Recent breakthroughs and large-scale profiling are unraveling TOR signaling in plant growth, proliferation, and metabolism.
Abstract
The target of rapamycin (TOR) kinase, a master regulator that is evolutionarily conserved among yeasts (Saccharomyces cerevisiae), plants, animals, and humans, integrates nutrient and energy signaling to promote cell proliferation and growth. Recent breakthroughs made possible by integrating chemical, genetic, and genomic analyses have greatly increased our understanding of the molecular functions and dynamic regulation of the TOR kinase in photosynthetic plants. TOR signaling plays fundamental roles in embryogenesis, meristem activation, root and leaf growth, flowering, senescence, and life span determination. The molecular mechanisms underlying TOR-mediated ribosomal biogenesis, translation promotion, readjustment of metabolism, and autophagy inhibition are now being uncovered. Moreover, monitoring photosynthesis-derived Glc and bioenergetics relays has revealed that TOR orchestrates unprecedented transcriptional networks that wire central metabolism and biosynthesis for energy and biomass production. In addition, these networks integrate localized stem/progenitor cell proliferation through interorgan nutrient coordination to control developmental transitions and growth.
The modulation of growth and development is a central process in all organisms, and an intimate relationship exists between nutrient availability, energy status, and cell growth rate, which are influenced by dynamic and diverse environmental stresses and challenges. The target of rapamycin (TOR), a Ser/Thr protein kinase, has emerged as a central coordinator of nutrient, energy, and stress signaling networks (Wullschleger et al., 2006; Laplante and Sabatini, 2012; Robaglia et al., 2012; Cornu et al., 2013; Dobrenel et al., 2013; Yuan et al., 2013). TOR was first identified in budding yeast (Saccharomyces cerevisiae) through genetic mutant screens for resistance to rapamycin, an immunosuppressant that blocks human T cell activation and proliferation (Heitman et al., 1991). Two TOR genes have been identified in yeast, but subsequent studies have identified only one TOR gene in Arabidopsis (Arabidopsis thaliana), Chlamydomonas reinhardtii, most animals, and humans. TOR is an atypical Ser/Thr protein kinase that belongs to the phosphoinositide 3-kinase-related kinase family and is structurally and functionally conserved among all eukaryotes (Wullschleger et al., 2006; Robaglia et al., 2012). Arabidopsis TOR shares high amino acid sequence similarity with human TOR, especially in the protein kinase domain (75% similarity; Fig. 1A), suggesting that these protein kinases potentially share similar properties and protein substrates (Menand et al., 2002; Ahn et al., 2011; Xiong and Sheen, 2012; Montané and Menand, 2013; Xiong et al., 2013). Understanding how a single protein kinase acts as a master regulator to modulate a myriad of cellular activities via multiple partners and effectors in complex signaling networks remains a fascinating challenge in both plants and animals. This review focuses on recent progress in unraveling the functions and regulatory mechanisms of TOR signaling networks in plant growth, proliferation, and metabolism.
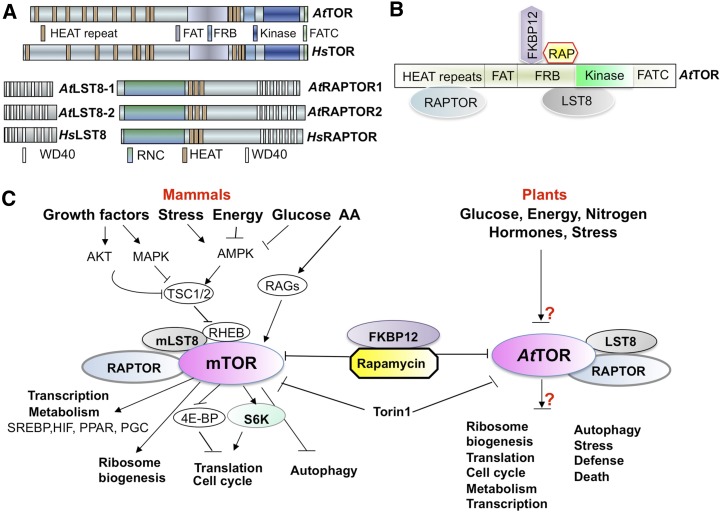
Figure 1.
TOR signaling in Arabidopsis and mammals. A, Conservation of domain organization in human (Hs) and Arabidopsis (At) TOR signaling components (Perry and Kleckner, 2003). B, Interaction map of Arabidopsis TOR domains and RAPTOR, LST8, FKBP12, and rapamycin (RAP). C, Plant and mammalian TOR signaling networks. AA, Amino acid; FAT, FRAP, ATM, and TRRAP domain; FATC, Carboxy-terminal FAT domain; HEAT repeats, Huntingtin, Elongation factor 3, subunit of protein phosphatase 2A and TOR1; HIF, hypoxia-inducible factor; PGC, peroxisome proliferator-activated receptor-γ coactivator; PPAR, peroxisome proliferator-activated receptor; RNC, Raptor N-terminal Conserved/putative Caspase domain; SREBP, sterol regulatory element-binding protein; TSC1/TSC2, tuberous sclerosis1/tuberous sclerosis2; WD40, WD40 repeat domain.
EVOLUTIONARY CONSERVATION OF TOR KINASES
In yeast and mammals, TOR forms at least two structurally and functionally distinct complexes, target of rapamycin complex1 (TORC1) and TORC2. Each complex contains shared and distinct TOR interacting partners, which recruit and regulate diverse TOR kinase substrates to control a variety of biological processes that contribute to cell growth, metabolism, and proliferation (Wullschleger et al., 2006; Laplante and Sabatini, 2012; Cornu et al., 2013; Kang et al., 2013). The rapamycin-sensitive mechanistic/mammalian target of rapamycin C1 (mTORC1), which contains mechanistic/mammalian target of rapamycin (mTOR), mammalian lethal with sec-13 protein8 (mLST8), and regulatory associate protein of target of rapamycin (RAPTOR) in its core, responds to amino acids, Glc, insulin, and growth factors to control proliferation and temporal cell growth by promoting anabolic processes (e.g. translation, transcription, and ribosome biogenesis) but negatively regulating autophagy (Fig. 1C). Although mTOR has been shown to enter the nucleus to directly regulate transcription (Cunningham et al., 2007), most of the functions of mTORC1 are mediated through translational control by its most well-characterized substrates, S6 kinase (S6K) and eukaryotic translation initiation factor 4E binding protein1 (4E-BP1; Fig. 1, B and C; Wullschleger et al., 2006; Düvel et al., 2010; Laplante and Sabatini, 2012; Cornu et al., 2013). A recent study identified lipin1 as a novel TOR substrate with complex phosphorylation sites that modulates nuclear eccentricity, lipin1 localization, and lipo- and sterol-genic transcription (Peterson et al., 2011). By contrast, mTORC2, which is insensitive to rapamycin and contains mTOR, mLST8, and rapamycin-insensitive companion of mTOR (RICTOR) as core components, controls spatial cell growth by regulating cytoskeleton structure and polarity, as well as glycolysis, glycogenesis, lipogenesis, and gluconeogenesis, via Akt phosphorylation (Wullschleger et al., 2006; Laplante and Sabatini, 2012; Cornu et al., 2013).
The precise compositions of the TOR kinase complexes have not been systematically characterized in plants. Some of the mTORC1 components and downstream effectors have been identified in photosynthetic eukaryotes through sequence similarity searches from C. reinhardtii to Arabidopsis, including Arabidopsis RAPTOR1/RAPTOR2, LST8-1/LST8-2, S6K1/S6K2, ribosome protein small subunit6 (RPS6A/B), type 2A-phosphatase-associated protein 46 kD (TAP46), and ErbB-3 epidermal growth factor receptor binding protein (EBP1; Fig. 1A; Table I; Turck et al., 2004; Anderson et al., 2005; Deprost et al., 2005; Horváth et al., 2006; Mahfouz et al., 2006; Díaz-Troya et al., 2008; Creff et al., 2010; Ahn et al., 2011; Moreau et al., 2012; Ren et al., 2012; Xiong and Sheen, 2012). Arabidopsis RAPTOR1 interacts with the Huntingtin, Elongation factor 3, subunit of protein phosphatase 2A, and TOR1 (HEAT) repeat domain, and LST8-1 directly binds to the FKBP-rapamycin binding (FRB) and kinase domains of TOR, indicating that they are functional TOR signaling components in plants (Fig. 1B; Mahfouz et al., 2006; Moreau et al., 2012). In addition to its similar rapamycin sensitivity and its role in modulating endogenous S6K phosphorylation and nutrient-dependent growth, endogenous Arabidopsis TOR kinase (which is pulled down by a specific TOR antibody) directly phosphorylates human 4E-BP1, strongly indicating the existence of a conserved and functional TORC1 in plants (Fig. 1C; Xiong and Sheen, 2012; Xiong et al., 2013). By contrast, there is no evidence of a plant TORC2 to date, because specific components of this complex, such as RICTOR and stress-activated mitogen-activated protein kinase-interacting protein1, seem to be absent from the genomes of photosynthetic organisms. However, plants may still possess a functional equivalent of TORC2, although its complex components may differ from those in its yeast and mammalian counterparts. It is also possible that TOR forms unique complexes that serve specialized functions; future research may aid in the discovery of novel TOR complexes.
Table I. Putative Arabidopsis TOR signaling components.
| Gene/Protein Name | Mammals | Arabidopsis | Arabidopsis Gene Initiative identifier | References |
|---|---|---|---|---|
| Target of rapamycin | mTOR | TOR | At1g50030 | Menand et al., 2002 |
| Deprost et al., 2007 | ||||
| Liu and Bassham, 2010 | ||||
| Ren et al., 2011 | ||||
| Ren et al., 2012 | ||||
| Xiong and Sheen, 2012 | ||||
| Caldana et al., 2013 | ||||
| Xiong et al., 2013 | ||||
| Montané and Menand, 2013 | ||||
| Regulatory associate protein of TOR | RAPTOR | RAPTOR1 | At3g08850 | Anderson et al., 2005 |
| RAPTOR2 | At5g01770 | Deprost et al., 2005 | ||
| Mahfouz et al., 2006 | ||||
| Lethal with sec-13 protein8 | LST8 | LST8-1 | At3g18140 | Moreau et al., 2012 |
| LST8-2 | At2g22040 | |||
| S6 kinase | S6K | S6K1 | At3g08730 | Turck et al., 2004 |
| S6K2 | At3g08720 | Mahfouz et al., 2006 | ||
| Henriques et al., 2010 | ||||
| Schepetilnikov et al., 2011 | ||||
| Shin et al., 2012 | ||||
| Xiong and Sheen, 2012 | ||||
| Schepetilnikov et al., 2013 | ||||
| Ribosome protein small subunit6 | RPS6 | RPS6A | At4g31700 | Mahfouz et al., 2006 |
| RPS6B | At5g01360 | Creff et al., 2010 | ||
| Ren et al., 2012 | ||||
| AMP-activated protein kinase | AMPK | KIN10 | At3g01090 | Baena-González et al., 2007 |
| KIN11 | At3g29160 | |||
| FK506-binding protein 12 | FKBP12 | FKBP12 | At5g64350 | Mahfouz et al., 2006 |
| Sormani et al., 2007 | ||||
| Leiber et al., 2010 | ||||
| Ren et al., 2012 | ||||
| Xiong and Sheen, 2012 | ||||
| Type 2A-phosphatase-associated protein 46 kD | α4 | TAP46 | At5g53000 | Ahn et al., 2011 |
| E2Fa transcription factor | E2F3 | E2Fa | At2g36010 | Xiong et al., 2013 |
| Repressor of LRR-extensin1 | ATPBD3 | ROL5 | At2g44270 | Leiber et al., 2010 |
| Translationally controlled tumor protein | TCTP | TCTP | At3g16640 | Berkowitz et al., 2008 |
| ErbB-3 epidermal growth factor receptor binding protein | EBP1 | EBP1 | At3g51800 | Horváth et al., 2006 |
In mammals, mTOR operates as a hub of the signal transduction network that coordinates translation, proliferation, and growth with diverse signals including nutrient availability, energy sufficiency, stress, hormones, and mitogens, whereas the molecular functions and dynamic regulatory mechanisms of the TOR kinase in plants have remained largely unclear until recent breakthroughs (Fig. 1C). This is mainly because of the lack of molecular and biochemical assays for endogenous TOR kinase activity, the embryo lethality of null Arabidopsis tor mutants, and the prevailing perceived rapamycin resistance of land plants (Menand et al., 2002; Ren et al., 2011). Based on antibody detection of the conserved and specific phosphorylation of T499 in S6K1 (Reyes de la Cruz et al., 2004; Hong et al., 2008; Schepetilnikov et al., 2011) and T455 in S6K2 (Xiong and Sheen, 2012), a sensitive and reliable in vivo cellular assay was recently established for monitoring endogenous TOR activity in Arabidopsis (Xiong and Sheen, 2012). Unexpectedly, this assay revealed that rapamycin can effectively inhibit Arabidopsis TOR activity when added at high concentrations (10 μM) to liquid cultures of Arabidopsis mesophyll protoplasts or seedlings. Rapamycin exerts TOR inhibition by inducing a direct interaction between TOR-FRB domain and FK506-binding protein12 (FKBP12) in plant cells, which was not previously detected by heterologous yeast two-hybrid assays (Fig. 1B). Interestingly, overexpression of either human, yeast, or Arabidopsis FKBP12 can enhance rapamycin sensitivity more than 100-fold in Arabidopsis (Sormani et al., 2007; Ren et al., 2012; Xiong and Sheen, 2012). Variable endogenous FKBP12 protein levels may offer a molecular explanation for the varied rapamycin resistance observed at low (nanomolar) rapamycin concentrations, and changing the levels of this protein may provide a feasible strategy for improving rapamycin efficacy to clarify the roles of TOR and to advance TOR research in plants, various mammalian cell lines, and diverse animal cell types and model systems (Xiong and Sheen, 2012; Kang et al., 2013). The discovery of the effectiveness of rapamycin in Arabidopsis opens up new possibilities for combined chemical and molecular dissection of the TOR signaling networks in plants. To circumvent the embryo lethality of the tor mutants in Arabidopsis (Menand et al., 2002; Ren et al., 2011), ethanol- and estradiol-inducible RNA interference (RNAi) lines have been generated to allow conditional silencing of the TOR gene to varying degrees (Deprost et al., 2007; Xiong and Sheen, 2012; Caldana et al., 2013). These Arabidopsis lines, together with rapamycin, provide invaluable genetic and chemical tools that have been used to begin deciphering the TOR signaling networks in plants. Genetic and physiological studies combined with large-scale transcript and metabolite profiling have revealed that TOR regulates plant growth, development, flowering, senescence, and life span by modulating translation, transcription, autophagy, and primary and secondary metabolism (Deprost et al., 2007, Ahn et al., 2011; Ren et al., 2011, 2012; Moreau et al., 2012; Xiong and Sheen, 2012; Caldana et al., 2013; Xiong et al., 2013).
ROLES OF TOR SIGNALING IN PLANT GROWTH AND DEVELOPMENT
Despite their distinct body plans and growth strategies, the function of TOR in coupling nutrient and energy availability with other environmental signals to coordinate growth, development, and survival appears to be conserved in yeast, plants, and animals. In general, Arabidopsis TOR expression levels correlate with root and shoot growth, cell size, and seed yield (Deprost et al., 2007). Manipulating TOR kinase activity or TOR expression levels leads to changes in growth and development from embryogenesis to senescence (Fig. 2; Menand et al., 2002; Deprost, et al., 2007; Ren et al., 2011, 2012; Xiong and Sheen, 2012; Caldana et al., 2013; Xiong et al., 2013).
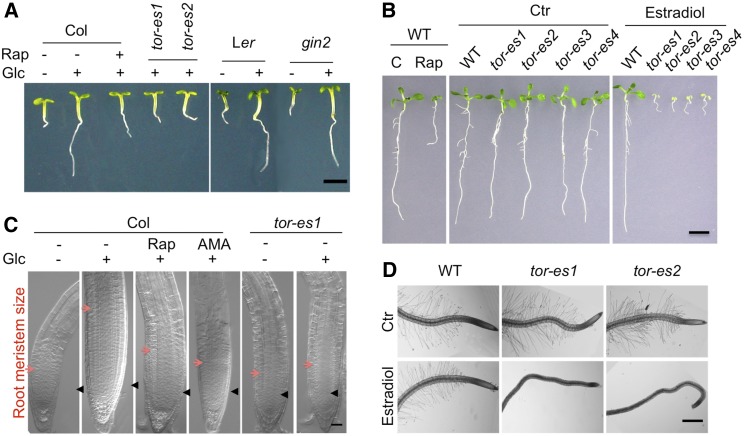
Figure 2.
TOR signaling functions in plant growth and development. A, Glc activation of Arabidopsis root growth is TOR dependent but gin2 independent. Wild-type (WT; ecotype Columbia of Arabidopsis or Ler), estradiol-inducible tor-es mutants (tor-es1, tor-es2), and HEXOKINASE1 mutant (gin2) seedlings at 3 d after germination (DAG) were incubated with or without Glc (15 mM) or rapamycin (Rap; 10 µM) for 24 h. B, Retarded seedling growth by rapamycin and in tor-es mutants. Rapamycin (10 µM) or estradiol was added at the time of germination, and the seedlings were incubated for 9 d. C, TOR controls root meristem activation. Seedlings at 3 DAG were incubated with or without Glc (15 mM), rapamycin (10 µM), or antimycin A (AMA; 5 µM) for 24 h. Arrowheads, Quiescent center; red arrow, transition zone. D, Retardation of root hair growth in tor-es mutants. Seedlings were germinated in the presence of 30 mm Glc for 4 D. A and C are adapted from Xiong et al. (2013). The Authors acknowledge first and reference publication in the Journal. B and D were originally published in Journal of Biological Chemistry. Xiong and Sheen, Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem. 2012; 287, 2836–2842. © the American Society for Biochemistry and Molecular Biology Bar in A = 2 mm; bar in B = 5 mm; bar in C = 25 µm; bar in D = 500 µm.
Arabidopsis TOR is highly expressed in embryos and endosperm, and the null tor mutant exhibits growth arrest at the 16- to 32-cell embryo stages, indicating that TOR signaling is involved in early embryo development. Although TOR is a large protein with multiple domains (Fig. 1, A and B), its kinase domain alone can partially rescue early embryo lethality of the null tor mutant from the early globular stage to the late heart stage. However, the N-terminal HEAT-repeat domain of TOR is critical for full complementation (Ren et al., 2011). Control of embryo development by TOR likely requires its interaction partner RAPTOR1, because mutation of this gene has also been reported to arrest embryo development (Deprost et al., 2005).
TOR signaling is also indispensible for postembryonic development in plants. Repression of TOR activity by rapamycin in wild-type and FKBP12 overexpression plants and induced down-regulation of TOR expression strongly retard many key aspects of organ growth in seedlings after germination, including cotyledon expansion, true leaf development, petiole elongation, and primary and lateral root growth (Fig. 2, A and B; Ren et al., 2012; Xiong and Sheen, 2012; Caldana et al., 2013; Xiong et al., 2013). However, specific plant growth conditions and different methods used to reduce TOR activity can lead to opposite results in senescence. For example, accelerated senescence was observed in the lst8 mutant and ethanol-inducible tor-RNAi lines (Deprost et al., 2007; Moreau et al., 2012), whereas delayed flowering and senescence with greatly extended life span was reported in rapamycin-treated transgenic Arabidopsis plants overexpressing yeast FKBP12 (Ren et al., 2012). Moderate increases in TOR expression levels (less than 2-fold) have been found to increase root and shoot growth, cell size, and seed yield without visibly affecting plant morphology (Deprost et al., 2007). By contrast, strong overexpression of full-length TOR results in developmental abnormalities including severe shoot and inflorescence meristem defects, shorter hypocotyls, compact architecture with short petioles, and early senescence of rosette leaves. Intriguingly, overexpression of the TOR kinase domain alone produces some distinct phenotypes such as changes in the proximal-distal axis, dark green and asymmetric leaves, tuberous roots, loss of apical dominance, delayed flowering and senescence, and bent siliques (Ren et al., 2011). These results suggest that precise regulation of both temporal and spatial TOR expression is crucial for plant growth and development. In addition to TOR, disruption of the TORC1 signaling components RAPTOR1, RAPTOR2, LST8-1, and RPS6A/B also results in delayed vegetative growth, reduced apical dominance, and abnormal flower development (Anderson et al., 2005; Deprost et al., 2005; Moreau et al., 2012; Ren et al., 2012), further reinforcing the indispensible role of TOR signaling in orchestrating plant growth and development.
The plant TOR kinase also acts as a gatekeeper, gauging and linking the nutrient status of the plant to regulate the development of the root system and associated functions (Fig. 2, C and D). Shoot photosynthesis-derived Glc is essential for the activation of TOR signaling (through the glycolysis and mitochondrial bioenergetics energy relay) for root meristem activation and establishment (Xiong et al., 2013). However, this TOR signaling activation is decoupled from direct Glc sensing via the hexokinase1 Glc sensor (Fig. 2A; Moore et al., 2003), primary growth hormone signaling, and stem cell maintenance (Müller and Sheen, 2008; Aichinger et al., 2012). Root hairs are long, tubular-shaped extensions from single epidermal cells that help plants absorb nutrients and water and interact with microorganisms, and they also help physically anchor plants to the soil. Several studies have suggested that TOR signaling plays an essential role in root hair growth. Rapamycin or the estradiol-inducible tor mutant completely abolishes Glc-promoting root hair growth associated with reactive oxygen species signaling (Fig. 2D; Ren et al., 2012; Xiong and Sheen, 2012). Interestingly, TOR kinase activity is also linked to modification of cell walls for root hair development. A mutant screen for suppressors of the root hair cell wall formation mutant LRR-extensin1 (lrx1) led to the identification of the repressor of lrx1 (rol5) mutant. The yeast ortholog of ROL5, needs Cla4 to survive6, is a downstream component of the yeast TOR pathway, indicating that TOR signaling is involved in cell wall formation in Arabidopsis (Leiber et al., 2010). Root hair development provides an excellent platform for integrating the molecular functions of TOR into one of the best-characterized gene regulatory networks governing root hair growth and development (Bruex et al., 2012).
PROMOTION OF RIBOSOMAL BIOGENESIS AND PROTEIN TRANSLATION
TOR controls protein synthesis at multiple levels, functioning in ribosomal biogenesis, polysome accumulation, and various protein translational processes (Fig. 3). The ribosome is a large and complex molecular machine comprising ribosomal RNAs (rRNAs) and ribosome proteins that supports protein synthesis (Ben-Shem et al., 2011). Arabidopsis tor mutants show reduced expression of 5.8S, 18S, and 25S rRNA (derived from the 45S rRNA precursor) in embryos, whereas overexpression of full-length TOR or the kinase domain alone significantly elevates the expression of rRNA (Ren et al., 2011). TOR regulates rRNA transcription through its kinase domain. Interestingly, Arabidopsis TOR-GFP fusion protein is observed in the nucleus of transiently transfected onion epidermal cells. Chromatin immunoprecipitation studies have revealed that Arabidopsis TOR directly binds to the 45S rRNA promoter and the 5′ untranslated region via the Leu zipper sequence within the HEAT repeat domain of TOR to directly regulate 45S rRNA transcription (Ren et al., 2011). It will be important to determine how the TOR kinase regulates transcription factors and RNA polymerase I during rRNA transcription.
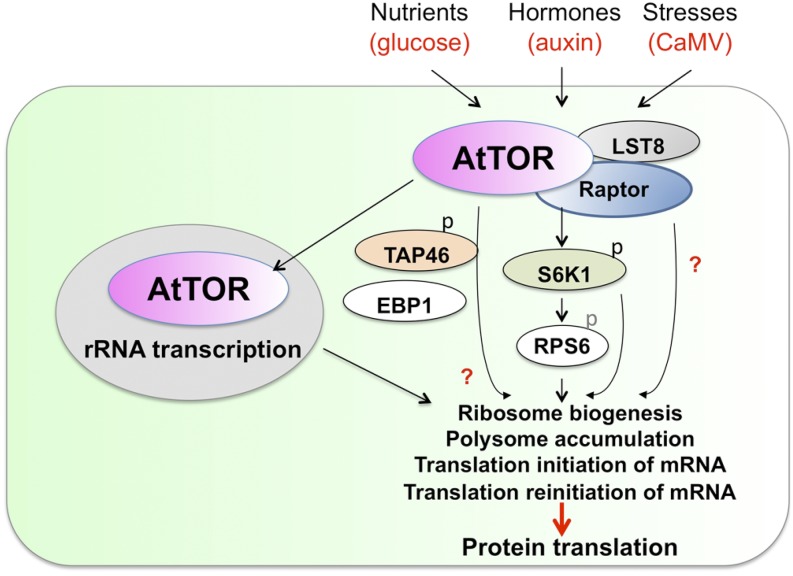
Figure 3.
Model of the role of plant TOR signaling in promoting protein translation. The upstream signals and downstream effectors of plant TOR signaling in controlling protein translation processes is shown. TOR controls protein synthesis at multiple levels, including rRNA transcription, ribosomal biogenesis, polysome accumulation, and various protein translational processes. AtTOR, Arabidopsis TOR; CaMV, Cauliflower mosaic virus.
Recent studies have revealed that the transcription of a large number of genes encoding ribosomal proteins (RPs) are similarly regulated by TOR in yeast, Arabidopsis, and mouse, suggesting another conserved TOR function that is central to ribosome biogenesis (Martin et al., 2004; Huber et al., 2009; Chauvin et al., 2013; Xiong et al., 2013). Advances in integrated functional genomic and computational screens will enable the identification of common cis-elements and transcription factors mediating TOR regulation of RP genes in plants. It will also be important to determine whether plant S6Ks play similar critical roles in RP gene regulation as mammalian S6Ks and yeast SCH9 protein kinase, which are activated by TOR (Huber et al., 2009; Chauvin et al., 2013).
Partial silencing of Arabidopsis TOR by ethanol-induced tor-RNAi or partial repression of TOR kinase activity by rapamycin in transgenic Arabidopsis plants expressing yeast FKBP12 decreases the accumulation of polysomes (Deprost et al., 2007; Sormani et al., 2007). The protein synthesis rate increases in transgenic Arabidopsis lines overexpressing TOR or RPS6A/B, as indicated by GUS activity measurements (Ren et al., 2012). Moreover, the expression of an Arabidopsis gene encoding the putative ortholog of human (EBP1) is strongly correlated with TOR expression in wild-type plants and in TOR silencing or overexpression transgenic plants. Because EBP1 is a regulator of ribosome assembly and translation, AtEBP1 may function downstream of TOR kinase in the mRNA translation machinery in plants (Fig. 3; Horváth et al., 2006; Deprost et al., 2007). It will be interesting to determine the molecular mechanism underlying TOR regulation of EBP1.
In mammals, mTORC1 directly phosphorylates the key translational regulators 4E-BP1 and S6K1 to promote protein synthesis. A plant ortholog of 4E-BP1 has not been identified based on sequence conservation. The TOR-S6K1 signaling pathway appears to be conserved in plants, because TOR activity directly affects plant S6K1 phosphorylation (Xiong and Sheen, 2012; Xiong et al., 2013). Interestingly, two reports suggest an essential role of the TOR-S6K1 pathway in translation reinitiation. Upon virus infection, transactivator-viroplasmin protein from the Cauliflower mosaic virus directly interacts with TOR to trigger TOR hyperactivation and S6K1 phosphorylation, which in turn phosphorylates reinitiation-supporting-protein to promote the translation reinitiation of the viral 35S mRNA (Schepetilnikov et al., 2011). Moreover, the TOR-S6K1 pathway can also be activated by the growth hormone auxin to facilitate the translation reinitiation of plant mRNAs containing upstream open reading frames at their 5′ untranslated regions (Schepetilnikov et al., 2013). These data indicate a common role for the TOR-S6K1 signaling pathway in the translation of mRNAs that contain upstream small open reading frames, whose expression is often affected by internal nutrient signals and external stress environments.
Protein phosphatase 2A (PP2A) is a major Ser/Thr protein phosphatase that regulates many key cellular processes, and a regulatory subunit of PP2A, Tap42/α4, has been found to function downstream of TOR in yeast (Urban et al., 2007). The Arabidopsis ortholog TAP46 interacts with TOR and can be phosphorylated in vitro by the Arabidopsis TOR kinase expressed in human embryonic kidney 293 cells. Silencing of TAP46 causes a dramatic decrease in the protein synthesis rate, indicating that TAP46 may act as a positive effector of the plant TOR signaling pathway (Ahn et al., 2011). In addition, TAP46 silencing also reduces polysome accumulation and global protein translation efficiency in Nicotiana benthamiana, Nicotiana tabacum BY-2 cells, and Arabidopsis plants. Future studies will help determine how the TOR kinase regulates TAP46 and PP2A and will help identify the substrates of PP2A that regulate translation, autophagy, nutrient recycling, and programmed cell death (Ahn et al., 2011).
INHIBITION OF AUTOPHAGY
Autophagy is a major catabolic process in which cells enclose their cytoplasmic components in double membrane structures, called autophagosomes, and send them to vacuoles or lysosomes for degradation (for review, see Li and Vierstra, 2012; Liu and Bassham, 2012; Jewell et al., 2013). In Arabidopsis, down-regulation of TOR expression leads to constitutive activation of autophagy (Liu and Bassham, 2010). In the green alga C. reinhardtii, rapamycin-mediated inhibition of TOR activity and nutrient limitation similarly induce autophagy (Pérez-Pérez et al., 2010). These results indicate that TOR is a conserved negative regulator of autophagy in photosynthetic species, as it is in yeast and mammals.
The autophagy related1 (ATG1)/ATG13/ATG17 kinase complex plays an essential role in the onset of autophagy and is a direct TORC1 substrate. Yeast TORC1 mediates the phosphorylation of Atg13, and this phosphorylation prevents its association with Atg17 and thus blocks autophagy. In mammals, mTORC1 inhibits autophagy by phosphorylating both the ATG13L (ATG13-like) and ULK1 (UNC51-like kinase1; the ortholog of yeast ATG1) subunits to repress ULK1 kinase activity (Jewell et al., 2013). In Arabidopsis, three putative ATG1 and two putative ATG13 homologs have been identified, and their roles in the regulation of autophagy in response to nutrient starvation have been confirmed (Suttangkakul et al., 2011). However, whether plant TOR represses autophagy by similar mechanism via ATG1/ATG13 phosphorylation requires further study.
Other universally conserved signaling pathways, such as TAP46-PP2A and the energy sensor AMP-activated protein kinase (AMPK) pathway, are also involved in autophagy regulation (Baena-González and Sheen, 2008; Ahn et al., 2011). Transient overexpression of Arabidopsis PROTEIN KINASE10 (KIN10) energy sensor kinase, the ortholog of the catalytic subunit of mammalian AMPK, strongly induces several autophagy genes, suggesting that KIN10 also positively activates autophagy via transcriptional control in plants (Baena-González et al., 2007). Interestingly, AMPK is an upstream negative regulator of mTORC1 in mammals (Fig. 1C), whereas TORC1 inhibits SUC NON-FERMENTATION1, the yeast ortholog of AMPK (Orlova et al., 2006; Kim et al., 2011). The precise regulatory relationship between KIN10 and TOR in different biological contexts and their opposite molecular actions in autophagy regulation represent interesting future research directions.
MODULATION OF METABOLISM
Studies incorporating technical advances in large-scale metabolite profiling have revealed profound and complex roles of TOR in the regulation of many aspects of primary and secondary metabolism in plants. Long-term and partial disruption of TOR signaling, either by reduction of TOR expression or kinase activity, or via mutation of LST8-1, usually leads to the accumulation of high levels of starch, triacylglycerides (TAGs), amino acids, TCA intermediates, and several secondary metabolites (Deprost et al., 2007; Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013).
Starch is the most widespread form of carbon storage in plants, and the abundance of starch is negatively correlated with plant growth (Ramon et al., 2008; Sulpice et al., 2009). The starch accumulation phenotype in the partially silenced tor-RNAi lines, lst8 mutants, and estradiol-inducible artificial microRNA amiR-tor plants (Deprost et al., 2007; Moreau et al., 2012; Caldana et al., 2013; Dobrenel et al., 2013) is reminiscent of the finding that disruption of TORC1 in yeast and animals leads to carbon storage through the accumulation of glycogen (Schmelzle et al., 2004; Cornu et al., 2013). Contrary to starch accumulation, raffinose and galactinol levels are reduced in the ethanol-inducible tor-RNAi and lst8 mutants and rapamycin-treated Arabidopsis plants overexpressing yeast FKBP12 but are elevated in the estradiol-inducible amiR-tor plants (Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013; Dobrenel et al., 2013). The reason for these opposite consequences is unclear, because raffinose and galactinol usually accumulate under stress conditions such as high light, starvation, cold, drought, and high salt (Taji et al., 2002; Nishizawa et al., 2008; Dobrenel et al., 2013). The accumulation of starch and TAGs in various TOR-deficient plants may reflect the presence of hierarchical controls in metabolism, which prevent the carbon flux required for the synthesis of inositol, raffinose, and galactinol. However, long-term TOR inactivation to varying degrees may trigger the redirection of carbon fluxes, depending on the metabolic circuit connections, physiological and nutrient status, and environmental conditions (Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013; Dobrenel et al., 2013). Future research is required to better understand the complexity of TOR-mediated metabolic changes.
Interestingly, although TORC1 promotes lipid synthesis and storage in mammals, long-term reduction of TOR expression results in the accumulation of the lipids TAGs, especially those with long-chain poly-unsaturated fatty acids, in Arabidopsis leaves (Caldana et al., 2013). Similar lipid accumulation is also observed in stressed plants (e.g. in response to cold treatment; Degenkolbe et al., 2012). However, during the heterotrophic-to-photoautotrophic transition in Arabidopsis seedlings, TOR plays a key role in repressing the metabolic programs involved in β-oxidation of lipids and oil stored in the seeds. For instance, the expression of genes encoding TAG lipase and acyl-CoA oxidase in germinating seeds is repressed by TOR, which is activated via Glc derived from photosynthesis (Graham, 2008; Xiong et al., 2013). Therefore, depending on the developmental stage and physiological context, TOR may redirect the storage of carbon accumulation to starch or lipids to adjust plant growth, developmental transitions, or stress tolerance in different organs. Understanding and manipulating the TOR-mediated TAG and lipid accumulation process may offer a technical innovation for the conversion of photosynthetic products to biofuels (Caldana et al., 2013).
The lst8 mutants, and plants with partial repression of TOR expression or activity, also accumulate high levels of branched chain (Leu, Ile, and Val), aromatic (Tyr and Trp) and other (Lys, His, and Pro) amino acids as well as γ-aminobutyrate. This amino acid accumulation pattern may be the result of the activation of autophagy, which is used for rapid protein degradation and recycling, or it may be attributable to decreased amino acid incorporation caused by the slowing of the new protein translation process (Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013). Because the expression of some nitrogen assimilation genes is elevated in the ethanol-inducible tor-RNAi plants and in the lst8 mutants, changes in nitrogen assimilation and nutrient remobilization rates may also affect amino acid levels (Deprost et al., 2007; Moreau et al., 2012). Moreover, the levels of TCA cycle intermediates, including citrate, α-ketoglutarate, succinate, fumarate, and malate, are significantly higher in the mutant plants after long-term TOR inhibition (Ren et al., 2012; Caldana et al., 2013). These TCA intermediates may arise from the reorganized metabolism required for cellular respiration and maintenance despite the restricted plant growth, which may also lead to complex changes in nitrogen metabolism and usage (depending on the experimental conditions) in the lst8-1 and tap46 mutants and in estradiol-inducible amiR-tor transgenic plants with different degrees of downstream deficiency in TOR signaling (Ahn et al., 2011; Moreau et al., 2012; Caldana et al., 2013).
The changes in secondary metabolites associated with the flavonoid, phenylpropanoid, and glucosinolate pathways are the most significant in estradiol-inducible amiR-tor transgenic plants grown in nutrient-rich conditions (Caldana et al., 2013). These results suggest that nutrient remobilization in secondary metabolism pathways may occur for cellular adjustment and stress adaptation when growth is restricted but Suc is available in the medium (Caldana et al., 2013). Using quiescent seedlings, transcript profiling of the wild type and the estradiol-inducible null tor-es mutant at the photoautotrophic transition checkpoint has provided evidence that TOR plays a critical role in the Glc activation of genes involved in the glucosinolate synthesis pathways, including MYB28 and MYB34, encoding key transcription factors (Xiong et al., 2013). These findings provide an exciting and previously unexpected opportunity to elucidate the precise molecular, biochemical, and genetic links between TOR kinase activity and the regulation of a large set of genes and enzymes involved in diverse metabolic pathways in plants. However, the modes of gene regulation and the levels of metabolite changes may vary depending on the physiological and developmental states of the plant, the method used for manipulating a range of TOR activities, and the plant growth conditions (Deprost et al., 2007; Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013; Xiong et al., 2013).
ALTERED GENE EXPRESSION BY LONG-TERM TOR INHIBITION
Genome-wide expression profiling experiments suggest that TOR controls diverse genes in plants. Long-term repression of TOR signaling has been achieved using multiple strategies in Arabidopsis plants grown in a nutrient-rich environment, including silencing TOR expression in estradiol-inducible amiR-tor mutants for 3 to 6 d, rapamycin treatment of transgenic lines overexpressing yeast FKBP12 for 3 d, and the use of lst8-1 mutants (Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013). Functional over-representation analysis revealed that TOR signaling activates major anabolic and biosynthetic pathways, including pathways involved in cell wall modification, the cell cycle, carbon and nitrogen utilization, photosynthesis, and nutrient transport. By contrast, genes associated with catabolic processes, including autophagy, senescence, and protein and lipid metabolism, are negatively regulated by TOR signaling. In addition to its opposite regulation of anabolism and catabolism, TOR inhibition also triggers altered expression of genes involved in chromatin structure, hormone metabolism, signaling, and stress-related processes.
These broad transcriptome changes closely match the roles of TOR signaling in plant growth and development. Consistent with its role in growth promotion, TOR signaling tightly controls genes involved in cell wall modification to promote cell growth and elongation (Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013). Disruption of TOR signaling also causes strong defects in the development of root hairs and primary and secondary roots (Fig. 2). Consistent with this notion, many root-related genes are significantly down-regulated by rapamycin treatment (Ren et al., 2012; Xiong et al., 2013), including MORPHOGENESIS OF ROOT HAIR6, which has been linked to root hair development (Jones et al., 2006). Down-regulation of TOR expression also strongly activates some genes encoding heat shock proteins, pathogenesis-related proteins (PRs), and disease resistance proteins (Caldana et al., 2013). Interestingly, silencing of TAP46 leads to programmed cell death in N.benthamiana, N. tabacum BY-2 cells, and Arabidopsis plants, as well as the activation of cell death- and defense-related genes, such as PR1, PR2, PR5, and HAIRPIN-INDUCED-GENE1, in Nicotiana benthamiana leaves (Ahn et al., 2011). How TOR-TAP46-PP2A signaling regulates programmed cell death and transcription in the processes of stress adaption and tolerance remains to be elucidated.
The transcriptome changes associated with long-term TOR inhibition also corroborate well with some of the observed changes in metabolites that have been revealed by large-scale profiling (Deprost et al., 2007; Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013). Key genes involved in the raffinose synthesis pathway, including GALACTINOL SYNTHASE1, GALACTINOL SYNTHASE2, and GALACTINOL SYNTHASE3 and MYO-INOSITOL1-PHOSPHATE SYNTHASE1 and MYO-INOSITOL1-PHOSPHATE SYNTHASE2 are strongly repressed by inhibiting TOR activity and in the lst8-1 mutants, which is consistent with reduced levels of galactinol and raffinose (Moreau et al., 2012; Ren et al., 2012). Increased polyamine biosynthesis in response to long-term rapamycin treatment in transgenic Arabidopsis plants overexpressing yeast FKBP12 is accompanied by the up-regulation of S-ADENOSYLMETHIONINE DECARBOXYLASE (Ren et al., 2012), which encodes a key enzyme in the polyamine biosynthesis pathway (Mehta et al., 2002). In the lst8-1 mutants, a group of genes involved in the nitrate or sulfur assimilation pathway are up-regulated, including ADENOSINE 5′-PHOSPHOSULFATE KINASE1, NITRATE REDUCTASE, NITRITE REDUCTASE, ASN SYNTHETASE2, and UROPORPHYRIN METHYLASES, which may be associated with the observed accumulation of high levels of Gln and Pro (Moreau et al., 2012). These data indicate that metabolic adjustments seem to largely follow gene regulatory programs, and TOR may play a central role in coordinating metabolic and transcriptional networks.
DYNAMIC Glc-TOR-MEDIATED TRANSCRIPTIONAL NETWORK
Although genome-wide expression profiling analyses of plants with long-term conditional TOR silencing (3 to 6 d) and of lst8-1 plants have revealed an important role for TOR signaling in the regulation of gene expression, limited evidence supports the direct role of TOR signaling in transcriptional control (Deprost et al., 2007; Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013). To better understand the molecular landscape of the TOR signaling networks and the dynamic links between TOR signaling and transcriptional regulation, rapid global changes in the transcriptome in response to a 2-h Glc (15 mM) treatment were investigated in wild-type and estradiol-inducible null tor-es seedlings at the photoautotrophic transition checkpoint (Xiong et al., 2013). The use of seedlings (3 d after germination) at the heterotrophic-to-photoautotrophic transition checkpoint, with normal growth and development in the absence of photosynthesis or exogenous sugars, offered a low TOR signaling background in which to detect rapid Glc induction. The minimal endogenous Glc levels of these seedlings maximized the detection sensitivity upon Glc induction, which led to the identification of 1,318 up-regulated and 1,050 down-regulated genes that were differentially affected by a physiological level of Glc. Strikingly, this swift, Glc-induced global transcriptional reprogramming is completely blocked in the tor-es mutant, which had not been reported previously in extensive mammalian or plant studies (Kim and Guan, 2011; Laplante and Sabatini, 2012; Ren et al., 2012; Caldana et al., 2013). Importantly, TOR senses and transduces shoot photosynthesis-derived Glc signals through the glycolysis and mitochondria energy relay to specifically control root meristem cell proliferation. Other sugars (Fru, Xyl, and Gal) and amino acids (mixed and Gln) are unable to substitute for Glc in TOR signaling, reinforcing the notion that Glc signal is the main nutrient mediator derived from source leaf photosynthesis that functions in systematic gene regulation and root growth (Xiong et al., 2013).
The sensitivity of seedlings 3 d after germination at the heterotrophic-to-photoautotrophic transition checkpoint facilitated the discovery of previously unknown primary Glc-TOR target genes, especially genes that function in the cell cycle and in DNA synthesis, transcription, and RNA synthesis/processing (among Glc-TOR activated genes) and genes involved in modulating transcription, protein degradation, and signaling (among Glc-TOR repressed genes). Remarkably, Glc-TOR target genes stratify into a myriad of regulatory and metabolic functional categories (Fig. 4A; Xiong et al., 2013). A total of 105 genes important for the cell cycle and DNA synthesis are highly represented among the Glc-TOR activated genes, and most of these genes have not been identified as primary TOR target genes. By comparison, only nine cell cycle-related genes have been identified in a previous study after long-term (6 d) TOR repression under nutrient-rich conditions (Caldana et al., 2013).
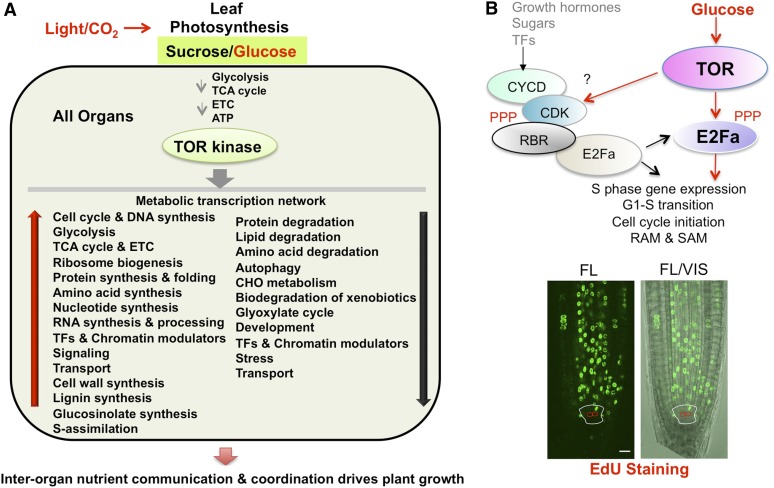
Figure 4.
Glc-TOR-mediated transcription networks in plants. A, Glc-TOR transcription networks mediate interorgan dialogs to drive plant growth. Glc activates the TOR kinase (via glycolysis and mitochondrial bioenergetics relays) to orchestrate global transcriptional reprogramming, which integrates central and secondary carbon metabolism with bioenergetics, biosynthesis, signaling, TFs, chromatin modulators, transporters, autophagy, and cell cycle regulation. B, E2Fa phosphorylation by Glc-TOR signaling is a key step in cell cycle S-phase entry. Glc-TOR signaling activates E2Fa, bypassing or acting downstream of the conventional CYC-CDK-RBR cascade, and activates the cell cycle in most cell types in the primary root meristem. S-phase entry is visualized by EdU staining. Red circles indicate quiescent center cells and the white outline marks the stem cell initials. Scale bar, 10 µm. EdU, 5-ethynyl-2′-deoxyuridine; ETC, electron transport chain; FL, fluorescent; RAM, root apical meristem; SAM, shoot apical meristem; TF, transcription factor; VIS, light. A and B (bottom) are adapted from Xiong et al. (2013). The authors acknowledge first and reference publication in the Journal.
Glc-TOR signaling activates genes involved in key, evolutionarily conserved anabolic processes, including amino acid, lipid, and nucleotide synthesis and the oxidative pentose phosphate pathway, which are essential for rapid growth. However, the Glc-TOR signaling pathway represses genes that function in mediating the degradation of proteins, amino acids, and lipids as well as xenobiotic and autophagy regulation, which are critical for survival under starvation (Baena-González et al., 2007; Miyashita and Good, 2008). In addition, Glc-TOR signaling activates genes involved in the entire Arabidopsis glycolysis pathway and the TCA cycle, mitochondria, and the electron transport chain, as well as RPs and protein synthesis machinery, suggesting a universal and conserved function for TOR in controlling translation and central carbon and energy metabolism (Urban et al., 2007; Laplante and Sabatini, 2012; Robaglia et al., 2012; Xiong et al., 2013). Although TOR was first discovered in yeast, the atypical fermentation lifestyle of this organism evolved in an opposite transcriptional regulation of glycolytic- and TCA-cycle-related genes by yeast TOR1 (Hardwick et al., 1999). It will be exciting to explore and elucidate the shared and distinct transcriptional programs in yeast, mammals and plants.
Unique to plant Glc-TOR signaling is its pivotal roles in repressing the expression of metabolic genes encoding enzymes involved in β-oxidation (TAG lipase and acyl-CoA oxidase) and the glyoxylate cycle (malate synthase and isocitrate lyase), which are required in the germination program of Arabidopsis seeds (Graham, 2008), and suppressing catabolic programs for plant survival in prolonged darkness, including dark-inducible 6/Asn synthase, phosphoenolpyruvate dikinase, phosphoenolpyruvate carboxykinase, Glu dehydrogenase, and trehalose metabolism and sensing (Baena-González et al., 2007; Miyashita and Good, 2008). Glc-TOR signaling also controls plant-specific genes, promoting the synthesis of cell wall polymers/proteins, as well as secondary metabolic pathways that are uniquely required for plant growth, defense, or communication to promote adaptation, fitness, and survival (Keurentjes et al., 2006). Significantly, genes that function in the entire glutathione, indolic/benzoic and aliphatic glucosinolate, lignin, and flavonoid synthesis pathways are all activated by Glc-TOR signaling (Xiong et al., 2013), which appears to be different from the results of studies involving long-term and partial repression of TOR activity in plants grown in the presence of Suc (Caldana et al., 2013). The plant Glc-TOR signaling networks also integrate a large number of chromatin modulators, transcription factors, signaling regulators, and growth- and stress-related proteins that are either unique to plants or conserved in eukaryotes. These findings suggest a previously unanticipated central role of the TOR kinase in Glc and energy signaling through transcriptome reprogramming, which goes beyond the conventional emphasis on the role of translational control for the mTOR kinase (Dowling et al., 2010; Hsieh et al., 2012; Laplante and Sabatini, 2012; Ben-Sahra et al., 2013).
TOR PHOSPHORYLATION OF E2FA CONTROLS THE CELL CYCLE
TOR senses and transduces Glc signals to activate root meristems by orchestrating global transcriptional reprogramming (Xiong and Sheen, 2013; Xiong et al., 2013). Many Glc-TOR-activated genes match typical G1- and S-phase genes and are highly overlapping (95%) with putative Arabidopsis E2 promoter binding factor (E2Fa) target genes including ORIGIN RECOGNITION COMPLEX2/6, MINOCHROMOSOME MAINTENANCE3/5/7, CELL DIVISION CYCLE6, E2F TARGET GENE1, and PROLIFERATING CELL NUCLEAR ANTIGEN1 (Vandepoele et al., 2005; de Jager et al., 2009; Naouar et al., 2009). Significantly, TOR directly phosphorylates the E2Fa transcription factor to activate these S-phase genes. This finding uncovers a previously unrecognized role for TOR in the direct transcriptional regulation of the cell cycle and provides a compelling novel mechanism for how Glc-TOR signaling controls the transcription of S-phase genes and the cell cycle to promote root meristem activation and growth, which differs from the conventional, well-known role of TOR in stimulating the translation of proteins involved in cell cycle progression through the activities of S6K1 and 4E-BP (Dowling et al., 2010; Ben-Sahra et al., 2013).
E2Fs are well-established targets of the universal CYCLIN (CYC)-CYCLIN-DEPENDENT KINASE (CDK)-RETINOBASTOMA-RELATED PROTEIN (RBR) cascade that initiates the cell cycle (Fig. 4B; De Veylder et al., 2007; Xiong et al., 2013). Functional analysis has suggested that the presence or absence of the RBR binding domain at the C terminus of E2Fa (Shen, 2002) does not affect TOR phosphorylation or the roles of E2Fa in S-phase gene activation and target gene promoter binding (Xiong et al., 2013). These results indicate that direct E2Fa protein phosphorylation by the TOR kinase may be a key step in Glc activation of S-phase genes, which bypasses or acts downstream of the conventional CYC-CDK-RBR cascade (Fig. 4B; Xiong et al., 2013).
Glc-TOR signaling activates S-phase entry in most cell types in the primary root meristem, except in the quiescent center (Fig. 4B; Xiong et al., 2013). Importantly, in the absence of Glc-TOR signaling, growth hormones (auxin, cytokinin, gibberellins, and brassinosteroid) fail to activate entry into the cell cycle despite normal transcriptional activation by auxin and cytokinin in the root meristem (Xiong et al., 2013). Therefore, Glc-TOR signaling provides essential energy, metabolites, biomass, and cell cycle machinery through dynamic transcriptional activation. This concept may explain the prerequisite, fundamentally indispensible, and global roles of Glc-TOR signaling in proliferation and growth. Plant hormones may modulate specific cell cycle regulators in specific cells and contexts and bring cell cycle connections to patterning and developmental programs when nutrients and Glc-TOR signaling are available (Santner et al., 2009; Aichinger et al., 2012; Hwang et al., 2012).
Recent studies have indicated that Arabidopsis S6K is also involved in cell cycle regulation (Henriques et al., 2010; Shin et al., 2012). Intriguingly, the regulation of the cell cycle by S6K is quite complex because TOR is not the sole regulator of S6K. Osmotic and salt stress and 3-phosphoinositide-dependent protein kinase1 also regulate S6K (Mahfouz et al., 2006). Previous studies have focused on the negative role of S6K in cell cycle regulation induced by stress, but not by TOR or nutrients (Mahfouz et al., 2006; Henriques et al., 2010; Shin et al., 2012). S6K can act as either a positive or negative regulator of the cell cycle in a context-dependent manner. In the presence of 1 µM naphthylacetic acid (which strongly inhibits primary root growth), S6K1 moderately represses or activates some cell cycle-related genes (CDKB1;1, RNR2, CYCB1;1, and CYCD3;1) in different cellular contexts, and these cell cycle-related genes are not E2Fa target genes (Shin et al., 2012; Xiong et al., 2013). In Arabidopsis suspension culture cells, the down-regulation of S6K1 and S6K2 leads to increased levels of E2Fb, Differentiation regulated transcription factor Protein a (DPa), and CDKB1;1 proteins and increased CDK activity. S6K1 is found to interact with RBR1 and regulate its nuclear localization and to negatively regulate chromosome numbers and stability (Henriques et al., 2010). These studies suggest that Arabidopsis cells grown under nutrient-limiting conditions require S6K for repression of cell proliferation and for the maintenance of chromosome stability and ploidy. It remains unclear whether this negative role of S6K is linked to the role of TOR signaling in the regulation of root meristem activity. It is possible that different upstream regulators modulate S6Ks to exert distinct functions with diverse partners in different subcellular compartments.
PERSPECTIVE AND FUTURE DIRECTION
In contrast with the significant progress made in discovering the downstream cellular and molecular functions of plant TOR signaling, the upstream regulators remain poorly defined. Small guanosine 5′‑triphosphatases (GTPases): Ras homolog enriched in brain (RHEB), and Rag guanosine 5′‑triphosphatases (RAGs) play crucial roles in sensing nutrients, growth factors, and amino acid signals and in activating mTOR (Fig. 1C; Laplante and Sabatini, 2012; Yuan et al., 2013), but plants seem to lack orthologs of RHEB and RAGs. In Drosophila melanogaster, the translationally controlled tumor protein (TCTP) has been shown to control TOR activity as a guanine exchange factor of RHEB (Hsu et al., 2007). Interestingly, a TCTP homolog has been identified in plants, and silencing Arabidopsis TCTP slows plant growth, reduces cell size, and impairs lateral root formation and root hair development, implying a potential role of TCTP in plant TOR signaling (Berkowitz et al., 2008). These results strongly indicate that plants may have developed alternative small GTPase signaling pathways to mediate TOR activity. Interestingly, plants posses a unique Rho-like small GTPase family called ROP/RAC, which function as hubs in signaling networks that control fundamental cellular processes (Nagawa et al., 2010). It will be interesting to test whether these ROPs/RACs may have functions analogous to RHEB and RAGs in plants. AMPK is another key upstream regulator of mTOR. It will be important to determine whether and how the plant energy sensor protein kinases KIN10 and KIN11 affect TOR kinase activity, and how these two evolutionarily conserved nutrient and energy sensing systems interact with each other in various biological processes modulated by TOR and KIN10/KIN11 protein kinases (Baena-González et al., 2007; Baena-González and Sheen, 2008; Ren et al., 2012; Caldana et al., 2013; Xiong et al., 2013).
A new generation of ATP-competitive inhibitors of the mTOR kinase (e.g. PP242, Torin1, Torin2, and AZD-8055) has recently been developed (Feldman et al., 2009; Thoreen et al., 2009; Liu et al., 2012, 2013). These inhibitors directly target the ATP site of the mTOR kinase and have been shown to be effective in suppressing animal cell growth and proliferation (Liu et al., 2012). Because the Arabidopsis TOR kinase domain shares high amino acid sequence similarity with the human TOR kinase domain, these chemical inhibitors represent promising new tools for probing plant TOR functions. A very recent study has shown that many ATP-competitive inhibitors of mTOR cause growth defects that are similar to those induced by rapamycin treatment and down-regulation of TOR expression (Montané and Menand, 2013). Because the causes of seedling growth suppression are complex, it will be important to show that these inhibitors indeed specifically block plant TOR kinase activity in vivo and in vitro with minimal off-target effects (Xiong and Sheen, 2012; Xiong et al., 2013). Unlike rapamycin, which is suggested to be specific for TORC1, these ATP-competitive inhibitors suppress a broader spectrum of mTOR functions through inactivation of both TORC1 and TORC2. It will be informative to combine and compare the effective range and specificity of these distinct inhibitors in future molecular studies of plant TOR signaling networks.
TOR signaling is mediated through a plethora of phosphorylation substrates and effectors. Identification and analysis of additional and novel TOR kinase substrates will provide a more comprehensive view of TOR kinase phosphorylation and its role in plant growth and development. Integrating large-scale phosphoproteomics analyses through quantitative mass spectrometry, screening of positional scanning peptide libraries using plant TOR complexes, and improving algorithms for computational prediction of TOR kinase substrates will greatly facilitate the identification of additional TOR kinase substrates (Hsu et al., 2011; Yu et al., 2011; Kang et al., 2013; Robitaille et al., 2013). The successful identification of transcription factors as direct TOR kinase substrates offers another innovative approach to the discovery of unconventional TOR kinase substrates with complex and combinatorial phosphorylation sites, especially for dynamic and low-abundance regulatory proteins, which may escape detection by current phosphoproteomics techniques (Xiong et al., 2013). The availability of versatile epitope-tagged protein-based artificial microRNA screens (Li et al., 2013a) and powerful genome editing tools (Feng et al., 2013; Li et al., 2013b) is also vital for systematic genetic characterization and functional analyses of novel and conserved TOR signaling components, which will open a new window to understanding the multifaceted regulatory roles of TOR in metabolism, growth, and senescence.
It is conceivable that Glc-TOR signaling in interorgan nutrient coordination, illustrated between shoots and roots (Xiong et al., 2013), may also be critical to supporting the meristem activity and growth of other sink organs, such as stems, tubers, flowers, fruits, and seeds, which are all targets of agronomic trait improvement. Plants are the source of renewable biomass and bioenergy used to feed and fuel the world, and they provide environmental protection by sustaining the ecosystem. Understanding the central growth regulatory mechanisms that function through the TOR signaling networks may lead to future exploration of the power of photosynthesis-driven interorgan nutrient coordination in manipulating specialized organ growth and associated metabolic circuits. Such studies may get at the core of agricultural and bioenergy productivity, which can be further explored to help reveal the amazingly diverse chemical synthesis capacity of plants to produce polymers, nutritional supplements, insecticides, and medicines. These findings on Glc-TOR signaling reveal a missing link in the nutritional regulation of meristems during plant growth, and they may aid in the elucidation of transcriptional networks that couple metabolism and proliferation of stem/progenitor cells, which are central to health, longevity, and disease development in animals and humans.
Acknowledgments
We thank Patrick Giavalisco and Camila Caldana for helpful discussions.
Glossary
- rRNA
ribosomal RNA
- RP
ribosomal proteins
- TAG
triacylglycerides
- PR
pathogenesis-related protein
References
- Ahn CS, Han JA, Lee HS, Lee S, Pai HS. (2011) The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 23: 185–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E, Kornet N, Friedrich T, Laux T. (2012) Plant stem cell niches. Annu Rev Plant Biol 63: 615–636 [DOI] [PubMed] [Google Scholar]
- Anderson GH, Velt B, Hanson MR. (2005) The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J. (2008) Convergent energy and stress signaling. Trends Plant Sci 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD. (2013) Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339: 1323–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. (2011) The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529 [DOI] [PubMed] [Google Scholar]
- Berkowitz O, Jost R, Pollmann S, Masle J. (2008) Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell 20: 3430–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, Kang YH, Bernhardt C, Xia Y, Zheng X, Wang JY, Lee MM, Benfey P, et al. (2012) A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P. (2013) Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J 73: 897–909 [DOI] [PubMed] [Google Scholar]
- Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O, et al. (January 14, 2013) Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene http//dx..org/ [DOI] [PubMed] [Google Scholar]
- Cornu M, Albert V, Hall MN. (2013) mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev 23: 53–62 [DOI] [PubMed] [Google Scholar]
- Creff A, Sormani R, Desnos T. (2010) The two Arabidopsis RPS6 genes, encoding for cytoplasmic ribosomal proteins S6, are functionally equivalent. Plant Mol Biol 73: 533–546 [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. (2007) mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450: 736–740 [DOI] [PubMed] [Google Scholar]
- de Jager SM, Scofield S, Huntley RP, Robinson AS, den Boer BG, Murray JA. (2009) Dissecting regulatory pathways of G1/S control in Arabidopsis: common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol Biol 71: 345–365 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D. (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Degenkolbe T, Giavalisco P, Zuther E, Seiwert B, Hincha DK, Willmitzer L. (October 13, 2012) Differential remodeling of the lipidome during cold acclimation in natural accessions of Arabidopsis thaliana. Plant J http//dx..org/ [DOI] [PubMed] [Google Scholar]
- Deprost D, Truong HN, Robaglia C, Meyer C. (2005) An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem Biophys Res Commun 326: 844–850 [DOI] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C. (2007) The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Troya S, Florencio FJ, Crespo JL. (2008) Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga Chlamydomonas reinhardtii. Eukaryot Cell 7: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Marchive C, Azzopardi M, Clément G, Moreau M, Sormani R, Robaglia C, Meyer C. (2013) Sugar metabolism and the plant target of rapamycin kinase: a sweet operaTOR? Front Plant Sci 4: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. (2010) mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328: 1172–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. (2010) Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39: 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. (2009) Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol 7: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, et al. (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23: 1229–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA. (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. (1999) Rapamycin modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA 96: 14866–14870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hiestand PC, Hall MN. (1991) FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 88: 1948–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bögre L. (2010) Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J 29: 2979–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Pan X, Welti R, Wang X. (2008) Phospholipase Dalpha3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth BM, Magyar Z, Zhang Y, Hamburger AW, Bakó L, Visser RG, Bachem CW, Bögre L. (2006) EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J 25: 4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. (2012) The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485: 55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332: 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Chern JJ, Cai Y, Liu M, Choi KW. (2007) Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature 445: 785–788 [DOI] [PubMed] [Google Scholar]
- Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. (2009) Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev 23: 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63: 353–380 [DOI] [PubMed] [Google Scholar]
- Jewell JL, Russell RC, Guan KL. (2013) Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 14: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N. (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45: 83–100 [DOI] [PubMed] [Google Scholar]
- Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM. (2013) mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341: 1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M. (2006) The genetics of plant metabolism. Nat Genet 38: 842–849 [DOI] [PubMed] [Google Scholar]
- Kim J, Guan KL. (2011) Amino acid signaling in TOR activation. Annu Rev Biochem 80: 1001–1032 [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. (2012) mTOR signaling in growth control and disease. Cell 149: 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiber RM, John F, Verhertbruggen Y, Diet A, Knox JP, Ringli C. (2010) The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 22: 1898–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD. (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Li JF, Chung HS, Niu Y, Bush J, McCormack M, Sheen J. (2013a) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25: 1507–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. (2013b) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31: 688–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kirubakaran S, Hur W, Niepel M, Westover K, Thoreen CC, Wang J, Ni J, Patricelli MP, Vogel K, et al. (2012) Kinome-wide selectivity profiling of ATP-competitive mammalian target of rapamycin (mTOR) inhibitors and characterization of their binding kinetics. J Biol Chem 287: 9742–9752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Xu C, Kirubakaran S, Zhang X, Hur W, Liu Y, Kwiatkowski NP, Wang J, Westover KD, Gao P, et al. (2013) Characterization of Torin2, an ATP-competitive inhibitor of mTOR, ATM, and ATR. Cancer Res 73: 2574–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. (2010) TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DP. (2006) Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN. (2004) TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119: 969–979 [DOI] [PubMed] [Google Scholar]
- Mehta RA, Cassol T, Li N, Ali N, Handa AK, Mattoo AK. (2002) Engineered polyamine accumulation in tomato enhances phytonutrient content, juice quality, and vine life. Nat Biotechnol 20: 613–618 [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita Y, Good AG. (2008) NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J Exp Bot 59: 667–680 [DOI] [PubMed] [Google Scholar]
- Montané MH, Menand B. (2013) ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J Exp Bot 64: 4361–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Moreau M, Azzopardi M, Clément G, Dobrenel T, Marchive C, Renne C, Martin-Magniette ML, Taconnat L, Renou JP, Robaglia C, et al. (2012) Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Sheen J. (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa S, Xu T, Yang Z. (2010) RHO GTPase in plants: Conservation and invention of regulators and effectors. Small GTPases 1: 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naouar N, Vandepoele K, Lammens T, Casneuf T, Zeller G, van Hummelen P, Weigel D, Rätsch G, Inzé D, Kuiper M, et al. (2009) Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J 57: 184–194 [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S. (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova M, Kanter E, Krakovich D, Kuchin S. (2006) Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell 5: 1831–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Florencio FJ, Crespo JL. (2010) Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 152: 1874–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Kleckner N. (2003) The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112: 151–155 [DOI] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. (2011) mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146: 408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M, Rolland F, Sheen J. (2008) Sugar sensing and signaling. The Arabidopsis Book 6: e0117, /10.1199/tab.0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Qiu S, Venglat P, Xiang D, Feng L, Selvaraj G, Datla R. (2011) Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol 155: 1367–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Venglat P, Qiu S, Feng L, Cao Y, Wang E, Xiang D, Wang J, Alexander D, Chalivendra S, et al. (2012) Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24: 4850–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes de la Cruz H, Aguilar R, Sánchez de Jiménez E. (2004) Functional characterization of a maize ribosomal S6 protein kinase (ZmS6K), a plant ortholog of metazoan p70(S6K). Biochemistry 43: 533–539 [DOI] [PubMed] [Google Scholar]
- Robaglia C, Thomas M, Meyer C. (2012) Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr Opin Plant Biol 15: 301–307 [DOI] [PubMed] [Google Scholar]
- Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. (2013) Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 339: 1320–1323 [DOI] [PubMed] [Google Scholar]
- Santner A, Calderon-Villalobos LI, Estelle M. (2009) Plant hormones are versatile chemical regulators of plant growth. Nat Chem Biol 5: 301–307 [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA. (2013) TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J 32: 1087–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Kobayashi K, Geldreich A, Caranta C, Robaglia C, Keller M, Ryabova LA. (2011) Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO J 30: 1343–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Beck T, Martin DE, Hall MN. (2004) Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol 24: 338–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WH. (2002) The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci 7: 505–511 [DOI] [PubMed] [Google Scholar]
- Shin YJ, Kim S, Du H, Choi S, Verma DP, Cheon CI. (2012) Possible dual regulatory circuits involving AtS6K1 in the regulation of plant cell cycle and growth. Mol Cells 33: 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormani R, Yao L, Menand B, Ennar N, Lecampion C, Meyer C, Robaglia C. (2007) Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol 7: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD. (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Zilbermann F, Kozma SC, Thomas G, Nagy F. (2004) Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol 134: 1527–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26: 663–674 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inzé D, De Veylder L. (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. (2013) Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. (2012) Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem 287: 2836–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. (2013) Moving beyond translation: glucose-TOR signaling in the transcriptional control of cell cycle. Cell Cycle 12: 1989–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332: 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan HX, Xiong Y, Guan KL. (2013) Nutrient sensing, metabolism, and cell growth control. Mol Cell 49: 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]