A nodule-specific lipid transfer protein contributes to transport to the symbiosome membrane and is required for effective legumerhizobium symbiosis.
Abstract
Rhizobia in legume root nodules fix nitrogen in symbiosomes, organelle-like structures in which a membrane from the host plant surrounds the symbiotic bacteria. However, the components that transport plant-synthesized lipids to the symbiosome membrane remain unknown. This study identified and functionally characterized the Chinese milk vetch (Astragalus sinicus) lipid transfer protein AsE246, which is specifically expressed in nodules. It was found that AsE246 can bind lipids in vitro. More importantly, AsE246 can bind the plant-synthesized membrane lipid digalactosyldiacylglycerol in vivo. Immunofluorescence and immunoelectron microscopy showed that AsE246 and digalactosyldiacylglycerol localize in the symbiosome membrane and are present in infection threads. Overexpression of AsE246 resulted in increased nodule numbers; knockdown of AsE246 resulted in reduced nodule numbers, decreased lipids contents in nodules, diminished nitrogen fixation activity, and abnormal development of symbiosomes. AsE246 knockdown also resulted in fewer infection threads, nodule primordia, and nodules, while AsE246 overexpression resulted in more infection threads and nodule primordia, suggesting that AsE246 affects nodule organogenesis associated with infection thread formation. Taken together, these results indicate that AsE246 contributes to lipids transport to the symbiosome membrane, and this transport is required for effective legume-rhizobium symbiosis.
Legume crops can act as hosts for nitrogen-fixing soil Rhizobium spp. bacteria, which induce and occupy a specialized organ, the root nodule (Limpens et al., 2009). This endosymbiotic relationship is mutualistic for both the host plant and the Rhizobium spp.; the plant receives a crucial supply of reduced nitrogen from the bacteria and the nodule bacteria receive reduced carbon and other nutrients (Held et al., 2010).
Symbiosis requires specialized host-symbiont communication and cellular development. Plant roots are exposed to various microorganisms in the soil, but their strong protective barriers, including cell walls, prevent the entry of most harmful species. To bypass these barriers, the invasion of plant roots by rhizobia begins with a reciprocal exchange of signals that allow the bacteria to enter through the plant root hair cells (Jones et al., 2007). The rhizobia enter the root hair and underlying cells via an infection thread (IT), from which they are eventually released into cortical cells via endocytosis. Each bacterial cell is endocytosed by a target cell into an individual, unwalled membrane compartment that originates from the IT. The bacteria are surrounded by a membrane of plant origin; this membrane is variously termed the endocytic, peribacteroid, or symbiosome membrane. Also, the entire unit is known as the symbiosome (Verma and Hong, 1996; Jones et al., 2007), and the space between the bacteria and the membrane is called the peribacteroid space.
The symbiosome membrane forms the structural and functional interface between the host plant and the rhizobia. First, it prevents direct contact between the host plant cell cytoplasm and the invading prokaryote, which may otherwise interfere with host cell metabolism and may provoke host defense responses. Second, the symbiosome membrane controls the exchange of substrate and signal molecules between host plant cell and the bacteria (Verma and Hong, 1996; Gaude et al., 2004). The host plant makes the symbiosome membrane, which has similar properties to the plant vacuolar membrane, but contains several nodule-specific proteins. For example, the syntaxin MtSYP132, a Medicago truncatula homolog of Arabidopsis (Arabidopsis thaliana) SYNTAXIN OF PLANTS132, occurs on M. truncatula symbiosomes throughout their development (Verma and Hong, 1996; Whitehead and Day, 1997; Catalano et al., 2007). The symbiosome membrane also contains a nonphosphorus galactoglycerolipid, digalactosyldiacylglycerol (DGDG), which is also found in chloroplast, extraplastidic, tonoplast, and plasma membranes (Gaude et al., 2004; Benning, 2009). The symbiosome membrane also contains saturated (16:0, palmitic acid; 18:0, stearic acid) and unsaturated fatty acids (16:1Δ3trans, palmitoleic acid; 18:1Δ9cis, oleic acid 18:2Δ9,12, linoleic acid; 18:3Δ9,12,15, α-linolenic acid), all of which are typically found in higher plants (Whitehead and Day, 1997; Gaude et al., 2004).
During nodule development, the number of rhizobia in infected cells increases dramatically. This increase requires membrane biosynthesis in the bacteria as part of cell division and also requires membrane biosynthesis by the plant to produce symbiosome membrane to enclose the bacteria (Verma, 1992; Gaude et al., 2004). Thus, symbiosis requires large amounts of lipid, but how plant-synthesized lipids are transported to the symbiosome membrane remains unclear.
In eukaryotic cells, vesicular and nonvesicular transport mechanisms mediate intracellular lipid trafficking (Voelker, 1990; Lev, 2010). Large amounts of lipids were thought to be transported between organelles only by vesicular transport. However, lipid transport can occur even when vesicular transport is blocked by ATP depletion, by reduction in temperature, or by treatment with specific drugs such as brefeldin A and colchicine (Kaplan and Simoni, 1985; Vance et al., 1991). Lipid transport can also occur between organelles that are not connected to the vesicular transport machinery, e.g. chloroplasts, mitochondria, and peroxisomes (Levine, 2004; Holthuis and Levine, 2005; Benning, 2009). These observations suggest that nonvesicular transport mechanisms have an important role in intracellular lipid trafficking. However, to date, no symbiosome-targeting plant lipid transport proteins have been characterized.
Nonvesicular lipid transport could occur, in principle, by spontaneous desorption of a lipid monomer from a bilayer and free diffusion through the cytosol, but this process is slow and insufficient to support substantial transport of most lipids. Lipid transfer proteins (LTPs) can facilitate lipid transport between membranes in vitro (Kader et al., 1984; Shin et al., 1995; Lee et al., 1998; Lascombe et al., 2008). LTPs were initially discovered as soluble factors that accelerate the exchange or net transfer of lipid species between membranes in vitro. Since then, many LTPs have been isolated, characterized, and crystallized. LTPs have been identified in eukaryotes and in bacteria and have been subdivided into different families according to their protein sequences and structures (D’Angelo et al., 2008).
Plant LTPs are cationic peptides, with molecular masses of around 7 to 10 kD and can reversibly bind and transport hydrophobic molecules in vitro. Plant LTPs are subdivided into two families, LTP1 and LTP2 (Carvalho et al., 2007). Both families possess conserved patterns of eight Cys residues, and their three-dimensional structure reveals an internal hydrophobic cavity that forms the lipid binding site. NMR data have demonstrated that a wheat (Triticum aestivum) LTP forms a complex with prostaglandin B2, suggesting that LTPs can accommodate large hydrophobic compounds (Tassin-Moindrot et al., 2000). Also, an LTP from Arabidopsis, LTPg1, is required for normal export of wax to the cuticle (Debono et al., 2009).
Chinese milk vetch (Astragalus sinicus) is a winter-growing green manure legume. It can establish a specific endosymbiosis with Mesorhizobium huakuii 7653R and form indeterminate-type N2-fixing root nodules, which are cylindrical and consist of a gradient of developmental zones with a persistent apical meristem (zone I), an infection zone (zone II), and a fixation zone (zone III). In mature nodules, a senescence zone (zone IV) is established proximal to zone III. As the nodule ages, this zone gradually moves in a proximal-distal direction until it reaches the apical part and the nodule degenerates (Monahan-Giovanelli et al., 2006; Van de Velde et al., 2006). Chinese milk vetch is mainly distributed in China, Japan, and Korea, where it is widely planted in rice fields to increase soil fertility (Li et al., 2008).
To investigate whether LTPs function in symbiosis membrane deposition and nodule organogenesis in Chinese milk vetch, we previously identified two candidate LTP genes, AsE246 and AsIB259, via suppressive subtractive hybridization. AsE246 is specifically expressed in nodules, but AsIB259 is expressed in both uninfected roots and nodules, indicating that only AsE246 is specific to nodulation (Chou et al., 2006). Here, we report the functional characterization of AsE246, a nodule-specific lipid transfer protein that localizes on the symbiosome membrane and plays an essential role in symbiosome membrane biogenesis and effective symbiosis.
RESULTS
Phylogenetic Characterization of AsE246
AsE246 has a 399-nucleotide open reading frame that encodes a putative protein of 132 amino acids, containing a 23-amino acid-long N-terminal signal peptide for the secretory pathway. According to a classification scheme based on primary structure (Boutrot et al., 2008), comparison of the deduced sequence of mature AsE246 to LTPs from Arabidopsis, M. truncatula, Lotus japonicus, and Glycine max showed that AsE246 belongs to the Type I LTPs. A further search of the Pfam database (Finn et al., 2010) showed that the Type I LTPs belong to the LTP1 family, also called the “Tryp α amyl” family (Supplemental Fig. S1; Supplemental Data Set S1). By contrast, MtN5 and AsIB259 belong to Type IV and Type V LTPs, respectively, and both are members of the LTP2 family (Supplemental Fig. S1).
AsE246 Has Lipid Binding Activity in Vitro
Protein sequences that contain the conserved eight Cys motif may be annotated as LTPs (Chou et al., 2006), but this is not a sufficient predictor of lipid-binding activity, as the conserved eight-Cys motif and α-helix structure occur in LTPs and in several plant protease and amylase inhibitors, as well as in proteins of unknown function (José-Estanyol et al., 2004).
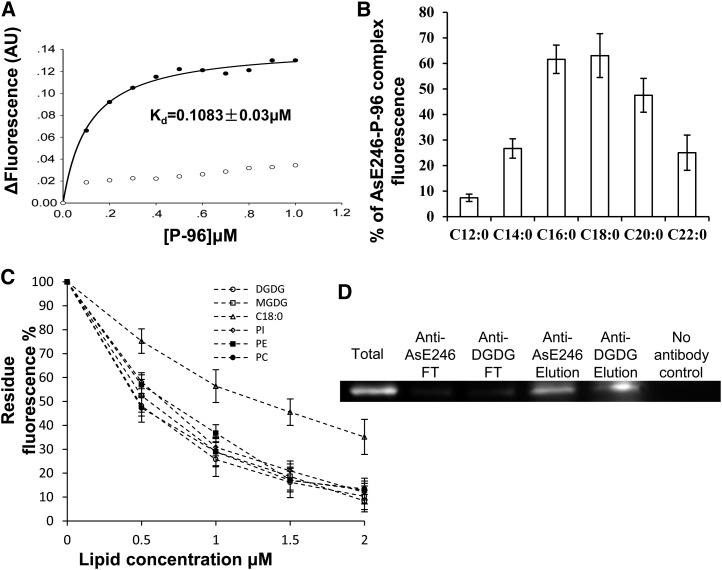
To confirm the lipid-binding capacity of AsE246, we used an in vitro lipid binding assay with purified AsE246 and a fluorescent lipid substrate. To produce a processed, soluble version of AsE246, which we termed ΔAsE246, the complementary DNA lacking the N-terminal signal sequence was expressed in Escherichia coli BL21 and purified using a glutathione S-transferase (GST) tag. To assess potential lipid-binding activity, ΔAsE246 was incubated with the fluorescent lipophilic probe 1-pyrenedodecanoic acid (P-96), which fluoresces weakly in aqueous solution but fluoresces intensely when bound in a hydrophobic environment. We found that, in binding assays with increasing amounts of P-96, fluorescence intensity increased initially and plateaued at greater than 0.5 μmol L–1 P-96 (Fig. 1A). A control experiment with an empty GST tag gave P-96 fluorescence at close to background levels. This result supports the hypothesis that AsE246 binds lipids.
Figure 1.
Lipid binding activity assay of AsE246. A, Association of P-96 with ΔAsE246. Purified ΔAsE246 (final concentration, 0.5 μm) was incubated in 10 mm MOPS, pH 7.2. Increasing amounts of P-96 (from a 30 μm solution in ethanol) were added, and the fluorescence intensity was measured at 378 nm (excitation at 343 nm). ΔAsE246, incubated with increasing concentrations of P-96, exhibited saturation binding defined by Kd = 0.1083 ± 0.03 μm (circles). No saturation binding was observed (squares) when P-96 was incubated with GST in the control experiment. B, Binding competition between P-96 and various unlabeled fatty acids. Purified ΔAsE246 (final concentration 0.5 μm) was incubated in 10 mm MOPS, pH 7.2, with 1 μm P-96 (from a 30 μm solution in ethanol). Various unlabeled fatty acids were added from concentrated solutions in ethanol (final concentration, 10 μm). Fluorescence intensity was measured at 378 nm (excitation at 343 nm) after its stabilization. C, Binding ability competition between P-96 and natural lipid, PC, PE, PI, DGDG, and its precursor MGDG. Purified ΔAsE246 (final concentration, 0.5 μm) was incubated in 10 mm MOPS, pH 7.2, with 1 μm P-96 (from a 30 μm solution in ethanol). Increasing amounts of unlabeled lipids were added from concentrated solutions. D, Co-IP of AsE246 with DGDG. The wild-type Chinese milk vetch nodules inoculated with M. huakuii extracts were incubated first with anti-DGDG antibody and then with immobilized Protein G beads. Proteins bound to the beads were separated on SDS-PAGE and immunoblotted with anti-AsE246 antibody.
We next used a competition test to determine whether common fatty acids bind to ΔAsE246. Addition of a fatty acid that competes with P-96 for ΔAsE246 binding will result in lower fluorescence of P-96 compared with the control with P-96 alone. Several lipids (each at 10 μm) were used as competitors: saturated fatty acids with C12 to C22 chain length reduced the signal from 1 μm P-96 (Fig. 1B). These fatty acids displayed different efficiencies in competing with P-96, depending on their chain length (Fig. 1B). Molecules with a 16- to 18-carbon chain length showed higher competition efficiency, but shorter (lauric and myristic) and longer (arachidic and behenic) fatty acids showed lower competition efficiency.
AsE246 Binds to Biological Membrane Lipid in Vitro and in Vivo
We next used the competition assay to test the capacity of AsE246 to bind to biological membrane lipids, phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), DGDG, and its precursor monogalactosyldiacylglycerol (MGDG; Fig. 1C). Increasing amounts of natural lipid (each to a maximum of 2 μm) were added to a solution of 1 μm P-96 plus 1 μm ΔAsE246. Stearic acid at the same concentration was used as a positive control. We found that the fluorescence decreased with the addition of natural lipid (Fig. 1C), showing that they can compete with the fluorescent lipophilic probe P-96 and indicating that ΔAsE246 can bind to PC, PE, PI, DGDG, and MGDG in vitro.
DGDG was reported to accumulate in the symbiosome membranes of G. max and L. japonicus (Gaude et al., 2004). More importantly, unlike other biological membrane lipids (PC, PE, or PI), DGDG was not present in the free-living rhizobia cells (Gaude et al., 2004). To achieve direct evidence to reveal the functional association and interaction between LTP and plant-synthesized lipids, we chose DGDG to perform coimmunoprecipitation (Co-IP) assay and subcellular localization and such therefore to further confirm the interaction between DGDG and AsE246. To conduct the designed experiment, anti-DGDG antibody was prepared as described in Botté et al. (2008). A nodule extract of wild-type Chinese milk vetch nodules was precipitated with immobilized anti-DGDG antibody or with anti-AsE246 antibody as a positive control. The captured immune complexes were resolved by SDS-PAGE and detected by immunoblotting with anti-AsE246 antibody. It was found that AsE246 was detected in the Co-IP products but not in the negative control without antibody (Fig. 1D). These results show that the nodule-specific lipid transfer protein AsE246 binds the biological membrane lipid DGDG in vivo.
AsE246 Localizes to the Infected Nodule Cells and Symbiosome Membrane
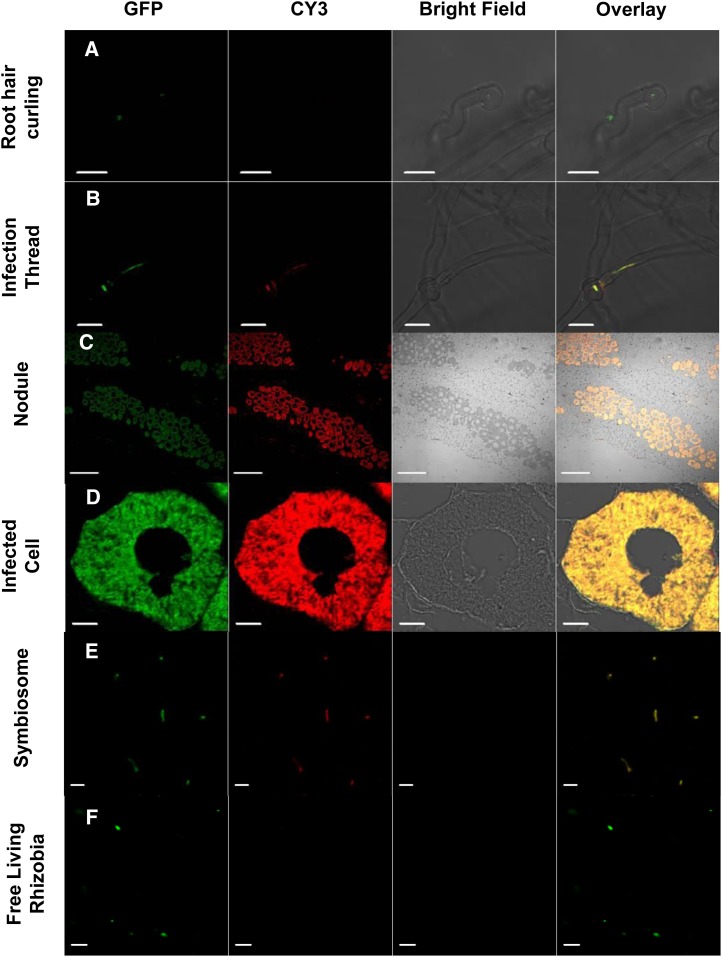
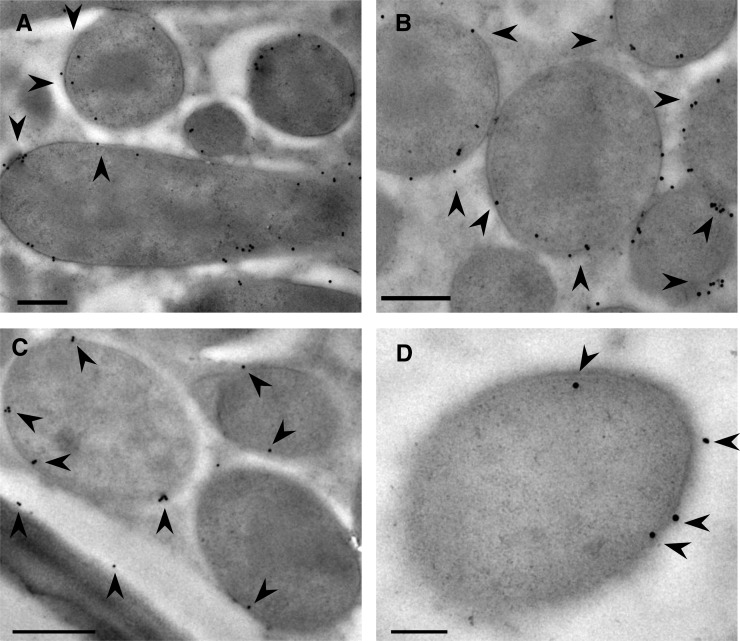
To determine the subcellular location of AsE246 under symbiotic conditions, an immunofluorescence analysis was carried out with wild-type Chinese milk vetch inoculated with GFP-labeled M. huakuii 7653R. The anti-AsE246 antibody is specific for AsE246 (Supplemental Fig. S2). AsE246 was expressed in ITs and infected cells in the nodule but was not detected in root hair curling (Fig. 2, A–D). AsE246 colocalized with symbiosomes (Fig. 2E) but was not detected in free-living rhizobia (Fig. 2F). The secondary antibody controls did not show any labeling in nodules (Supplemental Fig. S3A). Considering that AsE246 could bind to DGDG, a major constituent of the symbiosome membrane (Gaude et al., 2004), it is possible that AsE246 is present on the symbiosome membrane. Immunoelectron microscopy detected AsE246 on the symbiosome membrane (Fig. 3, A and B). The secondary antibody controls for immunoelectron microscopy also did not show any labeling (Supplemental Fig. S3B). These results are consistent with our findings that AsE246 is expressed in nodule cells containing symbiosomes.
Figure 2.
AsE246 is colocalized with symbiosomes in nodules. Immunolocalization of AsE246 in the infected roots of Chinese milk vetch, induced by M. huakuii 7653R, shows colocalization with GFP-labeled 7653R in ITs and nodules but not in root hair curls. A, Immunofluorescence of AsE246 in wild-type hair curling; the rhizobia are expressing GFP, but no CY3 signal for AsE246 is detected. B, Immunolocalization of AsE246 in the IT. The signal for AsE246 is revealed as red dots (secondary antibody tagged with CY3), showing colocalization with rhizobia (yellow color; in the overlay). C, Immunolocalization of anti-AsE246 on nodules. The signal shows only in the infected cells and colocalizes with rhizobia. D, Immunolocalization of AsE246 in infected cells. The signal shows specific colocalization with the nodule symbiosomes. E, Immunolocalization of AsE246 in crushed nodules, which were fixed in 4% (w/v) freshly depolymerized paraformaldehyde in 1× PBS, shows that AsE246 is localized on the symbiosome. F, Immunolocalization of AsE246 in free-living M. huakuii rhizobia as a negative control. Bars = 20 μm (A and B), 200 μm (C), 10 μm (D), and 5 μm (E and F). [See online article for color version of this figure.]
Figure 3.
Ultrastructural localization of AsE246 and DGDG on the symbiosome membrane. A and B, Immunogold detection of anti-AsE246 (immunogold signal appears as black dots; indicated by black arrowheads) in wild-type nodules. The 10-nm gold particles are present over the symbiosome membranes. C and D, Immunogold detection of anti-DGDG (immunogold signal appears as black dots; indicated by black arrowheads) in wild-type nodules. The 10-nm gold particles are present over the symbiosome membranes. Bars = 200 nm (A, B, and D) and 500 nm (C).
The Plant-Synthesized Glycolipid DGDG Occurs on the Chinese Milk Vetch Nodule Symbiosome Membrane
DGDG accumulates in the symbiosome membranes of G. max and L. japonicus (Gaude et al., 2004), both of which form determinate-type root nodules, while Chinese milk vetch forms indeterminate-type root nodules. Therefore, we analyzed the DGDG content of the Chinese milk vetch symbiosome by using liquid chromatography-mass spectrometry (LC-MS) and confirmed its presence occurring on the symbiosome membrane through immunolocalization methods.
The symbiosomes were isolated from Chinese milk vetch nodules, and LC-MS confirmed that the total lipids extracted from the symbiosomes and nodules contained DGDG (Supplemental Figs. S4 and S5). The LC-MS data also showed the DGDG species in the symbiosome were different from those in nodules and leaves. The result showed that in Chinese milk vetch nodules, the symbiosome contains the plant-synthesized glycolipid DGDG.
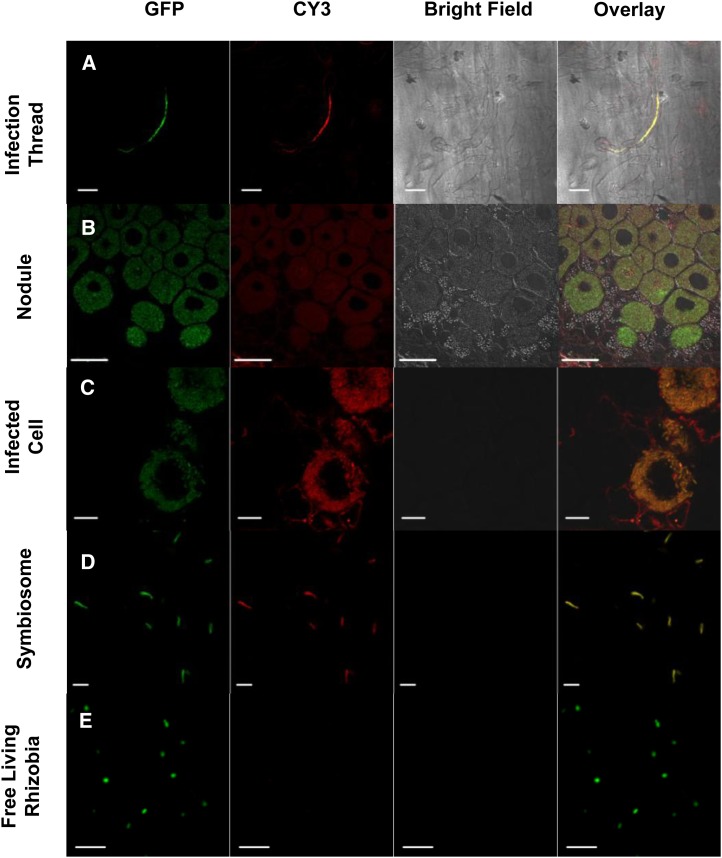
The immunofluorescence was carried out in the wild-type Chinese milk vetch inoculated with GFP-labeled M. huakuii 7653R and showed a weak anti-DGDG signal on the plasma membrane of the host plant cell (Fig. 4A) and in ITs and nodules. The signal was present in both infected and uninfected cells and colocalized with rhizobia in the infected cells (Fig. 4, B and C). The anti-DGDG signal was also detected on the symbiosomes. To examine the symbiosomes, crushed nodules were fixed with 1% (w/v) freshly depolymerized paraformaldehyde; immunolocalization showed that the anti-DGDG signal colocalized with the symbiosomes (Fig. 4D) but was not detected in the free-living M. huakuii control (Fig. 5E). To determine whether DGDG localized on the symbiosome membrane, we examined the ultrastructural localization of DGDG by electron microscopy (EM). EM immunogold labeling with an anti-DGDG antibody was used to localize DGDG in wild-type nodules (Fig. 3, C and D), and the result showed that plant-synthesized glycolipid DGDG occurs on the Chinese milk vetch nodule symbiosome membrane.
Figure 4.
DGDG is localized on the plasma membrane and colocalized with the symbiosome in infected cells. Immunolocalization of DGDG over the infected roots of Chinese milk vetch induced by M. huakuii 7653R shows colocalization with GFP-labeled 7653R in ITs and nodules but not with free-living M. huakuii. A, Immunofluorescence of DGDG on the IT; the rhizobia are expressing GFP, and the signal for DGDG is revealed as red dots (secondary antibody tagged with CY3). CY3 signal is detected both on root hair plasma membrane and colocalized with rhizobia (yellow color; in the overlay). B and C, Immunolocalization of DGDG on nodules. The signal shows in both infected and uninfected cells and colocalizes with rhizobia in the infected cells. D, Immunolocalization of DGDG in crushed nodules, which were fixed in 4% (w/v) of freshly depolymerized paraformaldehyde in 1× PBS. The signal shows that DGDG is localized on the symbiosome. E, Immunolocalization of DGDG on free-living M. huakuii rhizobia as a negative control shows that the free-living rhizobia do not contain DGDG. Bars = 20 μm (A and C), 50 μm (B), and 5 μm (D and E). [See online article for color version of this figure.]
Figure 5.
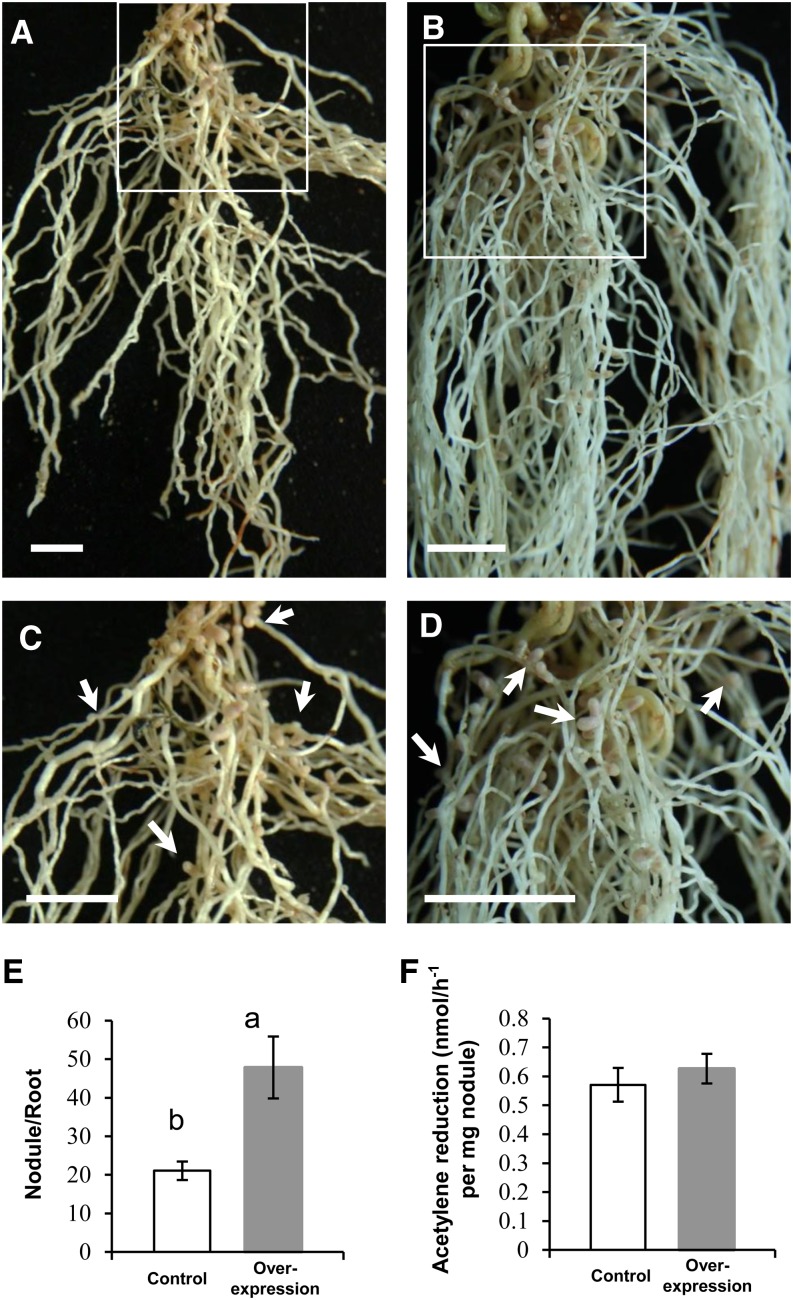
Overexpression of AsE246 results in increased numbers of nodules. A, The empty vector control hair root, inoculated with M. huakuii 7653R and grown in the absence of nitrogen. The photographs were taken 4 weeks after inoculation. B, AsE246-overexpressing hairy root inoculated with M. huakuii 7653R, showing the increase in nodule numbers. C, Close-up of boxed area in A. The nodules are indicated by white arrows. D, Close-up of boxed area in B. E, Numbers of nodules per root. The nodule number of AsE246-overexpressing roots (n = 17) is more than that of the empty vector control. F, The nitrogenase activity of AsE246 overexpressing and control nodules showed no remarkable difference. Different letters above bars indicate significant differences (P < 0.05, Student’s t test) between pairwise comparisons. Bars = 1 cm. [See online article for color version of this figure.]
Overexpression of AsE246 Results in Increased Numbers of Root Nodules
To investigate the role of AsE246 in symbiosis, we overexpressed AsE246 under the control of the 35S promoter. Expression levels of AsE246 in transgenic roots and nodules were examined by real-time PCR (Supplemental Fig. S6A). The level of AsE246 transcripts in transgenic nodules was about 2 to 3 times that in control nodules. AsE246 transcripts were also expressed in transgenic roots but were not detected in wild-type and control roots. The AsE246-overexpressing transgenic roots formed more root nodules (an average of 47.8, n = 17) than the empty vector control (an average of 21.1, n = 13; Fig. 5, A–E). Nodules in AsE246-overexpressing transgenic roots and empty vector controls showed similar nitrogenase activities (Fig. 5F).
Knockdown of AsE246 Impairs Nodulation and Symbiosome Development
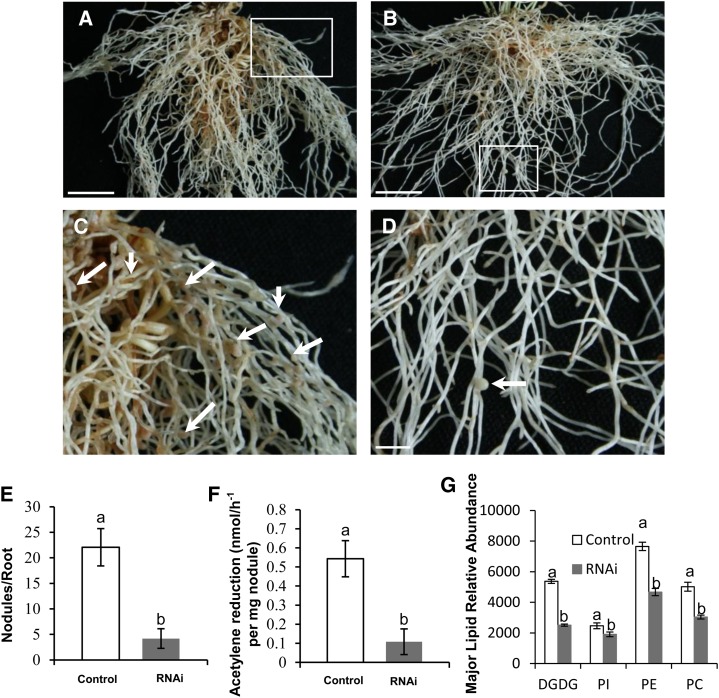
To further investigate the role of AsE246 in symbiosome development, we knocked down the expression of AsE246 using RNA interference (RNAi). Nodule RNA sequencing studies showed that the RNAi fragment used in this construct was unique (data not shown), indicating that the RNAi construct would specifically silence AsE246. Real-time PCR showed that AsE246 transcript levels in the transgenic nodules were 16.2% of levels in the control nodules (Supplemental Fig. S6B). The average number of nodules formed on control hairy roots was 22.1 (n = 35) but that on the RNAi plants was 4.2 (n = 47; Fig. 6, A–E). In addition, 14.9% of RNAi roots failed to develop any nodules. Compared with the empty vector control, the RNAi nodules were smaller and whiter (Fig. 6D), with much lower nitrogenase activity (Fig. 6F).
Figure 6.
Nodulation phenotype of AsE246-RNAi hair root. A, The empty vector control hair root, inoculated with M. huakuii 7653R and grown in the absence of nitrogen. The photographs were taken 4 weeks after inoculation. B, AsE246 RNAi hair root inoculated with M. huakuii 7653R formed fewer root nodules compared with control, and the nodules are also white colored. C, Close-up of boxed area in A. The nodules are indicated by white arrows. D, Close-up of boxed area in B. The nodules are indicated by white arrows. E, Numbers of nodules per root. There are fewer nodules in RNAi root (n = 47) compared with the empty control (n = 35). F, The nitrogenase activity of the AsE246-RNAi nodule is lower than that of the control nodule. Different letters above bars indicate significant differences (P < 0.05, Student’s t test) between pairwise comparisons. G, Lipid PC, PE, PI, and DGDG contents of transgenic nodule. Different letters above bars indicate significant differences (P < 0.05, Student’s t test) between pairwise comparisons. Bar = 1 cm. [See online article for color version of this figure.]
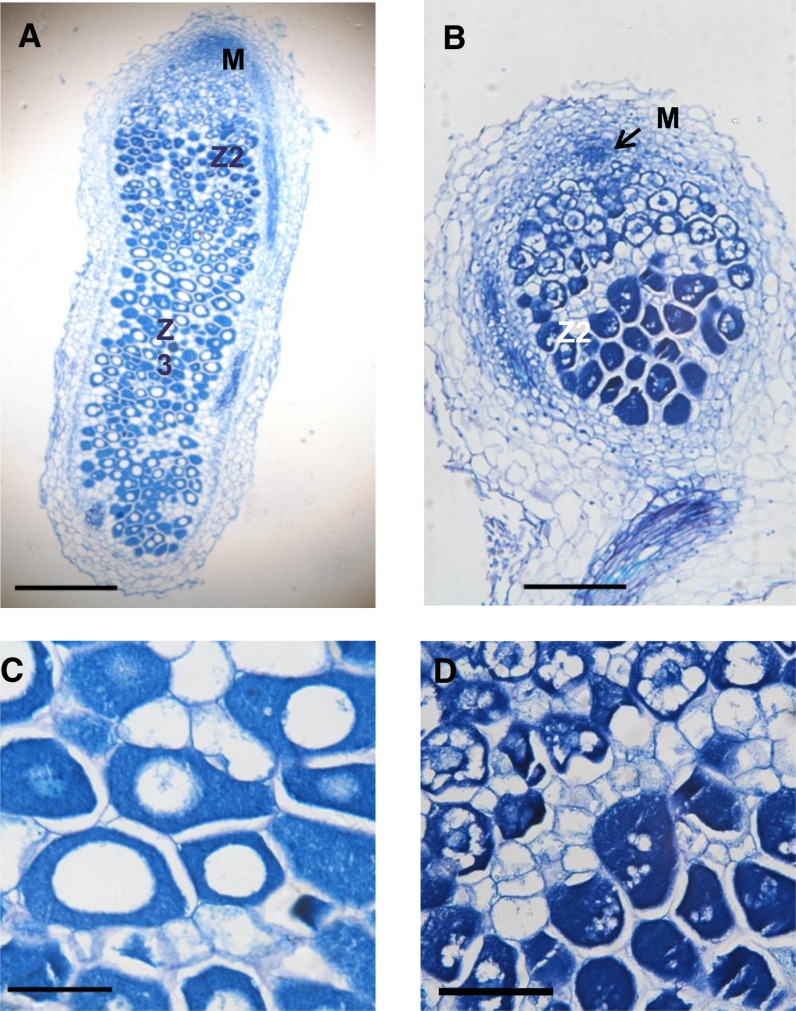
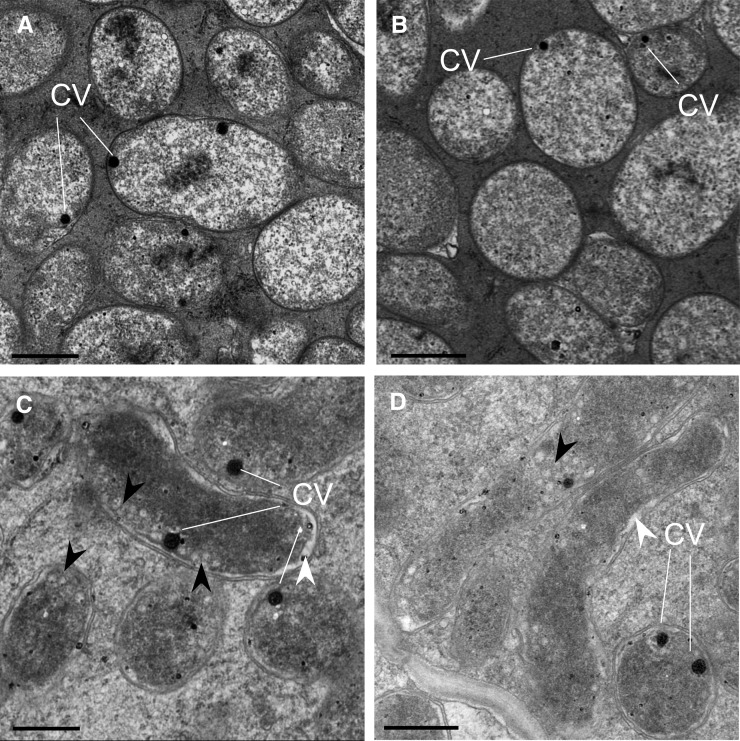
We also examined the structure of the nodules formed in AsE246 RNAi plants. Paraffin sections showed that RNAi nodules at 21 d after infection had fewer matured infected cells, as infected cell development in these nodules remained in zone II (infection zone; Fig. 7, B and D), and the mature fixation zone III was not observed. Furthermore, transmission electron microscopy (TEM) showed obvious shrinkage and peribacteroid space enlargement in the symbiosomes of RNAi nodules (Fig. 8, C and D, white arrows). There were many more unstained granules of poly-β-hydroxybutyrate (PHB) in the symbiosomes of RNAi nodules (Fig. 8, C and D, black arrows). By contrast, the symbiosomes of control nodules developed normal cellular morphology with a smoothly rounded boundary and more homogeneous cytoplasm (Fig. 8, A and B). TEM also showed that, for both the nodules formed in the empty vector and RNAi plants, there were coated vesicles over the symbiosomes.
Figure 7.
AsE246-RNAi impairs nodule development. A, Longitudinal section of a control (empty vector) 21-d-old nodule. B, Longitudinal section of a 21-d-old RNAi nodule, showing the infected cells remain in the meristem and infection zone. C, Magnification of nitrogen fixation zone in A, showing developed nitrogen-fixing symbiosomes. D, Magnification in B. M, Meristem; Z2, infection zone; Z3, nitrogen fixation zone. Bars = 500 μm (A), 200 μm (B), and 100 μm in (C and D). [See online article for color version of this figure.]
Figure 8.
AsE246-RNAi impairs symbiosome development. A and B, TEM images of symbiosomes of the control nodule. CV, Coated vesicle. C and D, TEM images of symbiosomes of the AsE246-RNAi nodule. There is obvious shrinkage and peribacteroid space enlargement in the symbiosomes of the RNAi nodule (white arrowheads) and PHB accumulation (black arrowheads). Bars = 500 nm.
Knockdown of AsE246 Affects Lipids Abundance in the Nodule
The lipid abundance in transgenic nodules was measured using LC-MS. The results showed that PC, PE, PI, and DGDG contents were significantly lower in AsE246 RNAi nodules than in controls, manifested by their corresponding contents in the RNAi nodules, which were 60.62%, 61.00%, 77.28%, and 46.74% of that in the control nodules, respectively (Fig. 6G; Supplemental Data Set S2). These data support the idea that AsE246 is involved in lipids transport to the nodule symbiosome membrane.
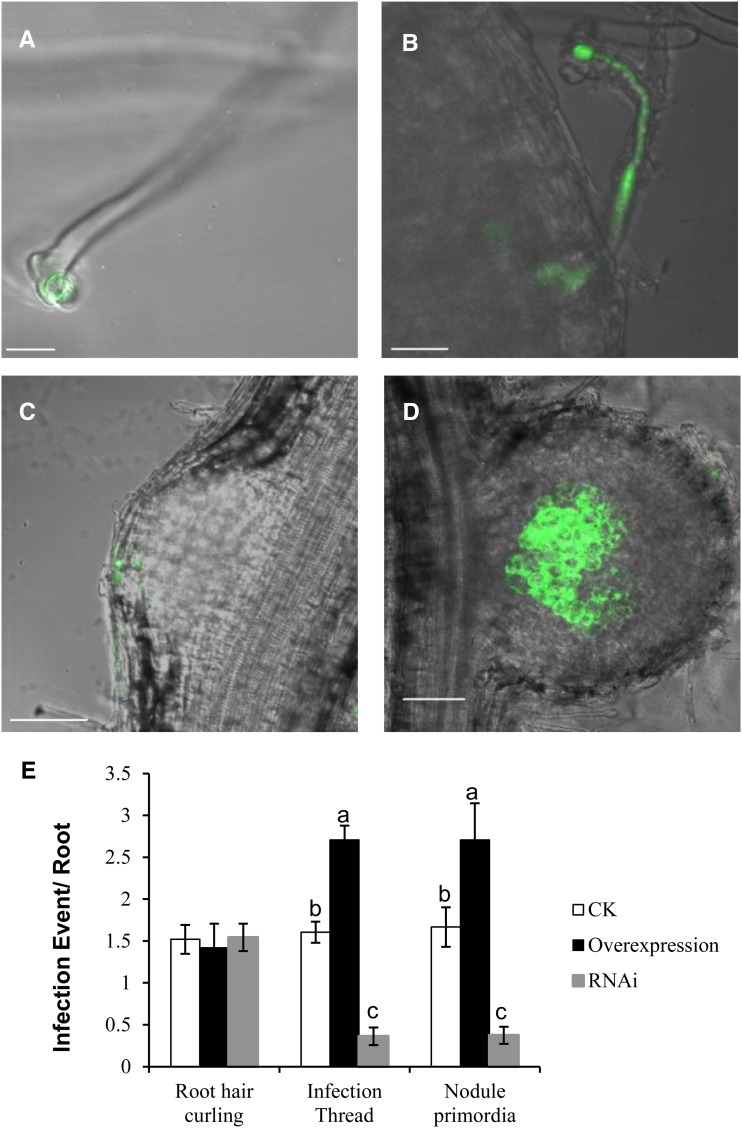
AsE246 Is Required for IT Formation
Rhizobial infection of transgenic roots was also monitored and quantified (Fig. 9) using GFP-labeled M. huakuii 7653R to visualize formation of ITs at 9 d after infection. In AsE246 RNAi roots, ITs initiated from curled root hair tips (Fig. 9F), grew through well-elongated root hairs, and formed ITs (Fig. 9B). These further penetrated into the root cortex and formed nodule primordia (Fig. 9C) and then developed into nodules (Fig. 9D). However, there were significantly fewer ITs and nodule primordia in RNAi roots compared with control roots; also in the overexpression roots, there were more ITs and nodule primordia (Fig. 9E). These data indicated that AsE246 participates in IT formation but not in root hair curling.
Figure 9.
Reduction of AsE246 affects rhizobia infection. A to D, Infection event observations of GFP-labeled M. huakuii 7653R strain to track the infection events at 9 d after infection. Representative root hair curling observed in (empty vector) control roots: root hair curling (A), IT (B), nodule primordia (C), and nodule (D). E, Frequencies of infection events per root. The data are presented as 16 individual transgenic plants for each construct. Different letters above bars indicate significant differences (P < 0.05, Student’s t test) between pairwise comparisons. Bars = 20 µm. [See online article for color version of this figure.]
DISCUSSION
AsE246 is specifically expressed in Chinese milk vetch nodules (Chou et al., 2006) and encodes a protein with a conserved eight-Cys motif characteristic of LTPs. Further phylogenetic analysis showed AsE246 belongs to the LTP1 family, also called the “Tryp α amyl” family, and is not homologous to another symbiosis-associated LTP, MtN5, a member of the LTP2 family (Chou et al., 2006; Pii et al., 2009). Various in vivo biological roles have been proposed for plant LTPs, including defense against pathogens and modulation of plant development.
In our search for the lipid transport machinery required for symbiosome development in legume plant nodules, we examined the involvement of LTPs. LTPs are abundantly expressed in epidermal cells, secreted to the extracellular matrix (Carvalho et al., 2007), and capable of binding and carrying lipids in vitro (Kader et al., 1984). These characteristics provide the rationale for the proposal that LTPs are potential carriers of lipid constituents to the nodule symbiosomes. In this study, we provide evidence that one member of the LTP family in Chinese milk vetch, AsE246, participates in symbiosome membrane lipid transport and is required for successful legume-rhizobium symbiosis, including infection and bacteroid maintenance.
The expression of AsE246 without a signal peptide in an E. coli system allowed us to purify AsE246 and assess its lipid binding capacity. As reported for several other LTPs (Buhot et al., 2004; Debono et al., 2009; Pii et al., 2009), AsE246 can bind to the fluorescent lipid probe P-96. P-96 binding to ΔAsE246 could be displaced by saturated fatty acids and biological glycolipids, including PC, PE, PI, DGDG, and its precursor MGDG, suggesting that lipids could compete with P-96 for the same hydrophobic binding cavity. The Co-IP experiment revealed that AsE246 could bind with DGDG to form a complex in vivo.
The immunofluorescence and immunoelectron microscopy images showed that AsE246 was expressed in the IT and nodule-infected cells and localized on the symbiosome membrane. This is quite similar to the symbiosome membrane-specific protein SNARE SYP132, which is present on the symbiosome membrane from the start of symbiosome formation and throughout its development (Catalano et al., 2007; Limpens et al., 2009). These results indicate that AsE246 might be involved in the targeting of plant-synthesized lipid during IT and symbiosome development, which require reorganization and de novo formation of membranes. Therefore, AsE246 can affect lipid homeostasis and the diverse cellular processes associated with it, such as signal transduction, membrane trafficking, and lipid metabolism, by capturing and responding to symbiosome membrane modifications.
In contrast with the empty vector control, the AsE246 RNAi roots formed fewer and smaller nodules, and there were fewer infected cells in the RNAi nodules. TEM showed obvious symbiosome shrinkage and peribacteroid space enlargement in the symbiosomes of the RNAi nodule. This suggests that the reduction of AsE246 led to changes in membrane permeability and caused symbiosomes to be more sensitive to the osmotic pressure in nodules; this abnormal development of the symbiosome membrane led to a significant decrease in nitrogen fixation ability of RNAi nodules. TEM also showed some coated vesicles over the symbiosome membrane, both in the control and RNAi nodules. This indicates that vesicular transport to the symbiosome was not affected by the reduction of AsE246.
The accumulation of PHB in AsE246-RNAi nodules is also of interest for analysis. As an indeterminate-type nodule formation legume plant, Chinese milk vetch (similar to alfalfa [Medicago sativa] interacting with Sinorhizobium meliloti [Hirsch et al., 1983]) does not usually accumulate PHB during symbiosis with M. huakuii 7653R. PHB synthesis does occur but is presumably accompanied by an equivalent rate of degradation during normal bacteroid differentiation, which leads to only a very small accumulation of PHB (Kim and Copeland, 1996). This amount is insufficient to allow for the formation of granules that are visible by EM. A study of Rhizobium leguminosarum bv viciae mutations in two genes encoding broad-specificity amino acid transporters, mutations that block the amino acid cycling pathway between the plant and the symbiosomes, showed that these mutants possessed PHB granules in mature symbiosomes (Lodwig et al., 2003; Trainer and Charles, 2006). We speculate that the knockdown of AsE246 led to changes in symbiosome membrane structure and function and also may have blocked the amino acid cycling pathway between the plant and symbiosomes.
The knockdown of AsE246 resulted in reduced PC, PE, PI, and DGDG contents of transgenic nodules. This result supported the idea that AsE246 participates in the transport of plant-synthesized lipid to the symbiosome membrane. In this case, the lipid contents in the symbiosomes of transgenic nodules was not examined because of technical difficulties caused by the low number of nodules formed in the RNAi plants.
AsE246 also plays an important role in rhizobia infection. The knockdown of AsE246 resulted in fewer ITs, nodule primordia, and nodules; overexpression of AsE246 resulted in more ITs and nodule primordia. Moreover, immunofluorescence showed that AsE246 colocalized with rhizobia in the IT. Considering that the symbiosome membrane is derived from the IT (Whitehead and Day, 1997), we conclude that AsE246 is also required for IT formation.
M. truncatula MtN5 encodes an LTP and is required for the symbiotic association between M. truncatula and the nitrogen-fixing rhizobial bacteria S. meliloti (Pii et al., 2009). However, the LTP mechanisms that underlie the symbiotic interaction have not been fully unraveled. Our results showing that a nodule-only LTP transports the plant-synthesized lipid including DGDG and targets to the symbiosome membranes, even in the ITs, raise deep questions concerning the DGDG biological functions associated with progression of symbiosis. Accumulation of DGDG, instead of phospholipids, in the symbiosome membrane might have two major functions. First, it might be a host plant protection strategy to facilitate rhizobia entry, differentiation, and functional maintenance inside the target host plant cells by avoiding host defense responses, which otherwise may interfere with normal metabolism of the host cell and provoke host defense responses. For example, there is a significant linear relationship between the amounts of MGDG and DGDG in noninoculated wheat leaves and the level of resistance to tan spot, which is caused by the fungus Pyrenophora triticirepentis (Kim et al., 2013). These findings suggest that the amount of DGDG in plant cells affects the resistance response to pathogens. The infection of rhizobia and their subsequent existence within the host plant cell are quite similar to phytopathogen infection and even to infection with some intracellular mammalian pathogens (i.e. Mycobacterium tuberculosis and Salmonella spp.; Schumpp and Deakin, 2010). Thus, DGDG in the symbiosome membrane (Figs. 5–7; Gaude et al., 2004) might be an alternative defense response to invading rhizobia and facilitate the intracellular symbiosis between the plant and the rhizobia.
Second, the accumulation of the nonphosphorus galactolipid DGDG, instead of phospholipids, in the symbiosome membrane might conserve phosphate for essential cellular processes during nodulation. Plants and algae synthesize DGDG in plastid envelope membranes (Maréchal et al., 1997, 2000; Awai et al., 2001; Jouhet et al., 2004, 2007; Benning and Ohta, 2005). Upon phosphate deprivation, DGDG is exported to the plasma membrane (Andersson et al., 2003) and the mitochondria (Jouhet et al., 2004). During nodule development, the number of rhizobia in infected cells increases dramatically. This cell division requires membrane biosynthesis by the bacteria and also by the plant for synthesis of the symbiosome membrane to enclose the bacteria in symbiosomes (Verma, 1992). As a consequence, nodulation requires the uptake of large amounts of phosphate for the synthesis of phospholipids, nucleic acids, phosphorylated proteins, and sugar phosphates. It is possible that cells infected with bacteroids might suffer from local phosphate limitation. Previous studies have demonstrated that phosphate availability is crucial for normal nodulation (Israel, 1987; Leidi and Rodriguez-Navarro, 2000). Thus, the presence of the nonphosphorus galactolipid DGDG might conserve phosphate for host plant metabolism (Gaude et al., 2004).
In conclusion, the novel LTP AsE246 may participate in the transport of the plant-synthesized lipid to the symbiosome membrane and is involved in legume-rhizobia symbiosis. These results increase our understanding of the molecular and physiological mechanisms of legume nodule organogenesis and symbiosome development.
MATERIALS AND METHODS
Phylogenetic Analysis
The Medicago truncatula genome database (assembly release 3.5; Young et al., 2011) was searched for nsLTP gene sequences using the gene annotations. The entire Glycine max and Lotus japonicus (release 2.5) proteome was searched for proteins having the Hidden Markov Model Pfam domain PF00234 (Plant lipid transfer/seed storage/trypsin-α amylase inhibitor).
Protein sequences of all nsLTPs were analyzed for the presence of potential signal peptide cleavage sites using the SignalP 4.0 program (Finn et al., 2010). Amino acid sequences were efficiently aligned to the Pfam profile Hidden Markov model defined from the protease inhibitor/seed storage/LTP family (Finn et al., 2010) using HMMalign from the HMMER package (Eddy, 1998).
Deduced M. truncatula, L. japonicus, G. max, and Arabidopsis (Arabidopsis thaliana) amino acid sequences were aligned to the Pfam glocal model using HMMalign from the HMMER package (Eddy, 1998). Phylogenetic trees were built from the protein alignment with the maximum-likelihood method using the PHYML program (Guindon and Gascuel, 2003). Maximum-likelihood inference analyses were conducted under the Le and Gascuel model (Le and Gascuel, 2008) with estimation of the proportion of invariant sites and estimation of variation rate among the remaining sites according to a γ distribution. The confidence level of each node was estimated by bootstrap procedure using 1,000 resampling repetitions of the data. The unrooted phylogenetic trees were visualized using Mega 5 (Tamura et al., 2011).
Plant Materials
Seeds of Chinese milk vetch (Astralagus sinicus; collected from Xinyang, Henan Province, China) were surface sterilized in 70% (v/v) alcohol for 3 min, then in 2% (w/v) sodium hypochlorite for 20 min, followed by five washes in distilled water. After immersion in distilled water for several hours, the seeds were placed on medium with 0.5% Suc and 1.2% (w/v) agar (Cho and Widholm, 2002).
Isolation of Symbiosomes from Chinese Milk Vetch
Approximately 10 g of fresh nodules from Chinese milk vetch plants was used for the isolation of symbiosomes via three-step Percoll density gradient centrifugation (Price et al., 1987; Panter et al., 2000). Intact symbiosomes were harvested from the 60%/80% Percoll interface. After washing with washing buffer (350 mm mannitol, 25 mm MES-KOH, 3 mm MgSO4, pH 7.0), the suspension was centrifuged at 4,000g for 5 min at 4°C. The pellet containing symbiosomes was resuspended in washing buffer and vortexed vigorously for 5 min.
Lipid Extraction and Quantification
Lipids were isolated from frozen plant material, rhizobia (isolated from liquid culture), or symbiosomes (Bligh and Dyer, 1959). To determine the lipid composition changes of transgenic nodule, the 35-d-after-infection transgenic nodules were harvested (100 mg) for lipid extraction and profiling. Samples were homogenized using liquid nitrogen and resuspended in 50 mL chloroform and 100 mL methanol, which was then sonicated two to three times. Then, to the mixture is added 50 mL chloroform and 50 mL distilled, deionized water, then transferred to a separating funnel, and after a few minutes for complete separation and clarification. The lower chloroform phase was dried using rotary evaporation, and the lipid extract was dissolved in methanol. The lipid extracts were introduced into the Quadrupole Time-of-Flight (Q-TOF) mass spectrometer (Agilent 6540 Accurate Mass Q-TOF LC-MS unit) operated in the positive mode. Lipids were supplied to the mass spectrometer in methanol. Q-TOF LC-MS was performed using the Agilent 6540 UHD accurate-mass Q-TOF LC-MS system with a ZORBAX Eclipse Plus C18 column (2.1 × 100 mm, 3.5 μm). The 1-μL sample was loaded with a flow rate of 0.3 mL min–1 A (water plus 5% methanol plus 10 mm NH4AcOA), B (methanol); solution B was at 25% (v/v) at the start, 100% (v/v) at 5 min, 100% (v/v) at 10 min, and 25% (v/v) again at 15 min. The acquisition range (m/z) was 400 to 1,100; nebulizer pressure, 40 pounds per square inch gauge; drying gas, N2 350°C 9 L min–1; electrospray ionization voltage of capillary, 4,000 V; fragment or 214v; skimmer, 65 V; and octopole radio frequency programming mode voltage, 750 V. MGDG and DGDG were analyzed as [M+NH4]+ and [M+Na]+ ions (Wu and Xue, 2010).
Nitrogenase Activity Measurement
Nitrogenase activity was measured by acetylene reduction activity (Hardy et al., 1973). For each sample at each time point, nine hairy root lines were analyzed. Hypogeal parts of hairy root plants (including nodules and roots) were incubated in 15-mL glass bottles with rubber seals containing 2 mL of acetylene (C2H2) for 1 h at 28°C. The C2H2 was measured in a Hitachi 163 gas chromatograph.
Protein Expression and Purification
AsE246 coding sequence (accession no. DQ199648) was amplified from complementary DNA obtained by reverse transcribing mRNA extracted from Chinese milk vetch nodules. The upstream primer was 5′-GGATCCAAACAATTAGTTGCCGT-3′ and the downstream primer 5′-CTCGAGTTAGACCAACAAAGTTTG-3′. The PCR product corresponding to the coding sequence of mature AsE246 was double digested with XhoI and BamHI, cloned into pGEX-6p-1 (GE), and checked by sequencing. The recombinant vector pGEX-246 was mobilized into the host strain Escherichia coli BL21 DE3. AsE246 protein was purified from E. coli lysate using a GST cartridge (Bio-Rad).
Lipid Binding Assay in Vitro
Lipid binding capacity of AsE246 was assayed by monitoring the displacement of the fluorescent probe P-96. Fluorescence experiments were performed at 25°C in a Shimadzu RF 5301 PC spectrofluorimeter, as previously described (Mikes et al., 1998; Osman et al., 2001), with minor modifications. The excitation and emission wavelengths were set at 343 and 378 nm, respectively.
P-96, with or without fatty acids, was incubated for 1 min in a stirred cuvette containing 2 mL of measurement buffer (10 mm MOPS, pH 7.2) before the fluorescence was recorded (F0). Then, AsE246 was added, and after 2 min, fluorescence was recorded at equilibrium (F). Results are expressed either as percentage of AsE246-P-96 complex fluorescence according to (F–Fc)/F, where Fc is the fluorescence of the AsE246-P-96 complex in absence of fatty acid, or as fluorescence (F, F0, F), in arbitrary units.
Antibody Preparation
AsE246 protein was purified from inclusion bodies using strong denaturing conditions (20 mm Tris HCl, pH 8.0; 0.5 m NaCl; 5 mm imidazole; and 6 m guanidine hydrochloride) and loaded on an immobilized metal affinity chromatography cartridge. AsE246 was refolded, applying a linear gradient from 6 to 0 m guanidine hydrochloride, and eluted using 250 mm imidazole. The recombinant protein was used to produce polyclonal antibodies in rabbit. Pure DGDG (Avanti) used for immunization in New Zealand white rabbits as described Botté et al. (2008).
Immunofluorescence Experiments
Nodules were hand sectioned using a double-edged razor blade. Nodule sections or roots were fixed in 1% (w/v) freshly depolymerized paraformaldehyde in 1× phosphate-buffered saline (PBS), pH 7.4, for 30 min at 4°C. Nodule sections were blocked in 3% (w/v) bovine serum albumin and further incubated in primary antibody overnight at 4°C in 1× PBS containing 0.3% (v/v) Triton X-100. The secondary antibodies CY3 (a red fluorescence dye)-conjugated goat anti-Rabbit were used. Controls were carried out in the absence of primary antibodies. Nodule sections containing GFP-labeled Mesorhizobium huakuii 7653R were examined by confocal microscopy. Primary antibody dilutions were prepared as follows: anti-AsE246 rabbit, 1:1,000; goat anti-rabbit (CY3), 1:1,000.
Sample Preparation for EM
Nodules were fixed in 4% (w/v) of freshly depolymerized paraformaldehyde, 0.4% (v/v) glutaraldehyde in 1× PBS, pH 7.4, for 1 h at 4°C. Nodules were treated with a graded ethanol series for dehydration, embedded using LR White embedding kit (Fluka), and polymerized at 50°C for 24 h.
EM Immunodetection
Thin sections (60 nm) were cut using a Leica Ultracut microtome. Nickel grids with sections were blocked in 2% (w/v) bovine serum albumin in PBS. Grids were incubated overnight at 4°C with the primary antibody according to dilutions given above. Goat anti-rabbit coupled with 10-nm gold (1:1,000 dilution) was used as the secondary antibody. The sections were examined using TEM (Hitachi, H-7650).
Construction of the Overexpression Plasmid
The open reading frame of AsE246 was amplified by reverse transcription-PCR using the following primers: 5′-GGATCCATGAAATTTGCATATGTGGTTGTG-3′, containing a BamHI site at the 5′ end, and 5′-CCCGGGCTAGACCAACAAAGTTTGAAGT-3′, containing a SmaI site at the 5′ end. The amplification product was digested with BamHI-SmaI and ligated into the pBI121 vector. This AsE246 construct was placed behind the Cauliflower mosaic virus 35S promoter. The construct was transferred into Agrobacterium rhizogenes K599 by electroporation.
Construction of the RNAi Plasmid
A 225-bp fragment of the 3′ untranslated region with a short part of the coding region of AsE246 was amplified by reverse transcription-PCR using the following primers: 5′-GAAGCAATTAAGAGTTTCAAAGTTGT-3′, containing a SmaI or PstΙ site at the 5′ end, and 5′-CCAAATCATTTATTTAATGGCATG-3′, containing a BamHI or SalΙ site at the 5′ end. The amplification products were digested with SmaI-BamHI or with PstΙ-SalΙ and ligated into the pCAMBIA1301-35S-int-T7 plasmid vector (gifted by Dr. Da Luo, School of Life Science, Sun Yat-sen University), in which the sense and antisense AsE246 RNA sequences were located in tandem with the Arabidopsis actin11 intron between them, and this RNAi construct was placed behind the Cauliflower mosaic virus 35S promoter. The constructs were transferred into A. rhizogenes K599 by electroporation.
Co-IP
The Chinese milk vetch nodules were extracted using native extraction buffer. The native extraction buffer contains 50 mm HEPES, 70 mm potassium acetate, 1 mm sodium fluoride, 20 mm β-glycerophosphate, 5 mm magnesium acetate, 0.3% (v/v) Triton X-100, 10% (v/v) glycerol, pH 7.4, and protease inhibitor cocktail Complete Mini Tablets (Roche). Nodules were ground with liquid nitrogen to a fine powder, and then 6 mL extraction buffer was added for each 1-g nodule sample, which was then sonicated one to two times. The samples were centrifuged at 10,000g for 10 min at 4°C, and the supernatant was removed and filtered through a 0.45 μm filter before addition of antibody. Anti-DGDG antibodies were added to the extract (10 μg mL–1 proteins), which was incubated with rotation overnight at 4°C. Protein G agarose beads (GE) were added to the mixture at 20 mg mL–1 to immobilize the immune complex. After gentle shaking at 4°C for 3 h, the agarose beads were recovered by centrifugation at 4,000 rpm for 1 min and washed three times with cold PBS.
Plant Transformation
A. rhizogenes-mediated plant transformation was carried out as described by Cho and Widholm (2002). Briefly, A. rhizogenes K599 lines containing pRNAi, pOVEREXPRESSION, empty pBI121, or pCAMBIA1301-35S-int-T7 were cultured in 20 mL of Luria-Bertani medium containing 100 μg mL–1 kanamycin until the culture reached an optical density at 600 nm of approximately 1.0. Sterilized 5-d-old seedlings were cut in the middle of the hypocotyl and incubated in the bacterial suspension culture for 10 min. After blotting with autoclaved filter paper, the seedlings were placed on Murashige and Skoog basal medium and grown in daily conditions of 16-h/8-h light/darkness at 24°C/20°C, respectively, for cocultivation. Three days later, the explants were transferred to fresh Murashige and Skoog medium with 500 mg L–1 carbenicillin (Duchefa) and 30 mg L–1 kanamycin (Duchefa) and grown for 10 more days until hairy roots developed from hypocotyls. For selection of transgenic hairy roots, root tips (2 to 3 mm) were excised for GUS staining overnight at 37°C in 100 mm sodium phosphate, pH 7.0, 0.1% (v/v) Triton X-100, 0.1% (w/v) N-laurylsarcosine, 10 mm EDTA, 1 mm K3Fe(CN)6, 1 mm K4Fe(CN)6, and 0.5 mg mL–1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid. The remaining portion of the hairy root (20 to 30 mm long) attached to the seedling was labeled. If its tip was GUS negative, the whole hairy root was discarded. If the hairy root tip was GUS positive, the remaining portion of the hairy root was saved. One to three transgenic hairy roots were saved for each seedling. Plants harboring transgenic hairy roots were transferred to pots filled with vermiculite and sand (1:1) and grown in a chamber in a 16-h/8-h day/night cycle at 22°C. After 5 to 7 d, plants were inoculated with M. huakuii 7653R. Nodule number was scored 4 weeks after inoculation.
Nodulation of Hairy Roots
Three weeks after transformation, plants with well-developed hairy roots were transferred to aeroponics chambers with sterile sand for nodulation tests. Transgenic plantlets were grown under the same growth conditions as described earlier. The plantlets were watered with Fahraeus nitrogen-free nutrient solution. After being starved of nitrogen for 7 d, the plantlets were inoculated with M. huakuii 7653R. Nodules were collected at various times for paraffin-embedded section slides, EM, and nitrogenase activity detection.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ199648.1 (AsE246) and DQ199649.1 (AsIB259).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. AsE246 phylogenetic relationships.
Supplemental Figure S2. Immunoblot analysis of anti-AsE246 antibody.
Supplemental Figure S3. Immunolocalization control in wild-type plant nodule.
Supplemental Figure S4. The symbiosome membrane contains DGDG.
Supplemental Figure S5. Electrospray ionization quadrupole MS precursor ion spectra acquired on extracts of A. sinicus.
Supplemental Figure S6. Quantification of AsE246 expression levels in transgenic roots and nodules.
Supplemental Data Set S1. Multiple sequence alignment of LTPs from genome-sequenced legume and Arabidopsis.
Supplemental Data Set S2. Different lipids species content in transgenic A. sinicus nodules.
Acknowledgments
We thank Andy Johnston (University of East Anglia) for critical comments and reediting of the manuscript.
Glossary
- DGDG
digalactosyldiacylglycerol
- GST
glutathione S-transferase
- P-96
1-pyrenedodecanoic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- MGDG
monogalactosyldiacylglycerol
- Co-IP
coimmunoprecipitation
- LC-MS
liquid chromatography-mass spectrometry
- EM
electron microscopy
- RNAi
RNA interference
- TEM
transmission electron microscopy
- IT
infection thread
- PHB
poly-β-hydroxybutyrate
- Q-TOF
Quadrupole Time-of-Flight
- PBS
phosphate-buffered saline
- CY3
cyanine-3
References
- Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AS. (2003) Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett 537: 128–132 [DOI] [PubMed] [Google Scholar]
- Awai K, Maréchal E, Block MA, Brun D, Masuda T, Shimada H, Takamiya K, Ohta H, Joyard J. (2001) Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 10960–10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C. (2009) Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu Rev Cell Dev Biol 25: 71–91 [DOI] [PubMed] [Google Scholar]
- Benning C, Ohta H. (2005) Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem 280: 2397–2400 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Botté C, Saïdani N, Mondragon R, Mondragón M, Isaac G, Mui E, McLeod R, Dubremetz JF, Vial H, Welti R, et al. (2008) Subcellular localization and dynamics of a digalactolipid-like epitope in Toxoplasma gondii. J Lipid Res 49: 746–762 [DOI] [PubMed] [Google Scholar]
- Boutrot F, Chantret N, Gautier MF. (2008) Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics 9: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhot N, Gomès E, Milat ML, Ponchet M, Marion D, Lequeu J, Delrot S, Coutos-Thévenot P, Blein JP. (2004) Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Mol Biol Cell 15: 5047–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AdeO, Gomes VM. (2007) Role of plant lipid transfer proteins in plant cell physiology-a concise review. Peptides 28: 1144–1153 [DOI] [PubMed] [Google Scholar]
- Catalano CM, Czymmek KJ, Gann JG, Sherrier DJ. (2007) Medicago truncatula syntaxin SYP132 defines the symbiosome membrane and infection droplet membrane in root nodules. Planta 225: 541–550 [DOI] [PubMed] [Google Scholar]
- Cho HJ, Widholm JM. (2002) Improved shoot regeneration protocol for hairy roots of the legume Astragalus sinicus. Plant Cell Tissue Organ Cult 69: 259–269 [Google Scholar]
- Chou MX, Wei XY, Chen DS, Zhou JC. (2006) Thirteen nodule-specific or nodule-enhanced genes encoding products homologous to cysteine cluster proteins or plant lipid transfer proteins are identified in Astragalus sinicus L. by suppressive subtractive hybridization. J Exp Bot 57: 2673–2685 [DOI] [PubMed] [Google Scholar]
- D’Angelo G, Vicinanza M, De Matteis MA. (2008) Lipid-transfer proteins in biosynthetic pathways. Curr Opin Cell Biol 20: 360–370 [DOI] [PubMed] [Google Scholar]
- Debono A, Yeats TH, Rose JK, Bird D, Jetter R, Kunst L, Samuels L. (2009) Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21: 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude N, Tippmann H, Flemetakis E, Katinakis P, Udvardi M, Dörmann P. (2004) The galactolipid digalactosyldiacylglycerol accumulates in the peribacteroid membrane of nitrogen-fixing nodules of soybean and Lotus. J Biol Chem 279: 34624–34630 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Hardy RW, Burns RC, Holsten RD. (1973) Applications of the acetylene-ethylene assay for measurement of nitrogen fixation. Soil Biol Biochem 5: 47–81 [Google Scholar]
- Held M, Hossain MS, Yokota K, Bonfante P, Stougaard J, Szczyglowski K. (2010) Common and not so common symbiotic entry. Trends Plant Sci 15: 540–545 [DOI] [PubMed] [Google Scholar]
- Hirsch AM, Bang M, Ausubel FM. (1983) Ultrastructural analysis of ineffective alfalfa nodules formed by nif:Tn5 mutants of Rhizobium meliloti. J Bacteriol 155: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JC, Levine TP. (2005) Lipid traffic: floppy drives and a superhighway. Nat Rev Mol Cell Biol 6: 209–220 [DOI] [PubMed] [Google Scholar]
- Israel DW. (1987) Investigation of the role of phosphorus in symbiotic dinitrogen fixation. Plant Physiol 84: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5: 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- José-Estanyol M, Gomis-Rüth FX, Puigdomènech P. (2004) The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol Biochem 42: 355–365 [DOI] [PubMed] [Google Scholar]
- Jouhet J, Maréchal E, Baldan B, Bligny R, Joyard J, Block MA. (2004) Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J Cell Biol 167: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Maréchal E, Block MA. (2007) Glycerolipid transfer for the building of membranes in plant cells. Prog Lipid Res 46: 37–55 [DOI] [PubMed] [Google Scholar]
- Kader JC, Julienne M, Vergnolle C. (1984) Purification and characterization of a spinach-leaf protein capable of transferring phospholipids from liposomes to mitochondria or chloroplasts. Eur J Biochem 139: 411–416 [DOI] [PubMed] [Google Scholar]
- Kaplan MR, Simoni RD. (1985) Intracellular transport of phosphatidylcholine to the plasma membrane. J Cell Biol 101: 441–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Jeannotte R, Welti R, Bockus WW. (2013) Lipid profiles in wheat cultivars resistant and susceptible to tan spot and the effect of disease on the profiles. Phytopathology 103: 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Copeland L. (1996) Enzymes of poly-β-hydroxybutyrate metabolism in soybean and chickpea bacteroids. Appl Environ Microbiol 62: 4186–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascombe MB, Bakan B, Buhot N, Marion D, Blein JP, Larue V, Lamb C, Prangé T. (2008) The structure of “defective in induced resistance” protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein. Protein Sci 17: 1522–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Lee JY, Min K, Cha H, Shin DH, Hwang KY, Suh SW. (1998) Rice non-specific lipid transfer protein: the 1.6 A crystal structure in the unliganded state reveals a small hydrophobic cavity. J Mol Biol 276: 437–448 [DOI] [PubMed] [Google Scholar]
- Leidi EO, Rodriguez-Navarro DN. (2000) Nitrogen and phosphorus availability limit N2 fixation in bean. New Phytol 147: 337–346 [Google Scholar]
- Lev S. (2010) Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol 11: 739–750 [DOI] [PubMed] [Google Scholar]
- Levine T. (2004) Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions. Trends Cell Biol 14: 483–490 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhou L, Li Y, Chen D, Tan X, Lei L, Zhou J. (2008) A nodule-specific plant cysteine proteinase, AsNODF32, is involved in nodule senescence and nitrogen fixation activity of the green manure legume Astragalus sinicus. New Phytol 180: 185–192 [DOI] [PubMed] [Google Scholar]
- Limpens E, Ivanov S, van Esse W, Voets G, Fedorova E, Bisseling T. (2009) Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell 21: 2811–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodwig EM, Hosie AH, Bourdès A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS. (2003) Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422: 722–726 [DOI] [PubMed] [Google Scholar]
- Maréchal E, Awai K, Block MA, Brun D, Masuda T, Shimada H, Takamiya K, Ohta H, Joyard J. (2000) The multigenic family of monogalactosyl diacylglycerol synthases. Biochem Soc Trans 28: 732–738 [PubMed] [Google Scholar]
- Maréchal E, Block MA, Dorne AJ, Douce R, Joyard J. (1997) Lipid synthesis and metabolism in the plastid envelope. Physiol Plant 1: 65–77 [Google Scholar]
- Mikes V, Milat ML, Ponchet M, Panabières F, Ricci P, Blein JP. (1998) Elicitins, proteinaceous elicitors of plant defense, are a new class of sterol carrier proteins. Biochem Biophys Res Commun 245: 133–139 [DOI] [PubMed] [Google Scholar]
- Monahan-Giovanelli H, Pinedo CA, Gage DJ. (2006) Architecture of infection thread networks in developing root nodules induced by the symbiotic bacterium Sinorhizobium meliloti on Medicago truncatula. Plant Physiol 140: 661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman H, Mikes V, Milat ML, Ponchet M, Marion D, Prangé T, Maume BF, Vauthrin S, Blein JP. (2001) Fatty acids bind to the fungal elicitor cryptogein and compete with sterols. FEBS Lett 489: 55–58 [DOI] [PubMed] [Google Scholar]
- Panter S, Thomson R, de Bruxelles G, Laver D, Trevaskis B, Udvardi M. (2000) Identification with proteomics of novel proteins associated with the peribacteroid membrane of soybean root nodules. Mol Plant Microbe Interact 13: 325–333 [DOI] [PubMed] [Google Scholar]
- Pii Y, Astegno A, Peroni E, Zaccardelli M, Pandolfini T, Crimi M. (2009) The Medicago truncatula N5 gene encoding a root-specific lipid transfer protein is required for the symbiotic interaction with Sinorhizobium meliloti. Mol Plant Microbe Interact 22: 1577–1587 [DOI] [PubMed] [Google Scholar]
- Price GD, Day DA, Gresshoff PM. (1987) Rapid isolation of intact peribacteroid envelopes from soybean nodules and demonstration of selective permeability to metabolites. J Plant Physiol 130: 157–164 [Google Scholar]
- Schumpp O, Deakin WJ. (2010) How inefficient rhizobia prolong their existence within nodules. Trends Plant Sci 15: 189–195 [DOI] [PubMed] [Google Scholar]
- Shin DH, Lee JY, Hwang KY, Kim KK, Suh SW. (1995) High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings. Structure 3: 189–199 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin-Moindrot S, Caille A, Douliez JP, Marion D, Vovelle F. (2000) The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2. Eur J Biochem 267: 1117–1124 [DOI] [PubMed] [Google Scholar]
- Trainer MA, Charles TC. (2006) The role of PHB metabolism in the symbiosis of rhizobia with legumes. Appl Microbiol Biotechnol 71: 377–386 [DOI] [PubMed] [Google Scholar]
- Van de Velde W, Guerra JC, De Keyser A, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S. (2006) Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol 141: 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE, Aasman EJ, Szarka R. (1991) Brefeldin A does not inhibit the movement of phosphatidylethanolamine from its sites for synthesis to the cell surface. J Biol Chem 266: 8241–8247 [PubMed] [Google Scholar]
- Verma D. (1992) Signals in root nodule organogenesis and endocytosis of Rhizobium. Plant Cell 4: 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma DP, Hong Z. (1996) Biogenesis of the peribacteroid membrane in root nodules. Trends Microbiol 4: 364–368 [DOI] [PubMed] [Google Scholar]
- Voelker DR. (1990) Lipid transport pathways in mammalian cells. Experientia 46: 569–579 [DOI] [PubMed] [Google Scholar]
- Whitehead LF, Day DA. (1997) The peribacteroid membrane. Physiol Plant 100: 30–44 [Google Scholar]
- Wu GZ, Xue HW. (2010) Arabidopsis β-ketoacyl-[acyl carrier protein] synthase I is crucial for fatty acid synthesis and plays a role in chloroplast division and embryo development. Plant Cell 22: 3726–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]