Two desaturase genes respond to drought and hypoxia stress in Arabidopsis crown galls.
Abstract
Agrobacterium tumefaciens-derived crown galls of Arabidopsis (Arabidopsis thaliana) contain elevated levels of unsaturated fatty acids and strongly express two fatty acid desaturase genes, ω3 FATTY ACID DESATURASE3 (FAD3) and STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 (SAD6). The fad3-2 mutant with impaired α-linolenic acid synthesis developed significantly smaller crown galls under normal, but not under high, relative humidity. This strongly suggests that FAD3 plays a role in increasing drought stress tolerance of crown galls. SAD6 is a member of the SAD family of as yet unknown function. Expression of the SAD6 gene is limited to hypoxia, a physiological condition found in crown galls. As no sad6 mutant exists and to link the function of SAD6 with fatty acid desaturation in crown galls, the lipid pattern was analyzed of plants with constitutive SAD6 overexpression (SAD6-OE). SAD6-OE plants contained lower stearic acid and higher oleic acid levels, which upon reduction of SAD6 overexpression by RNA interference (SAD6-OE-RNAi) regained wild-type-like levels. The development of crown galls was not affected either in SAD6-OE or SAD6-OE-RNAi or by RNA interference in crown galls. Since biochemical analysis of SAD6 in yeast (Saccharomyces cerevisiae) and Escherichia coli failed, SAD6 was ectopically expressed in the background of the well-known suppressor of salicylic acid-insensitive2 (ssi2-2) mutant to confirm the desaturase function of SAD6. All known ssi2-2 phenotypes were rescued, including the high stearic acid level. Thus, our findings suggest that SAD6 functions as a Δ9-desaturase, and together with FAD3 it increases the levels of unsaturated fatty acids in crown galls under hypoxia and drought stress conditions.
Plant tumors, also referred to as crown galls, develop upon infection of susceptible plants with Agrobacterium tumefaciens. A DNA fragment, the transfer DNA (T-DNA) of the tumor-inducing plasmid of virulent A. tumefaciens strains, is randomly integrated into the genome of a host plant (Thomashow et al., 1980; Kim et al., 2007; Pitzschke and Hirt 2010). Expression of the T-DNA-encoded oncogenes drives increased production of auxin and cytokinin, thereby promoting cell proliferation. Plant tumor growth causes disruption of the epidermal cell layer that is covered by a cuticle and thus is permanently endangered to lose water (Schurr et al., 1996). In order to prevent desiccation and wilting, rescue processes appear to be constitutively activated (Veselov et al., 2003). Thereby, ethylene and abscisic acid trigger the expression of drought stress-responsive genes, the accumulation of osmoprotectants, and suberization of the surface cell layers (Efetova et al., 2007). In addition, cell membrane lipids are the major targets of environmental stresses, and tolerance to drought stress is dependent on high levels of polyunsaturated fatty acids (PUFAs) and the ability to maintain fatty acid (FA) desaturation activity (Berberich et al., 1998; Mikami and Murata, 2003; Torres-Franklin et al., 2009). In Arabidopsis (Arabidopsis thaliana) crown gall tumors, 27% of the genes involved in lipid metabolism are differentially regulated (Deeken et al., 2006). Under drought stress, Arabidopsis increases the ratio of digalactosyl diglyceride (DGDG) to monogalactosyl diglyceride (MGDG) and FA unsaturation (Gigon et al., 2004). An increase in α-linolenic acid levels (18:3, where x:y denotes an FA with x carbons and y double bonds) by overexpression of the ω3 fatty acid desaturases FAD3 and FAD7 has been shown to enhance tolerance to drought stress in Nicotiana tabacum cells (Zhang et al., 2005), whereas nontolerant plants decline their fraction of 18:3 (Monteiro de Paula et al., 1993; Dakhma et al., 1995; Upchurch, 2008).
Developing crown gall tumors also face permanent oxidative stress. Reactive oxygen species (ROS) are produced in tumors, and glutathione S-transferases and peroxidases are strongly up-regulated (Jia et al., 1996; Lee et al., 2009). Due to a reduced respiratory and photosynthetic capacity in crown gall tumors, ATP production is predominantly derived from glycolysis and alcoholic fermentation (Deeken et al., 2006). In other words, the hypoxia physiology of the Arabidopsis tumor is governed by heterotrophic metabolism (Deeken et al., 2006). Since dioxygen is a cofactor of desaturases, its depletion limits the de novo synthesis of unsaturated FAs and thus membrane lipids (Brown and Beevers, 1987). In addition, hypoxia appears to be associated with ROS production, peroxidation of PUFAs, and finally, loss of membrane integrity (Blokhina et al., 2003; Upchurch, 2008).
The biosynthesis of PUFAs is initiated by introduction of the first double bond into stearic acid (18:0) by STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE (SAD). SAD genes exhibit a tissue-specific expression profile, and the encoding enzymes regulate the pools of oleic acid (18:1), a monounsaturated fatty acid (MUFA; Shanklin and Somerville 1991; Thompson et al., 1991; Cahoon et al., 1996, 1998; Whittle et al., 2005). In Arabidopsis, five out of seven members of the SAD gene family (SAD1, SAD3, SAD4, SAD5, and SUPPRESSOR OF SALICYLIC ACID INSENSITIVE2 [SSI2]) are capable of desaturating 18:0 and contribute to the 18:1 pool (Kachroo et al., 2007). A mutation in the Arabidopsis ssi2 locus results in the accumulation of 18:0, whereas the 18:1 content is reduced. Furthermore, the salicylic acid-mediated defense signaling pathway is constitutively active, resulting in lesion formation and increased expression of the PATHOGENESIS-RELATED (PR) genes. The 18:1 MUFAs are incorporated into membrane lipids by two glycerolipid biosynthesis pathways. Phospholipids and galactolipids of photosynthetic membranes in plastids are synthesized by the prokaryotic pathway, while lipids of extraplastidic membranes are produced in the endoplasmic reticulum (ER) by the eukaryotic pathway (Ohlrogge and Browse, 1995). MUFAs are further desaturated to PUFAs by two sets of membrane-bound FADs. These enzymes are either located in plastids or the ER (Ohlrogge and Browse, 1995). In the ER, conversion of the unsaturated phospholipid FAs 18:1 to 18:2 and of 18:2 to 18:3 is carried out by the ω6 desaturase FAD2 and the ω3 desaturase FAD3, respectively (Browse et al., 1993; Okuley et al., 1994; Los and Murata, 1998).
This study focuses on the role of desaturases in Arabidopsis crown galls in the context of drought and hypoxia stress. We document that crown galls produce increased levels of α-linolenic acid and strongly express the two FAD genes FAD3 and SAD6. In contrast to the well-known ω3 desaturase FAD3, the function of SAD6, a putative SAD, is unknown, and mutants for this gene are not available. However, the ability of SAD6 to replace the well-characterized SSI2 functionally in the ssi2-2 mutant suggests that SAD6 is a functional SAD. Overexpression of SAD6 decreased stearic acid and increased oleic acid levels. Down-regulation of SAD6 overexpression by RNA interference (RNAi) restored wild-type-like FA levels. The ability of SAD6 to influence the oleic acid levels together with the finding that SAD6 gene expression is restricted to hypoxia suggest that SAD6 catalyzes FA desaturation under hypoxic conditions. Unlike SAD6, the results obtained with the fad3-2 mutant impaired in α-linolenic acid biosynthesis indicate a role of FAD3 in increasing the drought stress tolerance of crown galls. Thus, both desaturases shape the pool of unsaturated FAs in drought stress- and oxidative stress-challenged Arabidopsis crown gall tumors.
RESULTS
Arabidopsis crown galls induced by the A. tumefaciens strain C58 are known to encounter hypoxia, drought stress, and ROS during their development (Deeken et al., 2006; Efetova et al., 2007). This raises the question of how plants in general and crown galls in particular cope with these abiotic stress conditions. In crown gall tumors, a high percentage of the genes involved in lipid metabolism are differentially regulated (Deeken et al., 2006), illustrating that crown gall development requires a unique lipid profile. To test this assumption, we determined the lipid profile of stem-derived crown galls and compared it with that of tumor-free stems. The crown gall-specific lipid pattern, which was dominated by PUFAs, was correlated with differentially expressed genes involved in lipid metabolism.
ER-Derived Unsaturated Phospholipids Dominate the Lipid Pool of Crown Gall Tumors
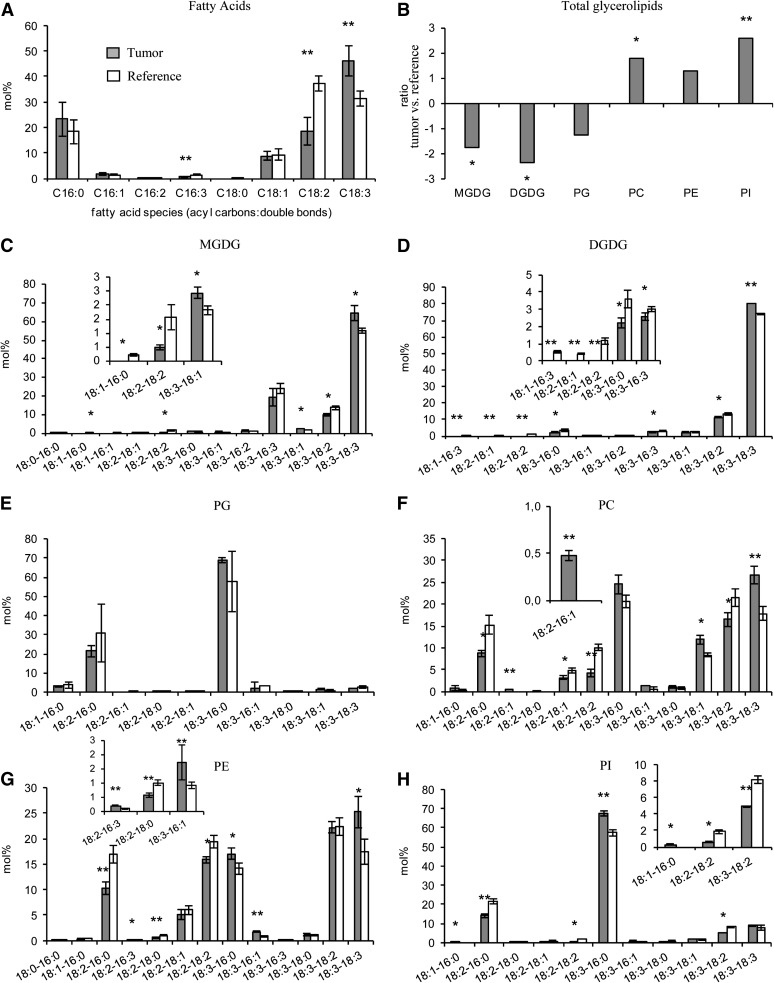
Determination of total FAs revealed that the relative amount of α-linolenic acid (18:3) was significantly higher in crown gall tumors (46 mol %) compared with reference stems (32 mol %) injected with buffer without agrobacteria (Fig. 1A). Coincident with this enrichment, two other PUFAs, roughanic acid (16:3) and linoleic acid (18:2), were 2-fold reduced, while all others remained unchanged. In order to trace back the origin of 18:3, the major lipid classes of plastids, MGDG, DGDG, and phosphatidylglycerol (PG), as well as those of the ER, phosphytidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI), were analyzed. In accordance with the reduced number of chloroplasts (Gee et al., 1967; Deeken et al., 2006), the total amounts of MGDG (−1.76-fold, P = 0.022), DGDG (−2.36-fold, P = 0.032), and PG (−1.25-fold, P = 0.153) were decreased in crown gall tumors, and again, the ER-specific phospholipid pools, PC (1.79-fold, 0.05), PE (1.31-fold, P = 0.29), and PI (2.61-fold, P = 0.002), were increased (Fig. 1B). Within all classes of the ER-derived lipids, the levels of those containing 18:3 were significantly (P ≤ 0.05) higher in tumors compared with reference stems (Fig. 1, C–H). Thus, the relatively high levels of 18:3 mainly derive from the phospholipid PC. Based on the relative distribution of FAs, α-linolenic acid was again significantly higher in each of the ER lipid classes of crown gall tumors. Furthermore, the unsaturation index, which represents the average number of double bonds per FA in a lipid pool, was significantly (P ≤ 0.05) increased in the tumor of all glycerolipids (Supplemental Table S1).
Figure 1.
Lipid profile of crown gall tumors in comparison with inflorescence stems of Arabidopsis (Ws-2). A, Relative amounts of C16 to C18 saturated and unsaturated FA species. B, Ratios of the total amount of each of the polar lipid classes MGDG, DGDG, PG, PC, PE, and PI in crown gall tumor versus inflorescence stem tissue. The ratios were calculated from the absolute amount of each lipid species in the respective galactolipid and phospholipid classes of three to four independent experiments. C to H, Lipid species of the polar head group classes MGDG (C), DGDG (D), PG (E), PC (F), PE (G), and PI (H). Insets show enlargements of the lipid species below 10 mol % with significant differences (P ≤ 0.05) between tumor and reference tissue. Tumor development was induced by inoculating A. tumefaciens (strain C58) into wounded inflorescence stems. Inflorescence stems of the same age inoculated with buffer served as a reference. FAs and polar lipid species were determined by LC-MS/MS, and mean values (mol % ± sd) were calculated from three to four independent experiments. Statistical analysis was performed by using an unpaired two-tailed Student’s t test: *P ≤ 0.05, **P ≤ 0.01.
High Expression Levels of Two Desaturase Genes in Crown Galls
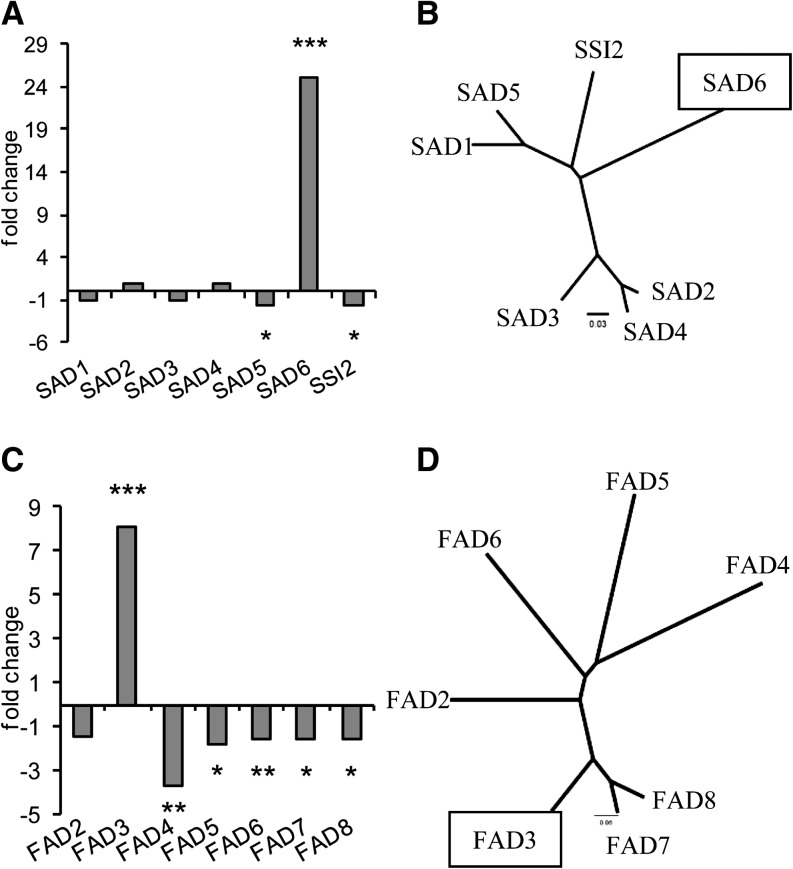
The higher proportion of unsaturated FAs in crown gall lipids compared with that of tumor-free stem tissue suggested that genes involved in FA desaturation must be differentially expressed. According to microarray analyses (Deeken et al., 2006), two genes involved in the biosynthesis of unsaturated FAs, the putative SAD6 and FAD3, were strongly up-regulated in Arabidopsis crown gall tumors compared with the reference stem. The transcription of FAD3 appeared 8-fold and that of SAD6 25-fold higher (Fig. 2, A and C). In contrast, FAD4, encoding a desaturase known to generate Δ3-trans-hexadecenoic acid (Gao et al., 2009), was almost 4-fold down-regulated. The expression of all other genes of the two desaturase families was either reduced or remained unchanged. Quantitative real-time PCR measurements confirmed the very high transcript levels of SAD6 and FAD3 in tumors as well as the significant down-regulation of the SAD gene SSI2 (Supplemental Fig. S1, A–C). SAD6 exhibited an overall very low transcript level in tissues of leaves, roots, flowers, and inflorescence stems, except in the tumor (Fig. 3A). The gene expression patterns suggest that SAD6 and FAD3 may play a unique role in the lipid metabolism of crown galls.
Figure 2.
SAD6 and FAD3 are the desaturase genes strongly up-regulated in Arabidopsis crown gall tumors. A, Fold changes of gene expression levels of SADs in crown gall versus stem tissues injected with buffer. B, Phylogenetic tree based on the amino acid sequences of the seven members constituting the family of Arabidopsis SAD proteins. C, Fold changes of gene expression of membrane-bound ω3/ω6 FADs. D, Phylogenetic tree of the five membrane-bound ω3/ω6 FAD proteins of Arabidopsis. Fold changes of gene expression were calculated as described (Deeken et al., 2006). Statistical analysis was performed using an unpaired two-tailed Student’s t test: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Phylograms were generated with ClustalX 2.0.12 (Larkin et al., 2007) and drawn by FigTree version 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
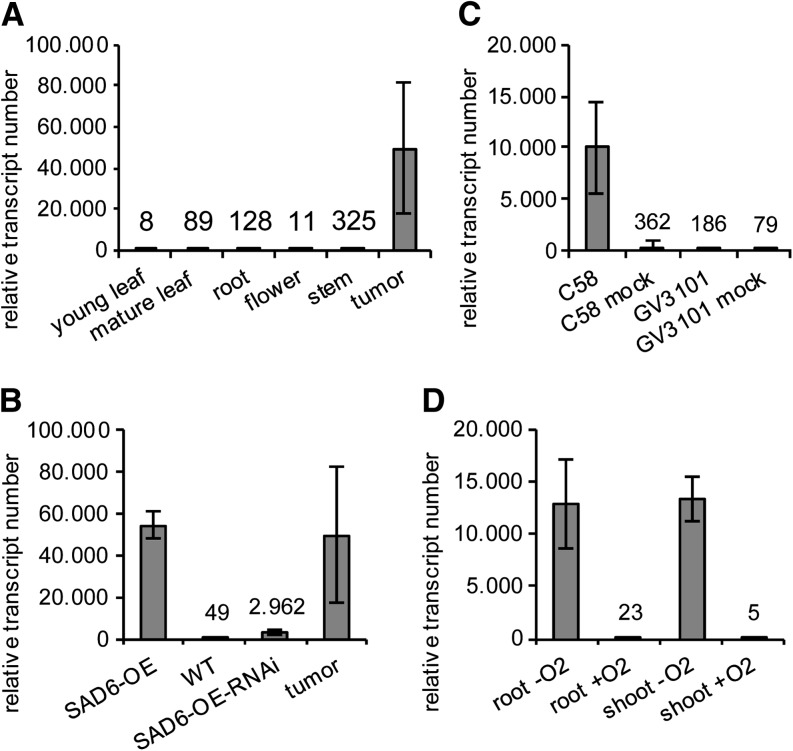
Figure 3.
SAD6 gene expression levels in different Arabidopsis tissues and after various treatments. Relative SAD6 transcript numbers are shown for different organs and the crown gall tumors induced by A. tumefaciens (strain C58) on Arabidopsis (Ws-2) stems (A), in young leaves of SAD6-OE plants, the Col-0 wild type (WT), SAD6-OE-RNAi plants, as well as crown gall tumors (B), in stems 6 d post injection of the oncogenic (C58) and nononcogenic A. tumefaciens strain GV3101 and stems injected with the respective cultivation medium serving as a mock treatment (C), and in shoots and roots treated with low (0.5% oxygen; −O2) and ambient (+O2) oxygen concentrations for 24 h (D). Relative SAD6 transcript numbers were determined and normalized to 10,000 ACTIN2/ACTIN8 molecules by applying quantitative real-time PCR. Bars and numbers above the bars represent mean values ± sd of three to four independent experiments.
SAD6 Overexpression Correlates with an Increase in FA Unsaturation
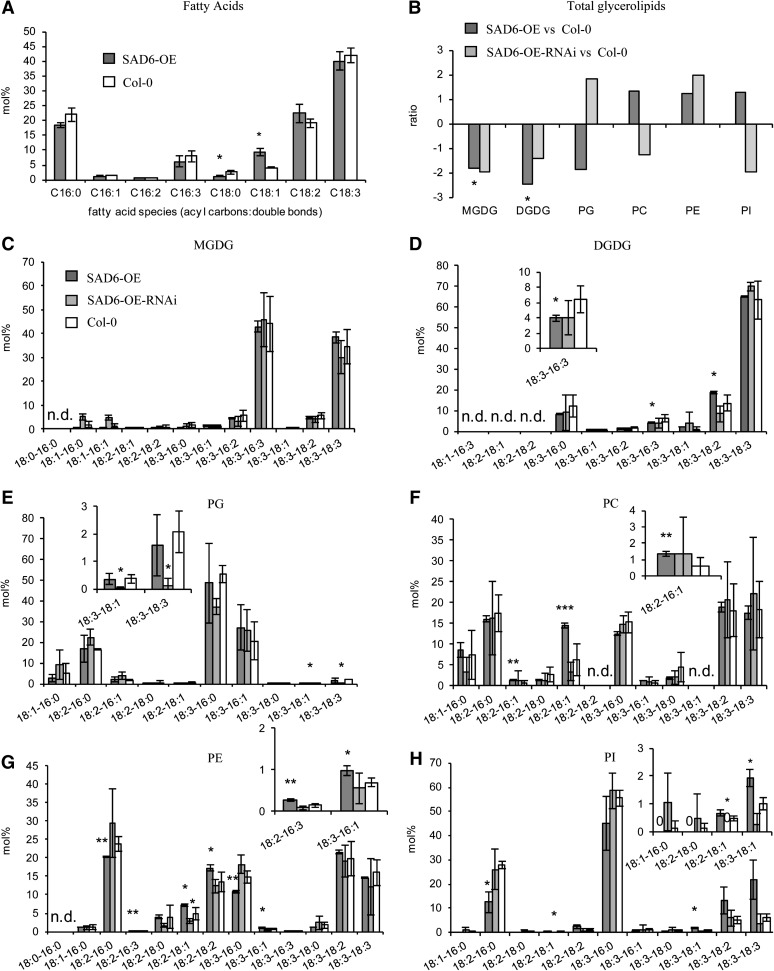
In order to study the function of the SAD6 gene and its role in crown gall physiology, we searched for a knockout mutant in the T-DNA express database SIGnAL (http://signal.salk.edu/cgi-bin/tdnaexpress). One line with a T-DNA insertion in the SAD6 locus existed. This line from the GABI-Kat population of T-DNA-mutagenized Arabidopsis harbored a T-DNA in the promoter close to the transcription start site (gk30d04; Supplemental Fig. S2, A and B). However, this line turned out to be a transactivation-tagged line that expressed high numbers of SAD6 transcripts (49,634 ± 6,781) compared to the wild type (Fig. 3B). A loss-of-function mutant was not available in any of the Arabidopsis stock centers. Since the expression levels of the transactivation-tagged line were similar to that found in crown gall tumors (54,438 ± 32,132; Fig. 3B), we decided to examine the lipid profile of this SAD6-overexpressing line (SAD6-OE). It should be mentioned that the strong expression of SAD6 had no impact on the transcript levels of the SSI2 gene (Supplemental Fig. S1D), a functionally well-described SAD enzyme and the nearest homolog to SAD6 (Fig. 2B). Determination of the total FA contents in leaves revealed that the relative amount of stearic acid (18:0) was significantly reduced in SAD6-OE compared with wild-type ecotype Columbia (Col-0) plants, whereas that of oleic acid (18:1) was increased (Fig. 4A). A similar result was obtained when the FA composition of the polar lipids in SAD6-OE and Col-0 was compared (Fig. 4, C–H). The relative amounts primarily of 18:0 were decreased and those of 18:1 significantly increased specifically in the ER-specific phospholipid pools (Fig. 4, F–H). All other FA species remained more or less unchanged between SAD6-OE and Col-0. Furthermore, the amount of the ER-specific phospholipids was increased whereas that of the plastid-derived galactolipids was reduced in leaves of SAD6-OE compared with wild-type Col-0 (Fig. 4B). In line with this finding, the desaturation index was higher in several polar lipid classes, similar to that in crown galls (Supplemental Table S1). However, the capability of SAD6-OE to increase unsaturated FA had no impact on crown gall development.
Figure 4.
Relative amounts of lipids in Arabidopsis leaves from SAD6-OE and SAD6-OE-RNAi in comparison with Col-0. A, Saturated and unsaturated FAs of C16 to C18 species. B, Ratios of the total amount of each of the polar lipid classes MGDG, DGDG, PG, PC, PE, and PI. The ratios were calculated from the absolute amounts of each lipid species in the respective galactolipid and phospholipid class of three to four independent experiments. C to H, Lipid species of the polar head group classes MGDG (C), DGDG (D), PG (E), PC (F), PE (G), and PI (H). Insets show enlargements of the lipid species below 10 mol % with significant differences (P ≤ 0.05) between SAD6-OE or SAD6-OE-RNAi and the wild-type Col-0. FAs and polar lipid species were determined by LC-MS/MS, and mean values (mol % ± sd) were calculated from three to four independent experiments. Statistical analysis was performed by using an unpaired two-tailed Student’s t test: *P ≤ 0.05, **P ≤ 0.01 ***P ≤ 0.001; n.d., not determined.
To confirm the notion that strong expression of SAD6 has an impact on FA desaturation, the RNAi technology was applied to posttranscriptionally reduce the high SAD6 transcript levels in the activation-tagged SAD6-OE line. RNAi plants were generated in the SAD6-OE background, because in wild-type plants, SAD6 expression was very low (Fig. 3A). In leaves of homozygous SAD6-OE lines, the expression of an artificial microRNA directed against SAD6 resulted in an 18-fold down-regulation of SAD6 transcription (SAD6-OE-RNAi; Fig. 3B). This strong suppression of SAD6 gene expression resulted in a reduction of the relative amount of unsaturated FAs of the galactolipid and phospholipid species (Fig. 4B). The differences observed between SAD6-OE and wild-type plants in the FA profiles of the polar lipid classes vanished in SAD6-OE-RNAi plants (Fig. 4, C–H). Thus, the lipid pattern in SAD6-OE-RNAi transgenics was wild type like, while the elevated levels of unsaturated FAs of ER-derived phospholipids in SAD6-OE plants reflected the situation in tumors with high SAD6 levels. Since other SAD genes may also be affected by the RNAi construct directed against SAD6, it cannot be excluded that the lipid profile of SAD6-OE-RNAi plants is the result of an altered expression of several SAD genes. Nevertheless, tissues with high levels of SAD6 transcripts (SAD6-OE and crown galls) have increased levels of unsaturated FAs in glycerolipids but not in sphingolipids (data not shown).
Due to the lack of a sad6 mutant and to prove the impact of SAD6 on crown gall development, the RNAi construct directed against SAD6 was expressed in crown galls to repress SAD6 expression on the side where it is induced. To do so, crown galls were induced by the virulent A. tumefaciens strain C58, harboring, in addition to the T-DNA of the tumor-inducing plasmid, a T-DNA with the RNAi construct. The RNAi construct reduced the level of SAD6 transcripts in crown galls 10-fold, from 22,715 ± 12,835 to 2,323 ± 1,286. The weight of crown galls per centimeter of Arabidopsis stems inoculated with the wild-type A. tumefaciens strain C58 was 20 ± 2.5 mg, and that induced by the RNAi-containing C58 reached 18 ± 2.5 mg. Despite a strong reduction of SAD6 gene expression, the transcript numbers never reached a level close to zero but were always high enough to support normal crown gall development.
SAD6 Is Located in Chloroplasts and Compensates for the Loss of ssi2 Function
Stearoyl-acyl carrier protein (S-ACP) desaturases like SSI2 or FATTY ACID BIOSYNTHESIS2 (FAB2) of Arabidopsis are soluble enzymes that are located in the chloroplast stroma and catalyze the desaturation of stearic acid to oleic acid (Shanklin and Somerville, 1991; Kachroo et al., 2007). To get a first hint if SAD6 is also operating as a plastidic S-ACP desaturase, database searches were performed (WoLF PSORT [http://wolfpsort.org/] and MultiLoc2 [http://www-bs.informatik.uni-tuebingen.de/Services/MultiLoc2]). Both databases were in favor of a chloroplast localization of SAD6. To verify this indirect evidence, western-blot analyses were performed with a V5 epitope/6x histidin tag-tagged SAD6 fusion protein transiently overexpressed in Nicotiana benthamiana leaves. Indeed, leaf protein extracts from the Light Harvesting Complex Protein-containing chloroplast-specific protein fraction exhibited a strong signal of the SAD6 protein (“C” in Supplemental Fig. S3). A weak SAD6-related signal was also observed in the total extract, which most likely is due to a chloroplast contamination (“L” in Supplemental Fig. S3).
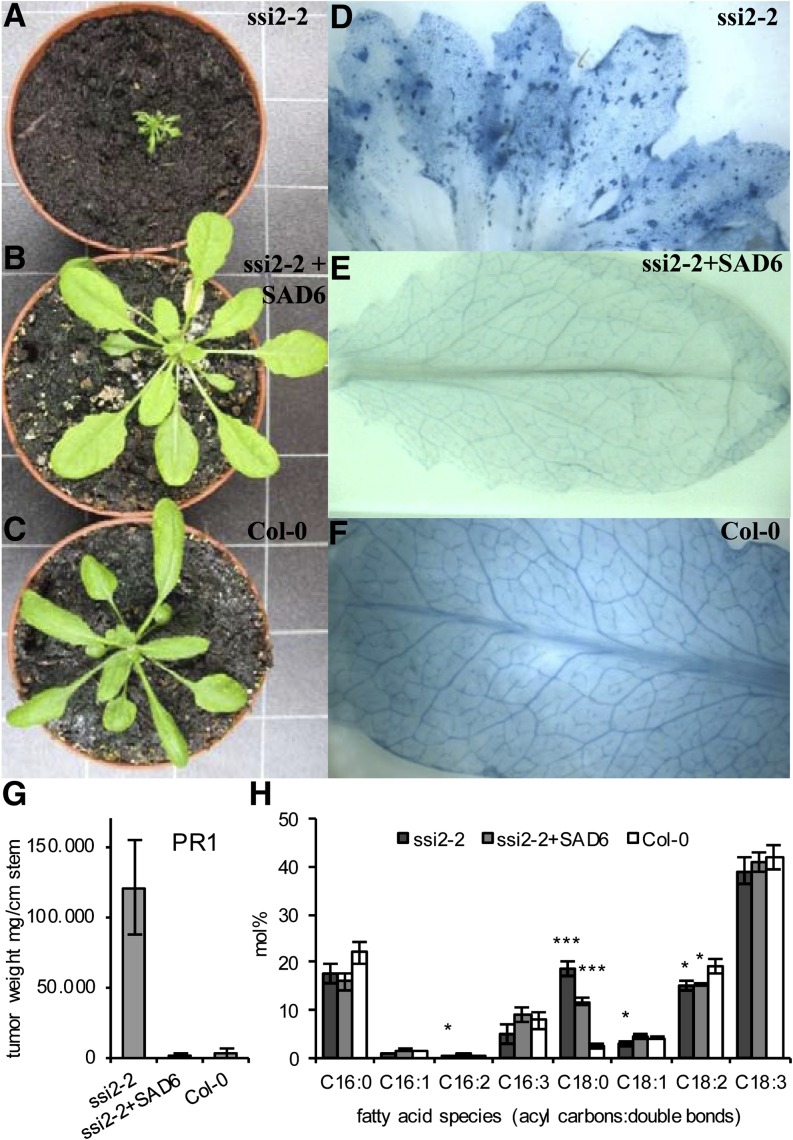
The chloroplast localization of the SAD6 protein and the increased levels of 18:1 in SAD6-OE supported the notion that SAD6 operates as an S-ACP desaturase. To test the enzymatic function of SAD6, the protein was expressed in different yeast (Saccharomyces cerevisiae) mutants and in Escherichia coli cells, as summarized in Supplemental Table S2. Despite its expression, the SAD6 protein neither rescued the growth of the yeast mutants to complement the loss of desaturase function nor altered the unsaturated FA profile of either the yeast strains or E. coli. As a consequence, we characterized the role of SAD6 by an in planta approach. The salk039682 line, which harbored a T-DNA insertion in the SSI2/FAB2 locus (AT2G43710; Supplemental Fig. S2, C and D), was chosen as a native expression system for SAD6. SSI2/FAB2 is the closest homolog of SAD6, and its enzymatic function as an S-ACP desaturase is well documented (Lightner et al., 1994; Kachroo et al., 2001). The SALK line with the T-DNA inserted into the SSI2/FAB2 locus displayed exactly the same phenotypes as the fab2 and ssi2 mutants, such as dwarf-like growth (Fig. 5A), formation of lesions (Fig. 5D, compare with E and F), and elevated levels of PR1 transcripts (Fig. 5G). Most importantly, the level of stearic acid (18:0) was found to be increased compared with wild-type Col-0 and that of oleic acid (18:1) was reduced (Fig. 5H), similar to those known for the ssi2 mutant. To distinguish the SALK line from the ssi2 and fab2 mutants, this mutant allele was named ssi2-2. Under normal growth conditions, leaves of ssi2-2 plants did not express noteworthy numbers of SAD6 transcripts according to real-time reverse transcription (RT)-PCR (9 ± 12 transcripts). Therefore, the ssi2-2 mutant was stably transformed with SAD6 copy DNA (cDNA) expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter. This ubiquitously active promoter was chosen, because the native SAD6 promoter was only weekly active in all Arabidopsis organs (Fig. 3A). Three different transformed ssi2-2 lines (ssi2-2+SAD6) expressing high levels of SAD6 transcripts in leaves (line 17, 173,765 transcripts; line 21, 52,776 transcripts; line 22, 53,732 transcripts) showed a similar growth phenotype as the wild-type Col-0 (Figure 5, compare B and C). Furthermore, SAD6 overexpression in the ssi2-2 mutant background prevented lesion formation (Figure 5, compare D with E and F) and PR1 gene expression (Fig. 5G). The levels of stearic acid (18:0) were markedly reduced in ssi2-2+SAD6 compared with ssi2-2 but did not reach the low levels of Col-0 (Fig. 5H). Oleic acid (18:1) levels, however, were wild type like. These results demonstrate that SAD6 seems to restore the ssi2/fab2 loss of function.
Figure 5.
Expression of SAD6 in the Arabidopsis ssi2-2 mutant restores a wild-type-like phenotype. A to C, Growth phenotypes of 8-week-old plants of the salk039682 line with a T-DNA insertion in the SSI2 locus (ssi2-2; A), an ssi2-2 T-DNA insertion line transformed with CaMV 35S::SAD6 cDNA (ssi2-2+SAD6; B), and the wild-type Col-0 (C). One representative plant of each genotype is shown. D to F, Trypan blue staining of leaves to visualize lesion formation in the ssi2-2 mutant (D), the ssi2-2 line transformed with CaMV 35S::SAD6 cDNA (E), and the wild-type Col-0 (F). G, Determination of expression levels of the PR1 gene in leaves of the ssi2-2 mutant, ssi2-2+SAD6, and Col-0 plants by applying real-time quantitative PCR. Relative transcript numbers were normalized to 10,000 molecules of ACTIN2/ACTIN8. H, Relative amounts of C16 to C18 saturated and unsaturated FAs in the total FA pool of leaves from the ssi2-2 mutant, ssi2-2+SAD6, and the wild-type Col-0. Mean values (mol % ± sd) of each FA species were calculated from three independent experiments. Statistical analysis was performed using an unpaired two-tailed Student’s t test: ***P ≤ 0.001, *P ≤ 0.05.
Hypoxia Induces SAD6 Gene Expression in Crown Gall Tumors
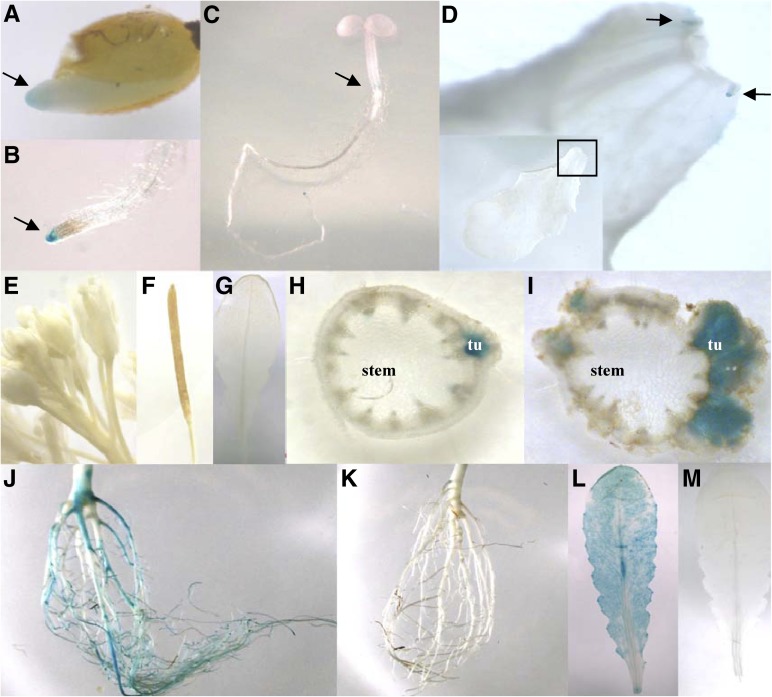
In order to localize the sites of SAD6 expression in Arabidopsis and to identify the stimulus that drives the pronounced expression of the SAD6 gene in crown gall tumors, SAD6 promoter-driven GUS expression was analyzed in stably transformed Arabidopsis lines. Three independent transgenic GUS reporter lines localized the SAD6 promoter activity in root tips of germinating seeds, seedlings, stipules (Fig. 6, A–G), and the crown gall throughout all developmental stages (Fig. 6, H and I). Thus, the SAD6 promoter-GUS lines confirmed the real-time PCR results that the SAD6 activity is very weak throughout the whole Arabidopsis plant but is high in crown gall tumors (Fig. 3A). SAD6 promoter-driven GUS expression appeared induced 6 d post inoculation of the virulent T-DNA harboring A. tumefaciens strain C58 (Fig. 6H), and SAD6 transcript numbers increased significantly (Fig. 3C). This increased SAD6 gene activity was not observed when the disarmed A. tumefaciens strain GV3101 was injected. GV3101 is a derivative of the A. tumefaciens strain C58 that lacks only the T-DNA but not the virulence factors, such as VirD2, VirE2, VirE3, and VirF, or any other effector proteins (Vergunst et al., 2000, 2003). Thus, agrobacterial effector proteins do not have an influence on SAD6 gene expression. This gene regulation pattern suggests that the developmental switch from stem to tumor cell physiology, but not agrobacteria, drives the up-regulation of SAD6.
Figure 6.
Expression pattern of the SAD6 gene in Arabidopsis. A to D, Histochemical GUS assay indicated by the blue color in a 7-d-old seedling. Arrows point to GUS-derived blue color in the root apex of a germinating seedling (A), the root tip (B), the developing zone of the hypocotyl (C), and stipules at the base of the leaf petiole shown in closeup (D). E to I, GUS expression in a flowering plant: flowers (E), silique (F), and leaf (G) show no GUS activity; GUS-derived blue staining is seen in cross sections of an inflorescence stem with a young tumor (tu) 6 d post inoculation (H) and a mature tumor 21 d post inoculation (I) with A. tumefaciens strain C58. J to M, GUS expression in adult nonflowering plants treated with low oxygen (0.5% oxygen) or ambient oxygen concentrations for 24 h: strong GUS activity in a root (J) and a leaf (L) of a plant exposed to hypoxia and no GUS activity in a root (K) and a leaf (M) from a plant grown in normoxia. Representative images of three independent transgenic GUS-expressing lines of the T3 generation are presented.
Since agrobacterial effectors appear not to be involved in SAD6 gene induction, other crown gall-relevant stimuli must have an impact on SAD6 gene expression. Analysis of the SAD6 promoter sequence provided a hint that the partial pressure of oxygen might affect SAD6 gene regulation. The motif database PLACE (http://www.dna.affrc.go.jp/PLACE/index.html) revealed seven hypoxia-related cis-acting regulatory DNA elements in the SAD6 promoter (Supplemental Fig. S4). When Arabidopsis was grown under hypoxia with 0.5% oxygen for 24 h, increases in relative SAD6 transcription of more than 500-fold (from 23 ± 15 to 12,977 ± 4,205) and 2,500-fold (from 5 ± 7 to 13,424 ± 2,169) were measured in roots and shoots, respectively (Fig. 3D). This real-time PCR result was again confirmed by histochemical GUS staining experiments with the transgenic SAD6 promoter::GUS reporter lines (Fig. 6, J–M). Under hypoxia, strong GUS staining was observed in roots and leaves (Fig. 6, J and L), which was completely absent under normoxia (Fig. 6, K and M). In addition, waterlogging, which causes hypoxia in underground plant tissue, was found to induce SAD6 transcription in roots, from 355 ± 301 to 34,846 ± 5,727, as well (Supplemental Fig. S1E). Taken together, hypoxia that is associated with Arabidopsis crown gall development drives SAD6 gene expression.
FAD3 Supports Crown Gall Growth under Low Relative Humidity
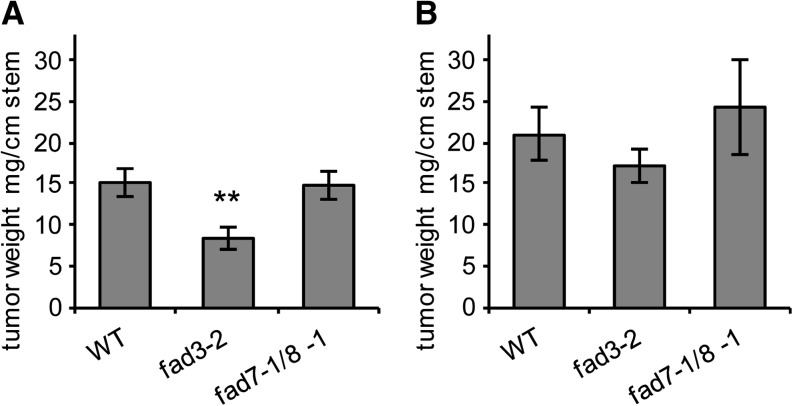
In contrast to SAD6, the role of FAD3 in crown gall physiology is more obvious. Arabidopsis crown galls are known to also cope with increased water loss and to express drought stress-protective mechanisms (Efetova et al., 2007). In order to prove if the elevated levels of FAD3 expression and 18:3 in crown galls add to the drought stress-protective mechanisms, tumor growth of the Arabidopsis desaturase mutants fad3-2 and fad7-1/fad8-1 and their respective wild-type Col-0 was analyzed under two different relative humidity (RH) conditions. The fad3-2 mutant of the ER-bound FAD is known to contain reduced amounts of trienoic FAs of extraplastidic phospholipids (James and Dooner, 1990; Browse et al., 1993), whereas the double mutant of the plastid-localized and most closely to FAD3 related desaturases, FAD7 and FAD8 (Fig. 2D), produces less plastidic galactolipid trienoic FAs compared with the wild type (Gibson et al., 1994; McConn et al., 1994). When these mutants and wild-type plants were inoculated with virulent agrobacteria (strain C58) and grown at normal RH (45%–50%), crown gall tumors from the fad3-2 plants were 45% smaller compared with those of wild-type Col-0 (Fig. 7A). No difference in tumor weight was observed between the fad7-1/fad8-1 lacking plastidic desaturases and the wild type. At high RH of 75% to 100%, crown gall tumors were larger on all three Arabidopsis genotypes (Col-0, fad3-2, and fad7-1/fad8-1) compared with those at 45% to 50% RH (Fig. 7B). The difference in tumor growth between Col-0 and the fad3-2 mutant, evident at 45% to 50% RH, was abolished when plants were grown at 75% to 100% RH (Fig. 7). The fad7-1/fad8-1 mutant again exhibited no significant change in tumor size in comparison with Col-0 under high-RH conditions. According to quantitative real-time PCR, the change in RH did not affect the transcriptional regulation of FAD3 in Col-0 (Supplemental Fig. S1F). This finding strongly suggests that a normal level of FAD3 expression and a normal amount of 18:3 are sufficient to support crown gall growth at low RH. Taken together, the tumor growth assays suggest that 18:3, synthesized by FAD3 in the ER, plays an important role in drought stress signaling in tumors.
Figure 7.
The Arabidopsis fad3-2 mutant develops smaller tumors only under low RH. Tumor weights from fad3-2 (Browse et al., 1993) and fad7-1/8-1 (McConn et al., 1994) mutants and the respective wild-type Col-0 (WT) grown at RH of 45% to 50% (A) and 75% to 100% (B). Tumor weights were determined per centimeter of infected stems 3 weeks after injection of A. tumefaciens strain C58. Mean values ± se were calculated from 22 tumors of fad3-2, 18 tumors of fad7-1/8-1, and 28 tumors of Col-0 (A) and from 22 tumors of fad3-2, six tumors of fad7-1/8-1, and 24 tumors of Col-0 (B). Statistical analysis was performed using an unpaired two-tailed Student’s t test: **P ≤ 0.01.
DISCUSSION
This study presents a comprehensive analysis of the lipid profile of Arabidopsis crown galls, considering the role of two major desaturases in tumor physiology. The lipid profile of the crown gall tumor shows strong similarity to that of heterotrophic tissues such as roots, calli, and other nongreen tissues. In heterotrophic tissues, the eukaryotic phospholipid biosynthesis pathway of the ER is dominating (Somerville and Browse, 1991). As a matter of fact, in galls of Quercus palustris or Solidago altissima, the overall level of unsaturated FAs of the ER-derived phospholipid class was higher than in leaf tissue (Bayer, 1991; Tooker and De Moraes, 2009). Accordingly, we found unsaturated FAs of the eukaryotic pathway enriched in crown gall tumors, while glycerolipids of the prokaryotic pathway appeared reduced. This result comes as no surprise, since it reflects the considerably decreased number of chloroplasts in Arabidopsis crown galls (Deeken et al., 2006), which was also reported for sunflower (Helianthus annuus) tumors (Gee et al., 1967). Consequently, the chlorophyll content and activity of genes involved in photosynthetic light reactions are also reduced in Arabidopsis crown galls (Deeken et al., 2006).
SAD6 and FAD3 Shape the Lipid Profile of Crown Galls
The reduced photosynthesis in the crown gall tumor gives rise to a heterotrophic metabolism and results in hypoxia (Deeken et al., 2006). Despite the limited oxygen availability, oxygen-dependent FA desaturation is increased in crown galls. The expression of two desaturase genes, FAD3 and SAD6, was strongly induced, and 18:3 levels were elevated. The functional unknown SAD6 gene has highest homology to SSI2/FAB2 from Arabidopsis. SSI2/FAB2 belongs to a class of soluble enzymes of the chloroplast stroma that introduces the first double bond at the Δ9 position of stearic acid to form oleic acid (Lightner et al., 1994; Kachroo et al., 2001). In order to test SAD6 for desaturase function, the gene was expressed in Saccharomyces cerevisiae as well as E. coli (Supplemental Table S2). In contrast to SSI2, the SAD6 protein was active in neither of the two heterologous expression systems tested (Cao et al., 2010). Since plastidic desaturation is dependent on ferredoxin as an electron donor and distinct ferredoxin isoforms (heterotrophic and autotrophic) influence the activity of SAD enzymes differentially (Schultz et al., 2000), it seems likely that heterologous expression systems lack the correct version of this cofactor. Consequently, we analyzed SAD6 function via an in planta approach. A knockout mutant was not available, and application of RNAi, despite several attempts, never resulted in the complete inhibition of SAD6 expression. These findings may suggest that SAD6 is an essential gene during the plant life cycle.
This situation forced us to study the impact of SAD6 on FA desaturation with plants of a transactivation-tagged line (SAD6-OE). These plants expressed SAD6 to a similar level as crown galls and contained increased levels of oleic acid, whereas those of stearic acid, the substrate of SAD enzymes, were considerably reduced. A similar shift, but not that prominent, was also observed for palmitoleic acid (16:1) and palmitic acid (16:0) in these plants. As 16:0 is a substrate for the SSI2 desaturase (Kachroo et al., 2007), we cannot exclude that SAD6 can desaturate this unsaturated FA as well. It should be noted that a decrease in 18:0 was also reported for rice (Oryza sativa) overexpressing the rice ortholog of SSI2, OsSSI2 (Jiang et al., 2009). Down-regulation of SAD6 in the SAD6-OE line by RNAi resulted in SAD6-OE-RNAi plants that were characterized by a wild-type-like FA profile. A final confirmation of the involvement of SAD6 in FA desaturation was provided by the ssi2-2 mutant transformed with the SAD6 coding sequence under the control of the CaMV 35S promoter because the native promoter was only active under hypoxia. Strong SAD6 expression in the ssi-2-2 background rescued all mutant phenotypes, including wild-type levels of oleic acid and partial suppression of the high ssi2-2-associated stearic acid levels. For crown gall development, the absolute level of SAD6 expression seems not important, because SAD6-OE as well as SAD6-OE-RNAi developed wild-type-like crown galls. This finding suggests that the transcript levels of SAD6 are not important for crown gall development as long as the gene is expressed at all.
In crown galls, the relative amount of C18:1 was not altered to the extent found in SAD6-OE. Nevertheless, the average number of double bonds per FA in the tumor lipid pool was comparable to that found in SAD6-OE. This may be due to PUFA levels that likely result from the desaturation of MUFAs. Crown galls with strong SAD6 expression contained increased amounts of the ER-derived 18:3-containing glycerolipids. These elevated α-linolenic acid levels correlated with the strong induction of the FAD3 gene. Previously, it was shown that FAD3 synthesizes 18:3 in the ER and that transcript numbers and 18:3 levels correlate positively (Browse et al., 1993; Collados et al., 2006). Thus, FAD3 very likely is responsible for the elevated 18:3 contents in Arabidopsis tumors, as this tissue contains fewer chloroplasts and increased levels of lipids of the ER pathway. The finding that the fad3-2 mutant developed significantly smaller tumors supports the notion that 18:3 of the extraplastidic membranes are required for tumor growth. In contrast to FAD3, the plastidic desaturases FAD7 and FAD8 do not have an impact on the PUFA profile of crown galls. The FAD7 and FAD8 genes are down-regulated in tumors, and the double mutant, impaired in plastidic biosynthesis of 18:3, allows wild-type-like tumor growth. From the results obtained with the transgene and mutant approaches, we conclude that SAD6 and FAD3 shape the unsaturated FA profile in Arabidopsis tumors.
FAD3 and SAD6 Respond to Drought and Hypoxia in Crown Galls
Arabidopsis crown gall tumor development is accompanied by hypoxia and hydrogen peroxide production. Coincidentally, several genes encoding enzymes known to protect against oxidative stress, such as glutathione S-transferases and peroxidases, are strongly up-regulated (Deeken et al., 2006; Lee et al., 2009). Hypoxia has been shown to also affect the expression of desaturase genes (Upchurch, 2008). We found that SAD6 gene expression is induced in tissues experiencing hypoxic conditions, whereas transcripts were almost undetectable under normoxia. A hypoxia-dependent expression is also known for OLEIC ACID-REQUIRING1, an ER-localized Δ9-FAD from yeast (Kwast et al., 1998; Nakagawa et al., 2001; Vasconcelles et al., 2001). The promoters of the OLEIC ACID-REQUIRING1 and SAD6 genes share low-oxygen-responsive cis-activation elements. The apparent low partial oxygen pressure in crown gall tumors most likely causes the strong induction of SAD6 gene expression. From this observation, it is tempting to assume that SAD6 is operating in plastids of the root that more often experience hypoxia rather than the shoot.
Arabidopsis microarray data sets concerning hypoxia documented the induction of SAD6 under oxygen deprivation and also of the FAD3 gene (Liu et al., 2005; Loreti et al., 2005; Mustroph et al., 2009). Thus, both desaturases seem to come into play when oxygen is limited. Hypoxia and the recovery from hypoxia are usually associated with a ROS burst and damage of PUFAs (Crawford, 1992; Blokhina et al., 2003; Mène-Saffrané et al., 2009; Blokhina and Fagerstedt, 2010). Hypoxia/ROS tolerance of cells is linked to the capacity of cells to increase FA unsaturation in order to maintain the polar lipid content (Gigon et al., 2004). As the levels of desaturase gene expression and of 18:3 are significantly elevated in hypoxic and ROS-producing crown gall tumors, we assume an involvement of unsaturated FAs in hypoxia/ROS signaling.
In contrast to other plant organs, crown galls face permanent water stress in addition to hypoxia and oxidative stress (Schurr et al., 1996). To circumvent dehydration, abscisic acid levels are elevated and processes of drought stress adaption are active (Veselov et al., 2003; Efetova et al., 2007). Evaporation from the tumor is dependent on RH in the environment (Grantz, 1990), and with a drop in the water content of the air the abscisic acid content increases (Okamoto et al., 2009). In addition, water stress-tolerant plants up-regulate ω3-FAD genes and increase PUFA biosynthesis during drought stress adaptation (Gigon et al., 2004; Torres-Franklin et al., 2009). We found 45% smaller tumors on fad3-2 plants with a reduced trionic FA content when the relative water content of the atmosphere dropped below 50%. This finding indicates that the induction of FAD3 gene expression provides a key for drought stress signaling and guarantees long-term increased production of 18:3 FA.
In conclusion, this study provides experimental support for the hypothesis that an oxygen-consuming desaturase system, increasing the levels of unsaturated FAs, is involved in hypoxia and water stress signaling.
MATERIALS AND METHODS
Plant Material and Transformation
In these studies, the Arabidopsis (Arabidopsis thaliana) ecotypes Wassilewskija (Ws-2) and Col-0 were used and compared with the following mutants: (1) fad3-2 (AT2G29980, N8034; James and Dooner, 1990; Browse et al., 1993) and (2) fad7-1/fad8-1 (AT3G11170/AT5G05580, N8036; Gibson et al., 1994; McConn et al., 1994). T-DNA insertion lines for the SAD6 (AT1G43800) gene locus were ordered from GABI-Kat (gk30d04) and from the Nottingham Arabidopsis Stock Centre (salk70018, N70018). Also from the Nottingham Arabidopsis Stock Centre was the salk039862 line (N539862) with a T-DNA insertion in the gene locus AT2G43710 (SSI2). Since the latter T-DNA insertion line is a different mutant allele of the fab2 (Lightner et al., 1994) and ssi2 (Kachroo et al., 2001) alleles, it was named ssi2-2. The genotypes of the T-DNA insertion lines were confirmed by PCR with the following gene-specific primers: (1) gk30d04, left genomic primer 5′-CCATCCATCTAAAGA-3′ and right genomic primer 5′-CTCCATACATGATAAGAA-3′; and (2) salk039862, left genomic primer 5′-GTCGGAAGTGCTTCTTCTGTG-3′ and right genomic primer 5′-ATAAGAATGGGCCATCCTCTG-3′. The T-DNA-specific left border region primer for the GABI-Kat line was 5′-ATATTGACCATCATACTCATTGC-3′ and that for the SALK lines was 5′-ATTTTGCCGATTTCGGAAC-3′.
Arabidopsis was grown in soil (Einheitserde type P; http://www.einheitserde.de) and cultivated in growth chambers under short-day conditions with 120 µE of cool-white light (TL70 [Philips] and Osram 25W [Osram]) at 22°C (8 h) and 16°C during the dark period (16 h) with an RH of 45% to 55%. Nicotiana benthamiana, supplied by Karl Oparka (Scottish Crop Research Institute), was cultivated in a greenhouse with a day and night interval of 16 h (26°C) to 8 h (22°C) and a photon flux density of 600 µE of light.
Tumor development was induced by injecting a bacterial suspension with a syringe into the bases of Arabidopsis inflorescence stems. The bacterial suspension with the virulent nopaline-utilizing Agrobacterium tumefaciens strain C58 (no. 584; Max Planck Institute for Plant Breeding) in autoinducer bioassay (AB) minimal medium (50 mL of 20× AB salts [20 g L−1 NH4Cl, 6 g L−1 MgSO4·7H2O, 3 g L−1 KCl, 0.2 g L−1 CaCl2, and 15 mg L−1 FeSO4AB 7H2O], 2.4 mL L−1 0.5 m phosphate buffer [60 g L−1 K2HPO4 and 40 g L−1 NaH2PO4], and 3.9 g L−1 MES, pH 5.5). Tumor tissue was separated from inflorescence stems with a scalpel 3 weeks after infection together with inflorescence stems of the same age, only injected with AB minimal medium (mock injected).
Stable transformation of Arabidopsis was carried out by applying the floral dipping technique as described by Clough and Bent (1998). For transient transformation, N. benthamiana was used by applying the agroinfiltration technique according to Latz et al. (2007). N. benthamiana leaves were infiltrated with a mixture of the disarmed A. tumefaciens strain GV3101 (pMP90; Holsters et al., 1980; Koncz and Schell, 1986) and the 19K strain in order to prevent gene silencing. A. tumefaciens was cultivated overnight at 28°C and 150 rpm.
Generation of DNA Constructs for Arabidopsis Transformation
For tissue-specific expression studies, a promoter fragment of 1,303 bp 5′ upstream of the SAD6 coding sequence of Arabidopsis (Ws-2) was PCR amplified by using the following primers: forward, 5′-TTGTGTGTGAATCATTTGTTTGGTG-3′; reverse, 5′-TTTTTATGGTGAAGTAGAGAAGACTTTAATTTAC-3′. The SAD6 promoter PCR product was inserted into the entry vector pCR8/GW-Topo using the TOPO-TA cloning strategy according to the manufacturer’s instructions (Invitrogen). Applying the Gateway cloning strategy (Curtis and Grossniklaus, 2003), the SAD6 promoter fragment was transferred from the entry clone into the binary destination vector pMDC164 to be inserted 5′ upstream of the GUS coding sequence.
For protein localization studies, the coding region of the SAD6 gene was PCR amplified using the following primers (forward, 5′-CACCATGCTTGCGCACAAGTCTCT-3′; reverse, 5′-CACACTAATCTGCTTATCGA-3′) by omitting the stop codon. In order to obtain a translational fusion of the SAD6 protein with the V5 epitope and 6 histidins, the PCR product was inserted into the vector pcDNA3.1-D-V5/HIS-TOPO using the directional TOPO cloning strategy (Invitrogen). The SAD6::V5::HIS6 fusion was PCR amplified using the following primers (forward, 5′-GGCTTAAUCGGATCCAGTAC-3′; reverse, 5′-GGTTTAAUTCAATGGTGATGG-3′) and finally cloned into the binary pCAMBIA2300 vector (Cambia) by applying uracil excision-based cloning as described by Nour-Eldin et al. (2006).
For posttranscriptional silencing of the SAD6 gene, the vector pRS400 containing the Arabidopsis stem-loop-forming precursor microRNA319a (Schwab et al., 2006) was used. Three fragments of the microRNA319a with 21 SAD6-specific nucleotides (5′-TAATGTTCGAATAGCATCGGC-3′) forming the stem were PCR amplified by applying site-specific mutagenesis according to Schwab et al. (2006) with the following primers: fragment 1, forward primer A, 5′-CTGCAAGGCGATTAAGTTGGGTAAC-3′, and microRNA* reverse primer, 5′-GATAATGTTCCAATAGCATCGTCTCTACATATATATTCCT-3′; fragment 2, microRNA* forward primer, 5′-GAGACGATGCTATTGGAACATTATCACAGGTCGTGATATG-3′, and microRNA reverse primer, 5′-GAGCCGATGCTATTCGAACATTATCAAAGAGAATCAATGA-3′; fragment 3, microRNA forward primer, 5′-GATAATGTTCGAATAGCATCGGCTCTCTCTTTTGTATTCC-3′, and reverse primer B, 5′-GCGGATAACAATTTCACACAGGAAACAG-3′. The three overlapping PCR fragments were fused in a PCR using the primers A1 (5′-GGCTTAAUCTGCAAGGCGATTAAGTTGGGTAAC-3′) and A2 (5′-GGTTTAAUGCGGATAACAATTTCACACAGGAAACAG-3′) and inserted into the binary vector pCAMBIA2300 (Cambia) 3′ downstream of the CaMV 35S promoter applying the uracil excision-based cloning strategy as described by Nour-Eldin et al. (2006). For complementation of the salk039862 line with a T-DNA insertion in the SSI2/FAB2 locus, the coding region of the SAD6 gene was PCR amplified with the forward primer 5′-GGCTTAAUATGCTTGCGCACAAGTCTC-3′ and the reverse primer 5′-GGTTTAAUTTACACACTAATCTGCTTATCGAAGATC-3′. The PCR product was inserted into the binary vector pCAMBIA3300 (Cambia) 3′ downstream of the CaMV 35S promoter applying the uracil excision-based cloning strategy as described by Nour-Eldin et al. (2006). The binary plasmids were finally transformed into the disarmed A. tumefaciens strain GV3101 (pMP90; Koncz and Schell, 1986) by electroporation.
Quantitative Real-Time RT-PCR
Poly(A)-mRNA was isolated by applying the Dynabead technology according to the protocol of the Dynabeads mRNA Direct Kit (Invitrogen). In order to eliminate contaminations with genomic DNA, each sample was treated with Dynabeads twice. First-strand cDNA synthesis and quantitative real-time RT-PCR experiments were performed as described previously (Szyroki et al., 2001) using the LIGHTCYCLER 3.1 (Roche). The following primers were used: SAD6 forward, 5′-ATCTAATGACAGACGG-3′, and reverse, 5′-TGCTTATCGAAGATCCA-3′; SSI2 forward, 5′-CAGTTGGATACACGAC-3′, and reverse, 5′-GCTAAAAAGCCACATGA-3′; FAD3 forward, 5′-TCACGACATTGGAACT-3′, and reverse, 5′-ATTTGTCAGAAGCGTA-3′. The relative number of transcripts was normalized to 10,000 molecules of ACTIN2/ACTIN8 mRNA (An et al., 1996).
Lipid Extraction
Total lipid extraction was carried out by chloroform/methanol extraction from 100 mg of plant material according to Bligh and Dyer (1959). Membrane lipids were extracted from 150 mg of fresh plant material with 1.5 mL of 2-isopropanol at 80°C in 15 min. Samples were centrifuged for 10 min at 14,000 rpm, and the supernatant was collected. The remaining plant material was reextracted twice with 1.5 mL of chloroform:2-isopropanol (2:1, v/v) and 1.5 mL of chloroform:methanol (1:2, v/v). Samples were centrifuged after each extraction step, and the supernatants were combined. Extracts were dried under a stream of nitrogen at 60°C and reconstituted in 100 µL of methanol. Total FAs were analyzed after alkaline hydrolysis as described (Triantaphylidès et al., 2008).
Liquid Chromatography-Mass Spectrometry and Liquid Chromatography-Tandem Mass Spectrometry Analyses
Liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analyses were performed using an Acquity ultra-high-performance liquid chromatograph coupled to a Micromass Quattro Premier triple quadrupole mass spectrometer (Waters) with an electrospray interface. System operation and data acquisition were controlled using MassLynx version 4.1 software. LC-mass spectrometry analyses of FAs were performed by using an Agilent 1200 LC system coupled to the mass spectrometer mentioned above.
Chromatographic Conditions
Chromatographic separations of phospholipids, galactolipids, and arabidopsides were carried out using an Acquity BEH column (2.1 × 50 mm, 1.7-µm particle size with a guard column; Waters) with the following solvent systems: eluent A was 1 mm aqueous ammonium acetate and eluent B was 1 mm ammonium acetate in methanol. For phospholipid analysis, a gradient elution was performed on a BEH C18 column at a flow rate of 0.3 mL min–1 at 40°C as follows: 75% to 100% B in 10 min, followed by 100% B for 2 min, and reconditioning at 75% B for 3 min. For galactolipids and arabidopsides, a linear step gradient (0.3 mL min–1 flow rate, 30°C) on a BEH C8 column, starting from 75% B for 1 min, followed by 75% to 100% B in 10 min, 100% B for 1 min, and 75% B for 4 min, was used.
For the separation of FAs, a Purospher Star RP-18ec column (2 × 125 mm, 5-µm particle size with a 4- × 4-mm guard column; Merck) was used with the following solvent system: solvent A was 1 mm aqueous ammonium acetate and solvent B was acetonitrile. A gradient elution was performed at a flow rate of 0.2 mL min–1 from 70% to 100% B within 10 min, followed by 100% B for 5 min, and reconditioning at 75% B for 10 min.
Mass Spectrometric Conditions
For the analysis of galactolipids, the electrospray source was operated in the negative electrospray mode at 120°C and a capillary voltage of 2.25 kV. Nitrogen was used as desolvation and cone gas with flow rates of 850 L h–1 at 450°C and 50 L h–1, respectively. The cone voltage (CV) was adjusted to 40 V. Galactolipids were analyzed by multiple reaction monitoring (MRM) using argon as collision gas at a pressure of approximately 3 × 10−3 bar and a collision energy (CE) of 26 eV. The characteristic formation of FA fragments ([M − H]− → [FA]−) for each individual compound during collision-induced dissociation was monitored for a scan time of 0.025 s per transition. Quantification was carried out using 3 µg of MGDG-C18:0-C18:0 and 3 µg of DGDG-18:0-C18:0-DGDG as internal standards.
The determination of PC, PGs, and PIs was carried out in the negative electrospray mode. The capillary voltage was 3.25 kV, and the flow rates were adjusted to 800 L h–1 at 400°C for the desolvation gas and 50 L h–1 for the cone gas. The CV and CE were optimized for each class of phospholipids with the following parameters: CV of 40 V and CE of 26 eV for PCs; CV of 48 V and CE of 40 eV for PGs; and CV of 38 V and CE of 42 eV for PIs. The quantification of lipids was performed by monitoring specific transitions of lipids (MRM) using 2 µg of PC-17:0-17:0 and 1 µg of PG-17:0-17:0 and PI-16:0-16:0 as internal standards. Lipids with two different acyl chains in the sn-1 and sn-2 positions were determined by integration of the signals of both product ions derived from the loss of an acyl group from the precursor ion ([M − H+]− → [FA]−).
PEs were analyzed in the positive ionization mode. The parameters for ionization and collision-induced dissociation were optimized as follows: capillary voltage, 3.25 kV; cone voltage, 32 V; collision energy, 22 eV. The determination of PE was carried out by monitoring a specific MRM transition for each individual compound, resulting from a loss of PE ([M + H]+ → [M − P − EA]+), using 1 µg of PE-18:0-18:0 as an internal standard.
For FA analyses, the electrospray source was operated in the negative ionization mode at a capillary voltage of 3.5 kV and a source temperature of 120°C. The optimal mass spectrometry parameters for the detection of FAs were as follows: desolvation gas, 800 L h–1 at 400°C; cone gas, 100 L h–1; cone voltage, 30 V. Quantification was performed using selected ion recording for FA carboxylate anions with a scan time of 0.1 s for each compound. Heptadecanoid acid was used as an internal standard (250 ng).
Protein Extraction from Chloroplasts and Western-Blot Analysis
Isolation of chloroplasts from transiently transformed N. benthamiana leaves was carried out as described by Pineda et al. (2010) with minor modifications. Chloroplasts were pelleted with 7,000g at 4°C for 1 min and then shock frozen in liquid nitrogen in order to break the chloroplasts.
Total soluble proteins were extracted from the chloroplast pellet or N. benthamiana leaf tissue, also frozen in liquid nitrogen. The frozen material was resuspended in an equal volume of extraction buffer (10% (v/v) glycerol, 10 mm dithiothreitol, 5 mm EDTA, and 100 mm HEPES, pH 7.2) containing a protease inhibitor cocktail (Complete Mini EGTA-free; Roche) and centrifuged at 15,000g for 10 min at 4°C. The supernatant with the soluble protein fraction (10 µg) was size fractionated in an SDS-PAGE gradient (8%–16%, w/v). Protein bands were stained with 0.006% (w/v) Coomassie Brilliant Blue R-250 in 10% (v/v) acetic acid. Proteins were electrotransferred to a polyvinylidene difluoride membrane (Amersham Hybond-P, GE Healthcare Life Sciences). Polyvinylidene difluoride membranes were blocked by incubation in Tris-buffered saline buffer (150 mm NaCl and 50 mm Tris, pH 7.8) containing 3% (w/v) low-fat milk powder at room temperature for 0.5 h. After three washings for 5 min in Tris-buffered saline buffer, membranes were incubated with mouse anti-V5 monoclonal antibodies (1:5,000 dilution; Invitrogen) in Tris-buffered saline plus Tween 20 (TBST; 150 mm NaCl, 50 mm Tris, pH 7.8, and 0.05% (v/v) Tween 20) for 1 h at room temperature. Blots were washed in TBST three times for 5 min and incubated with the goat anti-mouse secondary antibody conjugated to horseradish peroxidase (1:20,000 dilution; Pierce) for 1 h at room temperature. Excess secondary antibody was removed by three washings in TBST buffer for 5 min. Peroxidase activity was detected by using supersignal chemiluminescent substrate luminol (Pierce) and exposure to x-ray films (Kodak-Biomax Light).
Histochemical GUS Assay
GUS activity was assayed in Arabidopsis (Ws-2) homozygous for the T-DNA containing GUS under the control of the SAD6 promoter (AT1G43800). GUS-expressing plant tissue was vacuum infiltrated (100 mbar, 10 min) with GUS staining buffer (100 mm sodium phosphate, pH 7.0, 10 mm EDTA, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, and 0.1% [v/v] Triton X-100) containing 1 mm 5-bromo-4-chloro-3-indolyl glucuronide (Duchefa) and incubated at 37°C in the dark for 0.5 h. Three independent transgenic plant lines were analyzed. GUS staining was inspected and documented with a digital microscope (VHX-100; Keyence) and a fluorescence stereomicroscope (MZ FLIII; Leica) equipped with a CCD camera (Spot Insight Color 3.2.0; Diagnostic Instruments).
Application of Hypoxia and Waterlogging Stress
Arabidopsis plants were placed in a desiccator and flushed with a mixture of 2 mL min−1 air and 80 mL min−1 nitrogen (Messer Griesheim) containing 0.5% oxygen for 24 h. Flow controllers (FC-260; Tylan General) were used to adjust the gas flows. Control plants were flushed with 82 mL min−1 air. Waterlogging stress was applied in a water-filled basin by flooding the roots but not the shoots of Arabidopsis plants grown in pots filled with soil for 24 h. Pots with control plants were placed in a basin without water for the same time.
Expression and Analysis of SAD6 in Yeast and Escherichia coli Cells
The analysis of SAD6 in different yeast (Saccharomyces cerevisiae) strains was performed as described previously (Ahmann et al., 2011). For SAD6 protein expression in E. coli cells, strain Rosetta DE3 pLysS and the vector pET28a (both from Novagen) were applied according to the protocol described previously by Cao et al. (2010).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcriptional activity of three FAD genes under varying environmental stress conditions in different Arabidopsis tissues and genotypes.
Supplemental Figure S2. Two different Arabidopsis lines (Col-0) with T-DNA insertions in the SAD6 (AT1G43800) and SSI2 (AT2G43710) loci.
Supplemental Figure S3. The SAD6 protein is in the chloroplast fraction.
Supplemental Figure S4. Hypoxia-responsive cis-acting elements in the SAD6 promoter.
Supplemental Table S1. Relative changes in the unsaturation indexes of FAs.
Supplemental Table S2. Heterologous systems used for SAD6 protein expression in yeast and E. coli strains.
Acknowledgments
We thank the following colleagues from the University of Wuerzburg for their support: Dietmar Geiger and Chil-Woo Lee for molecular cloning, Stefan Ruemer for hypoxia experiments, Pablo Tarrazona Corrales, Cornelia. Herrfurth, and Kirstin Feussner for glycerolipid and sphingolipid analysis, Katharina Ahmann, Ellen Hornung, and Veronika Behnen for SAD6 analysis in yeast, and Werner Kaiser for critically reading the manuscript. We thank Detlef Weigel (Max Planck Institute for Developmental Biology, Tuebingen) for providing the vector PSR300 as well as the GABI-Kat, the Max Planck Institute for Plant Breeding, the Genomic Analysis Laboratory of the SALK Institute for Biological Studies, and the European Arabidopsis Stock Center for the sequence-indexed Arabidopsis T-DNA insertion mutants. The lipid analysis was performed in cooperation with the Metabolomics Core Facility of the University of Wuerzburg.
Glossary
- T-DNA
transfer DNA
- PUFA
polyunsaturated fatty acid
- FA
fatty acid
- DGDG
digalactosyl diglyceride
- MGDG
monogalactosyl diglyceride
- ROS
reactive oxygen species
- MUFA
monounsaturated fatty acid
- ER
endoplasmic reticulum
- RNAi
RNA interference
- PG
phosphatidylglycerol
- PC
phosphytidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- Col-0
ecotype Columbia
- S-ACP
stearoyl-acyl carrier protein
- RT
reverse transcription
- cDNA
copy DNA
- RH
relative humidity
- CaMV
cauliflower mosaic virus
- Ws-2
ecotype Wassilewskija
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- CV
cone voltage
- MRM
multiple reaction monitoring
- CE
collision energy
- TBST
Tris-buffered saline plus Tween 20
References
- Ahmann K, Heilmann M, Feussner I. (2011) Identification of a Δ4-desaturase from the microalga Ostreococcus lucimarinus. Eur J Lipid Sci Technol 113: 832–840 [Google Scholar]
- An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 [DOI] [PubMed] [Google Scholar]
- Bayer MH. (1991) Fatty acid composition of galactolipids and phospholipids in neoplasmatic plant tissues (cecidia) and normal leaf tissue. Physiol Plant 81: 313–318 [Google Scholar]
- Berberich T, Harada M, Sugawara K, Kodama H, Iba K, Kusano T. (1998) Two maize genes encoding omega-3 fatty acid desaturase and their differential expression to temperature. Plant Mol Biol 36: 297–306 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Blokhina O, Fagerstedt KV. (2010) Oxidative metabolism, ROS and NO under oxygen deprivation. Plant Physiol Biochem 48: 359–373 [DOI] [PubMed] [Google Scholar]
- Blokhina O, Virolainen E, Fagerstedt KV. (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot (Lond) 91: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DJ, Beevers H. (1987) Fatty acids of rice coleoptiles in air and anoxia. Plant Physiol 84: 555–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, McConn M, James D, Jr, Miquel M. (1993) Mutants of Arabidopsis deficient in the synthesis of alpha-linolenate: biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J Biol Chem 268: 16345–16351 [PubMed] [Google Scholar]
- Cahoon EB, Mills LA, Shanklin J. (1996) Modification of the fatty acid composition of Escherichia coli by coexpression of a plant acyl-acyl carrier protein desaturase and ferredoxin. J Bacteriol 178: 936–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Shah S, Shanklin J, Browse J. (1998) A determinant of substrate specificity predicted from the acyl-acyl carrier protein desaturase of developing cat’s claw seed. Plant Physiol 117: 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Xian M, Yang J, Xu X, Liu W, Li L. (2010) Heterologous expression of stearoyl-acyl carrier protein desaturase (S-ACP-DES) from Arabidopsis thaliana in Escherichia coli. Protein Expr Purif 69: 209–214 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collados R, Andreu V, Picorel R, Alfonso M. (2006) A light-sensitive mechanism differently regulates transcription and transcript stability of omega3 fatty-acid desaturases (FAD3, FAD7 and FAD8) in soybean photosynthetic cell suspensions. FEBS Lett 580: 4934–4940 [DOI] [PubMed] [Google Scholar]
- Crawford R. (1992) Oxygen availability as an ecological limit to plant distribution. Adv Ecol Res 23: 93–185 [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakhma WS, Zarrouk M, Cherif A. (1995) Effects of drought-stress on lipids in rape leaves. Phytochemistry 40: 1383–1386 [Google Scholar]
- Deeken R, Engelmann JC, Efetova M, Czirjak T, Müller T, Kaiser WM, Tietz O, Krischke M, Mueller MJ, Palme K, et al. (2006) An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell 18: 3617–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efetova M, Zeier J, Riederer M, Lee CW, Stingl N, Mueller M, Hartung W, Hedrich R, Deeken R. (2007) A central role of abscisic acid in drought stress protection of Agrobacterium-induced tumors on Arabidopsis. Plant Physiol 145: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Ajjawi I, Manoli A, Sawin A, Xu C, Froehlich JE, Last RL, Benning C. (2009) FATTY ACID DESATURASE4 of Arabidopsis encodes a protein distinct from characterized fatty acid desaturases. Plant J 60: 832–839 [DOI] [PubMed] [Google Scholar]
- Gee MM, Sun CN, Dwyer JD. (1967) An electron microscope study of sunflower crown gall tumor. Protoplasma 64: 195–200 [Google Scholar]
- Gibson S, Arondel V, Iba K, Somerville C. (1994) Cloning of a temperature-regulated gene encoding a chloroplast omega-3 desaturase from Arabidopsis thaliana. Plant Physiol 106: 1615–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigon A, Matos AR, Laffray D, Zuily-Fodil Y, Pham-Thi AT. (2004) Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (ecotype Columbia). Ann Bot (Lond) 94: 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantz DA. (1990) Plant response to atmospheric humidity. Plant Cell Environ 13: 667–679 [Google Scholar]
- Holsters M, Silva B, Van Vliet F, Genetello C, De Block M, Dhaese P, Depicker A, Inzé D, Engler G, Villarroel R, et al. (1980) The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid 3: 212–230 [DOI] [PubMed] [Google Scholar]
- James DW, Dooner HK. (1990) Isolation of EMS-induced mutants in Arabidopsis altered in seed fatty acid composition. Theor Appl Genet 80: 241–245 [DOI] [PubMed] [Google Scholar]
- Jia SR, Kumar PP, Kush A. (1996) Oxidative stress in Agrobacterium-induced tumors on Kalanchoe plants. Plant J 10: 545–551 [Google Scholar]
- Jiang CJ, Shimono M, Maeda S, Inoue H, Mori M, Hasegawa M, Sugano S, Takatsuji H. (2009) Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol Plant Microbe Interact 22: 820–829 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Shanklin J, Whittle E, Lapchyk L, Hildebrand D, Kachroo P. (2007) The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol Biol 63: 257–271 [DOI] [PubMed] [Google Scholar]
- Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF. (2001) A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc Natl Acad Sci USA 98: 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Veena, Gelvin SB. (2007) Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J 51: 779–791 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kwast KE, Burke PV, Poyton RO. (1998) Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J Exp Biol 201: 1177–1195 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics. 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Latz A, Ivashikina N, Fischer S, Ache P, Sano T, Becker D, Deeken R, Hedrich R. (2007) In planta AKT2 subunits constitute a pH- and Ca2+-sensitive inward rectifying K+ channel. Planta 225: 1179–1191 [DOI] [PubMed] [Google Scholar]
- Lee CW, Efetova M, Engelmann JC, Kramell R, Wasternack C, Ludwig-Müller J, Hedrich R, Deeken R. (2009) Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell 21: 2948–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner J, Wu J, Browse J. (1994) A mutant of Arabidopsis with increased levels of stearic acid. Plant Physiol 106: 1443–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Vantoai T, Moy LP, Bock G, Linford LD, Quackenbush J. (2005) Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol 137: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los DA, Murata N. (1998) Structure and expression of fatty acid desaturases. Biochim Biophys Acta 1394: 3–15 [DOI] [PubMed] [Google Scholar]
- McConn M, Hugly S, Browse J, Somerville C. (1994) A mutation at the fad8 locus of Arabidopsis identifies a second chloroplast [omega]-3 desaturase. Plant Physiol 106: 1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mène-Saffrané L, Dubugnon L, Chételat A, Stolz S, Gouhier-Darimont C, Farmer EE. (2009) Nonenzymatic oxidation of trienoic fatty acids contributes to reactive oxygen species management in Arabidopsis. J Biol Chem 284: 1702–1708 [DOI] [PubMed] [Google Scholar]
- Mikami K, Murata N. (2003) Membrane fluidity and the perception of environmental signals in cyanobacteria and plants. Prog Lipid Res 42: 527–543 [DOI] [PubMed] [Google Scholar]
- Monteiro de Paula F, Pham-Thi A, Zuily-Fodil Y, Ferrari-Iliou R, Vieira da Silva J, Mazliak P. (1993) Effect of water stress on the biosynthesis and degradation of polyunsaturated lipid molecular species in leaves of Vigna unguiculata. Plant Physiol Biochem 31: 707–715 [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, Sugioka S, Kaneko Y, Harashima S. (2001) O2R, a novel regulatory element mediating Rox1p-independent O2 and unsaturated fatty acid repression of OLE1 in Saccharomyces cerevisiae. J Bacteriol 183: 745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Nørholm MH, Jensen JK, Halkier BA. (2006) Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Tanaka Y, Abrams SR, Kamiya Y, Seki M, Nambara E. (2009) High humidity induces abscisic acid 8′-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis. Plant Physiol 149: 825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda M, Sajnani C, Barón M. (2010) Changes induced by the Pepper mild mottle tobamovirus on the chloroplast proteome of Nicotiana benthamiana. Photosynth Res 103: 31–45 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Hirt H. (2010) New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J 29: 1021–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DJ, Suh MC, Ohlrogge JB. (2000) Stearoyl-acyl carrier protein and unusual acyl-acyl carrier protein desaturase activities are differentially influenced by ferredoxin. Plant Physiol 124: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr U, Schuberth B, Aloni R, Pradel KS, Schmundt D, Jaehne B, Ullrich CI. (1996) Structural and functional evidence for xylem-mediated water transport and high transpiration in Agrobacterium tumefaciens-induced tumors of Ricinus communis. Bot Acta 109: 405–411 [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, Somerville C. (1991) Stearoyl-acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proc Natl Acad Sci USA 88: 2510–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Browse J. (1991) Plant lipids: metabolism, mutants, and membranes. Science 252: 80–87 [DOI] [PubMed] [Google Scholar]
- Szyroki A, Ivashikina N, Dietrich P, Roelfsema MR, Ache P, Reintanz B, Deeken R, Godde M, Felle H, Steinmeyer R, et al. (2001) KAT1 is not essential for stomatal opening. Proc Natl Acad Sci USA 98: 2917–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF, Nutter R, Montoya AL, Gordon MP, Nester EW. (1980) Integration and organization of Ti plasmid sequences in crown gall tumors. Cell 19: 729–739 [DOI] [PubMed] [Google Scholar]
- Thompson GA, Scherer DE, Foxall-Van Aken S, Kenny JW, Young HL, Shintani DK, Kridl JC, Knauf VC. (1991) Primary structures of the precursor and mature forms of stearoyl-acyl carrier protein desaturase from safflower embryos and requirement of ferredoxin for enzyme activity. Proc Natl Acad Sci USA 88: 2578–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooker JF, De Moraes CM. (2009) A gall-inducing caterpillar species increases essential fatty acid content of its host plant without concomitant increases in phytohormone levels. Mol Plant Microbe Interact 22: 551–559 [DOI] [PubMed] [Google Scholar]
- Torres-Franklin ML, Repellin A, Huynh VB, d’Arcy-Lameta A, Zuily-Fodil Y, Pham-Thi AT. (2009) Omega-3 fatty acid desaturase (FAD3, FAD7, FAD8) gene expression and linolenic acid content in cowpea leaves submitted to drought and after rehydration. Environ Exp Bot 65: 162–169 [Google Scholar]
- Triantaphylidès C, Krischke M, Hoeberichts FA, Ksas B, Gresser G, Havaux M, Van Breusegem F, Mueller MJ. (2008) Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol 148: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upchurch RG. (2008) Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol Lett 30: 967–977 [DOI] [PubMed] [Google Scholar]
- Vasconcelles MJ, Jiang Y, McDaid K, Gilooly L, Wretzel S, Porter DL, Martin CE, Goldberg MA. (2001) Identification and characterization of a low oxygen response element involved in the hypoxic induction of a family of Saccharomyces cerevisiae genes: implications for the conservation of oxygen sensing in eukaryotes. J Biol Chem 276: 14374–14384 [DOI] [PubMed] [Google Scholar]
- Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuïnk TJ, Hooykaas PJ. (2000) VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290: 979–982 [DOI] [PubMed] [Google Scholar]
- Vergunst AC, van Lier MC, den Dulk-Ras A, Hooykaas PJ. (2003) Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol 133: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselov D, Langhans M, Hartung W, Aloni R, Feussner I, Götz C, Veselova S, Schlomski S, Dickler C, Bächmann K, et al. (2003) Development of Agrobacterium tumefaciens C58-induced plant tumors and impact on host shoots are controlled by a cascade of jasmonic acid, auxin, cytokinin, ethylene and abscisic acid. Planta 216: 512–522 [DOI] [PubMed] [Google Scholar]
- Whittle E, Cahoon EB, Subrahmanyam S, Shanklin J. (2005) A multifunctional acyl-acyl carrier protein desaturase from Hedera helix L. (English ivy) can synthesize 16- and 18-carbon monoene and diene products. J Biol Chem 280: 28169–28176 [DOI] [PubMed] [Google Scholar]
- Zhang M, Barg R, Yin M, Gueta-Dahan Y, Leikin-Frenkel A, Salts Y, Shabtai S, Ben-Hayyim G. (2005) Modulated fatty acid desaturation via overexpression of two distinct omega-3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J 44: 361–371 [DOI] [PubMed] [Google Scholar]