The microRNA miR156 is a potential graft-transmissible signal regulating potato development.
Abstract
MicroRNA156 (miR156) functions in maintaining the juvenile phase in plants. However, the mobility of this microRNA has not been demonstrated. So far, only three microRNAs, miR399, miR395, and miR172, have been shown to be mobile. We demonstrate here that miR156 is a potential graft-transmissible signal that affects plant architecture and tuberization in potato (Solanum tuberosum). Under tuber-noninductive (long-day) conditions, miR156 shows higher abundance in leaves and stems, whereas an increase in abundance of miR156 has been observed in stolons under tuber-inductive (short-day) conditions, indicative of a photoperiodic control. Detection of miR156 in phloem cells of wild-type plants and mobility assays in heterografts suggest that miR156 is a graft-transmissible signal. This movement was correlated with changes in leaf morphology and longer trichomes in leaves. Overexpression of miR156 in potato caused a drastic phenotype resulting in altered plant architecture and reduced tuber yield. miR156 overexpression plants also exhibited altered levels of cytokinin and strigolactone along with increased levels of LONELY GUY1 and StCyclin D3.1 transcripts as compared with wild-type plants. RNA ligase-mediated rapid amplification of complementary DNA ends analysis validated SQUAMOSA PROMOTER BINDING-LIKE3 (StSPL3), StSPL6, StSPL9, StSPL13, and StLIGULELESS1 as targets of miR156. Gel-shift assays indicate the regulation of miR172 by miR156 through StSPL9. miR156-resistant SPL9 overexpression lines exhibited increased miR172 levels under a short-day photoperiod, supporting miR172 regulation via the miR156-SPL9 module. Overall, our results strongly suggest that miR156 is a phloem-mobile signal regulating potato development.
Long-distance transport of signaling molecules is known to be a major component in regulating plant growth and development as well as their adaptation to changing environmental conditions. This transport is implemented by the plant’s vascular system, especially through the complex of companion cells and sieve elements present in the phloem. Recent evidence has established the movement of macromolecules like proteins, mRNAs, and microRNAs (miRNAs) through the phloem. It is now clear that these entities act as long-distance signals for development and stress response pathways (Kehr and Buhtz, 2008; Atkins et al., 2011). A well-established example is the movement of FLOWERING TIME protein from leaves to the shoot apex in Arabidopsis (Arabidopsis thaliana) as a long-distance signal for the regulation of flowering time (Corbesier et al., 2007). Similarly, the movement of transcripts such as GIBBERELLIC ACID INSENSITIVE (Haywood et al., 2005), BELL1 LIKE TRANSCRIPTION FACTOR5 (Banerjee et al., 2006a; Lin et al., 2013), TOMATO KNOTTED2 (Kim et al., 2001), and POTATO HOMEOBOX1 TRANSCRIPTION FACTOR (Mahajan et al., 2012), acting as long-distance signals for plant developmental processes such as leaf development, tuberization, and root growth, has been demonstrated. The movement of small interfering RNAs is also reported in a few cases where the induction of posttranscriptional gene silencing against viruses has been well studied (Waterhouse et al., 2001). In addition, small interfering RNAs were also demonstrated to be mobile and to exert epigenetic changes in recipient cells (Molnar et al., 2010). However, very limited information is available on the mobility of plant miRNAs, another group of small noncoding RNAs. Recent reviews have summarized the miRNAs found in phloem exudates of different plant species, but little information is available on their mobility (Kehr and Buhtz, 2008, 2013; Chuck and O’Connor, 2010).
The cellular movement of microRNA165/166 (miR165/166) in root patterning, where mature miR165/166 appears to move from its site of biogenesis to adjacent cell layers, is an example of the short-distance movement of miRNA (Carlsbecker et al., 2010; Miyashima et al., 2011). Although some reports (Yoo et al., 2004; Buhtz et al., 2008; Varkonyi-Gasic et al., 2010) have demonstrated the presence of numerous miRNAs in phloem tissues, so far, only three miRNAs (miR399, miR395, and miR172) have been shown to move long distance in plants. miR399 acts as a long-distance mobile signal that regulates phosphate homeostasis in Arabidopsis (Pant et al., 2008), whereas miR395 was shown to move from wild-type scions to rootstocks of the miRNA-processing mutant hen1-1 under sulfate stress (Buhtz et al., 2010). In another study, Martin and coworkers (2009) proposed that miR172 functions as a long-distance mobile signal for potato (Solanum tuberosum) tuberization. Later, Kasai et al. (2010) showed that miR172 molecules can move systemically from source to sink tissues in Nicotiana benthamiana. Earlier studies have shown that miR172, along with another miRNA (miR156), regulate phase transitions and flowering in Arabidopsis. miR172 has been demonstrated to promote adult phase and flowering, whereas miR156 is involved in juvenile stage development (Wu et al., 2009). Similar roles of miR156 and miR172 were also reported in rice (Oryza sativa; Xie et al., 2006) and maize (Zea mays; Chuck et al., 2007). Sequential action of both these miRNAs appears to be pivotal for phase transition and flowering in plant development. Flowering and tuberization are reproductive strategies that bear similar environmental cues and molecular players (Jackson, 2009). With the evidence of miR172 being involved in both these pathways, we hypothesize that miR156 could be involved in the potato tuberization pathway acting in concert with miR172.
miR156 is a well-conserved miRNA present in all land plants (Axtell and Bowman, 2008). It targets the transcripts of SQUAMOSA PROMOTER BINDING-LIKE (SPL) transcription factors and acts as a master regulator of plant development (Schwab et al., 2005). In Arabidopsis, miR156 overexpression results in a prolonged juvenile phase and a delay in flowering, with increased branching and production of a large number of leaves (Huijser and Schmid, 2011). Similar phenotypes of miR156 overexpression were also observed in rice (Xie et al., 2006), maize (Chuck et al., 2007), switchgrass (Panicum virgatum; Fu et al., 2012), and tomato (Solanum lycopersicum; Zhang et al., 2011b). In a recent study, Eviatar-Ribak et al. (2013) overexpressed the Arabidopsis miR156 gene in potato (cv Desiree), where miR156-overexpressing lines exhibited suppressed leaf complexity and produced aerial tubers, indicating a role of miR156 in tuberization. In addition to these functions, miR156 and its targets, SPL transcription factors, have also been shown to regulate embryonic patterning (Nodine and Bartel, 2010), anthocyanin biosynthesis (Gou et al., 2011), and male fertility (Xing et al., 2010). Interestingly, miR156 has also been detected in phloem sap of pumpkin (Cucurbita maxima; Yoo et al., 2004), Arabidopsis, apple (Malus domestica; Varkonyi-Gasic et al., 2010), and Brassica napus (Buhtz et al., 2008, 2010). miRNAs present in phloem exudates are proposed to be mobile, with a putative role as long-distance regulators of development and stress pathways by acting on target genes (Marín-González and Suárez-López, 2012). Although miR156 is known to interact with the transcripts of SPL transcription factors, the mobility of miR156 in plants has not yet been investigated.
In this study, we have identified and validated a miR156a precursor from potato. To understand the role of miR156 and its target genes in potato development, we employed a number of strategies, including target gene validations, transgenic analysis, assays of miR156 abundance, high-resolution mass spectrometry (HR-MS)-based hormone quantification, phloem sap analysis, and grafting. Our results suggest that miR156 is a graft-transmissible signal that affects plant architecture and tuber development in potato. It is present in the phloem of wild-type plants, and it accumulates in short-day (SD)-induced stolons to facilitate tuber formation. In addition, miR156 overexpression (OE) lines show multiple morphological changes and produce aerial tubers under inductive conditions. Although the formation of aerial tubers was recently demonstrated by Eviatar-Ribak et al. (2013), our study reveals additional novel functions of miR156 in potato. Based on its accumulation in phloem sap of wild-type plants and its graft-transmissible effect, our results suggest that miR156 moves through the phloem and regulates development in potato.
RESULTS
Identification, Validation, and Expression Analysis of miR156 in Potato
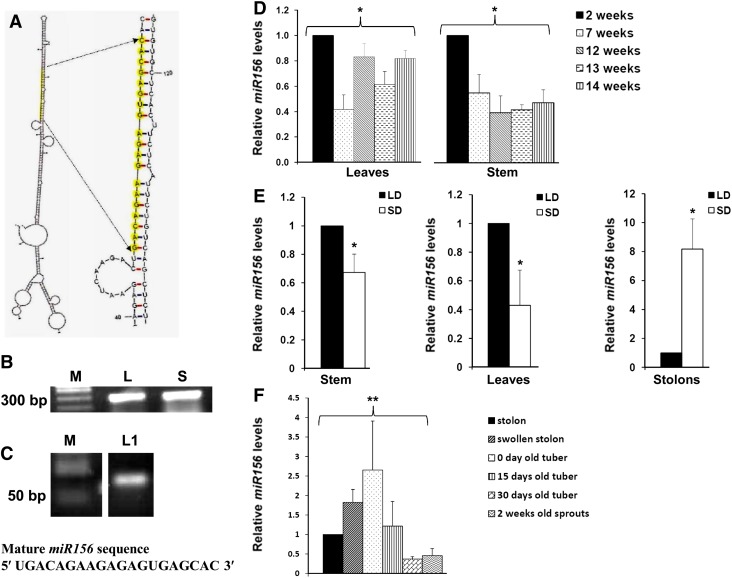
miR156 was predicted to be present in potato by an in silico analysis reported earlier (Zhang et al., 2009). The MFold-predicted secondary structure (Zuker, 2003) of the miR156a precursor sequence (BI432985.1) had a hairpin loop with mature miR156 in its stem region, a characteristic of miRNA precursors (Fig. 1A). To validate the presence of the miR156a precursor in potato, reverse transcription (RT)-PCR was performed, and the amplified fragment was sequence confirmed (Fig. 1B). A 20-bp mature miR156 was detected by stem-loop end-point PCR in leaves and was verified by sequencing (Fig. 1C), demonstrating that miR156 is expressed in potato.
Figure 1.
Identification, validation, and expression analysis of miR156 in potato. A, Secondary structure of miR156a precursor as predicted by MFold (Zuker, 2003). Mature miR156 sequence is highlighted in yellow. B, RT-PCR of miR156a precursor from leaf (L) and shoot (S). M represents a DNA marker. C, Stem-loop RT-PCR of mature miR156 from leaves of LD-grown plants (L1). D, Age-specific miR156 abundance in leaves and stem of wild-type potato grown under LD photoperiod. Error bars indicate sd of two biological replicates each with three technical replicates. Asterisks indicate one-factor ANOVA (*P < 0.05). E, miR156 abundance in stem, leaves, and stolons of wild-type potato grown under LD and SD photoperiods for 15 dpi. Error bars indicate sd of three biological replicates each with three technical replicates. Asterisks indicate Student’s t test (*P < 0.05). F, Relative abundance of miR156 in different developmental stages of tuber formation and dormancy. Error bars indicate sd of three biological replicates each with three technical replicates. Asterisks indicate one-factor ANOVA (**P < 0.01). [See online article for color version of this figure.]
The relative levels of miR156 were analyzed by stem-loop quantitative reverse transcription (qRT)-PCR in potato plants of different age groups. Two-week-old plants showed higher accumulation of miR156 in stem, and their levels decreased as the plant aged. However, miR156 levels varied in mature leaves of plants of different ages (Fig. 1D). To determine whether miR156 expression is regulated by the photoperiod, plants were grown under long-day (LD; tuber-noninductive) and SD (tuber-inductive) conditions. Stem-loop qRT-PCR analysis demonstrated a higher accumulation of miR156 in leaves and stem under LD conditions as compared with the plants under the SD photoperiod (Fig. 1E). However, in stolons, miR156 levels were found to be approximately 8-fold higher under SD as compared with LD photoperiod (Fig. 1E). Our analysis showed a range of miR156 abundance in swollen stolons, tubers stored postharvest for different time periods (0, 15, and 30 d), and 2-week-old sprouts. Zero-day-old tubers (postharvest) showed an approximately 2.5-fold higher accumulation than in stolons harvested from SD-induced plants, whereas miR156 levels in juvenile tuber sprouts were almost half the level in stolons (Fig. 1F). Overall, our expression analysis suggests that miR156 shows tissue-specific accumulation with respect to the age of the plant and the photoperiod.
Overexpression of miR156 Affects Multiple Morphological Traits
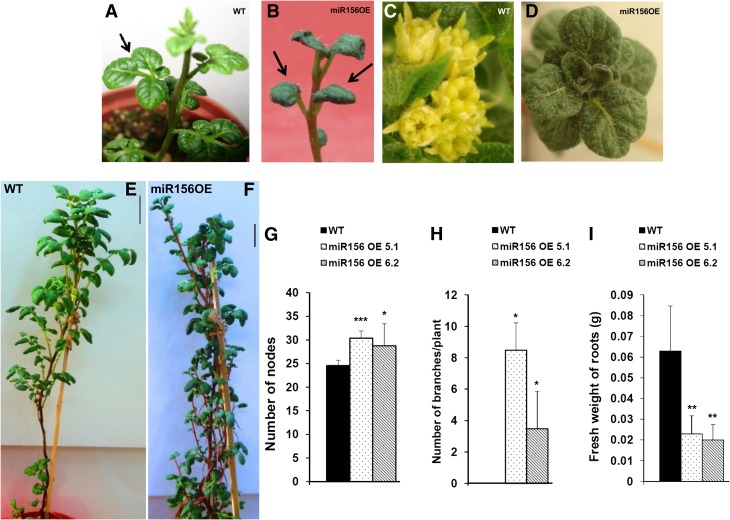
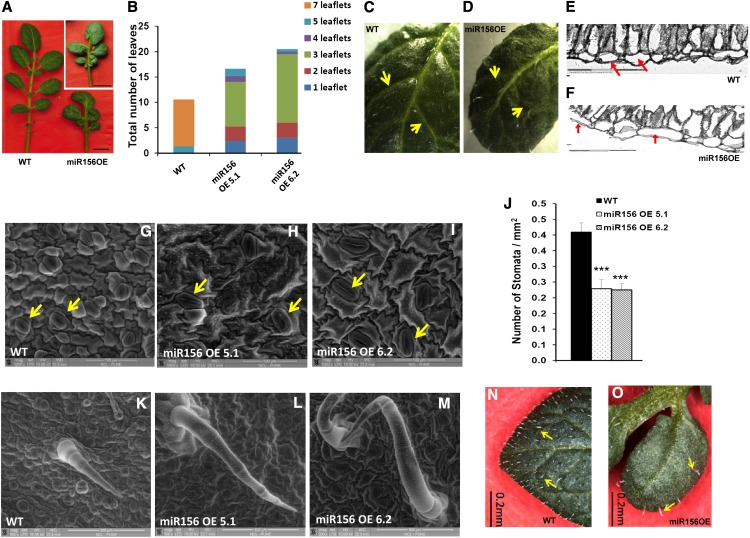
The level of miR156 in miR156 OE lines (miR156 OE 5.1 and 6.2) was determined by stem-loop qRT-PCR (Supplemental Fig. S1, A and B). Two-week-old OE lines exhibited a drastic change in leaf phenotype (Fig. 2, A and B), and as they matured, they did not form an inflorescence compared with wild-type plants (Fig. 2, C and D). These plants did not flower even after 18 weeks of growth, whereas wild-type plants produced inflorescences in 12 weeks. miR156 OE plants also exhibited enhanced branching from axillary buds and an increased number of nodes, resulting in a bushy appearance (Fig. 2, E–H). The fresh weight of roots in OE lines was also significantly reduced (Fig. 2I). The leaf architecture of miR156 OE lines was dramatically affected. miR156 OE plants produced smaller leaves with reduced leaflet number (Fig. 3, A and B). The venation pattern was found to be altered such that the side veins of transgenic leaves were less prominent (Fig. 3, C and D). Transverse sections of leaves showed disoriented cell arrangement as well as the presence of large epidermal cells in the miR156 OE 5.1 line (Fig. 3, E and F). We also observed a reduction in stomatal density in both OE lines as opposed to the wild type (Fig. 3, G–J). In addition, trichome number was reduced with an increase in trichome length in OE lines (Fig. 3, K–O). To gain more insight into the function of miR156, 35S::miR156 tobacco (Nicotiana tabacum) plants were generated. All the tobacco OE plants showed a similar phenotype to that observed in miR156 OE potato plants (Supplemental Fig. S2).
Figure 2.
Overexpression of miR156 affects multiple morphological traits in potato. A and B, Two-week-old plants of the wild type (WT; A) and miR156 OE 5.1 (B). C and D, Inflorescence produced at the apical tip of 12-week-old wild-type potato plants (C), while miR156 OE 5.1 plants of the same age produced leaves (D). E and F, Twelve-week-old plants of wild-type (E) and miR156 OE 5.1 (F) lines of potato. Bars = 5 cm. G to I, Number of nodes (G; n = 5), number of axillary branches (H; n = 4), and fresh weight of roots (I; n = 6) of wild-type and miR156 OE plants. Error bars indicate sd. Asterisks indicate statistical differences as determined using Student’s t test (***P < 0.001, **P < 0.01, *P < 0.05). [See online article for color version of this figure.]
Figure 3.
Effect of miR156 overexpression on leaf development of potato. A, Leaves of 8-week-old wild-type (WT) and miR156 OE 5.1 and 6.2 (inset) plants. Bars = 1 cm. B, Distribution of the number of leaflets per leaf in 8-week-old wild-type and miR156 OE 5.1 and 6.2 plants. C and D, Venation pattern of wild-type leaf (C) and miR156 OE 5.1 leaf (D). Arrows indicate veins. E and F, Transverse sections of leaves (20×) of wild-type (E) and miR156 OE 5.1 (F) plants showing differences in leaf architecture. The epidermal cells are marked by arrows. G to I, eSEM images of the leaf surface showing differences in the size of epidermal cells and stomata (marked by arrows) for wild-type (G) and miR156 OE 5.1 (H) and miR156 OE 6.2 (I) plants. Bars = 100 μm. J, Stomatal density of wild-type and miR156 OE 5.1 and 6.2 plants (n = 5). Error bars indicate sd. Asterisks indicate statistical differences as determined using Student’s t test (***P < 0.001). K to M, eSEM images of trichomes for wild-type (K) and miR156 OE 5.1 (L) and miR156 OE 6.2 (M) plants. Bars = 300 μm. N and O, Trichome phenotype of wild-type leaf (N) and miR156 OE 5.1 leaf (O). Bars = 0.2 mm. [See online article for color version of this figure.]
miR156 Regulates Potato Tuberization
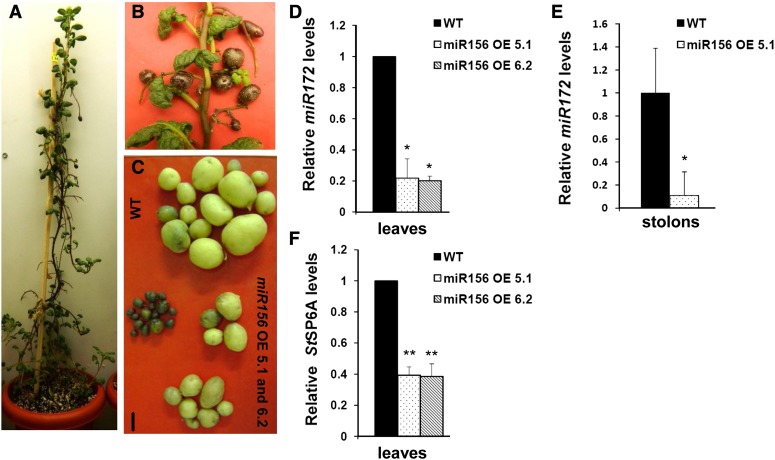
To examine whether an increase in miR156 levels in OE lines could have any impact on tuber development, we examined the tuberization phenotype of the miR156 OE 5.1 and 6.2 lines. OE line 5.1 produced aerial and underground tubers after 4 weeks of SD induction, whereas wild-type plants grown under an SD photoperiod only produced underground tubers (Fig. 4, A–C). Line 6.2 produced underground tubers and showed a delayed formation of aerial tubers. None of the plants produced tubers under the LD photoperiod.
Figure 4.
miR156 regulates potato tuberization. A, miR156 OE 5.1 plant incubated for 30 d under SD conditions. B, Aerial tubers developed on miR156 OE 5.1. C, Tubers of representative wild-type (WT) and miR156 OE line 5.1 and 6.2 plants. Bar = 1 cm. D to F, Levels of tuberization markers: miR172 in 8-d post SD-induced leaves of wild-type and miR156 OE line 5.1 and 6.2 plants (D); miR172 in 15-d post SD-induced stolons of wild-type and miR156 OE line 5.1 plants (E); and StSP6A in 8-d post SD-induced leaves of wild-type and miR156 OE line 5.1 and 6.2 plants (F). For miR172 in leaves (D), error bars indicate sd of two biological replicates each with three technical replicates; for miR172 in stolons (E; 15 dpi in SD conditions), error bars indicate sd of one biological replicate with three technical replicates; for StSP6A (F), semiquantitative analysis was performed with three independent replicates. Error bars indicate sd of three replicates. Asterisks indicate statistical differences as determined using Student’s t test (*P < 0.05, **P < 0.01). [See online article for color version of this figure.]
Overall, miR156 OE lines (5.1 and 6.2) developed fewer underground tubers and showed reduced tuber yields (Table I). Previous reports have shown miR172 and the Flowering Locus T-like paralog StSP6A to act as positive regulators of tuberization (Martin et al., 2009; Navarro et al., 2011). Since miR156 OE lines exhibited reduced tuber yield, we investigated the levels of StSP6A and miR172 (tuberization markers) in leaves of OE plants. Also, miR172 levels were quantified in SD-induced stolons. Our results showed a reduction in the levels of the tuberization markers miR172 and StSP6A in miR156 OE lines. miR172 levels were reduced by approximately 80% in leaves and stolons, while StSP6A levels were reduced by approximately 60% in leaves (Fig. 4, D–F).
Table I. Tuber yields (tuber number and weight) of wild-type and miR156 OE 5.1 and 6.2 plants incubated under SD conditions for 30 d.
Means of three plants each were calculated.
Zeatin Riboside and Orobanchyl Acetate Levels Are Affected by miR156 Overexpression
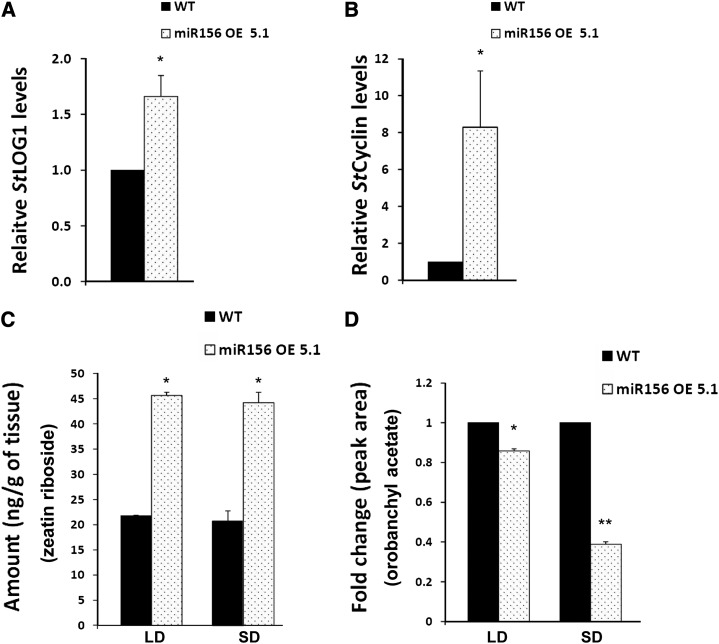
miR156 overexpression in potato resulted in a drastic phenotype of increased branching, a higher number of leaves with reduced leaflets, and a delay in flowering, a phenotype that was also recently described for tomato LONELY GUY1 (TLOG1) overexpression in tomato (Eviatar-Ribak et al., 2013). LONELY GUY1 (LOG1) is a cytokinin biosynthetic gene that converts cytokinin ribosides to biologically active cytokinin (Kurakawa et al., 2007). Considering the role of cytokinins in branching (Domagalska and Leyser, 2011), we investigated the effect of miR156 on the cytokinin pathway. miR156 OE plants showed approximately 1.8-fold increased expression of StLOG1 in the axillary meristems as compared with wild-type plants (Fig. 5A). Also, the levels of StCyclin D3.1, a cytokinin-responsive gene, were increased up to approximately 8-fold as compared with wild-type plants (Fig. 5B). To determine the amount of cytokinin (zeatin riboside), HR-MS analysis demonstrated increased levels (more than 2-fold) in miR156 OE plants as compared with the wild type in both SD and LD conditions (Fig. 5C; Supplemental Figs. S3 and S4). As strigolactones are also considered to be branching hormones (Domagalska and Leyser, 2011), we investigated the levels of one such strigolactone: orobanchyl acetate. HR-MS analysis demonstrated reduced levels of orobanchyl acetate (approximately 20% under LD conditions and approximately 60% under SD conditions) in miR156 OE plants as compared with the wild type (Fig. 5D; Supplemental Figs. S5 and S6). The changes in these hormone amounts correlated with the branching phenotype observed in miR156 OE lines.
Figure 5.
Zeatin riboside and orobanchyl acetate levels are affected by miR156 overexpression. A and B, qRT-PCR analysis of StLOG1 (A) and StCyclin D3.1 (B) in axillary meristems of wild-type (WT) and miR156 OE 5.1 plants incubated for 15 d under SD conditions. Error bars indicate sd of three biological replicates each with three technical replicates. Asterisks indicate statistical differences as determined using Student’s t test (*P < 0.05). C and D, HR-MS analysis of wild-type and miR156 OE 5.1 plants for zeatin riboside (C) and orobanchyl acetate (D). The tissues were axillary meristems of wild-type and miR156 OE 5.1 plants incubated for 15 d under both SD and LD conditions. Error bars indicate sd of two biological replicates. Asterisks indicate statistical differences as determined using Student’s t test (*P < 0.05, **P < 0.01).
miR156 Targets StSPL Transcription Factors
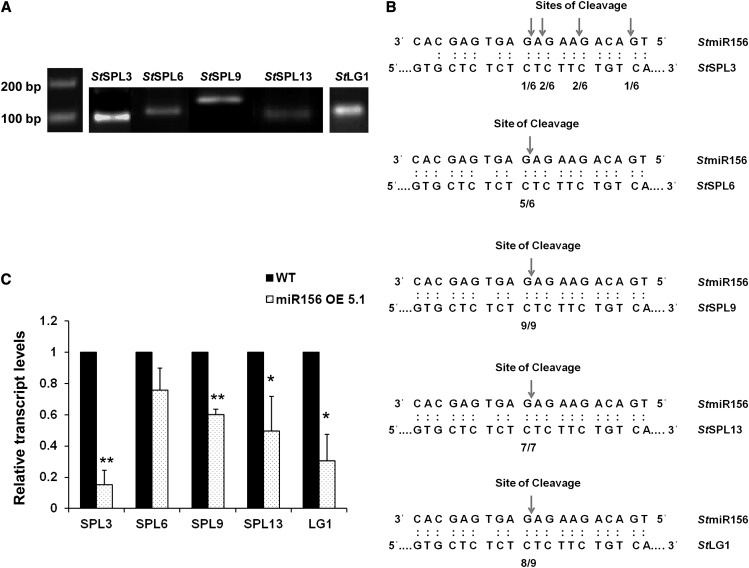
Our in silico analysis with psRNATarget software (plantgrn.noble.org/psRNATarget; Dai and Zhao, 2011) predicted 12 potential target genes for miR156 in potato (Supplemental Table S1). Further analysis of these target genes revealed that nine out of the 12 genes (including LIGULELESS1 [LG1]) belong to the SPL family of transcription factors. Two belong to the DNA topoisomerase family of proteins, while one target is of unknown function. Additionally, we have also predicted all these target genes by TargetAlign software. Finally, based on their scores and consistency of analysis in both softwares, we short listed five miR156 target genes, StSPL3, StSPL6, StSPL9, StSPL13, and StLG1, for further analysis. To determine if these genes are the targets of miR156 in potato, modified RNA ligase-mediated (RLM) 5′ RACE was performed. RNA sequences with 5′ termini corresponding to the 10th/11th nucleotides of miR156 were consistently detected, demonstrating that StSPL6, StSPL9, StSPL13, and StLG1 are targeted by miR156 in vivo (Fig. 6, A and B). However, StSPL3 was cleaved at sites other than the 10th/11th nucleotides of miR156, which is not a common observation in plant miRNAs. Levels of these targets were also quantified in miR156 OE plants. As expected, transcript levels of these targets showed different degrees of reduction (StSPL3, approximately 80%; StLG1, 70%; StSPL13, 60%; StSPL9, 40%; and StSPL6, 30% in miR156 OE 5.1 plants [Fig. 6C]). Our results are consistent with previous studies on the miR156-SPL interaction (Schwab et al., 2005).
Figure 6.
miR156 targets in potato. A and B, miR156 cleavage site mapping in miR156 targets as determined by modified RLM-RACE. A, Nested PCR products were cloned and sequenced. B, Frequency of 5′ RACE clones showing cleavage site (arrows) and fractions indicating proportions of clones showing these cleavage sites. C, Expression pattern of StSPL3, StSPL6, StSPL9, StSPL13, and StLG1 in wild-type (WT) and miR156 OE 5.1 plants by qRT-PCR. Error bars indicate sd of three biological replicates each with three technical replicates. Asterisks indicate statistical differences as determined using Student’s t test (*P < 0.05, **P < 0.01).
Regulation of miR172 by the miR156-SPL Module
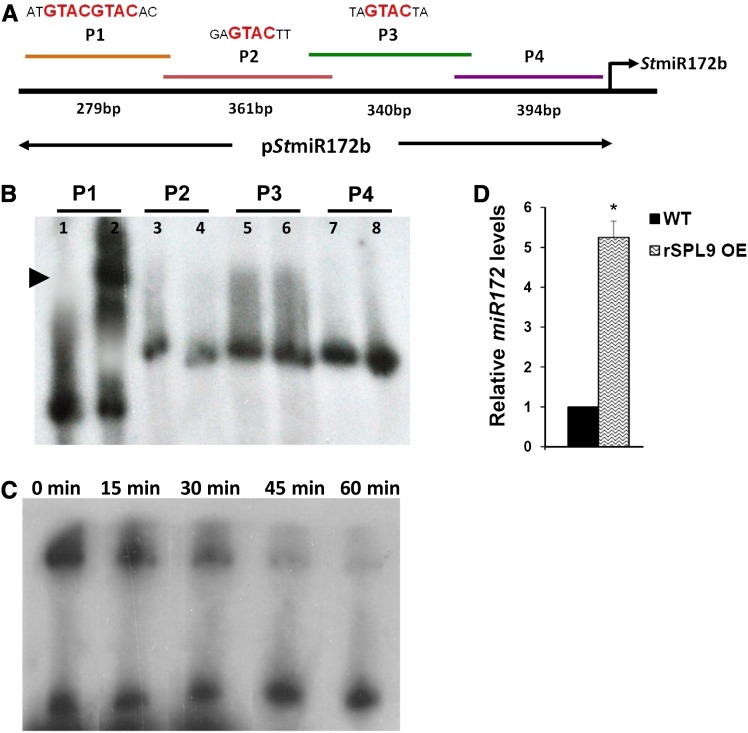
Overexpression of miR156 in potato resulted in lower tuber yields and reduced levels of miR172 and SPLs as mentioned above. Our bioinformatic analysis of the StMIR172b promoter showed the presence of multiple GTAC motifs, characteristic of SPL binding (Birkenbihl et al., 2005). We chose to continue our investigation with StSPL9, since the miR156-SPL9 interaction has previously been demonstrated in Arabidopsis (Wu et al., 2009) and rice (Jiao et al., 2010), suggesting that a similar interaction module might also be conserved in potato. To examine if StSPL9 binds to the StMIR172b promoter, gel retardation assays were performed. The StMIR172b promoter was analyzed in four fragments (P1–P4; Fig. 7A), having two binding motifs in the P1 fragment, a single motif each in P2 and P3, while P4 served as a negative control. Recombinant StSPL9 protein (42 kD) retarded the mobility of the P1 promoter sequence, whereas the other three promoter fragments (P2–P4) remained unaffected (Fig. 7B). Competition gel retardation assays were performed with 32P-labeled and unlabeled P1 fragment. With increased unlabeled P1, the P1-SPL9 complex was diminished over time (Fig. 7C). Our analysis demonstrated StSPL9-MIR172 promoter interactions in vitro, with StSPL9 binding to a promoter region with two binding sites.
Figure 7.
StSPL9 binds to the StMIR172b promoter. A, Schematic representation of StMIR172b promoter sequence showing SPL binding motifs and lengths of four fragments. B, Gel retardation assay of StMIR172b promoter fragments P1 to P4 with StSPL9. The lanes are alternate for free probe and probe + protein. C, Cold competition retardation assay of P1 with StSPL9. Labeled P1 was incubated with StSPL9 for 30 min at 25°C, and then a 100-fold molar excess of unlabeled P1 was added and aliquots were analyzed after the indicated times (0–60 min). D, Relative levels of miR172 in 15-d post SD-induced leaves of wild-type (WT) and rSPL9 OE plants. Error bars indicate sd of two biological replicates each with three technical replicates. The asterisk indicates a statistical difference as determined using Student’s t test (*P < 0.05). [See online article for color version of this figure.]
To further validate the miR156-StSPL9 interaction, we generated miR156-resistant StSPL9 OE potato plants (rSPL9 OE lines) driven by the cauliflower mosaic virus (CaMV) 35S promoter (Supplemental Fig. S7, A and B). rSPL9 transgenics were generated by introducing silent mutations in the microRNA recognition element (MRE), so that the mutated transcript is no longer recognized by miR156. Stem-loop qRT-PCR analysis revealed an approximately 5-fold increase in levels of miR172 under SD conditions compared with the wild type (Fig. 7D). This increase in miR172 levels under SD conditions, however, was not reflected by the tuberization phenotype of the rSPL9 OE line (Supplemental Fig. S7C).
Detection of miR156 in Phloem of Wild-Type Potato
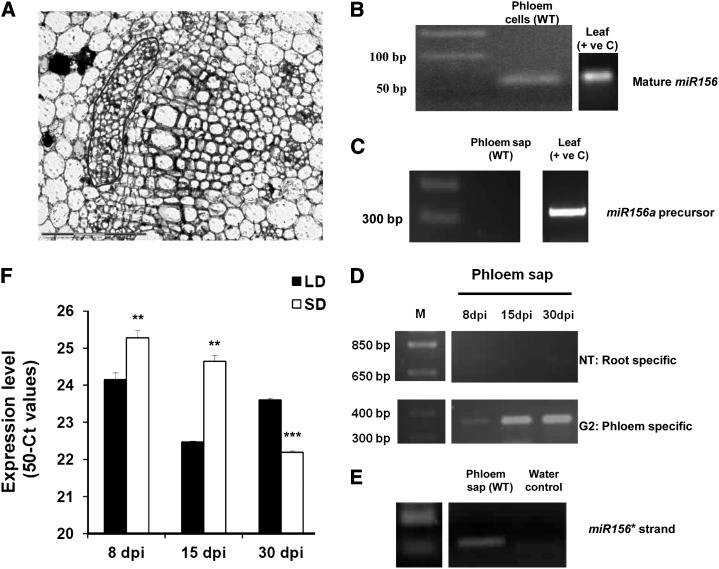
In order to investigate the presence of miR156 in phloem of potato plants, we harvested phloem cells by laser microdissection pressure catapulting (LMPC) and tested for the presence of miR156 in phloem cells of wild-type potato (Fig. 8, A and B). While miR156 was detected in phloem cells, we did not detect the miR156a precursor in phloem sap harvested from wild-type plants (Fig. 8C). The purity of phloem sap (phloem-enriched exudate) was confirmed by detecting the phloem-specific transcript G2-like transcription factor and the absence of root-specific transcript nitrate transporter (Fig. 8D). The miR156* strand, however, was detected in phloem sap exudates of wild-type plants by stem-loop qRT-PCR (Fig. 8E). To understand if photoperiod has any role in miR156 accumulation in the phloem, we also carried out a stem-loop qRT-PCR analysis of phloem sap harvested from wild-type plants incubated for 8, 15, and 30 d post induction (dpi) under both SD and LD conditions. Higher accumulation of miR156 was observed in phloem sap harvested from 8- and 15-dpi SD-induced plants, indicating that miR156 accumulation increased under SD conditions in phloem sap of potato. This pattern changed in plants incubated for longer times (30 dpi; Fig. 8F).
Figure 8.
Detection of miR156 in phloem. A, Phloem tissue in a wild-type stem section (marked in red). This tissue was harvested by LMPC. B, Detection of miR156 (mature) in phloem of wild-type plants (WT) and leaf tissue of wild-type plants (positive control [+ ve C]) by stem-loop RT-PCR. C, Absence of miR156a precursor (300 bp) in wild-type phloem sap and its presence in wild-type leaf, acting as a positive control, by RT-PCR analysis. D, RT-PCR analysis of nitrate transporter (NT; root-specific transcript) and G2-like transcription factor (G2; phloem-specific transcript) of potato phloem sap of the wild type (phloem-enriched exudate) to assess its purity (Banerjee et al., 2006a). E, Detection of the miR156* strand in phloem sap of the wild type by stem-loop RT-PCR. F, Differential accumulation of miR156 (mature) under SD and LD photoperiods in phloem sap of wild-type plants harvested after 8, 15, and 30 dpi. miR156 accumulation is plotted as 50 minus Ct (for cycle threshold; 50-Ct) values as described previously (Pant et al., 2008). Error bars indicate sd of one biological replicate with three technical replicates. Asterisks indicate statistical differences as determined using Student’s t test (**P < 0.01, ***P < 0.001).
miR156 Is Potentially a Graft-Transmissible Signal in Potato
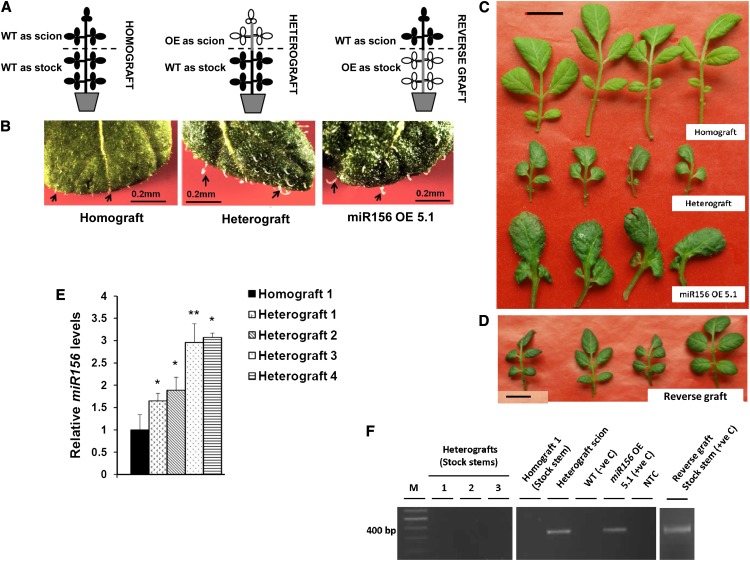
In our study, we detected miR156 in LMPC-harvested phloem cells, and it exhibited an SD-induced accumulation pattern in phloem sap. Considering this observation, we tested whether miR156 is a phloem-mobile signal in potato. Grafting experiments (homografts, heterografts, and reverse grafts) were performed to demonstrate the mobility of miR156 (Fig. 9A). After 4 weeks of SD induction, analysis of morphological changes in grafts as well as the quantitative analysis of miR156 were performed. Overall, the leaf shape and trichome morphology of stocks from the heterografts (miR156 OE plants as scion and the wild type as stock) were affected. The newly emerging leaves from the axillary shoots on the stock of heterografts had more prominent but fewer trichomes (Fig. 9B) and exhibited small and thick lamina along with reduced numbers of leaflets (Fig. 9C). On the other hand, newly emerging leaves from wild-type scions of reverse grafts did not show any phenotype similar to miR156 OE plants (Fig. 9D). All the heterografts had less tuber yield as compared with homografts, while reverse grafts did not form any tubers (Table II).
Figure 9.
miR156 is a potential graft-transmissible signal. A, Pictorial representation of the grafts. WT, Wild type. B, Trichomes of homograft (stock leaves), heterograft (stock leaves), and leaves from miR156 OE 5.1 plants, where trichomes in heterografts (stock leaves) are less in number and more in length, as observed for miR156 OE 5.1 plants. Bars = 0.2 mm. C and D, Leaves of homograft (stock), heterograft (stock), miR156 OE 5.1 plants, and reverse grafts (scion), where heterograft stock leaves mimic the phenotype of miR156 OE 5.1 leaves (C), while reverse graft scion leaves mimic the phenotype of homograft leaves (stock; D) Bars = 1 cm. E, Relative levels of mature miR156 in stock stems of four representative heterografts (1–4) and homograft incubated under SD conditions and harvested after 30 dpi were measured by stem-loop qRT-PCR. Error bars indicate sd of one biological replicate with three technical replicates. Asterisks indicate statistical differences as determined using Student’s t test (*P < 0.05, **P < 0.01). F, Detection of miR156a precursor transgene in stock stems of homograft, three representative heterografts (1–3), and reverse graft by RT-PCR analysis. Stem tissue of heterograft scion, reverse graft stock, and a miR156 OE 5.1 plant served as positive controls (+ve C), with the wild type as a negative control (−ve C). NTC, No template control. [See online article for color version of this figure.]
Table II. Number of tubers of homografts, heterografts, and reverse grafts incubated under SD conditions for 30 d.
Means of three grafted plants were calculated.
| Graft | No. of Tubers |
|---|---|
| Homografts | 9.5 ± 2.12 |
| Heterografts | 2 ± 1.41 |
| Reverse grafts | 0 ± 0 |
The morphological changes in the stock stems of heterografts could be due to (1) the transport of mature miR156 itself, (2) the transport of the overexpressed miR156a precursor transgene, or (3) a miR156-mediated up-regulated mobile factor activating miR156 transcription in stock stems. To analyze if mature miR156 is transported, we carried out a quantitative analysis by stem-loop qRT-PCR. In stock stems of all four heterografts, a higher accumulation of mature miR156 was observed, as opposed to homografts (Fig. 9E). On the other hand, absence of the miR156a precursor transgene in heterograft stock stems confirmed that the overexpressed transgene is not moving from scion to stock (Fig. 9F). Also, a comparative analysis of mature miR156 and miR156a precursor levels in both wild-type and grafted plants clearly demonstrated that mature miR156 had a higher accumulation than its precursor form (Supplemental Fig. S8, A–D). These findings make the possibility of miR156a precursor transgene movement as well as activated localized transcription of miR156 in stock stems of heterografts unlikely. Instead, the higher accumulation of mature miR156 in heterograft stock stems supports the preferential transport of mature miR156 itself, from scion to stock in grafted plants. Overall, our results suggest that miR156 is a graft-transmissible phloem-mobile signal that affects tuberization and plant architecture in potato.
DISCUSSION
miR156 in Potato
Several reports have previously demonstrated the function of miR156 in various plant species like maize (Chuck et al., 2007), Arabidopsis (Huijser and Schmid, 2011), switchgrass (Fu et al., 2012), rice (Xie et al., 2006), tomato (Zhang et al., 2011b), and poplar hybrid (Populus trichocarpa; Wang et al., 2011). Earlier studies (Zhang et al., 2009; Yang et al., 2010; Xie et al., 2011) have predicted miR156 in potato through a bioinformatic approach. The recent study by the Tomato Genome Consortium (2012) reported 13 members of the miR156 family in potato, whereas the miR156 family has 12 members in Arabidopsis, 12 in rice, and 11 in poplar hybrid (Griffiths-Jones et al., 2008). A very recent report (Eviatar-Ribak et al., 2013) has shown that overexpression of miR156 in the potato cv Desiree (a day-neutral cultivar) resulted in stolon-borne aerial mini tubers from almost all of the distal buds. miR156 OE plants had exhibited late flowering, suppression of leaf complexity, and a profuse branching phenotype. We had similar observations for miR156 overexpression in potato, but in the photoperiod-sensitive subspecies andigena 7540. We wanted to investigate the following. (1) Does miR156 affect multiple morphological traits other than what was already observed in potato? (2) What could be the target genes of miR156 and their roles in potato? (3) What is the role of miR156 in tuberization under different photoperiods? (4) Knowing its presence in phloem of other plant species, our hypothesis was to test if miR156 acts as a potential mobile signal in potato development.
To answer these questions, we started with validating the presence of one member of the miR156 family, miR156a in potato. This is different from the approach followed in a previous report (Eviatar-Ribak et al., 2013), where the Arabidopsis miR156a precursor was overexpressed in potato. The sequence of mature potato miR156 was found to be identical to that of miR156 of Arabidopsis, rice, maize, and sorghum (Sorghum bicolor), suggesting its conserved nature. In our analysis, miR156 exhibited an age-dependent expression pattern in potato stem tissues, and its levels decreased as the plant aged (Fig. 1D). This observation was consistent with the earlier studies in Arabidopsis and rice (Wu et al., 2009; Xie et al., 2012). Both of these reports showed a gradual decrease of the expression of miR156 in shoots as the plant aged. However, in developing leaves of rice, an opposite expression pattern of miR156 was demonstrated. In our study, no significant pattern of miR156 expression was observed in leaves (Fig. 1D). All of the previous reports mentioned above demonstrate decreased miR156 expression with respect to plant age, suggesting its role in juvenile phase maintenance. However, photoperiod-mediated expression and function of miR156 have not been documented earlier. Because tuberization in potato is a photoperiod-regulated process, we investigated the effect of the photoperiod on miR156 expression and its function. Our analysis suggests that miR156 is differentially expressed under SD/LD conditions in a tissue-specific manner (Fig. 1, E and F). As we have validated the miR156a precursor in potato, we carried out bioinformatic analysis of the upstream sequence of the MIR156a gene using the PLACE online tool (Higo et al., 1999). The MIR156a upstream sequence exhibited a number of light regulatory motifs (Supplemental Table S2; Waksman et al., 1987; Vorst et al., 1990; Vauterin et al., 1999), indicating a putative light-mediated regulation of this miRNA along with an endogenous, age-mediated regulation.
Overexpression of miR156 Affects Plant Architecture in Potato and Tobacco
In our study, miR156 OE lines (potato and tobacco) exhibited phenotypes like profuse axillary branching, altered leaf and trichome morphology, and delayed or no flowering, documented earlier in a number of plants such as Arabidopsis (Huijser and Schmid, 2011), rice (Xie et al., 2006), tomato (Zhang et al., 2011b), and potato (Eviatar-Ribak et al., 2013). In addition to these phenotypes, an altered venation pattern in leaves, disoriented cell organization with larger epidermal cells, and reduced stomatal density and root biomass were also observed in both the OE lines (Figs. 2 and 3), indicating several new functions for miR156 in potato. Overall, this suggests that miR156 acts as a master regulator involved in the regulation of different plant developmental traits. The altered leaf morphology in OE plants can possibly be a result of reduced levels of SPLs (Fig. 6C). A number of previous reports (Wu and Poethig, 2006; Shikata et al., 2009; Usami et al., 2009; Chen et al., 2010) have described the role of SPLs in leaf development in Arabidopsis, suggesting that StSPLs might control leaf size and shape, altered venation, and reduced leaflet number in potato as well. LG1 is a well-characterized SPL protein whose function in leaf development has previously been reported in maize (Harper and Freeling, 1996) and rice (Lee et al., 2007). It was shown to be involved in controlling ligule and auricle development and the formation of a laminar joint between leaf blade and leaf sheath. In our study, reduced StLG1 expression in miR156 OE plants could possibly explain the aberrant leaf morphology (reduced leaf lamina and curled leaf margins). To understand the cause of the profuse branching phenotype of miR156 OE plants in potato, we quantified the amounts of cytokinin and strigolactone, hormones that are known to play an important role in branching (Domagalska and Leyser, 2011). Cytokinins act antagonistically to auxins and promote branching from axillary meristems, leading to the loss of apical dominance (Domagalska and Leyser, 2011). miR156 OE plants exhibited increased levels (more than 2-fold) of zeatin riboside under both SD and LD photoperiods (Fig. 5C), which is consistent with the bushy phenotype of these OE plants. Increased cytokinin amount was also accompanied by increased expression of the cytokinin biosynthesis gene StLOG1 and the cytokinin-responsive gene StCyclin D3.1 (Fig. 5, A and B). This increase in the activity of cytokinin might have caused the profuse branching phenotype. On the other hand, in Arabidopsis, strigolactone mutants show increased branching (Gomez-Roldan et al., 2008). In our study as well, miR156 OE plants contained reduced amounts of orobanchyl acetate (Fig. 5D). The absence of flowering in both potato and tobacco OE plants supports the role of miR156 in controlling phase transitions. In Arabidopsis, AtSPL2, AtSPL3, AtSPL9, AtSPL10, and AtSPL11 are shown to act as positive regulators in promoting floral meristem identity by directly regulating genes like LEAFY, FRUITFUL, and APETALA1 (Chen et al., 2010). A similar mechanism might also be conserved in potato, since StSPL3 and StSPL9 are found to be reduced in miR156 OE plants (Fig. 6C).
miR156 Regulates Potato Tuberization
Flowering and tuberization are different reproductive strategies, both of which are photoperiod-mediated mechanisms (Jackson, 2009). Several molecular components like phytochrome B (Jackson et al., 1998), CONSTANS (Martinez-Garcia et al., 2002), and StSP6A (Navarro et al., 2011) have previously been shown to play roles in flowering and potato tuberization. In addition, two other miRNAs (miR156 and miR172) were shown to control developmental timing and flowering in Arabidopsis (Wu et al., 2009). The positive role of miR172 in tuberization was reported earlier (Martin et al., 2009), while a very recent report (Eviatar-Ribak et al., 2013) demonstrated the role of miR156 in tuber formation. In Arabidopsis, miR156 regulates miR172 expression via AtSPL9 (Wu et al., 2009) during phase transitions. Similarly, we observed reductions of miR172 and StSPL9 in miR156 OE lines (Figs. 4, D and E, and 6C) in potato. Although in our analysis, StSPL9 was found to be reduced by 40%, there is a possibility of miR156 acting on StSPL9 by translational arrest as well (Gandikota et al., 2007). Gel retardations assays confirmed the regulatory role of the miR156-SPL-miR172 module in potato. However, control of miR172 by SPLs other than StSPL9 cannot be ruled out. This regulatory module is likely to be active in leaves induced under LD conditions, as there are high levels of miR156 but reduced levels of SPL9 and miR172, whereas an increased accumulation of miR156 and miR172 in SD-induced stolons reflects a lack of regulation of miR172 by miR156, possibly due to the tissue-specific action of miR156 or spatial exclusion.
Several interesting observations regarding the effect of miR156 on tuberization were noted in our study. In stolons (the tissue destined to form a tuber) harvested from SD-induced wild-type plants, an approximately 8-fold increase in the level of miR156 was detected (Fig. 1E). Also, miR156 OE lines, when incubated under SD conditions, produced aerial tubers, as reported in a recent work (Eviatar-Ribak et al., 2013). However, in our study, miR156 OE lines exhibited a reduction in overall tuber yield and the levels of the tuberization markers miR172 and StSP6A (Fig. 4, D–F). Considering these observations, should miR156 be termed as an activator or a repressor of tuberization? If miR156 acts as an activator, miR156 OE lines would have produced tubers under LD (noninductive) conditions, as observed previously for StBEL5 (Banerjee et al., 2006a), miR172 (Martin et al., 2009), and StSP6A (Navarro et al., 2011) OE lines. In our study, miR156 OE lines produced aerial tubers in SD conditions (Fig. 4B). This rules out the possibility of miR156 functioning as a repressor. The reduced levels of tuberization markers in miR156 OE lines can possibly be due to the prolonged juvenile phase of these plants, which in turn reduced the overall tuber yield. In potato, all axillary meristems have the capacity to form tubers, and under permissive conditions any meristem can produce aerial tubers. However, this potential is suppressed except in stolons (Xu et al., 1998). We propose that under tuber-inductive conditions, a threshold level of miR156 facilitates tuber formation from a meristem. Overexpression of miR156 in potato results in levels above threshold in all the axillary meristems; hence, the plant produces aerial tubers under SD conditions. The recent work by Eviatar-Ribak and coworkers (2013) demonstrated that TLOG1 OE tomato plants produced sessile tubers only in basal meristems, whereas TLOG1-miR156 double OE plants produced sessile tubers from all axillary meristems. In our study, we used potato subspecies andigena, which is sensitive to photoperiod for tuberization. When miR156 is overexpressed in this background, the OE plants produced aerial tubers only under SD conditions, whereas in LD conditions, the axillary meristems produced only branches. This observation clearly established that increased levels of miR156 in OE plants alone are not sufficient for tuber formation but that tuber-inductive conditions are required for aerial tuber formation.
miR156 as a Potential Phloem-Mobile Long-Distance Signal
The detection of miR156 in LMPC-harvested phloem cells and an increased accumulation in phloem exudates under SD photoperiod suggest that miR156 could possibly act as a long-distance signal in potato development. Our grafting assays support this hypothesis, as increased levels of mature miR156 could be detected in the stock stems of SD-induced heterografts (Fig. 9E). Further molecular analysis ruled out the possibility of miR156a precursor transgene movement (Fig. 9F; Supplemental Fig. S8, A–D). In addition, morphological changes in leaves and trichome phenotypes further support miR156 transport. Reverse graft assays showed that the mobility of miR156 was restricted to a shoot-to-root direction. Questions could be raised. Is miR156 transported as a double-stranded or single-stranded form in the phloem? The presence of the miR156* strand in potato phloem sap indicates that it is possibly transported as a miR156/miR156* duplex. Similar to our results, previous studies (Buhtz et al., 2008, 2010; Pant et al., 2008; Hsieh et al., 2009) also reported the presence of a star strand along with the mature miRNA in the phloem stream. Another explanation for miR156* in potato phloem could be its association with the RNA-induced silencing complex to target different genes. miRNA* species are reported to be associated with Argonaut proteins and to have inhibitory effects on target gene expression in Drosophila species (Okamura et al., 2008). A recent report by Devers and coworkers (2011) described a similar phenomenon in Medicago truncatula roots. Although a handful of miRNAs have now been detected in phloem of several plant species such as pumpkin (Yoo et al., 2004), Arabidopsis (Varkonyi-Gasic et al., 2010), and Brassica (Buhtz et al., 2008), only three miRNAs, miR399, miR395, and miR172 (Pant et al., 2008; Buhtz et al., 2010; Kasai et al., 2010), were demonstrated to act as long-distance mobile signals. We show that miR156 is involved in the regulation of plant architecture and tuberization and might be another miRNA to be transported via the phloem over long distances. The availability of techniques to differentiate between mature endogenous miRNAs from transgenic miRNAs would perhaps provide the final evidence for miRNA mobility.
Based on our results, we propose a model for the regulation of tuberization by miR156. We hypothesize that under tuber-inductive (SD) conditions, miR156 is transported to stolons through the phloem, accumulates in underground stolons (which in turn reduces the miR156 accumulation in leaves and stems), and facilitates underground tuber formation. Reduced miR156 accumulation in aerial organs inhibits the formation of aerial tubers, whereas in LD conditions, increased levels of miR156 in leaves and stems assist the vegetative growth of the plant. miR156 exerts this effect presumably through a miR156-SPL9-miR172 regulatory module and possibly arrests tuberization under LD conditions. It appears that miR156 has a different function in SD and LD photoperiods. We also cannot rule out the possibility that the high accumulation of miR156 in SD-induced stolons is associated with controlling tuber transitions, the maintenance of the juvenile phase, or even tuber dormancy. Future work will help to elucidate the additional functions of miR156 in potato.
MATERIALS AND METHODS
Plant Material and Growth Conditions
In this study, potato (Solanum tuberosum subspecies andigena 7540) was used. This is a photoperiod-responsive plant that tuberizes under SD conditions (8 h of light) and does not produce tubers under LD conditions (16 h of light). In vitro plants were grown under LD conditions at 25°C in a growth incubator (Percival Scientific). Soil plants were grown at 22°C under LD photoperiod in environmental chambers (Percival Scientific). For age-specific expression studies of miR156, tissue culture-raised plants were transferred to soil and incubated up to 14 weeks. Tissues (fully expanded mature leaves and stem) were collected after specific time intervals (2, 7, 12, 13, and 14 weeks) and stored at −80°C until further use. For photoperiod-dependent expression studies, plants were induced under both SD and LD conditions in environmental chambers for 15 d. Different tissues (leaf, stem, stolon, and swollen stolon) were harvested after 15 dpi. For quantifying miR156 levels in 0-, 15-, and 30-d-old tubers stored post harvest (tuber dormancy), tuber eyes were isolated and stored at −80°C. In the case of tobacco (Nicotiana tabacum cv Petit Havana), plants were grown under LD conditions in environmental chambers.
Validation of miR156
Total RNA was harvested from mature leaves and stem tissue of potato, and StmiR156a (BI432985.1) precursor was amplified by RT-PCR (details of all the primers and accession numbers are given in Supplemental Tables S3 and S4). The amplicon was then sequence confirmed. Mature miR156 was detected by stem-loop RT-PCR as described earlier (Varkonyi-Gasic et al., 2007). Total RNA was isolated by TRIzol reagent (Invitrogen) following the manufacturer’s instructions. RT was carried out using stem-loop primer miR156STP. End-point PCR was performed using miR156FP and universal reverse primer (univRP). The 61-bp amplicon was cloned in the subcloning vector pGEM-T Easy (Promega) and was confirmed by sequencing.
Analysis of miRNA Levels
In the entire study, levels of miRNAs (miR156 and miR172) were determined by stem-loop qRT-PCR. One microgram of total RNA was used for all RT reactions except for the quantification of miRNAs from phloem sap, where 100 ng of RNA was used. Stem-loop reverse primers miR156STP and miR172STP were used for miR156 and miR172, respectively. RT was carried out as per a previous protocol (Varkonyi-Gasic et al., 2007). Quantitative PCR (qPCR) for miR156 (miR156FP and univRP) and miR172 (miR172FP and univRP) was performed in a Mastercycler ep realplex (Eppendorf). For normalization, 5S ribosomal RNA was reverse transcribed by stem-loop primer 5S rRNASTP and amplified by 5S rRNAFP and univRP. All the PCRs were incubated at 95°C for 5 min followed by 40 cycles of 95°C for 5 s, 60°C for 10 s, and 68°C for 8 s. PCR specificity was checked by melting curve analysis, and data were analyzed using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Construct Design and Plant Transformation
To generate miR156 OE lines of potato, the precursor sequence (StmiR156a; BI432985.1) of miR156 was amplified from total RNA harvested from leaves using the primers miR156preFP and miR156preRP. The PCR product was digested with XbaI-SacI and cloned into the binary vector pBI121 under the control of the CaMV 35S promoter. This construct was then mobilized into Agrobacterium tumefaciens strain GV2260. Transgenic plants were generated following the protocol by Banerjee et al. (2006b). miR156 OE lines of tobacco were raised as described by Horsch et al. (1985). Kanamycin-resistant transgenic plants were selected for further analysis and were maintained in Murashige and Skoog basal medium (Murashige and Skoog, 1962) until further use, whereas rSPL9 transgenic lines were generated by introducing silent mutations in the MRE. Mutations were incorporated by site-directed mutagenesis using Turbo DNA polymerase (Stratagene). The primers used for site-directed mutagenesis in MRE were rSPL9FP and rSPL9RP. Amplification of rSPL9 was carried out by using primer pair SPL9FP and SPL9RP3, and it was cloned in binary vector pBI121 downstream of the CaMV 35S promoter. Transgenic plants of rSPL9 were generated and maintained as described above.
Leaf and Stem Histology
For histology, a modified protocol of Cai and Lashbrook (2006) was followed. Briefly, leaves and stems of 8-week-old plants (the wild type and miR156 OE lines 5.1 and 6.2) grown under LD conditions were fixed in chilled ethanol:acetic acid (3:1; Merck). The tissues were vacuum infiltrated (400 mm of mercury) for 30 min and then stored at 4°C overnight. Fixed tissues were then dehydrated at room temperature in a graded series of ethanol (75% [v/v], 95% [v/v], and 100% [v/v] ethanol) followed by washes of a combination of ethanol and xylene series. Tissue blocks were prepared with molten Paraplast (Leica). Ten-micrometer sections were cut by a microtome (Leica) and placed on glass slides. Dried slides were deparaffinized by washing twice in 100% xylene and were observed with a microscope.
Environmental Mode Scanning Electron Microscopy of Leaves
Leaves of 8-week-old plants (the wild type and miR156 OE lines 5.1 and 6.2) grown under LD conditions were used for scanning electron microscopy in the environmental mode (eSEM) with a Quanta 200 3D eSEM apparatus (FEI), and leaf morphology was documented.
Analysis of Tuberization
Both wild-type and miR156 OE lines were grown in soil at 22°C under LD conditions in environmental chambers for 3 weeks. Thereafter, 10 plants each were shifted to SD and LD conditions and were incubated further for 4 weeks. To analyze StSP6A and miR172 levels in these plants, leaf tissues were harvested 8 dpi from both of these lines. miR172 levels were also quantified in 15-d post SD-induced stolons of wild-type and miR156 OE plants (line 5.1). The tuberization phenotype was scored after 4 weeks of induction.
Analysis of Zeatin Riboside and Orobanchyl Acetate by HR-MS
Axillary meristems were harvested from wild-type and miR156 OE 5.1 plants induced for 15 d in LD and SD conditions and ground in liquid nitrogen. For HR-MS analysis, a modified protocol of Forcat et al. (2008) was followed. One hundred milligrams of tissue was used for extraction in 400 µL of 10% (v/v) methanol and 1% (v/v) glacial acetic acid. This mixture was vigorously vortexed and stored on ice for 2 h, followed by centrifugation to obtain the supernatant. This was repeated three times, and the volume of the supernatant was adjusted to 2 mL in a volumetric flask. Samples were resolved through a Thermo Scientific Hypersil Gold column of particle size 5 μm with a flow rate of 0.5 mL min−1 and a gradient solvent program of 25 min (0 min, 10% methanol-water; 0.5 min, 10% methanol-water; 3 min, 45% methanol-water; 20 min, 50% methanol-water; 22 min, 90% methanol-water; 23 min, 10% methanol-water; 25 min, 10% methanol-water). Formic acid (0.1%; liquid chromatography-mass spectrometry grade) was also added to methanol and water. Mass spectrometry and tandem mass spectrometry experiments were performed in electrospray ionization-positive ion mode using the tune method as followed: sheath gas flow rate, 45 units N2; auxiliary gas flow rate, 10 units N2; sweep gas flow rate, 2 units N2; spray voltage, 3.60 kV; spray current, 3.70 μA; capillary temperature, 320°C; source lens RF level, 50; heater temperature, 350°C. Electrospray ionization-mass spectrometry data were recorded in full scan mode within the mass-to-charge ratio range 100 to 1,000. A standard curve for quantification was prepared using zeatin riboside (Sigma). Orobanchyl acetate was identified based on mass spectrometry analysis, and quantification was performed considering the peak areas.
Analysis of StSP6A, StLOG1, and StCyclin D3.1
One microgram of total RNA was used for StSP6A analysis from 8-d-old SD-induced leaves of wild-type and miR156 OE line plants. 18S ribosomal RNA (50 ng) was used for normalization. For RT-PCR, the SuperScript III one-step RT-PCR system with platinum Taq DNA polymerase (Invitrogen) was used as per the manufacturer’s instructions. Semiquantitative RT-PCR for StSP6A was performed using the following primers: SP6AFP and SP6ARP. RT-PCR conditions were as follows: 50°C for 30 min, 94°C for 2 min, followed by 25 cycles of 94°C 15 s, 55°C for 15 s, and 68°C for 1 min. The cycle number for 18S RNA was restricted to 10, while the program remained the same as for StSP6A. For analysis of StLOG1 and StCyclin D3.1, total RNA was isolated from axillary meristems of wild-type and miR156 OE 5.1 plants grown in SD photoperiod for 15 d by TRIzol reagent. One microgram of total RNA was used for complementary DNA (cDNA) synthesis by SuperScript III reverse transcriptase (Invitrogen) using an oligo(dT) primer. qPCR was performed on a Mastercycler ep realplex with LOG1FP-LOG1RP and CyclinFP-CyclinRP. The reactions were carried out using KAPA SYBR green master mix (Kapa Biosystems) and incubated at 95°C for 2 min followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. Glyceraldehyde 3 phosphate dehydrogenase was used for normalization for all the reactions. PCR specificity was checked by melting curve analysis, and data were analyzed using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Prediction of miR156 Targets
miR156 targets in potato were predicted using bioinformatic tools. To increase the efficiency of target prediction, psRNATarget (plantgrn.noble.org/psRNATarget/; Dai and Zhao, 2011) and TargetAlign (leonxie.com/targetAlign.php; Xie and Zhang, 2010) online tools were used. Based on their score and consistency of results, five sequences were short listed as potential miR156 targets. These targets showed homology (40%–80%) with AtSPL3, AtSPL6, AtSPL9, AtSPL13, and RcoLG1 (for Ricinus communis LIGULELESS1) and were termed StSPL3, StSPL6, StSPL9, StSPL13, and StLG1, respectively. Their coding sequences were retrieved from the Online Resource for Community Annotation of Eukaryotes (http://bioinformatics.psb.ugent.be/orcae/; Sterck et al., 2012) and the Database of Plant Transcription Factors (http://planttfdb.cbi.edu.cn/; Zhang et al., 2011a).
Cleavage Site Mapping
To validate the candidate targets of miR156 in planta, modified RLM 5′ RACE was performed using the First Choice RLM-RACE kit (Ambion). Total RNA was extracted from wild-type potato leaves by TRIzol reagent and was directly ligated to RNA adaptor without any enzymatic pretreatments. cDNA synthesis was performed using the respective gene-specific reverse primers. Two rounds of PCR were conducted with adaptor-specific forward primers and gene-specific reverse primers (SPL3, SPL3RP1-SPL3RP2; SPL6, SPL6RP1-SPL6RP2; SPL9, SPL9RP1-SPL9RP2; SPL13, SPL13RP1-SPL13RP2; LG1, LG1RP1-LG1RP2). Amplicons were then cloned into the subcloning vector pGEM-T Easy and were sequenced to identify the miRNA cleavage sites.
Analysis of StSPLs
Total RNA from wild-type and miR156 OE plants was isolated by TRIzol reagent as per the manufacturer’s instructions. One microgram of total RNA was reverse transcribed using gene-specific primers by Moloney murine leukemia virus reverse transcriptase (Promega). For normalization, GAPDH was reverse transcribed. The primers used for reverse transcription were SPL3RP2, SPL6RP2, SPL9RP2, SPL13RP2, LG1RP2, and GAPDHRP. qPCR was performed on a Mastercycler ep realplex with the same reverse primers mentioned above. Forward primers were SPL3qFP, SPL6qFP, SPL9qFP, SPL13qFP, LG1qFP, and GAPDHFP. The reactions were carried out using the KAPA SYBR green master mix and incubated at 95°C for 2 min followed by 40 cycles of 95°C for 15 s, 52°C for 15 s, and 60°C for 20 s. For GAPDH, all conditions were similar, but the annealing temperature was 55°C. For StLG1, all conditions were similar except that the extension time was 10 s. PCR specificity was checked by melting curve analysis, and data were analyzed using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Gel Retardation Assay
A 6× His-tagged fusion construct was generated by introducing the 1,152-bp coding sequence of StSPL9 in frame into the pET28a expression vector and transformed into Escherichia coli BL21 (DE3) cells. Cells were grown at 37°C until the optical density at 600 nm reached 0.6, induced with 1.0 mm isopropyl-β-d-thiogalactopyranoside, and cultured for 3 h at 37°C. The cells were lysed by sonication. The tagged protein was purified using nickel-nitrilotriacetic acid agarose beads. Purified StSPL9 protein aliquots were frozen in liquid N2 and stored at −80°C. Four overlapping fragments of the MIR172b promoter were used for gel mobility shift assays. Promoter fragments were PCR amplified from potato genomic DNA and were purified on columns. The respective primer sequences are provided in Supplemental Table S3. The 5′ ends of the fragments were then labeled with γ-32P using the KinaseMax kit (Ambion). The DNA-binding reactions were set up at 24°C in 20 μL containing 10 mm Tris-HCl (pH 7.5), 5% glycerol, 0.5 mm EDTA, 0.5 mm dithiothreitol, 0.05% Nonidet P-40, 50 mm NaCl, 50 mg L−1 poly(dG-dC), 250 ng of protein, and 1 fmol of labeled DNA. After incubation at 24°C for 60 min, the reactions were resolved on a 6% native polyacrylamide gel in 1× Tris-borate-EDTA buffer. The gel was dried and exposed to x-ray film. In the cold competition assays, 100-fold more unlabeled double-stranded DNA fragment (P1) was added to the reaction and loaded onto the gel every 15 min.
Detection of miR156 in the Phloem
Stem sections of 12-week-old wild-type plants were fixed as described above in the histology section. LMPC-mediated harvest of phloem cells (Carl Zeiss PALM laser micro beam) and RNA extraction from these cells were done as per Yu et al. (2007). Total RNA was extracted using TRIzol reagent. Mature miR156 and miR156* were detected by stem-loop RT-PCR as described earlier. Primer sequences for miR156* are provided in Supplemental Table S3.
For analysis of the differential accumulation of miR156 in phloem sap of SD- and LD-grown wild-type plants, sap extraction and RNA isolation were done as per Campbell et al. (2008) with a minor modification (the phloem exudate was harvested at 18°C). Sap collection was performed at 8, 15, and 30 dpi. To assess the purity of phloem sap, RT-PCR was performed for nitrate transporter (root-specific transcript) and G2-like transcription factor (phloem-specific transcript) using 150 ng of RNA as mentioned before (Banerjee et al., 2006a). The SuperScript III one-step RT-PCR system with platinum Taq DNA polymerase was used as per the manufacturer’s instructions. For nitrate transporter, RT-PCR conditions were as follows: 55°C for 30 min, 94°C for 2 min, followed by 40 cycles of 94°C for 15 s, 50°C for 30 s, and 68°C for 1 min, with a final extension at 68°C for 5 min. For G2-like transcription factor, all conditions were similar except that annealing was at 56°C and extension was for 30 s. To quantify miR156 levels in phloem sap, 100 ng of total RNA was used for miR156-specific stem-loop qRT-PCR as described above, except that the cycle number was increased to 50. qRT-PCR cycle threshold (Ct) value differences were calculated for miR156 accumulation and plotted as described previously (Pant et al., 2008).
Soil-Grown Heterografts
Wild-type and miR156 OE lines were maintained in an environmental chamber until grafting was performed. Grafts were made with wild-type and miR156 OE transgenic potato plants as per our previous protocol (Mahajan et al., 2012). miR156 OE lines were used as scions and wild-type plants as stock (heterografts), while for reverse grafting, miR156 OE plants served as stock and wild-type plants as scion (reverse grafts). Homografts (wild type on wild type) were used as controls in both cases. Equal numbers (10 each) of heterografts, reverse grafts, and homografts were made and maintained in environmental chambers for hardening for 4 weeks. Hardened grafts were further incubated in SD conditions for 4 weeks. Scion and stock samples (devoid of graft union) were harvested, and phenotypes such as leaf number, trichomes, and axillary branches were scored. qRT-PCR was performed for miR156 accumulation in both heterograft stock samples and reverse graft scion samples with respective tissues from homografts as controls. Tuberization phenotypes were scored for all grafts.
For miR156a precursor transgene detection, RT-PCR was performed by the SuperScript III one-step RT-PCR system with platinum Taq DNA polymerase using 250 ng of total RNA. The primers used were miR156pre FP and transgene-specific NOST RP. The RT-PCR conditions were as follows: 50°C for 30 min, 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, 50°C for 15 s, and 68°C for 1 min, with a final extension of 68°C for 5 min.
For comparative analysis of miR156 (mature) and miR156a precursor levels in wild-type and grafted plants, 500 ng of RNA was used. Stem-loop qRT-PCR of miR156 (mature) was performed as described above. For miR156a precursor quantification, cDNA synthesis was performed using oligo(dT) and SuperScript III reverse transcriptase enzyme. qPCR was performed on a Mastercycler ep realplex with miR156preFP and miR156preRP primers. The reactions were carried out using KAPA SYBR green master mix and incubated at 95°C for 2 min followed by 40 cycles of 95°C for 15 s, 50°C for 10 s, and 72°C for 18s. GAPDH was used for normalization. PCR specificity was checked by melting curve analysis, and data were analyzed using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers BI432985.1 (miR156a precursor), AC237992 (MIR172b promoter), CK267169.1 (NT), CK853924.1 (G2), in the Online Resource for Community Annotation of Eukaryotes (ORCAE) under accession numbers sotub10g009340 (SPL3), sotub12g015890 (SPL6), sotub10g020210 (SPL9), sotub05g016640 (SPL13), sotub05g016440 (LG1), and in the Potato Genome Sequencing Consortium under accession numbers: PGSC0003DMG400023365 (SP6A), PGSC0003DMT400009551 (LOG1), PGSC0003DMT400064307 (Cyclin D3.1), PGSC chr07:1782100..1784700 (MIR156a promoter).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. miR156 overexpression in potato.
Supplemental Figure S2. Overexpression of miR156 in tobacco.
Supplemental Figure S3. HR-MS of zeatin riboside.
Supplemental Figure S4. Mass spectrum of zeatin riboside.
Supplemental Figure S5. HR-MS of orobanchyl acetate.
Supplemental Figure S6. Mass spectrum of orobanchyl acetate.
Supplemental Figure S7. Overexpression of rSPL9 in potato.
Supplemental Figure S8. Comparative analysis of miR156 (mature) and miR156a precursor levels in wild-type and grafted plants.
Supplemental Table S1. Detailed analysis of miR156 targets in potato.
Supplemental Table S2. Potential light regulatory elements present in the upstream sequence of miR156a.
Supplemental Table S3. List of primers.
Supplemental Table S4. List of accession numbers.
Acknowledgments
We thank David J. Hannapel (Iowa State University) and Julia Kehr (University of Hamburg) for their critical reading of the manuscript, the director, National Chemical Laboratory, for providing us the eSEM facilities, Sanjeev Galande (Indian Institute of Science Education and Research) for his help in the gel retardation assays, and Saikat Haldar (National Chemical Laboratory) for his technical help in carrying out HR-MS analysis.
Glossary
- miRNA
microRNA
- HR-MS
high-resolution mass spectrometry
- SD
short-day
- OE
overexpression
- RT
reverse transcription
- qRT
quantitative reverse transcription
- LD
long-day
- RLM
RNA ligase-mediated
- CaMV
cauliflower mosaic virus
- MRE
microRNA recognition element
- LMPC
laser microdissection pressure catapulting
- dpi
days post induction
- univRP
universal reverse primer
- qPCR
quantitative PCR
- eSEM
scanning electron microscopy (environmental mode)
- cDNA
complementary DNA
Footnotes
This work was supported by the Indian Institute of Science Education and Research, the Council of Scientific and Industrial Research, India, and the Department of Biotechnology, India.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Atkins CA, Smith PMC, Rodriguez-Medina C. (2011) Macromolecules in phloem exudates: a review. Protoplasma 248: 165–172 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL. (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13: 343–349 [DOI] [PubMed] [Google Scholar]
- Banerjee AK, Chatterjee M, Yu Y, Suh SG, Miller WA, Hannapel DJ. (2006a) Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18: 3443–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Prat S, Hannapel DJ. (2006b) Efficient production of transgenic potato (S. tuberosum L. ssp. andigena) plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci 170: 732–738 [Google Scholar]
- Birkenbihl RP, Jach G, Saedler H, Huijser P. (2005) Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352: 585–596 [DOI] [PubMed] [Google Scholar]
- Buhtz A, Pieritz J, Springer F, Kehr J. (2010) Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol 10: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J. (2008) Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J 53: 739–749 [DOI] [PubMed] [Google Scholar]
- Cai S, Lashbrook CC. (2006) Laser capture microdissection of plant cells from tape-transferred paraffin sections promotes recovery of structurally intact RNA for global gene profiling. Plant J 48: 628–637 [DOI] [PubMed] [Google Scholar]
- Campbell BA, Hallengren J, Hannapel DJ. (2008) Accumulation of BEL1-like transcripts in solanaceous species. Planta 228: 897–906 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang Z, Liu D, Zhang K, Li A, Mao L. (2010) SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J Integr Plant Biol 52: 946–951 [DOI] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S. (2007) The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet 39: 544–549 [DOI] [PubMed] [Google Scholar]
- Chuck G, O’Connor D. (2010) Small RNAs going the distance during plant development. Curr Opin Plant Biol 13: 40–45 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dai X, Zhao PX. (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39: W155–W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devers EA, Branscheid A, May P, Krajinski F. (2011) Stars and symbiosis: microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol 156: 1990–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. (2011) Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 12: 211–221 [DOI] [PubMed] [Google Scholar]
- Eviatar-Ribak T, Shalit-Kaneh A, Chappell-Maor L, Amsellem Z, Eshed Y, Lifschitz E. (2013) A cytokinin-activating enzyme promotes tuber formation in tomato. Curr Biol 23: 1057–1064 [DOI] [PubMed] [Google Scholar]
- Forcat S, Bennett MH, Mansfield JW, Grant MR. (2008) A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods 4: 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Sunkar R, Zhou C, Shen H, Zhang JY, Matts J, Wolf J, Mann DGJ, Stewart CN, Jr, Tang Y, et al. (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol J 10: 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P. (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49: 683–693 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L, Freeling M. (1996) Interactions of liguleless1 and liguleless2 function during ligule induction in maize. Genetics 144: 1871–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood V, Yu TS, Huang NC, Lucas WJ. (2005) Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J 42: 49–68 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Rogers SG, Fraley RT. (1985) Transgenic plants. Cold Spring Harb Symp Quant Biol 50: 433–437 [DOI] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ. (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M. (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Jackson SD. (2009) Plant responses to photoperiod. New Phytol 181: 517–531 [DOI] [PubMed] [Google Scholar]
- Jackson SD, James P, Prat S, Thomas B. (1998) Phytochrome B affects the levels of a graft-transmissible signal involved in tuberization. Plant Physiol 117: 29–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42: 541–544 [DOI] [PubMed] [Google Scholar]
- Kasai A, Kanehira A, Harada T. (2010) miR172 can move long distance in Nicotiana benthamiana. The Open Plant Sci J 4: 1–6 [Google Scholar]
- Kehr J, Buhtz A. (2008) Long distance transport and movement of RNA through the phloem. J Exp Bot 59: 85–92 [DOI] [PubMed] [Google Scholar]
- Kehr J, Buhtz A. (2013) Endogenous RNA constituents of the phloem and their possible roles in long-distance signaling. In GA Thompson, AJE van Bel, eds, Phloem: Molecular Cell Biology, Systemic Communication, Biotic Interactions. Wiley-Blackwell Publishing, Oxford, pp 186–208 [Google Scholar]
- Kim M, Canio W, Kessler S, Sinha N. (2001) Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293: 287–289 [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Lee J, Park JJ, Kim SL, Yim J, An G. (2007) Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65: 487–499 [DOI] [PubMed] [Google Scholar]
- Lin T, Sharma P, Gonzalez DH, Viola IL, Hannapel DJ. (2013) The impact of the long-distance transport of a BEL1-like messenger RNA on development. Plant Physiol 161: 760–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mahajan A, Bhogale S, Kang IH, Hannapel DJ, Banerjee AK. (2012) The mRNA of a Knotted1-like transcription factor of potato is phloem mobile. Plant Mol Biol 79: 595–608 [DOI] [PubMed] [Google Scholar]
- Marín-González E, Suárez-López P. (2012) “And yet it moves”: cell-to-cell and long-distance signaling by plant microRNAs. Plant Sci 196: 18–30 [DOI] [PubMed] [Google Scholar]
- Martin A, Adam H, Díaz-Mendoza M, Zurczak M, González-Schain ND, Suárez-López P. (2009) Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136: 2873–2881 [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Virgos-Soler A, Prat S. (2002) Control of photoperiodregulated tuberization in potato by the Arabidopsis flowering-time gene CONSTANS. PNAS 99: 15211–15216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S, Koi S, Hashimoto T, Nakajima K. (2011) Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138: 2303–2313 [DOI] [PubMed] [Google Scholar]
- Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. (2010) Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Navarro C, Abelenda JA, Cruz-Oró E, Cuéllar CA, Tamaki S, Silva J, Shimamoto K, Prat S. (2011) Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478: 119–122 [DOI] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP. (2010) MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev 24: 2678–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. (2008) The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol 15: 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Shikata M, Koyama T, Mitsuda N, Ohme-Takagi M. (2009) Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol 50: 2133–2145 [DOI] [PubMed] [Google Scholar]
- Sterck L, Billiau K, Abeel T, Rouzé P, Van de Peer Y. (2012) ORCAE: online resource for community annotation of eukaryotes. Nat Methods 9: 1041. [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami T, Horiguchi G, Yano S, Tsukaya H. (2009) The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136: 955–964 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Gould N, Sandanayaka M, Sutherland P, MacDiarmid RM. (2010) Characterisation of microRNAs from apple (Malus domestica ‘Royal Gala’) vascular tissue and phloem sap. BMC Plant Biol 10: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauterin M, Frankard V, Jacobs M. (1999) The Arabidopsis thaliana dhdps gene encoding dihydrodipicolinate synthase, key enzyme of lysine biosynthesis, is expressed in a cell-specific manner. Plant Mol Biol 39: 695–708 [DOI] [PubMed] [Google Scholar]
- Vorst O, van Dam F, Oosterhoff-Teertstra R, Smeekens S, Weisbeek P. (1990) Tissue-specific expression directed by an Arabidopsis thaliana pre-ferredoxin promoter in transgenic tobacco plants. Plant Mol Biol 14: 491–499 [DOI] [PubMed] [Google Scholar]
- Waksman G, Lebrun M, Freyssinet G. (1987) Nucleotide sequence of a gene encoding sunflower ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (rbcs). Nucleic Acids Res 15: 7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Park MY, Wang LJ, Koo Y, Chen XY, Weigel D, Poethig RS. (2011) miRNA control of vegetative phase change in trees. PLoS Genet 7: e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Lough T. (2001) Gene silencing as an adaptive defence against viruses. Nature 411: 834–842 [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Frazier TP, Zhang B. (2011) Identification, characterization and expression analysis of microRNAs and their targets in the potato (Solanum tuberosum). Gene 473: 8–22 [DOI] [PubMed] [Google Scholar]
- Xie F, Zhang B. (2010) Target-align: a tool for plant microRNA target identification. Bioinformatics 26: 3002–3003 [DOI] [PubMed] [Google Scholar]
- Xie K, Shen J, Hou X, Yao J, Li X, Xiao J, Xiong L. (2012) Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol 158: 1382–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Wu C, Xiong L. (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Salinas M, Höhmann S, Berndtgen R, Huijser P. (2010) miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22: 3935–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Vreugdenhil D, van Lammeren AAM. (1998) Cell division and cell enlargement during potato tuber formation. J Exp Bot 49: 573–582 [Google Scholar]
- Yang W, Liu X, Zhang J, Feng J, Li C, Chen J. (2010) Prediction and validation of conservative microRNAs of Solanum tuberosum L. Mol Biol Rep 37: 3081–3087 [DOI] [PubMed] [Google Scholar]
- Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ. (2004) A systemic small RNA signaling system in plants. Plant Cell 16: 1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YY, Lashbrook CC, Hannapel DJ. (2007) Tissue integrity and RNA quality of laser microdissected phloem of potato. Planta 226: 797–803 [DOI] [PubMed] [Google Scholar]
- Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J. (2011a) PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res 39: D1114–D1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Luo Y, Gong X, Zeng W, Li S. (2009) Computational identification of 48 potato microRNAs and their targets. Comput Biol Chem 33: 84–93 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zou Z, Zhang J, Zhang Y, Han Q, Hu T, Xu X, Liu H, Li H, Ye Z. (2011b) Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett 585: 435–439 [DOI] [PubMed] [Google Scholar]
- Zuker M. (2003) MFold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]