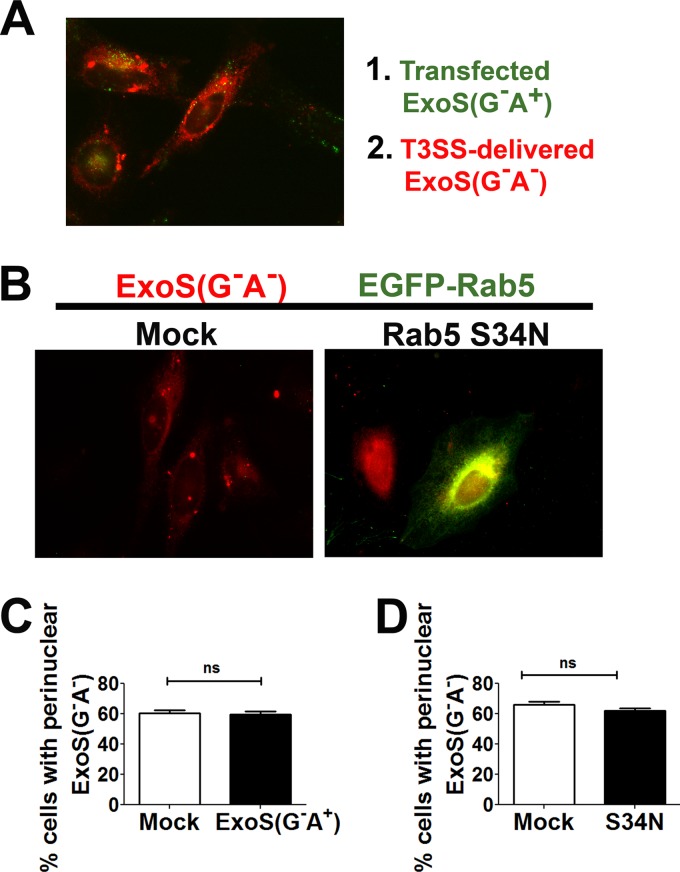
To determine if ExoS requires Rab5 activity for intracellular trafficking to the perinuclear region, HeLa cells were mock transfected or transfected with ExoS(G−A+)-HA for 20 h to block endocytosis and then intoxicated by PA103(ΔexoU exoT::Tc)[pUCP-ExoS(G−A−)-FLAG] as an indicator for the trafficking of T3SS-delivered ExoS. Cells were probed for the HA epitope to identify cells containing ExoS(G−A+) and the FLAG epitope to identify cells containing T3SS-delivered ExoS(G−A−). In cells either mock transfected or expressing ExoS(G−A+), T3SS-delivered ExoS(G−A−) trafficked to a perinuclear location (Fig. 6A). Quantification of the percentage of cells displaying perinuclear-localized T3SS-delivered ExoS(G−A−) showed no significant differences between the ExoS(G−A+) and mock-transfected cells (Fig. 6C). Controls showed that the experimental conditions used resulted in blockade of EGFR trafficking in cells expressing ExoS(G−A+) (data not shown).
FIG 6.
Intracellular trafficking of T3SS-delivered ExoS does not require functional Rab5. (A) HeLa cells were mock transfected or transfected with pExoS(G−A+)-HA for 20 h and then intoxicated by PA103(ΔexoU exoT::Tc)[pUCP-ExoS(G−A−)-FLAG] at a multiplicity of infection of 16 at 37°C for 1 h. Cells were fixed and probed for HA (transfected ExoS) and FLAG (T3SS-delivered ExoS), and the merged image is shown. Red, ExoS(G−A−)-FLAG; green, ExoS(G−A+)-HA. (B) Expression of a dominant negative Rab5 construct does not block ExoS trafficking to the perinuclear region. HeLa cells mock transfected or transfected with Rab5(Ser34Asn) for 20 h were intoxicated with PA103(ΔexoU exoT::Tc)[pUCP-ExoS(G−A−)-FLAG] for 1 h. The cells were then fixed, permeabilized, and stained for the FLAG epitope. (C) Percentage of cells displaying perinuclear localization of T3SS-delivered ExoS(G−A−) in pExoS(G−A+)- or mock-transfected cells. Approximately 100 cells were counted under each transfection condition in four independent experiments. Columns were compared by unpaired Student's t test. (D) Percentage of cells displaying perinuclear localization of T3SS-delivered ExoS(G−A−) in pEGFP-Rab5(Ser34Asn)-transfected cells. Approximately 100 cells were counted under each transfection condition in four independent experiments. Columns were compared by unpaired Student's t test.
A second approach was used to test if Rab5 contributed to T3SS-delivered ExoS intracellular trafficking. HeLa cells were transfected with dominant negative Rab5 [Rab5(Ser34Asn)], and after 20 h, cells were intoxicated with PA103(ΔexoU exoT::Tc)[pUCP-ExoS(G−A−)-FLAG]. In cells mock transfected and in cells expressing dominant negative Rab5, ExoS(G−A−) localized to the perinuclear region by 1 h postintoxication (Fig. 6B). Cells mock transfected and cells expressing Rab5(Ser34Asn) showed no significant difference in the percentage of cells displaying perinuclear localization of T3SS-delivered ExoS(G−A−) (Fig. 6D), indicating that ExoS does not require a functional Rab5 signaling pathway for intracellular trafficking to the perinuclear region.
DISCUSSION
ExoS is an active bacterial toxin, with multiple mechanisms to modify host cell function, including a RhoGAP activity and an ADP-ribosyltransferase activity. Previous research has identified Rab5 as a target of ADP-ribosylation, resulting in inhibition of fluid-phase and receptor-mediated endocytic pathways. The mechanism behind this inhibition was previously undefined. The current study identifies six arginine residues that are ADP-ribosylated by ExoS. While arginines at positions 195 and 197 are ADP-ribosylated by ExoS, they are lower-affinity targets and do not appear to play a role in ExoS ADPr-mediated endocytosis inhibition. While it was hypothesized that their location near the prenylation-site CAAX box would result in interference with prenylation and cellular localization abnormalities, no apparent localization differences have been identified in this study. ADP-ribosylation of arginines located at positions 81, 91, 110, and 120 appears to be the mechanism by which ExoS inhibits endocytosis. Mutation of all four of these residues rescues ExoS-induced trafficking inhibition in epithelial cells and prevents sequestration of plasma membrane protein receptors in clathrin-coated vesicles at the plasma membrane. Mutation of all 6 arginine residues (6×) also rescues EGFR trafficking; however, mutation of just the EF residues is sufficient to restore proper trafficking of EGFR, indicating that the EF residues are the primary residues responsible for ExoS-induced trafficking inhibition during intoxication. Of note, the point mutations utilized to characterize Rab5 at Arg81 and Arg91 were Arg→Lys mutations, since we observed that Rab5(Arg81Ala) and Rab5(Arg91Ala) did not localize like wild-type Rab5 (not shown). This is consistent with previous studies that reported that Rab5(Arg81Ala) is not prenylated and was mislocalized in cells (48).
The four arginines within the EF region are located within or are proximal to the switch II region of Rab5. In particular, Arg81 and Arg91 are contained on an α-helix of switch II that is implicated in interactions with downstream effectors, like EEA1 and Rabaptin5, or RabGEFs, like Rabex5 (49–52). Analysis of Rab5-effector protein cocrystal structures suggests that ADP-ribosylation at Arg81 or Arg91 could directly inhibit residue contacts required for protein-protein interactions, especially those at Rab5 Tyr82 (EEA1, Rabex5, and Rabaptin5), Tyr90 (Rabex5), or Arg91 (EEA1) (49–51). Disruption of these contacts would then result in loss of Rab5 activation through GEF activity or in an inability to interact with EEA1 to promote vesicle tethering and fusion (Fig. 1D) (52). ADP-ribosylation of Arg110 or Arg120 may also affect protein-protein interactions, although less directly, through displacement of the switch I loop or switch II α-helix, altering Rab5 structural organization through steric hindrance or charge repulsion from ADP-ribose. Disorder of either switch region structure results in loss of effector binding (50). An inability to interact with downstream effectors or nucleotide exchange factors as a result of ExoS ADP-ribosylation of the effector region arginines is likely the mechanism behind ExoS inhibition of Rab5 activity, which is required for phagocytosis of P. aeruginosa (43). However, it is unclear whether ExoS ADP-ribosylation blocks GDP-GTP exchange and Rab5 activation in vivo or merely inhibits the Rab5-EEA1 interaction (28). ADP-ribosylation of the effector region arginines may contribute to the inability of certain RabGEFs to rescue ExoS ADPr-induced inhibition of phagocytosis of P. aeruginosa, as the switch II region is required for interactions with GEF Rabex5, which was unable to rescue phagocytosis (43, 51).
The observation that ExoS does not ADP-ribosylate sites in Rab5 homologous to the sites ADP-ribosylated in Ras suggests that ExoS utilizes a specific mechanism of substrate recognition and does not simply modify all surface-exposed arginine residues. While the protein crystal structures of Ras and Rab5 are highly homologous (root mean square value of 1.71 Å for 160 α-carbon atoms), the residues ADP-ribosylated are in discrete locations. This suggests that ExoS may utilize a substrate-targeting mechanism to allow for the greatest inhibitory effect on the eukaryotic substrate. Additionally, despite the similarities between Ras and Rab5, the mechanisms of toxin activity are somewhat different. ExoS ADP-ribosylation of Ras at Arg41 inhibits Ras signal transduction through blocking nucleotide exchange factor binding to switch I, resulting in Ras inactivation. ADP-ribosylation of Rab5 in the switch II region blocks interactions with downstream effectors, as shown for EEA1 (28), which can prevent vesicle tethering and CCV fusion with early endosomes. The effects of ExoS ADP-ribosylation are clearly more varied and intricate than previously appreciated and suggest that closer examination of ExoS effects on other small GTPases may be beneficial.
Additionally, the discovery that ExoS does not require active Rab5 cycling to traffic from the plasma membrane to the perinuclear region where most of the cytotoxic effects are initiated is intriguing. Neither ExoS(G−A+) expression nor expression of dominant negative Rab5(Ser34Asn) inhibited T3SS-delivered ExoS trafficking to the perinuclear region. This suggests that ExoS may be utilizing a unique pathway or compartment to traffic in the cell, which warrants further evaluation.
ACKNOWLEDGMENT
This study was supported by NIH grant R01 AI-031062.
Footnotes
Published ahead of print 7 October 2013
REFERENCES
- 1.Mendelson MH, Gurtman A, Szabo S, Neibart E, Meyers BR, Policar M, Cheung TW, Lillienfeld D, Hammer G, Reddy S, Choi K, Hirschman SZ. 1994. Pseudomonas aeruginosa bacteremia in patients with AIDS. Clin. Infect. Dis. 18:886–895. 10.1093/clinids/18.6.886 [DOI] [PubMed] [Google Scholar]
- 2.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7:654–665. 10.1038/nrmicro2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659–2669 [DOI] [PubMed] [Google Scholar]
- 4.Yahr TL, Mende-Mueller LM, Friese MB, Frank DW. 1997. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 179:7165–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato H, Frank DW, Hillard CJ, Feix JB, Pankhaniya RR, Moriyama K, Finck-Barbancon V, Buchaklian A, Lei M, Long RM, Wiener-Kronish J, Sawa T. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959–2969. 10.1093/emboj/cdg290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson DM, Schmalzer KM, Sato H, Casey M, Terhune SS, Haas AL, Feix JB, Frank DW. 2011. Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol. Microbiol. 82:1454–1467. 10.1111/j.1365-2958.2011.07904.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yahr TL, Vallis AJ, Hancock MK, Barbieri JT, Frank DW. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. U. S. A. 95:13899–13904. 10.1073/pnas.95.23.13899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J, Barbieri JT. 2004. ExoS Rho GTPase-activating protein activity stimulates reorganization of the actin cytoskeleton through Rho GTPase guanine nucleotide disassociation inhibitor. J. Biol. Chem. 279:42936–42944. 10.1074/jbc.M406493200 [DOI] [PubMed] [Google Scholar]
- 9.Pederson KJ, Vallis AJ, Aktories K, Frank DW, Barbieri JT. 1999. The amino-terminal domain of Pseudomonas aeruginosa ExoS disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol. Microbiol. 32:393–401. 10.1046/j.1365-2958.1999.01359.x [DOI] [PubMed] [Google Scholar]
- 10.Krall R, Sun J, Pederson KJ, Barbieri JT. 2002. In vivo Rho GTPase-activating protein activity of Pseudomonas aeruginosa cytotoxin ExoS. Infect. Immun. 70:360–367. 10.1128/IAI.70.1.360-367.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henriksson ML, Sundin C, Jansson AL, Forsberg A, Palmer RH, Hallberg B. 2002. Exoenzyme S shows selective ADP-ribosylation and GTPase-activating protein (GAP) activities towards small GTPases in vivo. Biochem. J. 367:617–628. 10.1042/BJ20020714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrity-Ryan L, Kazmierczak B, Kowal R, Comolli J, Hauser A, Engel JN. 2000. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect. Immun. 68:7100–7113. 10.1128/IAI.68.12.7100-7113.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coburn J, Gill DM. 1991. ADP-ribosylation of p21ras and related proteins by Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 59:4259–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coburn J, Wyatt RT, Iglewski BH, Gill DM. 1989. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J. Biol. Chem. 264:9004–9008 [PubMed] [Google Scholar]
- 15.Radke J, Pederson KJ, Barbieri JT. 1999. Pseudomonas aeruginosa exoenzyme S is a biglutamic acid ADP-ribosyltransferase. Infect. Immun. 67:1508–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knight DA, Barbieri JT. 1997. Ecto-ADP-ribosyltransferase activity of Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 65:3304–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coburn J, Kane AV, Feig L, Gill DM. 1991. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J. Biol. Chem. 266:6438–6446 [PubMed] [Google Scholar]
- 18.Fu H, Coburn J, Collier RJ. 1993. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc. Natl. Acad. Sci. U. S. A. 90:2320–2324. 10.1073/pnas.90.6.2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu S, Yahr TL, Frank DW, Barbieri JT. 1997. Biochemical relationships between the 53-kilodalton (Exo53) and 49-kilodalton (ExoS) forms of exoenzyme S of Pseudomonas aeruginosa. J. Bacteriol. 179:1609–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J, Barbieri JT. 2003. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. J. Biol. Chem. 278:32794–32800. 10.1074/jbc.M304290200 [DOI] [PubMed] [Google Scholar]
- 21.Deng Q, Sun J, Barbieri JT. 2005. Uncoupling Crk signal transduction by Pseudomonas exoenzyme T. J. Biol. Chem. 280:35953–35960. 10.1074/jbc.M504901200 [DOI] [PubMed] [Google Scholar]
- 22.Maresso AW, Baldwin MR, Barbieri JT. 2004. Ezrin/radixin/moesin proteins are high affinity targets for ADP-ribosylation by Pseudomonas aeruginosa ExoS. J. Biol. Chem. 279:38402–38408. 10.1074/jbc.M405707200 [DOI] [PubMed] [Google Scholar]
- 23.Maresso AW, Deng Q, Pereckas MS, Wakim BT, Barbieri JT. 2007. Pseudomonas aeruginosa ExoS ADP-ribosyltransferase inhibits ERM phosphorylation. Cell. Microbiol. 9:97–105. 10.1111/j.1462-5822.2006.00770.x [DOI] [PubMed] [Google Scholar]
- 24.Vincent TS, Fraylick JE, McGuffie EM, Olson JC. 1999. ADP-ribosylation of oncogenic Ras proteins by Pseudomonas aeruginosa exoenzyme S in vivo. Mol. Microbiol. 32:1054–1064. 10.1046/j.1365-2958.1999.01420.x [DOI] [PubMed] [Google Scholar]
- 25.Fraylick JE, Rucks EA, Greene DM, Vincent TS, Olson JC. 2002. Eukaryotic cell determination of ExoS ADP-ribosyltransferase substrate specificity. Biochem. Biophys. Res. Commun. 291:91–100. 10.1006/bbrc.2002.6402 [DOI] [PubMed] [Google Scholar]
- 26.Ganesan AK, Vincent TS, Olson JC, Barbieri JT. 1999. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor-catalyzed nucleotide exchange. J. Biol. Chem. 274:21823–21829. 10.1074/jbc.274.31.21823 [DOI] [PubMed] [Google Scholar]
- 27.Rocha CL, Rucks EA, Vincent DM, Olson JC. 2005. Examination of the coordinate effects of Pseudomonas aeruginosa ExoS on Rac1. Infect. Immun. 73:5458–5467. 10.1128/IAI.73.9.5458-5467.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbieri AM, Sha Q, Bette-Bobillo P, Stahl PD, Vidal M. 2001. ADP-ribosylation of Rab5 by ExoS of Pseudomonas aeruginosa affects endocytosis. Infect. Immun. 69:5329–5334. 10.1128/IAI.69.9.5329-5334.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordens I, Marsman M, Kuijl C, Neefjes J. 2005. Rab proteins, connecting transport and vesicle fusion. Traffic 6:1070–1077. 10.1111/j.1600-0854.2005.00336.x [DOI] [PubMed] [Google Scholar]
- 30.Nikolova L, Soman K, Nichols JC, Daniel DS, Dickey BF, Hoffenberg S. 1998. Conformationally variable Rab protein surface regions mapped by limited proteolysis and homology modelling. Biochem. J. 336:461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes AQ, Ali BR, Ramalho JS, Godfrey RF, Barral DC, Hume AN, Seabra MC. 2003. Membrane targeting of Rab GTPases is influenced by the prenylation motif. Mol. Biol. Cell 14:1882–1899. 10.1091/mbc.E02-10-0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. 1992. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70:715–728. 10.1016/0092-8674(92)90306-W [DOI] [PubMed] [Google Scholar]
- 33.Christoforidis S, McBride HM, Burgoyne RD, Zerial M. 1999. The Rab5 effector EEA1 is a core component of endosome docking. Nature 397:621–625. 10.1038/17618 [DOI] [PubMed] [Google Scholar]
- 34.Rubino M, Miaczynska M, Lippé R, Zerial M. 2000. Selective membrane recruitment of EEA1 suggests a role in directional transport of clathrin-coated vesicles to early endosomes. J. Biol. Chem. 275:3745–3748. 10.1074/jbc.275.6.3745 [DOI] [PubMed] [Google Scholar]
- 35.van der Bliek AM. 2005. A sixth sense for Rab5. Nat. Cell Biol. 7:548–550. 10.1038/ncb0605-548 [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Deng Q, Barbieri JT. 2007. Intracellular localization of type III-delivered Pseudomonas ExoS with endosome vesicles. J. Biol. Chem. 282:13022–13032. 10.1074/jbc.M606305200 [DOI] [PubMed] [Google Scholar]
- 37.Deng Q, Zhang Y, Barbieri JT. 2007. Intracellular trafficking of Pseudomonas ExoS, a type III cytotoxin. Traffic 8:1331–1345. 10.1111/j.1600-0854.2007.00626.x [DOI] [PubMed] [Google Scholar]
- 38.Krall R, Zhang Y, Barbieri JT. 2004. Intracellular membrane localization of Pseudomonas ExoS and Yersinia YopE in mammalian cells. J. Biol. Chem. 279:2747–2753. 10.1074/jbc.M301963200 [DOI] [PubMed] [Google Scholar]
- 39.Deng Q, Barbieri JT. 2008. Modulation of host cell endocytosis by the type III cytotoxin, Pseudomonas ExoS. Traffic 9:1948–1957. 10.1111/j.1600-0854.2008.00808.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barbieri MA, Roberts RL, Gumusboga A, Highfield H, Alvarez-Dominguez C, Wells A, Stahl PD. 2000. Epidermal growth factor and membrane trafficking: Egf receptor activation of endocytosis requires Rab5a. J. Cell Biol. 151:539–550. 10.1083/jcb.151.3.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ceresa BP, Bahr SJ. 2006. rab7 activity affects epidermal growth factor:epidermal growth factor receptor degradation by regulating endocytic trafficking from the late endosome. J. Biol. Chem. 281:1099–1106. 10.1074/jbc.M504175200 [DOI] [PubMed] [Google Scholar]
- 42.Merrifield CJ, Feldman ME, Wan K, Almers W. 2002. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 4:691–698. 10.1038/ncb837 [DOI] [PubMed] [Google Scholar]
- 43.Mustafi S, Rivero N, Olson JC, Stahl PD, Barbieri MA. 2013. Regulation of Rab5 function during phagocytosis of live Pseudomonas aeruginosa in macrophages. Infect. Immun. 81:2426–2436. 10.1128/IAI.00387-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganesan AK, Frank DW, Misra RP, Schmidt G, Barbieri JT. 1998. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J. Biol. Chem. 273:7332–7337. 10.1074/jbc.273.13.7332 [DOI] [PubMed] [Google Scholar]
- 45.Chen P-I, Kong C, Su X, Stahl PD. 2009. Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J. Biol. Chem. 284:30328–30338. 10.1074/jbc.M109.034546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganesan AK, Mende-Mueller L, Selzer J, Barbieri JT. 1999. Pseudomonas aeruginosa exoenzyme S, a double ADP-ribosyltransferase, resembles vertebrate mono-ADP-ribosyltransferases. J. Biol. Chem. 274:9503–9508. 10.1074/jbc.274.14.9503 [DOI] [PubMed] [Google Scholar]
- 47.Knight DA, Finck-Barbancon V, Kulich SM, Barbieri JT. 1995. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 63:3182–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li G, Barbieri MA, Colombo MI, Stahl PD. 1994. Structural features of the GTP-binding defective Rab5 mutants required for their inhibitory activity on endocytosis. J. Biol. Chem. 269:14631–14635 [PubMed] [Google Scholar]
- 49.Mishra A, Eathiraj S, Corvera S, Lambright DG. 2010. Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of early endosomal autoantigen 1 (EEA1). Proc. Natl. Acad. Sci. U. S. A. 107:10866–10871. 10.1073/pnas.1000843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu G, Zhai P, Liu J, Terzyan S, Li G, Zhang XC. 2004. Structural basis of Rab5-Rabaptin5 interaction in endocytosis. Nat. Struct. Mol. Biol. 11:975–983. 10.1038/nsmb832 [DOI] [PubMed] [Google Scholar]
- 51.Delprato A, Merithew E, Lambright DG. 2004. Structure, exchange determinants, and family-wide Rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell 118:607–617. 10.1016/j.cell.2004.08.009 [DOI] [PubMed] [Google Scholar]
- 52.Merithew E, Stone C, Eathiraj S, Lambright DG. 2003. Determinants of Rab5 interaction with the N terminus of early endosome antigen 1. J. Biol. Chem. 278:8494–8500. 10.1074/jbc.M211514200 [DOI] [PubMed] [Google Scholar]