A “functional hypothesis” model is presented for the extensive functional pleiotropy of a poplar class III homeodomain-leucine zipper transcription factor in modulating extensive phenotypic variability.
Abstract
In plants, genes may sustain extensive pleiotropic functional properties by individually affecting multiple, distinct traits. We discuss results from three genome-wide association studies of approximately 400 natural poplar (Populus trichocarpa) accessions phenotyped for 60 ecological/biomass, wood quality, and rust fungus resistance traits. Single-nucleotide polymorphisms (SNPs) in the poplar ortholog of the class III homeodomain-leucine zipper transcription factor gene REVOLUTA (PtREV) were significantly associated with three specific traits. Based on SNP associations with fungal resistance, leaf drop, and cellulose content, the PtREV gene contains three potential regulatory sites within noncoding regions at the gene’s 3′ end, where alternative splicing and messenger RNA processing actively occur. The polymorphisms in this region associated with leaf abscission and cellulose content are suggested to represent more recent variants, whereas the SNP associated with leaf rust resistance may be more ancient, consistent with REV’s primary role in auxin signaling and its functional evolution in supporting fundamental processes of vascular plant development.
The spectrum of genetic control underlying the expression of phenotypic characteristics ranges from epistasis (governed by interactions of multiple genes) to pleiotropy (i.e. single genes or mutations affecting multiple unrelated traits). Phenotypic traits, albeit subjectively defined, typically outnumber genes by orders of magnitude in complex organisms, supporting the presence of pleiotropy (Wagner and Zhang, 2011). Knowledge about the molecular basis of pleiotropy is important for our understanding of evolvability (i.e. the capacity of an organism for adaptive evolution; Wang et al., 2010; Wagner and Zhang, 2011; Hill and Zhang, 2012a). Hodgkin (1998) defined seven types of pleiotropy depending on different underlying molecular mechanisms, including alternative splicing as a source of functional pleiotropy at a gene locus. Ongoing debate centers on whether pleiotropy is limited and modular (Wagner and Zhang, 2011) or is widespread, which would support the hypothesis of universal pleiotropy (Hill and Zhang, 2012a), and whether higher pleiotropy constrains or facilitates the evolution of organismal complexity (Wagner and Zhang, 2011; Hill and Zhang, 2012b). However, current measurements may not satisfactorily disclose the pleiotropic level for a given polymorphism (Wagner and Zhang, 2012). Studies of mutant phenotypes often overestimate gene pleiotropy (Wagner and Zhang, 2011) and thus do not reflect the effects of naturally occurring mutations, which are of particular interest to our understanding of normal gene function (Hodgkin, 1998). Quantitative trait locus data may also overestimate pleiotropy due to the large linkage intervals in pedigree studies of quantitative trait loci (Gardner and Latta, 2007). Quantitative trait nucleotide (QTN) functional variants can provide important insights into the genetics of evolution (Streisfeld and Rausher, 2011; Rockman, 2012), but only large-effect QTNs are typically accessible (Rockman, 2012). Thus, causal variants may not be the commonly detected QTNs; instead, such variants may be present in multiple, largely small-effect genes that, therefore, likely remain undetected (Rockman, 2012). Although generally less studied than exonic variants, mutational events within introns, untranslated regions (UTRs), and/or untranscribed gene regulatory regions could be key drivers of evolution (Lynch and Katju, 2004). This noncoding DNA is enriched with functional sequences that can arise spontaneously (Rockman, 2012).
The class III homeodomain leucine zipper (HD ZIP) gene REVOLUTA (REV) is involved in auxin-mediated adaxial-abaxial patterning of plant leaves in general and patterning of secondary vascular tissues in woody species in particular (Emery et al., 2003; Robischon et al., 2011) and represents an interesting example of how phenotypic variation can be mediated through interference with microRNA (miRNA) binding rather than nonsynonymous mutations in the coding sequence (Emery et al., 2003; Ong and Wickneswari, 2012). Here, we discuss specific results obtained from genotype-phenotype association studies in poplar (Populus trichocarpa; La Mantia et al., 2013; Porth et al., 2013a; A.D. McKown, unpublished data), a tree species of significant ecological, economic, and scientific importance (Tuskan et al., 2006; Geraldes et al., 2013), that highlight putative pleiotropic properties of REV.
RESULTS AND DISCUSSION
We conducted genome-wide association studies (GWAS) that employed approximately 400 phenotyped and genotyped natural poplar accessions grown in common gardens to elucidate the genetic architecture of wood quality as well as ecophysiological, phenological, biomass, and rust fungus resistance trait variation in three separate studies (La Mantia et al., 2013; Porth et al., 2013a; A.D. McKown, unpublished data). In total, we examined single-nucleotide polymorphism (SNP) polymorphisms in over 3,500 broad-based candidate genes using an Illumina 34K SNP genotyping array (Geraldes et al., 2013). The SNP discovery panel for the array (Geraldes et al., 2013) involved 20 accessions from the Pacific Northwest for mRNA sequencing (Geraldes et al., 2011) and 16 accessions taken from the southern distribution range of poplar for whole-genome sequencing (Slavov et al., 2012). Phenotypic trait variation was investigated for approximately 60 quantitative characteristics (La Mantia et al., 2013; McKown et al., 2013; Porth et al., 2013b). All three studies employed the generalized linear mixed-model approach to correct for any genetic structure present in the population. The optimal model was selected on a trait-by-trait basis using the Bayesian information criterion (Yu et al., 2006).
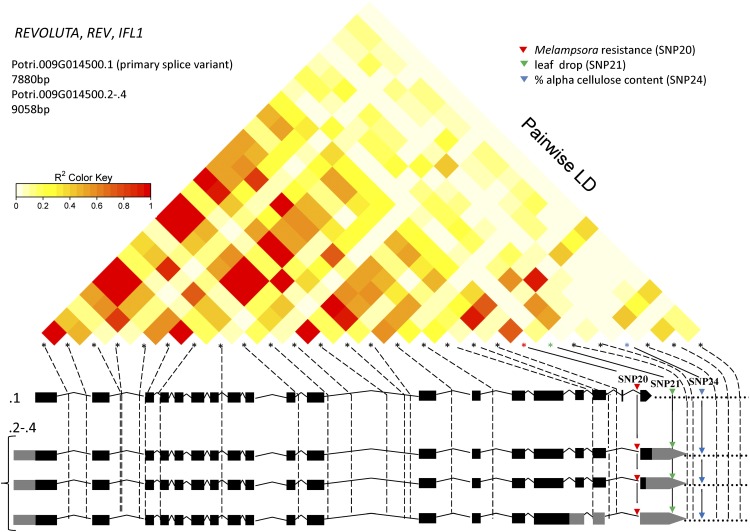
Among many SNP-trait associations, these studies revealed genetic associations of wood cellulose content and leaf phenology traits with SNPs in noncoding regions of distinct splice variants of the poplar REV gene Potri.009G014500 (PtREV; Fig. 1; Table I), consistent with fundamental functions for REV in regulating the architecture of leaf and vascular systems (Zhong et al., 1997; Zhong and Ye, 1999; Otsuga et al., 2001; Emery et al., 2003; Robischon et al., 2011; Stirnberg et al., 2012). An additional putatively functional polymorphism common to all splice variants of REV is associated with Melampsora spp. rust fungus resistance (Fig. 1; Table I), suggesting the involvement of REV with this trait also and providing a striking example of the extensive pleiotropic properties that plant genes can sustain.
Figure 1.
PtREV gene structure, LD plot, and SNP locations. The gene structures for splice variants/transcriptional variants of REV (Phytozome version 3) are presented schematically as exon (black boxes), intron (lines), UTR (gray boxes), and noncoding (dots) at the bottom. Locations of 27 genotyped SNPs are shown as asterisks above the gene models; the solid lines connect to the genetic associations of tag SNPs within the specific splice variants with Melampsora spp. rust fungus resistance (SNP20), leaf drop (SNP21), and percentage of α-cellulose content (SNP24); dashed lines indicate SNPs with no significant association with phenotype. The LD relationships of the SNPs are shown at the top and are color coded to show the extent of LD between genotyped SNPs. r2, Squared correlation coefficient. Principal component analysis was used to adjust for population structure in the analyses of wood traits and leaf rust resistance (La Mantia et al., 2013; Porth et al., 2013a); the “area under the disease curve” resistance measure was adjusted for date of bud set prior to association analysis (La Mantia et al., 2013); Q matrix population structure correction was applied for phenology traits that covary with latitudinal population structure (A.D. McKown, unpublished data).
Table I. Features of 27 SNPs in PtREV (Potri.009G014500) genotyped in an association population.
| Tag SNP (Geraldes et al., 2013)a | Location | Location Feature | HEb | FSTb | MAF | Detected Associations |
|---|---|---|---|---|---|---|
| scaffold_9_2556002 | Splice variants 1 to 4 | Intron | 0.504193 | 0.036061 | 0.47691 | |
| scaffold_9_2556257 | Splice variants 1 to 4 | Intron | 0.504857 | 0.033881 | 0.49884 | |
| scaffold_9_2556661 | Splice variants 1 to 4 | Intron | 0.428818 | 0.028909 | 0.31235 | |
| scaffold_9_2556664 | Splice variants 1 to 4 | Intron | 0.226606 | 0.008931 | 0.12587 | |
| scaffold_9_2556886 | Splice variants 1 to 4 | Intron | 0.453198 | 0.036915 | 0.34642 | |
| scaffold_9_2556960 | Splice variants 1 to 4 | Intron | 0.504843 | 0.033775 | 0.49769 | |
| scaffold_9_2557200 | Splice variants 1 to 4 | Intron | 0.504844 | 0.033773 | 0.49769 | |
| scaffold_9_2557957 | Splice variants 1 to 4 | Intron | 0.262775 | 0.010365 | 0.16051 | |
| scaffold_9_2558943 | Splice variants 1 to 4 | Intron | 0.395116 | 0.027997 | 0.27252 | |
| scaffold_9_2559321 | Splice variants 1 to 4 | Intron | 0.11118 | 0.008393 | 0.06134 | |
| scaffold_9_2559505 | Splice variants 1 to 4 | Intron | 0.504236 | 0.037609 | 0.47564 | |
| scaffold_9_2560010 | Splice variants 1 to 4 | Intron | 0.504859 | 0.033876 | 0.49883 | |
| scaffold_9_2560210 | Splice variants 1 to 4 | Intron | 0.263309 | 0.010729 | 0.16088 | |
| scaffold_9_2560374 | Splice variants 1 to 4 | Intron | 0.504283 | 0.036495 | 0.47801 | |
| scaffold_9_2560840 | Splice variants 1 to 4 | Intron | 0.44196 | 0.037899 | 0.31597 | |
| scaffold_9_2561340 | Splice variants 1 to 4 | Intron | 0.45543 | 0.030888 | 0.35082 | |
| scaffold_9_2562685 | Splice variants 1 to 4 | Intron | 0.458848 | 0.037309 | 0.35267 | |
| scaffold_9_2562888 | Splice variants 1 to 4 | Intron | 0.183453 | −0.00455 | 0.10233 | |
| scaffold_9_2562964 | Splice variants 1 to 4 | Intron | 0.470355 | 0.040831 | 0.37153 | |
| scaffold_9_2563210 | Splice variants 1 to 4 | Intron | 0.511584 | 0.08915 | 0.48956 | Melampsora × columbiana resistancec |
| scaffold_9_2563600 | Splice variants 2 to 4 | Exon, 3′ UTR | 0.24597 | 0.116677 | 0.13741 | Leaf dropd |
| scaffold_9_2563682 | 3′ flanking | Intergenic | 0.502272 | 0.040688 | 0.46846 | |
| scaffold_9_2563761 | 3′ flanking | Intergenic | 0.193176 | −0.00458 | 0.10855 | |
| scaffold_9_2563977 | 3′ flanking | Intergenic | 0.272897 | 0.020284 | 0.16051 | Percentage α-cellulosee |
| scaffold_9_2564040 | 3′ flanking | Intergenic | 0.352358 | 0.006295 | 0.23248 | |
| scaffold_9_2564643 | 3′ flanking | Intergenic | 0.4637 | 0.010006 | 0.36706 | |
| scaffold_9_2565072 | 3′ flanking | Intergenic | 0.331155 | 0.001465 | 0.20882 |
SNPs in boldface did not conform to Hardy-Weinberg equilibrium. bHE (expected heterozygosity) and FST (fixation index) were calculated using Fdist2 implemented in the LOSITAN software package (Antao et al., 2008) on the basis of 433 poplar individuals and subgrouping according to climatic variables (I. Porth, unpublished data). cArea under the disease curve resistance measure adjusted for date of bud set (La Mantia et al., 2013). dAn adaptive phenology trait (McKown et al., 2013; A.D. McKown, unpublished data). ePercentage of relative α-cellulose content in dry wood (Porth et al., 2013a, 2013b).
The wood association study also identified, among others, SNP associations within various auxin-related genes related to variation in fiber properties, including fiber length. Fiber development is tightly linked to the action of the phytohormone auxin (indole acetic acid) in vascular tissue formation (Schuetz et al., 2013; Ursache et al., 2013), as auxin signaling regulates molecular master switches during the initiation of fiber development (Gorshkova et al., 2012). Since indole acetic acid plays a role in regulating cambium, xylem, and fiber development as well as fiber secondary cell wall thickening and lignification (Gorshkova et al., 2012; Schuetz et al., 2013), the candidate SNPs within auxin-related genes identified in our study are potentially important in the upstream regulation of plant xylem and fiber formation. In particular, REV (synonymous with INTERFASCICULAR FBERLESS1 and AMPHIVASAL VASCULAR BUNDLES1 [AVB1]), encoding one of five Arabidopsis (Arabidopsis thaliana) class III HD ZIP transcription factors, regulates meristem function, polarity of lateral organs, vascular architecture and development, and interfascicular fiber differentiation in Arabidopsis and other angiosperms (Zhong et al., 1999; Zhong and Ye, 1999; Ratcliffe et al., 2000, Emery et al., 2003; Zhong and Ye, 2004). In poplar, the REV ortholog PtREV influences cambium initiation and patterning of woody stems (Robischon et al., 2011; Hu et al., 2012). REV is part of a complex regulatory system involving auxin, KANADI (KAN1) transcription factors, and miRNA (Emery et al., 2003; Ilegems et al., 2010; Brandt et al., 2012; Schuetz et al., 2013). The direct targets of regulation involve several transcription factors, some of which are regulated antagonistically to support the antagonistic influence by REV and KAN1 on a suite of gene promoters (Reinhart et al., 2013). REV mutants are highly pleiotropic across different species. In Arabidopsis, REV mutants exhibit differences compared with the wild type in thicker inflorescence stems with less lignified interfascicular cells, reduced stem strength, reduced number of branches (defect in paraclade growth) and fewer stem leaves, enlarged and downwardly curled (i.e. revolute) cauline leaves, a change in leaf color to dark green, and delayed leaf senescence (Talbert et al., 1995; Zhong et al., 1997), while the semidominant REV avb1 mutant exhibits aberrant cauline branches and leaves, abnormal floral tissues, and amphivasal instead of collateral vascular bundles (Emery et al., 2003; Zhong and Ye, 2004). In poplar, gain-of-function REV mutants show severe anatomical phenotypes related to leaf architecture, reduced internode length, callus formation on the stem surface, abnormal cambial growth, and cambial polarity defects correlated with the positions of leaves and axillary buds (Robischon et al., 2011). These consistent observations of phenotypic aberrations among REV gain-of-function mutants across different plant species (Talbert et al., 1995; Emery et al., 2003; Magnani and Barton, 2011; Robischon et al., 2011) suggest that the functionality of wild-type REV is indeed (1) pleiotropic and (2) a major hub gene (i.e. a key regulatory gene) regulating plant morphogenesis. Thus, it is anticipated that wild-type REV controls both stem and leaf growth (Talbert et al., 1995), and recently, it was also suggested that REV controls light-mediated elongation growth (Brandt et al., 2012).
In contrast to mutant phenotypes generated by loss of function or overexpression, forward genetics studies such as GWAS have the potential to elucidate normal gene functions by correlating differences in the observed natural phenotypic variation with the natural allelic variations. Our data from wild accessions of poplar support REV function in secondary growth and xylem development in poplar based on the genetic association between SNP24 in PtREV and wood α-cellulose content. This indicates a possible regulatory function for REV in coordinating cellulose biosynthesis during the differentiation of cells with secondary cell wall such as vessels and fibers, a process about which little is known (Ambavaram et al., 2011). Our poplar GWAS also revealed associations between an SNP in REV and natural variation in date of leaf abscission (SNP21; A.D. McKown, unpublished data). Together, these results suggest a putative auxin-meditated mechanism. In addition to vascular differentiation, auxin regulates leaf senescence and abscission (Ellis et al., 2005). Furthermore, an additional function for REV in stress and pathogen resistance was revealed in our studies by the association of SNP20 in REV with resistance to the poplar rust fungus Melampsora × columbiana (La Mantia et al., 2013), which causes yellow leaf rust and represents a serious pest to commercial poplar plantations. We reliably inferred haplotypes for the second alternative splice variant in 435 unrelated individuals with complete genotype data. We identified one “recessive” haplotype, AGACGTTGTAAAACGCCTAAA (based on the 21 genotyped SNPs for variant 2; Fig. 1), related to leaf rust susceptibility. This haplotype is prevalent in poplar accessions from the northern extent of the species’ range that set bud early compared with southern populations. Weaker selection pressure from the pathogen in more northerly located populations than southerly populations (Chandrashekar and Heather, 1980) is likely responsible for the maintenance of these sensitive alleles within the population. A proposed mechanism of resistance could be mediated through the suppression of axillary bud outgrowth by auxin and repressed leaf growth (Stirnberg et al., 2012), as REV function is implicated in polar auxin transport (Zhong and Ye, 1999). Auxin may also regulate defense signaling via cross talk with the GA3 negative regulators of the DELLA protein family. Proper polar auxin transport is necessary for degradation of DELLA and derepression of growth (Fu and Harberd, 2003). DELLA loss-of-function Arabidopsis mutants have primed salicylic acid with earlier, more robust expression of pathogenesis-related PR1 and PR2 genes (Navarro et al., 2008), yet for poplar (and Salicaceae spp. in general), a different mechanism may apply, as these plant systems can sustain constitutively high amounts of salicylic acid (Xue et al., 2013). Furthermore, AUXIN F-BOX genes positively regulate auxin signaling and are up-regulated in incompatible (resistant) poplar-leaf rust interactions (J. La Mantia, unpublished data). The observation that REV regulates genes encoding auxin biosynthetic genes (Brandt et al., 2012) helps to explain the nature of its pleiotropy, as this gene seems to regulate multiple fundamental plant developmental processes, such as basic plant organ patterning, all of which are tightly linked to auxin signaling (Otsuga et al., 2001; Emery et al., 2003; Ellis et al., 2005; Ilegems et al., 2010; Robischon et al., 2011; Brandt et al., 2012; Schuetz et al., 2013; Ursache et al., 2013).
How did REV obtain its pleiotropic functionality as evidenced by the association of REV variants with variation in multiple diverse traits? Class III HD ZIP genes, such as REV, that contain a steroidogenic acute regulatory protein-related lipid transfer (START) domain are plant specific and conserved across plants (Schrick et al., 2004). In animal systems, proteins containing the START domain, a protein module of about 210 amino acid residues that binds lipids such as sterols, show an overlap between developmental and disease-related gene functions. In mammals, besides being important for lipid metabolism and fertility, START domain proteins are also involved in atherosclerosis, autoimmune disease, and cancer and, therefore, are suggested targets for drug development against such diseases (Soccio and Breslow, 2003; Clark, 2012).
Class III HD ZIP genes predate the evolution of vascular plants (Prigge et al., 2005). REV represents the most recent class III HD ZIP in land plants (Magnani and Barton, 2011). The initial ancestral role for class III HD ZIP genes may have been auxin signaling (Prigge et al., 2005). New functions appear to have been acquired in parallel with the development of major body-plan innovations (Floyd et al., 2006). As suggested elsewhere (Hu et al., 2012), these functions were retained with relatively low divergence under purifying selection. PtREV exhibits a distinctive gene structure with four alternative transcriptional variants of its gene sequence, with the shortest transcriptional variant (2,568 bp) devoid of the 3′ UTR sequence (Fig. 1; Supplemental Table S1). This transcriptional variant is prevalent in xylem (based on RNA-Seq expression profiling; Geraldes et al., 2011; Bao et al., 2013). It is noteworthy that all detected phenotype-genotype associations are represented by distinct SNPs (Geraldes et al., 2013) that are all localized toward the 3′ end of the gene, where alternative splicing and alternative transcript processing occur (Fig. 1; Table I; Supplemental Table S1).
Interestingly, SNP21 (leaf drop association) is localized in the 3′ UTR present only in splice variants 2 to 4, suggesting that this polymorphism might affect mRNA stability. SNP24 (associated with cellulose content) is located in the 3′ nontranscribed flanking regions of all gene variants (and SNP21 is downstream of the 3′ UTR of variant 1) and, therefore, could be in a regulatory region affecting transcription of the gene. Thus, both SNPs may affect PtREV gene expression, with important potential consequences on REV function and the traits it affects (Kuersten and Goodwin, 2003). Among 27 genotyped SNPs in REV, 41% did not conform to Hardy-Weinberg equilibrium, including all SNPs genetically associated with the studied phenotypes (Table I). However, none of these polymorphisms were identified within the miRNA target region, which represents a protein-coding sequence important for maintaining normal REV gene function (Emery et al., 2003; Zhong and Ye, 2004; Floyd et al., 2006; Robischon et al., 2011).
Overall, the investigated SNPs within REV exhibited little to moderate genetic differentiation in the populations studied, when climate-related population grouping was employed, with the exception of the SNP associated with variation in the timing of leaf abscission; this SNP (SNP21) is thus a candidate for an adaptive QTN to different local climate regimes (see fixation index values in Table I). In brief, we used distinct climate partitions to identify adaptive traits due to population differentiation of quantitative traits based on local climate of origin. The estimated population differentiation values for leaf drop and rust resistance traits (both traits were also validated over time; La Mantia et al., 2013; A.D. McKown, unpublished data) deviated from neutral expectations (I. Porth, unpublished data). Then, we used the same subgroup partitioning to determine such QTNs associated with adaptive traits that also identified as fixation index outliers. The SNP within REV associated with leaf drop is suggested to be adaptive to different temperature regimes. However, we note here that the selection pressure driven by climate represents only one aspect of adaptive evolution in poplar; phenology traits, in particular, are strongly adapted to latitudinal origins, while soil composition (aridity and moisture) also contribute to plant adaptation.
Comparing the three associated SNPs, the polymorphism associated with Melampsora spp. resistance is found in relatively high linkage disequilibrium (LD) with several SNPs throughout the REV gene region. This increased LD might be due to diversifying selection evident for this SNP along the north-south geographic cline of the poplar distribution (A. Geraldes, unpublished data). In contrast, the leaf abscission- and cellulose-associated SNPs are not in LD with any other SNP within the REV gene region and may have evolved at different rates based on the observed recombination patterns (Fig. 1; Table I). The LD pattern across the REV gene is likely incomplete due to a possible ascertainment bias (rare SNPs in the population and/or nongenotyped SNPs), which is related to the employed SNP discovery panel for the genotyping platform. Unfortunately, we cannot confidently use this LD information to make inferences about the age of the mutations, although minor allele frequencies (MAFs) hint at the relative ages of mutations. In this context, the MAF for variants associated with leaf abscission and cellulose were much lower than the MAF for the rust resistance SNP (Table I). This is consistent with an initial role for REV in auxin signaling and subsequent functional evolution in supporting fundamental processes of vascular plant development (Prigge et al., 2005), and it is particularly consistent with auxin as a regulator in plant defense with effects on plant growth and development (Kazan and Manners, 2009). The complex regulatory network governing REV expression and the potentially profound effects of its misexpression in poplar (Robischon et al., 2011; Schuetz et al., 2013) suggest that pleiotropic phenotypic variability associated with SNPs in REV 3′ noncoding regions may be related to variation in the splice variants through four transcript levels that are conditioned by regulatory SNPs.
It is interesting that no SNPs were detected in exons (Table I). This could indicate low allelic frequency of exonic SNPs in the population (with allele frequencies lower than 5%), probably due to purifying selection. This is specifically suggested for the miRNA-targeted protein-coding sequence that is important for maintaining normal REV gene function. There is evidence from REV gain-of-function experiments by Magnani and Barton (2011) that the C terminus of REV (specifically, the Per-ARNT-Sim-like MEKHLA domain at the C terminus) is particularly important in regulating REV activity, in a mechanism that is sequence independent but involves steric masking of the Leu zipper domain such that homodimerization is prevented and REV cannot function as a transcription activator. MEKHLA, therefore, is considered a negative regulator in the normal function of REV (Magnani and Barton, 2011).
We detected several other examples of pleiotropy in poplar (A.D. McKown, unpublished data; I. Porth, unpublished data). This involves pleiotropy within trait categories (phenology, ecophysiology, or growth/biomass) and, importantly, also across these field trait categories (A.D. McKown, unpublished data). In addition, three association genetics studies (Porth et al., 2013a; La Mantia et al., 2013; A.D. McKown, unpublished data) revealed pleiotropy across completely different phenotypic traits (wood properties, phenology, and leaf rust resistance), of which the apparent pleiotropic functions of REV were the most striking example. REV has been intensively studied in Arabidopsis using reverse genetics functional approaches, in which this transcription factor was identified as an important regulator of multiple aspects of plant development. These data support our findings from GWAS in poplar that underscore the functional integration of plant developmental traits related to the molecular mechanisms governing cellulose fiber production (secondary growth) and leaf life span. Another example of considerable pleiotropy in forest trees involved the recent discovery of several genomic regions in spruce (Picea spp.) associated with growth and herbivory resistance traits (Porth et al., 2012).
The association of distinct SNP variants in the PtREV gene with phenotypic variation in several diverse traits underscores the importance of the REV transcription factor in regulating the expression of different developmental processes and provides a striking example of a pleiotropic gene with variants that impact these traits. Alternative splicing that generates different protein isoforms and gene expression-level variation may both facilitate pleiotropic gene functions. PtREV variants associated with variation in distinct traits are located in the last intron at the 3′ end, the 3′ UTR, and the untranscribed downstream region. Thus, functional pleiotropy of the PtREV gene in modulating extensive phenotypic variability may be best explained by gene expression and/or mRNA processing.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Sequences of PtREV alternative splice variants (Phytozome version 3).
Glossary
- QTN
quantitative trait nucleotide
- UTR
untranslated region
- HD ZIP
homeodomain leucine zipper
- miRNA
microRNA
- GWAS
genome-wide association studies
- SNP
single-nucleotide polymorphism
- START
steroidogenic acute regulatory protein-related lipid transfer
- LD
linkage disequilibrium
- MAF
minor allele frequency
References
- Ambavaram MM, Krishnan A, Trijatmiko KR, Pereira A. (2011) Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol 155: 916–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. (2008) LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics 9: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao H, Li E, Mansfield SD, Cronk QC, El-Kassaby YA, Douglas CJ. (2013) The developing xylem transcriptome and genome-wide analysis of alternative splicing in Populus trichocarpa (black cottonwood) populations. BMC Genomics 14: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R, Salla-Martret M, Bou-Torrent J, Musielak T, Stahl M, Lanz C, Ott F, Schmid M, Greb T, Schwarz M, et al. (2012) Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J 72: 31–42 [DOI] [PubMed] [Google Scholar]
- Chandrashekar M, Heather WA. (1980) Temperature sensitivity of reactions of Populus spp. to races of Melampsora larici-populina. Phytopathology 71: 421–424 [Google Scholar]
- Clark BJ. (2012) The mammalian START domain protein family in lipid transport in health and disease. J Endocrinol 212: 257–275 [DOI] [PubMed] [Google Scholar]
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW. (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132: 4563–4574 [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Floyd SK, Zalewski CS, Bowman JL. (2006) Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics 173: 373–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Harberd NP. (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743 [DOI] [PubMed] [Google Scholar]
- Gardner KM, Latta RG. (2007) Shared quantitative trait loci underlying the genetic correlation between continuous traits. Mol Ecol 16: 4195–4209 [DOI] [PubMed] [Google Scholar]
- Geraldes A, Difazio SP, Slavov GT, Ranjan P, Muchero W, Hannemann J, Gunter LE, Wymore AM, Grassa CJ, Farzaneh N, et al. (2013) A 34K SNP genotyping array for Populus trichocarpa: design, application to the study of natural populations and transferability to other Populus species. Mol Ecol Resour 13: 306–323 [DOI] [PubMed] [Google Scholar]
- Geraldes A, Pang J, Thiessen N, Cezard T, Moore R, Zhao Y, Tam A, Wang S, Friedmann M, Birol I, et al. (2011) SNP discovery in black cottonwood (Populus trichocarpa) by population transcriptome resequencing. Mol Ecol Resour (Suppl 1) 11: 81–92 [DOI] [PubMed] [Google Scholar]
- Gorshkova T, Brutch N, Chabbert B, Deyholos M, Hayashi T, Lev-Yadun S, Mellerowicz EJ, Morvan C, Neutelings G, Pilate G. (2012) Plant fiber formation: state of the art, recent and expected progress, and open questions. Crit Rev Plant Sci 31: 201–228 [Google Scholar]
- Hill WG, Zhang XS. (2012a) On the pleiotropic structure of the genotype-phenotype map and the evolvability of complex organisms. Genetics 190: 1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Zhang XS. (2012b) Assessing pleiotropy and its evolutionary consequences: pleiotropy is not necessarily limited, nor need it hinder the evolution of complexity. Nat Rev Genet 13: 296. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. (1998) Seven types of pleiotropy. Int J Dev Biol 42: 501–505 [PubMed] [Google Scholar]
- Hu R, Chi X, Chai G, Kong Y, He G, Wang X, Shi D, Zhang D, Zhou G. (2012) Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa). PLoS ONE 7: e31149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilegems M, Douet V, Meylan-Bettex M, Uyttewaal M, Brand L, Bowman JL, Stieger PA. (2010) Interplay of auxin, KANADI and class III HD-ZIP transcription factors in vascular tissue formation. Development 137: 975–984 [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2009) Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci 14: 373–382 [DOI] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB. (2003) The power of the 3′ UTR: translational control and development. Nat Rev Genet 4: 626–637 [DOI] [PubMed] [Google Scholar]
- La Mantia J, Klápště J, El-Kassaby YA, Azam S, Guy RD, Douglas CJ, Mansfield SD, Hamelin R. (2013) Association analysis identifies Melampsora × columbiana poplar leaf rust resistance SNPs. PLoS ONE 8: e78423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Katju V. (2004) The altered evolutionary trajectories of gene duplicates. Trends Genet 20: 544–549 [DOI] [PubMed] [Google Scholar]
- Magnani E, Barton MK. (2011) A per-ARNT-sim-like sensor domain uniquely regulates the activity of the homeodomain leucine zipper transcription factor REVOLUTA in Arabidopsis. Plant Cell 23: 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown AD, Guy RD, Azam MS, Drewes EC, Quamme LK. (2013) Seasonality and phenology alter functional leaf traits. Oecologia 172: 653–665 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JD. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Ong SS, Wickneswari R. (2012) Characterization of microRNAs expressed during secondary wall biosynthesis in Acacia mangium. PLoS ONE 7: e49662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Porth I, Klápště J, Skyba O, Hannemann J, McKown AD, Guy RD, DiFazio S, Muchero W, Ranjan P, Tuskan GA, et al. (2013a) Genome-wide association mapping for wood characteristics in Populus identifies an array of candidate single nucleotide polymorphisms. New Phytol 200: 710–726 [DOI] [PubMed] [Google Scholar]
- Porth I, Klápště J, Skyba O, Lai BS, Geraldes A, Muchero W, Tuskan GA, Douglas CJ, El-Kassaby YA, Mansfield SD. (2013b) Populus trichocarpa cell wall chemistry and ultrastructure trait variation, genetic control and genetic correlations. New Phytol 197: 777–790 [DOI] [PubMed] [Google Scholar]
- Porth I, White R, Jaquish B, Alfaro R, Ritland C, Ritland K. (2012) Genetical genomics identifies the genetic architecture for growth and weevil resistance in spruce. PLoS ONE 7: e44397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Riechmann JL, Zhang JZ. (2000) INTERFASCICULAR FIBERLESS1 is the same gene as REVOLUTA. Plant Cell 12: 315–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Liu T, Newell NR, Magnani E, Huang T, Kerstetter R, Michaels S, Barton MK. (2013) Establishing a framework for the ad/abaxial regulatory network of Arabidopsis: ascertaining targets of class III HOMEODOMAIN LEUCINE ZIPPER and KANADI regulation. Plant Cell 25: 3228–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robischon M, Du J, Miura E, Groover A. (2011) The Populus class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol 155: 1214–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV. (2012) The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Nguyen D, Karlowski WM, Mayer KF. (2004) START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol 5: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz M, Smith R, Ellis B. (2013) Xylem tissue specification, patterning, and differentiation mechanisms. J Exp Bot 64: 11–31 [DOI] [PubMed] [Google Scholar]
- Slavov GT, DiFazio SP, Martin J, Schackwitz W, Muchero W, Rodgers-Melnick E, Lipphardt MF, Pennacchio CP, Hellsten U, Pennacchio LA, et al. (2012) Genome resequencing reveals multiscale geographic structure and extensive linkage disequilibrium in the forest tree Populus trichocarpa. New Phytol 196: 713–725 [DOI] [PubMed] [Google Scholar]
- Soccio RE, Breslow JL. (2003) StAR-related lipid transfer (START) proteins: mediators of intracellular lipid metabolism. J Biol Chem 278: 22183–22186 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Zhao S, Williamson L, Ward S, Leyser O. (2012) FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J 71: 907–920 [DOI] [PubMed] [Google Scholar]
- Streisfeld MA, Rausher MD. (2011) Population genetics, pleiotropy, and the preferential fixation of mutations during adaptive evolution. Evolution 65: 629–642 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Adler HT, Parks DW, Comai L. (1995) The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Ursache R, Nieminen K, Helariutta Y. (2013) Genetic and hormonal regulation of cambial development. Physiol Plant 147: 36–45 [DOI] [PubMed] [Google Scholar]
- Wagner GP, Zhang J. (2011) The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat Rev Genet 12: 204–213 [DOI] [PubMed] [Google Scholar]
- Wagner GP, Zhang J. (2012) Universal pleiotropy is not a valid null hypothesis: reply to Hill and Zhang. Nat Rev Genet 13: 296. [DOI] [PubMed] [Google Scholar]
- Wang Z, Liao B-Y, Zhang J. (2010) Genomic patterns of pleiotropy and the evolution of complexity. Proc Natl Acad Sci USA 107: 18034–18039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue LJ, Guo W, Yuan Y, Anino EO, Nyamdari B, Wilson MC, Frost CJ, Chen HY, Babst BA, Harding SA, et al. (2013) Constitutively elevated salicylic acid levels alter photosynthesis and oxidative state but not growth in transgenic Populus. Plant Cell 25: 2714–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, et al. (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Zhong R, Taylor JJ, Ye ZH. (1997) Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9: 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Taylor JJ, Ye ZH. (1999) Transformation of the collateral vascular bundles into amphivasal vascular bundles in an Arabidopsis mutant. Plant Physiol 120: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. (1999) IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11: 2139–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. (2004) Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol 45: 369–385 [DOI] [PubMed] [Google Scholar]