MAKER-P annotates the entire Arabidopsis and maize genomes in less than 3 h with comparable quality to the current TAIR10 and maize V2 annotation builds.
Abstract
We have optimized and extended the widely used annotation engine MAKER in order to better support plant genome annotation efforts. New features include better parallelization for large repeat-rich plant genomes, noncoding RNA annotation capabilities, and support for pseudogene identification. We have benchmarked the resulting software tool kit, MAKER-P, using the Arabidopsis (Arabidopsis thaliana) and maize (Zea mays) genomes. Here, we demonstrate the ability of the MAKER-P tool kit to automatically update, extend, and revise the Arabidopsis annotations in light of newly available data and to annotate pseudogenes and noncoding RNAs absent from The Arabidopsis Informatics Resource 10 build. Our results demonstrate that MAKER-P can be used to manage and improve the annotations of even Arabidopsis, perhaps the best-annotated plant genome. We have also installed and benchmarked MAKER-P on the Texas Advanced Computing Center. We show that this public resource can de novo annotate the entire Arabidopsis and maize genomes in less than 3 h and produce annotations of comparable quality to those of the current The Arabidopsis Information Resource 10 and maize V2 annotation builds.
Because high-throughput genome sequencing technology has become widely available, many genome projects are now carried out by small groups with little prior experience in genome annotation. A major challenge for these researchers is the generation and dissemination of high-quality gene structure annotations for downstream applications. This is especially true for plant genomics researchers, given that plant genomes can be difficult targets for annotation: they are unusually rich in transposable elements (Feschotte et al., 2002; Schnable et al., 2009; Kejnovsky et al., 2012), have high rates of pseudogenization (Thibaud-Nissen et al., 2009; Zou et al., 2009; Hua et al., 2011), and contain many novel protein-coding and noncoding RNA (ncRNA) genes as revealed through RNA-Seq and proteomics studies (Campbell et al., 2007; Hanada et al., 2007; Jiang et al., 2009; Yang et al., 2009; Li et al., 2010; Lin et al., 2010; Donoghue et al., 2011; Garg et al., 2011; Boerner and McGinnis, 2012; Moghe et al., 2013). Plant genomes are also relatively large compared with other eukaryotes, representing some of the largest genomes in existence (Pellicer et al., 2010; Birol et al., 2013; Nystedt et al., 2013), meaning that the time required to annotate a large plant genome can be measured in months rather than hours. Moreover, different plant genomes, and in some cases even the same plant genome, have been annotated using very different procedures and to very different levels of accuracy. The plant genomics community is thus in need of an annotation engine that will scale to extremely large data sets; can produce accurate annotations in a repeat- and ncRNA-rich genomic landscape; integrate computational predictions and transcriptome data; and compare, evaluate, merge, and update legacy annotations. Most importantly, this software must be easy to use, as many of today’s plant genome sequencing groups have only limited bioinformatics expertise and computational resources.
To achieve these goals, we have optimized and extended an established genome annotation engine, MAKER (Holt and Yandell, 2011), for the plant genome research community. Not only is MAKER portable and easy to use, it is already in wide use by the animal and fungal research communities (Kumar et al., 2012; Amemiya et al., 2013; Eckalbar et al., 2013; Schardl et al., 2013; Smith et al., 2013). MAKER, unlike existing pipelines, can produce accurate annotations even in the absence of training data (Holt and Yandell, 2011). Importantly, MAKER generates a set of quality-control measures to compare, evaluate, merge, and update legacy annotations (Cantarel et al., 2008; Eilbeck et al., 2009; Holt and Yandell, 2011).
We have extended MAKER for better performance on plant genomes, developing means for the annotation of pseudogenes and ncRNAs, and optimized its parallelization for maximal performance on large, repeat-rich plant genomes. The resulting software is available for download, and a MAKER-P module is installed at the Texas Advanced Computing Center (TACC) using the iPlant Cyberinfrastructure (Goff et al., 2011).
Here, we benchmark MAKER-P’s accuracy and speed using two previously annotated plant genomes: Arabidopsis (Arabidopsis thaliana) and maize (Zea mays). Our Arabidopsis results demonstrate that MAKER-P can be used to manage and improve the annotations of what is arguably the best-annotated plant genome. Using a massively parallel version of MAKER-P on the TACC, we also show that MAKER-P can de novo annotate the Arabidopsis and maize genomes in less than 3 h and that the resulting annotations are of comparable quality to the current The Arabidopsis Information Resource 10 (TAIR10) and maize V2 annotation builds. Collectively, these results demonstrate that MAKER-P provides the plant genomics community with a very rapid and effective means for both de novo annotation of new plant genomes and the management of existing plant genome annotations.
RESULTS AND DISCUSSION
Choice of Target Species
We chose to benchmark MAKER-P using Arabidopsis because it has a well-assembled reference genome and its genome annotations have been subject to extensive computational and manual curation (Lamesch et al., 2012). In addition, there is a large pool of experimental evidence available to aid the annotation of the Arabidopsis genome, including traditional ESTs, full-length complementary DNAs (cDNAs), and vast amounts of RNA-Seq data (Rounsley et al., 1996; Paz-Ares, 2002; Seki et al., 2002; Yamada et al., 2003). Moreover, The Arabidopsis Information Resource (TAIR; Lamesch et al., 2012) has put great effort into assigning evidence-based quality values to each annotation via its five-star rating system (The Arabidopsis Information Resource, 2009) in the current release of the Arabidopsis annotation set (TAIR10; Lamesch et al., 2012). Thus, the Arabidopsis genome provides a perfect opportunity to benchmark the performance of MAKER-P.
Gene-Level Accuracies
We first used the TAIR10 annotations as a gold standard with which to determine gene-level accuracies of the ab initio gene finders Semi Hidden Markov model [HMM]-Based Nucleic Acid Parser (SNAP; Korf, 2004) and Augustus (Stanke and Waack, 2003; Stanke et al., 2008). To do so, we ran SNAP and Augustus trained for Arabidopsis both with and without MAKER-P. When run within, MAKER-P can pass SNAP and Augustus additional information regarding protein, EST, and RNA-Seq evidence, allowing these programs to modify their predictions based on the evidence (Holt and Yandell, 2011). The results of this analysis are reported in Table I. As can be seen, all three approaches achieve similar gene-level accuracies. These results demonstrate an established fact of gene finding: given sufficient training data, good gene-level accuracies are relatively easy to obtain (Guigó et al., 2006; Yandell and Ence, 2012). However, often no training data are available for novel genomes. In such cases, ab initio gene finders perform poorly, requiring an evidence-driven means of genome annotation (Yandell and Ence, 2012). This phenomenon is illustrated by the penultimate column in Table I, wherein we have run SNAP using the maize HMM as a surrogate for a poorly trained gene finder. In this case, the gene-level accuracy is much poorer: 70% compared with 82% using the Arabidopsis HMM. This demonstrates that attempts to leverage training data from other plants, maize in this example, are fraught with difficulty, a fact that is well established (Korf, 2004; Holt and Yandell, 2011; Yandell and Ence, 2012). The last column of Table I reports the impact of running the same version of SNAP trained for maize within the MAKER software harness along with the RNA-Seq, EST/cDNA, and protein evidence data sets, as described in “Materials and Methods.” This column of Table I demonstrates that MAKER-P’s evidence-driven functions allow it to achieve high gene-level accuracies even using poorly trained ab initio gene finders, an observation consistent with previous work using animal genomes (Holt and Yandell, 2011) and one that demonstrates the utility of MAKER-P as a means to annotate novel plant genomes.
Table I. Effects of MAKER-P’s supervision of gene finders on genome-level sensitivity and specificity.
MAKER default, standard, and max refer to different MAKER gene-build options (see “Materials and Methods” and Supplemental Fig. S5).
| Parameter | MAKER Default | MAKER Standard | MAKER Max | Augustus Trained for Arabidopsis Run outside of MAKER | SNAP Trained for Arabidopsis Run outside of MAKER | SNAP Trained for Maize Run outside of MAKER | SNAP Trained for Maize Run inside of MAKER |
|---|---|---|---|---|---|---|---|
| Sensitivity | 0.88 | 0.91 | 0.93 | 0.91 | 0.84 | 0.47 | 0.67 |
| Specificity | 0.93 | 0.91 | 0.81 | 0.93 | 0.81 | 0.92 | 0.94 |
| Accuracy | 0.90 | 0.91 | 0.87 | 0.92 | 0.82 | 0.70 | 0.80 |
Using Annotation Edit Distance to Measure Exon-Level Accuracy
Gene-level accuracy is only the first step toward producing a well-annotated genome. Gene annotations must do more than simply overlap genes, as downstream applications require that their intron-exon structures and predicted protein sequences also be correct. The accuracy of intron-exon structures is usually assessed by means of exon-level or nucleotide-level accuracy calculations using gold standard annotations (for review, see Yandell and Ence, 2012). One question that naturally arises in such analyses is how to assess the accuracy of the gold standard annotations themselves. MAKER-P, like its parent application MAKER (Holt and Yandell, 2011), provides an automated means for addressing both these questions. MAKER-P uses Annotation Edit Distance (AED; Cantarel et al., 2008; Eilbeck et al., 2009; Holt and Yandell, 2011) to measure the goodness of fit of an annotation to the evidence supporting it. AED is a number between 0 and 1, with an AED of 0 denoting perfect concordance with the available evidence and a value of 1 indicating a complete absence of support for the annotated gene model (Eilbeck et al., 2009). AED can be calculated relative to any specific sort of evidence: EST and protein alignments, ab initio gene predictions, or RNA-Seq data. In each case, the AED score provides a measure of each annotation’s congruency with a particular type or types of evidence. By plotting the cumulative distribution function (CDF) of AED across all annotations (Holt and Yandell, 2011), a genome-wide perspective of how well the annotations and/or ab initio gene predictions reflect the EST, protein, and RNA-Seq evidence can be obtained. Importantly, this can be done even in the absence of a gold standard set of reference annotations for that genome (for an example comparing gene models produced by the ab initio gene finder Augustus run with and without MAKER supervision, see Supplemental Fig. S1). Similarly, the same procedure can be used to evaluate the goodness of fit between a gold standard annotation data set and the evidence used to produce it. For additional information on AED, see Eilbeck et al. (2009), Holt and Yandell (2011), and Yandell and Ence (2012).
Cross-Genome Validation
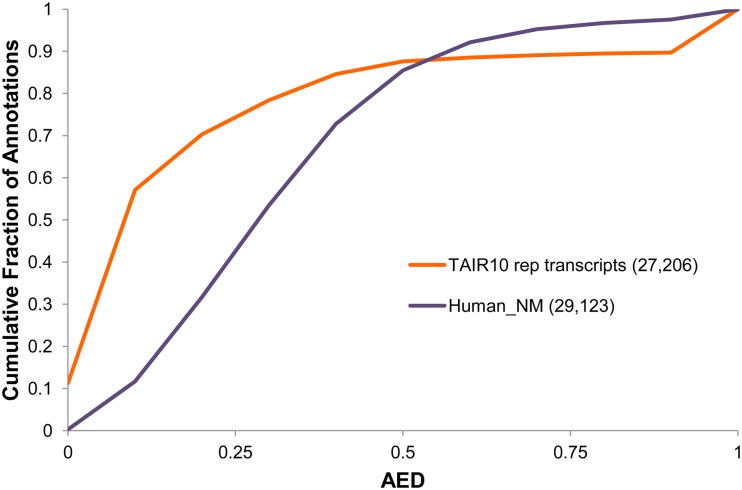
AED also makes possible cross-genome assessments of annotation data sets in the context of each genome’s own supporting evidence (Eilbeck et al., 2009; Holt and Yandell, 2011). An example is shown in Figure 1, which provides a genome-wide overview of the goodness of fit of the TAIR10 annotations to the evidence data sets used for our benchmarking analyses (for evidence data set details, see “Materials and Methods”). As can be seen, Arabidopsis is a very well-annotated genome; overall, the congruency of the TAIR10 annotations with this evidence is roughly equivalent to that of the human RefSeq annotations, in that greater than 85% of annotations have an AED score less than 0.5 when compared with a previously published analysis of human RefSeq annotations (Lander et al., 2001; Venter et al., 2001; for details of the data set, see “Materials and Methods”). Figure 1 also demonstrates that our evidence set provides support for 90% of the annotated genes in the TAIR10 data set.
Figure 1.
AED CDF for TAIR10 annotations compared with human RefSeq annotations. AED can be used to assess how well an annotation set agrees with its associated evidence. When plotted as a cumulative AED distribution, multiple annotation sets can be visualized on the same plot. Here, we have included the AED CDF for the TAIR10 (orange line) annotation of Arabidopsis and the human RefSeq (purple line) annotations of human for purposes of comparison.
Comparison of AED and TAIR’s Five-Star System
One advantage of using the TAIR10 annotations to benchmark MAKER-P is that each TAIR10 annotation has already been assigned a quality score via TAIR’s five-star ranking system (The Arabidopsis Information Resource, 2009), whereby the best-supported genes are afforded five stars or four stars, with less well-supported annotations assigned three-, two-, and one-star status. Annotations with no external support are classified as “no star.” Table II provides a breakdown of TAIR10 annotations by their star rating in the context of their supporting evidence using the evidence data sets used for our benchmarking analyses. Also shown in Table II is the cumulative support for the TAIR10 annotations in total and for the MAKER standard annotation build produced using the same evidence (for details, see “Materials and Methods”). Importantly, these results demonstrate that (1) MAKER-P can automatically produce a de novo genome annotation data set of very similar quality to the highly curated TAIR10 annotations and (2) there is good concordance between the TAIR10 star rating and the degree of evidence support.
Table II. Breakdown of evidence types supporting TAIR10 and MAKER-P annotations.
The percentage of MAKER standard and TAIR10 annotations are broken down by star rating with Pfam domains, homology to eukaryotes in RefSeq, or various combinations of RNA-Seq/EST/cDNA evidence.
| Star Rating | Fraction of Annotations with Pfam Domains | Fraction of Annotations with Eukaryotic RefSeq Protein Homology | Fraction of Annotations with Spliced RNA-Seq Support | Fraction of Annotations with RNA-Seq Support | Fraction of Annotations with Any RNA Support (mRNA-Seq, EST, cDNA) |
|---|---|---|---|---|---|
| Five stars (n = 7,880) | 0.76 | 0.81 | 0.42 | 0.79 | 0.96 |
| Four stars (n = 12,654) | 0.87 | 0.84 | 0.94 | 0.95 | 0.99 |
| Three stars (n = 2,087) | 0.80 | 0.80 | 0.53 | 0.71 | 0.85 |
| Two stars (n = 2,188) | 0.81 | 0.79 | 0.64 | 0.69 | 0.80 |
| One star (n = 1,788) | 0.54 | 0.61 | 0.06 | 0.22 | 0.40 |
| No star (n = 604) | 0.14 | 0.22 | 0.02 | 0.04 | 0.07 |
| TAIR10 representative transcripts (n = 27,206) | 0.79 | 0.79 | 0.65 | 0.80 | 0.90 |
| MAKER standard (n = 25,956) | 0.79 | 0.72 | 0.66 | 0.82 | 0.93 |
Next, we sought to determine the ability of MAKER-P to revise and improve upon the preexisting TAIR10 annotations when fed new evidence. We first used MAKER-P’s update functionality (Holt and Yandell, 2011) to automatically update each of the TAIR10 annotations, bringing each gene model into better agreement with the available evidence, by means of extending and modifying the exon coordinates of each existing TAIR10 gene annotation in light of RNA-Seq-based transcript assembly data, EST, cDNA, and protein evidence (for details, see “Materials and Methods”). Then we ran MAKER-P as we would to annotate a novel genome using the same evidence data set, allowing MAKER-P to create a new or de novo set of gene annotations based upon the same evidence that we used to update the TAIR10 annotations.
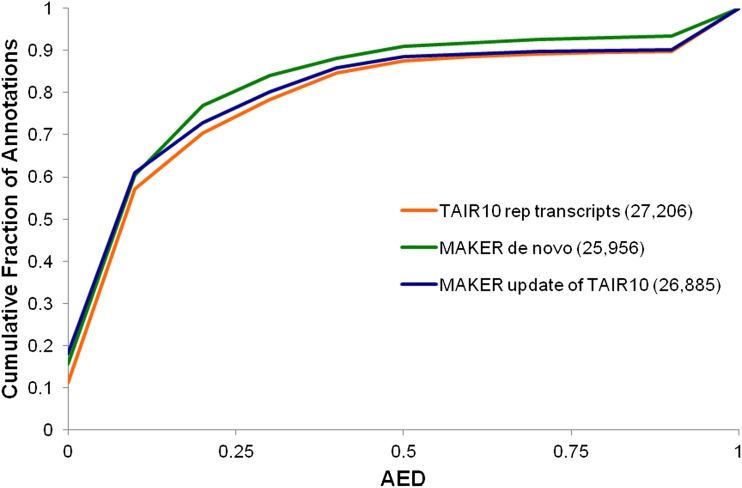
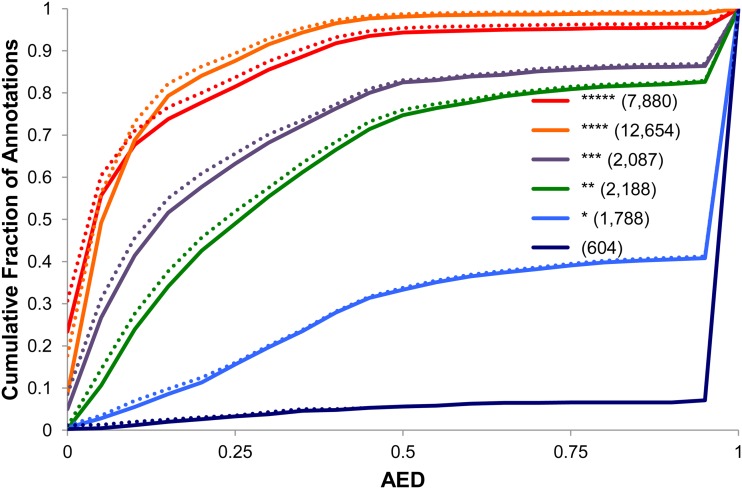
Figure 2 displays the cumulative AED distributions for the MAKER de novo, the MAKER-updated TAIR10 annotations, and the original TAIR10 Arabidopsis annotations as a reference. As can be seen, both the updated and the de novo MAKER-P data sets are in better agreement with supporting evidence than the original TAIR10 annotations. Much of the improvement, especially in the case of the MAKER-P de novo annotations, is due to the absence of poorly supported TAIR10 genes in the MAKER-P de novo gene build. The MAKER-P de novo gene build, for example, contains 1,250 fewer genes than the TAIR10 data set. In total, there are 2,368 genes present in TAIR10 that are absent from the MAKER de novo gene build. Sixty percent of the absent models are single-exon genes; 53% are one- or no-star gene-models; but 96% of all TAIR five-, four-, three-, and two-star transcripts are present. We also evaluated MAKER-P’s performance using a subset of genes with a one-to-one relationship between the TAIR10 and MAKER-P de novo annotations shown in Figure 2 and allowed MAKER-P to update the TAIR10 annotations. These results are shown in Supplemental Figure S2 and demonstrate that MAKER-P’s improvements to the TAIR10 gene models are not solely due to having culled the unsupported TAIR10 gene models; rather, the improvements are made across the entire TAIR10 data set. Figure 3 demonstrates this fact quite clearly. There is excellent agreement between the TAIR10 manually curated evidence classifications and MAKER’s automatic AED-based quality-control scheme, cross validating both MAKER-P’s AED and TAIR10’s star rating approaches to assigning confidence levels to individual annotations. For five-star TAIR10 genes, 94% have AED scores of less than 0.5, whereas only 33% of one-star genes have an AED less than 0.5. Note that the four- and five-star genes’ AED curves are very similar. This is because under the TAIR system, genes supported entirely by a single piece of evidence (usually a single full-length cDNA) are afforded five-star status, whereas an annotation completely supported by tiled evidence is afforded four-star status. MAKER-P’s AED calculation makes no such distinction; hence, the two curves are quite similar.
Figure 2.
MAKER-P de novo annotation and update of TAIR10 annotations. AED CDF curves are shown for MAKER-P run as a de novo plant annotation engine (green curve) and when used to update the existing TAIR10 gene annotation data set (blue curve), bringing it into better agreement with the evidence. Both MAKER-P data sets improve upon the existing TAIR10 annotations (orange curve).
Figure 3.
MAKER-P improvements in AED are distributed across the entire TAIR10 data set. The cumulative AED distributions for the TAIR10 representative transcripts are broken down by the TAIR star rating system. Note the excellent agreement between the TAIR10 manually curated evidence classifications and MAKER’s automatic AED-based quality-control scheme. The dotted lines denote the AED curves for the MAKER-P-updated TAIR10 annotations.
Figure 3 also demonstrates another important point: the greatest improvements are made to the highest confidence TAIR10 gene models. The dotted lines denote the AED curves for the MAKER-updated TAIR10 annotations. Note that the greatest MAKER-P-mediated improvements to the TAIR10 gene models are seen for two-star through five-star genes. While this may seem a paradoxical result, it is wholly expected. Single-star and no-star genes by definition have little supporting evidence; hence, there is little raw material available to MAKER-P with which to effect revisions. In contrast, the better supported genes (two-star through five-star annotations) have correspondingly more evidence, some supporting, some contradicting, the TAIR10 models. It is thus to the best-supported gene models under the TAIR10 classification system that MAKER-P is able to make the most positive changes. This is an important point, and it demonstrates a key strength of MAKER-P. Highly supported, highly expressed genes often have some data that strongly support a given transcript model. A single full-length cDNA, for example, may confirm the entire exon-intron structure of the annotated transcript, affording that model five-star status. Contradictory evidence is not considered under the TAIR scheme; however, it is considered by MAKER-P. This means that the resulting MAKER-P transcript structure is not necessarily a perfect match to any given piece of evidence but rather reflects the best-possible gestalt of all of the evidence for that gene. Consequently, no matter how well supported a gene model, it will have an AED greater than 0 if other evidence contradicts that model. The ability of AED to take into account both confirming and contradictory evidence is a key strength of the MAKER-P approach. The fact that MAKER-P is able to effect positive revisions to what would appear to be the best-annotated genes in the TAIR10 data sets (five- and four-star genes) demonstrates the strength of the AED approach to quality control. Further insight into the nature of these revisions is provided in Table III, which focuses on gene models with alternatively spliced transcripts
Table III. Features of alternatively spliced genes in the MAKER-P de novo annotation of Arabidopsis, TAIR10, and a MAKER-P update of TAIR10.
Comparison is shown for structural features between alternatively spliced genes generated by MAKER run de novo, TAIR10, and MAKER updating TAIR10.
| Feature | MAKER-P | TAIR10 | MAKER-P Update of TAIR10 |
|---|---|---|---|
| No. of alternatively spliced genes | 3,024 | 5,804 | 5,726 |
| No. of alternatively spliced transcripts | 7,190 | 13,774 | 13,590 |
| Average exons per transcript | 10.18 | 7.79 | 7.82 |
| Total transcripts with 5′ UTR | 5,708 | 12,714 | 12,352 |
| Total transcripts with 3′ UTR | 6,195 | 13,148 | 13,198 |
| Average nucleotides per transcript | 2,029.87 | 1,737.20 | 1,788.76 |
| Average nucleotides per coding sequence | 1,617.68 | 1,333.73 | 1,333.26 |
| Average 5′ UTR length | 169.18 | 160.13 | 179.41 |
| Average 3′ UTR length | 243 | 243.34 | 275.22 |
| Fraction of transcripts with AED less than 0.2 | 0.81 | 0.79 | 0.82 |
Alternative Splicing
MAKER-P annotates only the most certain of alternatively spliced transcripts, those with clear support for differential internal exon (cassette splicing); hence, the number of alternatively spliced transcripts is very limited compared with TAIR10. MAKER-P’s update functionality, on the other hand, provides a means to update individual alternatively spliced transcripts. MAKER-P deleted or merged 184 alternatively spliced transcripts and added an average of 19 5′ untranslated region (UTR) nucleotides and 32 3′ UTR nucleotides per transcript genome wide. The cumulative effects of the revisions are shown in the last column of Table III; prior to revision, 79% of TAIR10 transcripts had an AED less than 0.2. After revision, the proportion of gene models with AED less than 0.2 has climbed to 82%. MAKER-P thus provides a rapid and automated means to improve even intensively manually curated alternatively spliced gene models.
Repeats
Plant genomes can be difficult targets for annotation because they can be unusually rich in transposable elements (Bennetzen, 2005; Schnable et al., 2009), have high rates of pseudogenization (Zou et al., 2009; Hua et al., 2011), and contain many novel ncRNA genes as revealed through RNA-Seq (Fahlgren et al., 2007; Sunkar et al., 2008). We have attempted to address these points with the MAKER-P project. Although MAKER-P employs RepeatMasker (A.F. Smit, R. Hubley, and P. Green, unpublished data) as well as its own internal repeat-finding method (Cantarel et al., 2008), novel genomes, especially plant genomes, often contain new classes of repeats absent from both RepBase (Jurka et al., 2005) and from MAKER’s internal repeat library (Cantarel et al., 2008). Failure to identify, annotate, and mask repeats during the gene-finding stages of annotation can result in spurious gene calls and lead to the creation of gene models containing portions of transposons and retrotransposons in the form of exons derived from transposon sequences fused to legitimate protein-coding genes. Although there exist several packages to identify repeats and to construct repeat libraries for new genomes (for discussion, see Lerat, 2010), many MAKER users report that these tools are difficult to use. Moreover, the resulting output of existing packages often contains nontransposon genes or gene fragments, which may lead to the masking of bona fide genes. To address this point, the MAKER-P tool kit now contains two guided tutorials, walking users through a series of steps necessary to create their own custom repeat library. The basic tutorial describes the process of generating a species-specific repeat library suitable for repeat masking prior to protein-coding gene annotation with MAKER or MAKER-P. The advanced tutorial explains how to classify repeats identified using the basic tutorial into families. For the Web addresses for both tutorials, see Table IV. We used the approach outlined in the basic tutorial to construct a novel Arabidopsis repeat library and then assayed the impact of using it for de novo annotation of Arabidopsis, using AED to evaluate the results. These data are shown in Supplemental Figure S3. In this case, we found little difference in MAKER-P’s performance. However, Arabidopsis is not an ideal genome to demonstrate the effect of repeats on gene annotation, because the Arabidopsis genome contains the fewest repeats among all the sequenced plant genomes with the exception of the carnivorous bladderwort plant Utricularia gibba (Arabidopsis Genome Initiative, 2000; Slotkin et al., 2012; Ibarra-Laclette et al., 2013).
Table IV. Locations of all software and data sets.
Pseudogenes
With MAKER-P, we have also extended MAKER to include means for the annotation of pseudogenes and ncRNAs. These tools are included in the MAKER-P tool kit (see “Materials and Methods”). We benchmarked them on the Arabidopsis genome. The MAKER-P pseudogene tools define pseudogenes as unannotated genomic regions with significant resemblance to annotated protein sequences from the genome in question (e.g. Arabidopsis; see “Materials and Methods”). In total, we identified 4,204 pseudogenes. Among these presumed pseudogenes, 2,277 have at least one premature stop and/or frame shift (referred to as disabling substitutions). Although the rest are without disabling substitutions, the median pseudogene length is 175 bp (Supplemental Fig. S4), significantly shorter than those of TAIR10 genes and annotated pseudogenes. Thus, they are severely truncated genes that likely have no function. Because our method relied on the use of annotated protein-coding genes, all pseudogene annotations have significant similarities to known Arabidopsis proteins. Nonetheless, 18% have RNA-Seq coverage. If the analysis pipeline is applied to the whole genome, 2.5% and 0.6% of currently annotated protein-coding genes are identified as pseudogenes due to the presence of misidentified stops and frame shifts, respectively, indicating that the false-positive rate of our pipeline is 3.1%. Assuming that the pseudogene and its most closely related functional gene are paralogous, we found that the most commonly occurring domains in progenitors that gave rise to pseudogenes are F-box and related domains, RNase H, and protein kinase. Although the size of a domain family with annotated genes generally correlates with the number of pseudogenes, families differ significantly in their pseudogene:gene ratio. For example, the pseudogene:gene ratios differ significantly between F-box (152:567) and protein kinases (54:1,021; P < 2.2 × e-16), demonstrating that these families differ greatly in their loss rates.
ncRNAs
Using nine small RNA-Seq data sets of Arabidopsis (Supplemental Tables S1 and S2), the MAKER-P ncRNA tools identified 807 ncRNAs in total. The intersections of our predictions and TAIR10 annotations are summarized in Table V for tRNA, ribosomal RNA, small nucleolar RNA (snoRNA), microRNA (miRNA), and other types of ncRNA genes. It is worth noting that the number of identified ncRNAs, especially miRNAs, heavily depends on the RNA-Seq data. Some previously annotated ncRNAs are not transcribed or have extremely low transcription levels (e.g. one mapped read) in the RNA-Seq data we used for our analyses.
Table V. ncRNA annotations.
The numbers of ncRNA annotations broken down by type in the TAIR10 and MAKER-P annotation sets are shown. The last column shows the number of each type of ncRNA annotated in both sets.
Community Availability
Web addresses, download sites, and passwords (where applicable) for all tools, data sets, and online documentation described in this report are listed in Table IV. MAKER-P, like its parent package MAKER, is a multithreaded, fully message passing interface-compliant annotation engine (Holt and Yandell, 2011). MAKER-P was specifically optimized for improved functionality on the iPlant infrastructure relative to MAKER and is packaged with the necessary launch scripts to ensure optimal performance. MAKER-P also includes integrated means for tRNA and snoRNAs. MAKER-P is available to iPlant users as a supported module on the TACC Lonestar cluster (for usage instructions [specifically “iPlant MAKER-P documentation”], see Table IV). The MAKER-P tool kit is freely available for academic use; for download information, see Table IV.
Speed Benchmarks
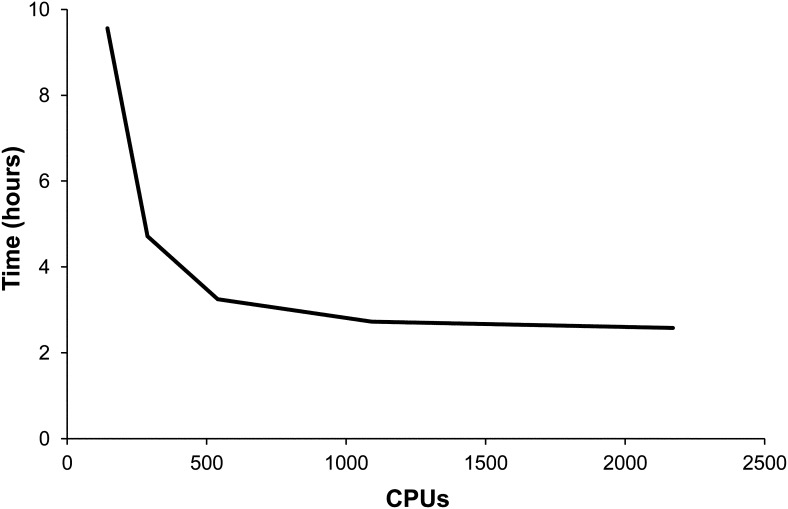
We first used the Arabidopsis genome to benchmark MAKER-P’s performance on the TACC, which hosts the iPlant compute infrastructure. Using 600 central processing units (CPUs), we were able to complete the entire de novo annotation of the Arabidopsis assembly (approximately 120 Mb) in 2 h and 44 min. Even faster compute times can be achieved using additional CPUs and/or by launching multiple instances of MAKER-P (e.g. chromosome by chromosome). By doing so, we were able to perform the same annotation in 1 h and 27 min on 1,500 CPUs. An additional benchmarking analysis using the maize assembly (approximately 2 Gb) and 2,172 CPUs finished in 2 h and 53 min (Fig. 4). Run times are both a function of the evidence data set presented for alignment as well as the gene density of a genome, but the observed throughput of greater than 500 Mb h−1 demonstrates that even the largest of plant genomes could be annotated in a reasonable time frame by leveraging MAKER-P’s scalability. Supplemental Figure S5 compares the resulting MAKER-P maize annotations with those of the current chromosome 10 V2 annotations available at MaizeGDB. As can be seen, the MAKER-P results compare favorably with the V2 annotations, with MAKER-P generating 3,059 gene annotations on this chromosome, an additional 365 gene annotations compared with the current V2 build. All of the 365 additional MAKER-P annotations are supported by RNA-Seq, EST, protein, or Pfam domain evidence and have overall better AED scores (Supplemental Fig. S6). Moreover, MAKER-P’s annotation of alternatively spliced transcripts (Supplemental Table S3) mirrors its performance on the Arabidopsis genome (Table III), further demonstrating that MAKER-P can produce highly accurate maize annotations and that it can systematically improve upon the quality of the existing V2 annotation build. Collectively, these results demonstrate that, using MAKER-P, a single investigator can carry out the de novo annotation of a grass genome and/or update its existing genome annotations with new RNA-Seq data in a few hours.
Figure 4.
MAKER-P run times on the entire maize V2 genome assembly versus the number of processors used. Increasing the number of processors given to MAKER-P decreases the run time. Run time is less than 4 h using fewer than 500 CPUs, decreasing to less than 3 h with 1,092 CPUs.
Redistribution of Annotations
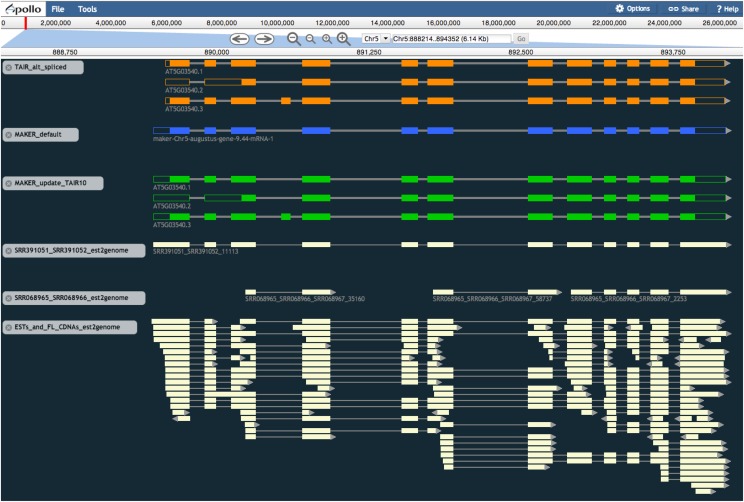
Dissemination of genome annotations, especially those of novel genomes, to the wider biological community is often a bottleneck for genome annotation projects. To remedy this problem, we have worked with the WebApollo project (Lee et al., 2013) to provide MAKER and MAKER-P users with easy means to distribute their annotation data sets to the wider community. MAKER-P’s outputs are fully WebApollo ready; thus, a WebApollo database can be constructed and placed online within hours of finishing an annotation run using ether the downloadable version of MAKER-P run locally on a user’s machine or using the community iPlant version installed on the TACC. As proof of principle, we constructed a WebApollo database containing the TAIR10, MAKER-P de novo, and MAKER-P updated annotations, the pseudogene and ncRNA annotations, and their associated protein and RNA-Seq evidence described in this report. This database is available online at http://weatherby.genetics.utah.edu:8080/WebApollo_A_thaliana (username, MAKER-P; password, marksentme). For example, click the edit button on the first page, then drag and drop any data set shown on the left-hand panel into the JBrowse central frame. For additional details and data set download locations, see Table IV. WebApollo has many features that will benefit the plant genomes community. For example, WebApollo provides functionality for remote editing of the annotations and supports concurrent users, meaning that it can be easily deployed in the classroom for purposes of hands-on instruction and rapidly deployed in support of distributed genome jamborees that aim to rapidly curate all or a specific subset of the gene annotations. Figure 5 shows a screen shot for the TAIR10 AT5G03540 gene from the database. Note that this TAIR10 gene has three annotated transcripts, two four-star and one two-star transcripts; as expected, the MAKER-P default model summarizes these with a single consensus transcript (minus the fourth exon of AT5G03540.3, for which there is no RNA-Seq, EST, or cDNA evidence). The MAKER-P update of the TAIR10 gene model maintained all three transcripts, each containing additional 5′ and 3′ UTR sequences, as suggested by the RNA-Seq data, improving the overall AED of this gene model to 0.04 compared with the AED of 0.06 of the original TAIR10 gene model.
Figure 5.
MAKER-P annotations can be easily visualized using WebApollo. This view from WebApollo shows the original TAIR10 AT5G03540 gene transcripts (orange), the MAKER-P de novo gene annotation at that locus (blue), and the MAKER-P-updated AT5G03540 gene transcripts (green). A subset of the mRNA-Seq and EST/cDNA data are shown in beige.
CONCLUSION
Today, the evidence for genome annotations evolves more rapidly than the annotations. In many cases, annotations fall out of synchronization with the available evidence almost as soon as they are created. MAKER-P provides a solution to this problem, providing a means to rapidly update a genome’s annotations, bringing them into synchronization with the latest data sets. As we have demonstrated, the greatest revisions are accomplished for those genes with the most evidence. In such cases, the quantity and complexity of RNA-Seq data supporting and contradicting even the most established gene models can confound attempts by human annotators to produce consistent, coherent gene models. MAKER-P, in contrast, guarantees a constant, complete analysis of these data, resulting in demonstrable improvements to the annotations of even the well-annotated Arabidopsis genome. Moreover, our time trials using the maize genome demonstrate that even large, complex plant genomes can be annotated in only a few hours using the version of MAKER-P installed on the iPlant resources at TACC. The availability of MAKER-P within the iPlant Cyberinfrastructure will grant independent plant genome researchers the ability to rapidly annotate new plant genomes, to revise and manage existing ones, and to create online databases for the distribution of their results. MAKER-P thus provides the plant genome research community with a basic resource that democratizes genome annotation.
MATERIALS AND METHODS
Evidence Sources and Assembly
Sequence evidence used for annotation by MAKER-P consisted of SwissProt protein data, EST and cDNA sequences from Arabidopsis (Arabidopsis thaliana), and transcript assemblies derived from publicly available RNA-Seq data sets. A SwissProt data file containing only protein sequences from plants was obtained from UniProt (release 2011_12). All Arabidopsis proteins were removed from this file, and only the non-Arabidopsis plant proteins were used when running MAKER-P. A file of Arabidopsis EST sequences (ATH_EST_sequences_20101108.fas) was obtained from TAIR (Lamesch et al., 2012). Full-length Arabidopsis cDNA sequences were downloaded from the National Center for Biotechnology Information (NCBI) Nucleotide database (Benson et al., 2013). Forty-seven RNA-Seq data sets derived from different Arabidopsis tissues and/or grown under different conditions were collected from the NCBI Short Read Archive (Supplemental Table S4; Wheeler et al., 2008). The reads from each file were cleaned using programs from the FASTX tool kit (version 0.0.13; http://hannonlab.cshl.edu/fastx_toolkit/). Fastx_clipper removed Illumina adapter sequences, and fastx_artifacts_filter removed any aberrant reads. Finally, fastx_quality_trimmer removed nucleotides with Phred scores less than 30 and discarded reads less than 20 bases long. The Trinity transcript assembly package (r2011-11-26) was used to generate transcript assemblies with lengths of 150 nucleotides or longer (Grabherr et al., 2011). The 47 RNA-Seq data sets were from 17 Short Read Archive studies and were thus assembled into 17 different transcript assemblies (Supplemental Table S4). All RNA-Seq data were treated as single-end reads in order to avoid aligning transcripts with stretches of Ns. The same procedures were used for the maize (Zea mays) data sets detailed in Supplemental Table S5.
Human annotations for release 37.2 were downloaded from the NCBI. AED metrics were computed using all mouse proteins from release 37.1, all UniProt/SwissProt proteins minus human proteins, and all human ESTs in dbEST.
Repeat Library
In this study, we established two protocols to satisfy the demands of different users. For the basic protocol (for the Web address of the tutorial, see Table IV), RepeatModeler (http://www.repeatmasker.org/RepeatModeler.html) was used to process the genomic sequences with all Arabidopsis repeats excluded from the RepeatMasker repeat library so that the Arabidopsis genome would act as a “novel” genome. Among the repetitive sequences generated by RepeatModeler, some are classified, and they are considered as transposable elements. Sequences with unknown identity from RepeatModeler were searched against a transposase database (without Arabidopsis transposase), and sequences matching transposases were considered as transposons belonging to the relevant superfamily. Many transposable elements carry genes or gene fragments. To exclude gene fragments, all repeats were searched against a plant protein database with transposon proteins excluded. Sequences matching plant proteins as well as 50 bp of flanking sequence were excluded. After the exclusion, if the remaining portion of the sequence was shorter than 50 bp, the entire sequence was excluded.
For the advanced protocol (for the Web address of the advanced tutorial, see Table IV), we used a combination of structure-based and homology-based approaches to maximize the opportunity for repeat collection. Briefly, sequences of miniature inverted repeat transposable elements were collected using MITE-Hunter (Han and Wessler, 2010) with all default parameters. Long terminal repeat retrotransposons were collected using LTR-harvest and LTR-digest (Ellinghaus et al., 2008; Steinbiss et al., 2009), followed by a filtering to exclude false positives. To reduce redundancy, representative sequences (exemplars) were chosen as described previously (Schnable et al., 2009). To collect other repetitive sequences, the genomic sequence was then masked using the long terminal repeat and miniature inverted repeat transposable element sequences. The unmasked sequence was extracted and processed by RepeatModeler. The gene fragments contained in all repetitive sequences were excluded as described above. More details can be found in the advanced repeat library construction tutorial; its Web location is given in Table IV. The libraries made through different protocols masked different percentages of the genome (Supplemental Table S2); however, the use of the basic protocol versus the advanced protocol did not significantly affect the overall AED distribution or gene-level accuracy. The resulting annotation with the basic transposable element library is a possible exception, generating a slightly lower accuracy and slightly higher overall AED scores (Supplemental Fig. S3).
MAKER-P de Novo Annotation of Arabidopsis
MAKER-P 2.27 r1020 was run on Arabidopsis (TAIR10 assembly) using the assembled Arabidopsis mRNA-Seq data, a set of traditional ESTs and full-length cDNAs, and a set of plant proteins from UniProt/SwissProt as evidence. Repetitive regions were masked using a custom repeat library. The details surrounding evidence and repeat library generation were described above. Additional areas of low complexity were soft masked (Korf et al., 2003) using RepeatMasker to prevent seeding of evidence alignments in those regions but still allowing the extension of evidence alignments through them (Korf et al., 2003; Cantarel et al., 2008). Genes were predicted using SNAP (Korf, 2004) and Augustus (Stanke and Waack, 2003; Stanke et al., 2008) trained for Arabidopsis or maize using MAKER-P in an iterative fashion as described for MAKER by Cantarel et al. (2008).
Generating MAKER-P Default, Standard, and Max Builds
When using MAKER-P to generate de novo annotations for a genome, users can choose from three different options to produce their final annotation data set: default, standard, and max. The MAKER-P default build consists only of those gene models that are supported by the evidence (i.e. AED less than 1.0). The default build is thus very conservative. The MAKER-P standard build (which was used in Fig. 2 and Tables I and II) includes every gene model in the default build, plus every ab initio gene prediction that (1) encodes a Pfam domain as detected by InterProScan (Quevillon et al., 2005) and (2) does not overlap an annotation in the MAKER default set. The MAKER-P max build includes every gene model in the default build plus every ab initio gene prediction that does not overlap an annotation in the MAKER default set, regardless of whether it encodes a Pfam domain. When using TAIR10 as a gold standard, the MAKER-P default build had the highest specificity, the MAKER-P max build had the highest sensitivity, and the MAKER-P standard build balances sensitivity and specificity to give the highest overall accuracy, which is why we used it for the comparisons in this paper (Supplemental Fig. S5). MAKER-P annotation of alternative transcripts was not evoked unless specified in the text.
Generating AED Scores for TAIR10 and Gene Finders Only
AED scores for the TAIR10 annotation set were generated using MAKER-P 2.27 r1020. The TAIR10 annotations were passed to MAKER-P as gene models in a GFF file and evaluated against the same evidence and repeat library used for the MAKER-P de novo annotation. This allowed MAKER-P to calculate AED scores for each of the TAIR10 annotations without allowing MAKER-P to modify the annotation in any way. This same procedure was used to generate AED scores for the ab initio gene predictions generated without MAKER-P supervision.
MAKER-P Update of TAIR10
The TAIR10 gene models were passed to MAKER-P as gene predictions with the same evidence and repeat library used for the MAKER-P de novo annotation. This allows MAKER-P to update the TAIR10 annotations to better match the evidence.
Pseudogene Identification
We adapted a previously published pseudogene pipeline for use with MAKER-P (Zou et al., 2009). To identify genomic regions likely to be pseudogenes, we first searched the Arabidopsis genome using all Arabidopsis annotated protein sequences as queries. The output was filtered based on the following thresholds: E value < 1e-5, identity greater than 40%, match length greater than 30 amino acids, and coverage greater than 5% of the query sequence. The filtered matches provide pseudoexon definitions. These pseudoexons that are less than 457 bp (95th percentile of the intron length distribution) from each other and having matches to the same protein are concatenated together to form putative pseudogenes. Pseudogenes overlapping with annotated protein-coding regions were removed from the data set. Finally, pseudogenes with significant similarity to known Viridiplantae repeats (cutoff = 300, divergence = 30; RepeatMasker 3.3.0) were discarded. This MAKER-P pseudogene identification pipeline is available for download at the location given in Table IV.
tRNA and snoRNA Annotation
MAKER-P features integrated means for the annotation of tRNAs and snoRNAs. tRNAs are identified using tRNAScan-SE (Lowe and Eddy, 1997) and snoRNAs with snoscan (Lowe and Eddy, 1999). Both tools are now supported and integrated within the MAKER-P software harness, and their outputs are included in MAKER-P’s GFF outputs, where they are described using the sequence ontology terms tRNA and snoRNA, respectively.
miRNA Annotation
Our ncRNA annotation pipeline uses multiple ncRNA homology search tools (described below) and small RNA RNA-Seq data to identify transcribed ncRNAs. There are three major components in the pipeline. First, we employ Infernal (Nawrocki et al., 2009), a stochastic context-free grammar-based general ncRNA search tool to identify ncRNA homologs to annotated ncRNA families in Rfam (Gardner et al., 2009). The output of this step provides candidate ncRNA genes. However, it is known that genome-scale stochastic context-free grammar searches can incur high false-positive rates. In order to discard false predictions, we evaluate the expression levels of the candidate ncRNAs in the second step. As the expression of many types of ncRNAs is condition and tissue specific, we quantified the expression levels of these putative ncRNAs in multiple small RNA-Seq data sets (Supplemental Tables S1, S2, and S7), which were sequenced from different tissues and conditions. All ncRNAs that were expressed in at least one RNA-Seq data set were validated using family-specific properties. tRNAScan-SE (Lowe and Eddy, 1997) and snoscan (Lowe and Eddy, 1999) were applied to candidate tRNAs and snoRNAs, respectively. For miRNAs, we used our own miRNA identification tool, miR-PREFeR. miR-PREFeR and its documentation are available for download at https://github.com/hangelwen/miR-PREFeR. When running this tool on Arabidopsis, we used the properties that are associated with the biogenesis of miRNA maturation as features and trained an Alternating-Decision-tree-based classification model to distinguish true from false stem loops. The features we examined include the expression pattern of the mature miRNA and miRNA* (for the RNA strand that does not go on to become the active miRNA), 3′ overhang, secondary structure, minimum free energy, existence of the regulation target (miRNA target finding), number of samples in which the miRNA is expressed, and expression-level change across multiple RNA-Seq samples. All ncRNAs that pass the three-step pipeline are reported in Table V. The total run time for miR-PREFeR on Arabidopsis was 12 h and 21 min using four processing cores and nine RNA-Seq samples.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Performance of an ab initio gene finder improves when supervised by MAKER.
Supplemental Figure S2. MAKER-P's improvements to the TAIR-10 gene models are not limited to culling of poorly supported gene models or merging gene models.
Supplemental Figure S3. Basic versus advanced repeat library generation has little effect on overall AED in Arabidopsis.
Supplemental Figure S4. Length distributions of genic and pseudogene features.
Supplemental Figure S5. Benchmarks of MAKER-P using the TAIR10 annotation dataset.
Supplemental Figure S6. Maize chromosome 10 analysis of V2 gene models.
Supplemental Table S1. RNA-Seq data sources used for miRNA identification from the NCBI's Sequence Read Archive.
Supplemental Table S2. RNA-Seq data sources used for miRNA identification from Massively Parallel Signature Sequencing Database.
Supplemental Table S3. Features of alternatively spliced genes in the MAKER-P de novo annotation of maize chromosome 10.
Supplemental Table S4. RNA-Seq data sources used for Arabidopsis benchmarks.
Supplemental Table S5. RNA-Seq data sources used for maize benchmarks.
Supplemental Table S6. Percentage of genomic sequences masked by different repeat libraries.
Supplemental Table S7. RNA-Seq data sources used for small RNA identification from the NCBI's Gene Expression Omnibus.
Acknowledgments
We gratefully acknowledge the TACC support personnel. We also acknowledge and thank Chris Towne of J. Craig Venter Institute for helpful discussion and feedback as well as Suzie Lewis at the University of California, Berkeley, and the rest of the WebApollo team for their efforts to ensure WebApollo compatibility with MAKER-P outputs.
Glossary
- ncRNA
noncoding RNA
- TACC
Texas Advanced Computing Center
- TAIR10
The Arabidopsis Information Resource 10
- cDNA
complementary DNA
- TAIR
The Arabidopsis Information Resource
- AED
Annotation Edit Distance
- CDF
cumulative distribution function
- UTR
untranslated region
- miRNA
microRNA
- CPUs
central processing units
- NCBI
National Center for Biotechnology Information
References
- Amemiya CT, Alföldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, et al. (2013) The African coelacanth genome provides insights into tetrapod evolution. Nature 496: 311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. (2005) Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev 15: 621–627 [DOI] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. (2013) GenBank. Nucleic Acids Res 41: D36–D42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birol I, Raymond A, Jackman SD, Pleasance S, Coope R, Taylor GA, Yuen MM, Keeling CI, Brand D, Vandervalk BP, et al. (2013) Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 29: 1492–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerner S, McGinnis KM. (2012) Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS ONE 7: e43047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MA, Zhu W, Jiang N, Lin H, Ouyang S, Childs KL, Haas BJ, Hamilton JP, Buell CR. (2007) Identification and characterization of lineage-specific genes within the Poaceae. Plant Physiol 145: 1311–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Alvarado AS, Yandell M. (2008) MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue MT, Keshavaiah C, Swamidatta SH, Spillane C. (2011) Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol Biol 11: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckalbar WL, Hutchins ED, Markov GJ, Allen AN, Corneveaux JJ, Lindblad-Toh K, Di Palma F, Alföldi J, Huentelman MJ, Kusumi K. (2013) Genome reannotation of the lizard Anolis carolinensis based on 14 adult and embryonic deep transcriptomes. BMC Genomics 14: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K, Moore B, Holt C, Yandell M. (2009) Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics 10: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U. (2008) LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR. (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3: 329–341 [DOI] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al (2009) Rfam: updates to the RNA families database. Nucleic Acids Res 37: D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Patel RK, Jhanwar S, Priya P, Bhattacharjee A, Yadav G, Bhatia S, Chattopadhyay D, Tyagi AK, Jain M. (2011) Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and Web resource development. Plant Physiol 156: 1661–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, Gessler D, Matasci N, Wang L, Hanlon M, Lenards A, et al. (2011) The iPlant Collaborative: cyberinfrastructure for plant biology. Front Plant Sci 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigó R, Flicek P, Abril JF, Reymond A, Lagarde J, Denoeud F, Antonarakis S, Ashburner M, Bajic VB, Birney E, et al. (2006) EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol (Suppl 1) 7: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wessler SR. (2010) MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res 38: e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH. (2007) A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res 17: 632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. (2011) MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Zou C, Shiu SH, Vierstra RD. (2011) Phylogenetic comparison of F-Box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS ONE 6: e16219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Laclette E, Lyons E, Hernández-Guzmán G, Pérez-Torres CA, Carretero-Paulet L, Chang TH, Lan T, Welch AJ, Juárez MJ, Simpson J, et al. (2013) Architecture and evolution of a minute plant genome. Nature 498: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SY, Christoffels A, Ramamoorthy R, Ramachandran S. (2009) Expansion mechanisms and functional annotations of hypothetical genes in the rice genome. Plant Physiol 150: 1997–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467 [DOI] [PubMed] [Google Scholar]
- Kejnovsky E, Hawkins J, Feschotte C (2012) Plant Transposable Elements: Biology and Evolution. In J Wendel, J Greilhuber, J Dolezel, IJ Leitch, eds, Plant Genome Diversity, Vol 1: Plant Genomes, Their Residents, and Their Evolutionary Dynamics. Springer, New York, pp 17–34 [Google Scholar]
- Korf I. (2004) Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I, Yandell M, Bedell J (2003) BLAST. O’Reilly, Sebastopol, CA [Google Scholar]
- Kumar S, Kushwaha H, Bachhawat AK, Raghava GPS, Ganesan K. (2012) Genome sequence of the oleaginous red yeast Rhodosporidium toruloides MTCC 457. Eukaryot Cell 11: 1083–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lee E, Helt GA, Reese JT, Munoz-Torres MC, Childers CP, Buels RM, Stein L, Holmes IH, Elsik CG, Lewis SE. (2013) Web Apollo: a web-based genomic annotation editing platform. Genome Biol 14: R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E. (2010) Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity (Edinb) 104: 520–533 [DOI] [PubMed] [Google Scholar]
- Li X, Wu HX, Southerton SG. (2010) Comparative genomics reveals conservative evolution of the xylem transcriptome in vascular plants. BMC Evol Biol 10: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Moghe G, Ouyang S, Iezzoni A, Shiu SH, Gu X, Buell CR. (2010) Comparative analyses reveal distinct sets of lineage-specific genes within Arabidopsis thaliana. BMC Evol Biol 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. (1999) A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171 [DOI] [PubMed] [Google Scholar]
- Moghe GD, Lehti-Shiu MD, Seddon AE, Yin S, Chen Y, Juntawong P, Brandizzi F, Bailey-Serres J, Shiu SH. (2013) Characteristics and significance of intergenic polyadenylated RNA transcription in Arabidopsis. Plant Physiol 161: 210–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR. (2009) Infernal 1.0: inference of RNA alignments. Bioinformatics 25: 1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584 [DOI] [PubMed] [Google Scholar]
- Paz-Ares J, Valencia A, Costantino P, Vittorioso P, Davies B, Gilmartin P, Giraudat J, Parcy F, Reindl A, Sablowski R, et al. (2002) REGIA, an EU project on functional genomics of transcription factors from Arabidopsis thaliana. Comp Funct Genomics 3: 102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicer J, Fay M, Leitch I. (2010) The largest eukaryotic genome of them all? Bot J Linn Soc 165: 10–15 [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33: W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley SD, Glodek A, Sutton G, Adams MD, Somerville CR, Venter JC, Kerlavage AR. (1996) The construction of Arabidopsis expressed sequence tag assemblies: a new resource to facilitate gene identification. Plant Physiol 112: 1177–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl CL, Young CA, Hesse U, Amyotte SG, Andreeva K, Calie PJ, Fleetwood DJ, Haws DC, Moore N, Oeser B, et al. (2013) Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet 9: e1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves T, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Slotkin R, Nuthikatuu S, Jaing N (2012) Plant Genome Diversity, Vol 1: Plant Genomes, Their Residents, and Their Evolutionary Dynamics. Springer, New York, pp 35–58 [Google Scholar]
- Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, et al. (2013) Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet 45: 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. (2008) Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644 [DOI] [PubMed] [Google Scholar]
- Stanke M, Waack S. (2003) Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics (Suppl 2) 19: ii215–ii225 [DOI] [PubMed] [Google Scholar]
- Steinbiss S, Willhoeft U, Gremme G, Kurtz S. (2009) Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res 37: 7002–7013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK. (2008) Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol 8: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Information Resource (2009) Documentation for the TAIR gene model and exon confidence ranking system. ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR_gene_confidence_ranking/DOCUMENTATION_TAIR_Gene_Confidence.pdf (June 22, 2012)
- Thibaud-Nissen F, Ouyang S, Buell CR. (2009) Identification and characterization of pseudogenes in the rice gene complement. BMC Genomics 10: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. (2001) The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. (2008) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 36: D13–D21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al. (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yandell M, Ence D. (2012) A beginner’s guide to eukaryotic genome annotation. Nat Rev Genet 13: 329–342 [DOI] [PubMed] [Google Scholar]
- Yang X, Jawdy S, Tschaplinski TJ, Tuskan GA. (2009) Genome-wide identification of lineage-specific genes in Arabidopsis, Oryza and Populus. Genomics 93: 473–480 [DOI] [PubMed] [Google Scholar]
- Zou C, Lehti-Shiu MD, Thibaud-Nissen F, Prakash T, Buell CR, Shiu SH. (2009) Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol 151: 3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]