A jasmonic acid biosynthesis enzyme enhances wheat salinity tolerance.
Abstract
One of the two branches of the α-linolenic acid metabolism pathway is catalyzed by 12-oxo-phytodienoic acid reductase I, and the other is involved in jasmonic acid (JA) synthesis. The former is known to be active in the response to salinity tolerance in wheat (Triticum aestivum), but the participation of the latter in this response has not been established as yet. Here, the salinity-responsive bread wheat gene TaAOC1, which encodes an allene oxide cyclase involved in the α-linolenic acid metabolism pathway, was constitutively expressed in both bread wheat and Arabidopsis (Arabidopsis thaliana). In both species, transgenic lines exhibited an enhanced level of tolerance to salinity. The transgenic plants accumulated a higher content of JA and developed shorter roots. Both the shortened roots and the salinity tolerance were abolished in a background lacking a functional AtMYC2, a key component of the JA and abscisic acid signaling pathway, but were still expressed in a background deficient with respect to abscisic acid synthesis. We provide the first evidence, to our knowledge, suggesting that JA is also involved in the plant salinity response and that the α-linolenic acid metabolism pathway has a regulatory role over this response.
High levels of soil salinity impose both osmotic stress and ion toxicity on plants, leading to cell damage and growth arrest. Exhaustive investigation of the Arabidopsis (Arabidopsis thaliana) response to salinity stress has uncovered a number of participating pathways, some dependent and others independent of abscisic acid (ABA; Zhu, 2001; Xiong and Zhu, 2002; Bartels and Sunkar, 2005). Activation of the Salt Overly Sensitive (SOS) pathway helps to maintain ionic homeostasis, while both ABA-independent and -dependent signaling pathways regulate an array of genes (Zhu, 2002; Wang et al., 2003; Munns and Tester, 2008). In addition to osmotic stress and ion toxicity, salinity also induces the production of reactive oxygen species (ROS), which damage lipid layers as a result of the production of oxylipins, the presence of which is a reliable indicator of membrane oxidation (Mittler, 2002; Miller et al., 2010). Oxylipins also serve as signaling molecules, for example, controlling stomatal closure in an ABA-independent manner to enhance pathogen resistance (Farmer et al., 2003; Montillet et al., 2013).
Among the lipids that can be peroxidized via the action of ROS, α-linolenic acid has attracted particular attention because it represents a substrate in the synthesis of the phytohormone jasmonic acid (JA; Hause et al., 2009; Acosta and Farmer, 2010; Wasternack and Hause, 2013). The pathway begins with the peroxidization of α-linolenic acid by lipoxygenase, the product of which is then converted to 12,13(S)-epoxy-octadecatrienoic acid (12,13-EOT) by the enzyme allene oxide synthase (AOS). 12,13-EOT is then processed to 12-oxo-phytodienoic acid (OPDA) by the action of allene oxide cyclase (AOC), and OPDA is reduced by OPDA reductase (OPR; Creelman and Mullet, 1997; Mueller, 1997; Stenzel et al., 2003a; Schaller and Stintzi, 2009). OPDA exists as four distinct isomers, namely cis-(+), cis-(–), trans(+), and trans(–); of these, cis-(+)-OPDA is predominant in most plants. OPR enzymes are present in two forms, OPRI and OPRII (Mueller, 1997; Beynon et al., 2009; Schaller and Stintzi, 2009). OPRII reduces cis-(+)-OPDA to form 3-oxo-2-(2′-[Z]-pentenyl)cyclopentane-1-octanoic acid, which is finally oxidized to produce JA (Schaller et al., 1998). OPRI enzymes do not interact with cis-(+)-OPDA; however, they are known to interact with trans(+)- and cis-(–)-OPDA in vitro (Biesgen and Weiler, 1999). The AOC-catalyzed enolization of 12,13-EOT is also responsible for the slow conversion of cis- to trans-OPDA. Hence, it is believed that in addition to its involvement in JA synthesis, the α-linolenic acid metabolism pathway also has an OPRI-catalyzed branch.
A growing body of evidence confirms that α-linolenic acid metabolism is responsive to abiotic stress (Lenka et al., 2011). A global transcriptomic analysis of bread wheat (Triticum aestivum) has shown that the genes involved in α-linolenic acid metabolism are induced by salinity stress (Liu et al., 2012). As applied similarly to genes encoding OPRI enzymes, these genes are characteristically up-regulated by pathogen invasion, wounding, and oxidative stress (Biesgen and Weiler, 1999; Agrawal et al., 2003). When expressed heterologously in Arabidopsis, the product of the bread wheat OPRI gene TaOPR1 has been shown to enhance salinity tolerance by the up-regulation of AtMYC2, thereby triggering an ABA-dependent signaling pathway in a JA-independent manner (Dong et al., 2013). Whether the JA synthesis branch of α-linolenic acid metabolism is also involved in the response to salinity stress remains to be resolved.
Here, we describe the isolation of the bread wheat AOC gene TaAOC1 (the first AOC gene to be isolated from wheat), which, when the plants are subjected to salinity stress, is more strongly transcribed in the salinity tolerant cv SR3 (derived from a somatic hybrid between cv JN177 and tall wheat grass; Xia et al., 2003; Xia, 2009) than in the parental cv JN177. We show that its constitutive expression in both wheat and Arabidopsis induces a JA-sufficient phenotype (Hause et al., 2009) and improved the plant salinity tolerance. The experiments demonstrate that the JA synthesis branch of the α-linolenic acid metabolism pathway enhances salinity tolerance, and the presence of AtMYC2 is required for this effect to be expressed.
RESULTS
TaAOC1 Was Induced in Wheat Seedlings Subjected to Stress or Exposed to Hormones
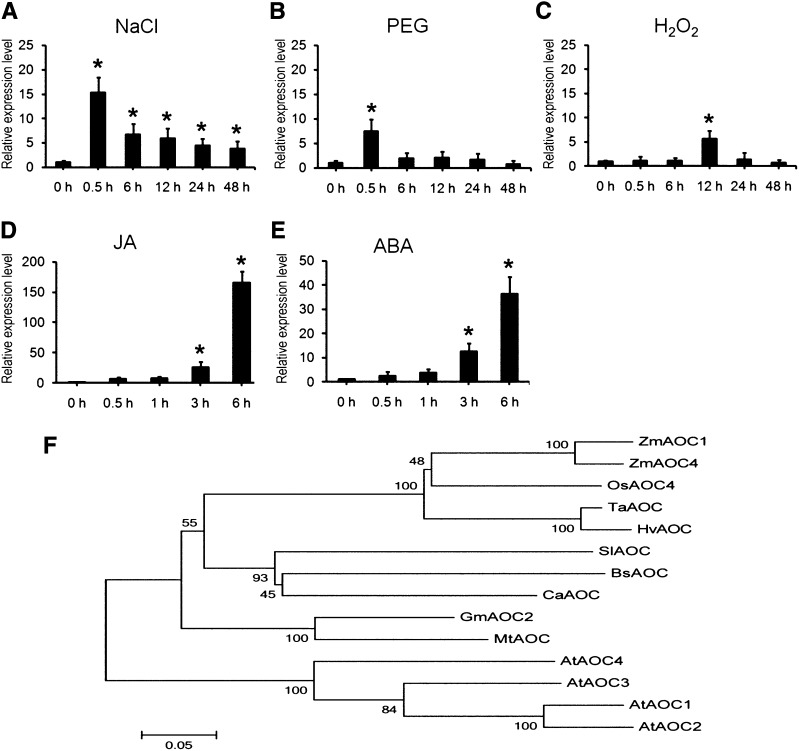
In wheat cv SR3 plants stressed with either NaCl or polyethylene glycerol (PEG), TaAOC1 transcript abundance rose by, respectively, about 15- and 8-fold, with the first signs of induction already detectable within 0.5 h of the imposition of the stress (Fig. 1, A and B). The consequences of exposure to H2O2 were similar, although less intense (Fig. 1C). Because JA and ABA are both important for regulating the plant’s response to abiotic stress, the effect on TaAOC1 transcription of exogenously supplied JA and ABA was then investigated. JA treatment up-regulated TaAOC1 transcription very early, with transcript abundance rising by more than 150-fold over time (Fig. 1D). The effect of the ABA treatment was similar in its timing, although the extent of the induction was less (Fig. 1E). TaAOC1 was also induced in ‘JN177’ wheat plants under these stresses and hormone treatment, although the induction was relatively lower than that in ‘SR3’ (Supplemental Fig. S1).
Figure 1.
TaAOC1 expression under treatments in wheat ‘SR3’ plants and phylogenetic analysis of AOCs from different plant species. A to E, The expression levels of TaAOC1 in wheat ‘SR3’ plants under 200 mm NaCl treatment (A), 20% (w/v) PEG treatment (B), 10 mm H2O2 treatment (C), 100 µm JA treatment (D), and 100 µm ABA treatment (E) were determined by qPCR using wheat actin gene (AB181991) as the internal control. The expression levels at 0 h were set at 1.0. Error bars represent the sd from three biological replicates. Each replicate contained at least 30 plants. F, Phylogenetic tree. Zm represents maize, Os represents rice, Ta represents wheat, Hv represents barley, Sl represents Solanum lycopersicum, Bs represents Bruguiera sexangula, Ca represents C. acuminate, Gm represents Glycine max, Mt represents Medicago truncatula, and At represents Arabidopsis.
TaAOC1 Encodes a Putative Wheat AOC Enzyme
The cv SR3 copy of TaAOC1 was 720 bp in length, and its predicted product was a 239-residue protein carrying an allene oxide cyclase domain at its C terminus. A multialignment with 14 plant homologs showed that the most conserved region of the sequence was the enzyme’s functional domain (Supplemental Fig. S2). A phylogenetic analysis clustered the TaAOC1 product with its homologs in maize (Zea mays), rice (Oryza sativa), and barley (Hordeum vulgare), with the sequences from dicotyledonous species forming a distinct second clade (Fig. 1F). An analysis of the spatial-temporal pattern of TaAOC1 transcription in cv SR3 plants demonstrated that the gene was ubiquitously transcribed, but that the level of transcript abundance was variable from organ to organ. The highest levels recorded obtained when the plant was in the booting stage, particularly in the root, but also in the stem, sheath, and ear. A substantial level of transcription was also detected in the awn and ear at anthesis (Supplemental Fig. S2).
The Constitutive Expression of TaAOC1 in Wheat Enhanced Salinity Tolerance
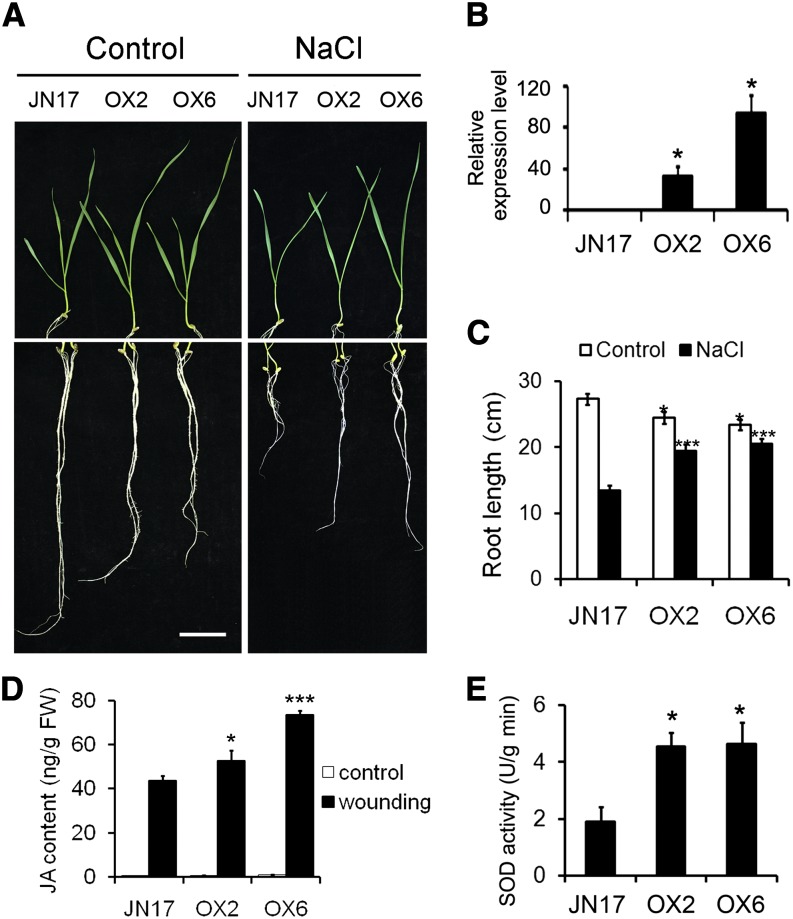
Among the 44 transgenic constitutive expression lines generated, lines OVEREXPRESSION2 (OX2) and OX6 showed the highest level of TaAOC1 transcript (Fig. 2B). The relative root growth of 10-d-old OX2, OX6, and ‘JN17’ seedlings exposed to salinity stress for 8 d was then compared. In the absence of the stress, OX2 and OX6 roots were shorter than those of ‘JN17’ (Fig. 2, A and C), but in its presence, root growth in ‘JN17’ seedlings was more inhibited than in OX2 and OX6 seedlings (Fig. 2, A and C). Because TaAOC1 encodes a putative AOC enzyme involved in JA synthesis, the JA levels in vivo were quantified in OX2, OX6, and ‘JN17’ seedlings. As shown in Figure 2D, wounding induced higher levels of JA accumulation in two transgenic lines with constitutive expression of TaAOC1. Moreover, superoxide dismutase (SOD) activity in OX2 and OX6 plants was about double the level detected in ‘JN17’ plants (Fig. 2E).
Figure 2.
Constitutively expressing the TaAOC1 enhanced salinity tolerance and JA levels in wheat. A, Seedlings of ‘JN17’ and two transgenic lines constitutively expressing the TaAOC1 (OX2 and OX6) under NaCl treatment for 8 d. Bar = 1 cm. B, The expression levels of TaAOC1 relative to the internal control actin gene in ‘JN17,’ OX2, and OX6 determined by qPCR. C, Root lengths of ‘JN17’ and two transgenic lines under NaCl treatment. Error bars represent the sd from 30 plants. D, Measurement of JA levels in 2-week-old ‘JN17,’ OX2, and OX6 seedlings, with or without wounding. E, SOD activity in ‘JN17,’ OX2, and OX6 plants. Data in B, D, and E are presented as means ± sd, and columns marked with asterisks indicate significant differences from ‘JN17’ using Student’s t test (* indicates P < 0.05, *** indicates P < 0.001). Error bars represent the sd from three biological replicates. Each replicate contained at least 30 plants. [See online article for color version of this figure.]
The Constitutive Expression of TaAOC1 in Arabidopsis Enhanced Salinity Tolerance and JA Accumulation in Wounded Leaves
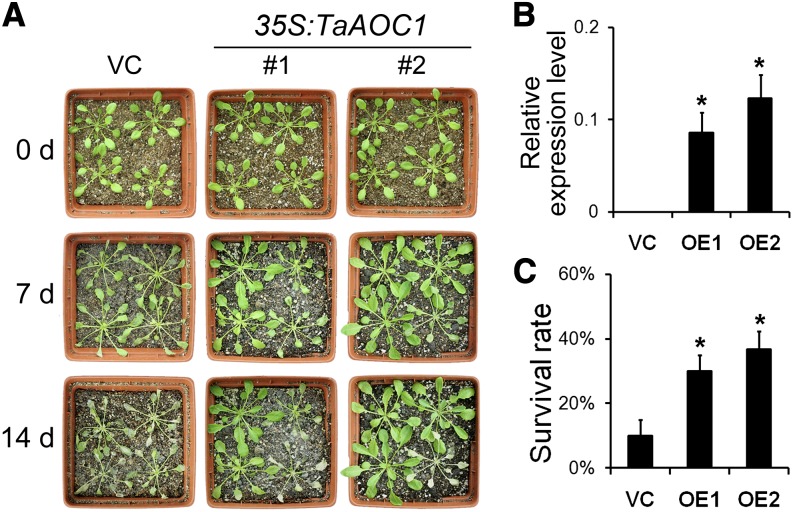
Two independent transgenic Arabidopsis lines (OVEREXPRESSING1 [OE1] and OE2), each showing a high level of TaAOC1 transcription, were selected (Fig. 3B). Measuring the root growth of 4-d-old seedlings not exposed to stress showed that the vector control (VC) plants were more vigorous than either of the OE plants (Supplemental Fig. S3A). However, in the presence of salinity stress, the root growth of VC plants was more noticeably suppressed than that of the OE plants (Supplemental Fig. S3, B–D). In the presence of 100 mm NaCl, the length of the VC roots was 80% of those of nonstressed plants, whereas the equivalent for the OE roots was greater than 95%, and at 200 mm NaCl, the respective relative root growths were 40% and greater than 60% (Supplemental Fig. S3E). At the adult stage (4-week-old plants), VC plants’ rosette leaves showed severe chlorosis, while the OE leaves remained green (Fig. 3A). Later in the experiment, almost all the VC plants died, but greater than 30% of the OE plants were still viable (Fig. 3C).
Figure 3.
Constitutively expressing the TaAOC1 enhanced the salinity tolerance in Arabidopsis adult plants. A, Phenotype of 4-week-old VC and two transgenic lines (OE1 and OE2) at 0, 7, and 14 d after NaCl treatment. B, The expression levels of TaAOC1 relative to the internal control gene AtUBIQUITIN10 (AtUBQ10) in VC, OE1, and OE2 plants determined by qPCR. C, Survival rates of 4-week-old VC, OE1, and OE2 plants at 14 d after NaCl treatment. Data in B and C are presented as means ± sd, and columns marked with asterisks indicate significant differences from the VC line using Student’s t test (P < 0.05). Error bars represent the sd from three biological replicates. Each replicate contained at least 30 plants. [See online article for color version of this figure.]
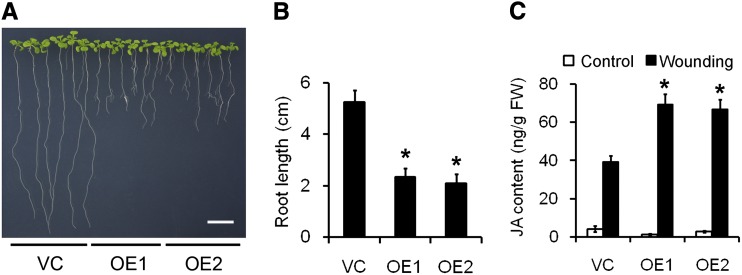
The phenotype of these lines differed from that of the control line carrying only an empty vector. Root growth in 10-d-old OE plants was retarded compared with that in VC plants (Fig. 4, A and B), resembling the shortened roots produced by Arabidopsis seedlings challenged with JA. When the endogenous level of JA was quantified in rosette leaves harvested from nonwounded 4-week-old OE and VC plants, there was little difference between the OE and VC plants. By contrast, in wounded leaves, the JA content rose more strongly in the two OE lines than in VC (Fig. 4C).
Figure 4.
JA-sufficient phenotypes in transgenic Arabidopsis constitutively expressing the TaAOC1. A, Two-week-old VC and two transgenic lines (OE1 and OE2). Bar = 1 cm. B, Root length of 2-week-old VC, OE1, and OE2 seedlings. Error bars represent the sd from at least 30 plants. C, Measurement of JA levels in the rosette leaves of VC, OE1, and OE2 plants, with or without wounding. Data are presented as means ± sd, and columns marked with asterisks indicate significant differences from the VC line using Student’s t test (P < 0.05). Error bars represent the sd from three biological replicates. Each replicate contained at least 30 plants. [See online article for color version of this figure.]
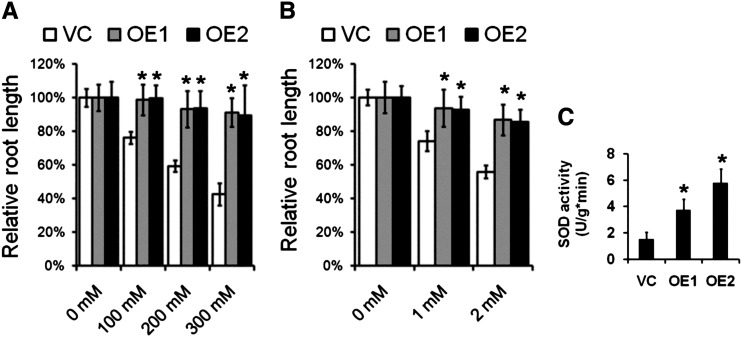
The Constitutive Heterologous Expression of TaAOC1 in Arabidopsis Enhanced Tolerance to Osmotic and H2O2 Stress
A similar set of experiments compared the response of the OE and VC plants to stress imposed by either mannitol (osmotic stress) or H2O2 (Fig. 5). In the presence of 100 mm mannitol, root growth in OE1 and OE2 plants was hardly affected (98% and 99%, respectively), whereas that of VC plants fell to 76% (Fig. 5A; Supplemental Fig. S4, A and B). Under higher concentrations of mannitol, the differential performance of the OE and VC lines was even more pronounced (Fig. 5A; Supplemental Fig. S4). The effect of exposure to 1 mm H2O2 was to only slightly reduce the relative root length of OE1 and OE2 plants (94% and 93%, respectively), while that of VC plants was reduced to 74% (Fig. 5B); the effect was greater when the stress level was doubled (Fig. 5B; Supplemental Fig. S5). The OE plants also expressed a higher level of SOD activity than VC plants (Fig. 5C).
Figure 5.
Constitutively expressing the TaAOC1 enhanced the osmotic and H2O2 tolerance in Arabidopsis. A, Relative root length of the VC and two transgenic lines (OE1 and OE2) under mannitol treatments. The root length of each line on MS plates without mannitol was set at 100%. B, Relative root length of VC, OE1, and OE2 seedlings under H2O2 treatment. The root length of each line on MS plates without H2O2 was set at 100%. C, SOD activity in the VC, OE1, and OE2 lines. Data in A, B, and C are presented as means ± sd, and columns marked with asterisks indicate significant differences from the VC line using Student’s t test (P < 0.05). Error bars represent the sd from three biological replicates. Each replicate contained at least 30 plants.
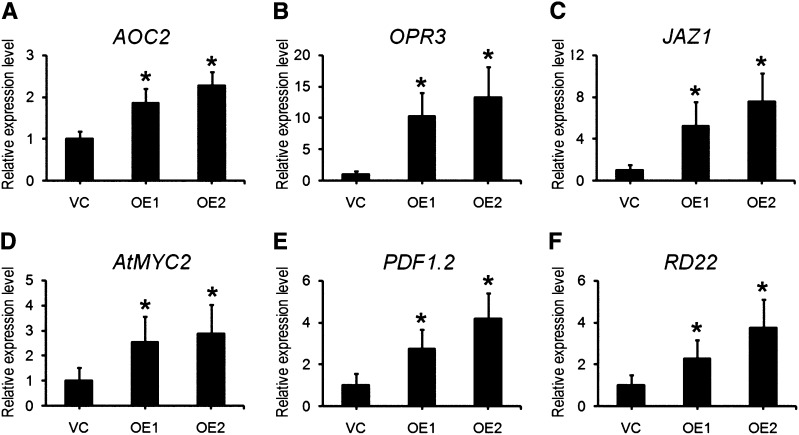
The Constitutive Heterologous Expression of TaAOC1 in Arabidopsis Up-Regulated the JA Pathway
The effect of constitutively expressing TaAOC1 in Arabidopsis on the transcription of genes in the JA pathway was investigated by monitoring the abundance of AOC2 (an Arabidopsis homolog of TaAOC1 that functions in JA synthesis), OPR3, which encodes a JA synthesis enzyme downstream of AOC, JASMONATE-ZIM-DOMAIN PROTEIN1 (JAZ1) and AtMYC2 (JA signaling genes), PLANT DEFENSIN1.2 (PDF1.2, a downstream gene in the JA signaling pathway), and RESPONSIVE TO DESSICATION22 (RD22, a drought-responsive element containing an AtMYC2 binding site in its promoter) transcript. AOC2 transcript was more abundant in the OE lines than in VC (Fig. 6A), as was also the case for the OPR3, JAZ1, AtMYC2, PDF1.2, and RD22 (Fig. 6, C–F). The data were consistent with the JA-sufficient phenotypes and higher JA accumulation shown by the OE plants (Fig. 4).
Figure 6.
The expression levels of AOC2 (A), OPR3 (B), JAZ1 (C), AtMYC2 (D), PDF1.2 (E), and RD22 (F) in 2-week-old VC and transgenic lines (OE1 and OE2) determined by qPCR using AtUBQ10 as the internal control. The expression levels in the VC line were set at 1.0. The columns marked with asterisks indicate significant differences from the VC line using Student’s t test (P < 0.05). Error bars represent the sd from three biological replicates. Each replicate contained at least 30 plants.
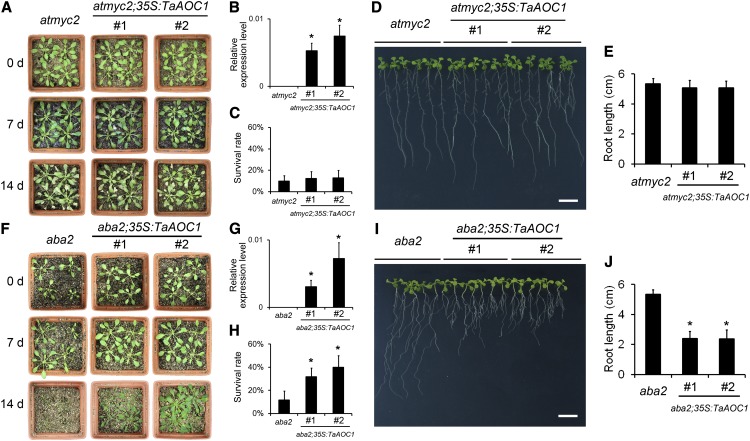
The Improved Salinity Tolerance of the OE Lines Relies on the Presence of AtMYC2, But Not of ABA2
The effect of the constitutive expression of TaAOC1 in the absence of AtMYC2 function was studied by introducing TaAOC1 into the atmyc2-2 mutant. The two selected transgenic lines showing a high level of constitutive TaAOC1 transcription (Fig. 7B) exhibited a similar response to salinity stress and plant survival rate as the nontransformed atmyc2-2 mutants (Fig. 7, A and C). Seedling root lengths were also comparable (Fig. 7, D and E). Thus, the JA-related phenotype and salinity tolerance conferred by the constitutive expression of TaAOC1 required the presence of a functional copy of AtMYC2. A similar experiment was based on the expression of TaAOC1 in a background deficient for ABA2 (Fig. 7G), a gene involved in ABA synthesis. The survival rate of the transgenic plants was higher than that of the nontransformed aba2 mutant (Fig. 7, F and H), and the former produced shorter roots than the latter (Fig. 7, I and J). Thus, the enhanced salinity tolerance conferred by the constitutive expression of TaAOC1 was unaffected by a deficiency in ABA synthesis capacity.
Figure 7.
The enhanced salinity tolerance and shortened seedling roots conferred by constitutively expressing the TaAOC1 relied on AtMYC2. A, Phenotypes of atmyc2, atmyc2;35S:TaAOC1#1, and atmyc2;35S:TaAOC1#2 plants at 0, 7, and 14 d after NaCl treatment. B, The expression levels of TaAOC1 relative to the internal control gene AtUBQ10 in atmyc2, atmyc2;35S:TaAOC1#1, and atmyc2;35S:TaAOC1#2 determined by qPCR. C, Survival rates of atmyc2, atmyc2;35S:TaAOC1#1, and atmyc2;35S:TaAOC1#2 plants at 14 d after NaCl treatment. D, Two-week-old atmyc2, atmyc2;35S:TaAOC1#1, and atmyc2;35S:TaAOC1#2 seedlings. Bar = 1 cm. E, Root length of 2-week-old atmyc2, atmyc2;35S:TaAOC1#1, and atmyc2;35S:TaAOC1#2 seedlings. F, Phenotypes of aba2, aba2;35S:TaAOC1#1, and aba2;35S:TaAOC1#2 at 0, 7, and 14 d after NaCl treatment. G, The expression levels of TaAOC1 relative to the internal control gene AtUBQ10 in aba2, aba2;35S:TaAOC1#1, and aba2;35S:TaAOC1#2 determined by qPCR. H, Survival rates of aba2, aba2;35S:TaAOC1#1, and aba2;35S:TaAOC1#2 plants at 14 d after NaCl treatment. I, Two-week-old aba2, aba2;35S:TaAOC1#1, and aba2;35S:TaAOC1#2 seedlings. Bar = 1 cm. J, Root length of 2-week-old aba2, aba2;35S:TaAOC1#1, and aba2;35S:TaAOC1#2 seedlings. Error bars in B, C, G, and H represent the sd from three biological replicates. Each replicate contained at least 30 plants. Error bars in E and J represent the sd from at least 30 plants. The columns marked with asterisks indicate significant differences from atmyc2 or aba2, respectively (P < 0.05). [See online article for color version of this figure.]
DISCUSSION
TaAOC1 Elevated the JA Synthesis Branch of the α-Linolenic Acid Metabolism Pathway
We have shown that the constitutive expression of TaAOC1 enhanced the accumulation of JA and resulted in a JA-sufficient phenotype (Figs. 2 and 4). JA synthesis is regulated by positive feedback, in a way that the genes encoding the enzymes involved in JA synthesis are JA inducible (Delker et al., 2006; Wasternack, 2007; Wasternack and Hause, 2013). Consistent with this, we were able to show that OPR3 was induced by the constitutive expression of TaAOC1 (Fig. 6B). The OPR3 product is involved in the catalysis of the JA precursor cis-(+)-OPDA (Stintzi and Browse, 2000). The Arabidopsis genome includes four AOC loci; as yet, the distinctness (if any) of their functions has not been fully defined, and no clear correlation has been established between their transcription and JA synthesis (Stenzel et al., 2003b, 2012). AOC2 is known to be inducible by the exogenous supply of JA (Ziegler et al., 2000; Stenzel et al., 2003b) and was induced by the constitutive expression of TaAOC1 (Fig. 6A). Moreover, JA synthesis is dependent on substrate availability, and the process is highly tissue specific (Browse, 2009; Wasternack and Hause, 2013). In Arabidopsis, the constitutive expression of AOS genes elevates JA levels in wounded but not in unwounded leaves (Laudert et al., 2000), and the same result was obtained by the constitutive expression of TaAOC1 (Fig. 4C). The roots of the TaAOC1 constitutive expression plants were significantly shorter than those of the control plants (Fig. 4, A and B). Given the phenotype of the constitutive expression of TaAOC1 in both wheat and Arabidopsis with respect to JA, a reasonable hypothesis is therefore that the TaAOC1 plays a key role in JA synthesis.
The Elevation of JA Synthesis Branch Enhances Salinity Tolerance beyond Just Regulating Ionic Homeostasis
The constitutive expression of TaAOC1 enhanced the salinity tolerance of both wheat and Arabidopsis (Figs. 2 and 3; Supplemental Fig. S3), just as does the heterologous expression of either a mangrove (Bruguiera sexangula) or a Camptotheca acuminate AOC in tobacco (Nicotiana tabacum) cells (Yamada et al., 2002; Pi et al., 2009). It is also well established that exposure to JA can improve the salinity tolerance of a number of plant species (Walia et al., 2007; del Amor and Cuadra-Crespo, 2011; Ismail et al., 2012). Hence, it is likely that JA does play a positive role in the salinity stress response. High levels of salinity induce both ionic and osmotic stress, as well as ROS-induced oxidative toxicity (Hasegawa et al., 2000; Munns and Tester, 2008). The constitutive expression of TaAOC1 enhanced tolerance to both osmotic and oxidative stress (Fig. 5), suggesting that the beneficial effect of JA is achieved via a systemic physiological alteration rather than merely by controlling ionic homeostasis. The neutralization of ROS has been proposed as a possible component of stress tolerance; if so, then a key enzyme ought to be SOD, which provides the first line of defense against the toxic effects of elevated levels of ROS (Mittler, 2002; Apel and Hirt, 2004; Mittler et al., 2004; Gill and Tuteja, 2010). SOD activity was increased in both wheat and Arabidopsis plants constitutively expressing TaAOC1 (Figs. 2E and 5C), indicating that one of the roles that JA may play in salinity tolerance surrounds the regulation of ROS homeostasis.
The JA Synthesis Branch Confers Salinity Tolerance via a Different Route Than Used by the OPRI-Catalyzed Branch
In a previous study, the elevation of the OPRI-catalyzed branch, achieved by the constitutive expression of TaOPR1, enhanced salinity tolerance via promoting activity in an ABA-dependent pathway (Dong et al., 2013). The contribution of TaOPR1 to the salinity tolerance of the ABA synthesis mutant aba2-2 was partially impaired, even though the higher transcript abundance of TaOPR1 had no effect on either JA content or the transcription level of JAZ1, known to be a key upstream component of the JA signaling pathway (Dong et al., 2013). Here, TaAOC1 constitutive expression increased JA content (Figs. 2D and 4C) as well as the transcript abundance of JAZ1 and the JA-responsive gene PDF1.2 (Fig. 6, C and E), while in the ABA synthesis mutant aba2-1, TaAOC1 made similar contribution to salinity tolerance (Fig. 7, F and H). Thus, unlike the situation with the OPRI-catalyzed branch, the JA synthesis branch achieved an improvement in salinity tolerance by promoting the JA pathway.
AtMYC2 is a key component of both the JA signaling pathway and the ABA-dependent abiotic stress responsive signaling pathway in Arabidopsis (Abe et al., 1997, 2003; Dombrecht et al., 2007). The loss of AtMYC2 function impairs the level of sensitivity to both ABA and JA (Abe et al., 2003). The constitutive expression of TaOPR1 up-regulated AtMYC2 and enhanced the plant’s sensitivity to ABA (Dong et al., 2013). Similarly, the constitutive expression of TaAOC1 also induced AtMYC2, while the loss of this gene in the atmyc2-2 mutant abolished the salinity tolerance promotive effect of TaAOC1 (Fig. 7, A and C). Therefore, the two branches of the α-linolenic acid metabolism pathway regulate AtMYC2 via distinct pathways, both acting to enhance the level of salinity tolerance.
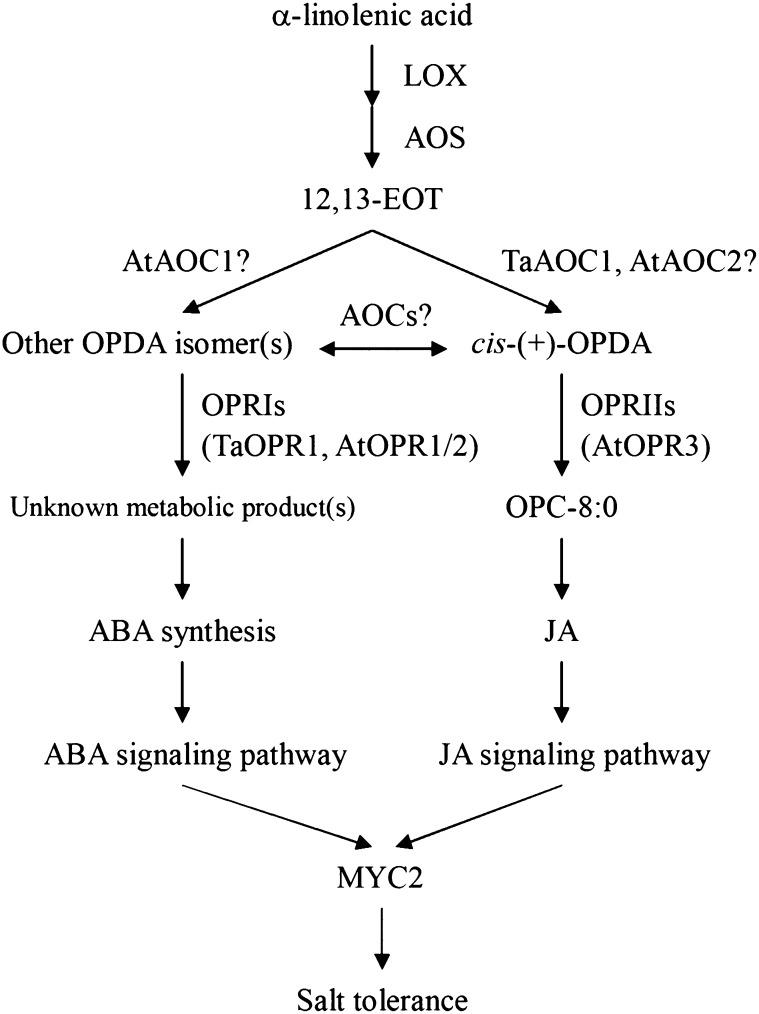
A working model of the α-linolenic acid metabolism pathway is outlined in Figure 8. The network comprises both an OPRI-catalyzed and a JA synthesis branch in which 12,13-EOT provides a common substrate leading to the formation of different OPDA isomers through the action of distinct AOCs. The two branches do not interact with one another, although a slow conversion between cis- and trans- isomers does occur, because the OPDA pool is well buffered. The metabolic output of the OPRI-catalyzed branch is unknown, but it provides signaling molecules to stimulate ABA synthesis and thereby up-regulate the ABA-dependent abiotic stress responsive signaling pathway. The JA synthesis branch performs a similar role via JA signaling. Both routes regulate AtMYC2, which is suggested therefore to be the key component linking the ABA- and JA-mediated salinity response pathways. Our results provide the first experimental evidence, to our knowledge, for the existence of a JA-related pathway responsible for salinity tolerance. The linkage between the α-linolenic acid metabolism pathway and the response to abiotic stress means that genetic manipulation directed at either (or both) branches could be beneficial in the area of breeding for improved salinity tolerance in crops.
Figure 8.
The proposed model of α-linolenic acid metabolism pathway in salinity tolerance. LOX, Lipoxygenase; OPC-8:0, 3-oxo-2-(2′-[Z]-pentenyl)cyclopentane-1-octanoic acid. Arrowed lines mean promotion effect.
MATERIALS AND METHODS
Wheat Growing Conditions and Abiotic Stress Treatments
‘SR3’ plants were grown at 22°C in one-half-strength Hoagland’s liquid medium under a 16-h photoperiod. Gene transcription assays were derived from seedlings at the three-leaf stage, following the addition to the medium of 200 mm NaCl, 20% (w/v) PEG 6000, 10 mm H2O2, 100 µm JA, or 100 µm ABA. After a 48-h exposure to each of the stress agents, RNA was extracted from root tissue and processed for quantitative reverse transcription-PCR (qPCR). The phenotypic effects of salinity stress were noted at 4 d after adding 50 mm NaCl in 4 consecutive days (Dong et al., 2013).
qPCR
RNA extracted with the TRIzol reagent (Invitrogen) represented the template for complementary DNA (cDNA) synthesis using SuperScript II reverse transcriptase (Invitrogen; conducted following the manufacturer’s protocols). Three biological replicates were run for each qPCR, and the reactions were based on FastStart Universal SYBR Green Master (Roche) run in an iCycler thermal cycler machine (Bio-Rad). The reference sequences comprised segments of the wheat (Triticum aestivum) actin (AB181991) or the Arabidopsis (Arabidopsis thaliana) tubulin (At1g04820) genes (Livak and Schmittgen, 2001). The relevant primers are listed in Supplemental Table S1.
TaAOC1 Isolation and Plant Transformation
The sequence of the TaAOC1 fragment identified in a previous microarray experiment (Liu et al., 2012) was used as a search query against a database of wheat expressed sequence tags (http://www.ncbi.nlm.nih.gov/nucest/?term=wheat). Matching expressed sequence tags, assembled using CAP3 software, were used to design a pair of specific primers (5′-CCA AGC TTC AAG AAT ATC ATC ATC CGC T and 5′-CGG GAT CCA CCG ACA TTC ATT CAA CAC CA) able to amplify the full TaAOC1 cDNA sequence from a cv SR3 cDNA library (Xia et al., 2003; Xia, 2009). The resulting coding sequence was ligated into the pGA3626 vector (which includes the maize [Zea mays] ubiquitin promoter to drive expression of the transgene) and then transformed into cv JN17 using the shoot apical meristem method (Zhao et al., 2006). Transgenic Arabidopsis plants were produced by inserting the TaAOC1 coding sequence into the pSTART vector, which was then transformed into ecotype Columbia, as well as into the mutant atmyc2-2 (SALK_083483) and aba2-1. Transformants were selected as described previously (Zhao et al., 2012), and homozygous T3 offspring were used in the subsequent experiments.
Phenotypic Response of Transgenic and Nontransgenic Arabidopsis to Abiotic Stress
Transgenic and nontransgenic seed was surface sterilized by immersion in 0.1% (w/v) mercuric chloride and then planted on Murashige and Skoog (MS) solid medium. The plates were kept at 4°C in the dark for 3 d and then moved to a 16-h photoperiod (light intensity, 200 μm m–2 s–1) at 22°C and 70% relative humidity. Four-day-old seedlings were planted on MS solid medium containing 0, 100, 200, or 300 mm mannitol, 0, 100, 150, or 200 mm NaCl, or 0, 1, or 2 mm H2O2 for 10 d. For NaCl treatment, 50 mm NaCl was added to 4-week-old Arabidopsis plants in 4 d until it reached a final concentration of 200 mm. The plants were incubated for another 2 weeks and used for a survival test (Ren et al., 2010).
Quantification of JA and SOD Activity
The leaves from 2-week-old wheat seedlings and rosette leaves from 4-week-old Arabidopsis plants were wounded using a sterile pair of scissors, as previously described (Li et al., 2006). After 1 h, wounded and unwounded leaves were harvested and their JA content measured via liquid chromatography-tandem mass spectrometry, as described by Dong et al. (2013). SOD activity was assessed in wheat seedlings at the three-leaf stage and in 2-week-old Arabidopsis seedlings, following methods described previously (Sequeira and Mineo, 1966).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number KF573524 (TaAOC1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. TaAOC1 expression under treatments in wheat ‘JN177’ plants.
Supplemental Figure S2. Multialignment of AOCs from different plant species and TaAOC1 expression in different wheat ‘SR3’ tissues.
Supplemental Figure S3. Constitutively expressing the TaAOC1 enhanced the salinity tolerance in Arabidopsis seedlings.
Supplemental Figure S4. Constitutively expressing the TaAOC1 enhanced the osmotic tolerance in Arabidopsis.
Supplemental Figure S5. Constitutively expressing the TaAOC1 enhanced the H2O2 tolerance in Arabidopsis.
Supplemental Table S1. PCR primer sequences used for this research.
Acknowledgments
We thank Zhaojun Ding (Shandong University) for kindly providing the Arabidopsis mutant lines atmyc2-2 and aba2-1.
Glossary
- JA
jasmonic acid
- ABA
abscisic acid
- ROS
reactive oxygen species
- 12,13-EOT
12,13(S)-epoxy-octadecatrienoic acid
- OPDA
12-oxo-phytodienoic acid
- PEG
polyethylene glycerol
- SOD
superoxide dismutase
- VC
vector control
- OE
overexpressing
- qPCR
quantitative reverse transcription-PCR
- cDNA
complementary DNA
- MS
Murashige and Skoog
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta IF, Farmer EE. (2010) Jasmonates. Arabidopsis Book 8: e0129, 10.1199/tab.0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal GK, Jwa NS, Shibato J, Han O, Iwahashi H, Rakwal R. (2003) Diverse environmental cues transiently regulate OsOPR1 of the “octadecanoid pathway” revealing its importance in rice defense/stress and development. Biochem Biophys Res Commun 310: 1073–1082 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Bartels D, Sunkar R. (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24: 23–58 [Google Scholar]
- Beynon ER, Symons ZC, Jackson RG, Lorenz A, Rylott EL, Bruce NC. (2009) The role of oxophytodienoate reductases in the detoxification of the explosive 2,4,6-trinitrotoluene by Arabidopsis. Plant Physiol 151: 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesgen C, Weiler EW. (1999) Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta 208: 155–165 [DOI] [PubMed] [Google Scholar]
- Browse J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48: 355–381 [DOI] [PubMed] [Google Scholar]
- del Amor F, Cuadra-Crespo P. (2011) Alleviation of salinity stress in broccoli using foliar urea or methyl-jasmonate: analysis of growth, gas exchange, and isotope composition. Plant Growth Regul 63: 55–62 [Google Scholar]
- Delker C, Stenzel I, Hause B, Miersch O, Feussner I, Wasternack C. (2006) Jasmonate biosynthesis in Arabidopsis thaliana—enzymes, products, regulation. Plant Biol (Stuttg) 8: 297–306 [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Wang M, Xu F, Quan T, Peng K, Xiao L, Xia G. (2013) Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of abscisic acid signaling and reactive oxygen species scavenging. Plant Physiol 161: 1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Alméras E, Krishnamurthy V. (2003) Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr Opin Plant Biol 6: 372–378 [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48: 909–930 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499 [DOI] [PubMed] [Google Scholar]
- Hause B, Wasternack C, Strack D. (2009) Jasmonates in stress responses and development. Phytochemistry 70: 1483–1484 [DOI] [PubMed] [Google Scholar]
- Ismail A, Riemann M, Nick P. (2012) The jasmonate pathway mediates salt tolerance in grapevines. J Exp Bot 63: 2127–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudert D, Schaller F, Weiler EW. (2000) Transgenic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase. Planta 211: 163–165 [DOI] [PubMed] [Google Scholar]
- Lenka SK, Katiyar A, Chinnusamy V, Bansal KC. (2011) Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol J 9: 315–327 [DOI] [PubMed] [Google Scholar]
- Li CB, Zhao JH, Jiang HL, Wu XY, Sun JQ, Zhang CQ, Wang X, Lou YG, Li CY. (2006) The wound response mutant suppressor of prosystemin-mediated responses6 (spr6) is a weak allele of the tomato homolog of CORONATINE-INSENSITIVE1 (COI1). Plant Cell Physiol 47: 653–663 [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Wang M, Xia G. (2012) A transcriptomic analysis reveals the nature of salinity tolerance of a wheat introgression line. Plant Mol Biol 78: 159–169 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Montillet JL, Leonhardt N, Mondy S, Tranchimand S, Rumeau D, Boudsocq M, Garcia AV, Douki T, Bigeard J, Laurière C, et al. (2013) An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol 11: e1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller MJ. (1997) Enzymes involved in jasmonic acid biosynthesis. Physiol Plant 100: 653–663 [Google Scholar]
- Munns R, Tester M. (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Pi Y, Jiang K, Cao Y, Wang Q, Huang Z, Li L, Hu L, Li W, Sun X, Tang K. (2009) Allene oxide cyclase from Camptotheca acuminata improves tolerance against low temperature and salt stress in tobacco and bacteria. Mol Biotechnol 41: 115–122 [DOI] [PubMed] [Google Scholar]
- Ren Z, Zheng Z, Chinnusamy V, Zhu J, Cui X, Iida K, Zhu JK. (2010) RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA 107: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Stintzi A. (2009) Enzymes in jasmonate biosynthesis: structure, function, regulation. Phytochemistry 70: 1532–1538 [DOI] [PubMed] [Google Scholar]
- Schaller F, Hennig P, Weiler EW. (1998) 12-Oxophytodienoate-10,11-reductase: occurrence of two isoenzymes of different specificity against stereoisomers of 12-oxophytodienoic acid. Plant Physiol 118: 1345–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira L, Mineo L. (1966) Partial purification and kinetics of indoleacetic acid oxidase from tobacco roots. Plant Physiol 41: 1200–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Maucher H, Pitzschke A, Miersch O, Ziegler J, Ryan CA, Wasternack C. (2003a) Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato: amplification in wound signalling. Plant J 33: 577–589 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Miersch O, Kurz T, Maucher H, Weichert H, Ziegler J, Feussner I, Wasternack C. (2003b) Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol Biol 51: 895–911 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Otto M, Delker C, Kirmse N, Schmidt D, Miersch O, Hause B, Wasternack C. (2012) ALLENE OXIDE CYCLASE (AOC) gene family members of Arabidopsis thaliana: tissue- and organ-specific promoter activities and in vivo heteromerization. J Exp Bot 63: 6125–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Browse J. (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Close TJ. (2007) Large-scale expression profiling and physiological characterization of jasmonic acid-mediated adaptation of barley to salinity stress. Plant Cell Environ 30: 410–421 [DOI] [PubMed] [Google Scholar]
- Wang WX, Vinocur B, Altman A. (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218: 1–14 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot (Lond) 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G. (2009) Progress of chromosome engineering mediated by asymmetric somatic hybridization. J Genet Genomics 36: 547–556 [DOI] [PubMed] [Google Scholar]
- Xia GM, Xiang FN, Zhou AF, Wang HA, Chen HM. (2003) Asymmetric somatic hybridization between wheat (Triticum aestivum L.) and Agropyron elongatum (Host) Nevishi. Theor Appl Genet 107: 299–305 [DOI] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. (2002) Salt tolerance. Arabidopsis Book 1: e0048–e0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A, Saitoh T, Mimura T, Ozeki Y. (2002) Expression of mangrove allene oxide cyclase enhances salt tolerance in Escherichia coli, yeast, and tobacco cells. Plant Cell Physiol 43: 903–910 [DOI] [PubMed] [Google Scholar]
- Zhao TJ, Zhao SY, Chen HM, Zhao QZ, Hu ZM, Hou BK, Xia GM. (2006) Transgenic wheat progeny resistant to powdery mildew generated by Agrobacterium inoculum to the basal portion of wheat seedling. Plant Cell Rep 25: 1199–1204 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wei T, Yin KQ, Chen Z, Gu H, Qu LJ, Qin G. (2012) Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol 195: 450–460 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2001) Plant salt tolerance. Trends Plant Sci 6: 66–71 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Stenzel I, Hause B, Maucher H, Hamberg M, Grimm R, Ganal M, Wasternack C. (2000) Molecular cloning of allene oxide cyclase. The enzyme establishing the stereochemistry of octadecanoids and jasmonates. J Biol Chem 275: 19132–19138 [DOI] [PubMed] [Google Scholar]