Abstract
Virus diversity and escape from immune responses are the biggest challenges to the development of an effective vaccine against HIV-1. We hypothesized that T-cell vaccines targeting the most conserved regions of the HIV-1 proteome, which are common to most variants and bear fitness costs when mutated, will generate effectors that efficiently recognize and kill virus-infected cells early enough after transmission to potentially impact on HIV-1 replication and will do so more efficiently than whole protein-based T-cell vaccines. Here, we describe the first-ever administration of conserved immunogen vaccines vectored using prime-boost regimens of DNA, simian adenovirus and modified vaccinia virus Ankara to uninfected UK volunteers. The vaccine induced high levels of effector T cells that recognized virus-infected autologous CD4+ cells and inhibited HIV-1 replication by up to 5.79 log10. The virus inhibition was mediated by both Gag- and Pol- specific effector CD8+ T cells targeting epitopes that are typically subdominant in natural infection. These results provide proof of concept for using a vaccine to target T cells at conserved epitopes, showing that these T cells can control HIV-1 replication in vitro.
Introduction
A safe, effective, accessible preventive vaccine against HIV-1 infection will be key to any solution for the AIDS epidemic even in the context of the broader prevention landscape. Ideally, such a vaccine should induce broadly neutralizing antibodies and protective T cells at the same time,1 although either one of these arms alone, if effective, could have an impact on the epidemic.
The work presented here focuses on optimizing the anti–HIV-1 T-cell response. CD8+ T cells exert their protective function by killing virus-infected cells and producing soluble factors with antiviral activities. Despite strong evidence that HIV-1–specific CD8+ T cells contribute to the control of HIV-1 replication during both acute and chronic phases of infection, it has been difficult to identify a single phenotypic correlate of protection in humans.2 However, inhibition of HIV-1 replication in activated CD4+ T cells by autologous CD8+ T cells in vitro does correlate well with virus control in vivo.3,4,5,6
Vaccine-induced T cells and antibodies face the same two problems: trying to match the enormous variability of the infecting HIV-1 and its subsequent ability to escape immune control by mutation.7 Attempts have been made to tackle these major challenges for an HIV-1 vaccine by assembling cocktails of immunogens derived from multiple HIV-1 isolates8 or computing the most representative aa sequences7,9 Another rational approach is to design vaccine immunogens based on the most conserved proteins such as Gag,10,11,12 or better yet subprotein regions of the proteome, which are shared by the majority of HIV-1 variants and thus in principle provide a universal vaccine with potential for global deployment.13,14,15,16,17 It is known that escape mutations in many of these conserved regions will reduce the HIV-1 fitness.18,19,20,21 The challenge is that immune responses to these regions are typically subdominant and have to compete for recognition with rapidly changing (and therefore less protective), immunodominant epitopes.22 Furthermore, in virus infections, immunodomination can decrease the efficiency of T cells responding to subdominant epitopes both at the level of T-cell priming by antigen-presenting cells and recognition of HIV-1–infected targets by effectors.22
We have pioneered the targeting of conserved regions of the HIV-1 proteome by T-cell vaccines.16 We designed the immunogen HIVconsv based on the 14 most conserved subprotein domains of HIV-1 irrespective of known CD8+ T-cell epitopes. For each region, the consensus aa sequence alternates between the four major clades A, B, C, and D (Figure 1a). The gene coding for HIVconsv was inserted into plasmid DNA (pSG2.HIVconsv), nonreplicating chimpanzee adenovirus type 63 (ChAdV63.HIVconsv) and nonreplicating modified vaccinia virus Ankara (MVA.HIVconsv).23 Prior to clinical studies, we demonstrated safety24 and high immunogenicity of the HIVconsv vaccines in mice and macaques.16,23,25,26 Here, we report on the first evaluation of the three HIVconsv vaccines administered in heterologous prime-boost regimens to HIV-1/2–negative volunteers in phase I trial HIV-CORE 002. We show that the vaccines were safe and elicited strong CD4+ and CD8+ T-cell responses, and that the CD8+ T cells recognized HIV-1–infected autologous CD4+ cells and inhibited HIV-1 replication. These efficacy data encourage further clinical development of this approach to a T-cell stimulating vaccine against HIV-1.
Figure 1.
The HIVconsv immunogen and trial design. (a) A schematic representation of the conserved region immunogen HIVconsv. Capital letters above bars indicate the clade used for the derivation of the consensus aa sequence for each conserved segment.16 Below are the regions of the HIVconsv protein covered by overlapping 15/11 peptide pools used in enzyme linked immunoabsorbent spot (ELISPOT) and intracellular cytokine staining (ICS) immunological assays. HIV-1 protein origin of conserved regions are color-coded with navy blue indicating Mamu-A*01 and H-2Dd–restricted T-cell epitopes, and mAb Pk tag added to facilitate protein detection. (b) Trial design. Four immunization schedules used in the HIV-CORE 002 clinical trial are shown, whereby c and C are low and regular doses of ChAdV63.HIVconsv, respectively, D is pSG2.HIVconsv DNA, M is MVA.HIVconsv and P is placebo. The vaccine-to-placebo ratio was 4:1. Note that a longer gap of 8 weeks was used after the ChAdV63.HIVconsv administration before MVA.HIVconsv boost.50
Results
HIVconsv vaccines induced high frequencies of HIV-1–specific T cells
The humanized gene coding for the HIVconsv immunogen, which codes for the 14 most conserved regions of the consensus HIV-1 Gag, Pol, Vif, and Env proteins (Figure 1a), had previously been prepared for vaccination as plasmid DNA, pSG2.HIVconsv (D), and vectored by chimpanzee adenovirus 63, ChAdV63.HIVconsv (C) and modified vaccinia virus Ankara, MVA.HIVconsv (M).16,23 After manufacture to GMP standards, these vaccines were administered to healthy, HIV-1– and 2–uninfected, low-risk adults in Oxford, UK in a double-blind, block randomized phase I trial. In Group 1, 2 subjects received a single low dose of ChAdV63.HIVconsv (c; Figure 1b) to assess vaccine safety as this was the first time that an HIV-1 vaccine delivered by any ChAdV vector was administered to humans. Ten volunteers were recruited into each of Groups 2–4, and received heterologous CM, DDDCM, and DDDMC regimens or placebo (Figure 1b) at a vaccine-to-placebo ratio of 4:1. All vaccines were very well tolerated and induced only mild-to-moderate local or systemic reactions; there were no serious vaccine-related adverse events. The safety data will be reported in a separate manuscript.
As the primary immunological read out, HIVconsv-specific T cells were enumerated using a standardized interferon (IFN)-γ enzyme linked immunoabsorbent spot (ELISPOT) assay, whereby freshly isolated peripheral blood mononuclear cells (PBMC) were stimulated with 199 15-mer peptides overlapping by 11 aa (15/11) representing the entire HIVconsv immunogen, divided into 6 pools of 32–33 peptides (Figure 1a). All (100%) vaccine recipients responded to the c, CM, DDDCM, and DDDMC regimens with median (range) peak total, i.e., sum of pools 1–6, net frequencies of 443 (295–590), 5,150 (1,475–16,510), 5,790 (1,670–7,960), and 2,035 (380–2,550) spot-forming units (SFU)/106 PBMC, respectively (Figure 2). These were unprecedented high levels of postvaccination T-cell responses to HIV-1 peptides. None of the blinded placebo recipients responded. A number of other observations were made. First, the DDD priming before ChAdV63.HIVconsv increased the median (range) peak response from 630 (330–1,470) to 1,550 (390–4,345) SFU/106 PBMC (C versus DDDC: not significant). Second, the highest responses after CM were detected uniformly after 1 week, while after DDDCM varied between 1 and 8 weeks after the MVA.HIVconsv administration implying an effect of DNA priming on effector recruitment. The peak frequencies between DDDCM and DDDMC regimens were significantly different (P = 0.006 after Bonferroni correction).
Figure 2.

Vaccine-elicited frequencies of T cells recognizing conserved regions of HIV-1. Freshly isolated, unexpanded peripheral blood mononuclear cells (PBMC) were stimulated with 6 pools of overlapping 15/11 peptide pools derived from the HIVconsv immunogen (Figure 1a) in an IFN-γ enzyme linked immunoabsorbent spot (ELISPOT) assay and the sums of net peptide Pool 1–6 frequencies are plotted for individual volunteers for each time point. S, screening visit maximum 2 weeks before vaccination. Timing of vaccine administrations is indicated below. Volunteers are color-coded and boxed numbers identify placebo recipients. c and C are low and regular doses of ChAdV63.HIVconsv, respectively; D is pSG2.HIVconsv DNA; and M is MVA.HIVconsv.
The prevaccination mock-stimulated background mean frequency was 20 SFU/106 PBMC, elevated slightly because of two volunteers with increased backgrounds, and similar to the responses seen repeatedly in the blinded placebo controls (Supplementary Figure S1). For all volunteers, frozen samples at 1 week after the last vaccination were retested in the International AIDS Vaccine Initiative reference laboratory and yielded frequencies, which correlated very well with those obtained using the fresh samples (Spearman rank = 0.9156; P < 0.0001), but with a lower magnitude as expected for frozen and thawed samples (Supplementary Figure S2).
HIVconsv vaccine-elicited T cells were of broad specificities
We reasoned that a vaccine that targets multiple HIV-1 epitopes would provide the greatest barrier to immune escape, especially if these epitopes were derived from functionally conserved regions of the virus.12,27,28 Therefore, the breadth of HIVconsv-specific T cells was considered to be one of the critical parameters of the vaccine immunogenicity. Breadth was first indicated by the number of pools, to which individual volunteers responded in the ex vivo IFN-γ ELISPOT assay. Responses elicited by all three heterologous regimens were polyspecific and vaccine recipients in the CM and DDDCM groups recognized median (range) of 6 (5–6) HIVconsv pools (Figure 3). For these two regimens, a cultured IFN-γ ELISPOT assay on week-28 samples was employed to confirm proliferative capacity and expand T-cell numbers prior to testing individual peptides. An average of 13 stimulatory peptides covering an estimated 10 epitopes per vaccine recipient was found. 60% of these responses were to peptides that contained known CD8+ T-cell epitopes matching the volunteers' human leukocyte antigen (HLA) types (Figure 4), while ~40% had not been described previously. 20% of responses were specific for junctional peptides spanning two adjacent conserved regions. No T-cell responses to HIVconsv-derived peptides were identified in the blinded placebo recipients. Thus, the vaccines induced broadly specific T cells against conserved HIV-1 epitopes that are normally, i.e. in natural infection, immunologically subdominant.
Figure 3.
The breadth of vaccine-elicited T cells. Freshly isolated, unexpanded peripheral blood mononuclear cells (PBMC) were stimulated with Pools 1–6 of overlapping 15/11 peptides derived from the HIVconsv immunogen (Figure 1a) or the FEC (influenza virus, EBV, CMV) pool (color-coded) in an IFN-γ enzyme linked immunoabsorbent spot (ELISPOT) assay and the net frequencies of cells specific for each pool are shown. S is screening visit. Volunteer numbers are shown above the graphs and boxed numbers indicate placebo recipients.
Figure 4.
Breadth of vaccine-elicited responses in groups CM and DDDCM. Stimulatory peptides were identified using a cultured IFN-γ enzyme linked immunoabsorbent spot (ELISPOT) assay and are shown as a heat map for individual vaccines. Frozen peripheral blood mononuclear cells (PBMC) from week time points were cultured for 10 days using HIVconsv Pools 1–6 and assayed using individual peptides. Numbers in each box show SFU/106 of cultured cells. In the first column, peptide numbers are color-coded as for their origin: blue, Gag; pink, Pol; yellow, Vif; purple, Env. In the first and second columns, peptides containing a junction indication by a hyphen are shown on orange background. Black horizontal lines separate Pools 1–6. For volunteer no. 405, not enough cells were available for epitope mapping. Volunteers' HLA class I types are shown on the top of the figure.
HIVconsv vaccine-elicited CD4+ and CD8+ T cells were polyfunctional
Multicolor flow cytometry analysis was performed to assess the functionality of vaccine-elicited T cells in terms of production of IFN-γ, tumor necrosis factor (TNF)-α, and interleukin (IL)-2, and degranulation (CD107a). Samples were chosen from time points that were at or close to the peak of the ELISPOT response. Responses to individual peptide pools confirmed the broadly specific responses observed in the IFN-γ ELISPOT assay (Supplementary Figure S3a). For total IFN-γ and TNF-α, the medians varied significantly among the groups (P < 0.012) and some pairwise comparisons with control reached significance (Supplementary Figure S3b). Separately, frozen PBMC from the CM group were restimulated with HIVconsv peptide Pools 1–6 for 2 days and IFN-γ, TNF-α and β, GM-CSF, MIP-1β (CCL4), and IL-2, -4, -5, -9, -10, -13, and -17a were quantified in the culture supernatant using the Luminex assay. This analysis showed that the responses induced by the HIVconsv vaccines were more polyfunctional than the intracellular cytokine assay alone suggested. No responses were seen in samples from the blinded placebo recipients (Supplementary Table S1).
The ability to proliferate to recall antigens is an important feature of protective memory T cells. Vaccine-induced T cells proliferated and produced IFN-γ in the cultured ELISPOT assay employed in the peptide mapping experiments. The carboxyfluorescein succinimidyl ester (CFSE) dilution assay was used for the CM and DDDCM regimens to directly assess T-cell proliferation upon restimulation with HIVconsv peptides at the last time point of the clinical study, week 28. A robust proliferative capacity for both CD4+ and CD8+ T cells was detected, which was not statistically separable between the two regimens (Supplementary Figure S4).
HIVconsv vaccine-elicited T cells inhibited HIV-1 replication in vitro
There is a danger that T-cell responses elicited by artificial peptide-based immunogens might be specific for epitopes that are not naturally presented by HIV-1.29,30 This critical question was tested in a virus-inhibition assay (VIA), whereby vaccine-induced CD8+ T cells were tested for recognition of virus-infected autologous CD4+ T cells. PBMC were expanded in vitro for 7 days in the presence of bi-specific anti-CD3/CD4 antibodies to yield ~90% pure CD3+CD8+ cultures. Autologous target cells were prepared by incubation of PBMC with bi-specific anti-CD3/CD8 antibodies to yield 98% pure CD3+CD4+ cells, which were then infected with HIV-1. CD8+ T cells from every volunteer in the CM, DDDCM, and DDDMC groups were assessed at prevaccination and three postvaccination time points for inhibition of eight different HIV-1 isolates. These were ELI, CH077, CH106, 247Fv2, ZA97012, U455, Bal, and Nef-mutated IIIB chosen for their availability and coverage of clades A, B, and C. Volunteers nos. 411 (CM) and 418 (DDDCM) inhibited the growth of 8 and 6 out of the 8 tested viruses, respectively, with up to 105.79-fold inhibition (Figure 5a). These two vaccinees had protective HLA types B*51:01 and B*27:05, respectively, although several other volunteers with a narrower virus control had protective HLA alleles, too (Figure 4). Overall, 7/7, 3/8, and 5/8 vaccine recipients in the respective CM, DDDCM, and DDDMC groups inhibited both clade B (IIIB) and A (U455) viruses (Figure 5b). For many CM controllers, their suppressive activity was maintained for 20 weeks after vaccination. There was no obvious link between the replicative capacity of individual virus isolates in different volunteers' CD4+ cells (Figure 6) and the virus inhibition. No activity was detected in the prevaccination samples or placebo recipients. Therefore, the vaccination had induced CD8+ T cells capable of recognizing HIV-1–infected cells and inhibiting virus replication in vitro. This is the first evidence that in-natural-infection subdominant T cells recognizing conserved epitopes can control HIV-1 replication in vitro if they are induced strongly enough by a vaccine.
Figure 5.
In vitro HIV-1 inhibition by vaccine-elicited CD8+ T cells. (a) HIV-1 inhibition by two individuals with broad VIA activity. Eight available viruses employed in this assay are listed and color-coded on the right. Dotted lines show the highest background inhibition (1.3 log10). (b) HIV-1 inhibition by individual volunteers for the two most commonly inhibited viruses indicated above the graphs. Dotted lines show the highest background inhibition for each virus of all tested individuals. Boxed volunteer numbers indicate placebo recipients.
Figure 6.
Replicative capacity of HIV-1 isolates in autologous CD4+ cells. HIV-1 growth in expanded autologous CD4+ cells was measured by the p24 concentration in the tissue culture supernatant on day 13. Broad controllers numbers 411 and 418 are highlighted in red and green, respectively. Blue bars indicate the average concentration for individual viruses.
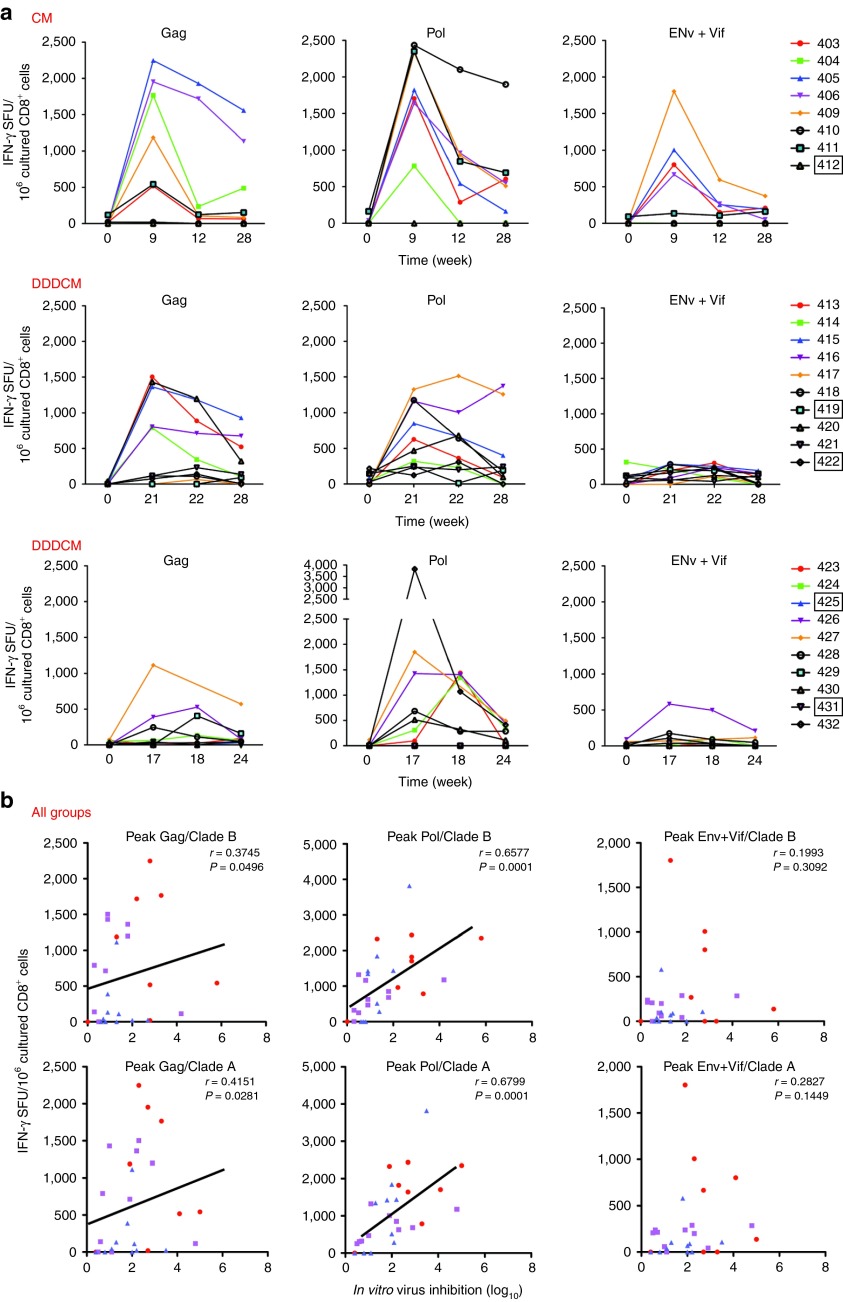
Both Gag- and Pol-specific CD8+ T-cell frequencies correlated with in vitro HIV-1 inhibition
Because protective CD8+ T-cell responses in infected patients are mostly specific for Gag,10,11,12 while Pol-specific responses are less common,31 we determined whether vaccine-induced Pol-specific T cells were as effective in the VIA as those specific for Gag. Therefore, the bi-specific antibody-expanded CD8+ cells were also tested in the IFN-γ ELISPOT assay using peptide pools assembled separately for Gag, Pol, and Env+Vif HIVconsv regions. This analysis revealed stronger responses to Gag and Pol compared to Env+Vif (Figure 7a), probably reflecting the relative representation of these proteins in the HIVconsv immunogen. For all three regimens combined together, there were strong correlations between virus inhibition and the net frequencies of Gag- and, more strongly, Pol-targeted CD8+ T cells from the peak-response time points (Figure 7b). Also the total ex vivo IFN-γ ELISPOT assay frequencies correlated with in vitro peak inhibition of HIV-1 replication for all CM, DDDCM, and DDDMC groups combined (clade A; r = 0.5124; P < 0.0053; Supplementary Figure S5). Thus, although Pol-specific responses are relatively uncommon in natural HIV-1 infection, CD8+ T cells with Pol specificity are perfectly capable of recognizing virus-infected cells and inhibiting HIV-1 replication.
Figure 7.
Correlation of frequencies of Gag- and Pol-specific CD8+ effector cells with virus inhibition. (a) Peripheral blood mononuclear cells (PBMC) from pre- and three postvaccination time points were nonspecifically expanded by incubation with anti-CD3/CD4 bi-specific antibody and subjected to an IFN-γ enzyme linked immunoabsorbent spot (ELISPOT) assay using 4 pools of overlapping 15/11 peptides derived from Gag, 2 pools of Pol and Env+Vif. (The same cultured cells were used for virus-inhibition assay (VIA) shown in Figure 5.) Individual volunteers' frequencies are color-coded with boxed volunteer numbers indicating placebo recipients. (b) For the two most commonly inhibited viruses indicated above the graphs, correlations of nonspecifically expanded CD8+ effector frequencies specific for Gag, Pol, and Env+Vif with overall, i.e., by cells specific for Gag, Pol, Env, and Vif together, virus in vitro inhibition are shown for the peak inhibition time points. Red dots, CM; purple squares, DDDCM; blue triangles, DDDMC. For each correlation, Spearman rank (r) and P (two-tailed) values are shown.
Discussion
The use of prophylactic vaccination to focus early CD8+ T cells on the most conserved regions of the HIV-1 proteome could provide a critical advantage in ensuring a good match between vaccine and infecting virus, thus suppressing the breakthrough infection transmitted/founder viruses and their rapidly emerging escape mutants. This is strongly supported by recent analysis of T-cell responses and escape in acute HIV-1 infection.31 Here, we tested for the first time a conserved immunogen HIVconsv for its T-cell immunogenicity in a phase I clinical trial in HIV-1/2–negative volunteers in the UK. This is a very important step forward, moving from promising preclinical observations in animals to detailed studies in humans, given that immune responses in experimental animals do not always predict how humans will respond.32,33,34 Furthermore, the study enabled us to ask whether vaccine-induced T cells could recognize HIV-1–infected autologous CD4+ T cells.
The gene coding for HIVconsv was delivered by combinations of three nonreplicating vaccine vectors DNA (D), ChAdV-63 (C), and MVA (M). The CM and DDDCM regimens appeared to be the most effective regimens and induced transgene-specific, IFN-γ–producing T cells, which (i) reached unprecedented high median total frequencies of 5.2 k and 5.8 k SFU/106 PBMC, respectively; (ii) included both CD8+ and CD4+ T-cell subpopulations; (iii) were broadly specific; (iv) produced multiple intercellular signaling molecules; (v) proliferated to recall antigens; and (vi) showed efficacy in vitro by inhibiting HIV-1 replication in cultured autologous cells. Therefore, there is no doubt that the conserved region construct in combination with the delivery used here is strongly immunogenic, opening up this approach to countering HIV-1 variability and propensity to escape.
All volunteers in the main three groups were tested at four time points for inhibition of a panel of eight viruses representing the major clades. The first important conclusion from these experiments is that chimeric immunogen HIVconsv made of consensus clade sequences, which may not be present in natural isolates, elicited T cells that recognized HIV-1–infected cells. Second, it was encouraging to detect an overall correlation of ex vivo IFN-γ ELISPOT assay frequencies with virus inhibition (Supplementary Figure S5). Third, and importantly for this construct, Pol-specific CD8+ T-cell responses correlated as well as or better than the Gag-specific responses with virus inhibition. This may be due to the larger representation of Pol relative to Gag in the HIVconsv protein. Preliminary experiments using short-term Pol peptide-expanded cell lines confirmed that these effectors can kill peptide-pulsed target cells (data not shown); assessment of killing of HIV-1–infected cells is under way. This finding appears to contrast with many studies emphasizing the protective association with mainly Gag-specific CD8+ T-cell responses and little or no protection associated with Pol-specific CD8+ T cells.10,11,12 The relative scarcity of Pol-specific responses in acute and chronic infections31 has been attributed to low expression of Pol epitopes on virus-infected cells, but our results suggest that this might affect T-cell priming more than target cell recognition and that Pol, which is relatively conserved, is a good target for vaccination. Indeed induction of strong Pol-specific responses would be a way in which a vaccine could improve on natural anti–HIV-1 responses.
It was intriguing that while T cells from two volunteers were able to inhibit most viruses, most of the others inhibited only the clade B and A viruses employed. The failure to inhibit all viruses could not be explained by differences in the virus replication potentials (Figure 6). For the HIV-1 IIIB isolate used here, inactivation of Nef may have contributed to higher MHC class I cell surface density, making the infected cells better CD8+ T-cell targets. Optimal epitope mapping is under way and this will allow for their sequence comparison with the viruses employed in VIA. Few previous studies have compared different viruses in this type of assay and it is possible that virus isolates differ in their susceptibility to inhibition by T-cell responses, possibly analogous to the tiers of virus sensitivity to antibody neutralization. Further studies will attempt to characterize the mechanism(s) of suppression in greater detail.
The level of virus inhibition in VIA we observed after vaccination is not as strong as that seen in PBMC taken ex vivo in HIV-1–infected people who control infection. This concurs with comparison of DNA/HAdV-5 vaccines with chronically infected patients.35 Given the good prospective correlations between strong virus suppression, delayed loss of CD4+ T cells and lower viral load set-point, VIA could be a highly clinically relevant assay for the assessment of the likely efficacy of CD8+ T-cell–inducing vaccines.6,35,36 Future vaccination strategies will focus on strengthening this effector function.
The frequencies of HIV-1–specific T cells induced by the vaccines were high and correlated with assays on the same samples by the International AIDS Vaccine Initiative reference laboratory. The level of response was considerably higher than the responses seen in the Step Study, which used a trivalent HAdV-5 vaccine delivering HIV-1 B clade Gag, Pol, and Nef, and failed to protect. Furthermore, the HIVconsv-specific responses were detected in 100% of vaccine recipients and recognized an average of eight naturally subdominant HIV-1 epitopes, which originated from conserved regions common to many circulating viruses. This contrasts to the responses seen in the Step Study where only 77% subjects responded, of whom 62% recognized only two to three epitopes, and the T-cell responses were often directed against more variable regions of the clade B virus resulting in a sieve effect that selected escape mutants in those subjects who were subsequently HIV-1 infected.37 Thus, HIV-CORE 002 responses appear to be both quantitatively and qualitatively different from those elicited by the vaccine used in the Step Study, which did not show any protective efficacy. Whether the responses elicited by HIVconsv will be enough to protect in vivo remains to be seen, but it is clear that any T-cell–inducing vaccine that is taken to an efficacy trial in the future will have to generate T-cell responses that demonstrably exceed those seen in the Step Study.
In contrast to the Step Study, the RV144 trial, which tested a combination of ALVAC-vectored Env prime and Env protein boost, gave 31% protection against acquisition, most likely through the generation of nonneutralizing anti-Env antibodies.38,39 No CD8+ T-cell responses were measurable after that vaccination. Currently, attempts are being made to improve the RV144 vaccine regimen and an improved outcome might be achievable if the vaccine induces T cells similar to those elicited by the ChAdV63.HIVconsv-MVA.HIVconsv regimen alongside anti-Env antibodies. Other vaccine designs tested in phase I trials have also given weaker T-cell responses, often in less than 100% of vaccine recipients and have not focused on conserved epitopes.8,40,41,42,43
The HIVconsv vaccine-induced T cells were of broad specificities, whereby each vaccine recipient in the CM and DDDCM groups recognized on average 10 epitopes, of which 10% and 59% originated in the Gag and Pol proteins, respectively. Based on the volunteers' HLA types, ~40% of these were not clearly identifiable in the Los Alamos National Laboratory HIV Immunology Database and were therefore novel CD8+ epitopes or epitopes recognized by CD4+ T cells. Given the HIVconsv epitope subdominance in natural infection, this broadly concurs with a detailed mapping of CD8+ T-cell responses during acute HIV-1 infection, which identified about 30% of previously undescribed epitopes, some of which were in variable regions and escaped early.44 The HIVconsv immunogen is a chimeric protein and contains junctional regions; these junctions can elicit responses against irrelevant epitopes not present in the HIV-1 proteome. This was the case for ~20% of estimated epitope responses. However, it is unlikely that these T-cell responses will interfere with the induction of relevant anti–HIV-1 responses any more that the antivector T cells45 and they will certainly not compete for recognition of HIV-1–infected cells. In BLAST searches on the human genome, the junctional regions had no difference in the number of matches compared with other regions of HIV-1.16 Furthermore, strong junctional responses can be avoided in future vaccine designs/regimens.
One of the vectors used here for the HIVconsv vaccine was simian adenovirus serotype 63. Following the Step, Phambili, and HVTN 505 trials, which all used HAdV-5 as a vector and showed trends of increased acquisition of HIV-1 in vaccine recipients, there is some concern that all adenoviruses may be precluded from use as prophylactic HIV-1 vaccines,32,46 although more qualified recommendations might be appropriate.47 ChAdV-63 is serologically distinct from HAdV-5, but there could be some CD4+ T-cell cross reactivity, which may relate to an increased risk. If that is shown to be the case, alternative delivery for the HIVconsv immunogen will be found. The important finding here concerns the insert, which is immunogenic and elicits T cells that suppress virus replication in vitro. In principle, a conserved region T-cell vaccine such as this, that elicits strong antiviral T-cell responses, should be a good candidate for progressing to a phase IIb efficacy trial, testing either on its own or better in a combination with vaccines that can elicit broadly neutralizing antibodies or antibodies to the V1/V2 regions of the gp120 Env, as in the RV144 trial.38,48 Also this platform, if immunogenic in HIV-1-positive individuals, would be very appropriate for studies aimed at eliminating HIV-1 reservoirs, in conjunction with antiretroviral treatment and reactivation of the latent virus.49 Stimulation of CD8+ T cells specific for conserved epitopes in such patients would be greatly preferable to simply reactivating the preantiretroviral treatment T-cell responses that had failed to control the virus.
In conclusion, we have tested for the first time in humans an HIV-1 vaccine, which focuses vaccine-elicited T cells on the conserved subprotein regions common to most HIV-1 isolates. Demonstration of strong anti–HIV-1 T-cell responses in these volunteers represents an important milestone in the translation of this platform towards clinical use. The current vaccine regimen showed unique potency and the induced T cells showed efficacy against HIV-1 in vitro. Some modification of the delivery regimen may be needed because of currently perceived problems with the HAdV-5 vector, but should be readily achievable. Then the next step will be to demonstrate protection of humans against HIV-1 infection, ideally in conjunction with a vaccine that elicits effective anti-Env antibodies.
Materials and Methods
Ethical and regulatory approvals. Approvals for HIV-CORE 002 were granted by the National Research Ethics Service Committee West London (Ref: 10/H0707/52) and the UK Medicines and Healthcare products Regulatory Agency (Ref: 21584/0271/001). The study was conducted according to the principles of the Declaration of Helsinki (2008) and complied with the International Conference on Harmonization Good Clinical Practice guidelines.
Subjects. The trial was conducted at the Centre for Clinical Vaccinology and Tropical Medicine, University of Oxford between March 2011 and August 2012. Healthy males and nonpregnant females aged 18–50 years were invited to participate in the study. All volunteers were at low risk of HIV-1 infection and all gave written informed consent before participation. There was no selection of volunteers on the basis of pre-existing neutralizing antibodies to ChAdV-63 or HAdV-5 before enrolment. Two volunteers in the CM group withdrew after the first immunization for reasons unrelated to the vaccine.
Vaccines. The pSG2.HIVconsv DNA vaccine was produced by the Clinical Biotechnology Centre, Bristol Institute for Transfusion Science, University of Bristol, UK and formulated in PBS pH 7.4 at 4.0 mg/ml. The ChAdV63.HIVconsv vaccine23 was produced at the Clinical Biomanufacturing Facility, University of Oxford and diluted in formulation buffer to 1.35 × 1011 vp/ml (36 vp/infectious unit). The MVA.HIVconsv vaccine23 was produced by IDT Biologika GmbH, Germany and diluted in a formulation buffer to 5.5 × 108 PFU/ml. All vaccines were stored below −70 °C until use.
Vaccinations. Vaccines were thawed no more then 30 minutes prior to injection and kept on ice. All vaccines were administered into the deltoid muscle of both arms by an i.m. needle injection at the following doses: pSG2.HIVconsv DNA: D—4 mg; ChAdV63.HIVconsv: c (low dose)—5 × 109 vp and C (regular dose)—5 × 1010 vp; MVA.HIVconsv: M—2 × 108 PFU (half of each dose delivered into each arm). The regimens are depicted in Figure 1b.
Peptides and antigens. HIVconsv peptides (Ana Spec, San Jose, CA; 95131) were reconstituted to 40 mg/ml in dimethyl sulfoxide and diluted to working stock solutions of 4 mg/ml in phosphate-buffered saline (PBS). For ELISPOT and intracellular cytokine staining (ICS) assays, peptides were combined into 6 pools, Pool 1–6, 32–36 peptides per pool in R10 (RPMI 1460 supplemented with 10% fetal bovine serum, 2 mmol/l L-glutamine, 1 mmol/l sodium pyruvate, 10 mmol/l HEPES and penicillin-streptomycin antibiotics; Sigma Aldrich, St. Louis, MO). For subsequent proliferation and inhibition studies, the peptides were combined into 4 pools covering Gag, Pol (2 pools) and Env+Vif. For ELISPOT assays, 2x-concentrated stocks of 3 µg/ml were prepared and aliquoted into peptide plates along with the positive and negative controls. For ICS, proliferation and inhibition assays, 10x-concentrated pools at 15 µg/ml were prepared in between 500-µl and 1-ml aliquots. To map responses of individual participants, 2x-concentrated stocks of individual peptides were prepared in peptide plates. A pool of FEC peptides consisting of 32 previously defined CD8+ T-cell epitopes from influenza virus, Epstein-Barr virus and cytomegalovirus (CEF; NIH AIDS Research and Reference Program) was reconstituted in dimethyl sulfoxide and used at a final concentration of 1 µg/ml as a positive control. All peptide stocks and plates were stored at −80 °C until use.
Isolation and cryopreservation of PBMC. Blood was drawn into heparinized vacutainers (Becton Dickinson, Franklin Lakes, NJ) and processed by the laboratory within 6 hours. Standard procedures were used for cryopreservation.
Ex vivo IFN-γ ELISPOT assay. Freshly isolated PBMC were used. ELISPOT plates (S5EJ044I10; Merck Millipore, Darmstadt, Germany) prewetted for 1 minute with 15 µl of 35% ethanol were coated overnight at +4 °C with anti–IFN-γ antibody (10 µg/ml in PBS; clone 1-D1K; Mabtech, Nacka Strand, Sweden). Prior to use, plates were washed with PBS and blocked with R10 for a minimum of 1 hour at 37 °C. The PBMC were plated out at 2 × 105 cells/well in 50 µl for the majority of time points and at 1 × 105 cells/well at peak time points. For HIVconsv, Pools 1–6 responses were detected in triplicate wells. Six negative no-peptide control wells were cells cultured in R10 supplemented with 0.45% dimethyl sulfoxide. Positive controls in triplicate wells were cells cultured with 10 µg/ml PHA (Sigma Aldrich, St Louis, MO) or a pool of FEC peptides at 1 µg/ml. As an external positive control, the cell line NKL was cultured in duplicate wells in the presence of PMA (4 µg/ml) plus ionomycin (1 µg/ml; both from Sigma Aldrich), The cells were incubated overnight at 37 °C in 5% CO2. Spots were visualized using biotin anti–IFN-γ combined with streptavidin/alkaline phosphate (both from Mabtech) and the color was developed using substrate BCIP/NBTPlus (Mabtech). The reaction was stopped after 5 minutes by washing under the tap. The plates were air dried overnight and the spots counted using an AID ELISpot Reader and version 5.0 software (AID GmbH, Stassberg, Germany). The median number of SFU in no-peptide wells were subtracted from test wells and the results were expressed as the median net SFU/106 PBMC. Electronic records for each plate were retained.
Peptide mapping. In order to map the responses of individual participants short-term cell lines were derived from frozen PBMC samples taken on week 28. The PBMC were thawed and an equal volume of R10 plus 50 U/ml benzonase nuclease (Novagen, Darmstadt, Germany) warmed to 37 °C was added dropwise. Cells were expanded using Pools 1–6 for 10 days, rested and tested an IFN-γ ELISPOT assay.
ICS assay. Cryopreserved PBMC were thawed and resuspended in R10 medium at 106 cells/ml. The cells were labeled with anti-CD107a-PE-Cy7 (BD Biosciences, San Jose, CA), stimulated with 1.5 µg/ml of peptide Pools 1–6 or 1 µg/ml SEB (positive control) or R10 (negative control) and 1 µg/ml of anti-CD28 and anti-CD49d for 2 hours at 37 °C, 5% CO2. Then, 10 µg/ml of GolgiPlug (BD Biosciences, San Jose, CA) and 0.7 µl/ml of GolgiStop (BD Biosciences) were added for a further 17 hours and the cells were stained with a dead-cell marker (LIVE/DEAD Fixable Aqua stain; Invitrogen, Carlsbad, CA) followed by incubation with anti-CD14-Qdot655 (Invitrogen, Paisley, UK), anti-CD4-BV570 (BioLegend), anti-CD3-ECD (Beckman Coulter), anti-CD8-APC-eFluor780 (eBioscience) antibodies, permeabilized with 200 µl of Cytofix/Cytoperm solution (BD Biosciences), stained with anti-IFN-γ-V450 (BD Biosciences), anti-TNF-α–FITC (eBioscience), anti-IL-2 PE (BD Pharmingen), fixed in 1% paraformaldehyde and acquired on a BD LSR II flow cytometer (BD Biosciences). Data analysis was performed using FlowJo software (Tree Star, Ashland, OR).
CFSE proliferation assay. Cryopreserved PBMC were thawed, resuspended in prewarmed PBS/0.1% bovine serum albumin at a final concentration of 106 cells/ml and labeled with 500 nmol/l CFSE (Molecular Probes) for 10 minutes at 37 °C, 5% CO2. The staining was quenched by adding five volumes of ice-cold R10 followed by a 5-minute incubation on ice. The cells were pelleted, washed, and plated in 96-well round bottom plates at a concentration of 5 × 105 cells/well. The CFSE-labeled cells were then stimulated with 1.5 µg/m of peptide Pools 1–4 or 1 µg/ml SEB (positive control) and R10 (negative control) for 5 days, stained with a dead cell marker (LIVE/DEAD Fixable Aqua stain; Invitrogen, Paisley, UK) and anti-CD4-BV570 (BioLegend), anti-CD3-ECD (Beckman Coulter, High Wycomb, UK) and anti-CD8-APC-eFluor780 (eBioscience), fixed and acquired on a BD LSR II flow cytometer. Data analysis was performed using FlowJo software (Tree Star, Ashland, OR).
Viral inhibition assay. Antiviral capacity of vaccine-elicited CD8+ T cells was assessed in VIA as described previously.5 Briefly, to generate CD4+ and CD8+ T cells, frozen PBMC were thawed, resuspended in R10 supplemented with IL-2 at 50 IU/ml, activated using either 0.5 µg/ml anti-CD3/8 or anti-CD3/4 bi-specific antibodies, respectively, (generously provided by J Wong, Harvard Medical School) and cultured for 7 days with the addition of fresh R10/IL-2 media on days 3 and 6. Viral infections were performed on autologous expanded CD4+ T cells at MOI 0.01 for Nef-mutated IIIB, ELI, CH07, CH106, 247Fv2, ZA97012, and U455 viruses, while BaL was used at MOI 0.005. CD4+ cells were incubated with viruses for 3.5–4 hours at 37 °C, 5% CO2, washed once in R10, resuspended at 106 cells/ml in R10 supplemented with 100 IU/ml IL-2 and 5 × 105 CD4+ cells were aliquoted into assay wells of 48-well tissue culture plates. 5 × 105 CD8+ effector cells were added to the matched infected CD4+ T cells and incubated at 37 °C, 5% CO2 for 13 days. On days 3, 6, 8, and 10 of coculture, half of the culture media was replaced with fresh R10 supplemented with 50 IU/ml IL-2. On day 13, supernatants were collected for analysis by p24 ELISA (PerkinElmer, Foster City, CA). The log10 reduction in the p24 content in the supernatants of CD8+/CD4+ coculture wells relative to infected CD4+ cells alone was calculated for each sample.
Multiplex bead array. Frozen PBMC samples from the CM group, week 12 were stimulated using the HIVconsv peptides Pools 1–6 for 48 hours and the supernatants were analyzed for intercellular signaling molecules using MILLIPLEX MAP Human High Sensitivity Cytokine/Chemokine Panel Kit. All samples were acquired on a Luminex 200 instrument (Millipore, Darmstadt, Germany) using the maximum likelihood method.
Statistics. Analyses of variance were performed using GraphPad Prism 5.0a (GraphPad Software, San Diego, CA). Results were assumed to be non-Gaussian in distribution, thus nonparametric tests were used throughout and medians (range) are shown. Variation among groups was assessed in the Kruskal-Wallis test and individual group means were compared either to control using Dunn's multiple comparison or among themselves in pairwise comparisons followed by the Bonferroni adjustment. For unpaired analyses, the Mann–Whitney U test was used. Correlations were made using Spearman rank test. Two-tailed P values were used and P value of less than 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. IFN-γ ELISPOT assay. Figure S2. Correlation of fresh and frozen ELISPOT assay frequencies of HIVconsv vaccine-elicited IFN-γ-producing T cells. Figure S3. A multicolor flow cytometry analysis of vaccine-elicited T cells. Figure S4. Proliferative capacity of vaccine-elicited T cells. Figure S5. Correlation of total ex vivo IFN-γ ELISPOT assay frequencies with in vitro peak inhibition of clade A HIV-1 replication in autologous CD4+ cells for all CM, DDDCM, and DDDMC groups combined. Table S1. Multifunctionality of T-cell responses induced by the CM regimen.
Acknowledgments
The authors thank the volunteers for making this study possible. The work was supported by Medical Research Council (MRC) UK and Department for International Development UK through an Experimental Medicine call II award G0701669 with contributions from the International AIDS Vaccine Initiative. HIV-1 infectious molecular clones were obtained from Dr George Shaw, University of Pennsylvania. The FEC Control Peptide Pool was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH (ref. no. 19626). A.N. and S.C. who were employees and shareholders of Okairos and Advent during the conduct of the study, and are inventors on patents WO 2005071093 (A3), WO 2006133911 (A3) and WO 03031588 (A2), A.J.McM. who reports grants from MRC and NIH, personal fees from International AIDS Vaccine Initiative SAB during the conduct of the study and is an inventor on patent WO 06123256, LC reports grants from MRC during the conduct of the study, and T.H. who reports grants from MRC and European and Developing Countries Clinical Trial Partnership obtained during the conduct of the study and is an inventor on patent WO 06123256. The other authors declare no conflict of interest.
Supplementary Material
References
- McMichael AJ, Haynes BF. Lessons learned from HIV-1 vaccine trials: new priorities and directions. Nat Immunol. 2012;13:423–427. doi: 10.1038/ni.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B, McMichael A. The T-cell response to HIV. Cold Spring Harb Perspect Med. 2012;2:a007054. doi: 10.1101/cshperspect.a007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Takiguchi M. HIV-1-specific CTLs effectively suppress replication of HIV-1 in HIV-1-infected macrophages. Blood. 2007;109:4832–4838. doi: 10.1182/blood-2006-07-037481. [DOI] [PubMed] [Google Scholar]
- Sáez-Cirión A, Lacabaratz C, Lambotte O, Versmisse P, Urrutia A, Boufassa F, et al. Agence Nationale de Recherches sur le Sida EP36 HIV Controllers Study Group HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci USA. 2007;104:6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spentzou A, Bergin P, Gill D, Cheeseman H, Ashraf A, Kaltsidis H, et al. Viral inhibition assay: a CD8 T cell neutralization assay for use in clinical trials of HIV-1 vaccine candidates. J Infect Dis. 2010;201:720–729. doi: 10.1086/650492. [DOI] [PubMed] [Google Scholar]
- Yang H, Wu H, Hancock G, Clutton G, Sande N, Xu X, et al. Antiviral inhibitory capacity of CD8+ T cells predicts the rate of CD4+ T-cell decline in HIV-1 infection. J Infect Dis. 2012;206:552–561. doi: 10.1093/infdis/jis379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- Churchyard GJ, Morgan C, Adams E, Hural J, Graham BS, Moodie Z, et al. NIAID HIV Vaccine Trials Network A phase IIA randomized clinical trial of a multiclade HIV-1 DNA prime followed by a multiclade rAd5 HIV-1 vaccine boost in healthy adults (HVTN204). PLoS ONE. 2011;6:e21225. doi: 10.1371/journal.pone.0021225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007;13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- Honeyborne I, Prendergast A, Pereyra F, Leslie A, Crawford H, Payne R, et al. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J Virol. 2007;81:3667–3672. doi: 10.1128/JVI.02689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- Rolland M, Heckerman D, Deng W, Rousseau CM, Coovadia H, Bishop K, et al. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS ONE. 2008;3:e1424. doi: 10.1371/journal.pone.0001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altfeld M, Allen TM. Hitting HIV where it hurts: an alternative approach to HIV vaccine design. Trends Immunol. 2006;27:504–510. doi: 10.1016/j.it.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Hanke T, McMichael AJ. HIV-1: from escapism to conservatism. Eur J Immunol. 2011;41:3390–3393. doi: 10.1002/eji.201190072. [DOI] [PubMed] [Google Scholar]
- Kunwar P, Hawkins N, Dinges WL, Liu Y, Gabriel EE, Swan DA, et al. Superior control of HIV-1 replication by CD8+ T cells targeting conserved epitopes: implications for HIV vaccine design. PLoS ONE. 2013;8:e64405. doi: 10.1371/journal.pone.0064405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Létourneau S, Im EJ, Mashishi T, Brereton C, Bridgeman A, Yang H, et al. Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS ONE. 2007;2:e984. doi: 10.1371/journal.pone.0000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland M, Nickle DC, Mullins JI. HIV-1 group M conserved elements vaccine. PLoS Pathog. 2007;3:e157. doi: 10.1371/journal.ppat.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS. 2003;17:2581–2591. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- Ferguson AL, Mann JK, Omarjee S, Ndung'u T, Walker BD, Chakraborty AK. Translating HIV sequences into quantitative fitness landscapes predicts viral vulnerabilities for rational immunogen design. Immunity. 2013;38:606–617. doi: 10.1016/j.immuni.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, et al. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Rosario M, Bridgeman A, Quakkelaar ED, Quigley MF, Hill BJ, Knudsen ML, et al. Long peptides induce polyfunctional T cells against conserved regions of HIV-1 with superior breadth to single-gene vaccines in macaques. Eur J Immunol. 2010;40:1973–1984. doi: 10.1002/eji.201040344. [DOI] [PubMed] [Google Scholar]
- Ondondo B, Brennan C, Nicosia A, Crome SJ, Hanke T. Absence of systemic toxicity changes following intramuscular administration of novel pSG2.HIVconsv DNA, ChAdV63.HIVconsv and MVA.HIVconsv vaccines to BALB/c mice. Vaccine. 2013;31:5594–5601. doi: 10.1016/j.vaccine.2013.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen ML, Mbewe-Mvula A, Rosario M, Johansson DX, Kakoulidou M, Bridgeman A, et al. Superior induction of T cell responses to conserved HIV-1 regions by electroporated alphavirus replicon DNA compared to that with conventional plasmid DNA vaccine. J Virol. 2012;86:4082–4090. doi: 10.1128/JVI.06535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario M, Borthwick N, Stewart-Jones GB, Mbewe-Mvula A, Bridgeman A, Colloca S, et al. Prime-boost regimens with adjuvanted synthetic long peptides elicit T cells and antibodies to conserved regions of HIV-1 in macaques. AIDS. 2012;26:275–284. doi: 10.1097/QAD.0b013e32834ed9b2. [DOI] [PubMed] [Google Scholar]
- Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidewind A, Brockman MA, Yang R, Adam RI, Li B, Le Gall S, et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone FR, Bevan MJ. Induction of ovalbumin-specific cytotoxic T cells by in vivo peptide immunization. J Exp Med. 1989;169:603–612. doi: 10.1084/jem.169.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone FR, Moore MW, Sheil JM, Bevan MJ. Induction of cytotoxic T lymphocytes by primary in vitro stimulation with peptides. J Exp Med. 1988;167:1767–1779. doi: 10.1084/jem.167.6.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MK, Hawkins N, Ritchie AJ, Ganusov VV, Whale V, Brackenridge S, et al. CHAVI Core B Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. J Clin Invest. 2013;123:380–393. doi: 10.1172/JCI65330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, et al. Step Study Protocol Team Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam G, Perry MR, Ward S, Brett SJ, Castello-Cortes A, Brunner MD, et al. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N Engl J Med. 2006;355:1018–1028. doi: 10.1056/NEJMoa063842. [DOI] [PubMed] [Google Scholar]
- Wilson NA, Watkins DI. Is an HIV vaccine possible. Braz J Infect Dis. 2009;13:304–310. doi: 10.1590/s1413-86702009000400013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel SA, Lamoreaux L, Chattopadhyay PK, Saunders K, Zarkowsky D, Overman RG, et al. Phenotypic and functional profile of HIV-inhibitory CD8 T cells elicited by natural infection and heterologous prime/boost vaccination. J Virol. 2010;84:4998–5006. doi: 10.1128/JVI.00138-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel SA, Picking RA, Ferrari G, Ding H, Ochsenbauer C, Kappes JC, et al. Initial HIV-1 antigen-specific CD8+ T cells in acute HIV-1 infection inhibit transmitted/founder virus replication. J Virol. 2012;86:6835–6846. doi: 10.1128/JVI.00437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland M, Tovanabutra S, deCamp AC, Frahm N, Gilbert PB, Sanders-Buell E, et al. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med. 2011;17:366–371. doi: 10.1038/nm.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. MOPH-TAVEG Investigators Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Barouch DH, Liu J, Peter L, Abbink P, Iampietro MJ, Cheung A, et al. Characterization of humoral and cellular immune responses elicited by a recombinant adenovirus serotype 26 HIV-1 Env vaccine in healthy adults (IPCAVD 001). J Infect Dis. 2013;207:248–256. doi: 10.1093/infdis/jis671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier JR, Ngauy V, de Souza MS, Ratto-Kim S, Cox JH, Polonis VR, et al. Phase I safety and immunogenicity evaluation of MVA-CMDR, a multigenic, recombinant modified vaccinia Ankara-HIV-1 vaccine candidate. PLoS ONE. 2010;5:e13983. doi: 10.1371/journal.pone.0013983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke T, Goonetilleke N, McMichael AJ, Dorrell L. Clinical experience with plasmid DNA- and modified vaccinia virus Ankara-vectored human immunodeficiency virus type 1 clade A vaccine focusing on T-cell induction. J Gen Virol. 2007;88 Pt 1:1–12. doi: 10.1099/vir.0.82493-0. [DOI] [PubMed] [Google Scholar]
- McCormack S, Stöhr W, Barber T, Bart PA, Harari A, Moog C, et al. EV02: a Phase I trial to compare the safety and immunogenicity of HIV DNA-C prime-NYVAC-C boost to NYVAC-C alone. Vaccine. 2008;26:3162–3174. doi: 10.1016/j.vaccine.2008.02.072. [DOI] [PubMed] [Google Scholar]
- Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, et al. CHAVI Clinical Core B The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm N, DeCamp AC, Friedrich DP, Carter DK, Defawe OD, Kublin JG, et al. Human adenovirus-specific T cells modulate HIV-specific T cell responses to an Ad5-vectored HIV-1 vaccine. J Clin Invest. 2012;122:359–367. doi: 10.1172/JCI60202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr A, Huang Y, Buchbinder S, Coombs RW, Sanchez J, del Rio C, et al. Step/HVTN 504 Study Team Extended follow-up confirms early vaccine-enhanced risk of HIV acquisition and demonstrates waning effect over time among participants in a randomized trial of recombinant adenovirus HIV vaccine (Step Study). J Infect Dis. 2012;206:258–266. doi: 10.1093/infdis/jis342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie TL, Villasante EF. Use of adenovirus serotype 5 vaccine vectors in seropositive, uncircumcised men: safety lessons from the step trial. J Infect Dis. 2013;207:689–690. doi: 10.1093/infdis/jis737. [DOI] [PubMed] [Google Scholar]
- Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DM, Hazuda DJ. Combined approaches for HIV cure. Curr Opin HIV AIDS. 2013;8:230–235. doi: 10.1097/COH.0b013e32835ef089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SC, Schneider J, Hannan CM, Hu JT, Plebanski M, Sinden R, et al. Enhanced CD8 T cell immunogenicity and protective efficacy in a mouse malaria model using a recombinant adenoviral vaccine in heterologous prime-boost immunisation regimes. Vaccine. 2002;20:1039–1045. doi: 10.1016/s0264-410x(01)00450-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.