Transcript and metabolite analysis of the methyl jasmonate-elicited sweet basil identified the genes involved in the biosynthesis of the medicinally important pentacyclic triterpenes.
Abstract
Sweet basil (Ocimum basilicum) is well known for its diverse pharmacological properties and has been widely used in traditional medicine for the treatment of various ailments. Although a variety of secondary metabolites with potent biological activities are identified, our understanding of the biosynthetic pathways that produce them has remained largely incomplete. We studied transcriptional changes in sweet basil after methyl jasmonate (MeJA) treatment, which is considered an elicitor of secondary metabolites, and identified 388 candidate MeJA-responsive unique transcripts. Transcript analysis suggests that in addition to controlling its own biosynthesis and stress responses, MeJA up-regulates transcripts of the various secondary metabolic pathways, including terpenoids and phenylpropanoids/flavonoids. Furthermore, combined transcript and metabolite analysis revealed MeJA-induced biosynthesis of the medicinally important ursane-type and oleanane-type pentacyclic triterpenes. Two MeJA-responsive oxidosqualene cyclases (ObAS1 and ObAS2) that encode for 761- and 765-amino acid proteins, respectively, were identified and characterized. Functional expressions of ObAS1 and ObAS2 in Saccharomyces cerevisiae led to the production of β-amyrin and α-amyrin, the direct precursors of oleanane-type and ursane-type pentacyclic triterpenes, respectively. ObAS1 was identified as a β-amyrin synthase, whereas ObAS2 was a mixed amyrin synthase that produced both α-amyrin and β-amyrin but had a product preference for α-amyrin. Moreover, transcript and metabolite analysis shed light on the spatiotemporal regulation of pentacyclic triterpene biosynthesis in sweet basil. Taken together, these results will be helpful in elucidating the secondary metabolic pathways of sweet basil and developing metabolic engineering strategies for enhanced production of pentacyclic triterpenes.
Plants are capable of synthesizing an amazing diversity of low molecular weight secondary metabolites that have a variety of applications as pharmaceuticals, flavors, fragrances, and pesticides (Bohlmann and Keeling, 2008; Wink, 2010a). Although some plant-derived secondary metabolites are directly used as drugs, many are leading models for the development of semisynthetic and synthetic drugs (Heilmann, 2010). To date, over 100,000 different structures of secondary metabolites or natural products are known; however, the real number is undoubtedly much higher because only 20% to 30% of the plant species are exploited (Wink, 2010a). Because of the insufficient supply of raw materials for extraction of many valuable secondary metabolites, increasing the production of these compounds in plants is a well-established target for genetic manipulation through traditional and biotechnological approaches (Goossens et al., 2003; Canter et al., 2005; Alfermann, 2010; Lange and Ahkami, 2013). However, this presents some severe challenges owing to the lack of in-depth knowledge on the secondary metabolite biosynthetic pathways and their regulation (Canter et al., 2005; Lange and Ahkami, 2013). A comprehensive understanding of the metabolic pathway is extremely important to overcome low product yield of the secondary metabolites in plants or plant cell cultures. Once the biosynthetic genes and their regulatory controls have been elucidated, pathway engineering can be employed to increase metabolite accumulation not only in the native plants but also in the organisms that do not even synthesize them (Yun et al., 1992; Chen et al., 2000; Zhang et al., 2004; Lange and Ahkami, 2013; Paddon et al., 2013).
Biologically active phytochemicals are synthesized in plants in tissue-specific, organ-specific, and developmentally specific ways that involve highly complex and sophisticated biosynthetic pathways (Facchini and De Luca, 2008; Murata et al., 2008; Besser et al., 2009; Wink, 2010a). Plants synthesize and accumulate these compounds in specific tissue types perhaps for autotoxicity avoidance, because many of the secondary metabolites are known to be phytotoxic (Reigosa and Malvido-Pazos, 2007; Ghosh et al., 2013). Moreover, the sites of biosynthesis may not necessarily be the sites of storage, and long-distance transport via the xylem, phloem, or apoplast is also possible (Wink, 2010b). Lipophilic metabolites are usually stored in the cuticle, resin ducts, laticifers, trichomes, or oil cells, whereas hydrophilic metabolites are stored in the vacuole. When plants experience various biotic and abiotic stresses, biosynthesis of the secondary metabolites is triggered that helps plants adapt to the challenging environment (Gershenzon and Dudareva, 2007; Hartmann, 2007; Reichling, 2010). Consequently, transcript and metabolite profiling of stress/elicitor-treated plants or cell cultures represents a powerful approach to determine gene function in the biosynthesis of the secondary metabolites (Aerts et al., 1994; Goossens et al., 2003; Suzuki et al., 2005; Zhao et al., 2005; Naoumkina et al., 2008; De Geyter et al., 2012; Lenka et al., 2012; Yu et al., 2012; Ee et al., 2013; Mishra et al., 2013; Sun et al., 2013).
The plant hormone methyl jasmonate (MeJA) acts as a conserved elicitor of secondary metabolite production across the plant kingdom, from angiosperms to gymnosperms (De Geyter et al., 2012). Several studies have demonstrated that MeJA treatment can trigger the biosynthesis of all three major classes of secondary metabolites (i.e. terpenoids, phenylpropanoids, and alkaloids) through an extensive transcriptional reprogramming of the plant metabolism (Gundlach et al., 1992; Cheong and Choi, 2003; Zhao et al., 2005; Pauwels et al., 2009; De Geyter et al., 2012; Sun et al., 2013). Here, we carried out transcriptome and selected metabolite analyses to identify the MeJA-responsive secondary metabolic pathways of sweet basil (Ocimum basilicum), which is a plant of the Lamiaceae family with highly valued aromatic and medicinal properties. Sweet basil has been extensively used for centuries in traditional medicine for the treatment of various ailments (Bilal et al., 2012). Several pharmacological studies have demonstrated that sweet basil contains biologically active constitutions that are analgesic, anti-inflammatory, antimicrobial, antioxidant, antiulcerogenic, anticarcinogenic, cardiac stimulant, chemomodulatory, central nervous system depressant, hepatoprotective, hypoglycemic, hypolipidemic, immunomodulatory, and larvicidal (Dasgupta et al., 2004; Muralidharan and Dhananjayan, 2004; Lee et al., 2005; Zeggwagh et al., 2007; Meera et al., 2009; Zhang et al., 2009; Arshad Qamar et al., 2010; Choudhury et al., 2010; Dashputre and Naikwade, 2010; Fathiazad et al., 2012).
Sweet basil essential oil contains numerous volatile compounds, including monoterpenes, sesquiterpenes, and phenylpropanoids (Iijima et al., 2004a; Zhang et al., 2009; Verma et al., 2012). In addition, various nonvolatile phytochemicals such as triterpenes, rosmarinic acid, and flavonoids that are present in considerable amounts in sweet basil are shown to have prominent biological properties (Siddiqui et al., 2007; Arshad Qamar et al., 2010; Strazzer et al., 2011; Fathiazad et al., 2012). Although efforts have been made to understand the biochemical and molecular basis of volatile terpene and phenylpropanoid biosynthesis in Ocimum species (Gang et al., 2001, 2002a, 2002b; Iijima et al., 2004a, 2004b; Louie et al., 2007; Rastogi et al., 2013), our knowledge of how nonvolatile compounds are biosynthesized has remained largely incomplete.
Ursane-type and oleanane-type pentacyclic triterpenes (C30 compounds) are known for a variety of pharmaceutical properties, including anti-inflammatory, hepatoprotective, antitumor, antihyperlipidemic, antiulcer, and antimicrobial benefits (Kunkel et al., 2011; Wilkinson et al., 2011; Kurek et al., 2012; da Silva Ferreira et al., 2013; Kong et al., 2013). However, there are limited data on the biosynthesis of these secondary metabolites in plants (Brendolise et al., 2011; Huang et al., 2012; Yu et al., 2013). Moreover, the regulatory controls of these secondary metabolic pathways are completely unknown. Ocimum species are known to accumulate these pentacyclic triterpenes, although the biosynthetic genes are yet to be identified and characterized (Silva et al., 2008).
Here we report the identification and characterization of the MeJA-responsive transcripts in sweet basil using a suppression subtractive hybridization (SSH) approach. A total of 388 candidate MeJA-responsive unique transcripts were isolated, which represent various secondary metabolic pathways (e.g. terpenoid, phenylpropanoid/flavonoid), in addition to transcripts associated with plant stress responses. Furthermore, transcript and metabolite analysis revealed MeJA responsiveness of ursane-type and oleanane-type pentacyclic triterpenes (ursolic acid and oleanolic acid), whose biosynthesis is spatiotemporally regulated in sweet basil. Moreover, cloning and functional characterization of two MeJA-responsive oxidosqualene cyclases revealed their involvement in the biosynthesis of the direct precursors (α-amyrin and β-amyrin) of these medicinally important triterpenes.
RESULTS
Cloning and Characterization of the MeJA-Responsive Transcripts
To identify sweet basil transcripts expressed in response to MeJA treatment, the SSH approach was followed because of its high efficiency in enriching low-expressing genes and normalization of the targeted sequences. An SSH library was generated from 2-month-old plants treated with 250 µm MeJA versus control plants, as described in the materials and methods. A total of 550 individual clones was randomly selected from the SSH library for sequencing using a vector-specific M13 forward primer. This resulted in the isolation of 509 high-quality ESTs with an average sequence length of 481.59 bp, after removing low-quality, vector, and short sequences (Table I). The average guanine-cytosine (GC) content of these high-quality ESTs was 43.92%, which was comparable with that observed for other plant species (Asamizu et al., 2000). EST sequences were deposited in the National Center for Biotechnology Information (NCBI) GenBank database (accession numbers JZ190506 to JZ191014). Furthermore, using the CAP3 program, ESTs were assembled into a total of 388 unique sequences, including 74 contigs and 314 singletons. The contigs consisted of ESTs ranging from 2 to 28, thereby suggesting a high level of MeJA-inducible transcription associated with the corresponding genes, such as mevalonate kinase, oxidosqualene cyclase, lectin, global transcription factor group E, putative ethylene-responsive element-binding protein-type transcription factor, and so forth. The detailed functional annotation of the unique ESTs is presented in Supplemental Table S1. BLASTN analysis of the unique sequences against the sweet basil EST database at the NCBI (http://www.ncbi.nlm.nih.gov) revealed that 70.87% of the unique ESTs are novel. To confirm the MeJA-induced expression, some of the transcripts were subjected to semiquantitative reverse transcription PCR (sqRT-PCR) and quantitative reverse transcription PCR (qRT-PCR) analysis (Figs. 1D and 2C).
Table I. An overview of the sweet basil MeJA SSH library.
| Description | No. |
|---|---|
| Total clones sequenced | 550 |
| High-quality ESTs obtained | 509 |
| Average length of ESTs (bp) | 481.59 |
| GC content of ESTs (%) | 43.92 |
| Unique sequences | 388 |
| Average length of unique sequence (bp) | 478.82 |
| Total contigs | 74 |
| Total singletons | 314 |
| Annotated unique sequencesa | 323 |
| Nonannotated unique sequences | 65 |
| Novel sweet basil unique ESTsb | 275 |
Figure 1.
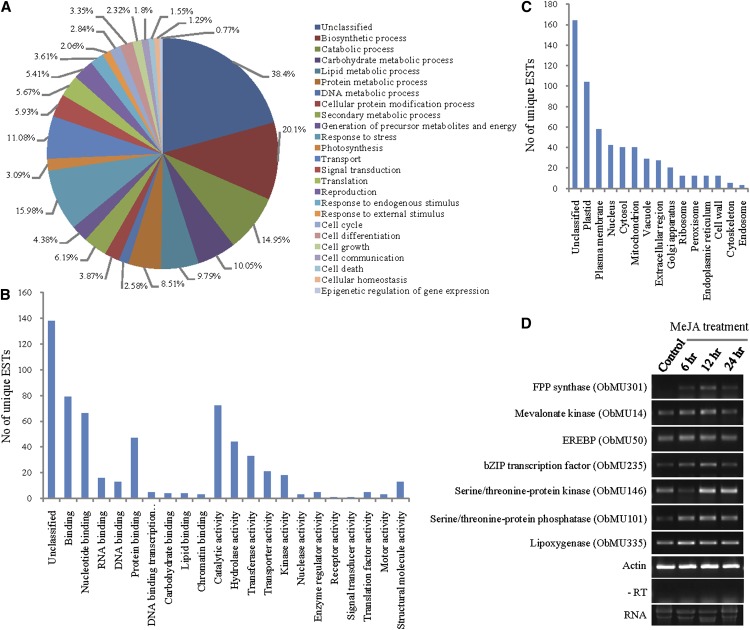
Functional categorization of the MeJA-responsive transcripts. A to C, GO classification of 388 MeJA-responsive transcripts on the basis of the biological process (A), molecular function (B), and cellular component (C). D, MeJA-inducible expression of the transcripts was detected by RT-PCR analysis. Actin amplification was used to normalize the amount of template. The negative control lacked reverse transcriptase in cDNA synthesis reaction (−RT). The experiment was repeated twice independently with similar results. bZIP, basic leucine zipper; EREBP, ethylene-responsive element-binding protein; FPP, farnesyl pyrophosphate. [See online article for color version of this figure.]
Figure 2.
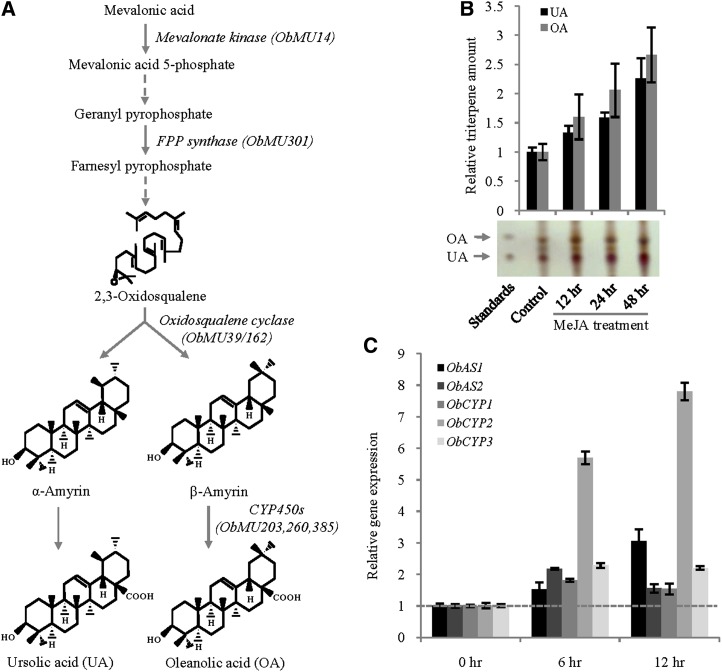
MeJA responsiveness of the pentacyclic triterpene biosynthetic pathway. A, Ursolic acid and oleanolic acid biosynthetic pathway. Transcripts identified in the MeJA SSH library and the corresponding biosynthetic steps are shown. Solid and dashed arrows denote single and multiple enzymatic steps, respectively. B, The relative levels of ursolic acid and oleanolic acid in sweet basil leaves were determined by TLC analysis after MeJA treatment for the indicated time periods. Data are the mean ± se from three to four independent experiments. The photograph of one representative TLC plate is presented. C, qRT-PCR expression analysis of the OSCs (ObAS1, ObAS2) and cytochrome P450s (ObCYP1, ObCYP2, ObCYP3) to show their MeJA-inducible expression. Data are mean ± se from two independent experiments (n = 5 to 6). FPP, farnesyl pyrophosphate; OA, oleanolic acid; UA, ursolic acid. [See online article for color version of this figure.]
Gene Ontology (GO) annotation was performed to assign putative function to the gene products (Fig. 1, A–C). Altogether, 73.97% of the unique sequences could be functionally categorized based on the GO biological process, cellular component, and molecular function (http://www.blast2go.com). The majority of the transcripts were predicted to be associated with the plastid (26.8%), plasma membrane (14.95%), nucleus (10.82%), cytosol (10.31%), mitochondria (10.31%), and vacuole (7.47%), suggesting that these cellular compartments play an important role in mediating MeJA-induced modulation of the cellular metabolic pathways. On the other hand, the majority of the unique sequences under the molecular function category were assigned to binding (20.36%) and catalytic activity (18.56%). Transferase, oxidoreductase, and hydrolase were found to be the major enzyme classes (Supplemental Table S2). Under the biological process, categories such as biosynthetic process (20.1%), response to stress (15.98%), catabolic process (14.95%), and transport (11.08%) were highly represented. This finding highlighted the key role of MeJA in regulating cellular metabolism and stress responses in plants (Cheong and Choi, 2003; Pauwels et al., 2009).
Few transcriptional regulators of APETALA2/Ethylene Response Factor (ERF), WRKY, Plant Homeo Domain (PHD), and Zinc Finger families were identified in the SSH library (Supplemental Table S3). Functional characterization of these MeJA-responsive transcription factors may lead to a better understanding of the underlying transcriptional regulation of MeJA-induced alteration in cellular metabolism. The SSH library was also enriched with the transcripts that presumably have no direct role in cellular metabolism. Some of them were related to the plant defense responses and are known to be induced by MeJA in other plant species as well (Supplemental Table S4; Cheong and Choi, 2003; Lenka et al., 2012; Ee et al., 2013).
Identification of the Metabolic Pathways
To gain an overview of the sweet basil metabolic pathways that are modulated by MeJA, unique ESTs were analyzed according to the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp), which provides an alternative functional annotation of the transcripts based on the associated biochemical pathways. The analysis revealed a total of 77 KEGG pathways (Supplemental Table S5). Interestingly, six transcripts were found to be associated with the α-linolenic acid metabolism that leads to jasmonic acid biosynthesis (Supplemental Fig. S1; Supplemental Table S5). These transcripts included 13S-lipoxygenase (EC:1.13.11.12), C-acyltransferase (EC:2.3.1.16), and oxidase (EC:1.3.3.6), with three, two, and one unique ESTs, respectively. These results support the existence of a positive feedback regulatory system for the biosynthesis of jasmonic acid (Sasaki et al., 2001).
A list of various secondary metabolic pathways represented by the unique ESTs is provided in Table II. Interestingly, among the secondary metabolic processes, terpenoid and phenylpropanoid/flavonoid biosynthesis pathways were found to be maximally represented (Table II; Supplemental Fig. S2). These secondary metabolite types were previously reported to accumulate in high levels in sweet basil after MeJA treatment; however, the genes involved in the biosynthesis of these compounds were not identified (Kim et al., 2006; Li et al., 2007).
Table II. MeJA-responsive unique ESTs of sweet basil related to secondary metabolite biosynthesis.
| Sl No | Contig ID | Length | Functional Annotation to GenBank Database | E- Value |
|---|---|---|---|---|
| bp | ||||
| Terpenoid | ||||
| 1 | ObMU14 | 893 | Mevalonate kinase (AEZ55674.1) | 7.00E-120 |
| 2 | ObMU39 | 771 | Mixed amyrin synthase (BAF63702.1) | 3.00E-160 |
| 3 | ObMU118 | 513 | Nerolidol synthase (AER36088.1) | 3.00E-59 |
| 4 | ObMU156 | 480 | Acetoacetyl-coenzyme A thiolase (ABV08820.1) | 9.33E-109 |
| 5 | ObMU162 | 673 | Mixed amyrin synthase (AFJ19235.1) | 2.00E-138 |
| 6 | ObMU203 | 442 | Cytochrome P450 CYP716A52v2 (AFO63032.1) | 9.00E-31 |
| 7 | ObMU260 | 130 | Cytochrome P450 CYP716A17 (NP_001268076.1) | 4.00E-15 |
| 8 | ObMU261 | 169 | Linalool synthase (Q5SBP3.1) | 7.00E-23 |
| 9 | ObMU262 | 726 | 4-Diphosphocytidyl-2-C-methyl-d-erythritol kinase (ABP96842.1) | 2.00E-18 |
| 10 | ObMU301 | 589 | Farnesyl diphosphate synthase (AAK63847.1) | 5.00E-128 |
| 11 | ObMU385 | 306 | Cytochrome P450 CYP716A52v2 (AFO63032.1) | 1.00E-36 |
| Phenylpropanoid/flavonoid | ||||
| 12 | ObMU78 | 526 | Cytochrome P450 82A3 (XP_002281995.2) | 4.00E-51 |
| 13 | ObMU89 | 356 | Isoflavone reductase homolog P3 (XP_002282110.1) | 3.00E-54 |
| 14 | ObMU96 | 375 | Dihydroflavonol reductase (XP_002276827.1) | 2.00E-37 |
| 15 | ObMU135 | 80 | Flavonol synthase (ABB53382.1) | 6.00E-06 |
| 16 | ObMU350 | 180 | Flavonoid O-methyltransferase 3 (AFU50297.1) | 2.00E-27 |
| 17 | ObMU355 | 312 | Cinnamoyl-CoA reductase (XP_002520066.1) | 3.00E-19 |
| 18 | ObMU374 | 187 | Eugenol synthase (Q15GI4.1) | 2.00E-25 |
| Alkaloid | ||||
| 19 | ObMU1 | 339 | Polyphenol oxidase (AAD53122.1) | 8.00E-05 |
| 20 | ObMU187 | 522 | Strictosidine synthase (XP_002273764.1) | 7.00E-80 |
| 21 | ObMU190 | 221 | Polyamine oxidase 2-like (XP_004251556.1) | 1.14E-33 |
| 22 | ObMU253 | 144 | Primary amine oxidase-like (XP_004298688) | 5.00E-25 |
| Tocopherol | ||||
| 23 | ObMU131 | 271 | Homogentisate phytyltransferase 2 (XP_002276728.1) | 9.00E-25 |
| Carotenoid | ||||
| 24 | ObMU314 | 331 | Cytochrome P450 CYP97C10 (ABD97103.1) | 3.00E-61 |
| Others | ||||
| 25 | ObMU92 | 877 | 3-Ketoacyl-CoA thiolase 1 (AGC59769.1) | 7.19E-163 |
| 26 | ObMU209 | 628 | Lipoxygenase (XP_002516771.1) | 2.00E-73 |
| 27 | ObMU272 | 879 | Lipoxygenase 6 (XP_003531186.1) | 1.00E-177 |
| 28 | ObMU306 | 408 | Acetoacetyl-CoA thiolase (ABB45810.1) | 8.75E-52 |
| 29 | ObMU335 | 408 | Lipoxygenase (NP_001233812.1) | 2.00E-53 |
MeJA Responsiveness of the Pentacyclic Triterpene Biosynthetic Pathway
In plants, all diverse types of terpene metabolites are synthesized from two 5C building blocks (dimethylallyl diphosphate and isopentenyl diphosphate) that are formed via two independent pathways: the mevalonic acid (MEV) pathway and the 2C-methyl-d-erythritol-4-phosphate (MEP) pathway (Tholl and Lee, 2011). Isopentenyl diphosphate and dimethylallyl diphosphate derived from the cytosolic MEV pathway serve as the precursors for the biosynthesis of the sesquiterpenes (C15) and triterpenes (C30), whereas the plastidial MEP pathway provides precursors for the biosynthesis of the monoterpenes (C10), diterpenes (C20), and tetraterpenes (C40) (Sawai and Saito, 2011). The identification of the transcripts similar to mevalonate kinase (three ESTs) and 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase (enzymes of the MEV and MEP pathways, respectively) suggests the up-regulation of these pathways in sweet basil after MeJA treatment. Moreover, MeJA induction of the pentacyclic triterpene biosynthesis was envisaged because the SSH library also represented one EST for putative farnesyl diphosphate synthase and three ESTs for putative oxidosqualene cyclase (OSC) with homology to amyrin synthases of other plant species (Fig. 2A). Furthermore, three cytochrome P450s of the CYP716A subfamily were identified in the SSH library (Fig. 2A). CYP716A subfamily cytochrome P450 enzymes were recently found to be involved in triterpene biosynthesis in Panax notoginseng, Medicago truncatula, and Catharanthus roseus (Carelli et al., 2011; Han et al., 2011; Huang et al., 2012). To confirm MeJA induction of the pentacyclic triterpene biosynthesis in sweet basil, the levels of ursolic acid and its isomer oleanolic acid were determined by thin-layer chromatography (TLC) analysis (Supplemental Fig. S3; Wójciak-Kosior, 2007). The analysis revealed 2.2- and 2.6-fold higher level accumulation of ursolic acid and oleanolic acid, respectively, in sweet basil leaves at 48 h after MeJA treatment (Fig. 2B). The MeJA-inducible expression of the candidate pentacyclic triterpene biosynthesis genes (amyrin synthases and CYP716A subfamily cytochrome P450 enzymes) of sweet basil was validated by qRT-PCR analysis (Fig. 2C). Altogether, these data suggest MeJA responsiveness of the ursane-type and oleanane-type pentacyclic triterpene biosynthetic pathways in sweet basil.
Cloning and Sequence Analysis of the OSCs
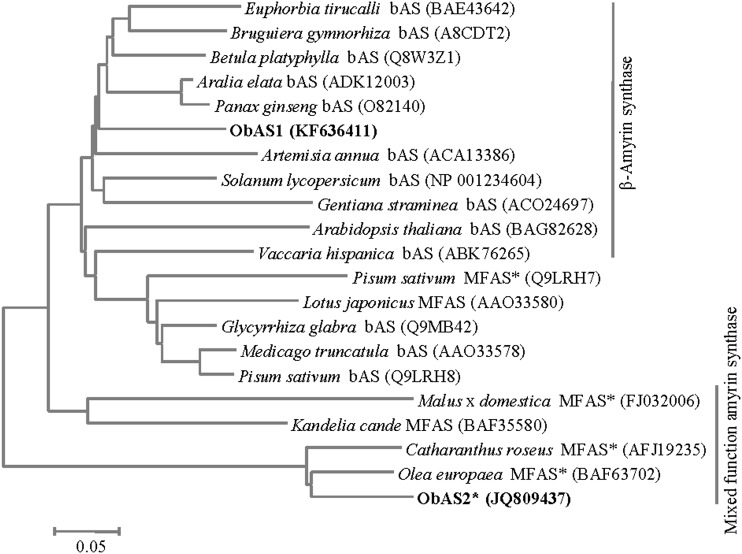
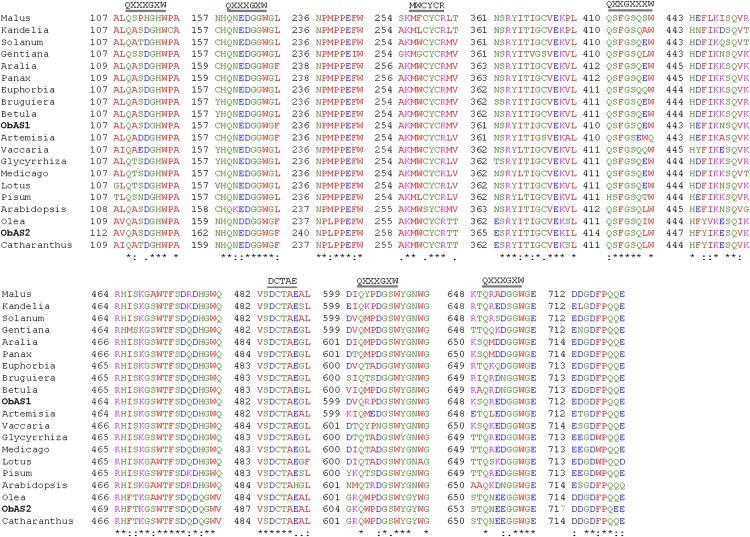
2,3-Oxidosqualene, the precursor of ursolic acid and oleanolic acid, serves as the endogenous substrate for the enzyme OSC that produces α-amyrin and/or β-amyrin, which then act as the substrates for the cytochrome P450 enzymes for the biosynthesis of the ursolic acid and oleanolic acid, respectively (Fig. 2A). To shed light on pentacyclic triterpene biosynthesis in sweet basil, two MeJA-responsive unique ESTs encoding putative OSCs (ObMU39 and ObMU162) were targeted for the functional analysis. The full-length complementary DNAs (cDNAs) specific to the OSCs were obtained from leaf total RNAs by a combination of reverse transcription PCR (RT-PCR) amplification of the unique sequences and rapid amplification of the cDNA ends (5′ and 3′ RACE). Analysis of the full-length sequences revealed that both of the unique sequences, ObMU39 and ObMU162, are the common gene products with a match to the 5′ and 3′ regions, respectively, of the complete cDNA sequence. To identify additional OSCs of sweet basil, we conducted RT-PCR using degenerate oligonucleotides followed by 5′ and 3′ RACE. Sequence analysis of the amplified cDNAs revealed one more OSC in addition to the previous one. Both of the complete cDNA sequences were submitted to the GenBank database under the accession numbers KF636411 (ObAS1) and JQ809437 (ObAS2). Sequence analysis revealed that ObAS1 and ObAS2 encode for 60.32% identical polypeptides of 761 and 765 amino acids, respectively, with calculated molecular masses of 87.73 kD and 87.49 kD. ObAS1 and ObAS2 were subjected to phylogenetic analysis with the OSCs of other plants (Fig. 3). At the amino acid sequence level, ObAS1 and ObAS2 shared 83.05% and 86.61% identities, respectively, with the Panax ginseng β-amyrin synthase and the Olea europaea mixed-function OSC that catalyzes the production of α-amyrin and β-amyrin as well as small amounts of ψ-taraxasterol and butyrospermol (Kushiro et al., 1998; Saimaru et al., 2007). On the contrary, ObAS1 and ObAS2 shared 61.37% and 61.07% identities with the O. europaea mixed-function OSC and the P. ginseng β-amyrin synthase, respectively. ObAS1 and ObAS2 have all of the conserved amino acid motifs characteristic of an OSC (Fig. 4). These include the DCTAE motif that is considered a part of the active site that binds the substrate 2,3-oxidosqualene, the MWCYCR motif, and few β-strand turn QW motifs that are known to be shared by the OSC superfamily (Poralla et al., 1994; Abe and Prestwich, 1995). These results suggest that ObAS1 and ObAS2 are the candidate OSCs that possibly provide the direct precursors for ursolic acid and oleanolic acid biosynthesis in sweet basil. Moreover, ObAS1 and ObAS2 exhibited MeJA-induced expression, suggesting their potential role in MeJA-induced biosynthesis of these pentacyclic triterpenes in sweet basil (Fig. 2, B and C).
Figure 3.
Phylogenetic tree of plant OSCs and their relationships to sweet basil ObAS1 and ObAS2. The complete amino acid sequences of OSCs obtained from the GenBank database, together with that of ObAS1 and ObAS2 were analyzed by MEGA5. The phylogenetic tree was built using the Neighbor Joining Method. MFASs with product preference for α-amyrin are marked with an asterisk. bAS, β-amyrin synthase; MFAS, mixed-function amyrin synthase.
Figure 4.
Amino acid sequence comparisons of the sweet basil ObAS1 and ObAS2 with plant OSCs. Multiple sequence alignment was carried out by ClustalW2 and manually edited to present the regions having high homology (complete alignment is presented in Supplemental Figure S4). Identical amino acid residues are depicted with an asterisk, whereas conserved and semiconserved substitutions are marked with a colon and period, respectively. [See online article for color version of this figure.]
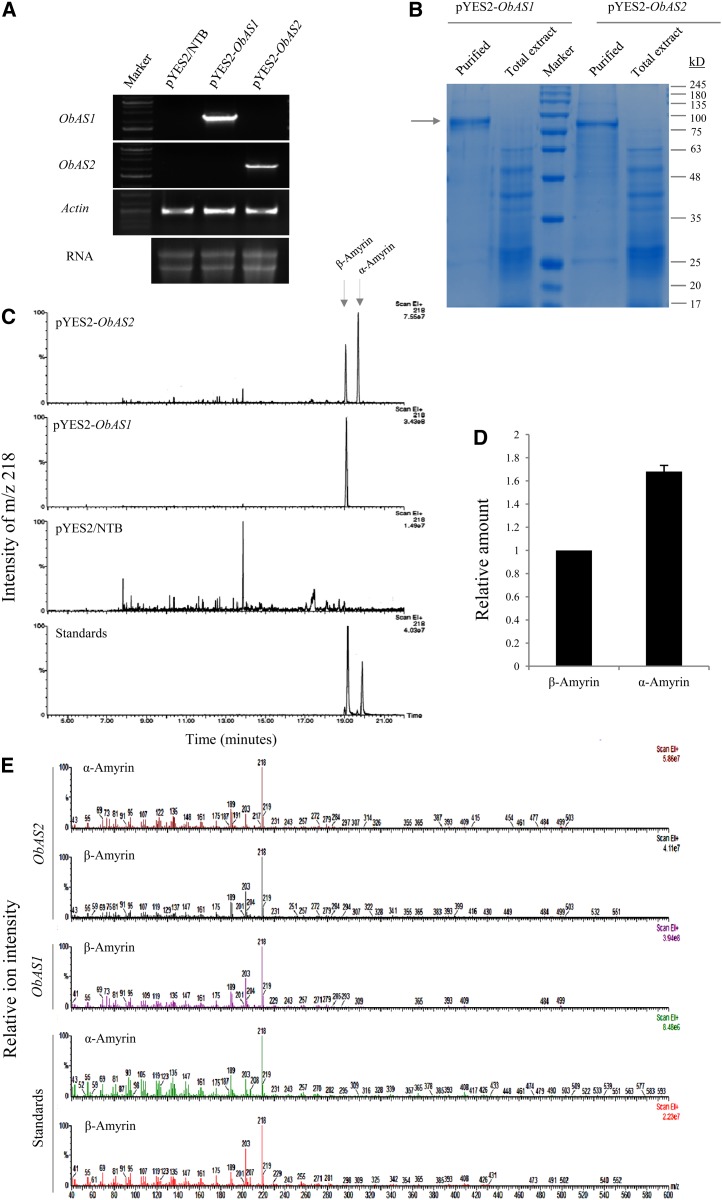
Functional Expression of ObAS1 and ObAS2 in Yeast
To determine the product specificities of ObAS1 and ObAS2, the full-length cDNAs (2286 and 2298 nucleotides, respectively) were expressed in yeast (Saccharomyces cerevisiae strain BY4741) under the control of the Gal-inducible GAL1 promoter. In yeast, endogenous 2,3-oxidosqualene can serve as an in vivo substrate for ObAS1 and ObAS2. The expression of ObAS1 and ObAS2 in transgenic yeasts was confirmed by RT-PCR analysis and by purifying poly(His) (6×His)-tagged proteins by using a nickel-nitrilotriacetic acid agarose column (Fig. 5, A and B). ObAS1 and ObAS2 were resolved as 97-kD recombinant proteins on 10% (w/v) SDS-PAGE (Fig. 5B). This was in agreement with the calculated molecular masses of 6×His-tagged recombinant proteins, indicative of the complete translation of the full-length cDNA sequences in yeast. To determine the enzyme function of ObAS1 and ObAS2, Gal-induced transgenic yeast cells were analyzed for the accumulation of pentacyclic triterpene metabolites. Yeast cells were extracted with hexane after alkaline treatment and metabolites were identified through gas chromatography-mass spectrometry (GC-MS) analysis. Compared with the vector control strain (S. cerevisiae transformed with pYES2/NTB empty vector), the yeast cells expressing ObAS1 and ObAS2 produced one and two new products, respectively (Fig. 5C). By comparing the retention time and mass spectra of those new products with those of the authentic standards (α-amyrin and β-amyrin), we found that ObAS1-expressing transgenic yeast accumulated β-amyrin, which was not present in the control yeast cells transformed with empty vector (Fig. 5, C and E). By contrast, ObAS2-expressing transgenic yeast accumulated both α-amyrin and β-amyrin as new products in a 5:3 ratio (Fig. 5, C–E). The yeast expression experiments were repeated three times, always with very similar results. These results clearly indicated that ObAS1 encodes for a β-amyrin synthase, whereas ObAS2 encodes a mixed-function amyrin synthase with a product preference for α-amyrin.
Figure 5.
Functional expression of ObAS1 and ObAS2 in yeast and identification of the amyrin products. A and B, The expression of the full-length cDNAs in S. cerevisiae was confirmed by RT-PCR analysis (A) and purifying 6×His-tagged recombinant proteins (B). C, GC-MS analysis of hexane extracts of yeast cells expressing ObAS1 and ObAS2. Yeast transformed with empty vector pYES2/NTB was used as the control. Products were monitored on the basis of intensity of the base peak (mass-to-charge ratio of 218) and retention time using a mixture of α-amyrin and β-amyrin as standards. The experiments were repeated thrice independently, always with identical patterns. Total ion chromatograms are presented in Supplemental Fig. S5. D, The relative comparison of α-amyrin and β-amyrin accumulated in the ObAS2-expressing yeast strain based on GC peak area. Data are the mean ± se from three biological replicates. E, Mass spectra of the peaks correspond to α-amyrin and β-amyrin in standards and ObAS1 and ObAS2 expressing yeast extracts are compared. [See online article for color version of this figure.]
Spatial and Temporal Regulation of Pentacyclic Triterpene Biosynthesis
To determine the involvement of ObAS1 and ObAS2 in the biosynthesis of ursane-type and oleanane-type pentacyclic triterpenes in sweet basil, the spatiotemporal expression patterns of these genes were correlated with in planta metabolite accumulation patterns. The relative transcript levels of ObAS1 and ObAS2 in different plant organs, tissue types, and leaf developmental ages were determined by qRT-PCR analysis and compared with the ursolic acid and oleanolic acid accumulation patterns (Figs. 6, 7, and 8). The level of these metabolites in sweet basil was determined by TLC analysis after iodine derivatization (Supplemental Fig. S3; Wójciak-Kosior, 2007).
Figure 6.
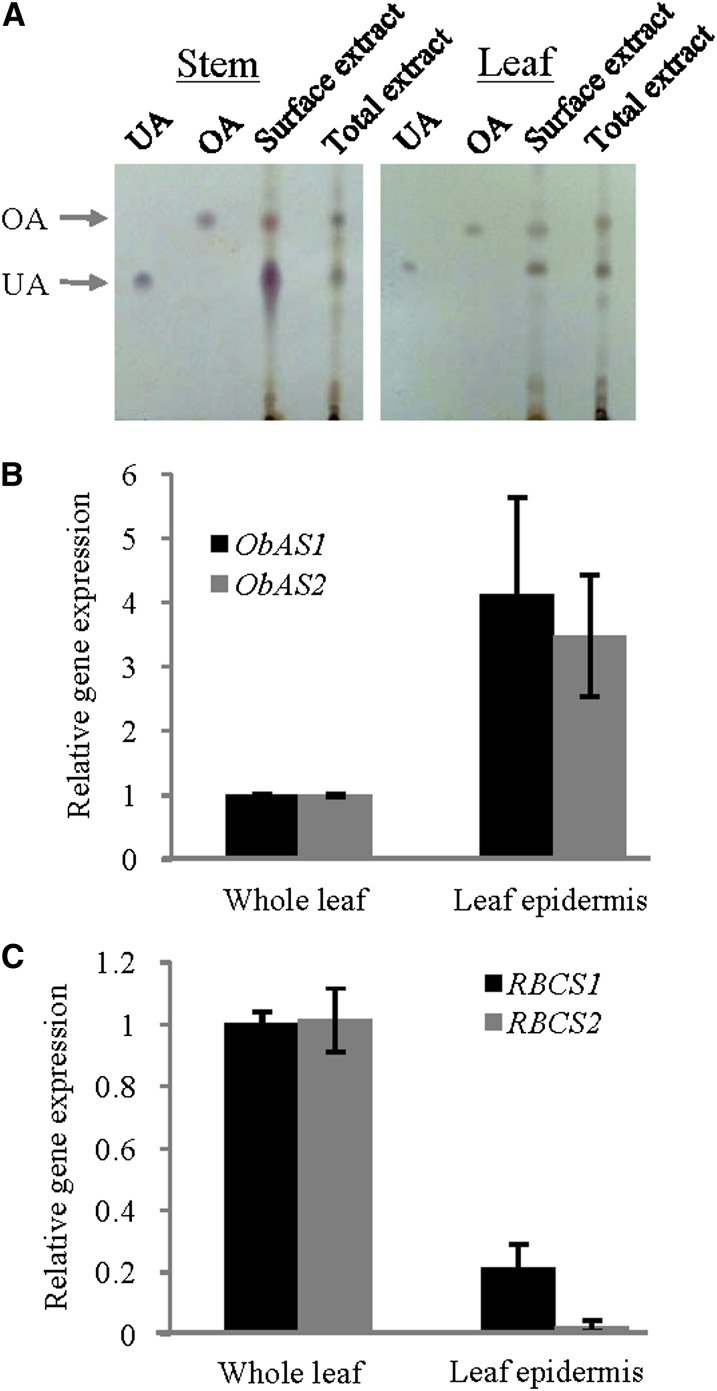
Epidermal cell localization of the pentacyclic triterpene biosynthesis. A, Stem and leaf surface extracts were obtained by dipping stem and leaves, respectively, into chloroform as described in the materials and methods. Total extracts were obtained after complete homogenization of stem and leaf tissues followed by chloroform extraction. Each extract was analyzed by TLC using 200 ng of ursolic acid (UA) and oleanolic acid (OA) as the standards. The analysis was repeated thrice independently with similar results. B, qRT-PCR expression analysis of ObAS1 and ObAS2 using whole leaf and epidermal-enriched RNAs. Data are the mean ± se from seven biological replicates. C, The expression levels of two unique transcripts of sweet basil Rubisco small subunit (RBCS) were determined using whole leaf and epidermal-enriched RNAs. Data are mean ± se (n = 4, from two biological replicates). GenBank accession numbers for RBCS1 and RBCS2 are DY342732 and DY336269, respectively. [See online article for color version of this figure.]
Figure 7.
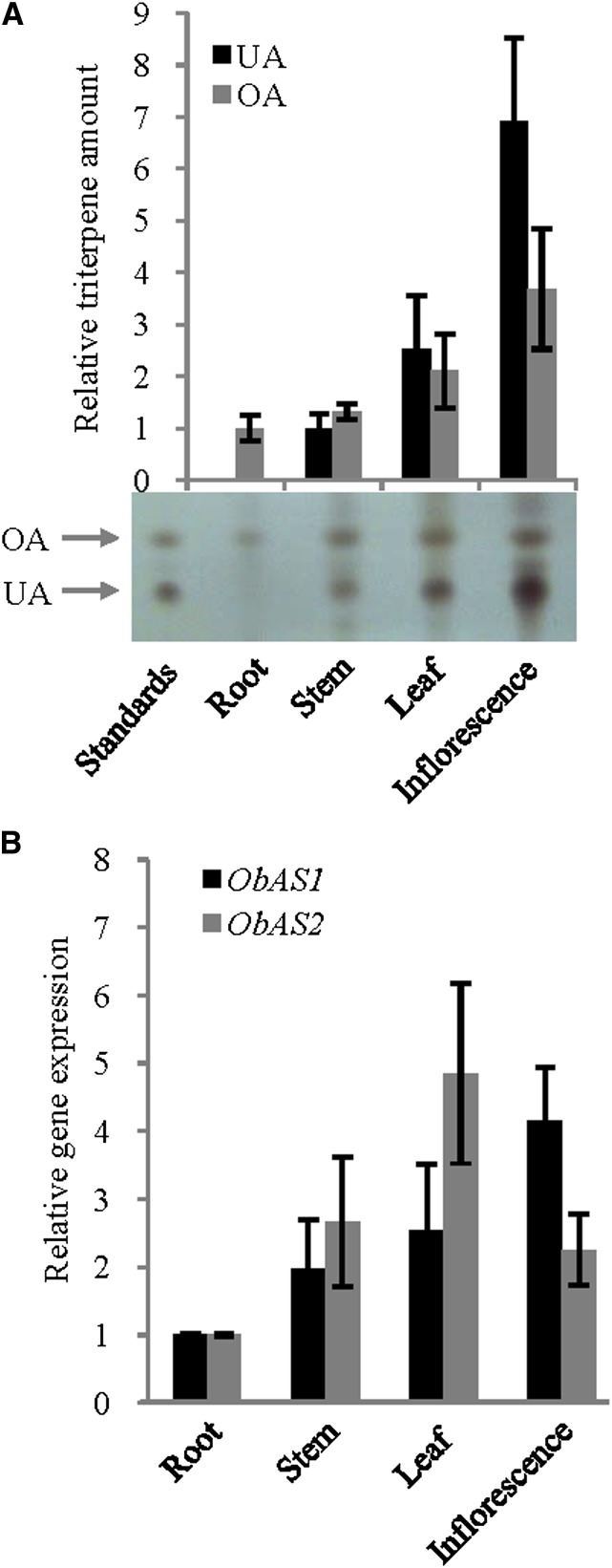
Tissue-specific expression profiles of ObAS1 and ObAS2 and differential accumulation of the pentacyclic triterpenes. A, The relative levels of ursolic acid (UA) and oleanolic acid (OA) in different tissues of sweet basil were determined by TLC analysis. Data are the mean ± se from six independent experiments and the photograph of one representative TLC plate is presented. B, The relative transcript level was determined by qRT-PCR analysis. Data are the mean ± se from four biological replicates. [See online article for color version of this figure.]
Figure 8.
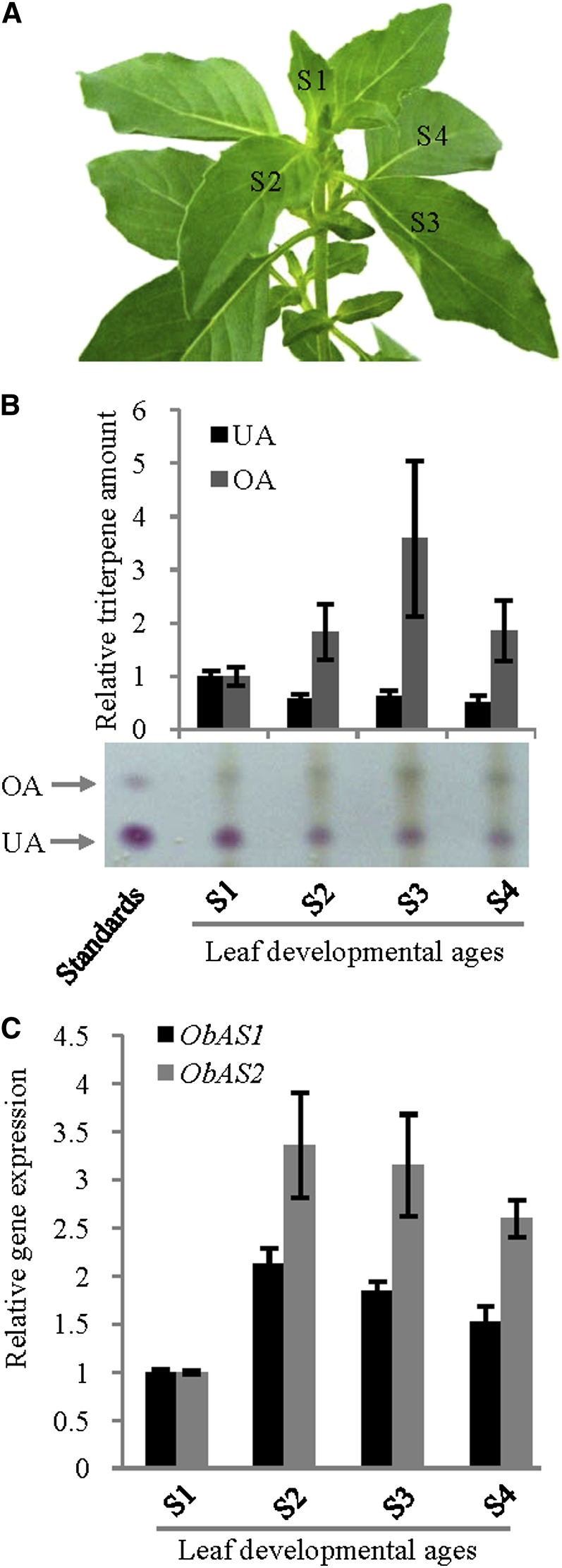
Differential expression of ObAS1 and ObAS2 and differential accumulation of the pentacyclic triterpenes in leaves of different developmental ages. A, Four stages (S1 to S4) of leaf pairs used for the gene expression and metabolite analysis. B, The relative levels of ursolic acid (UA) and oleanolic acid (OA) in different leaf ages were determined by TLC analysis. Data are the mean ± se from five independent experiments and the photograph of one representative TLC plate is presented. C, The relative transcript levels of ObAS1 and ObAS2 were determined by qRT-PCR analysis. Data are the mean ± se from four biological replicates. [See online article for color version of this figure.]
In some plant species, pentacyclic triterpenes accumulate in a substantial amount in surface cuticular wax, possibly because of epidermal cell specialization of the biosynthetic pathways (Murata et al., 2008; Yeats and Rose, 2013). The cuticular wax component of sweet basil was selectively extracted from stems and leaves by brief dipping in chloroform and analysis was conducted with TLC (Fig. 6A). The presence of ursolic acid and oleanolic acid in surface extracts suggests epidermal cell specialization of their biosynthesis in sweet basil. To substantiate these results, the expression levels of ObAS1 and ObAS2 were determined in leaf epidermal cells using the small subunit of Rubisco, a marker for the mesophyll cells, as the negative control. The carborundum abrasion method (Levac et al., 2008) was followed to harvest leaf epidermis-enriched RNAs and the relative gene expression was determined by qRT-PCR analysis (Fig. 6, B and C). Compared with the whole leaf, ObAS1 and ObAS2 exhibited 4.1- and 3.4-fold higher expression levels in the leaf epidermis, respectively (Fig. 6B). By contrast, the expression level of Rubisco small subunit was very low in the leaf epidermis compared with the whole leaf (Fig. 6C). Together, these results suggest epidermal cell localization and possible involvement of ObAS1 and ObAS2 in the biosynthesis of ursolic acid and oleanolic acid.
The expression levels of ObAS1 and ObAS2 in different plant organs and leaves of different developmental ages were determined and correlated with the ursolic acid and oleanolic acid levels (Figs. 7 and 8). ObAS1 and ObAS2 showed the highest expression in inflorescences and leaves, respectively, and the lowest expression in roots (Fig. 7B). On the other hand, both genes exhibited higher expression levels in older leaves compared with the youngest leaves (Fig. 8C). These differential expression patterns of ObAS1 were quite well correlated with the accumulation profiles of oleanolic acid in plant organs and leaves of different developmental ages (Figs. 7A and 8B). Oleanolic acid accumulated 3.6 times more in inflorescences compared with the roots and 1.8 to 3.5 times more in older leaves compared with the youngest leaves. Therefore, enzyme function (Fig. 5) and gene expression patterns of ObAS1 (Figs. 6–8) strongly suggest that this β-amyrin synthase is involved in the biosynthesis of oleanolic acid in sweet basil. However, the level of ursolic acid was found to be 2.7 times higher in inflorescences compared with the leaves and the highest level was detected in the youngest leaves compared with the older leaves. The role of ObAS2 for in planta biosynthesis of the ursolic acid was envisaged based on the epidermal cell enrichment of the ObAS2 mRNA and accumulation of the ursolic acid in the plant surface (Fig. 6, A and B). However, it is also possible that the accumulation of ursolic acid in different plant organs and during leaf development is also influenced by its catabolism/metabolism and transport after biosynthesis. Moreover, the presence of the additional α-amyrin synthase that is involved in high-level biosynthesis of the ursolic acid in inflorescences and youngest leaves cannot be excluded, because very limited transcript information is available and the genome of sweet basil is yet to be sequenced.
DISCUSSION
Although a number of structurally and functionally diverse classes of secondary metabolites with important pharmacological traits are identified from sweet basil (Iijima et al., 2004a; Meera et al., 2009; Zhang et al., 2009; Arshad Qamar et al., 2010; Choudhury et al., 2010; Dashputre and Naikwade, 2010; Verma et al., 2012), their biosynthetic pathways remained largely unexplored. The plant endogenous signaling molecule MeJA, in addition to regulating the normal plant developmental processes, induces defense responses during herbivore and pathogen attacks by accumulating diverse secondary metabolites (Cheong and Choi, 2003). Because MeJA has been shown to be a powerful inducer of secondary metabolite production in diverse plants (Aerts et al., 1994; Suzuki et al., 2005; Kim et al., 2006; De Geyter et al., 2012; Lenka et al., 2012), we studied MeJA-elicited transcriptional changes with the goal to identify the candidate genes involved in the biosynthesis of secondary metabolites in sweet basil. SSH is a powerful genomics tool for identification of differentially expressed genes for nonmodel organisms such as sweet basil, in which genomic information is currently scarce (Ohlrogge and Benning, 2000). The SSH approach led to the isolation of 388 candidate MeJA-responsive transcripts that included several (70.87%) new transcripts of sweet basil. Many of these transcripts comprise the secondary metabolic pathways, including terpenoids and phenylpropanoids/flavonoids, in addition to the transcripts that are associated with MeJA biosynthesis and plant stress responses (Table II; Supplemental Figs. S1 and S2; Supplemental Tables S1 and S4).
Some of the MeJA up-regulated transcripts are potential candidates for regulation of jasmonic acid biosynthesis (Supplemental Fig. S1). Among these, lipoxygenases are known to catalyze the oxygenation of polyunsaturated fatty acids, such as linolenic acids, to produce fatty acid hydroperoxides that serve as intermediates in the formation of jasmonic acid (Sasaki et al., 2001). Moreover, additional transcripts of the jasmonic acid biosynthesis pathway, such as C-acyltransferases and oxidases, were also identified (Supplemental Fig. S1; Supplemental Table S5). These results suggest that MeJA controls its own biosynthesis by a positive feedback mechanism (Sasaki et al., 2001). Few MeJA up-regulated transcripts are the putative transcriptional regulators of MeJA responses in sweet basil (Supplemental Table S3). The APETALA2/ERF and WRKY transcription factors were previously shown as regulators of secondary metabolite biosynthesis in plants in response to MeJA (Yu et al., 2012; Mishra et al., 2013), suggesting their possible involvement in the regulation of secondary metabolite biosynthesis in sweet basil. Lectins, callose synthase, and thaumatin-like proteins that presumably do not have any role in secondary metabolite accumulation were also identified among the MeJA up-regulated transcripts (Supplemental Table S4). These genes function as plant defense components and were also found to be induced by MeJA in other plant species (Cheong and Choi, 2003; Lenka et al., 2012; Ee et al., 2013). The data also suggest MeJA responsiveness of the MEP and MEV pathways that provide 5C building blocks for the biosynthesis of the diverse terpene metabolites (Table II). The MeJA-induced expression of the linalool synthase, eugenol synthase, and polyphenol oxidase substantiates the earlier reports of MeJA-induced biosynthesis of the corresponding metabolites in sweet basil (Table II; Kim et al., 2006; Li et al., 2007).
Our transcript and metabolite analysis revealed MeJA induction of the ursane-type and oleanane-type pentacyclic triterpene biosynthesis pathways of sweet basil (Fig. 2, A to C). OSCs (ObAS1 and ObAS2) and CYP716A subfamily cytochrome P450s (ObCYP1, ObCYP2, and ObCYP3) that showed induced expression after MeJA treatment could be involved in MeJA-induced biosynthesis of ursane-type and oleanane-type pentacyclic triterpene skeletons in sweet basil (Fig. 2C). The OSC-mediated cyclization of 2,3-oxidosqualene is the initial diversifying step for the formation of a large group of structurally diverse triterpene compounds that are widespread throughout the plant kingdom (Abe and Prestwich 1995; Phillips et al., 2006). Hydroxylation by cytochrome P450 enzymes, alkylation by methyltransferases, and glycosylation by glycosyltransferases are among the further enzymatic modifications of the triterpene skeletons. In contrast with animals and fungi that generally have a single OSC (lanosterol synthase) for biosynthesis of the membrane sterol, the higher plants contain several OSCs for biosynthesis not only of the sterols (cycloartenol synthase and lanosterol synthase) but also of diverse triterpenes (Phillips et al., 2006; Sawai and Saito, 2011). The functional diversity of the OSCs and evolution of the mixed-function OSCs that produce more than one specific compound enable over 100 different carbon skeletons of naturally occurring triterpenes in plants (Sawai and Saito, 2011). The product specificities of ObAS1 and ObAS2 were determined by heterologous expression in yeast (Fig. 5). In yeast, ObAS1 formed β-amyrin as its sole product, whereas ObAS2 catalyzed the formation of both α-amyrin and β-amyrin, with α-amyrin as the major product. Moreover, phylogenetic analysis also indicated that ObAS1 is closely related to the β-amyrin synthases, whereas ObAS2 is more similar to the mixed-function OSCs (Fig. 3). Although OSCs that produce β-amyrin specifically are known from other plant species, no enzyme producing α-amyrin as a sole product has been identified to date. All of the identified mixed-function OSCs that catalyze the formation of α-amyrin also produce β-amyrin and few other minor triterpenes (Morita et al., 2000; Saimaru et al., 2007; Brendolise et al., 2011; Yu et al., 2013). Further studies on ObAS1 and ObAS2 can be directed to elucidate the molecular mechanisms and evolutionary perspectives of the amyrin product specificities for OSCs.
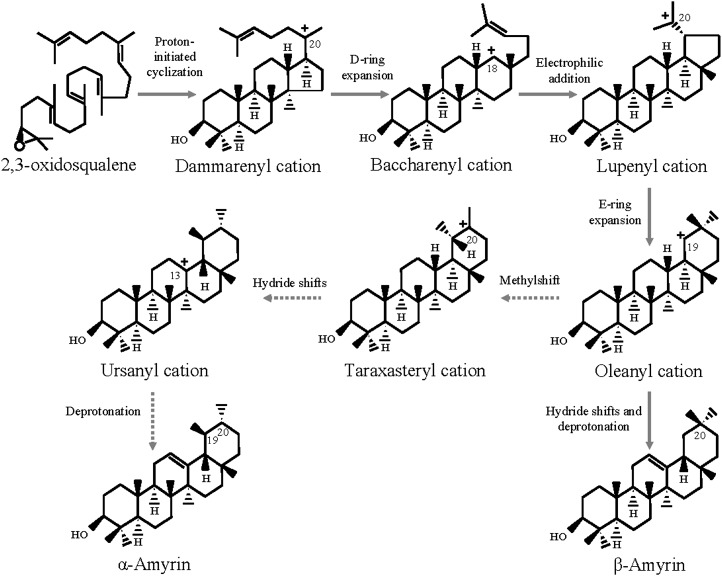
The conversion of 2,3-oxidosqualene into α-amyrin and β-amyrin involves an initial protonation step followed by a polyene addition cascade that leads to the formation of tetracyclic C-20 dammarenyl carbocation to pentacyclic C-20 lupenyl, C19 oleanyl, C-20 taraxasteryl, and C-13 ursanyl carbocation intermediates. Finally, a series of 1,2-shifts of hydride and/or methyl groups, followed by a deprotonation step, give a stable pentacyclic triterpene skeleton (Fig. 9; Phillips et al., 2006; Abe, 2007). The cyclization mechanism of the α-amyrin synthase and β-amyrin synthase is identical up to the C-19 oleanyl carbocation stage. The hydride shifts and deprotonation of the C-19 oleanyl carbocation result in the formation of β-amyrin. However, the migration of a methyl group from C-20 of oleanyl carbocation gives a C-20 taraxasteryl carbocation intermediate that is further converted to α-amyrin following 1,2-hydride shifts and deprotonation (Morita et al., 2000). The reaction mechanisms of α-amyrin synthase and β-amyrin synthase suggest that the conserved amino acid residues in the enzyme active sites take part in a common reaction up to the C-19 oleanyl carbocation stage. However, some specific amino acid residues of these enzymes determine which of two alternative paths to follow after the formation of the C-19 oleanyl carbocation intermediate (Fig. 9).
Figure 9.
The mechanism of ObAS1 and ObAS2-mediated cyclization of 2,3-oxidosqualene into pentacyclic triterpenes. The protonation of 2,3-oxidosqualene triggers a cyclization cascade that leads to the generation of different carbocation intermediates, which further undergo hydride and methyl shift rearrangement steps before being deprotonated. The differential stability of the C-19 oleanyl and C-13 ursanyl carbocations bound to the enzyme is likely to affect the product specificities of ObAS1 and ObAS2. The reaction steps are common for ObAS1 and ObAS2 up to C-19 oleanyl carbocation intermediate. After that, the reaction path favored by ObAS2 is depicted by dashed arrows.
ObAS2 produced more α-amyrin than β-amyrin; therefore, it must have characteristic amino acid residues that favor the formation of the ursane-type over the oleanane-type pentacyclic triterpene by directing the reaction toward C-13 ursanyl carbocation following a C-20 methyl shift, instead of completely stabilizing the C-19 oleanyl carbocation (Fig. 9). At the amino acid sequence level, ObAS1 and ObAS2 shared 60.32% identity and all of the conserved amino acids within the DCTAE, MWCYCR, and QG motifs were identical (Fig. 4). Therefore, the observed difference in their enzymatic functions could be due to the variation in amino acid residues that are outside these conserved motifs. On the basis of the amino acid sequence comparison of apple (Malus domestica) MdOSC1 with other OSCs, it was proposed earlier that substitution of Phe to Trp within the MWCYCR motif may possibly be responsible for the increased product specificity to α-amyrin (Brendolise et al., 2011), because site-directed mutagenesis of Trp residue led to loss of the product specificities for some OSCs (Kushiro et al., 2000). However, the presence of Trp in the MWCYCR motifs of both ObAS1 and ObAS2 suggests that α-amyrin product specificity is a result of the changes at other amino acid positions.
Sequence comparison of ObAS1 with ObAS2 highlighted some amino acid residues that are also conserved in two other mixed-function OSCs from C. roseus and O. europaea, which have α-amyrin product preference such as ObAS2 (Saimaru et al., 2007; Huang et al., 2012; Yu et al., 2013). Some of these residues include Tyr-173, Tyr-227, Leu-242, Tyr-255, ThrThr-266, TyrHis-279, Tyr-358, Glu-392, Tyr-439, PheTyr-448, PheThr-470, Val-485, Arg-516, Tyr-526, Phe-535, ProTyr-545, His-565, Trp-607, Tyr-615, LeuGlu-665, and MetTyr-694 (Supplemental Fig. S4). Aromatic amino acid substitutions are particularly interesting because they are thought to play an important role in stabilizing the high energy carbocation intermediates of the terpene cyclase reactions (Christianson, 2006; Phillips et al., 2006). To determine the functional significance of these amino acid substitutions in ObAS2, further studies using domain swapping/site-directed mutagenesis approaches are needed.
The tissue-specific expression profiles of ObAS1 and ObAS2 suggest that these enzymes are localized exclusively in the epidermal cells (Fig. 6B). Moreover, high levels of ursolic acid and oleanolic acid were also extracted as cuticular wax components coating the epidermal cells (Fig. 6A). This correlation in transcript and metabolite abundance implies that the biological role of ObAS1 and ObAS2 is to synthesize the triterpenes destined for the cuticular wax component of the plant surface. Taking together differential gene expression patterns and metabolite levels, it is very likely that ObAS1 and ObAS2 are the major enzymes responsible for the plant organ and leaf age-specific biosynthesis of oleanolic acid (Figs. 7 and 8). By contrast, plant organ and leaf age-specific accumulation of ursolic acid was somewhat different from ObAS2 expression patterns. Although the data suggest the involvement of ObAS2 in the biosynthesis of the ursolic acid in stems and leaves, the role of other OSCs in high-level biosynthesis in inflorescences cannot be excluded. Moreover, it is also possible that metabolite transport function determines ursolic acid accumulation in high levels to the actively growing parts and reproductive organs of the plants, as shown for other secondary metabolites (Wink, 2010b). This is quite likely because cuticular triterpenes are thought to play a role in plant defense, which signifies their accumulation in these organs (González-Coloma et al., 2011). Although the biological role of the pentacyclic triterpenes has not been clearly established, the potential insecticidal and antimicrobial activities of the pentacyclic triterpenes suggest a protective role against pests and pathogens (González-Coloma et al., 2011; Kurek et al., 2012).
Pentacyclic triterpenes are synthesized inside the epidermal cells but are accumulated in the cuticle (Fig. 6, A and B; Murata et al., 2008), suggesting the involvement of the transporter for secretion into the cuticle. The transport is possibly facilitated by the proteins, because it is unlikely that these lipophilic metabolites travel from the epidermal cell to the cuticle through diffusion. Although ATP binding cassette transporters that are involved in secretion of antifungal diterpene and cuticular lipid are known in plants, no triterpene transporter has been identified to date (Jasiński et al., 2001; Pighin et al., 2004). Moreover, long-distance transport of the secondary metabolites from the site of synthesis to the site of storage by means of the xylem and phloem or via the apoplast has been shown for some instances (Wink, 2010a, 2010b). It is also possible that sterol or lipid transporters that have broad substrate spectra are involved in the export of pentacyclic triterpenes (Buschhaus and Jetter, 2012).
In conclusion, this work led to the identification of several MeJA-responsive transcripts in sweet basil and functional characterization of two OSCs that are involved in the biosynthesis of the direct precursors of two medicinally important pentacyclic triterpenes, ursolic acid and oleanolic acid. This study will be useful in expanding our understanding of the molecular basis of the secondary metabolite accumulation in Ocimum species and in metabolic engineering of pentacyclic triterpene biosynthesis. Further work on the enzyme structure-function relationship using ObAS1 and ObAS2 could reveal the molecular mechanism of α-amyrin and β-amyrin product specificity of the OSCs.
MATERIALS AND METHODS
Plant Materials and MeJA Treatment
Sweet basil (Ocimum basilicum ‘CIM-Saumya’) seeds were obtained from the National Gene Bank for Medicinal and Aromatic Plants at the Central Institute of Medicinal and Aromatic Plants. Seeds were germinated in presterilized soil. Seedlings at the second true leaf stage were transplanted into earthen pots (15 cm height and internal diameter) containing a mixture of soil and farmyard manure in a 2:1 ratio. The plants were grown under natural light in a glass house at 24°C to 26°C and watered daily with application of Hoagland solution once a week. Roots, stems, matured green leaves, and inflorescences were collected from 90-d-old plants (average height, 63.71 ± 3.77 cm; n = 7), washed with reverse osmosis water, frozen immediately in liquid nitrogen, and stored at −80°C. MeJA (Sigma-Aldrich) treatment was given to 60 d-old plants (average height, 26.66 ± 3.7 cm, n = 9; number of matured green leaves, 48.66 ± 6.76, n = 9) with each group consisting of at least three plants. Plants were thoroughly sprayed until run-off with 250 µM of MeJA solution prepared in MilliQ water containing 0.2% (v/v) dimethyl sulfoxide and 0.05% (v/v) TritonX-100 and covered with polyethylene bags. After 2 h of treatment, the polyethylene bags were removed and the plants were left in open air for 2 h to completely remove the remaining MeJA and then were later transferred to the glass house. The control plants were sprayed with same solution without MeJA, bagged, and kept in isolation from the MeJA-treated plants, however, under the same treatment/growth conditions as mentioned for the MeJA-treated plants. After the specified time of treatment, leaves (stage S3; Fig. 8A) from the plants were harvested, frozen immediately in liquid nitrogen, and stored at −80°C for further metabolite and RNA isolation.
Construction of the Subtraction cDNA Library
The SSH library was generated according to the PCR-Select cDNA Substraction kit instruction manual (Takara) as previously described (Ghosh et al., 2013). In brief, poly(A+) RNA was isolated from the total RNAs using a mRNA purification kit (Qiagen) and 2 µg of purified poly(A+) RNA was reverse transcribed using avian myeloblastosis virus reverse transcriptase. To produce a cDNA library representative of MeJA-induced transcripts, sweet basil plants treated with 250 µM MeJA were used as the tester samples, whereas mock-treated plants were used as the driver samples. Double-stranded tester and driver cDNAs were prepared from poly(A+) RNA of leaf tissues (stage S3; Fig. 8A) harvested from the plants after treatment for 6, 12, and 24 h. Total RNAs were isolated individually from each time point and pooled together in equal ratios to prepare tester and driver RNA samples for poly(A+) RNA isolation. Both tester and driver cDNAs were digested with RsaI for producing blunt end cDNAs. Two populations of adapter-ligated tester cDNA samples were prepared and two rounds of hybridization between the driver and adaptor-linked tester were performed to remove common or noninduced transcripts. Tester-specific cDNAs were then PCR amplified (Advantage 2 PCR kit; Takara). PCR products were ligated into pTZ57R/T vector (Thermo Scientific) and transformed into Escherichia coli (DH5α) for propagation. Recombinant clones were selected based on blue-white selection on a Luria-Bertani agar plate containing 50 μg mL−1 ampicillin and grown in Luria-Bertani media at 37°C for plasmid isolation and glycerol stock preparation.
Sequencing and Analysis of ESTs
To obtain the sequences of ESTs, plasmid DNAs were isolated from the individual recombinant clones. Purified plasmid DNAs were analyzed on 0.8% (w/v) agarose gel before sequencing. Plasmid DNAs were sequenced using the BigDye Terminator kit (Applied Biosystems) with the M13 forward primer in an ABI Prism 3700 DNA analyzer (Applied Biosystems). A total of 550 independent recombinant clones were randomly picked; after sequencing, 509 high-quality sequences were obtained that were submitted to GenBank (accession numbers JZ190506 to JZ191014). These sequences were assembled into contigs after cleaning of vector sequences (Masoudi-Nejad et al., 2006). Functional annotation of the unigenes (contigs and singletons) was carried out by a homology search against the nonredundant protein databases of NCBI (http://www.blast2go.com) and UniProtKB/The Arabidopsis Information Resource (http://www.agbase.msstate.edu) and by following the GO scheme and KEGG pathways (http://www.blast2go.com).
Cloning and Sequence Analysis of ObAS1 and ObAS2
Gene-specific oligonucleotide primers were designed based on the sequences of two MeJA-responsive unique ESTs encoding for putative OSCs (ObMU39, ObMU162) and 5′ and 3′ end sequences were amplified as previously described (Ghosh et al., 2011). In addition, gene-specific degenerate oligonucleotides were also designed based on the conserved motifs of OSCs identified from the multiple sequence alignment. Total RNAs were isolated from the sweet basil leaves and reverse transcribed to generate cDNAs using a poly(A) tail-specific oligonucleotide (3′ RACE adapter primer; Invitrogen). The left primer corresponding to either peptide HQNEDGGWG (5′-CAYCAAAAYGARGAYGGIGGITGG-3′) or NPMPPEFW (5′-AAYCCIATGCCHCCIGARTTYTGG-3′) and the right primer corresponding to peptide DQDHGWQV (5′-ACTTGCCAICCRTGRTCYTGRTC-3′) were used to amplify partial sequences of OSCs that were later cloned into pTZ57R/T vector (Thermo Scientific) and sequenced. The remaining 5′ and 3′ regions were amplified using the RACE kit (Invitrogen/Takara). Related protein sequences from other species were taken and phylogenic analysis (Tamura et al., 2011) and sequence alignments (ClustalW2; http://www.ebi.ac.uk) were performed.
RNA Isolation, sqRT-PCR, and qRT-PCR
RNA was isolated according to the LiCl precipitation method (Meli et al., 2010), treated with DNase I (Thermo Scientific), and purified using the RNeasy Mini Kit (Qiagen). At least two independent isolations consisting of three plants for each group were performed. RNA quality was checked by running 1.2% (w/v) agarose gel and by determining the A260/280 ratio. Four micrograms of total RNA, quantified using a nanodrop (Thermo Scientific), was reverse transcribed using superscript II (Invitrogen) following the manufacturer’s instructions. cDNAs were diluted three times before use in the sqRT-PCR and qRT-PCR reactions. sqRT-PCR was performed in a 25-µl reaction volume with 200 nm of each primers using one unit of recombinant Taq DNA polymerase (Invitrogen) for 28 cycles of denaturation, annealing, and extension steps. qRT-PCR reactions (10 µl reaction volume) consisted of 5 µl 2X Power SYBR Green PCR Master Mix (Applied Biosystems), 0.5 µl of diluted cDNA, and 500 nm of each forward and reverse gene-specific primer. qRT-PCR was performed using 7900 HT Fast Real-Time PCR (Applied Biosystems). A list of the oligonucleotide primers used is shown in Supplemental Table S6. Melting curves were analyzed at the dissociation step to examine the specificity of amplification. Relative gene expression was analyzed following the 2−ΔΔCt (cycle threshold) method, using actin (DY329457) as the endogenous control.
Yeast Expression, Protein Purification, and GC-MS Analysis of Yeast Extracts
The ObAS1 and ObAS2 were expressed in yeast (Saccharomyces cerevisiae) as N-terminal poly(His)-tagged proteins under the control of Gal-inducible GAL1 promoter. The full-length coding sequences of the ObAS1 and ObAS2 were PCR amplified using Phusion high-fidelity DNA polymerase (Finnzymes) with gene-specific forward and reverse primers that contained NotI and XbaI restriction sites, respectively. The PCR products were digested with NotI/XbaI and ligated into the NotI/XbaI sites of the yeast expression vector pYES2/NTB to create pYES2-ObAS1 and pYES2-ObAS2. After verifying the integrity of the gene through sequencing, expression plasmids were transformed into S. cerevisiae (strain BY4741) following the standard lithium acetate method. Transformants were selected on synthetic dextrose medium without uracil after 3 d of incubation at 30°C and were confirmed by colony PCR.
To induce protein expression, yeast transformants were initially grown at 30°C in synthetic dextrose medium without uracil for 16 h and cells were collected by centrifugation. Cells were then washed twice with MilliQ water, resuspended in synthetic complete medium lacking uracil, containing 2% (w/v) Gal and 1% (w/v) raffinose, and incubated at 30°C for protein induction. After 12 h of growth, cells were collected by centrifugation, crushed with liquid nitrogen using a pestle and mortar, and resuspended in lysis buffer (50 mm Na2PO4, 300 mm sodium chloride, 20 mm imidazole, pH 8.0 with 0.1 mm phenylmethylsulfonyl fluoride and protease inhibitor cocktail; Sigma-Aldrich). After dialysis for 12 h in lysis buffer, poly(His)-tagged proteins were purified using nickel-nitrilotriacetic acid agarose beads according to the manufacturer’s protocol (Qiagen). The purity of the proteins was determined by resolving on 10% (w/v) SDS/PAGE gel.
For analysis of yeast products, cells were harvested by centrifugation after 48 h of Gal induction and saponified with 20% (w/v) KOH in 50% (v/v) ethanol at 80°C for 45 min with intermittent vortexing. After extraction with hexane twice, the hexane phase was dried and the residue was derivatized using bis-N,O-(trimethylsilyl) trifluoroacetamide (Sigma-Aldrich) at 80°C for 45 min prior to GC-MS analysis. The PerkinElmer Clarus 680 GC system equipped with an Elite-5MS column (30 m length, 0.25 mm i.d., 0.25-µm film thickness) and coupled with a PerkinElmer Clarus SQ 8C Mass Spectrometer was used for GC-MS analysis. The sample was injected into a split injection mode (1:25) and the carrier gas was helium with a flow rate of 1.2 mL/min. The injection temperature was 250°C. The GC oven temperature was programmed from 80°C (held for 2 min) to 290°C at 20°C/min and held for 20 min. For metabolite identification, full mass spectra were generated by scanning within the mass-to-charge ratio range of 40 to 600. Triterpene products in yeast were monitored on the basis of the intensity of the base peak (mass-to-charge ratio of 218), with pYES2/NTB empty vector as a negative control and a mixture of authentic standards, α-amyrin and β-amyrin (Sigma-Aldrich), as positive control. Triterpenes were confirmed by comparing both the retention time and mass spectra with the authentic standards.
Extraction and Analysis of Triterpenes
Fresh tissues from different plant parts were harvested separately, frozen in liquid nitrogen, and grinded into fine powder using a pestle and mortar. The ground tissue (500 mg) was extracted twice with 10 mL of chloroform. The obtained extract was evaporated to dryness and the residue was dissolved in methanol to a fixed volume (200 µl). To obtain surface extracts, tissues (500 mg) were dipped in 10 mL chloroform, vortexed for 2 min, and incubated for 5 min at room temperature. The surface extracts were dried and finally dissolved in 200 µl of methanol.
Triterpene analysis was carried out as described previously (Wójciak-Kosior, 2007) with few modifications. Ursolic acid and oleanolic acid stock solutions (0.2 mg mL−1) were prepared in methanol. For analysis of the plant extracts, TLC plates (silica gel 60 F254, 20×20 cm; Merck) were washed with methanol and dried before use. Standards and samples were derivatized with 1% (w/v) iodine solution in chloroform for 30 min at dark (30°C) and spotted onto a TLC plate. Excess iodine from the TLC plate was removed by drying. Further, the plates were developed with a mixture of petroleum ether:ethyl acetate: acetone (8:2:1, v/v/v) as a mobile phase. After drying, the plates were sprayed with 2% (w/v) ceric sulfate solution prepared in 5% (v/v) H2SO4, incubated at 37°C for 10 min, and heated to 110°C for 5 min. The visualized spots were documented with a digital camera in daylight. The quantification was carried out by densitometric scanning (Camag TLC Scanner 3) in absorbance/transmittance mode at λ = 530 nm.
Isolation of Leaf Epidermis-Enriched RNAs
Leaf epidermis-enriched RNAs were isolated as described previously (Levac et al., 2008), with few modifications. Sweet basil leaves (1 g fresh weight) of developmental stage S2 (Fig. 8A) were freshly harvested and mixed with 7.5 mL of Trizol reagent (Invitrogen) and 1g of carborundum powder (300 grit; Fisher Scientific) in a 50-mL conical bottom centrifuge tube (Tarsons). The tube was vortexed at maximum intensity for 1 min using a cyclomixer (REMI CM 101) to ensure that majority of the leaves in the tube were well abraded. The tube was then kept at room temperature for 5 min and abraded leaves were removed with forceps. The whole process was repeated once with an additional 1 g of fresh leaves. The extract was centrifuged at 3,000g for 5 min and the supernatant was transferred to a fresh centrifuge tube. To the supernatant 0.2 volume of 5 m sodium chloride was added and vortexed for 30 s. Further, a 0.6 volume of chloroform was added, vortexed for 30 s, and centrifuged at 3,000g for 30 min. The aqueous solution was transferred to 1.5-mL centrifuge tubes together with 0.9 volume of isopropanol and 10 µg of linear polyacrylamide and incubated at −20°C for 12 h. Samples were then centrifuged at 20,800g for 30 min to collect the RNA precipitate. The RNA pellet was washed with 70% (v/v) ethanol, air-dried, and dissolved with diethyl pyrocarbonate-treated MilliQ water. For RNA isolation from whole leaf tissues, 0.5 g of freshly harvested stage S2 leaves were ground into powder using liquid nitrogen and 7.5 mL of Trizol was added. The rest of the isolation steps were the same as the epidermal cell RNA isolation. RNA quality and quantity were determined using a nanodrop (Thermo Scientific) and resolving on 1.2% (w/v) agarose gel. Through carborundum extraction, approximately 2 µg of total RNAs were obtained from 1 g of fresh leaves.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JZ190506-JZ191014 (ESTs), KF636411 (ObAS1), JQ809437 (ObAS2), FJ032006 (Malus), BAF35580 (Kandelia), NP 001234604 (Solanum), ACO24697 (Gentiana), ADK12003 (Aralia), O82140 (Panax), BAE43642 (Euphorbia), A8CDT2 (Bruguiera), Q8W3Z1 (Betula), ACA13386 (Artemisia), ABK76265 (Vaccaria), Q9MB42 (Glycyrrhiza), AAO33578 (Medicago), AAO33580 (Lotus), Q9LRH7 (Pisum), BAG82628 (Arabidopsis), BAF63702 (Olea), and AFJ19235 (Catharanthus).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alpha-linolenic acid metabolism and jasmonic acid biosynthetic pathway obtained from the KEGG database (http://www.genome.jp/kegg/kegg2.html).
Supplemental Figure S2. A simplified representation of the phenylpropanoid/flavonoid pathway.
Supplemental Figure S3. TLC separation of the isomeric pentacyclic triterpene compounds ursolic acid (UA) and oleanolic acid (OA) after iodine derivatization.
Supplemental Figure S4. Amino acid sequence comparisons of sweet basil ObAS1 and ObAS2 with mixed-function OSCs of C. roseus and O. europaea.
Supplemental Figure S5. Total ion chromatograms obtained from GC-MS analysis of hexane extracts of yeast cells expressing ObAS1 and ObAS2.
Supplemental Table S1. Functional annotation of the MeJA-responsive unique ESTs of sweet basil.
Supplemental Table S2. List of the enzyme classes identified in the MeJA SSH library.
Supplemental Table S3. List of the sweet basil transcription factors identified in the MeJA SSH library.
Supplemental Table S4. Some of the plant defense-related transcripts identified in the MeJA SSH library.
Supplemental Table S5. KEGG pathway analysis of the MeJA-responsive unique ESTs.
Supplemental Table S6. List of the primers used in this study.
Acknowledgments
The authors gratefully acknowledge the Council of Scientific and Industrial Research Central Institute of Medicinal and Aromatic Plants for providing research facilities; Asis Datta (National Institute of Plant Genome Research) for valuable suggestions and extending laboratory facilities; Madan Mohan Gupta and Santosh Kumar Srivastava (Central Institute of Medicinal and Aromatic Plants) for the high-performance TLC facility and authentic standards, respectively; Anju Yadav for GC-MS analysis; and Rakesh Kumar Shukla (Central Institute of Medicinal and Aromatic Plants) for sharing laboratory space at the initial period of the research.
Glossary
- MeJA
methyl jasmonate
- SSH
suppression subtractive hybridization
- GC
gas chromatography
- NCBI
National Center for Biotechnology Information
- sqRT-PCR
semiquantitative reverse transcription PCR
- qRT-PCR
quantitative reverse transcription PCR
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MEV
mevalonic acid
- MEP
2C-methyl-d-erythritol-4-phosphate
- OSC
oxidosqualene cyclase
- TLC
thin-layer chromatography
- cDNA
complementary DNA
- RT-PCR
quantitative reverse transcription PCR
- GC-MS
gas chromatography-mass spectrometry
References
- Abe I. (2007) Enzymatic synthesis of cyclic triterpenes. Nat Prod Rep 24: 1311–1331 [DOI] [PubMed] [Google Scholar]
- Abe I, Prestwich GD. (1995) Identification of the active site of vertebrate oxidosqualene cyclase. Lipids 30: 231–234 [DOI] [PubMed] [Google Scholar]
- Aerts RJ, Gisi D, De Carolis E, De Luca V, Baumann TW. (1994) Methyl jasmonate vapor increases the developmentally controlled synthesis of alkaloids in Catharanthus and Cinchona seedlings. Plant J 5: 635–643 [Google Scholar]
- Alfermann AW (2010) Production of natural products by plant cell and organ cultures. In M Wink, ed, Functions and Biotechnology of Plant Secondary Metabolites, Annual Plant Reviews, Ed 2, Vol 39, Wiley–Blackwell, Oxford, pp 381–399 [Google Scholar]
- Arshad Qamar K, Dar A, Siddiqui BS, Kabir N, Aslam H, Ahmed S, Erum S, Habib S, Begum S. (2010) Anticancer activity of Ocimum basilicum and the effect of ursolic acid on the cytoskeleton of MCF-7 human breast cancer cells. Lett Drug Des Discov 7: 726–736 [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S. (2000) A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res 7: 175–180 [DOI] [PubMed] [Google Scholar]
- Besser K, Harper A, Welsby N, Schauvinhold I, Slocombe S, Li Y, Dixon RA, Broun P. (2009) Divergent regulation of terpenoid metabolism in the trichomes of wild and cultivated tomato species. Plant Physiol 149: 499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilal A, Jahan N, Ahmed A, Bilal SN, Habib S, Hajra S. (2012) Phytochemical and pharmacological studies on Ocimum basilicum. Int J Curr Res Rev 4: 73–83 [Google Scholar]
- Bohlmann J, Keeling CI. (2008) Terpenoid biomaterials. Plant J 54: 656–669 [DOI] [PubMed] [Google Scholar]
- Brendolise C, Yauk YK, Eberhard ED, Wang M, Chagne D, Andre C, Greenwood DR, Beuning LL. (2011) An unusual plant triterpene synthase with predominant α-amyrin-producing activity identified by characterizing oxidosqualene cyclases from Malus × domestica. FEBS J 278: 2485–2499 [DOI] [PubMed] [Google Scholar]
- Buschhaus C, Jetter R. (2012) Composition and physiological function of the wax layers coating Arabidopsis leaves: β-amyrin negatively affects the intracuticular water barrier. Plant Physiol 160: 1120–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canter PH, Thomas H, Ernst E. (2005) Bringing medicinal plants into cultivation: opportunities and challenges for biotechnology. Trends Biotechnol 23: 180–185 [DOI] [PubMed] [Google Scholar]
- Carelli M, Biazzi E, Panara F, Tava A, Scaramelli L, Porceddu A, Graham N, Odoardi M, Piano E, Arcioni S, et al. (2011) Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell 23: 3070–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ye H, Li G. (2000) Expression of a chimeric farnesyl diphosphate synthase gene in Artemisia annua L. transgenic plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci 155: 179–185 [DOI] [PubMed] [Google Scholar]
- Cheong JJ, Choi YD. (2003) Methyl jasmonate as a vital substance in plants. Trends Genet 19: 409–413 [DOI] [PubMed] [Google Scholar]
- Choudhury GB, Prabhat KJ, Nayak BS, Panda SK, Tripathy SK. (2010) Phytochemical investigation and evaluation of analgesic activity of leafy extracts of various Ocimum (tulsi) species. The Indian Pharmacist 8: 67–70 [Google Scholar]
- Christianson DW. (2006) Structural biology and chemistry of the terpenoid cyclases. Chem Rev 106: 3412–3442 [DOI] [PubMed] [Google Scholar]
- da Silva Ferreira D, Esperandim VR, Toldo MP, Kuehn CC, do Prado Júnior JC, Cunha WR, e Silva ML, de Albuquerque S. (2013) In vivo activity of ursolic and oleanolic acids during the acute phase of Trypanosoma cruzi infection. Exp Parasitol 134: 455–459 [DOI] [PubMed] [Google Scholar]
- Dasgupta T, Rao AR, Yadava PK. (2004) Chemomodulatory efficacy of basil leaf (Ocimum basilicum) on drug metabolizing and antioxidant enzymes, and on carcinogen-induced skin and forestomach papillomagenesis. Phytomedicine 11: 139–151 [DOI] [PubMed] [Google Scholar]
- Dashputre NL, Naikwade NS. (2010) Preliminary immunomodulatory activity of aqueous and ethanolic leaves extracts of Ocimum basilicum Linn. in mice. Int J Pharm Tech Res 2: 1342–1349 [Google Scholar]
- De Geyter N, Gholami A, Goormachtig S, Goossens A. (2012) Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci 17: 349–359 [DOI] [PubMed] [Google Scholar]
- Ee SF, Oh JM, Mohd Noor N, Kwon TR, Mohamed-Hussein ZA, Ismail I, Zainal Z. (2013) Transcriptome profiling of genes induced by salicylic acid and methyl jasmonate in Polygonum minus. Mol Biol Rep 40: 2231–2241 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, De Luca V. (2008) Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J 54: 763–784 [DOI] [PubMed] [Google Scholar]
- Fathiazad F, Matlobi A, Khorrami A, Hamedeyazdan S, Soraya H, Hammami M, Maleki-Dizaji N, Garjani A. (2012) Phytochemical screening and evaluation of cardioprotective activity of ethanolic extract of Ocimum basilicum L. (basil) against isoproterenol induced myocardial infarction in rats. Daru 20: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang DR, Beuerle T, Ullmann P, Werck-Reichhart D, Pichersky E. (2002a) Differential production of meta hydroxylated phenylpropanoids in sweet basil peltate glandular trichomes and leaves is controlled by the activities of specific acyltransferases and hydroxylases. Plant Physiol 130: 1536–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang DR, Lavid N, Zubieta C, Chen F, Beuerle T, Lewinsohn E, Noel JP, Pichersky E. (2002b) Characterization of phenylpropene O-methyltransferases from sweet basil: facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell 14: 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang DR, Wang J, Dudareva N, Nam KH, Simon JE, Lewinsohn E, Pichersky E. (2001) An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant Physiol 125: 539–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J, Dudareva N. (2007) The function of terpene natural products in the natural world. Nat Chem Biol 3: 408–414 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Meli VS, Kumar A, Thakur A, Chakraborty N, Chakraborty S, Datta A. (2011) The N-glycan processing enzymes α-mannosidase and β-D-N-acetylhexosaminidase are involved in ripening-associated softening in the non-climacteric fruits of capsicum. J Exp Bot 62: 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Singh UK, Meli VS, Kumar V, Kumar A, Irfan M, Chakraborty N, Chakraborty S, Datta A. (2013) Induction of senescence and identification of differentially expressed genes in tomato in response to monoterpene. PLoS ONE 8: e76029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Coloma A, Lopez-Balboa C, Santana O, Reina M, Fraga BM. (2011) Triterpene-based plant defences. Phytochem Rev 10: 245–260 [Google Scholar]
- Goossens A, Häkkinen ST, Laakso I, Seppänen-Laakso T, Biondi S, De Sutter V, Lammertyn F, Nuutila AM, Söderlund H, Zabeau M, et al. (2003) A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc Natl Acad Sci USA 100: 8595–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach H, Müller MJ, Kutchan TM, Zenk MH. (1992) Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc Natl Acad Sci USA 89: 2389–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JY, Kim HJ, Kwon YS, Choi YE. (2011) The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol 52: 2062–2073 [DOI] [PubMed] [Google Scholar]
- Hartmann T. (2007) From waste products to ecochemicals: fifty years research of plant secondary metabolism. Phytochemistry 68: 2831–2846 [DOI] [PubMed] [Google Scholar]
- Heilmann J (2010) New medical applications of plant secondary metabolites. In M Wink, ed, Functions and Biotechnology of Plant Secondary Metabolites, Annual Plant Reviews, Ed 2, Vol 39, Wiley-Blackwell, Oxford, pp 348–380 [Google Scholar]
- Huang L, Li J, Ye H, Li C, Wang H, Liu B, Zhang Y. (2012) Molecular characterization of the pentacyclic triterpenoid biosynthetic pathway in Catharanthus roseus. Planta 236: 1571–1581 [DOI] [PubMed] [Google Scholar]
- Iijima Y, Davidovich-Rikanati R, Fridman E, Gang DR, Bar E, Lewinsohn E, Pichersky E. (2004a) The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant Physiol 136: 3724–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y, Gang DR, Fridman E, Lewinsohn E, Pichersky E. (2004b) Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol 134: 370–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasiński M, Stukkens Y, Degand H, Purnelle B, Marchand-Brynaert J, Boutry M. (2001) A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 13: 1095–1107 [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Chen F, Wang X, Rajapakse NC. (2006) Effect of methyl jasmonate on secondary metabolites of sweet basil (Ocimum basilicum L.). J Agric Food Chem 54: 2327–2332 [DOI] [PubMed] [Google Scholar]
- Kong L, Li S, Liao Q, Zhang Y, Sun R, Zhu X, Zhang Q, Wang J, Wu X, Fang X, et al. (2013) Oleanolic acid and ursolic acid: novel hepatitis C virus antivirals that inhibit NS5B activity. Antiviral Res 98: 44–53 [DOI] [PubMed] [Google Scholar]
- Kunkel SD, Suneja M, Ebert SM, Bongers KS, Fox DK, Malmberg SE, Alipour F, Shields RK, Adams CM. (2011) mRNA expression signatures of human skeletal muscle atrophy identify a natural compound that increases muscle mass. Cell Metab 13: 627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek A, Nadkowska P, Pliszka S, Wolska KI. (2012) Modulation of antibiotic resistance in bacterial pathogens by oleanolic acid and ursolic acid. Phytomedicine 19: 515–519 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Shibuya M, Ebizuka Y. (1998) Beta-amyrin synthase—cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. Eur J Biochem 256: 238–244 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Shibuya M, Masuda K, Ebizuka Y. (2000) Mutational studies on triterpene synthases: engineering lupeol synthase into β-amyrin synthase. J Am Chem Soc 122: 6816–6824 [Google Scholar]
- Lange BM, Ahkami A. (2013) Metabolic engineering of plant monoterpenes, sesquiterpenes and diterpenes—current status and future opportunities. Plant Biotechnol J 11: 169–196 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Umano K, Shibamoto T, Lee KG. (2005) Identification of volatile components in basil (Ocimum basilicum L.) and thyme leaves (Thymus vulgaris L.) and their antioxidant properties. Food Chem 91: 131–137 [Google Scholar]
- Lenka SK, Boutaoui N, Paulose B, Vongpaseuth K, Normanly J, Roberts SC, Walker EL. (2012) Identification and expression analysis of methyl jasmonate responsive ESTs in paclitaxel producing Taxus cuspidata suspension culture cells. BMC Genomics 13: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levac D, Murata J, Kim WS, De Luca V. (2008) Application of carborundum abrasion for investigating the leaf epidermis: molecular cloning of Catharanthus roseus 16-hydroxytabersonine-16-O-methyltransferase. Plant J 53: 225–236 [DOI] [PubMed] [Google Scholar]
- Li Z, Wang X, Chen F, Kim HJ. (2007) Chemical changes and overexpressed genes in sweet basil (Ocimum basilicum L.) upon methyl jasmonate treatment. J Agric Food Chem 55: 706–713 [DOI] [PubMed] [Google Scholar]
- Louie GV, Baiga TJ, Bowman ME, Koeduka T, Taylor JH, Spassova SM, Pichersky E, Noel JP. (2007) Structure and reaction mechanism of basil eugenol synthase. PLoS ONE 2: e993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoudi-Nejad A, Tonomura K, Kawashima S, Moriya Y, Suzuki M, Itoh M, Kanehisa M, Endo T, Goto S. (2006) EGassembler: online bioinformatics service for large-scale processing, clustering and assembling ESTs and genomic DNA fragments. Nucleic Acids Res 34: W459–W462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meera R, Devi P, Kameswari B, Madhumitha B, Merlin NJ. (2009) Antioxidant and hepatoprotective activities of Ocimum basilicum Linn. and Trigonella foenum-graecum Linn. against H2O2 and CCL4 induced hepatotoxicity in goat liver. Indian J Exp Biol 47: 584–590 [PubMed] [Google Scholar]
- Meli VS, Ghosh S, Prabha TN, Chakraborty N, Chakraborty S, Datta A. (2010) Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc Natl Acad Sci USA 107: 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Triptahi V, Singh S, Phukan UJ, Gupta MM, Shanker K, Shukla RK. (2013) Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PLoS ONE 8: e52784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Shibuya M, Kushiro T, Masuda K, Ebizuka Y. (2000) Molecular cloning and functional expression of triterpene synthases from pea (Pisum sativum) new alpha-amyrin-producing enzyme is a multifunctional triterpene synthase. Eur J Biochem 267: 3453–3460 [DOI] [PubMed] [Google Scholar]
- Muralidharan A, Dhananjayan R. (2004) Cardiac stimulant activity of Ocimum basilicum Linn. extracts. Indian J Pharmacol 36: 163–166 [Google Scholar]
- Murata J, Roepke J, Gordon H, De Luca V. (2008) The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 20: 524–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina MA, He X, Dixon RA. (2008) Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol 8: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Benning C. (2000) Unraveling plant metabolism by EST analysis. Curr Opin Plant Biol 3: 224–228 [PubMed] [Google Scholar]
- Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, et al. (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496: 528–532 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Inzé D, Goossens A. (2009) Jasmonate-inducible gene: What does it mean? Trends Plant Sci 14: 87–91 [DOI] [PubMed] [Google Scholar]
- Phillips DR, Rasbery JM, Bartel B, Matsuda SPT. (2006) Biosynthetic diversity in plant triterpene cyclization. Curr Opin Plant Biol 9: 305–314 [DOI] [PubMed] [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL. (2004) Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Poralla K, Hewelt A, Prestwich GD, Abe I, Reipen I, Sprenger G. (1994) A specific amino acid repeat in squalene and oxidosqualene cyclases. Trends Biochem Sci 19: 157–158 [DOI] [PubMed] [Google Scholar]
- Rastogi S, Kumar R, Chanotiya CS, Shanker K, Gupta MM, Nagegowda DA, Shasany AK. (2013) 4-coumarate: CoA ligase partitions metabolites for eugenol biosynthesis. Plant Cell Physiol 54: 1238–1252 [DOI] [PubMed] [Google Scholar]
- Reichling J (2010) Plant-microbe interactions and secondary metabolites with antibacterial, antifungal and antiviral properties. In M Wink, ed, Functions and Biotechnology of Plant Secondary Metabolites, Annual Plant Reviews, Ed 2, Vol 39, Wiley–Blackwell, Oxford, pp 214–347 [Google Scholar]
- Reigosa MJ, Malvido-Pazos E. (2007) Phytotoxic effects of 21 plant secondary metabolites on Arabidopsis thaliana germination and root growth. J Chem Ecol 33: 1456–1466 [DOI] [PubMed] [Google Scholar]
- Saimaru H, Orihara Y, Tansakul P, Kang YH, Shibuya M, Ebizuka Y. (2007) Production of triterpene acids by cell suspension cultures of Olea europaea. Chem Pharm Bull (Tokyo) 55: 784–788 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, Awai K, Amagai M, Kuwata C, Tsugane T, Masuda T, et al. (2001) Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res 8: 153–161 [DOI] [PubMed] [Google Scholar]
- Sawai S, Saito K. (2011) Triterpenoid biosynthesis and engineering in plants. Front Plant Sci 2: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui BS, Aslam H, Ali ST, Begum S, Khatoon N. (2007) Two new triterpenoids and a steroidal glycoside from the aerial parts of Ocimum basilicum. Chem Pharm Bull (Tokyo) 55: 516–519 [DOI] [PubMed] [Google Scholar]
- Silva MG, Vieira IG, Mendes FN, Albuquerque IL, dos Santos RN, Silva FO, Morais SM. (2008) Variation of ursolic acid content in eight Ocimum species from northeastern Brazil. Molecules 13: 2482–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazzer P, Guzzo F, Levi M. (2011) Correlated accumulation of anthocyanins and rosmarinic acid in mechanically stressed red cell suspensions of basil (Ocimum basilicum). J Plant Physiol 168: 288–293 [DOI] [PubMed] [Google Scholar]
- Sun G, Yang Y, Xie F, Wen JF, Wu J, Wilson IW, Tang Q, Liu H, Qiu D. (2013) Deep sequencing reveals transcriptome re-programming of Taxus × media cells to the elicitation with methyl jasmonate. PLoS ONE 8: e62865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Reddy MS, Naoumkina M, Aziz N, May GD, Huhman DV, Sumner LW, Blount JW, Mendes P, Dixon RA. (2005) Methyl jasmonate and yeast elicitor induce differential transcriptional and metabolic re-programming in cell suspension cultures of the model legume Medicago truncatula. Planta 220: 696–707 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D, Lee S. (2011) Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143, 10.1199/tab.0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma RS, Padalia RC, Chauhan A. (2012) Variation in the volatile terpenoids of two industrially important basil (Ocimum basilicum L.) cultivars during plant ontogeny in two different cropping seasons from India. J Sci Food Agric 92: 626–631 [DOI] [PubMed] [Google Scholar]
- Wilkinson K, Boyd JD, Glicksman M, Moore KJ, El Khoury J. (2011) A high content drug screen identifies ursolic acid as an inhibitor of amyloid beta protein interactions with its receptor CD36. J Biol Chem 286: 34914–34922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink M (2010a) Introduction. In M Wink, ed, Functions and Biotechnology of Plant Secondary Metabolites, Annual Plant Reviews, Ed 2, Vol 39, Wiley–Blackwell, Oxford, pp 1–20 [Google Scholar]
- Wink M (2010b) Introduction: biochemistry, physiology and ecological functions of secondary metabolites. In M Wink, ed, Biochemistry of Plant Secondary Metabolism, Annual Plant Reviews, Ed 2, Vol 40, Wiley–Blackwell, Oxford, pp 1–19 [Google Scholar]
- Wójciak-Kosior M. (2007) Separation and determination of closely related triterpenic acids by high performance thin-layer chromatography after iodine derivatization. J Pharm Biomed Anal 45: 337–340 [DOI] [PubMed] [Google Scholar]
- Yeats TH, Rose JK. (2013) The formation and function of plant cuticles. Plant Physiol 163: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Thamm AM, Reed D, Villa-Ruano N, Quesada AL, Gloria EL, Covello P, De Luca V. (2013) Functional characterization of amyrin synthase involved in ursolic acid biosynthesis in Catharanthus roseus leaf epidermis. Phytochemistry 91: 122–127 [DOI] [PubMed] [Google Scholar]
- Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ, Chen XY. (2012) The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol Plant 5: 353–365 [DOI] [PubMed] [Google Scholar]
- Yun DJ, Hashimoto T, Yamada Y. (1992) Metabolic engineering of medicinal plants: transgenic Atropa belladonna with an improved alkaloid composition. Proc Natl Acad Sci USA 89: 11799–11803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggwagh NA, Sulpice T, Eddouks M. (2007) Anti-hyperglycaemic and hypolipidemic effects of Ocimum basilicum aqueous extract in diabetic rats. Am J Pharmacol Toxicol 2: 123–129 [Google Scholar]
- Zhang JW, Li SK, Wu WJ. (2009) The main chemical composition and in vitro antifungal activity of the essential oils of Ocimum basilicum Linn. var. pilosum (Willd.) Benth. Molecules 14: 273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ding R, Chai Y, Bonfill M, Moyano E, Oksman-Caldentey KM, Xu T, Pi Y, Wang Z, Zhang H, et al. (2004) Engineering tropane biosynthetic pathway in Hyoscyamus niger hairy root cultures. Proc Natl Acad Sci USA 101: 6786–6791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Davis LC, Verpoorte R. (2005) Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol Adv 23: 283–333 [DOI] [PubMed] [Google Scholar]