Abstract
Most models of gene duplication assume that the ancestral functions of the preduplication gene are independent and can therefore be neatly partitioned between descendant paralogs. However, many gene products, such as transcriptional regulators, are components within cooperative assemblies; here, we show that a natural consequence of duplication and divergence of such proteins can be competitive interference between the paralogs. Our example is based on the duplication of the essential MADS-box transcriptional regulator Mcm1, which is found in all fungi and regulates a large set of genes. We show that a set of historical amino acid sequence substitutions minimized paralog interference in contemporary species and, in doing so, increased the molecular complexity of this gene regulatory network. We propose that paralog interference is a common constraint on gene duplicate evolution, and its resolution, which can generate additional regulatory complexity, is needed to stabilize duplicated genes in the genome.
Gene duplications are an important source of new genes, and a variety of models have been developed to rationalize why certain gene duplicates have been maintained over evolutionary time (1-3). For instance, the neofunctionalization model posits that soon after duplication, one of the duplicates evolves a new function that can be selected for and, thereby, maintained over time (2, 3). Alternatively, subfunctionalization (via the duplication-degeneration-complementation model) holds that duplicates can be maintained in the genome by acquiring reciprocal loss-of-function mutations, such that both duplicates become necessary to perform the combined functions of the preduplication ancestor (1-3). Classically, these models have assumed that ancestral functions can be treated independently, making the partitioning of these functions among the descendant paralogs possible without detrimental effects (2). However, for the many gene products that participate in cooperative assemblies, the molecular interactions that underlie gene functions are not intrinsically independent (4). For example, many transcriptional regulators depend on a cooperative network of protein-protein and protein–nucleic acid interactions. In these instances, loss of one or more ancestral molecular interactions will often give rise to competitive interference between gene duplicates (paralog interference) (5). Although in some instances this competition may be advantageous, we suspect that paralog interference following gene duplication would typically have detrimental effects that must be evolutionarily bypassed for the paralogs to be maintained. Because many proteins form cooperative assemblies, resolution of paralog interference is likely to be a widespread phenomenon influencing the fate of duplicated genes.
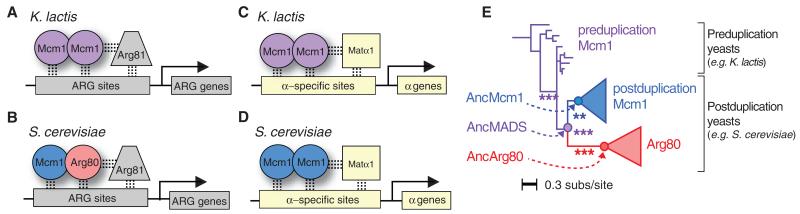
Mcm1 is a fungal MADS-box transcriptional regulator that binds DNA cooperatively with seven different partner transcriptional regulators (cofactors) to control the expression of many genes, including those coding for mating functions and arginine metabolic enzymes (6). The way in which Mcm1 assembles at the arginine metabolism (ARG) genes varies between fungal clades. In the yeasts Kluyveromyces lactis and Candida albicans, an Mcm1 homodimer regulates transcription of ARG genes by binding specifically to DNA with the cofactor Arg81 (Fig. 1A) (7, 8). In the lineage leading to baker’s yeast (Saccharomyces cerevisiae), a tandem gene duplication event introduced an extra copy of Mcm1 (called Arg80), such that the S. cerevisiae regulatory architecture is more complex. In S. cerevisiae, an Mcm1-Arg80 heterodimer regulates the transcription of ARG genes by binding DNA with the cofactor Arg81 (Fig. 1B) (9). Other Mcm1-regulated gene sets in S. cerevisiae did not experience an increase in regulatory complexity following gene duplication. For instance, the α-specific genes (genes that give α mating cells their specialized properties) are regulated by an Mcm1 homodimer that binds specifically to DNA with the cofactor Matα1 in species that branch before and after the gene duplication event (Fig. 1, C and D) (10-12). In all instances, gene regulation by Mcm1 and Arg80 depends on the formation of strong interactions with both cofactors and DNA.
Fig. 1. Function and evolution of MADS-box proteins in hemiascomycete yeasts.
(A) In K. lactis, an Mcm1 homodimer regulates the ARG genes by interacting with Arg81 and binding a specific DNA sequence. (B) In S. cerevisiae, an Mcm1-Arg80 heterodimer interacts with Arg81 to regulate ARG genes. (C) An Mcm1 homodimer interacts with Matα1 to regulate α-specific genes in K. lactis and (D) S. cerevisiae. (E) A maximum likelihood phylogeny of MADS-box domain proteins in hemiascomycete yeasts. A tandem gene duplication generated paralogs Mcm1 and Arg80 in the last shared common ancestor of Zygosaccharomyces rouxii and S. cerevisiae. Circles denote ancestral proteins reconstructed in this study. Asterisks on internal branches correspond to approximate-likelihood ratio support for the monophyly of the descendant clade: *** denotes support > 10.0; ** denotes support > 5.0. subs, substitutions.
To understand how the linked biochemical functions of DNA and cofactor binding diverged after Mcm1 duplicated, we reconstructed ancestral MADS-box proteins, characterized these ancestral proteins in vivo and in vitro, and identified the mutations through which their functions diversified [see supplementary materials and methods and (13)]. Specifically, we reconstructed the MADS-box domains of the most recent common shared ancestor of all postduplication Mcm1 paralogs (AncMcm1); all postduplication Arg80 paralogs (AncArg80); and the preduplication, most recent shared common ancestor of all Mcm1 and Arg80 paralogs (AncMADS) (Fig. 1E, fig. S1, and tables S1 and S2).
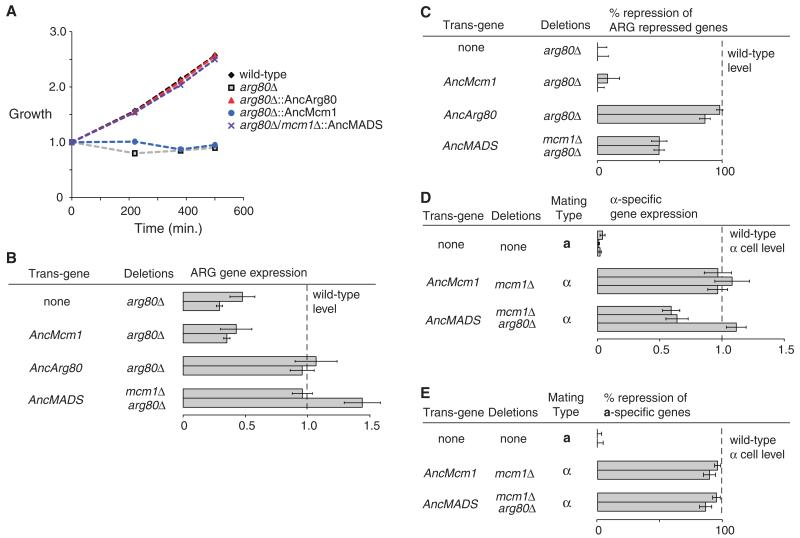
We integrated the reconstructed ancestral MADS-box proteins into S. cerevisiae and removed the modern copies of ARG80 and MCM1 to determine if the ancestral proteins could complement their deletion. Deletion of S. cerevisiae MCM1 is lethal, and deletion of S. cerevisiae ARG80 produces defects in arginine metabolism (14, 15). We found that the preduplication AncMADS protein complemented both defects: Replacement of Mcm1 or Arg80 with AncMADS had no impact on growth in either rich media or media with a precursor of arginine as a sole nitrogen source, a phenotype that depends on normal ARG gene regulation (Fig. 2A and fig. S2A). We measured the expression levels of a representative set of Mcm1 and Arg80/Mcm1 regulated genes and found that AncMADS restored activation and repression of these genes, and most genes showed the same dynamic range as in the wild type (Fig. 2, B to E, and fig. S2B). The exceptions were a diminished dynamic range of gene expression for the ARG repressed genes, a mildly diminished dynamic range of gene expression for the α-specific genes, and stronger activation than the wild type for the ARG activated gene CAR2 (Fig. 2, B to D). In contrast, the postduplication MADS-box proteins (AncArg80 and AncMcm1) failed to complement deletions of the sister paralogs. Specifically, AncMcm1 did not complement the deletion of the native ARG80 and, similarly, the presence of AncArg80 alone did not rescue the deletion of MCM1 (an essential gene) (Fig. 2, A to C, and fig. S2C). The capacity of the preduplication ancestral MADS-box protein to complement the functions of both daughter genes in a modern species, combined with the inability of the postduplication ancestors to do the same, shows that AncMcm1 and AncArg80 acquired degenerative mutations that necessitated the retention of both paralogs over evolutionary time.
Fig. 2. The preduplication ancestral gene complements both paralogs.
(A) Growth of ancestral MADS-box gene strains using ornithine as a sole nitrogen source. Ornithine is converted into arginine and then modified to produce the other essential amino acids. In the absence of a functional ARG gene regulatory complex, strains cannot use ornithine as a nitrogen source. The preduplication AncMADS can supply the function of the modern Arg80 paralog (purple), but the postduplication AncMcm1 paralog cannot (blue). “Growth” on the y axis is the ratio of optical density at 600 nm (OD600) at the indicated time point divided by OD600 at time zero. (B to E) Gene expression profiling of ancestral MADS-box proteins in S. cerevisiae quantified with NanoString (www.nanostring.com). (B) MADS-box activated ARG genes. Row 1, CAR1; row 2, CAR2. (C) MADS-box repressed ARG genes. Row 1, ARG3; row 2, ARG5,6. (D) MADS-box activated mating genes (α-specific genes). Row 1, SAG1; row 2, MFa1; row 3, STE3. (E) MADS-box repressed mating genes (a-specific genes). Row 1, STE2; row 2, STE6. In each experiment, mean and standard error (indicated by error bars) were determined using three replicates.
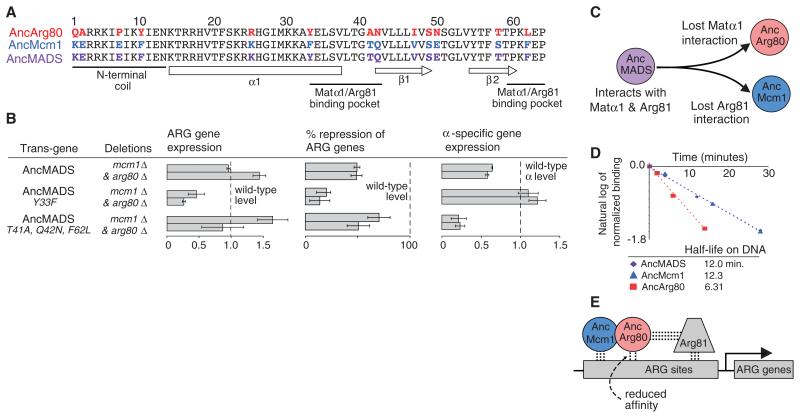
We next determined the mutations that underlie the diversification of AncMcm1 and AncArg80 following the duplication of AncMADS. The cofactors Matα1 and Arg81 both interact with the same portion of the MADS-box domain, which we refer to as the cofactor binding pocket (16, 17). We modeled the reconstructed protein sequences for AncMADS, AncMcm1, and AncArg80 onto the structure of S. cerevisiae Mcm1 in complex with DNA (18) and then compared the sequences within the cofactor binding pocket (fig. S3, A to C). On the lineage from AncMADS to AncMcm1, one substitution occurred in the pocket [Tyr33→Phe33 (Y33F); Y, Tyr; F, Phe], and it has been conserved in all Mcm1 descendants (Fig. 3A and table S1). On the lineage from AncMADS to AncArg80, three sequence substitutions occurred within the cofactor binding pocket (T41A, Q42N, F62L; T, Thr; A, Ala, Q, Gln; N, Asn; L, Leu), and each has been strongly conserved in postduplication Arg80 descendant sequences (Fig. 3A and table S1). Previous work has shown that these residues play a critical role in stabilizing the interactions between Arg81 and Arg80, as well as Matα1 and Mcm1 (16, 17). To assess the impact of these changes on the preduplication ancestor, we introduced these mutations into AncMADS and observed their effect on expression levels of ARG genes and α-specific genes. When we introduced the Mcm1 substitution (Y33F) into AncMADS, we observed a disruption of ARG gene regulation but no decrease in α-specific gene regulation; in fact, the dynamic range of α-specific gene regulation slightly expanded (Fig. 3B). When we introduced the Arg80 substitutions (T41A, Q42N, F62L) into AncMADS, we observed decreased α-specific gene regulation but no compromise in ARG gene regulation (Fig. 3B). (We note that CAR2 even returns to wild-type expression levels from its elevated state in the AncMADS background.) We observed similar effects of these mutations on AncMADS when we swapped in the cofactors Arg81 and Matα1 from the preduplication species K. lactis, indicating that changes to these two cofactors did not play a major role in partitioning the ancestral molecular interactions of AncMADS (fig. S3, D and E). Taken together, these results show that the preduplication AncMADS could form cofactor interactions with both Arg81 and Matα1 and that these interactions reciprocally degenerated in the descendant paralogs: AncMcm1 lost its ability to productively interact with Arg81, whereas AncArg80 lost its ability to interact with Matα1 (Fig. 3C).
Fig. 3. Divergence in cofactor and DNA-binding following gene duplication of ancestral MADS-box proteins.
(A) Alignment of the N-terminal 63 amino acids of the MADS-box domain with residues that changed identity between AncMcm1, AncArg80, and AncMADS in color (24). α1 denotes a long α helix; β1 and β2 signify an antiparallel β sheet. (B) Gene expression profiling to determine the impact of mutants on the function of preduplication AncMADS protein in S. cerevisiae. Gene expression quantified using NanoString. Panel 1: MADS-box activated ARG genes; row 1, CAR1; row 2, CAR2. Panel 2: MADS-box repressed ARG genes; row 1, ARG3; row 2, ARG5,6. Panel 3: MADS-box activated mating genes (α-specific genes); row 1, SAG1; row 2, MFa1. Mean and standard error (indicated by error bars) were determined from three replicates. (C) After duplication, AncArg80 lost the ability to form a strong interaction with Matα1, and AncMcm1 lost the ability to form a strong interaction with Arg81. These losses destroyed the abilities of AncArg80 and AncMcm1 to regulate α-specific genes and ARG genes, respectively. (D) Half-lives of MADS-box ancestors on the S. cerevisiae CAR2 cis-regulatory sequence (see supplementary materials and methods). Saturating levels of unlabeled DNA were added at time point zero. (E) After the duplication of AncMADS, α-specific genes are regulated by a homodimer of AncMcm1, whereas ARG genes are regulated by a heterodimer of AncMcm1 and AncArg80 due to the reduced affinity of AncArg80 for DNA.
We next examined the DNA-binding surfaces of the pre- and postduplication MADS-box proteins. S. cerevisiae Arg80 and Mcm1 have very closely related DNA-binding specificities (19, 20), indicating that, at most, a limited divergence in MADS-box DNA-binding specificity occurred following duplication. That the DNA-binding specificity did not change substantially is also supported by our observation that the preduplication AncMADS protein can complement the deletion of either postduplication gene (Fig. 2). However, S. cerevisiae Arg80 has a substantially lower affinity for DNA than Mcm1 (15, 16). To determine when this affinity change occurred, we compared the DNA-binding affinities of AncMADS, AncArg80, and AncMcm1 by measuring their half-lives on DNA. To make the comparisons meaningful, we used an endogenous S. cerevisiae MADS-box binding site (taken from an arginine metabolic gene) that closely resembles the consensus site for S. cerevisiae Mcm1 and Arg80 and that has been shown to support binding by both these proteins in vitro (8, 20). We observed that the half-life of AncArg80 was significantly lower than the half-lives of AncMADS and AncMcm1 (Fig. 3D). Thus, the difference in affinity between modern Mcm1 and Arg80 is due to a decrease in Arg80 DNA-binding affinity that occurred soon after the duplication (Fig. 3E).
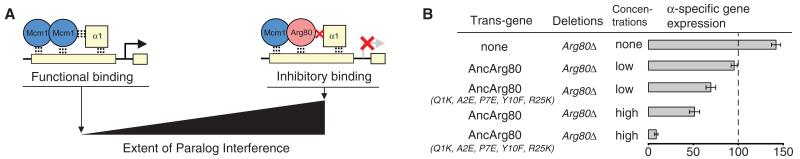
Next, we investigated the consequences of this reduction in Arg80 DNA-binding affinity on gene expression. We hypothesized that a version of Arg80 with full DNA-binding strength might interfere with Mcm1 by binding to α-specific gene regulatory sites and acting as a dominant negative mutant by preventing Mcm1 from binding cooperatively with Matα1 (Fig. 4A). If this were true, then the reduction of Arg80 DNA-binding affinity would minimize competition by weighting DNA-binding at the α-specific genes in favor of Mcm1. To test this idea, we increased the DNA-binding affinity of AncArg80 to that of the preduplication protein and measured the extent of competitive interference with Mcm1 in S. cerevisiae. We identified a total of five mutations (K1Q, E2A, E7P, F10Y, K25R; K, Lys; E, Glu; P, Pro; R, Arg) that occurred on the DNA-binding surface of AncArg80 after the AncMADS duplication (Fig. 3A). A subset of these residues are known to affect MADS-box DNA-binding affinity in S. cerevisiae (14, 16). We reversed these mutations in AncArg80, returning the DNA-binding region of AncArg80 to its preduplication, high-affinity state, and then measured α-specific gene expression in an S. cerevisiae strain lacking the native Arg80. As predicted by our hypothesis (paralog interference), the AncArg80 mutant significantly reduced α-specific gene expression (Fig. 4B). When overexpressed, the nonmutant, low-affinity AncArg80 protein dampened α-specific gene expression, and the overexpressed, high-affinity AncArg80 mutant blocked the expression of the α-specific genes almost entirely (Fig. 4B). [These effects were not an indirect consequence of altering MCM1 gene expression levels (fig. S4A).] The antagonism between Arg80 and Mcm1 persists in an attenuated form in contemporary species, as the deletion of ARG80 in S. cerevisiae slightly increases α-specific gene expression (Fig. 4B and fig. S4B). On the basis of these observations, we conclude that the historical reduction in Arg80 DNA-binding affinity limited the degree of paralog interference between Arg80 and Mcm1 at the α-specific genes.
Fig. 4. Paralog interference between Arg80 and Mcm1.
(A) The ratio of functional binding to inhibitory binding determines the extent of competitive interference between Mcm1 and Arg80. (B) Arg80, AncArg80, and AncArg80 mutant with DNA-binding surface changes reverted to ancestral state interfere with α-specific gene expression. α-specific gene expression was quantified by quantitative reverse-transcriptase fluorescence polymerase chain reaction using SAG1 transcript (normalized to URA6 transcript). Endogenous expression is driven by the native ARG80 promoter, and overexpression is driven by the TEF1 promoter. For gene expression experiments, mean and standard error (indicated by error bars) were determined from five replicates.
The diminished DNA-binding affinity of Arg80 also provides a simple explanation for the origins of the Arg80-Mcm1 heterodimer (as opposed to an Arg80 homodimer) at the ARG genes in S. cerevisiae. The cofactor Arg81 contacts only a single (proximal) subunit within the MADS-box dimer, and this interaction will favor Arg80 because of its strong interaction with Arg81. The energetics will favor Mcm1, rather than Arg80, as the second (distal) subunit because of its higher affinity for DNA (Fig. 3E).
By combining ancestral gene reconstructions with the biochemical and genetic tools available for yeasts, we have shown that competition between paralogs arose as an intrinsic consequence of the duplication and subfunctionalization of a deeply conserved transcriptional regulator. This interference was minimized by a set of historical amino acid substitutions, and we suggest that this was necessary for both paralogs to be maintained, as without the weakened affinity of Arg80 for DNA, gene regulation is severely compromised (Fig. 4B). The minimization of interference was accompanied by an increase in regulatory complexity: The increased number of distinct subunits needed to regulate the ARG genes in S. cerevisiae relative to the preduplication ancestor (three versus two) is necessary to compensate for the reduced DNA-binding affinity of Arg80. Although we do not know whether the mutations that affected protein-protein interactions occurred before, after, or in concert with those that affected DNA-binding, we have shown that each mutation is a loss-of-function (or at least a reduction-of-function) amino acid substitution (21, 22).
For the many gene products that form cooperative assemblies, exemplified by the proteins studied here, ancestral functions depend on a network of molecular interactions. Following gene duplication, the loss of ancestral interactions by such proteins, resulting in subfunctionalization, may unavoidably give rise to dominant negative effects between duplicates. Although in some cases, this interference can be exploited, for example, by using it to repress gene expression (5, 23), we propose that a more common outcome is the minimization of this interference in gene duplicates that persist over evolutionary time. Whether such minimization is generally accompanied by an increase in regulatory complexity, as seen here, remains to be determined.
Supplementary Material
Acknowledgments
We thank B. Tuch for assistance and comments; O. Homann for assistance on experimental techniques; and C. Nobile, C. Dalal, S. Devika, R. Bennett, L. Mera, and T. Sorrells for comments. This work was funded by grant RO1 GM057049 from the NIH.
Footnotes
Supplementary Materials
References and Notes
- 1.Force A, et al. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Innan H, Kondrashov F. Nat. Rev. Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 3.Wagner A. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-reviews1012. reviews1012.1-reviews1012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner A. Proc. Biol. Sci. 2003;270:457–466. doi: 10.1098/rspb.2002.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bridgham JT, Brown JE, Rodríguez-Marí A, Catchen JM, Thornton JW. PLOS Genet. 2008;4:e1000191. doi: 10.1371/journal.pgen.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messenguy F, Dubois E. Gene. 2003;316:1–21. doi: 10.1016/s0378-1119(03)00747-9. [DOI] [PubMed] [Google Scholar]
- 7.Boonchird C, Messenguy F, Dubois E. Mol. Gen. Genet. 1991;226:154–166. doi: 10.1007/BF00273599. [DOI] [PubMed] [Google Scholar]
- 8.Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. PLOS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Messenguy F, Dubois E. Mol. Cell. Biol. 1993;13:2586–2592. doi: 10.1128/mcb.13.4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bender A, Sprague GF., Jr. Cell. 1987;50:681–691. doi: 10.1016/0092-8674(87)90326-6. [DOI] [PubMed] [Google Scholar]
- 11.Tsong AE, Miller MG, Raisner RM, Johnson AD. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 12.Baker CR, Tuch BB, Johnson AD. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7493–7498. doi: 10.1073/pnas.1019177108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harms MJ, Thornton JW. Curr. Opin. Struct. Biol. 2010;20:360–366. doi: 10.1016/j.sbi.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acton TB, Mead J, Steiner AM, Vershon AK. Mol. Cell. Biol. 2000;20:1–11. doi: 10.1128/mcb.20.1.1-11.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amar N, Messenguy F, El Bakkoury M, Dubois E. Mol. Cell. Biol. 2000;20:2087–2097. doi: 10.1128/mcb.20.6.2087-2097.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamai A, Dubois E, Vershon AK, Messenguy F. Mol. Cell. Biol. 2002;22:5741–5752. doi: 10.1128/MCB.22.16.5741-5752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mead J, et al. Mol. Cell. Biol. 2002;22:4607–4621. doi: 10.1128/MCB.22.13.4607-4621.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan S, Richmond TJ. Nature. 1998;391:660–666. doi: 10.1038/35563. [DOI] [PubMed] [Google Scholar]
- 19.Hayes TE, Sengupta P, Cochran BH. Genes Dev. 1988;2:1713–1722. doi: 10.1101/gad.2.12b.1713. [DOI] [PubMed] [Google Scholar]
- 20.Messenguy F, Dubois E, Boonchird C. Mol. Cell. Biol. 1991;11:2852–2863. doi: 10.1128/mcb.11.5.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finnigan GC, Hanson-Smith V, Stevens TH, Thornton JW. Nature. 2012;481:360–364. doi: 10.1038/nature10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch M, Conery JS. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 23.Dennis MY, et al. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.