Arabidopsis plants with mutations in PECTIN METHYLESTERASEs are impaired in resistance to a bacterial pathogen.
Abstract
Pectins, major components of dicot cell walls, are synthesized in a heavily methylesterified form in the Golgi and are partially deesterified by pectin methylesterases (PMEs) upon export to the cell wall. PME activity is important for the virulence of the necrotrophic fungal pathogen Botrytis cinerea. Here, the roles of Arabidopsis PMEs in pattern-triggered immunity and immune responses to the necrotrophic fungus Alternaria brassicicola and the bacterial hemibiotroph Pseudomonas syringae pv maculicola ES4326 (Pma ES4326) were studied. Plant PME activity increased during pattern-triggered immunity and after inoculation with either pathogen. The increase of PME activity in response to pathogen treatment was concomitant with a decrease in pectin methylesterification. The pathogen-induced PME activity did not require salicylic acid or ethylene signaling, but was dependent on jasmonic acid signaling. In the case of induction by A. brassicicola, the ethylene response factor, but not the MYC2 branch of jasmonic acid signaling, contributed to induction of PME activity, whereas in the case of induction by Pma ES4326, both branches contributed. There are 66 PME genes in Arabidopsis, suggesting extensive genetic redundancy. Nevertheless, selected pme single, double, triple and quadruple mutants allowed significantly more growth of Pma ES4326 than wild-type plants, indicating a role of PMEs in resistance to this pathogen. No decreases in total PME activity were detected in these pme mutants, suggesting that the determinant of immunity is not total PME activity; rather, it is some specific effect of PMEs such as changes in the pattern of pectin methylesterification.
The plant cell wall determines cell shape, facilitates cell-cell interaction, and provides mechanical strength to plant cells. De Bary (1886) first observed that a plant pathogen, Sclerotina sclerotiorum, degraded host cell walls during infection. Later, it was concluded that plant cell walls act as preformed structural barriers against pathogen entry, because it was noticed that many plant pathogens produced various types of cell wall-degrading enzymes and that some of those were required for optimal infection of host plants (Albersheim et al., 1969).
Arabidopsis mesophyll cells are surrounded by primary cell walls consisting of three major components: cellulose, hemicelluloses, and pectins. Pectins make up approximately 50% of Arabidopsis leaf cell walls (Zablackis et al., 1995; Harholt et al., 2010). They are complex GalA-containing polysaccharides composed of homogalacturonan (HG), rhamnogalacturonan I and II, and xylogalacturonan (Mohnen, 2008). HG is typically the most abundant polysaccharide, constituting approximately 65% of the pectin (Mohnen, 2008; Harholt et al., 2010). HG is a linear homopolymer of 1,4-linked GalA and is synthesized in the Golgi in a highly methylesterified form (Caffall and Mohnen, 2009). Pectin methylesterases (PMEs) demethylesterify HG in the apoplast (Mohnen, 2008; Harholt et al., 2010). Demethylesterification of pectin is considered to be important for mechanical strength of cell walls, because blockwise deesterified pectin can form calcium bonds that promote the formation of so-called egg-box structures and thus supramolecular pectic gels (Jarvis, 1984; Liners et al., 1989). However, there is some controversy. The cell walls of the shoot apical meristem cells of plants overexpressing PME5 appear softer and cell walls of plants overexpressing a pectin methylesterase inhibitor (PMEI), such as PMEI3, appear harder than wild-type walls when measured using atomic force microscopy (Peaucelle et al., 2011). The degree and pattern of esterification influence the elasticity and porosity of pectic gels, making it likely that PMEs producing HG with varying deesterification patterns influence cell wall matrix properties in planta (Willats et al., 2001).
Pectolytic, necrotrophic pathogens, such as the fungus Botrytis cinerea, secrete pectin-degrading enzymes, including pectate lyases and polygalacturonases as well as PMEs early during infection (Espino et al., 2010). Moreover, the B. cinerea endopolygalacturonases Bcpg1 and Bcpg2 and the PME Bcpme1 are required for full virulence (ten Have et al., 1998; Valette-Collet et al., 2003; Kars et al., 2005), illustrating the importance of pectin degradation for the success of this pathogen.
In Arabidopsis, microarray experiments showed that the expression of plant PME genes is altered during infection with various pathogens, including the biotrophic pathogen Golovinomyces cichoracearum, the hemibiotroph Pseudomonas syringae, nonhost pathogens Phytophthora infestans and Blumeria graminis, as well as the necrotrophic pathogen B. cinerea (Lionetti et al., 2012). One PME, PME3, has been implicated in plant immune responses. Mutant pme3 is more resistant to B. cinerea and Pectobacterium carotovorum and total PME activity is reduced (Raiola et al., 2011). In addition, ectopic expression of PMEI genes rendered plants more resistant against various necrotrophic pathogens (Raiola et al., 2004; Lionetti et al., 2007; Volpi et al., 2011).
PME genes constitute a family of 66 members in Arabidopsis (Harholt et al., 2010). Many PMEs are encoded as preproproteins. The preregion contains a signal peptide and is required for protein targeting to the endoplasmic reticulum. Pro-PMEs are secreted to the apoplast, but only the mature part of the PME (without the proregion) is found in the cell wall (Micheli, 2001). Proregions of PMEs are homologous to PMEI genes from kiwifruit (Actinidia deliciosa) and Arabidopsis (Giovane et al., 2004). However, their function is not yet understood (Jolie et al., 2010). Type I PME genes contain the proregion, whereas type II PMEs do not (Pelloux et al., 2007; Jolie et al., 2010). PMEs identified in phytopathogenic organisms also lack a proregion (Markovic and Janecek, 2004).
At one time, plant PMEs were thought to demethylesterify pectin in a processive, blockwise pattern, whereas pathogen PMEs were thought to randomly deesterify pectin (Limberg et al., 2000a, 2000b; Willats et al., 2001). However, studies using monoclonal antibodies recognizing specific methylesterification patterns showed that plant cell wall PMEs can produce HG with nonblockwise and blockwise methylesters in discrete cell wall microdomains (Willats et al., 2001).
Plants recognize molecular patterns that are characteristic of microbes or are released from plant molecules during infection (Jones and Dangl, 2006; Monaghan and Zipfel, 2012), which are referred to as microbe-associated molecular patterns (MAMPs) and damage-associated molecular patterns (DAMPs), respectively. Well-studied MAMPs include flg22, a 22-amino acid peptide derived from bacterial flagellin, and elf18, an 18-amino acid peptide derived from bacterial elongation factor Tu (Felix et al., 1999; Kunze et al., 2004). Plant-derived peptides such as PEP1 , a 23-amino acid peptide isolated from Arabidopsis, are one group of molecular patterns produced during infection that act as DAMPs (Huffaker et al., 2006). Another group comprises small pectic fragments called oligogalacturonides (OGs) (Ferrari et al., 2013). The degree and pattern of pectin methylesterification are important for pectin degradation and hence generation of OGs by pectolytic enzymes (Berger and Reid, 1979; Limberg et al., 2000a, 2000b). The elicitor activity of OGs depends on their length and degree of esterification (Osorio et al., 2008; Ferrari et al., 2013). It has been shown that pectic fragments can be perceived by WALL-ASSOCIATED KINASE1 (WAK1) and WAK2 (Kohorn et al., 2009; Brutus et al., 2010). Recognition of OGs activates downstream defense responses including reactive oxygen species production, ethylene production, and callose deposition (Ferrari et al., 2013). Deesterified OGs show increased elicitor activity and binding to WAK1 when in the egg-box conformation (Cabrera et al., 2008).
Upon recognition of pathogen-derived patterns, plant immune responses are initiated and transduced through signaling events mediated by three plant hormones with major effects on immunity: salicylic acid (SA), jasmonate (JA), and ethylene (ET) (Robert-Seilaniantz et al., 2011; Pieterse et al., 2012). Other plant hormones such as abscisic acid (ABA), cytokinins, and auxins were recently shown to influence pathogen responses (Robert-Seilaniantz et al., 2011; Pieterse et al., 2012). SA and JA signaling processes inhibit each other (Robert-Seilaniantz et al., 2011; Pieterse et al., 2012).
Here, we found that pathogen-induced PME activity is affected by JA signaling. (+)-JA-l-Ile is the biologically active form of JA (Fonseca et al., 2009). It is produced by JASMONATE RESISTANT1, a JA-amino acid synthetase that conjugates the amino acid Ile to jasmonic acid (Staswick and Tiryaki, 2004). JA-Ile is recognized by a receptor complex constituted of the F-box protein CORONATINE INSENSITIVE1 (COI1) and JASMONATE ZIM (JAZ) transcriptional repressor proteins (Xie et al., 1998; Thines et al., 2007; Katsir et al., 2008; Sheard et al., 2010). After JA-Ile binding, JAZ proteins are ubiquitinated by the S-phase kinase-associated protein1 (Skp1), Cullin1, F-box containing (SCF)-COI1 complex and are thus targeted for degradation by the 26S proteasome (Chini et al., 2007; Thines et al., 2007). In the absence of JA, JAZ repressor proteins bind to the basic Helix-Loop-Helix transcription factors MYC2, MYC3, and MYC4 (Pauwels et al., 2010; Fernández-Calvo et al., 2011) and repress MYC2, MYC3, and MYC4-dependent gene expression. These MYC transcription factors have overlapping but distinct effects (Fernández-Calvo et al., 2011) and regulate expression of a subset of JA responsive genes together with ABA. Another branch of JA signaling incorporates ET-dependent responses and requires the ethylene response factor (ERF)-type transcription factors ERF1 and OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59; Lorenzo et al., 2003; Pré et al., 2008). ERF-type transcription factors have not been shown to interact with JAZ repressors. SA suppresses JA signaling downstream of the ET-dependent signaling branch in an ORA59-dependent manner (Van der Does et al., 2013).
PMEs control the esterification status of pectin and have been linked to pathogen success. Here, we aimed to study the role of PME genes in plant immunity using a genetics approach. We report that inoculation with the bacterial hemibiotroph P. syringae pv maculicola ES4326 (Pma ES4326) or the fungal necrotroph Alternaria brassicicola induces PME activity and decreases pectin methylesterification in Arabidopsis. PME activity is also induced after treatment with the MAMPs flg22 and elf18, but not the DAMP PEP1. We show that the pathogen-induced PME activity is host plant derived and dependent on JA signaling. Overexpression of ERF1 promotes PME activity induced by either pathogen. The MYC2 branch of JA signaling is not required for A. brassicicola-induced PME activity, but is required for Pma ES4326-induced PME activity. Plants with mutations in various PME genes are more susceptible to Pma ES4326. We did not detect measurable decreases in total PME activity in susceptible pme mutants, so distinct patterns of esterification produced by specific PMEs may be more relevant for immunity than total PME activity.
RESULTS
A. brassicicola, Pma ES4326, and MAMPs Treatment of Arabidopsis Induce Total PME Activity
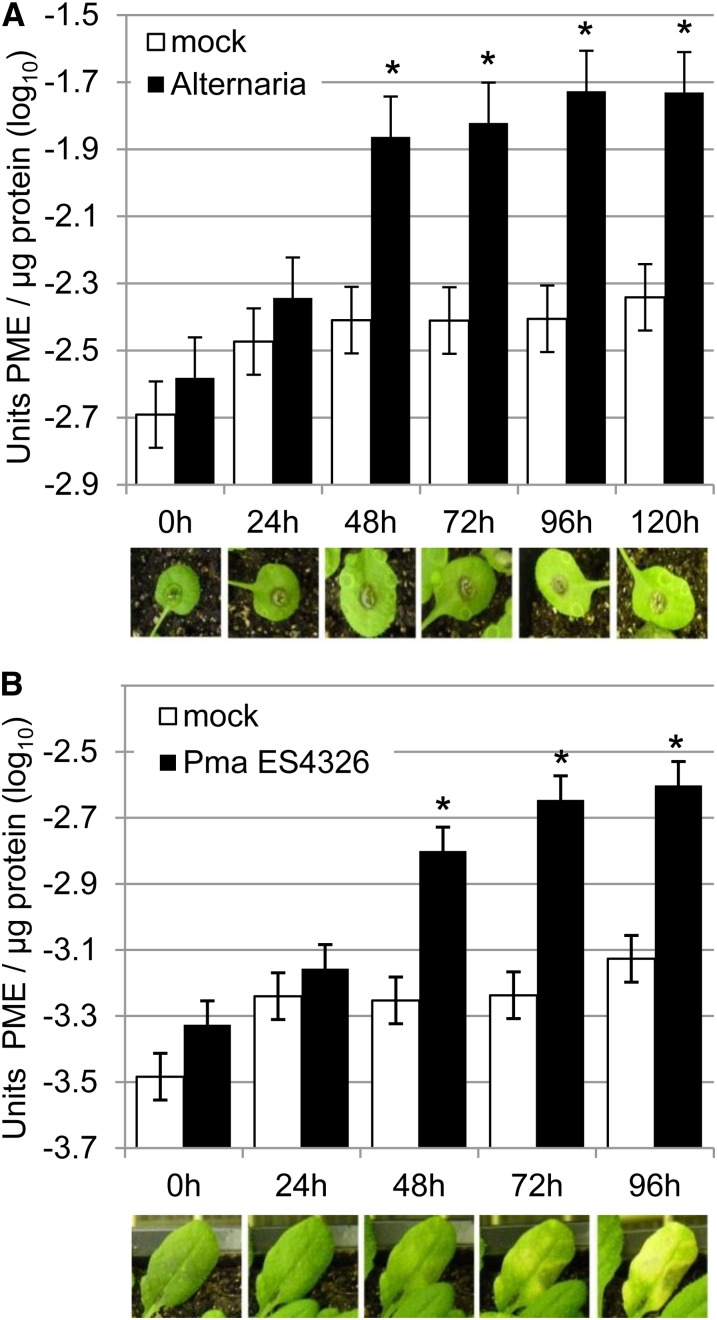
To test for effects of pathogens with different lifestyles on host PME activity, we monitored total PME activity in Arabidopsis after challenge by A. brassicicola, a fungal necrotroph to which wild-type accession ecotype Columbia 0 of Arabidopsis (Col-0) is resistant, or Pma ES4326, a hemibiotrophic bacterial pathogen to which Col-0 is susceptible. We further monitored total PME activity during pattern-triggered immunity initiated by treatment with the MAMPs flg22 or elf18 or the DAMP PEP1. PME activity was determined by extracting cytoplasmic and cell wall-bound proteins from homogenized plant samples and measuring PME activity in these samples using a gel diffusion assay (Downie et al., 1998; Supplemental Fig. S1). First, leaves of Col-0 plants were challenged with A. brassicicola or mock. Total PME activity was measured every 24 h for 5 d. Elevated PME activity was detected beginning 48 h after infection with A. brassicicola. No increase in PME activity was observed in mock-treated leaves (Fig. 1A). Second, Col-0 plants were inoculated with Pma ES4326 or mock. PME activity was measured every 24 h for 4 d. An increase in total PME activity was again detected beginning 48 h after inoculation with Pma ES4326 but not after mock treatment (Fig. 1B). Third, Col-0 plants were inoculated with elf18, flg22, PEP1, or mock. PME activity was measured after 2, 4, 8, 24, 32, and 48 h. Increased total PME activity was detected after treatment with the MAMPs at early time points but not after treatment with PEP1 (Supplemental Fig. S2). Evidently, total PME activity in Arabidopsis increases after challenge by A. brassicicola, Pma ES4326, flg22, or elf18.
Figure 1.
PME activity was induced after pathogen treatment. A, Total PME activity was induced upon challenge with A. brassicicola. Wild-type Col-0 plants were inoculated with A. brassicicola (Alternaria) or mock. Leaf tissue was harvested immediately (0 h) and after 24, 48, 72, 96, and 120 h. Total protein was extracted and PME activity determined using a gel diffusion assay. B, Total PME activity was induced upon challenge with Pma ES4326. Wild-type Col-0 plants were inoculated with Pma ES4326 (OD600 = 0.002) or mock. Experiments were performed as in A. Bars represent means and ses of data from four independent experiments, each with three technical replicates analyzed together using a mixed linear model. Asterisks indicate PME activity significantly higher than in mock-treated-plants at the same time point (q < 0.01). Photographs show representative leaves infected with A. brassicicola (A) or Pma ES4326 (B) at the indicated time points. [See online article for color version of this figure.]
Cell Wall Pectin Is Demethylesterified in Response to Pathogen Challenge
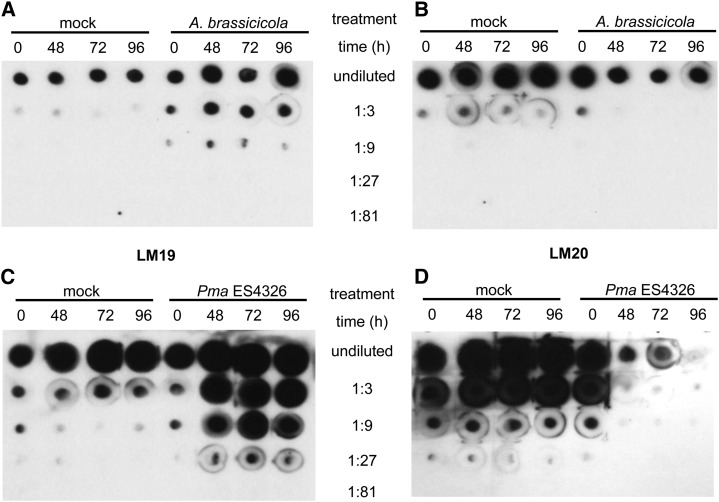
Increased PME activity might result in decreased pectin methylesterification in the cell wall. Thus, we tested for effects of pathogen challenge on pectin methylesterification by immunoblot analysis with the LM19 and LM20 antibodies (Verhertbruggen et al., 2009). LM19 binds more strongly to nonmethylesterified pectin than to methylesterified pectin, whereas LM20 requires methylester groups for binding to pectin and does not bind to nonmethylesterified pectin (Verhertbruggen et al., 2009). Plant cell wall pectins of Col-0 plants inoculated with A. brassicicola were enriched in nonmethylesterified pectin (Fig. 2A) and contained less methylesterified pectin (Fig. 2B) after infection compared with mock-inoculated plants. Similarly, cell wall pectins from Col-0 plants infected with Pma ES4326 contained more nonmethylesterified and less methylesterified pectin after infection (Fig. 2, C and D). These data show that challenge by either A. brassicicola or Pma ES4326 reduces the degree of methylesterification of pectins in the plant cell wall.
Figure 2.
Methylesterification of cell wall pectins was reduced after pathogen challenge. A and B, Nonmethylesterified pectin was enriched (A) and methylesterified pectin was reduced (B) after challenge with A. brassicicola. Wild-type Col-0 plants were inoculated with A. brassicicola (1 × 106 spores mL−1) or mock. Leaf tissue was harvested immediately (0 h) and after 48, 72, and 96 h. Pectins were extracted and samples were diluted to a final concentration equivalent to 1 nmol µL−1 GalA (undiluted). Samples were serially diluted (1:3 to 1:81) and 1 µL each was applied to a nitrocellulose membrane. Membranes were probed with LM19 (A) or LM20 (B) antibodies. C and D, nonmethylesterified pectin was enriched (C) and methylesterified pectin was reduced (D) after challenge with Pma ES4326. Experiments were performed as in A and B, but sample concentration was equivalent to 2 nmol µL−1 GalA for the undiluted sample. Two biological replicates were performed and yielded similar results.
The Pathogen-Induced PME Activity Is Plant Derived
PME activity was induced in MAMP-treated Arabidopsis samples, indicating that plants activate PMEs as part of their immune response. However, some pathogens, such as B. cinerea, are known to produce PMEs. In principle, the increase in PME activity in response to pathogen challenge could be the result of PMEs produced by the plant or PMEs produced by the pathogens. We carried out several experiments to distinguish these possibilities. We searched the pathogen genomes for PME genes that could encode PMEs responsible for the measured PME activity. A. brassicicola has four genes that have been annotated as PMEs: AB00162.1, AB01108.1, AB01671.1, and AB10201.1 (http://genome.jgi-psf.org; Cho et al., 2012; Ohm et al., 2012). Some of the A. brassicicola PMEs showed an increase in expression after infection (Supplemental Fig. S3). No PME activity could be detected in A. brassicicola grown in rich medium. We reasoned that if the PME activity in A. brassicicola-challenged leaves was derived from the fungus, then there should be more PME activity in plant genotypes that allow more growth of the fungus. Arabidopsis wild-type Col-0 plants are resistant to A. brassicicola but phytoalexin-deficient3 (pad3) plants lacking the phytoalexin camalexin (Zhou et al., 1999) or delayed dehiscence2 (dde2) plants lacking JA due to absence of allene oxide synthase activity (von Malek et al., 2002) are more susceptible (van Wees et al., 2003; Nafisi et al., 2007). We compared PME activity in A. brassicicola-challenged wild-type, pad3, and dde2 plants. The induction of PME activity in dde2 plants was reduced, whereas that in pad3 plants was similar to the wild type (Fig. 3A). We conclude that the enhanced PME activity observed during A. brassicicola infection is produced by the plant. Pma ES4326 does not contain any genes annotated as PMEs (Baltrus et al., 2011). PME activity could not be detected in Pma ES4326 grown in either rich King’s B medium or in hypersensitive response and pathogenicity-inducing minimal medium. Furthermore, pad4 and salicylic acid induction deficient2 (sid2) plants are more susceptible to Pma ES4326 and thus carry more bacteria 72 h after inoculation, but nevertheless show similar PME activity than wild-type Col-0 plants (Fig. 4A). Hence, the PME activity detected in Arabidopsis leaves after infection with Pma ES4326 is also likely of plant origin.
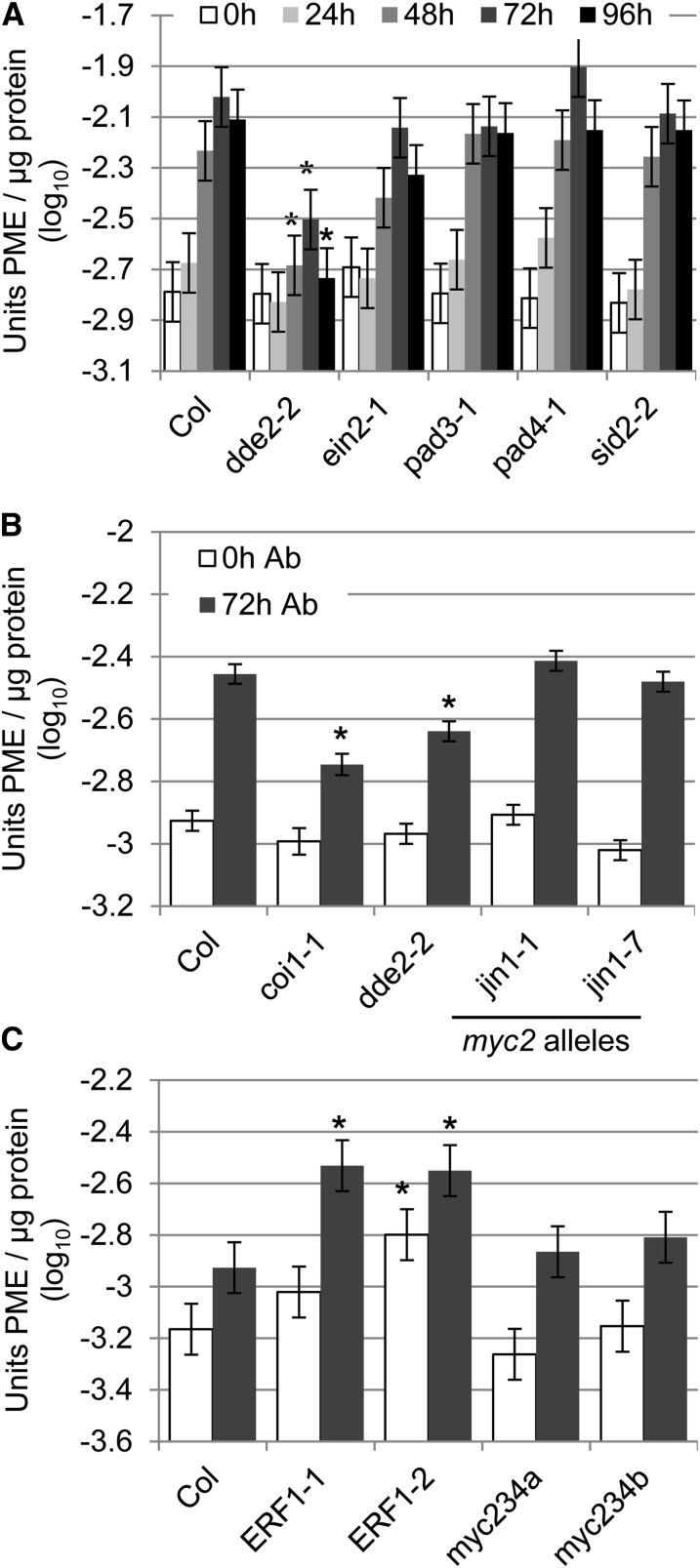
Figure 3.
Induction of PME activity by A. brassicicola required JA signaling and was promoted by ERF1. A, A. brassicicola-induced PME activity required DDE2. Wild-type Col-0 (Col) and the indicated mutants were inoculated with A. brassicicola (1 × 106 spores mL−1). PME activity was measured in tissue harvested immediately (0 h) and 24, 48, 72, and 96 h after inoculation. B, A. brassicicola-induced PME activity was unaltered in MYC2 mutants (jin1-1, jin1-7), but reduced in both a JA biosynthesis (dde2-2) and a JA coreceptor mutant (coi1-1). Experiments were performed as in A but samples harvested only after 0 and 72 h (0 h Ab and 72 h Ab). C, A. brassicicola-induced PME activity was unaltered in myc2 myc3 myc4 mutants (myc234a or b), but increased in two ERF1 overexpression lines (ERF1-1 and ERF1-2). Experiments were performed as in B. Bars represent means and ses of data from three (A and C) or four (B) independent biological replicates each with three technical replicates combined using a mixed linear model. Asterisks indicate significant differences from Col-0 at the indicated time point (q < 0.01).
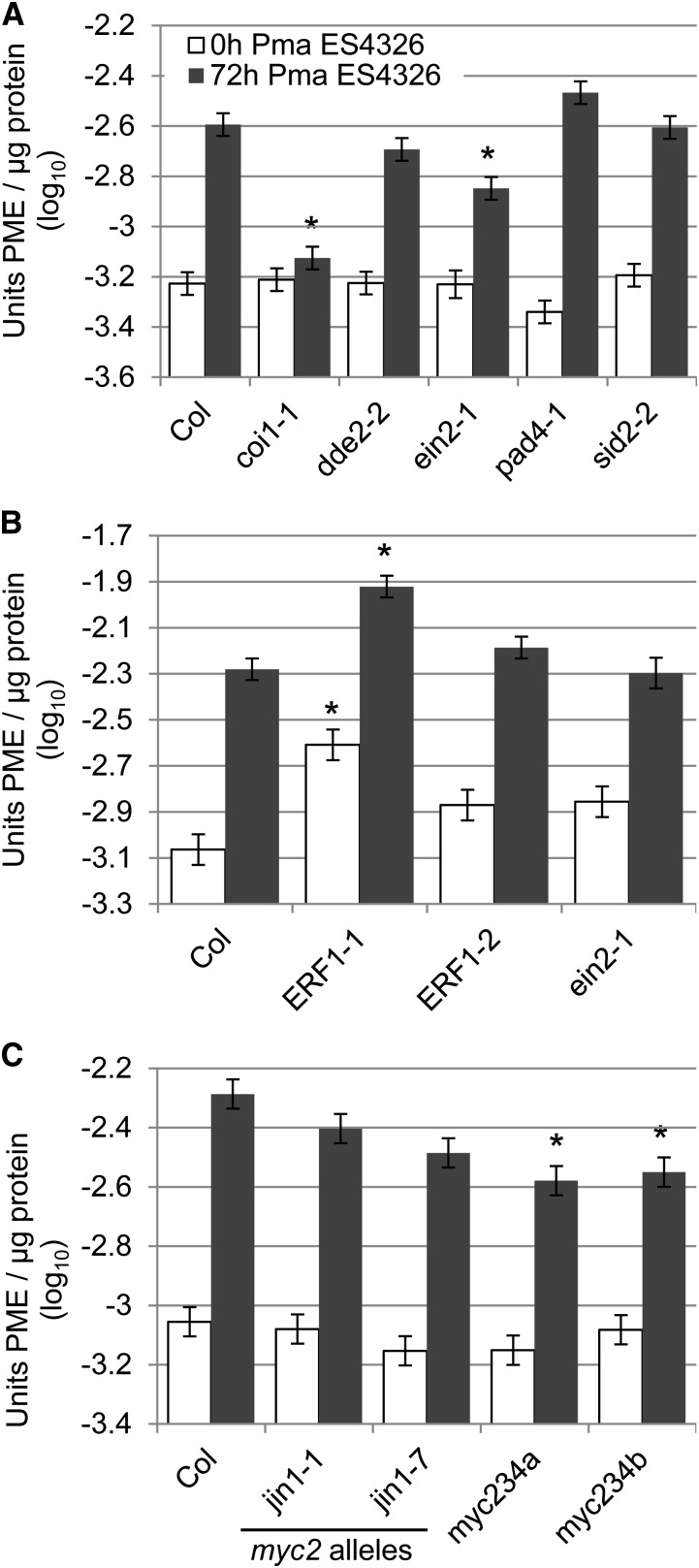
Figure 4.
Pma ES4326-induced PME activity involved both the ERF1- and the MYC2-dependent branch of JA signaling. A, Pma ES4326-induced PME activity was strongly reduced in coi1. Wild-type Col-0 (Col) plants and mutants impaired in JA perception (coi1-1), JA biosynthesis (dde2-2), ET signaling (ein2-1), PAD4-dependent signaling (pad4-1), and SA biosynthesis (sid2-2) were inoculated with Pma ES4326 (OD600 = 0.002). Leaves were harvested immediately (0 h) and after 72 h (72 h) and total PME activity was determined. B, Pma ES4326-induced PME activity was increased in the ERF1 overexpression line ERF1-1. Experiments were performed as in A. C, Pma ES4326-induced PME activity was unaltered in MYC2 single mutants jin1-1 and jin1-7 but reduced in two myc2 myc3 myc4 triple mutants. Experiments were performed as in A. Bars represent means and ses from two (A and C) or four (B) independent biological replicates with three technical replicates per sample, combined using a mixed linear model analysis. Asterisks indicate significant differences from Col-0 at each time point (q < 0.01).
Pathogen-Induced PME Activity Requires JA
The observed pathogen-induced increase in PME activity could be controlled as part of the plant immune response. To test this idea, we determined A. brassicicola-induced PME activity in plants with defects in major sectors of the immune signaling network, including dde2 (blocked in JA signaling), ethylene-insensitive2 (ein2) (blocked in ET signaling; Alonso et al., 1999), sid2 (blocked in SA signaling; Wildermuth et al., 2001), and pad4 (PAD4 is required for SA signaling and many SA-independent gene expression changes after infection by Pma ES4326; Jirage et al., 1999; Glazebrook et al., 2003; Wang et al., 2008). A. brassicicola-induced PME activity was reduced in dde2; however, some residual pathogen-inducible PME activity remained (Fig. 3A). The induction of PME activity in ein2, pad4, sid2, and pad3 plants was indistinguishable from the wild type (Fig. 3A). No difference from Col-0 could be detected for any genotype in mock-inoculated samples (Supplemental Fig. S4A). These data indicate that JA, but not ET, SA, or PAD4, is required for full A. brassicicola-induced PME activity.
To determine whether JA signaling is also required for the Pseudomonas-induced PME activity, a similar experiment was performed. Mutants dde2, ein2, pad4, and sid2 were inoculated with Pma ES4326 or mock. A coi1 mutant was also included, because Pma ES4326 produces the JA mimic coronatine and hence JA signaling in dde2, a biosynthetic mutant, is only slightly altered in Pma ES4326-infected plants (Wang et al., 2008). Pma ES4326-inducible PME activity was slightly reduced in ein2 and strongly reduced in coi1 72 h after infection (Fig. 4A). No significant difference in Pma ES4326-induced PME activity was found in dde2, pad4, or sid2. No difference from Col-0 could be detected for any genotype in the mock-inoculated samples (Supplemental Fig. S5A). These data indicate that similar to A. brassicicola-induced PME activity, JA signaling, but not SA or PAD4 signaling, is required for Pma ES4326-induced PME activity. Unlike A. brassicicola-induced PME activity, a small effect of ethylene signaling was detected in this particular set of experiments, but was not reproducible in independent sets of experiments (see below).
Both the MYC2- and the ERF1-Dependent Branches of JA Signaling Contribute to Pathogen-Induced PME Activity
Because JA signaling is required for the majority of the pathogen-inducible PME activity, we investigated the roles of the two JA signaling branches by analyzing additional JA signaling mutants, including jasmonate-insensitive1-1 (jin1-1) and jin1-7 (myc2 mutants; Lorenzo et al., 2004), two myc2 myc3 myc4 triple mutants (these lines are more completely blocked in JA signaling than myc2 single mutants; Fernández-Calvo et al., 2011), and two ERF1 overexpression lines (Solano et al., 1998; Berrocal-Lobo et al., 2002; Lorenzo et al., 2003). We used the ERF1 overexpression lines because there are 122 ERF genes in Arabidopsis (Nakano et al., 2006); ERF1 has many close homologs with redundant functions, and no phenotypes for any erf1 null mutants have been described.
We confirmed that A. brassicicola-induced PME activity measured 72 h after inoculation was reduced in dde2 (Fig. 3B). In addition, PME activity was reduced in the coi1 JA coreceptor mutant (Fig. 3B). No change in A. brassicicola-induced PME activity was found in the MYC2 mutants jin1-1 or jin1-7 (Fig. 3B). PME activity was increased in ERF1-2, one of two independent ERF1 overexpression lines tested, immediately after inoculation with mock or A. brassicicola (Fig. 3C; Supplemental Fig. S4C). Both ERF1 overexpression lines, ERF1-1 and ERF1-2, showed increased PME activity 72 h after treatment with mock or A. brassicicola (Fig. 3C; Supplemental Fig. S4C). No effect on PME activity could be detected for myc2 myc3 myc4 triple mutants (Fig. 3C). Collectively, the data show that the A. brassicicola-dependent induction of PME activity involves the JA signaling sector and is promoted by ERF1, but does not require MYC2, MYC3, and MYC4-dependent responses in the JA and ABA-dependent signaling sector.
Next, Pma ES4326-induced PME activity was examined in various JA mutants. No reduction in Pma ES4326-induced PME activity could be detected in ein2, jin1-1, or jin1-7 (Fig. 4, B and C). This result differs from Figure 4A. The reduction in Pma ES4326-induced PME activity in ein2 plants found there was smaller than the reduction in coi1, which was detected in both sets of experiments. The effect of ein2 might be too small to be reproducibly detected with the assay used. ERF1-1 showed elevated PME activity 0 and 72 h after inoculation with mock and Pma ES4326 (Fig. 4B; Supplemental Fig. S5B). No effect could be detected in the second ERF1 overexpression line, ERF1-2, possibly due to differences in ERF1 expression levels. Pma ES4326-dependent induction of PME activity was significantly reduced in myc2 myc3 myc4 triple mutants (Fig. 4C; Supplemental Fig. S5C). These data suggest that Pma ES4326-induced PME activity involves both branches of the JA signaling sector.
Expression of Arabidopsis PME Genes Is Up-Regulated upon Pathogen Challenge
We attempted to identify which of the 66 Arabidopsis PME genes are responsible for the observed pathogen-induced increases in PME activity. In publicly available microarray data, expression of three Arabidopsis PMEs (PME3, PME17, and PME41) was up-regulated upon inoculation with A. brassicicola. PME3 expression is up-regulated after infection with two other necrotrophic pathogens: B. cinerea and P. carotovorum (Raiola et al., 2011). Using quantitative reverse transcription PCR (qRT-PCR), we confirmed up-regulation of these genes in Col-0 and/or dde2 plants (Supplemental Fig. S6). PME3 is a good candidate for a plant PME responsible for the A. brassicicola-induced increase in PME activity, because its expression in Col-0 but not dde2 after A. brassicicola inoculation mirrors the PME activity measured in these genotypes.
We then measured A. brassicicola-induced PME activity in the corresponding mutants. Untreated pme3 plants were previously shown to have reduced total PME activity (Raiola et al., 2011). We confirmed this, but found that the increase in PME activity in pme3 plants 72 h after infection with A. brassicicola is indistinguishable from the increase in Col-0 (Supplemental Fig. S7A). Therefore, PME3 cannot be responsible for the change in total PME activity observed upon inoculation with A. brassicicola. We also measured A. brassicicola-induced PME activity in pme17 and pme41 mutants, which was not different from wild-type Col-0 (Supplemental Fig. S7B).
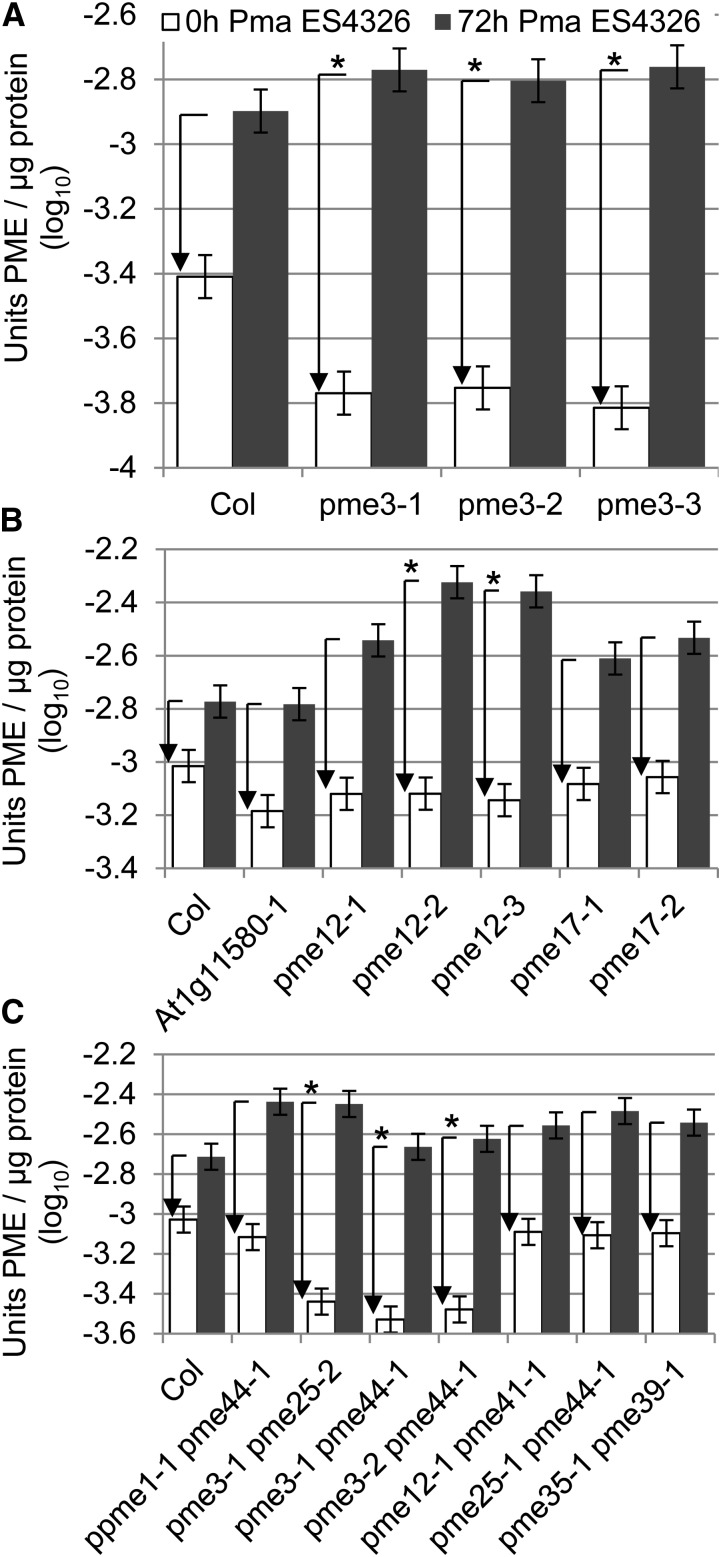
Microarray data showed that expression of three Arabidopsis PME genes (PME12, PME17, and At1g11580, which is one of three Arabidopsis PMEs that have not been assigned a number in the UniProt database; Supplemental Table S1) was up-regulated after inoculation with Pma ES4326 (Wang et al., 2008). We confirmed this by qRT-PCR (Supplemental Fig. S8). We also measured Pma ES4326-induced PME activity in the corresponding mutants and in pme3 plants. As in the A. brassicicola experiments, pme3 plants had lower PME activity in the absence of the pathogen (Supplemental Fig. S9A), but the induced level of PME activity 72 h after inoculation with Pma ES4326 was comparable to wild-type plants; thus, the induction of PME activity was increased in pme3 (Fig. 5A). Pma ES4326-induced PME activity in the At1g11580 transfer DNA (T-DNA) line and in pme17 was unaltered, but increased in two of three independent pme12 lines (Fig. 5B; Supplemental Fig. S9B).
Figure 5.
Pma ES4326-dependent induction in PME activity was not reduced but increased in some pme mutants. A, Pma ES4326-dependent induction of PME activity was enhanced in pme3 mutants. Wild-type Col-0 (Col) and pme3 plants were inoculated with Pma ES4326 (OD600 = 0.002). Leaves were harvested immediately (0 h Pma ES4326) and after 72 h (72 h Pma ES4326) and total PME activity was determined. B, Pma ES4326-dependent induction of PME activity was enhanced in pme12 mutants, but unaltered in pme17 mutants and a At1g11580 mutant. Wild-type Col-0 and pme mutant plants as specified were inoculated with Pma ES4326. Leaves were harvested immediately and after 72 h and total PME activity was determined. C, Pma ES4326-dependent induction of PME activity was enhanced in pme3 pme25 and pme3 pme44 mutants, but unaltered in ppme1 pme44, pme12 pme41, pme25 pme44, and pme35 pme39 mutants. Experiment was performed as in B. Bars represent means and ses from two independent biological replicates with three technical replicates per sample, combined using a mixed linear model. Arrows represent the change in PME activity between the 0 and the 72-h time point. Asterisks indicate significant differences to wild-type Col-0 (q < 0.01).
All together, we identified five Arabidopsis PME genes that are differentially expressed after pathogen challenge. No single PME mutant line showed a reduction in pathogen-induced PME activity, indicating that none of these genes are essential for the pathogen-induced increase in PME activity.
PME Genes Are involved in Immunity to Pma ES4326
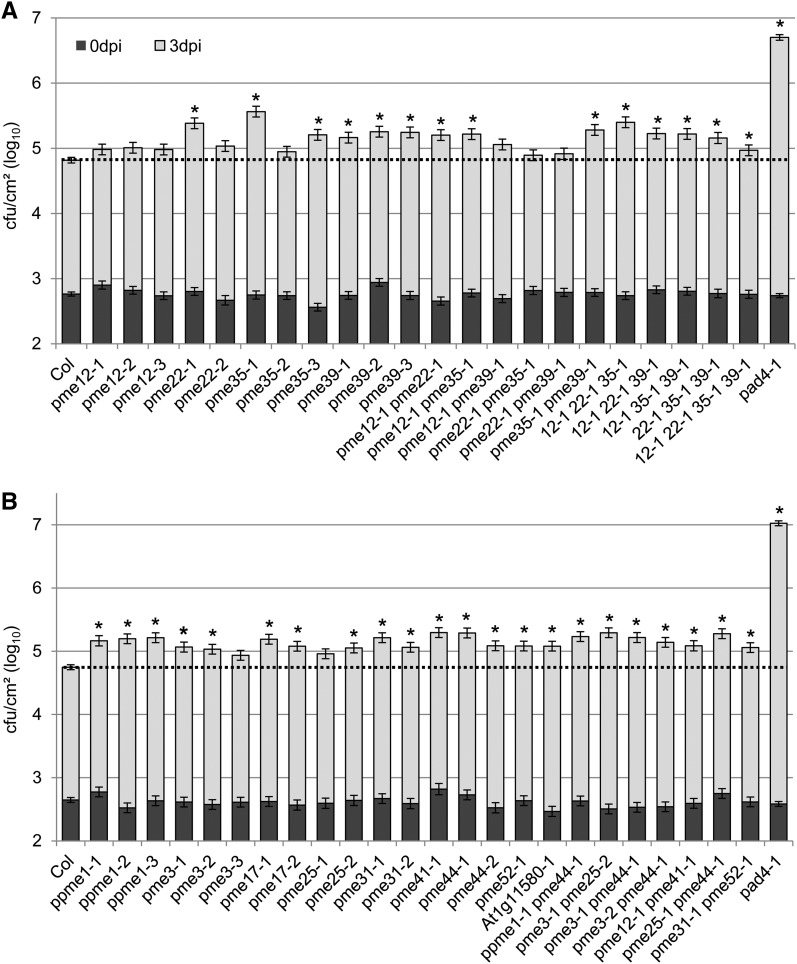
After establishing that total PME activity is enhanced upon pathogen challenge, we studied the role of PMEs in immunity by measuring pathogen growth in selected PME mutants. It was impractical to test all 66 PME genes, so we prioritized candidates using several criteria. First, we measured pathogen growth in mutants with T-DNA insertions in PME genes shown to be up-regulated after pathogen challenge. We measured growth of Pma ES4326 in pme3, an At1g11580 mutant, pme12, and pme17. None of three pme12 alleles tested showed significant changes in Pma ES4326 growth (Fig. 6A). However, two of three pme3 alleles tested, two of two pme17 alleles tested, and the At1g11580 mutant allowed significantly more growth of Pma ES4326 (Fig. 6B).
Figure 6.
Many pme mutants allowed enhanced growth of Pma ES4326. A, Single pme mutants and multiple mutants of the PME group D are more susceptible to Pma ES4326. Plants of the indicated genotypes were inoculated with Pma ES4326 (OD600 = 0.0002). For multiply mutant lines, numbers indicate PME gene and allele numbers; for example, 12-1 22-1 35-1 indicates pme12-1 pme22-1 pme35-1 triple mutant. Bacterial titers were determined immediately (0 dpi) and 3 d later (3 dpi). B, Mutants with defects in PME genes that show Pma ES4326-dependent expression and mutants with multiple mutations combined according to common cell wall alterations or expression patterns were more susceptible to Pma ES4326. Plants of the indicated genotypes were inoculated with Pma ES4326 by syringe infiltration. Bacterial titers were determined immediately (0 dpi) and 3 d later (3 dpi). Each bar represents the mean and se of three independent experiments, each with 4 or 12 biological replicates at 0 and 3 dpi, respectively, combined using a mixed linear model. Asterisks indicate significant differences from Col wild-type (q < 0.01). Susceptible pad4 plants were included as a positive control. Mutants have been plotted in numerical order. Col, Col-0; dpi, days post inoculation.
A variety of Arabidopsis T-DNA lines with insertions in genes implicated in cell wall biosynthesis or modification have been screened for cell wall alterations by Fourier transform infrared (FTIR) microspectroscopy (http://cellwall.genomics.purdue.edu). In that screen, 12 pme mutants displayed altered cell wall compositions (see Supplemental Table S1 for results of the FTIR analysis). This was our second criterion for selecting PME genes for pathogen assays. We tested Pma ES4326 growth in at least two independent T-DNA alleles of each of these PME genes, including the alleles used for FTIR analysis. For six PME genes, at least two independent mutations caused small but significant increases in susceptibility to Pma ES4326 (pme35 and pme39 in Fig. 6A; ppme1, pme17, pme31, and pme44 in Fig. 6B).
It is likely that there is some level of functional redundancy among PME family members. Phylogenetic analysis has grouped the PME genes in Arabidopsis into five clades (groups A to E; http://cellwall.genomics.purdue.edu/). We decided to create multiply mutant lines and to test pathogen growth. Because the cell wall composition of four PMEs in group A (PPME1, PME31, PME52, and PME55) and six PMEs in group D (PME12, PME22, PME32, PME35, PME39, and PME44) appeared altered in the FTIR screen, we combined T-DNA insertions according to the phylogenetic relationships of these PME genes. We combined two PMEs of group A (PME31 and PME52) and four PMEs of group D (PME12, PME22, PME35, and PME39), using the alleles tested in the FTIR screen. The double mutant derived from group A PMEs (pme31 pme52) was more susceptible to Pma ES4326 (Fig. 6B). Enhanced growth of Pma ES4326 was also detected in three group D double mutants (pme12 pme22, pme12 pme35, and pme35 pme39), all triple mutants, and the quadruple mutant (Fig. 6A).
We also combined PME mutations according to the type of alterations they caused in cell walls. We clustered all 12 pme mutants according to their FTIR spectrum differences from Col-0 using uncentered Pearson correlation with complete linkage, to determine which mutations caused similar effects. The results suggested combining pme35 pme39, as well as ppme1 and pme44 (Supplemental Fig. S10A). We have already stated that the pme35 pme39 double mutant was more susceptible to Pma ES4326 (Fig. 6A). Similarly, a ppme1 pme44 double mutant showed enhanced growth of Pma ES4326 (Fig. 6B).
Third, we selected PME genes that underwent changes in expression level in response to pathogen treatments. Expression data for selected PMEs were extracted from publicly available microarray experiments that profiled responses to a variety of biotrophic and necrotrophic pathogens and MAMPs. Hierarchical clustering identified PMEs with similar responses to infection at the gene expression level. We combined pme3, pme25, and pme44 on the basis of their shared suppression by pathogens and pme12 and pme41, which were both strongly up-regulated in response to pathogens and MAMPs (Supplemental Fig. S10B). We measured pathogen growth in pme3 pme25, pme3 pme44, pme25 pme44, and pme12 pme41 double mutants. Each double mutant allowed more growth of Pma ES4326 3 d after infection (Fig. 6B).
In summary, many PME single mutants and higher-order mutants were more susceptible to Pma ES4326 than wild-type Col-0 (Fig. 6). We tested a total of 17 PME genes and mutations in 7 of those caused increased susceptibility in at least two independent T-DNA insertion lines. None of 10 randomly selected SALK T-DNA insertion lines was more susceptible to Pma ES4326 (Supplemental Fig. S11). We conclude that many PME genes contribute to immunity against Pma ES4326.
By contrast, there were no PME genes for which two independent mutations caused altered susceptibility to A. brassicicola and none of the double, triple, or quadruple mutant lines tested showed any change in A. brassicicola growth (Supplemental Figs. S7C and S12). There is no evidence that PME genes contribute to resistance to A. brassicicola.
Total PME Activity Is Not Reduced in pme Mutants with Enhanced Susceptibility to Pma ES4326
To determine whether the enhanced susceptibility to Pma ES4326 is due to reduced total PME activity, we measured PME activity in a selection of pme mutants that showed enhanced susceptibility to Pma ES4326. We could not detect any differences in Pma ES4326-induced PME activity in any of the group D triple mutants or in the quadruple mutant that had been created according to the phylogenetic relationships of the PME genes (Supplemental Fig. S13). In addition, Pma ES4326-induced PME activity was unaltered in pme35 pme39 and ppme1 pme44 (Fig. 5C; Supplemental Fig. S9C). These double mutants had been created because the corresponding single mutants possessed similar changes in their cell wall, as indicated by clustered FTIR data. We also tested double mutants that had been created because PME genes appeared to be similarly expressed upon pathogen challenge. PME activity in pme12 pme41 and pme25 pme44 was similar to the wild type (Fig. 5C; Supplemental Fig. S9C). All double mutants containing pme3 (pme3 pme25 and pme3 pme44) showed reduced PME activity in mock-treated samples and immediately after Pma ES4326 inoculation, whereas induction of PME activity 72 h after inoculation with Pma ES4326 was higher than in Col-0 (Fig. 5C; Supplemental Fig. S9C). These results are similar to those for pme3 single mutants (Fig. 5A; Supplemental Fig. S9A). Evidently, enhanced susceptibility to Pma ES4326 is not correlated with reductions in total PME activity.
DISCUSSION AND CONCLUSION
We showed that two pathogens with very different lifestyles, the necrotrophic fungus A. brassicicola and the hemibiotrophic bacterial pathogen Pma ES4326, greatly induce PME activity in the host plants. PME activity is also increased during flg22- and elf18-induced pattern-triggered immunity, suggesting that Arabidopsis activates PME activity as part of its immune response. Increased PME activity might render the pectin in the cell wall more demethylesterified. Hence, pathogens producing pectolytic enzymes might trigger the production of OGs that activate plant defense responses resulting in increased plant immunity. The pathogen-induced increase in PME activity is the likely cause of decreased methylesterification of pectins in pathogen-challenged leaves. The observed increase in PME activity occurred rather late after pathogen challenge, between 24 and 48 h after inoculation in both cases. By this time, A. brassicicola is growing very slowly or not at all, whereas Pma ES4326 is growing rapidly, causing visible damage to infected leaves. This raises the question of the effect of induced PME activity on these plant-pathogen interactions. Perhaps the increased PME activity is a response to damage to the cell wall caused by pathogens. Degradation of pectin by pectin-degrading enzymes requires a certain level of pectin deesterification. Pectin fragments with a low degree of esterification and a size between 10 and 15 GalA residues show the highest elicitor activity (Cabrera et al., 2008; Ferrari et al., 2013). Perhaps increased PME activity triggers the production of OGs with more elicitor activity, resulting in increased induction of defense responses.
Pathogen-inducible PME activity required JA signaling, because induction by either pathogen required the JA-Ile coreceptor, COI1. Induction by A. brassicicola also required DDE2, a JA biosynthetic enzyme. As expected, DDE2 was not required for induction by Pma ES4326, because this strain produces the JA-Ile mimic, coronatine (Cui et al., 2005; Katsir et al., 2008). We studied diagnostic mutants to distinguish the roles of two branches of JA signaling. We used ERF1 overexpression lines to probe the ethylene branch of JA signaling because no loss-of-function mutants with defects specific to this branch are known. The ERF1-1 line showed higher induced levels of PME activity in response to either pathogen, whereas the ERF1-2 line did so only in experiments with A. brassicicola. Overexpression of ERF1 is known to be detrimental, causing plants to be dwarfed with elongated leaves (Solano et al., 1998; Lorenzo et al., 2003). Unsurprisingly, the ERF1 transgene is often silenced. We observed plants of a range of sizes in our experiments. We suspect that the difference in the PME levels between the lines is due to differences in the expression levels of ERF1. We conclude that the ERF-dependent branch of JA signaling, as reported by the effect of ERF1 overexpression, can promote PME activity.
We probed the effect of the MYC2-dependent branch of JA signaling using myc2 single mutants and myc2 myc3 myc4 triple mutants, which are blocked in this pathway (Dombrecht et al., 2007; Fernández-Calvo et al., 2011). We found that these mutants had reduced PME activity after Pma ES4326 infection, but did not affect activity in A. brassicicola-challenged plants. Apparently, both the JA/ET and JA/ABA branches contribute to induction of PME activity by Pma ES4326, but only the JA/ET branch contributes to induction by A. brassicicola. It seems unlikely that the mechanism underlying PME activation is different after challenge by different pathogens. Rather, the responses to the two pathogens may differ depending on which signaling pathways are activated. Pma ES4326 induces robust accumulation of SA, whereas A. brassicicola does not (De Vos et al., 2005; Wang et al., 2008). SA inhibits JA signaling by targeting GCC promoter elements in the ERF-dependent branch of JA signaling (Van der Does et al., 2013). Consistently, Pma ES4326 induces expression of the SA reporter gene PATHOGENESIS-RELATED1, the JA/ABA reporter gene VEGETATIVE STORAGE PROTEIN2, but not the JA/ET reporter gene PLANT DEFENSIN 1.2 (PDF1.2) (Wang et al., 2008). A. brassicicola does not induce PATHOGENESIS-RELATED1 or VEGETATIVE STORAGE PROTEIN2, but strongly induces PDF1.2 (De Vos et al., 2005). Thus, the likely reason that we did not observe MYC2 dependence of PME activation is that A. brassicicola does not activate the JA/ABA pathway. The ability to activate PMEs by either branch of the JA pathway makes this response robust to possible pathogen repression of one of the branches.
Although challenge with A. brassicicola induced PME activity, we did not detect any reproducible differences in disease severity of A. brassicicola in any pme mutant tested. Two single T-DNA insertion lines (pme12-3 and pme22-2) showed significantly increased disease severity, but this was not detected in allelic T-DNA lines. Thus, we have no evidence that individual PME genes are important for immunity to A. brassicicola. However, the A. brassicicola disease severity assay shows more variation than Pma ES4326 growth assays, and the feasible number of replicates is lower, resulting in less statistical power to resolve small differences. It therefore remains possible that some pme mutants have small effects on A. brassicicola disease severity.
Many of the pme mutants that we tested allowed modestly increased growth of Pma ES4326. For most of the pme single mutants that showed enhanced susceptibility phenotypes in preliminary bacterial growth assays, we tested additional independent alleles. Five PME genes were found to contribute to immunity based on findings that all of two or three alleles tested showed enhanced susceptibility (PPME1, PME17, PME31, PME39, and PME44), whereas mutations in a further three PME genes showed enhanced susceptibility, but only one allele was tested (PME41, PME52, and At1g11580). For four PME genes (PME3, PME22, PME25, and PME35), some alleles showed enhanced susceptibility, whereas others did not. There are two likely explanations. One is that the PME genes mutated in fact contribute to immunity, but with a small effect that is close to the limit of our statistical power to detect differences from the wild type, such that the effect was not detected in some alleles. Another possibility is that the effect observed in some mutants is due to secondary mutations elsewhere in the genome. This latter possibility explains pme35-1, which is very susceptible to Pma ES4326 (Fig. 6A). The allelic T-DNA line pme35-2 was no more susceptible than the wild type and pme35-3 was less susceptible than pme35-1 but more susceptible than the wild type (Fig. 6A). We noticed that the pme35-1 single mutant plants showed an altered rosette morphology (Supplemental Fig. S14, A and B). Reverse transcription PCR (RT-PCR) experiments showed that none of the pme35 alleles produce full-length transcripts, suggesting that they are all null alleles of PME35 (Supplemental Fig. S14, C and D). The altered leaf morphology and the enhanced susceptibility were lost in a backcrossed line (pme35-1_bc) and in various multiply mutant lines containing pme35-1 (Fig. 6A; Supplemental Fig. S14E). We conclude that secondary mutations in pme35-1 are responsible for the particularly high susceptibility and altered rosette morphology in this line. Importantly, the presence of secondary mutations compromising immunity is not very common in T-DNA lines, as we did not observe enhanced susceptibility in any of the 10 randomly chosen T-DNA lines that we tested. We conclude that there is a high probability that the five PME genes for which all alleles showed enhanced susceptibility in fact contribute to immunity, and it is likely that some of the PME genes for which one allele caused enhanced susceptibility also contribute to immunity. Most of the double or higher-order multiple pme mutants tested also showed enhanced susceptibility to Pma ES4326, strengthening our conclusion that some PMEs are important contributors to immunity.
Generally, PMEs can be classified as type I and type II PMEs, depending on the presence of PMEI domains (Micheli, 2001). The significance of these domains for PME activity is not clear (Micheli, 2001; Di Matteo et al., 2005; Pelloux et al., 2007). We did not detect any correlation between the type of PME mutated and disease susceptibility to Pma ES4326 (Supplemental Table S1). Mutants with defects in type I PMEs, including pme39 and pme44, as well as mutants with defects in type II PMEs, including ppme1 and pme31, exhibited significantly increased susceptibility to Pma ES4326.
Single or double mutants containing pme3 display lower basal levels of PME activity (Supplemental Figs. S7A and S9). A resulting increase in the degree of pectin methylesterification might affect Ca2+-binding and hence formation of egg-box structures in these plants. Such changes in the cell wall structure might render these plants more susceptible to Pma ES4326. However, other pme mutant plants with no detectable changes in basal PME activity show similar levels of Pma ES4326 growth, showing that changes in basal PME activity do not account for all increases in plant susceptibility.
None of the pme mutants that are more susceptible to Pma ES4326 showed reduced pathogen-induced PME activity. Curiously, pme3 and pme12 mutants displayed increased Pma ES4326-induced PME activity, possibly due to overcompensation for loss of these PMEs. The PME assay we used cannot detect small differences, so it is possible that total PME activity is slightly reduced in some pme mutants. For such undetectable differences in PME activity to explain the Pma ES4326 growth phenotypes, immunity to this pathogen would have to be very sensitive to reduced PME activity. On the other hand, some PMEs might have specific pH or substrate requirements, and may not be active with the commercial methylated pectin and the experimental conditions we used. Such issues could have prevented detection of differences in PME activities critical for disease phenotypes.
An alternative possibility is that the importance of PMEs lies not in total activity, but in their effects on the pattern of pectin methylation. We found that many pme mutants are susceptible to Pma ES4326. This indicates that the PMEs do not function redundantly, suggesting that different PMEs have specific functions. Certain PME genes are expressed after infection with various pathogens or in a tissue/developmental stage-specific manner (Pelloux et al., 2007; Lionetti et al., 2012). PME activity is affected by a variety of PMEIs, 69 in Arabidopsis, as well as pH and ionic strength in the cell wall (Lionetti et al., 2012). Specifically, overexpression of PMEI1 and PMEI2 in Arabidopsis has been shown to increase resistance to B. cinerea (Lionetti et al., 2007). Interestingly, other studies indicate that methylester distribution might be important for defense against some pathogens. For example, carrot cultivars with similar pectin content and degree of esterification show different amounts of pectin release by polygalacturonases and differences in susceptibility to the necrotrophic, pectolytic fungus Mycocentrospora acerina (LeCam et al., 1994). The difference in pectin release by polygalacturonases was diminished after PME treatment (LeCam et al., 1994), suggesting that differences in methylester distribution rather than total degree of esterification affected pathogen success. Moreover, in wheat (Triticum aestivum) near-isogenic lines, a blockwise distribution of methylesters was detected in lines susceptible to Puccinia graminis compared with a more random distribution in resistant lines (Wiethölter et al., 2003). In addition, the elicitor activity of OGs is dependent on the degree and distribution of methylesters (Cabrera et al., 2008; Osorio et al., 2008). Thus, it is likely that patterns of methylesterification affect plant-pathogen interactions. The PMEs required for full immunity to Pma ES4326 may affect patterns of methylesterification in a way that impacts immunity to this pathogen. Future studies should aim to elucidate distinct roles of individual PME genes such as by studying methylesterification patterns in pme mutant plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type Columbia (Col-0) Arabidopsis and all mutant plants (all in Col-0 background) were grown on autoclaved BM2 germinating mix (Berger Inc) in a controlled environment chamber (Conviron) at a 12-h photoperiod under 100 µm m−2 s−1 fluorescent illumination at 22°C and 75% relative humidity. Germplasm used includes dde2-2 (At5g42650; von Malek et al., 2002), ein2-1 (At5g03280; Alonso et al., 1999), pad3-1 (At3g26830; Zhou et al., 1999), pad4-1 (At3g52430; Jirage et al., 1999), sid2-2 (At1g74710; Wildermuth et al., 2001), coi1-1 (At2g39940; Xie et al., 1998), jin1-1 (At1g32640; Lorenzo et al., 2004), jin1-7 (SALK_040500), myc2 myc3 myc4 (MYC3, At5g46760; MYC4, At4g17880; Fernández-Calvo et al., 2011), and ERF1 overexpression lines (At3g23240; Solano et al., 1998; Lorenzo et al., 2003). PME mutants used are listed in Supplemental Table S1. They are T-DNA insertion lines of the SALK (Alonso et al., 2003), SAIL (Sessions et al., 2002), GABI-Kat (Kleinboelting et al., 2012), WiscDsLox (Woody et al., 2007), or SM_3 (John Innes Center) collections and were obtained from the Arabidopsis Biological Resource Center. One pme3 allele (GK002A10) has been studied previously (Raiola et al., 2011), and this allele was referred to as pme3-3 in this study.
Pathogen Strains, Growth Conditions, and Pathogen Growth Assays
Prior to each experiment Alternaria brassicicola strain ATCC96836 was grown on a modified potato dextrose agar medium (Flors et al., 2008) for 10 d at room temperature. The spores were washed from the surface of the plate with inoculation medium (1 × Gamborg B5 [Sigma], 10 mm Suc, 10 mm KH2PO4; Flors et al., 2008), and fungal hyphae were removed from the suspension by filtering through four layers of cheesecloth. The concentration of spores was determined using a hemacytometer and adjusted to 1 × 105 spores mL−1 for A. brassicicola growth assays and 1 × 106 spores mL−1 for PME activity and qRT-PCR assays. Leaves of 3- to 4-week-old Arabidopsis plants were inoculated by placing 10-µL droplets of A. brassicicola spore solution or inoculation medium (mock) on the adaxial leaf surface. Inoculated plants were kept at 100% relative humidity. Samples were harvested at various time points by cutting out the infection sites using a cork borer. For A. brassicicola growth assays, at least 15 leaf discs from five to six plants were pooled per biological sample. Disease severity, a proxy of fungal growth, was determined as previously described (Tsuda et al., 2009; Botanga et al., 2012). For PME assays, at least four leaf discs from one plant were combined for each biological sample.
Pma ES4326 was grown overnight at room temperature in King’s B medium supplemented with 50 µg mL−1 of streptomycin. The bacteria were harvested by centrifugation, washed twice, and diluted to the desired density (for bacterial growth assays, OD600 = 0.0002 for PME activity; for qRT-PCR assays, OD600 = 0.002) in 5 mm MgSO4. To measure bacterial growth, bacteria were inoculated into 4- to 5-week-old plants using a needleless syringe and bacterial titers determined immediately or 3 d after infection as previously described (Tsuda et al., 2008). To measure PME activity, plants were inoculated with bacteria or mock (5 mm MgSO4) and three leaves from one plant were collected at the indicated time points per biological replicate. To measure PME activity in culture grown bacteria, Pma ES4326 was grown overnight at room temperature in either King’s B or hypersensitive response and pathogenicity-inducing minimal medium (Kim et al., 2009) using 0.2% Fru as a carbon source.
PME Activity Assay
Leaf tissue was collected at the indicated time points, flash frozen, and pulverized and cytoplasmic and cell wall-bound proteins were extracted by vortexing in cell wall extraction buffer (0.1 m citrate to 0.2 m Na2HPO4, 1 m sodium chloride, pH 5.0). Protein concentration was measured using Bradford reagent (BioRad), diluted to equal concentration and used to perform a gel diffusion assay (Downie et al., 1998). For the gel diffusion assay, petri dishes (10-cm diameter) were filled with 13 mL of a gel made of highly esterified pectin (0.1% pectin ≥ 85% esterified [Sigma P9561], 1% agarose, 12.5 mm citric acid, 50 mm Na2HPO4, pH 7.0; Lionetti et al., 2007) and four holes per plate were cut out using a number 2 cork borer. Wells were filled with the protein extract (40 µL volume) and plates were incubated at 30°C for 16 h. Plates were then stained with 0.05% Ruthenium Red (MP Biomedicals) for 30 min and destained with water. Plates were scanned and darker colored areas that are indicative of deesterified pectin were measured using ImageJ software. At least two independent biological replicates with three technical replicates each were performed. Known amounts of commercially available PME from orange peel (Sigma P5400) were used as standards. Linear regression was fit to the darker colored area size versus log10PME units of the standards to obtain the standard curve, which was used to calculate the total PME activity of the sample extracts.
Cyclohexane Diamine Tetraacetic Acid Extraction of Cell Wall Pectin
Leaf tissue from inoculated plants was collected at the indicated time points, flash frozen, and pulverized, and cell wall pectin was extracted with extraction buffer (50 mm Trizma, 50 mm cyclohexane diamine tetraacetic acid, pH 7.2) at 95°C for 15 min (Siedlecka et al., 2008). Debris was precipitated by centrifugation for 10 min at 10,000g and the supernatant containing the pectin was divided in two aliquots per sample and freeze dried. One aliquot was used to measure total uronic acids (Filisetti-Cozzi and Carpita, 1991). The concentration of pectin as equivalent to GalA was calculated by comparison with known amounts of GalA. The second aliquot was resuspended in water to achieve equal concentrations of pectin and used for dot blot experiments.
Dot Blot Experiments
Pectin solutions were serially diluted and nitrocellulose membranes were spotted with 1 µL of the diluted pectin solutions. Membranes were dried overnight, blocked with 5% milk in 1 × phosphate-buffered saline (PBS; 8 g L−1 sodium chloride, 0.2 g L−1 KCl, 1.44 g L−1 Na2HPO4, 0.24 g L−1 KH2PO4, pH 7.4) and probed with LM19 or LM20 antibodies (Verhertbruggen et al., 2009). LM19 and LM20 were diluted 1:250 in 5% milk powder in 1 × PBS. A goat anti-rat horseradish peroxidase-conjugated antibody (Bethyl A110-105P) was used 1:5,000 diluted in 5% milk powder in 1 × PBS as secondary antibody, and membranes were washed with 1 × PBS. Dot blots were developed using the Amersham ECL Prime system (GE Healthcare).
Expression Analysis
Tissue from inoculated plants was harvested at the indicated time points, flash frozen, and pulverized, and RNA was extracted using Trizol (Invitrogen). qRT-PCR was performed as previously described (Truman and Glazebrook, 2012). For A. brassicicola gene expression, β-TUBULIN (AbTUB, AB07628.1) was used as the internal reference. To verify T-DNA insertion lines, RT-PCR was performed using the One-Step RT-PCR kit (Qiagen) according to the manufacturer’s instructions. Actin2 was used as stably expressed reference gene. The primer sets used are provided in Supplemental Table S2.
Clustering of Microarray Data
Affymetrix full genome (ATH1) microarray data were downloaded from a variety of sources detailed below. The data were normalized using the gcrma package of the Bioconductor suite of programs within the R statistical environment (Gentleman et al., 2004; Wu et al., 2004). Replicates for different treatments, if available, were averaged and the log2 ratios to control treatments were calculated. Clustering was performed using CLUSTER and visualized using TREEVIEW (Eisen et al., 1998). Prior to clustering, genes and treatments were ordered with the self-organizing maps algorithm and complete linkage hierarchical clustering was performed using an uncentered Pearson correlation metric.
Microarray data from this article can be found in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) data libraries under accession numbers GSE431, GSE5525, GSE5615, GSE5616, GSE5684, GSE12856, GSE13739, GSE18978, GSE22274, and GSE50526, and in the NASCArrays database under numbers 120 and 447. A list of accession numbers for all Arabidopsis PME genes described in this study is provided in Supplemental Table S1. Sequence data for all other Arabidopsis genes from this article can be found in The Arabidopsis Information Resource (http://www.arabidopsis.org) under accession numbers At5g42650 (DDE2, AOS), At5g03280 (EIN2), At3g26830 (PAD3, CYP71B15), At3g52430 (PAD4), At1g74710 (SID2, ICS1), At2g39940 (COI1), At1g32640 (MYC2), At5g46760 (MYC3), At4g17880 (MYC4), At3g23240 (ERF1), and At3g18780 (ACTIN2). Genome sequence data and gene information for A. brassicicola strain ATCC96836 can be found in the JGI database (http://genome.jgi-psf.org) using the following identifiers AB00162.1, AB01108.1, AB01671.1, AB10201.1, and β-TUBULIN (AB07628.1). Sequence data for larger contigs, each containing multiple genes, can be found in the GenBank data library under accession numbers ACIW01000050.1 (AB00162.1), ACIW01000290.1 (AB01108.1), ACIW01000452.1 (AB01671.1), ACIW01003035.1 (AB10201.1), and ACIW01002235.1 (β-TUBULIN).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PME activity of commercially available purified PME from orange peel.
Supplemental Figure S2. PME activity is induced by treatment with the MAMPs flg22 and elf18 but not the DAMP PEP1.
Supplemental Figure S3. A. brassicicola PME genes were differentially expressed during infection.
Supplemental Figure S4. PME activity in mock-inoculated samples corresponding to Fig. 3.
Supplemental Figure S5. PME activity in mock-inoculated samples corresponding to Fig. 4.
Supplemental Figure S6. Two Arabidopsis PME genes, other than PME3, showed increased expression after challenge with A. brassicicola.
Supplemental Figure S7. Arabidopsis PME genes up-regulated after inoculation with A. brassicicola were not required for A. brassicicola-induced PME activity or immunity against A. brassicicola.
Supplemental Figure S8. Expression of three Arabidopsis PME genes was up-regulated after challenge with Pma ES4326.
Supplemental Figure S9. PME activity in mock-inoculated samples corresponding to Figure 6.
Supplemental Figure S10. Clustering of pme mutants by cell wall differences and of PME genes by their pathogen-induced expression.
Supplemental Figure S11. Randomly selected T-DNA lines showed no difference in growth of Pma ES4326.
Supplemental Figure S12. Single mutants and multiply mutant lines with known cell wall alterations do not show reproducible differences in A. brassicicola disease severity.
Supplemental Figure S13. Pma ES4326-induced PME activity was unaltered in plants with multiple mutations in group D PMEs.
Supplemental Figure S14. Altered rosette morphology and enhanced susceptibility of pme35-1 were due to second site mutation(s).
Supplemental Table S1. List of Arabidopsis PME genes.
Supplemental Table S2. List of primers used.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for T-DNA insertion lines and ERF1 overexpression lines, Paul Knox’s laboratory for the LM19/LM20 antibodies, Roberto Solano’s laboratory for myc2 myc3 myc4 triple mutants, Barbara Kunkel for jin1-1 and jin1-7 seeds, and George Haughn’s laboratory for SALK_059908 and SALK_067447 seeds.
Glossary
- PME
pectin methylesterase
- HG
homogalacturonan
- PMEI
pectin methylesterase inhibitor
- MAMP
microbe-associated molecular pattern
- DAMP
damage-associated molecular pattern
- OG
oligogalacturonide
- SA
salicylic acid
- JA
jasmonate
- ET
ethylene
- ABA
abscisic acid
- ERF
ethylene response factor
- Col-0
ecotype Columbia 0 of Arabidopsis
- qRT-PCR
quantitative reverse transcription PCR
- T-DNA
transfer DNA
- FTIR
Fourier transform infrared
- RT-PCR
reverse transcription PCR
- PBS
phosphate-buffered saline (8 g L−1 sodium chloride, 0.2 g L−1 KCl, 1.44 g L−1 Na2HPO4, 0.24 g L−1 KH2PO4, pH 7.4)
References
- Albersheim P, Jones TM, English PD. (1969) Biochemistry of the cell wall in relation to infective processes. Annu Rev Phytopathol 7: 171–194 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog 7: e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger RK, Reid PD. (1979) Role of polygalacturonase in bean leaf abscission. Plant Physiol 63: 1133–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Botanga CJ, Bethke G, Chen Z, Gallie DR, Fiehn O, Glazebrook J. (2012) Metabolite profiling of Arabidopsis inoculated with Alternaria brassicicola reveals that ascorbate reduces disease severity. Mol Plant Microbe Interact 25: 1628–1638 [DOI] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera JC, Boland A, Messiaen J, Cambier P, Van Cutsem P. (2008) Egg box conformation of oligogalacturonides: The time-dependent stabilization of the elicitor-active conformation increases its biological activity. Glycobiology 18: 473–482 [DOI] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D. (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344: 1879–1900 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Cho Y, Srivastava A, Ohm RA, Lawrence CB, Wang KH, Grigoriev IV, Marahatta SP. (2012) Transcription factor Amr1 induces melanin biosynthesis and suppresses virulence in Alternaria brassicicola. PLoS Pathog 8: e1002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Bahrami AK, Pringle EG, Hernandez-Guzman G, Bender CL, Pierce NE, Ausubel FM. (2005) Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci USA 102: 1791–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bary A. (1886) Über einige Sclerotinien und Sclerotinienkrankheiten. Botanische Zeitung 44: 377–480 [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RM, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux JP, Van Loon LC, et al. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18: 923–937 [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Giovane A, Raiola A, Camardella L, Bonivento D, De Lorenzo G, Cervone F, Bellincampi D, Tsernoglou D. (2005) Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 17: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie B, Dirk LM, Hadfield KA, Wilkins TA, Bennett AB, Bradford KJ. (1998) A gel diffusion assay for quantification of pectin methylesterase activity. Anal Biochem 264: 149–157 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espino JJ, Gutiérrez-Sánchez G, Brito N, Shah P, Orlando R, González C. (2010) The Botrytis cinerea early secretome. Proteomics 10: 3020–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, Lorenzo GD. (2013) Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front Plant Sci 4: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filisetti-Cozzi TM, Carpita NC. (1991) Measurement of uronic acids without interference from neutral sugars. Anal Biochem 197: 157–162 [DOI] [PubMed] [Google Scholar]
- Flors V, Ton J, van Doorn R, Jakab G, García-Agustín P, Mauch-Mani B. (2008) Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J 54: 81–92 [DOI] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R. (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovane A, Servillo L, Balestrieri C, Raiola A, D’Avino R, Tamburrini M, Ciardiello MA, Camardella L. (2004) Pectin methylesterase inhibitor. Biochim Biophys Acta 1696: 245–252 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Chen WJ, Estes B, Chang HS, Nawrath C, Métraux JP, Zhu T, Katagiri F. (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34: 217–228 [DOI] [PubMed] [Google Scholar]
- Harholt J, Suttangkakul A, Vibe Scheller H. (2010) Biosynthesis of pectin. Plant Physiol 153: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis MC. (1984) Structure and properties of pectin gels in plant-cell walls. Plant Cell Environ 7: 153–164 [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Nat l Acad Sci USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolie RP, Duvetter T, Van Loey AM, Hendrickx ME. (2010) Pectin methylesterase and its proteinaceous inhibitor: a review. Carbohydr Res 345: 2583–2595 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kars I, Krooshof GH, Wagemakers L, Joosten R, Benen JA, van Kan JA. (2005) Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J 43: 213–225 [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Park JH, Park TH, Bronstein PA, Schneider DJ, Cartinhour SW, Shuler ML. (2009) Effect of iron concentration on the growth rate of Pseudomonas syringae and the expression of virulence factors in hrp-inducing minimal medium. Appl Environ Microbiol 75: 2720–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. (2012) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Johansen S, Shishido A, Todorova T, Martinez R, Defeo E, Obregon P. (2009) Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J 60: 974–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam B, Massiot P, Campion C, Rouxel F. (1994) Susceptibility of carrot cultivars to Mycocentrospora acerina and the structure of cell wall polysaccharides. Physiol Mol Plant Pathol 45: 139–151 [Google Scholar]
- Limberg G, Körner R, Buchholt HC, Christensen TM, Roepstorff P, Mikkelsen JD. (2000a) Analysis of different de-esterification mechanisms for pectin by enzymatic fingerprinting using endopectin lyase and endopolygalacturonase II from A. niger. Carbohydr Res 327: 293–307 [DOI] [PubMed] [Google Scholar]
- Limberg G, Körner R, Buchholt HC, Christensen TM, Roepstorff P, Mikkelsen JD. (2000b) Quantification of the amount of galacturonic acid residues in blocksequences in pectin homogalacturonan by enzymatic fingerprinting with exo- and endo-polygalacturonase II from Aspergillus niger. Carbohydr Res 327: 321–332 [DOI] [PubMed] [Google Scholar]
- Liners F, Letesson JJ, Didembourg C, Van Cutsem P. (1989) Monoclonal antibodies against pectin: recognition of a conformation induced by calcium. Plant Physiol 91: 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Cervone F, Bellincampi D. (2012) Methyl esterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases. J Plant Physiol 169: 1623–1630 [DOI] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D. (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic O, Janecek S. (2004) Pectin methylesterases: sequence-structural features and phylogenetic relationships. Carbohydr Res 339: 2281–2295 [DOI] [PubMed] [Google Scholar]
- Micheli F. (2001) Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci 6: 414–419 [DOI] [PubMed] [Google Scholar]
- Mohnen D. (2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J. (2007) Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA, Barry KW, Condon BJ, Copeland AC, Dhillon B, Glaser F, et al. (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog 8: e1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Castillejo C, Quesada MA, Medina-Escobar N, Brownsey GJ, Suau R, Heredia A, Botella MA, Valpuesta V. (2008) Partial demethylation of oligogalacturonides by pectin methyl esterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca). Plant J 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Höfte H. (2011) Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol 21: 1720–1726 [DOI] [PubMed] [Google Scholar]
- Pelloux J, Rustérucci C, Mellerowicz EJ. (2007) New insights into pectin methylesterase structure and function. Trends Plant Sci 12: 267–277 [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiola A, Camardella L, Giovane A, Mattei B, De Lorenzo G, Cervone F, Bellincampi D. (2004) Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett 557: 199–203 [DOI] [PubMed] [Google Scholar]
- Raiola A, Lionetti V, Elmaghraby I, Immerzeel P, Mellerowicz EJ, Salvi G, Cervone F, Bellincampi D. (2011) Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol Plant Microbe Interact 24: 432–440 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedlecka A, Wiklund S, Péronne MA, Micheli F, Lesniewska J, Sethson I, Edlund U, Richard L, Sundberg B, Mellerowicz EJ. (2008) Pectin methyl esterase inhibits intrusive and symplastic cell growth in developing wood cells of Populus. Plant Physiol 146: 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao QM, Ecker JR. (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I. (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Have A, Mulder W, Visser J, van Kan JAL. (1998) The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol Plant Microbe Interact 11: 1009–1016 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu GH, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Truman W, Glazebrook J. (2012) Co-expression analysis identifies putative targets for CBP60g and SARD1 regulation. BMC Plant Biol 12: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. (2008) Interplay between MAMP-triggered and SA-mediated defense responses. Plant J 53: 763–775 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valette-Collet O, Cimerman A, Reignault P, Levis C, Boccara M. (2003) Disruption of Botrytis cinerea pectin methylesterase gene Bcpme1 reduces virulence on several host plants. Mol Plant Microbe Interact 16: 360–367 [DOI] [PubMed] [Google Scholar]
- Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Körbes AP, Memelink J, Ritsema T, et al. (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees SC, Chang HS, Zhu T, Glazebrook J. (2003) Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol 132: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y, Marcus SE, Haeger A, Ordaz-Ortiz JJ, Knox JP. (2009) An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr Res 344: 1858–1862 [DOI] [PubMed] [Google Scholar]
- Volpi C, Janni M, Lionetti V, Bellincampi D, Favaron F, D’Ovidio R. (2011) The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol Plant Microbe Interact 24: 1012–1019 [DOI] [PubMed] [Google Scholar]
- von Malek B, van der Graaff E, Schneitz K, Keller B. (2002) The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- Wang L, Mitra RM, Hasselmann KD, Sato M, Lenarz-Wyatt L, Cohen JD, Katagiri F, Glazebrook J. (2008) The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: roles of major regulators and the phytotoxin coronatine. Mol Plant Microbe Interact 21: 1408–1420 [DOI] [PubMed] [Google Scholar]
- Wiethölter N, Graessner B, Mierau M, Mort AJ, Moerschbacher BM. (2003) Differences in the methyl ester distribution of homogalacturonans from near-isogenic wheat lines resistant and susceptible to the wheat stem rust fungus. Mol Plant Microbe Interact 16: 945–952 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Willats WG, Orfila C, Limberg G, Buchholt HC, van Alebeek GJ, Voragen AG, Marcus SE, Christensen TM, Mikkelsen JD, Murray BS, et al. (2001) Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J Biol Chem 276: 19404–19413 [DOI] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ. (2007) The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res 120: 157–165 [DOI] [PubMed] [Google Scholar]
- Wu ZJ, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. (2004) A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99: 909–917 [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Zablackis E, Huang J, Müller B, Darvill AG, Albersheim P. (1995) Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol 107: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Glazebrook J. (1999) Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]