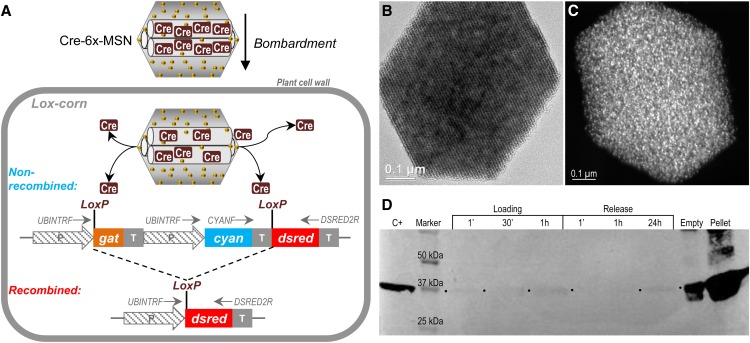
A recombinase protein loaded into mesoporous silica nanoparticles was delivered through the biolistic method to maize tissues, leading to site-specific recombination.
Abstract
The delivery of proteins instead of DNA into plant cells allows for a transient presence of the protein or enzyme that can be useful for biochemical analysis or genome modifications. This may be of particular interest for genome editing, because it can avoid DNA (transgene) integration into the genome and generate precisely modified “nontransgenic” plants. In this work, we explore direct protein delivery to plant cells using mesoporous silica nanoparticles (MSNs) as carriers to deliver Cre recombinase protein into maize (Zea mays) cells. Cre protein was loaded inside the pores of gold-plated MSNs, and these particles were delivered by the biolistic method to plant cells harboring loxP sites flanking a selection gene and a reporter gene. Cre protein was released inside the cell, leading to recombination of the loxP sites and elimination of both genes. Visual selection was used to select recombination events from which fertile plants were regenerated. Up to 20% of bombarded embryos produced calli with the recombined loxP sites under our experimental conditions. This direct and reproducible technology offers an alternative for DNA-free genome-editing technologies in which MSNs can be tailored to accommodate the desired enzyme and to reach the desired tissue through the biolistic method.
Introducing DNA-modifying enzymes rather than DNA-based expression cassettes is an attractive alternative for genetic engineering and genome-editing applications such as gene targeting or site-specific recombination. It offers a transient presence of the enzymes, and the process can be coordinated with high levels of enzymatic activity at the time and sites of the desired DNA recombination events. Many DNA-metabolizing enzymes (endonucleases, transposases, and topoisomerases), when delivered in an unrestrained manner, show adverse effects on cell viability. Delivery in the form of protein or RNA may help to mitigate these effects (Cui et al., 2011; Sander et al., 2011; Watanabe et al., 2012). In addition, by introducing proteins, one can avoid the need to remove the protein-encoding DNA fragments from the engineered plant genome. This may help shorten the time from laboratory to field for future improved germplasms.
Site-specific recombinases such as Cre or FLP have been widely used in genetic engineering applications (Sorrell and Kolb, 2005). The 38-kD Cre enzyme specifically binds to and recombines the 34-bp loxP sequences, allowing the removal, integration, or inversion of the DNA fragment flanked by these sequences (for review, see Wang et al., 2011). There are a number of established methodologies designed to provide the Cre recombinase activity for site-specific recombination in eukaryotic cells that do not involve the delivery of DNA. These methods include lipofection (Baubonis and Sauer, 1993), microinjection of protein or mRNA (de Wit et al., 1998; Luckow et al., 2009), electroporation of protein or mRNA (Kolb and Siddell, 1996; Ponsaerts et al., 2004), or using modified microorganisms for Cre delivery to their host cells (Vergunst et al., 2000; Koshy et al., 2010). Another strategy that has been used is the incubation or injection of tissues/cell cultures with cell-permeant Cre, a modified Cre protein fused to protein transduction domains or cell-penetrating peptides (Jo et al., 2001; Will et al., 2002; Lin et al., 2004; Nolden et al., 2006).
For biotechnological applications in plant sciences, protein delivery systems have been developed, including microinjection (Wymer et al., 2001), protein immobilization to gold particles (Wu et al., 2011), and protein transduction through cell-penetrating peptides (for review, see Chugh et al., 2010). The cell-penetrating peptides were shown to enable intracellular delivery of the Cre recombinase protein to rice (Oryza sativa) callus tissues (Cao et al., 2006). Nanobiotechnology is offering an attractive alternative, since nanoparticles can be precisely tailored to deliver a particular biomolecule to the cell, tissue, or organism of interest when needed (for review, see Du et al., 2012). Mesoporous silica nanoparticles (MSNs) are particularly suited for this purpose. These porous nanoparticles are formed by a matrix of well-ordered pores that confers high loading capacity of molecules like proteins (for review, see Popat et al., 2011). Additionally, surfaces of MSNs can be readily modified, permitting the customization of nanoparticles to particular experimental needs (for review, see Trewyn et al., 2007). In our previous studies, it was shown that MSNs can be used for the codelivery of DNA and chemicals (Torney et al., 2007) as well as DNA and proteins (Martin-Ortigosa et al., 2012a) to plant cells via biolistics. To improve MSN performance as a projectile, gold plating of MSN surfaces was performed, increasing nanoparticle density and, subsequently, the ability to pass through the plant cell wall upon bombardment (Martin-Ortigosa et al., 2012b).
In this work, the Cre recombinase enzyme was loaded into the pores of gold-plated MSNs and delivered through the biolistic method to maize (Zea mays) cells containing loxP sites integrated into chromosomal DNA (Lox-corn; Fig. 1A). Lox-corn expressed the glyphosate acetyltransferase gene (gat) and the Anemonia majano cyan fluorescent protein gene (AmCyan1) flanked by loxP sites. The MSN-released Cre enzyme recombined the loxP sites, thus removing the DNA fragment flanked by these sequences. Such excisions led to the expression of a variant of Discosoma sp. red fluorescent protein gene (DsRed2) and the loss of the selectable marker gene (Fig. 1A). Visual selection was used to recover the recombination events. Subsequently, fertile maize plants were regenerated from the recombined events and DNA analyses confirmed the recombination events. To our knowledge, this is the first time that MSNs have been used for the delivery of a functional recombinase into plant tissues, leading to successful genome editing.
Figure 1.
A, Schematic representation of the MSN-based bombardment technology. Cre protein is loaded into the pores of gold-plated MSN (Cre-6x-MSN) and subsequently bombarded onto immature embryos of a transgenic maize line carrying a loxP construct (Lox-corn). The parental transgenic Lox-corn tissues are blue fluorescence and herbicide resistant because they harbor a cassette with the glyphosate acetyltransferase (gat) selection gene and the AmCyan1 (cyan) marker gene flanked by the loxP sites. The DsRed2 (dsred) gene for the expression of a red fluorescent protein is placed downstream of the cassette. Once Cre recombinase is released inside the cell, it performs the recombination, excising gat-AmCyan1 genes and leading to the expression of the DsRed2 gene, switching the cell fluorescence pattern from blue to red. P, Promoter; T, terminator. UBINTRF, CYANF, and DSRED2R are primers for DNA analysis. B, Transmission electron microscope image showing the typical hexagonal shape and the well-ordered pore structure of a 6x-MSN. C, Scanning electron microscope image showing gold nanoparticle deposition (white dots) in all surfaces of 6x-MSN. D, Western blot showing Cre protein loading and release dynamics from 6x-MSN. The protein loading is almost immediate, even though some protein can be detected in the buffer even after 1 h of loading. For the release, some Cre protein can be observed after 24 h of incubation. Most of the protein remains in the 6x-MSN pellet. C+, 400 ng of Cre protein; Empty, a lane with no protein loading. The bands observed in the Empty lane were the spillover from the neighboring Pellet lane, which represents Cre-loaded 6x-MSN after the release experiment resuspended in Laemmli loading buffer (see “Materials and Methods”).
RESULTS
Cre Protein Loading onto Gold-Plated MSNs
MSNs are inorganic nanoparticles containing a uniform and structured network of well-ordered pores (Fig. 1B). The surfaces of these nanoparticles can be functionalized for specific purposes (Trewyn et al., 2007). In our previous work, 600-nm MSNs were synthesized with an average pore diameter of 10 nm, sufficient for loading enhanced GFP (28 kD) or bovine serum albumin (66.8 kD) model proteins (Martin-Ortigosa et al., 2012a). The preferred method for MSN introduction into cell-walled plant cells is the biolistic method. To improve MSN characteristics as projectiles, six rounds of gold plating were performed (6x-MSN; Fig. 1C). The amount of deposited gold increased 42.3% (w/w) in 6x-MSN (Supplemental Table S1). The deposition of the reduced gold nanoparticles on the surfaces of 6x-MSN can be observed as white dots in Figure 1C and was detected through energy-dispersive x-ray spectrum analysis (Supplemental Fig. S1A). Electron microscopy results indicate a uniform distribution of gold on the MSN surface. This amount of gold reduced the surface area and the pore volume of MSNs by 58% and 60%, respectively (Supplemental Table S1), but the values can be considered sufficient for surface functionalization and cargo loading.
The biolistic performance of 6x-MSN was tested on onion (Allium cepa) epidermis tissues using the MSN coating and bombardment protocols described previously (Martin-Ortigosa et al., 2012b). The plasmid ER-rk (double 35S promoter, endoplasmic reticulum-targeted mCherry, and nos 3′ terminator; Nelson et al., 2007) was used to coat 0x-MSN (non-gold-plated MSN) or 6x-MSN. One or 4 d after bombardment, the number of transiently fluorescent onion cells was counted on each sample as an indirect measurement of intracellular MSN delivery to plant cells. As can be seen in Supplemental Figure S1B, 6x-MSN (more than 60 fluorescent cells per sample) was 3 times more efficient as a projectile than 0x-MSN (approximately 20 fluorescent cells per sample).
Next, the loading and release of active Cre protein (Supplemental Fig. S2) into 6x-MSN were tested using time-course and western-blot analyses (Fig. 1D). The purified protein was loaded onto 6x-MSN by diffusion, and the loaded particles were lyophilized according to a previously developed protocol (Martin-Ortigosa et al., 2012a). For release experiments, the pellet was resuspended in buffer, and supernatant and pellet samples were analyzed. As shown on the western blot in Figure 1D (loading lanes), Cre protein loading onto 6x-MSN was immediate; only a small amount of Cre protein was detected in the supernatant after 1 min of incubation. Similar results were observed after 30 min and 1 h, suggesting that 30 µg of Cre protein per 1 mg of 6x-MSN saturated the particles. This immediate loading phenomenon could be due to the strong electrostatic interactions between positively charged Cre proteins (pI of 10.3; Ghosh and Van Duyne, 2002) and negatively charged MSNs (z-potential of −28.8 mV; Martin-Ortigosa et al., 2012b). These properties could facilitate Cre protein loading into MSN but could also impede its release. As shown in Figure 1D (release lanes), some Cre protein could be detected after overnight (24-h) incubation, suggesting slow release of Cre protein from MSNs into the supernatant. After overnight release experiments, the remaining MSN pellet was also loaded on the gel (Fig. 1D, pellet lane), showing that most Cre protein remained loaded in the nanoparticles.
In an attempt to improve the protein-release efficiency, a surface functionalization of 6x-MSN with amine groups was performed. The amine groups were incorporated via postsynthetically grafting 3-aminopropyltrimethoxysilane in toluene after gold deposition. These resultant MSNs (AP-6x-MSN) had a z-potential of +20.6 mV, which conferred an overall positive charge to MSNs. It was expected that the positive charges of both AP-6x-MSN and Cre protein would facilitate Cre release due to electrostatic repulsion. However, this modified surface functionalization did not achieve our intended results. Both loading and releasing processes appeared to be inefficient (Supplemental Fig. S1C).
Transient Analysis of MSN-Mediated Cre Protein Delivery to Plant Cells
The delivery of functional Cre protein to plant tissues was tested using transient fluorescence assays. Both onion epidermis cells and maize immature embryos were chosen as target tissues using the biolistic method. In our previous work, two coating/bombardment protocols for biomolecule delivery were established (Martin-Ortigosa et al., 2012b). The first protocol (MSN only) uses gold-plated MSN directly, while the second protocol (MSN/gold) uses a mixture of MSN and 0.6-µm gold microprojectiles for bombardment. The second approach was more efficient in the delivery of nanoparticles and biomolecules (Martin-Ortigosa et al., 2012b).
The transient experiments were carried out using the MSN/gold method. Cre-loaded MSNs (Cre-6x-MSN) were mixed with 0.6-µm gold particles and then coated with a plasmid DNA (PHP43619; Supplemental Fig. S3A) carrying a similar fragment for gat and AmCyan1 gene deletion and DsRed2 gene activation upon Cre-mediated recombination, as described in Figure 1A. This Cre-6x-MSN/gold-DNA mixture was bombarded into both onion epidermal tissues and maize immature embryos. One to 4 d after bombardment, red and blue fluorescent cells were scored using a microscope. Blue fluorescent cells (expressing the AmCyan1 gene) indicate successful plasmid DNA delivery. Red fluorescent cells (expressing the DsRed2 gene), on the other hand, indicate Cre protein-mediated intracellular recombination of the plasmid DNA.
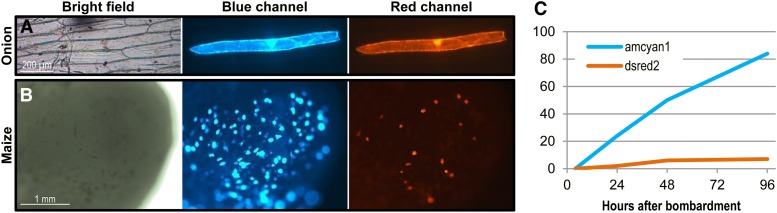
Figure 2 shows the transient fluorescence assay results for both onion and maize tissues. One day after bombardment, blue fluorescent cells were detected as a consequence of transient expression of the AmCyan1 gene on the plasmid PHP43619. Some of these blue fluorescent cells colocalized red fluorescence as well (Fig. 2, A and B) as a result of the in vivo Cre protein-mediated loxP recombination and the activation of the expression of DsRed2 on the plasmid PHP43619. No red fluorescent cells were detected on tissues where the plasmid PHP43619 was bombarded alone, with no Cre-6x-MSN, suggesting that no spontaneous recombination occurred when the Cre protein was not present.
Figure 2.
A and B, In planta Cre protein delivery. Microscope images are shown for onion epidermis tissue (A) or Hi-II maize immature embryo (B) bombarded with Cre-6x-MSN mixed with 0.6 µm of gold and coated with PHP43619 plasmid DNA. Red fluorescence is the result of Cre protein-mediated recombination in the cell. C, Graph showing the time course of the total number of fluorescent cells in an onion epidermis sample bombarded with Cre-6x-MSN and coated with PHP43619 plasmid.
In onion tissues, while the number of blue fluorescent cells increased steadily for 4 d (from 24 cells at 24 h to more than 80 cells at 96 h), the number of red cells increased only during the first 48 h after bombardment (from two cells at 24 h to seven cells at 96 h; Fig. 2C). It is possible that under the conditions tested, Cre protein release or its recombinase activity could only be detected up to 2 d. In maize tissues, the number of red fluorescent cells observed varied between different bombarded plates and individual embryos on the same target plates. In one experiment in which three target plates (25 embryos per plate) were bombarded, the total number of red fluorescent dots (Fig. 2B) on each plate was estimated around 170 to 450 (Supplemental Fig. S3C).
In the transient Cre protein delivery experiments reported in this study, the recombination events (measured as the number of red cells) achieved through the codelivery of plasmid DNA PHP43619 and Cre-6x-MSN were less frequent than those achieved through the codelivery of PHP43619 and Cre-encoding plasmid DNA PHP16072 (Supplemental Fig. S3, B and D). In Cre protein delivery experiments in onion samples, the percentage of red fluorescent cells ranged from 0% to 8.3% of the total number of blue fluorescent cells (Fig. 2C). In maize immature embryos, the number of red fluorescent cells ranged from one to 55 (Fig. 2B; Supplemental Fig. S3, C and D). However, when Cre-encoding plasmid DNA was delivered to maize immature embryos, the number of red fluorescent cells could exceed 100 (Supplemental Fig. S3D).
Maize Genome Recombination by MSN-Mediated Cre Protein Delivery
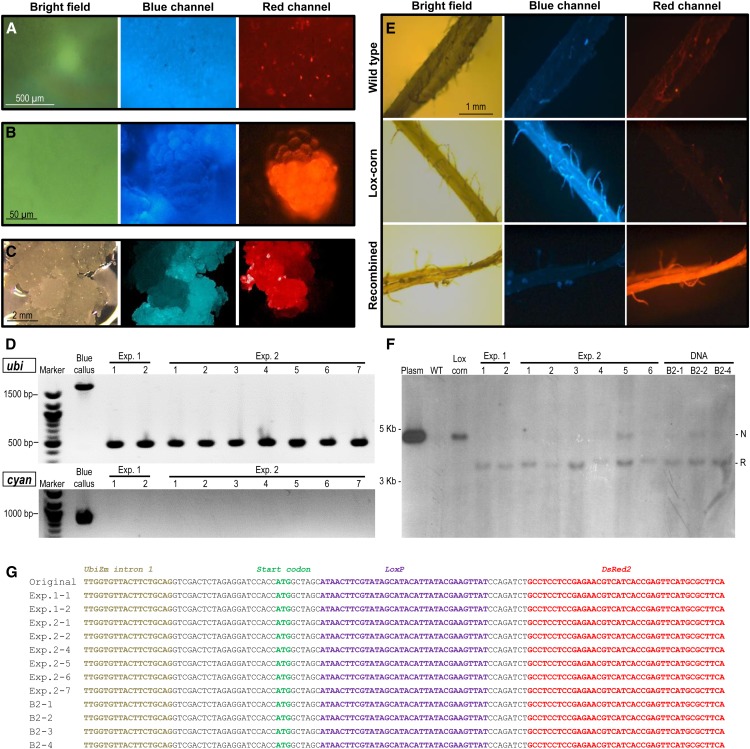
A transgenic Hi-II maize line (Lox-corn) was used for MSN-mediated Cre protein delivery experiments. The Lox-corn carried a transgene with the gat and AmCyan1 genes flanked by loxP sites; upon Cre-mediated recombination, activation of the DsRed2 marker gene could be achieved (Fig. 1A; Supplemental Fig. S3A). This transgenic Lox-corn line was a single-locus insertion event (Supplemental Fig. S4, A and B). The Lox-corn T2 plants were crossed using pollen from nontransgenic maize genotype Hi-II (Armstrong et al., 1991) to obtain immature embryos. Blue fluorescent immature embryos from this cross were bombarded with Cre-6x-MSN with or without 0.6 µm of gold. One day after bombardment, red fluorescent cells could be observed in the scutellum of the blue fluorescent embryos (Fig. 3A). This indicated that Cre protein had excised the sequences of the gat and AmCyan1 genes flanked by loxP sites (Fig. 1A), leading to DsRed2 gene expression driven by the ubiquitin promoter that once drove gat gene expression (Fig. 1A; Supplemental Fig. S3A).
Figure 3.
Selection and analysis of Cre recombinant events. A, Microscope images of a blue fluorescent Lox-corn embryo scutellum showing red fluorescent cells as a result of Cre protein-mediated recombination 4 d after bombardment. B, Early callus formation from a red fluorescent cell. C, Red fluorescent callus of 3 to 5 mm in size emerging from blue fluorescent tissue. D, PCR analysis results of callus from Lox-corn (blue callus) or red fluorescent calli events (Table I) obtained after Cre protein-mediated recombination in experiments 1 and 2 (Exp. 1 and Exp. 2 respectively). The top image shows ubi PCR using primers UBINTRF and DSRED2R (Fig. 1A; nonrecombined, 1,870 bp; recombined, 460 bp); the bottom image shows cyan PCR using the CYANF and DSRED2R primers (Fig. 1A; nonrecombined, 920 bp; recombined, 0 bp). E, Microscope images of ear silks from wild type Hi-II, Lox-corn, and the Cre protein recombinant event. F, Southern-blot analysis of T0 plants generated from Cre protein events (Exp. 1 and Exp. 2) and Cre-DNA callus events (DNA) using DsRed fragment as a probe (for recombination diagram, see Supplemental Fig. S4A). Lox-corn, Parental maize line DNA showing the original nonrecombined loxP fragment; N, nonrecombination band, 4.51 kb; Plasm, plasmid PHP43619; R, recombination band, 3.45 kb; WT, wild-type Hi-II DNA as a negative control. G, Recombination junction sequencing analysis of T0 plants generated from red fluorescent callus events. The PCR products were obtained by using primers UBINTRF and DSRED2R (Fig. 1A). The recombined loxP site (purple letters) was intact, as expected.
As a positive control, blue fluorescent Lox-corn embryos were bombarded with the Cre-encoding plasmid DNA PHP16072 (Supplemental Fig. S3B). More red fluorescent cells were observed in embryos bombarded with Cre-encoding plasmid DNA compared with those bombarded with Cre protein loaded onto 6x-MSN (Supplemental Fig. S5). It is likely that the cells bombarded with the Cre-encoding DNA produced more Cre recombinase protein than the cells bombarded with the Cre protein-loaded 6x-MSNs.
Delivery of Cre protein did not introduce any additional selectable marker genes. Therefore, visual selection of red fluorescent cells and tissues was used for the detection, separation, and multiplication of the recombination events. After 3 weeks, a few groups of spherical cell clusters could be detected in the newly formed type II callus from the embryos (Fig. 3B). These structures were small and formed by a few dozen cells. Seven to 10 weeks after bombardment, red fluorescent callus pieces (3–5 mm) were separated from the blue fluorescent calli background (Fig. 3C). Because there was no selection pressure, the growth of callus cultures was rapid. Some of the red fluorescent callus lines could have been neglected due to blue fluorescent callus overgrowth. Nevertheless, the selected callus lines were independently grown, separated from blue fluorescent cells, and multiplied.
PCR analysis was performed on both blue and red fluorescent callus lines. Primers were designed to verify the recombination events (Fig. 1A). Primer pair “ubi” (UBINTRF-DSRED2R) is expected to amplify a 1.8-kb band in blue fluorescent calli (no recombination) and a 460-bp band in any red fluorescent calli (recombination event; Fig. 1A). Primer pair “cyan” (CYANF-DSRED2R) should amplify a 920-bp band in blue fluorescent callus and no bands for red fluorescent callus analysis (Fig. 1A). As can be seen from Figure 3D, all nine independent red callus events produced the 460-bp band with the ubi primers and no bands with the cyan primers (Fig. 3D), as expected. The blue fluorescent callus control produced the expected bands of 1.8 kb with ubi primers and 920 bp with cyan primers (Fig. 3D).
A total of 310 Lox-corn embryos (four independent experiments) were bombarded with Cre-loaded MSNs using various bombardment conditions (Table I). In these experiments, the recombination efficiency (defined as the number of PCR-verified red fluorescent calli per 100 embryos bombarded) varied between 1.25% and 20%. A total of 46 red callus events were identified. The selected events were regenerated, and T0 plants were grown to maturity in the greenhouse. T0 plants were tested for fluorescence in silk tissues. The cyan fluorescence was not detectable in these tissues, while the red fluorescence was visible (Fig. 3E).
Table I. Summary of stable Cre protein delivery experiments.
| Experimenta | Gold | No. of Shots per Plate | No. of Bombarded Plates (No. of Embryos per Plate) | No. of Red Callus Events Confirmed by PCR | No. of Events Selected for Sequencing | Efficiencyb |
|---|---|---|---|---|---|---|
| µg shot−1 | ||||||
| 1 | 30 | 1 | 4 (30) | 24 | 2 | 20% |
| 2 | 0 | 1 | 2 (25) | 10 | 6 | 20% |
| 3 | 30 | 1 | 3 (20) | 12 | n/tc | 20% |
| 4 | 30 | 2 | 4 (20) | 1 | n/t | 1.25% |
Cre protein-loaded 6x-MSNs were used for bombarding blue fluorescent immature embryos of Lox-corn. bPercentage of red fluorescent callus that were recovered and PCR confirmed per number of bombarded embryos. cn/t, Not tested.
Southern-blot analyses were performed on T0 plants, including eight Cre protein events and three Cre DNA events randomly selected from the four bombardment experiments. As illustrated in Supplemental Figure S4A, if the Cre-mediated recombination is accomplished, a single band of 3.5 kb should be expected when the DsRed probe is used for hybridization. Otherwise, a 4.5-kb band representing nonrecombination should be detected (Supplemental Fig. S4A). As shown in Figure 3F, most Cre protein events have the 3.5-kb recombination band (indicated as R). Interestingly, some of the Cre protein events and Cre DNA events also showed the 4.5-kb band (indicated as N), suggesting the chimeric nature of these recombination events (Fig. 3F). We also noticed that the nonrecombined band in line Exp.2-5 (Fig. 3F) was not detected in the callus PCR analysis (Fig. 3D). One explanation could be that the callus sampled for PCR analysis consisted mainly of cells with complete excision. Because there was no growth selection on the cells with or without recombination, the regenerated plantlets could have germinated from chimeric somatic embryos consisting of both recombined and nonrecombined cells, as shown in the Southern analysis.
Regenerated events were also PCR verified and sequenced for the determination of recombination junctions. All events produced precise recombination junction sequences (Fig. 3G), indicating that the direct delivery of Cre protein was as effective as Cre DNA delivery in generating recombination on the loxP sites in the maize genome. In addition, when PCR was used to examine the presence of the Cre-encoding DNA, it was readily detected in the red callus events generated from DNA-bombarded Lox-corn embryos (Supplemental Fig. S6A) but not in the red callus events generated from Cre protein-bombarded embryos (Supplemental Fig. S6B), as expected.
DISCUSSION
In this work, a novel nanotechnology-based method for the delivery of an active Cre recombinase to plant tissues is demonstrated. Cre-mediated site-specific recombination has been extensively studied in many organisms. The published technologies rely on either transient/inducible expression of Cre or introducing Cre-expressing DNA through genetic crosses (Zuo et al., 2001; Wang et al., 2011). These technologies have been tested for the removal of marker genes in different plant species. The recombination success ranges from 0.25% to 82% for transient expression methodologies, from 13% to 100% for inducible Cre expression, and from 19.2% to 95% for cross pollination with a Cre-expressing line (for review, see Kopertekh and Schiemann, 2012). Because the presence of the Cre-encoding DNA is not desirable in the final products, subsequent analyses are needed to eliminate the DNA fragment in the genome. In this report, the maize genome has been modified by MSN-mediated Cre protein delivery through the biolistic method. This methodology could save time and expense for DNA analysis and is not target tissue or organism dependent. Additionally, unwanted genome rearrangements may be minimized, because a protein and its activity can be short lived in the cells.
Several transient Cre expression methodologies have been developed for the elimination of transgenes from plants, such as the use of inducible promoters, transient transformation, or even fusions of Cre protein to virulence proteins of Agrobacterium tumefaciens (for review, see Hare and Chua, 2002). Compared with these technologies, MSN-mediated Cre protein delivery is a straightforward method that does not require complex DNA constructs, analyses to detect unwanted DNA integration, or genetic engineering of microorganisms.
Most of the technologies for Cre protein delivery, such as microinjection (Luckow et al., 2009), electroporation (Van den Plas et al., 2003), lipofection (Baubonis and Sauer, 1993), or protein transduction through cell-penetrating peptides (Nolden et al., 2006), have been demonstrated in mammalian cell models. While studies have shown that native or His-tagged Cre protein can be taken up by mammalian cells via simple incubation (Will et al., 2002; Lin et al., 2004), there are no reports of the successful uptake of native Cre protein in plant cells or tissues. Cre protein transduction to plant cells has been achieved by using a Cre protein fused to a membrane translocation sequence and to a nuclear localization sequence (Cao et al., 2006). Rice callus explants were incubated with this protein, and the results varied depending on the age of the callus and maceration of the cell wall. The recombination efficiency of the different treatments, based on the initial number of explants, varied between 4% and 25.9% (Cao et al., 2006), similar to the frequencies achieved in our methodology. Cell-penetrating peptides are being used for protein delivery to plant tissues, but the technology has disadvantages, such as delivery dependence on cell type, physiology, permeability of the cell wall, and size of the cargo (for review, see Chugh et al., 2010). In addition, issues such as fusion protein stability and activity also need to be considered and evaluated when using protein transduction technology.
MSNs are versatile and customizable nanoparticles that have been used for intracellular biomolecule delivery (for review, see Slowing et al., 2010). When considering the loading of a protein into the pores of an MSN, not only the size but also the tertiary structure of the protein has to be taken into account. While no reports of structural dimensions or hydrodynamic radius for the 38-kD Cre monomer could be found, the complex of a Cre tetramer bound to loxP sites was shown to be 10.8 × 16.4 × 19.4 nm (Ennifar et al., 2003), which led to our deduction that the Cre monomer might be within a size allowable for the 10-nm diameter of MSN pores. Even though the pore size limitation can be perceived as a hindrance for loading larger proteins, methods for the synthesis of large-pore mesoporous silica materials are available (Hartono et al., 2010). Moreover, proteins as big as bovine serum albumin (67 kD) have already been loaded and released into plant cells following this method (Martin-Ortigosa et al., 2012a).
Loading the MSN particles with the Cre protein did not seem to be a limiting factor in this application. About 60 µg of Cre protein is rapidly loaded into 2 mg of 6x-MSNs after mixing (Fig. 1D). The loading process is mostly based on electrostatic interactions. Cre recombinase protein has a high pI, 10.3 (Ghosh and Van Duyne, 2002), and MSNs are negatively charged by nature, even after gold plating the surfaces (Martin-Ortigosa et al., 2012b). MSNs can be functionalized to promote loading/release of the cargo of interest (for review, see Popat et al., 2011). Amine functionalization of 6x-MSN resulted in positively charged AP-6x-MSN, which could, in theory, be more effective in the release due to electrostatic repulsion of the positive charges. However, no release of the Cre protein was observed when the amino groups were attached to the MSN particles. As demonstrated by data in Supplemental Figure S1C, no detectable Cre protein was loaded in AP-6x-MSN, and as a consequence, no protein release was detected. On the contrary, 6x-MSN Cre protein loading was fast and efficient. Through extensive nitrogen sorption and x-ray diffraction analyses in a previous study of MSN-loaded proteins, we are confident that the biomolecules diffuse into the pores (Slowing et al., 2007). However, due to the strong electrostatic interactions between the protein and the MSN materials, it is possible that some protein deposition occurred on the outer surface of the nanoparticle. After an overnight release experiment, some protein could be detected in the buffer, but most of the protein remained packaged inside MSN pores (Fig. 1D).
One concern could be that the Cre protein delivery described here may not be mediated by the MSN but rather by a direct introduction via gold microprojectiles in experiments where the 0.6-µm gold particles were used. Our previous work has shown that gold-plated MSN alone (without being associated with 0.6-µm gold microprojectiles) could be used as an effective protein delivery system for plant cells (Martin-Ortigosa et al., 2012a). In Table I, we have shown that recombination efficiency of 20% could be achieved using either Cre-MSN alone (experiment 2) or Cre-MSN combined with 0.6-µm gold microprojectiles (experiments 1 and 3). While we cannot dismiss the possibility that some of the Cre protein could somehow be released from the nanoparticle pores and be attached to the gold microprojectiles, we conclude that the Cre protein delivery described in this work was mediated mainly by MSN.
While the release process can be further optimized, there was an adequate amount of the Cre protein released for the intracellular recombination of loxP sites. Transient experiments in onion epidermis tissues and maize immature embryos showed that intracellular Cre protein delivery can be achieved using 6x-MSNs as carriers and that the enzyme is active to carry out plasmid DNA recombination (Fig. 2). In these experiments, Cre activity was detected in the first 48 h post bombardment (Fig. 2C). It is hypothesized that the bulk of the release is diffusion of the readily available Cre protein from the pores. Because Cre protein could have a limited lifespan and the rest of the packaged Cre might have a very slow release rate due to strong electrostatic interactions, no continuous Cre activity was detected (Fig. 2C). In these experiments, Cre protein was first loaded into 6x-MSN, and the Cre-loaded MSNs were then lyophilized and mixed with 0.6-µm gold before the mixture was coated with loxP-containing plasmid DNA. It is possible that premature loxP site recombination may have occurred prior to bombardment. If this were the case, red fluorescent cells (as a result of in vitro recombination) should have been detected in the same time frame as blue fluorescent cells. However, during the first 24 h, we only detected blue fluorescent cells in plant tissues. Most red fluorescent cells were detected after 24 h and colocalized with the blue fluorescence, suggesting that the recombination likely occurred in vivo.
In stable genome recombination experiments, 6x-MSN delivered active Cre recombinase protein to transgenic Lox-corn immature embryos, resulting in multiple red fluorescent single-cell recombined events. After visual selection and callus multiplication, these events showed the expected recombined DNA pattern, demonstrating, to our knowledge for the first time, the suitability of MSN-based protein delivery technology to edit a plant genome. To improve Cre protein delivery, different bombardment conditions, such as cobombardment with 0.6-µm gold particles or bombarding the same plate twice, were tested. A 20% recombination frequency was achieved using either MSN only or MSN/gold. However, cells bombarded twice seemed to yield fewer recombination events (1.25%; Table I). This could be due to excessive physical damage to the embryos from the double bombardment. At the current stage, the efficiency of the excision from Cre protein delivery was lower when compared with the Cre DNA delivery (Supplemental Figs. S3D and S5). However, no need for further removal of the Cre DNA fragment from the plant genome can compensate for the initial lower efficiency of the protein delivery system.
In this study, we demonstrate that it is feasible to deliver a recombinase (Cre) using a nanoparticle and biolistic approach to remove a loxP-defined DNA fragment from the maize genome. We selected the recombinant events by visual selection of the fluorescence pattern change (from blue fluorescence to red fluorescence). Because there was no growth advantage for successful recombinant events in this visual screening process, the regenerated somatic embryos could be chimeric (Armstrong and Phillips, 1988). Some of the T0 events showed a nonrecombined band in addition to the recombination band in the Southern-blot analysis (Fig. 3F). The coexistence of both recombined and nonrecombined patterns has been previously described in other Cre/lox system studies in which DNA fragment excision was accomplished using the Cre DNA delivery system (Zuo et al., 2001; Djukanovic et al., 2008). Because the chimerism was observed in both Cre protein events and Cre DNA events (Fig. 3F) in our study, we believe that this phenomenon is not due to the Cre delivery methods but rather to the visual selection procedure. This type of chimerism may be minimized if molecular analyses are performed on the red fluorescent callus to select for events lacking the nonrecombined pattern.
One important application of the technology described here can be the recombinase-mediated site-specific gene integration (Albert et al., 1995; Srivastava and Ow, 2002; Srivastava et al., 2004; Li et al., 2009). In our previous work, we have shown that MSN technology enables the intracellular codelivery of DNA and chemicals (Torney et al., 2007) and DNA and proteins (Martin-Ortigosa et al., 2012a). Therefore, it is feasible that this system can be used for site-specific integration by codelivering a loxP-bearing DNA molecule along with the Cre protein. Because the recombinase is delivered as a protein, there will be no integration of the undesired recombinase-expressing gene fragment. This can eliminate issues such as the reversibility of the integration/excision reactions. In addition, the transgene intended to be integrated can be selected if it is associated with a selectable marker gene.
MSN-mediated protein delivery to plant cells has advantages and disadvantages when compared with other methods such as protein transduction. MSN technology can be customized for each protein of interest, and it could protect proteins from degradation once loaded inside the pores. Since it is a direct physical method, it can be considered independent of the target tissue and its physiological status. Additionally, the pore openings of MSN can be capped for the controlled release of the cargo (Torney et al., 2007). Finally, multiple chemicals and biomolecules can be loaded and codelivered to plant cells (Torney et al., 2007; Martin-Ortigosa et al., 2012a). On the other hand, this technology requires adapting MSN synthesis to the size and nature of each particular protein. Moreover, release of the pore-packed protein can be inefficient, requiring further research to optimize the loading-release ratio for each particular protein.
Because direct protein delivery can bypass the integration of foreign DNA into the plant chromosomes, this method can provide a flexible means to edit chromosomal DNA without adding genetic elements not required in the final product. Therefore, this method has potential to be a powerful tool for both basic and applied research in plant science research.
MATERIALS AND METHODS
MSN Synthesis
The synthesis of 6x-MSN and AP-6x-MSN was modified from our previously published methods (Martin-Ortigosa et al., 2012b). To synthesize 10-nm pore MSN, P104 surfactant (7.0 g) was dissolved in 1.6 m HCl and stirred for 1 h at 55°C. Rapid addition of 10.64 g of tetramethyl orthosilicate was followed by 24 h of stirring. The suspension was transferred to a high-pressure vessel and placed in an oven at 150°C for 24 h. The MSN was filtered and washed with water and methanol. The surfactant was removed by thermally treating the sample to 550°C for 6 h. To modify the MSN surface with gold, 372 mg of Au(en)2Cl3 [for the detailed synthesis of Au(en)2Cl3 from HAuCl4·3H2O, see Martin-Ortigosa et al., 2012b] was dissolved in 150 mL of water, and the pH was adjusted to 10 using NaOH. After adding MSN (2.0 g), the pH was adjusted to 9.0 with NaOH and the mixture was stirred for 2 h at room temperature. The final product (6x-MSN) was filtered, washed with water and methanol, and dried under vacuum for 48 h. Surface-bound gold was reduced under H2 flow for 3 h at 150°C. This procedure was repeated six times to increase the quantity of gold on the MSN surface. Primary amine functionalization was achieved by refluxing 6x-MSN (100 mg) suspended in 3-aminopropyltrimethoxysilane (54 mg, 0.3 mmol) in anhydrous toluene (50 mL) overnight. The resulting materials were filtered, washed with toluene and methanol, and dried under vacuum overnight.
MSN Characterization Analysis
Zeta potential measurements were obtained using a Malvern Instruments Zetasizer. The surface area and porosity analyses were done using nitrogen sorption analysis on an ASAP 2020 BET surface analyzer (Micromeritics). The quantification of gold loading was accomplished through inductively coupled plasma-optical emission spectroscopy analysis using a Perkin-Elmer Optima 7300 spectrometer.
Electron Microscope Imaging
After air drying a small aliquot of 6x-MSN in aqueous solution over a lacey carbon film coated 400-mesh copper grid, images were taken for transmission electron microscopy with a Tecnai F2 microscope (FEI). For scanning electron microscopy, 10-kV accelerating voltage was used in an S4700 FE-SEM system (Hitachi). The presence of gold on the MSN material was verified by energy-dispersive x-ray analysis.
Plasmids
The PHP43619 plasmid provided a substrate for loxP recombination. The 34-bp loxP sites were integrated in frame at the 5′ ends of the glyphosate acetyltransferase gene gat4621 and the red fluorescent protein DsRed2 gene coding sequences. As a result, an additional 17 amino acids were fused to the gat4621 gene, whose expression was controlled by the maize (Zea mays) ubiquitin promoter. The AmCyan1 gene expression cassette was inserted behind the gat4621 gene but in front of the loxP site-containing DsRed2 coding sequence. The fluorescent protein genes were obtained from Clontech Laboratories (Matz et al., 1999). The vectors were built on the pSB11 plasmid backbone (Komari and Kubo, 1999). Subsequently, electroporation was used to integrate the pSB11-based PHP43619 vector into the superbinary vector pSB1 residing in Agrobacterium tumefaciens strain LBA4404, thus producing the PHP43635 plasmid used for stable transformation of the Hi-II immature maize embryos.
Construct PHP16072 (Supplemental Fig. S3B) was built for Cre recombinase expression in planta. The maize codon-optimized moCre recombinase gene containing the ST-LS1 intron (Vancanneyt et al., 1990) was driven by the maize ubiquitin promoter (Christensen and Quail, 1996), and the potato (Solanum tuberosum) proteinase inhibitor II gene 3′ end sequence was used as a terminator of transcription and a polyadenylation processing signal (An et al., 1989).
Cre Recombinase Protein Purification and in Vitro Activity Measurement
The Cre protein was supplied by Dr. Paul Sadowski (University of Toronto). It was stored at –80°C in 20 mm HEPES, pH 7.5, 0.71 m ammonium sulfate buffer at 1.0 mg mL−1. Its concentration was determined by amino acid analysis.
Cre enzyme activity was measured using the commercially available pLox2+ system from New England Biolabs. Cre recombinase from New England Biolabs was used as a positive control.
Plant Materials
Experiments with onion (Allium cepa) epidermis tissues were described previously (Martin-Ortigosa et al., 2012b). For maize immature embryos, the in vitro culture protocol described by Frame et al. (2000) was used, but with no herbicide selection. The T0 regenerated plants were self-pollinated, and then homozygous Lox-corn was pollinated with nontransgenic Hi-II pollen. The heterozygous immature embryos from this cross were used for stable Cre delivery experiments. For visual selection of recombined events, red fluorescent callus pieces of around 3 to 5 mm were separated using a fluorescence stereoscope.
MSN Protein Loading and DNA Coating
For MSN-Cre protein loading, a fresh 25 µg µL−1 MSN stock was prepared in Tris/NaCl buffer (15 mm Tris-HCl, pH 8, and 250 mm NaCl). Per shot, 50 µg of 6x-MSN was mixed with 1 or 2 µg of Cre (0.809 µg µL−1 stock, final concentration of 40 or 80 µg of Cre per 1 mg of MSN). Tris/NaCl buffer was added until a volume of 100 µL was reached; the mix was slowly shaken at 4°C for 30 min and then centrifuged at 5,000 rpm for 10 s (Spectrafuge 16M from Labnet). For experiments 1 and 2, the supernatant was removed; the pellet was frozen for 5 min in liquid nitrogen and then lyophilized for 1 h (Freezone 2.5 from Labconco). For experiment 2, the lyophilized pellet was resuspended in 5 µL of ice-cold ethanol and immediately poured over a macrocarrier. After the ethanol had evaporated, the macrocarrier was ready to be used for bombardment. For experiment 1, the MSNs were mixed with 0.6 µm of gold for better plant tissue penetration during bombardment. The lyophilized MSN pellet was mixed by pipetting with 2 µL of a 30 µg µL−1 stock of 0.6-µm gold particles (Bio-Rad) in water. After that, 5 µL of ice-cold ethanol was added and the mix was pipetted onto a macrocarrier. For experiments 3 and 4, once the supernatant was removed after centrifugation, no lyophilization was performed. Two microliters of 0.6-µm gold stock (30 µg µL−1) was mixed with the MSN pellet, and 5 µL of ice-cold ethanol was added. The mixture was immediately pipetted onto a macrocarrier. Bombardment was carried out once the ethanol was evaporated.
For transient Cre protein delivery into onion epidermis tissues and maize immature embryos, 6x-MSN loaded with Cre protein was mixed with 0.6 µm of gold and coated with plasmid PHP43619 as described previously (Martin-Ortigosa et al., 2012a). Per shot, 100 or 50 µg (for the onion or maize embryo experiment, respectively) of Cre-6x-MSN was mixed with 2 µL of a 60 µg µL−1 suspension of 0.6-µm gold particles; this mix was coated with 1 µg of plasmid DNA using a calcium chloride/spermidine-based coating protocol.
Bombardment Conditions
Onion epidermis tissues were bombarded as described previously (Martin-Ortigosa et al., 2012a, 2012b). Briefly, rectangular pieces of onion epidermis tissue were placed on solid medium with the peeled surface upward and bombarded at 1,100 p.s.i., 6-cm target distance, and 28 inches of mercury vacuum. For maize immature embryo bombardment, the protocol described by Frame et al. (2000) was followed, but no protective metallic screen was used between the plate and the launch assembly. Embryos were placed with the scutellum upward on osmoticum medium and bombarded at 650 p.s.i., 6-cm target distance, and 28 inches of mercury vacuum.
MSN-Cre Protein Loading and Release Experiments
For protein loading, 2 mg of 6x-MSN or AP-6x-MSN was mixed with 60 µg of purified Cre protein (0.809 µg µL−1 stock), and then Tris/NaCl buffer was added to reach a 100-µL final volume, followed by constant mixing for 1 h at 4°C. To monitor protein loading, supernatant samples were collected after 1, 30, or 60 min by spinning down the mixture at 5,000 rpm for 10 s (Spectrafuge 16M from Labnet). After the last centrifugation, all the supernatant was removed; the nanoparticle pellet was frozen in liquid nitrogen for 1 to 5 min and then lyophilized for 1 h. For the release experiment, 100 µL of Tris/NaCl buffer was added to the dry pellet. Samples of the supernatant were collected after centrifugation (10 s at 5,000 rpm) after 1 min, 1 h, or 24 h of incubation. Western-blot analyses were performed to detect Cre protein loading and release from MSNs. The supernatant samples taken for the time course, and the remaining nanoparticle pellet after the release experiment, were incubated in boiling water for 5 min in Laemmli sample buffer (Bio-Rad). The mouse monoclonal anti-Cre antibody (Sigma-Aldrich) was used at 1:1,000 dilution. As a secondary antibody, a goat anti-mouse IgG-horseradish peroxidase-conjugated antibody (Bio-Rad) was used at 1:3,000 dilution. The tetramethylbenzidine-stabilized substrate was used as a detection system (Promega).
Fluorescence Microscopy
Microscope images were taken using ProgRes Capture Pro 2.6 software and a ProgRes C3 digital camera from Jenoptik attached to an Axiostar plus microscope from Zeiss. Images were taken with the objectives A-Plan 5×/0.12 or 10×/0.25. DsRed filter (excitation, 545 nm; emission, 620 nm; beam splitter, 570 nm) and enhanced cyan fluorescent protein filter (excitation, 436 nm; emission, 480 nm; beam splitter, 455 nm) from Chroma Technology were used for fluorescence imaging. Scope images were taken using Spot 4.6 software and an RT color camera from Diagnostic Instruments attached to an Olympus ZSH10 research stereoscope. A red fluorescent protein filter (excitation, 540–580 nm; emission, 600–660 nm) and a GFP filter (excitation, 460–490 nm; emission, 510–550 nm) were used for fluorescence imaging. Green fluorescence images taken with the GFP filter were converted to blue using Adobe Photoshop software.
Recombination Analysis by PCR and Sequencing
Maize DNA was extracted from leaves or callus pieces using the cetyltrimethylammonium bromide procedure (Murray and Thompson, 1980). For ubi PCR, primers UBINTRF (5′-TTTAGCCCTGCCTTCATAGC-3′) and DSRED2R (5′-CCGTCCTCGAAGTTCATCAC-3′) were used. The bands were also purified and sequenced. For cyan PCR, CYANF primer (5′-CAGAACCGACCTGGACAAG-3′) and DSRED2R were used. Comparison with the original sequences was done using Blast2seq software (Tatusova and Madden, 1999). For Cre plasmid DNA detection experiments, primers CREX2F2 (5′-ACAGGTGCCAGGACATTAGG-3′) and CREX2R (5′-ACAATGTTCACGTTCGTCCA-3′) were used. Primer3 software was used for primer design (Rozen and Skaletsky, 2000). PCR procedures were done as follows: an initial step of 5 min at 95°C; 25 to 35 cycles of 30 s at 95°C denaturation step, 30 s at 60°C annealing step, and 2 min, 1 min, or 45 s at 72°C extension step for ubi, cyan, or Cre PCR, respectively; a final extension step of 10 min at 72°C was performed before cooling down the tubes to 4°C.
Southern Blotting
The recombination pattern of T0 plants was determined by Southern blot. Ten micrograms of genomic DNA was extracted from leaf tissue (Murray and Thompson, 1980) and digested with EcoRV enzyme. The digested DNA was separated on a 0.8% (w/v) agarose gel by electrophoresis, transferred to a membrane (BrightStar Plus; Ambion), cross linked, and prehybridized using Church’s buffer (Church and Gilbert, 1984) for 4 h at 65°C. The probe for the detection of the DsRed2 gene was amplified by PCR from the plasmid PHP43635 (Supplemental Fig. S3A) using primers RED_PROBE_F (5′-GCCTCCTCCGAGAACGTCATCA-3′) and RED_PROBE_R (5′-CGTCCTTCAGCTTCAGGGCCTT-3′). The 505-bp band was purified and labeled with [32P]dCTP using the Prime-it II kit (Agilent Technologies). The probe was purified using Illustra Probequant columns (GE Healthcare) and added for overnight incubation at 65°C. The membrane washing and film development were done according to standard protocols.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. MSN characterization analysis and MSN-Cre protein loading/release experiments.
Supplemental Figure S2. Cre protein and in vitro Cre activity test.
Supplemental Figure S3. Maps of the plasmids used for transformation and transient results of MSN-mediated transformation.
Supplemental Figure S4. Southern blot analysis.
Supplemental Figure S5. Microscope images of Lox-corn plant material after bombardment with MSN.
Supplemental Figure S6. PCR detection of Cre-DNA in callus samples.
Supplemental Table S1. Results of inductively coupled plasma-optical emission spectroscopy and nitrogen sorption analyses of a series of gold-plated MSN.
Acknowledgments
We thank Angela Nguyen, Qing Ji, Brian Lenderts, Lijuan Wang, and Christopher Scelonge for technical support, Bronwyn Frame and Katey Warnberg for providing maize materials and guidance in maize culture, and James Register, S. Carl Falco, and Todd J. Jones for scientific discussion and input.
Glossary
- MSN
mesoporous silica nanoparticle
References
- Albert H, Dale EC, Lee E, Ow DW. (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7: 649–659 [DOI] [PubMed] [Google Scholar]
- An G, Mitra A, Choi HK, Costa MA, An K, Thornburg RW, Ryan CA. (1989) Functional analysis of the 3′ control region of the potato wound-inducible proteinase inhibitor II gene. Plant Cell 1: 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CL, Green CE, Phillips RL (1991) Development and availability of germplasm with high type II culture formation response. Maize Genet Coop Newsl 65: 92–93 [Google Scholar]
- Armstrong CL, Phillips RL. (1988) Genetic and cytogenetic variation in plants regenerated from organogenic and friable, embryogenic tissue cultures of maize. Crop Sci 28: 363–369 [Google Scholar]
- Baubonis W, Sauer B. (1993) Genomic targeting with purified Cre recombinase. Nucleic Acids Res 21: 2025–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao MX, Huang JQ, Yao QH, Liu SJ, Wang CL, Wei ZM. (2006) Site-specific DNA excision in transgenic rice with a cell-permeable cre recombinase. Mol Biotechnol 32: 55–63 [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH. (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5: 213–218 [DOI] [PubMed] [Google Scholar]
- Chugh A, Eudes F, Shim YS. (2010) Cell-penetrating peptides: nanocarrier for macromolecule delivery in living cells. IUBMB Life 62: 183–193 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. (2011) Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol 29: 64–67 [DOI] [PubMed] [Google Scholar]
- de Wit T, Drabek D, Grosveld F. (1998) Microinjection of cre recombinase RNA induces site-specific recombination of a transgene in mouse oocytes. Nucleic Acids Res 26: 676–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djukanovic V, Lenderts B, Bidney D, Lyznik LA. (2008) A Cre:FLP fusion protein recombines FRT or loxP sites in transgenic maize plants. Plant Biotechnol J 6: 770–781 [DOI] [PubMed] [Google Scholar]
- Du J, Jin J, Yan M, Lu Y. (2012) Synthetic nanocarriers for intracellular protein delivery. Curr Drug Metab 13: 82–92 [DOI] [PubMed] [Google Scholar]
- Ennifar E, Meyer JEW, Buchholz F, Stewart AF, Suck D. (2003) Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation. Nucleic Acids Res 31: 5449–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame BR, Zhang HY, Cocciolone SM, Sidorenko LV, Dietrich CR, Pegg SE, Zhen SF, Schnable PS, Wang K. (2000) Production of transgenic maize from bombarded type II callus: effect of gold particle size and callus morphology on transformation efficiency. In Vitro Cell Dev Biol Plant 36: 21–29 [Google Scholar]
- Ghosh K, Van Duyne GD. (2002) Cre-loxP biochemistry. Methods 28: 374–383 [DOI] [PubMed] [Google Scholar]
- Hare PD, Chua NH. (2002) Excision of selectable marker genes from transgenic plants. Nat Biotechnol 20: 575–580 [DOI] [PubMed] [Google Scholar]
- Hartono SB, Qiao SZ, Liu J, Jack K, Ladewig BP, Hao Z, Lu GQM. (2010) Functionalized mesoporous silica with very large pores for cellulase immobilization. J Phys Chem C 114: 8353–8362 [Google Scholar]
- Jo D, Nashabi A, Doxsee C, Lin Q, Unutmaz D, Chen J, Ruley HE. (2001) Epigenetic regulation of gene structure and function with a cell-permeable Cre recombinase. Nat Biotechnol 19: 929–933 [DOI] [PubMed] [Google Scholar]
- Kolb AF, Siddell SG. (1996) Genomic targeting with an MBP-Cre fusion protein. Gene 183: 53–60 [DOI] [PubMed] [Google Scholar]
- Komari T, Kubo T (1999) Methods of genetic transformation: Agrobacterium tumefaciens. In I Vasil, ed, Molecular Improvement of Cereal Crops, Vol 5. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 43–82 [Google Scholar]
- Kopertekh L, Schiemann J (2012) Elimination of transgenic sequences in plants by Cre gene expression. In Y Özden Çiftçi, ed, Transgenic Plants: Advances and Limitations. InTech, Rijeka, Croatia, pp 449–468
- Koshy AA, Fouts AE, Lodoen MB, Alkan O, Blau HM, Boothroyd JC. (2010) Toxoplasma secreting Cre recombinase for analysis of host-parasite interactions. Nat Methods 7: 307–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Xing A, Moon BP, McCardell RP, Mills K, Falco SC. (2009) Site-specific integration of transgenes in soybean via recombinase-mediated DNA cassette exchange. Plant Physiol 151: 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Jo D, Gebre-Amlak KD, Ruley HE. (2004) Enhanced cell-permeant Cre protein for site-specific recombination in cultured cells. BMC Biotechnol 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckow B, Hänggli A, Maier H, Chilla S, Loewe RP, Dehmel S, Schlöndorff D, Loetscher P, Zerwes HG, Müller M. (2009) Microinjection of Cre recombinase protein into zygotes enables specific deletion of two eukaryotic selection cassettes and enhances the expression of a DsRed2 reporter gene in Ccr2/Ccr5 double-deficient mice. Genesis 47: 545–558 [DOI] [PubMed] [Google Scholar]
- Martin-Ortigosa S, Valenstein JS, Lin VSY, Trewyn BG, Wang K. (2012a) Gold functionalized mesoporous silica nanoparticle mediated protein and DNA codelivery to plant cells via the biolistic method. Adv Funct Mater 22: 3576–3582 [Google Scholar]
- Martin-Ortigosa S, Valenstein JS, Sun W, Moeller L, Fang N, Trewyn BG, Lin VSY, Wang K. (2012b) Parameters affecting the efficient delivery of mesoporous silica nanoparticle materials and gold nanorods into plant tissues by the biolistic method. Small 8: 413–422 [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17: 969–973 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nolden L, Edenhofer F, Haupt S, Koch P, Wunderlich FT, Siemen H, Brüstle O. (2006) Site-specific recombination in human embryonic stem cells induced by cell-permeant Cre recombinase. Nat Methods 3: 461–467 [DOI] [PubMed] [Google Scholar]
- Ponsaerts P, Brown JP, Van den Plas D, Van den Eeden L, Van Bockstaele DR, Jorens PG, Van Tendeloo VFI, Merregaert J, Singh PB, Berneman ZN. (2004) Messenger RNA electroporation is highly efficient in mouse embryonic stem cells: successful FLPe- and Cre-mediated recombination. Gene Ther 11: 1606–1610 [DOI] [PubMed] [Google Scholar]
- Popat A, Hartono SB, Stahr F, Liu J, Qiao SZ, Lu GQM. (2011) Mesoporous silica nanoparticles for bioadsorption, enzyme immobilisation, and delivery carriers. Nanoscale 3: 2801–2818 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In S Krawetz, S Misener, eds, Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. (2011) Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol 29: 697–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slowing II, Trewyn BG, Lin VSY. (2007) Mesoporous silica nanoparticles for intracellular delivery of membrane-impermeable proteins. J Am Chem Soc 129: 8845–8849 [DOI] [PubMed] [Google Scholar]
- Slowing II, Vivero-Escoto JL, Trewyn BG, Lin VSY. (2010) Mesoporous silica nanoparticles: structural design and applications. J Mater Chem 20: 7924–7937 [Google Scholar]
- Sorrell DA, Kolb AF. (2005) Targeted modification of mammalian genomes. Biotechnol Adv 23: 431–469 [DOI] [PubMed] [Google Scholar]
- Srivastava V, Ariza-Nieto M, Wilson AJ. (2004) Cre-mediated site-specific gene integration for consistent transgene expression in rice. Plant Biotechnol J 2: 169–179 [DOI] [PubMed] [Google Scholar]
- Srivastava V, Ow DW. (2002) Biolistic mediated site-specific integration in rice. Mol Breed 8: 345–350 [Google Scholar]
- Tatusova TA, Madden TL. (1999) BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174: 247–250 [DOI] [PubMed] [Google Scholar]
- Torney F, Trewyn BG, Lin VSY, Wang K. (2007) Mesoporous silica nanoparticles deliver DNA and chemicals into plants. Nat Nanotechnol 2: 295–300 [DOI] [PubMed] [Google Scholar]
- Trewyn BG, Slowing II, Giri S, Chen HT, Lin VSY. (2007) Synthesis and functionalization of a mesoporous silica nanoparticle based on the sol-gel process and applications in controlled release. Acc Chem Res 40: 846–853 [DOI] [PubMed] [Google Scholar]
- Van den Plas D, Ponsaerts P, Van Tendeloo V, Van Bockstaele DR, Berneman ZN, Merregaert J. (2003) Efficient removal of LoxP-flanked genes by electroporation of Cre-recombinase mRNA. Biochem Biophys Res Commun 305: 10–15 [DOI] [PubMed] [Google Scholar]
- Vancanneyt G, Schmidt R, O’Connor-Sanchez A, Willmitzer L, Rocha-Sosa M. (1990) Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Gen Genet 220: 245–250 [DOI] [PubMed] [Google Scholar]
- Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CMT, Regensburg-Tuïnk TJ, Hooykaas PJJ. (2000) VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290: 979–982 [DOI] [PubMed] [Google Scholar]
- Wang Y, Yau YY, Perkins-Balding D, Thomson JG. (2011) Recombinase technology: applications and possibilities. Plant Cell Rep 30: 267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Ochiai H, Sakuma T, Horch HW, Hamaguchi N, Nakamura T, Bando T, Ohuchi H, Yamamoto T, Noji S, et al. (2012) Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat Commun 3: 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will E, Klump H, Heffner N, Schwieger M, Schiedlmeier B, Ostertag W, Baum C, Stocking C. (2002) Unmodified Cre recombinase crosses the membrane. Nucleic Acids Res 30: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Du H, Liao X, Zhao Y, Li L, Yang L. (2011) Tn5 transposase-assisted transformation of indica rice. Plant J 68: 186–200 [DOI] [PubMed] [Google Scholar]
- Wymer CL, Fernández-Abalos JM, Doonan JH. (2001) Microinjection reveals cell-to-cell movement of green fluorescent protein in cells of maize coleoptiles. Planta 212: 692–695 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Møller SG, Chua NH. (2001) Chemical-regulated, site-specific DNA excision in transgenic plants. Nat Biotechnol 19: 157–161 [DOI] [PubMed] [Google Scholar]