microRNA-targeted BrpTCP genes modulate the head shape in B. rapa by differential cell division arrest in leaf regions.
Abstract
Leafy heads of cabbage (Brassica oleracea), Chinese cabbage (Brassica rapa), and lettuce (Lactuca sativa) are composed of extremely incurved leaves. The shape of these heads often dictates the quality, and thus the commercial value, of these crops. Using quantitative trait locus mapping of head traits within a population of 150 recombinant inbred lines of Chinese cabbage, we investigated the relationship between expression levels of microRNA-targeted Brassica rapa ssp. pekinensis TEOSINTE BRANCHED1, cycloidea, and PCF transcription factor4 (BrpTCP4) genes and head shape. Here, we demonstrate that a cylindrical head shape is associated with relatively low BrpTCP4-1 expression, whereas a round head shape is associated with high BrpTCP4-1 expression. In the round-type Chinese cabbage, microRNA319 (miR319) accumulation and BrpTCP4-1 expression decrease from the apical to central regions of leaves. Overexpression of BrpMIR319a2 reduced the expression levels of BrpTCP4 and resulted in an even distribution of BrpTCP4 transcripts within all leaf regions. Changes in temporal and spatial patterns of BrpTCP4 expression appear to be associated with excess growth of both apical and interveinal regions, straightened leaf tips, and a transition from the round to the cylindrical head shape. These results suggest that the miR319a-targeted BrpTCP gene regulates the round shape of leafy heads via differential cell division arrest in leaf regions. Therefore, the manipulation of miR319a and BrpTCP4 genes is a potentially important tool for use in the genetic improvement of head shape in these crops.
Chinese cabbage (Brassica rapa), cabbage (Brassica oleracea), and lettuce (Lactuca sativa) have uniform, tightly formed heads composed of yellow-green, crinkly leaves. These leaves serve as an important source of mineral nutrients, crude fiber, and vitamins in the human diet. Head shape can vary depending on the cultivar; the two most common shapes are round and cylindrical, with others often being oblong or cone like. Chinese cabbage is commonly sold by head shape and color rather than by individual varieties. The favorable shape often changes according to current consumer preference.
Leaf curvature is essential for the formation of the leafy head. During vegetative growth of plants, leaf growth occurs via primary growth to promote cell proliferation and secondary growth to increase cell size (Donnelly et al., 1999; Horiguchi et al., 2006). During this process, the cell division arrest front is convex and moves gradually from tip to base along the longitudinal axis, resulting in a flat leaf. The CINCINNATA (CIN) gene in Antirrhinum majus is a major regulator of cell division arrest in leaves. In leaves of the cin mutant, the arrest front becomes concave and moves down more slowly than in wild-type leaves. As a consequence, the excessive marginal cell proliferation produces a wrinkled leaf (Nath et al., 2003; Andriankaja et al., 2012). Among the 24 TCP family genes in Arabidopsis (Arabidopsis thaliana), only eight members belong to the CIN-class TCP genes. Due to the gene redundancy, a single mutation of the individual CIN-like TCP genes does not generate visible phenotypes, whereas multiple mutations (or the negative dominant mutants) exhibit wrinkled leaves in a dose-dependent manner. In addition to the transcriptional regulation of gene expression, five out of the eight CIN-class TCP genes are also under the posttranscriptional regulation of microRNAs (miRNAs; Palatnik et al., 2003). For example, the dramatically enhanced miR319 in the Arabidopsis jagged and wavy-D (jaw-D) mutants results in the suppression of its target genes TCP2, TCP3, TCP4, TCP10, and TCP24 and the development of a cin-like phenotype. By contrast, ectopic expression of microRNA319 (miR319)-resistant TCP genes induces leaf fusion and causes defects in the shoot apical meristem. According to the digital differentiation index, the primary role of CIN-class TCPs is as negative regulators for cell proliferation that cause premature leaf differentiation (Efroni et al., 2008). Thus, the temporal and spatial regulation of TCP genes is crucial for determining the total number of leaf cells and the final leaf shape. In Arabidopsis, appropriate miRNA accumulation depends on the activity of the nuclear protein HYPONASTIC LEAVES1 (HYL1; Han et al., 2004; Vaucheret et al., 2004). Nearly all null mutants of hyl1 mutants show leaf incurvature in the transverse direction (Wu et al., 2007; Liu et al., 2011).
In the previous study, we took advantage of recent advances in genome resequencing to perform quantitative trait locus (QTL) mapping using 150 recombinant inbred lines (RILs) derived from the cross between heading and nonheading Chinese cabbage (Yu et al., 2013). Among the 15 QTLs obtained for head traits, three QTLs were identified for head shape indices (head height-to-diameter ratio). Unfortunately, it would take some time to map the genes that control head shape using these QTLs. Thus, as an alternative approach, we divided the RIL population into several subpopulations according to head compactness and head shape. We then analyzed the expression of a subset of miRNA target genes involved in leaf curvature. Our analyses indicated that the cylindrical shape of the leafy head is associated with relatively low expression of the microRNA (miR) 319a-targeted Brassica rapa ssp. pekinensis TEOSINTE BRANCHED1, cycloidea, and PCF transcription factor4 (BrpTCP4) gene. The transgenic plants that overexpress the BrpMIR319a2 gene suggest that the miR319-targeted TCP4 genes regulate the shape and size of leafy heads.
RESULTS
Head Shape in Chinese Cabbage
The vegetative growth of Chinese cabbage goes through four stages: seedling, rosette, folding, and heading. Leafy heads can be divided into four shapes: round, oblong, cylindrical, and cone like. The two most common shapes are round and cylindrical. Overall head shape can vary depending on leaf shape, size, and curvature. To investigate the genetic basis of head shape, we used a population of 150 RILs developed from a cross between cv Bre (heading Chinese cabbage) and cv Wut (nonheading Chinese cabbage; Yu et al., 2013). Bre and Wut are the two representative cultivars of heading and nonheading Chinese cabbage (Supplemental Fig. S1). Compared with cv Wut leaves, the interior leaves inside the leafy heads of cv Bre plants are larger, incurved, and pale green, with relatively small inclination angles and short petioles.
A total of 150 RILs exhibited significant variation in head compactness and head shape. Of these, 58 RILs produced compact heads, whereas 92 RILs formed small, immature heads or failed to form any head (Supplemental Table S1). The compact heads differed in shape between the RILs. Within the RIL population, the proportion of the four shapes (cone like, cylindrical, oblong, and round) also differed.
We examined the relationship of head shape with forms of rosette leaves at the heading stage. Eighty percent of the RILs with round heads possessed flat rosette leaves (Fig. 1, A and E; Supplemental Table S1); 60% of the RILs with cylindrical heads possessed wavy margins of rosette leaves (Fig. 1, B and F); 100% of the RILs within cone-like heads possessed incurved rosette leaves (Fig. 1, C and G); and 74% of the RILs with oblong heads possessed shrinking rosette leaves (Fig. 1, D and H). Interestingly, the association of wavy leaf margins with the cylindrical head was reminiscent of cin mutants of Antirrhinum majus and jaw-D mutants of Arabidopsis (Nath et al., 2003; Palatnik et al., 2003).
Figure 1.
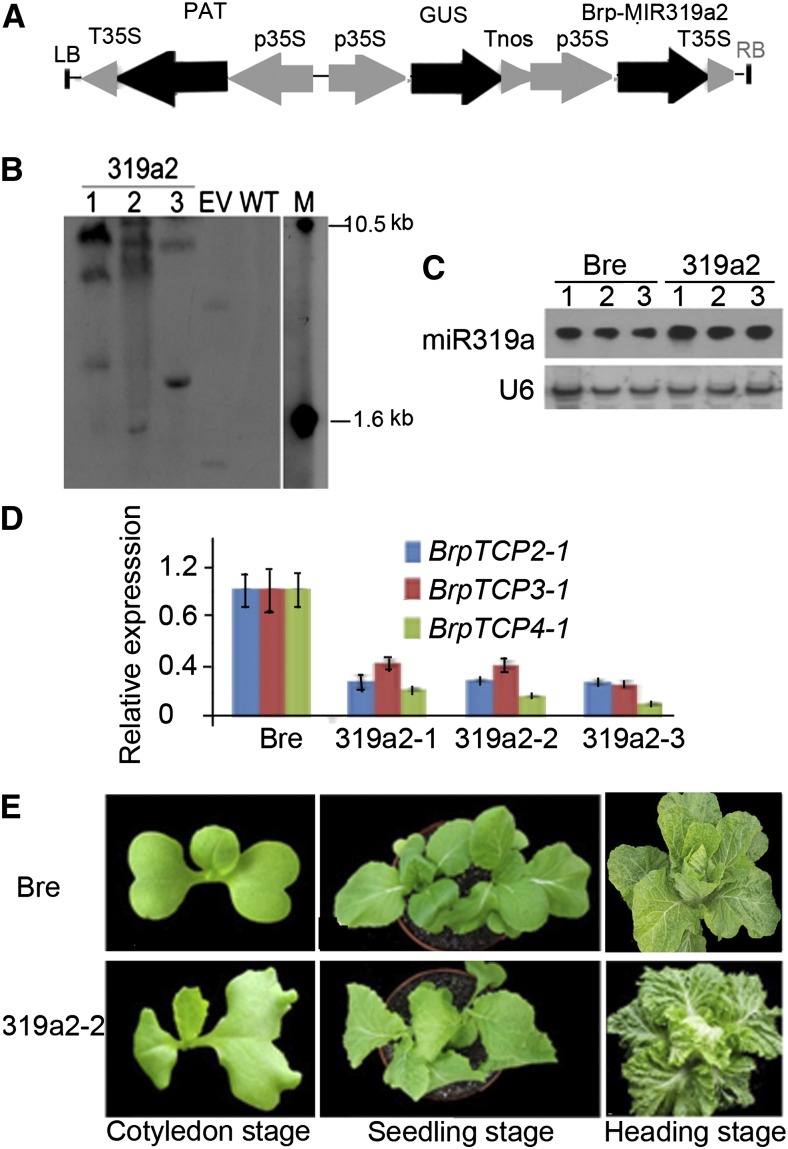
The rosettes and leafy heads of the representative RILs and the transgenic plants overexpressing the BrpMIR319a gene. A to D, The plants of RILs with round (A), oblong (B), cylindrical (C), and cone-like (D) heads at the rosette stage. E to H, The round (E), oblong (F), cylindrical (G), and cone-like (H) heads. I and J, The plant (I) and head (J) of cv Bre. K and L, The plant (K) and head (L) of 319a2-2. M and N, The plants of 319a2-1 (M) and miR319a2-3 (N). O and P, Cross sections of shoot apex of the wild type (O) and 319a2-2 (P) at the heading stage. hl, Head leaf; rl, rosette leaf; s, stalk; yhl, young head leaf. Bars = 5 cm in A to L and 0.5 mm in M and N. [See online article for color version of this figure.]
Sequences and Expression of BrpTCP Genes in Chinese Cabbage
In cin mutants of Antirrhinum majus and jaw-D mutants of Arabidopsis, silencing of either CIN genes and miR319-targeted genes, respectively, causes excess growth on marginal regions of leaves and the gradual introduction of negative leaf curvature and wavy leaf margins (Nath et al., 2003; Palatnik et al., 2003). To identify the possible mutation of miR319 and miR319-targeted BrpTCP genes that may affect leaf curvature and head shape, we analyzed the homology of the related genes between Chinese cabbage and Arabidopsis. In Arabidopsis, there were one miR319a gene and five TCP target genes (Palatnik et al., 2003). The alignment of Arabidopsis miR319a precursors with the published genomic sequences of Chinese cabbage (var Chiifu-401-42; Wang et al., 2011) revealed the existence of eight copies of MIR319 in Chiifu-401-42 (Fig. 2A). These copies were classified into three groups according to Arabidopsis homologs. The numbers of BraMIR319a, BraMIR319b, and BraMIR319c were three, two, and three, respectively. Within the BraMIR319a group of Chiifu-401-42, three miRNA genes were highly conserved (more than 99% homology). The alignment of AtTCP genes with the published genomic sequences of Chinese cabbage revealed two BraTCP2, one BraTCP3, three BraTCP4, one BraTCP10, and two BraTCP24 genes (Fig. 2B). These copies were classified into five subfamilies.
Figure 2.
Phylogenetic trees of BrpMIR319 and miR319a-targeted BrpTCP genes. A, Multiple sequence alignment of BrpMIR319 genes with Arabidopsis and Chiifu-401-42 homologous genes. B, Multiple sequence alignment of BrpTCP genes with Arabidopsis and Chiifu-401-42 homologs. C, Multiple sequence alignment and complementation of mature miRNAs in BrpMIR319a and the complementary sequences in BrpTCP genes. The phylogenetic trees are constructed using the maximum likelihood method based on the Tamura-Nei model by MEGA5. The sequences of AtMIR159a and AtTCP1 are designated as the out group.
Using the sequences of pre-miR319 and TCP genes in Arabidopsis and their homologs in Chiifu-401-42, we searched for homologous genomic sequences and transcripts of BrpMIR319a and BrpTCP genes within our genome resequencing data of cv Bre (Yu et al., 2013). Compared with AtMIR319a, BrpMIR319a2 in cv Bre contains one nucleotide polymorphism at the site of mature miR319a, which leads to an increase in sequence complementary to target genes. This change may reduce the expression levels of some BrpTCP genes in cv Bre plants. There were several nucleotide polymorphisms in sequences of BrpTCP genes relative to those in Chiifu-401-42. At the 14th nucleotide of the miRNA-binding site in BrpTCP4-3, there was a C-to-T substitution, increasing its sequence complementary to miR319 (Fig. 2C). This substitution may result in a change in the expression level of BrpTCP4-3 in cv Bre plants.
We designed primers specific for the BrpTCP4-1 gene and performed real-time PCR across 150 RIL lines using developing leaves (less than 1 cm in length) at the early heading stage. The BrpTCP4-1 gene in cv Bre (female parent) leaves was down-regulated 1.6 times relative to that of cv Wut (male parent) leaves (Supplemental Table S2). The expression levels of the BrpTCP4-1 gene varied greatly across the 150 RILs (Fig. 3A). To investigate the effects of BrpTCP genes on the formation of leafy heads, we divided the RIL population into two subpopulations according to head compactness. Fifty-eight RILs had compact heads and hence were classified as the “compact” subpopulation, whereas 92 RILs had loose heads or a nonheading phenotype and hence were classified as the “loose” subpopulation. Using a box plot (Hoffmann, 1995) and the Kruskal-Wallis test (Kruskal and Wallis, 1952), we demonstrated that the difference in the medians of BrpTCP4-1 expression between the compact (Sub1) and loose (Sub2) subpopulations was not significant (Fig. 3B), indicating that head compactness was not associated with BrpTCP4 expression. According to head shape, we divided the Sub1 subpopulation further into four subpopulations: Sub1-C (cylindrical), Sub1-N (cone like), Sub1-O (oblong), and Sub1-R (round). Real-time PCR indicated that the median of BrpTCP4-1 expression in the Sub1-C subpopulation was much lower compared with BrpTCP4-1 expression in cv Bre, whereas the medians of BrpTCP4-1 expression in the Sub1-N, Sub1-O, and Sub1-R subpopulations were higher (Fig. 3C; Supplemental Table S1). The Kruskal-Wallis test showed that the median of BrpTCP4-1 expression in the Sub1-C subpopulation was highly significantly different from those of the Sub1-N, Sub1-O, and Sub1-R subpopulations (Fig. 3D). Noticeably, the difference between the Sub1-C and Sub1-R subpopulations was extremely significant (P value threshold of 1 × 10−6). On the other hand, the differences in the medians of BrpTCP4-1 expression between the Sub1-N, Sub1-O, and Sub1-R subpopulations were not significant. These results suggest that the subpopulation with the cylindrical head shape is separated from those with the cone-like, oblong, and round shapes.
Figure 3.
Box plots and Kruskal-Wallis test for differences between the RIL subpopulations and BrpTCP4-1 expression. The data are from Supplemental Table S2. A, Composition of the RIL population and subpopulations. Sub1 and Sub2 represent the compact and loose subpopulations, respectively. Sub1-C, Sub1-N, Sub1-O, and Sub1-R represent the cylindrical, cone-like, oblong, and round subpopulations, respectively. The numbers of RILs are shown in parentheses. B and C, Box plots for the compact and loose subpopulations (B) and for the cylindrical, cone-like, oblong, and round subpopulations (C). On each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles, and the whiskers extend to the most extreme data points. Asterisks indicate the relative expression levels of BrpTCP4-1. D, P values using the Kruskal-Wallis test.
Silencing of BrpTCP Genes by BrpMIR319a in Round-Headed Chinese Cabbage
To examine the putative function of miR319a-targeted BrpTCP genes in head shape, we cloned the cv Bre BrpMIR319a2 gene and inserted it into a binary vector under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Fig. 4A). We modified the vacuum infiltration method of Clough and Bent (1998) and improved the in planta transformation via the vernalization-infiltration method (Bai et al., 2013). Use of this vernalization-infiltration method resulted in three lines transgenic for p35S::BrpMIR319a2. These transgenic lines were designated as 319a2-1, 319a2-2, and 319a2-3, and all were identified by Southern hybridization (Fig. 4B) using a CaMV 35S promoter probe and via sequencing of PCR products (Supplemental Fig. S2) obtained using the two pairs of the primers specific for the CaMV 35S promoter and for the transfer DNA (T-DNA) fragment containing BrpMIR319a2. Southern hybridization showed two or three bands in the transgenic lines 319a2-1, 319a2-2, and 319a2-3. Within the T2 population of 319a2-2, 42 phosphinothricin-resistant seedlings and 12 phosphinothricin-sensitive seedlings were identified, consistent with a 3:1 Mendelian ratio. These data indicated that one copy of p35S::BrpMIR319a2 was successfully integrated in the genome of 319a2-2. Segregation analysis showed that 319a2-1 and 319a2-3 had two and one copies of p35S::BrpMIR319a2, respectively.
Figure 4.
Overexpression of BrpMIR319a2 and silencing of BrpTCP genes in the plants transgenic for BrpMIR319a2. A, Schematic diagram showing the structure of T-DNA containing p35S::BrpMIR319a2. B, Southern hybridization of the three transgenic lines (F3 generation) with p35S::BrpMIR319a2 using CaMV 35S probes. C, Northern blotting showing the expression levels of BrpMIR319a2 in young rosette leaves (less than 1 cm long) of the three transgenic lines. D, Real-time PCR showing the expression levels of three BrpTCP genes in young rosette leaves of the three transgenic lines (n = 3; 1 cm long). E, Phenotypes of cv Bre and 319a2-2 transgenic plants at the cotyledon, seedling, and heading stages. EV, Transgenic plants with empty vectors; LB, left border of the T-DNA; M, molecular size markers; PAT, phosphinothricin acetyl transferase; p35S, promoter of CaMV 35S; RB, right border of the T-DNA; Tnos, terminator of the nopaline synthase gene; T35S, terminator of CaMV 35S; WT, wild type. [See online article for color version of this figure.]
BrpMIR319a2 was overaccumulated in all three transgenic lines (Fig. 4C). In contrast, BrpTCP4-1, BrpTCP2-1, and BrpTCP3-1 were significantly down-regulated (Fig. 4D), indicating that the exogenous BrpMIR319a2 reduced expression levels of BrpTCP genes in the transgenic plants.
Changes in Head Shape and Size
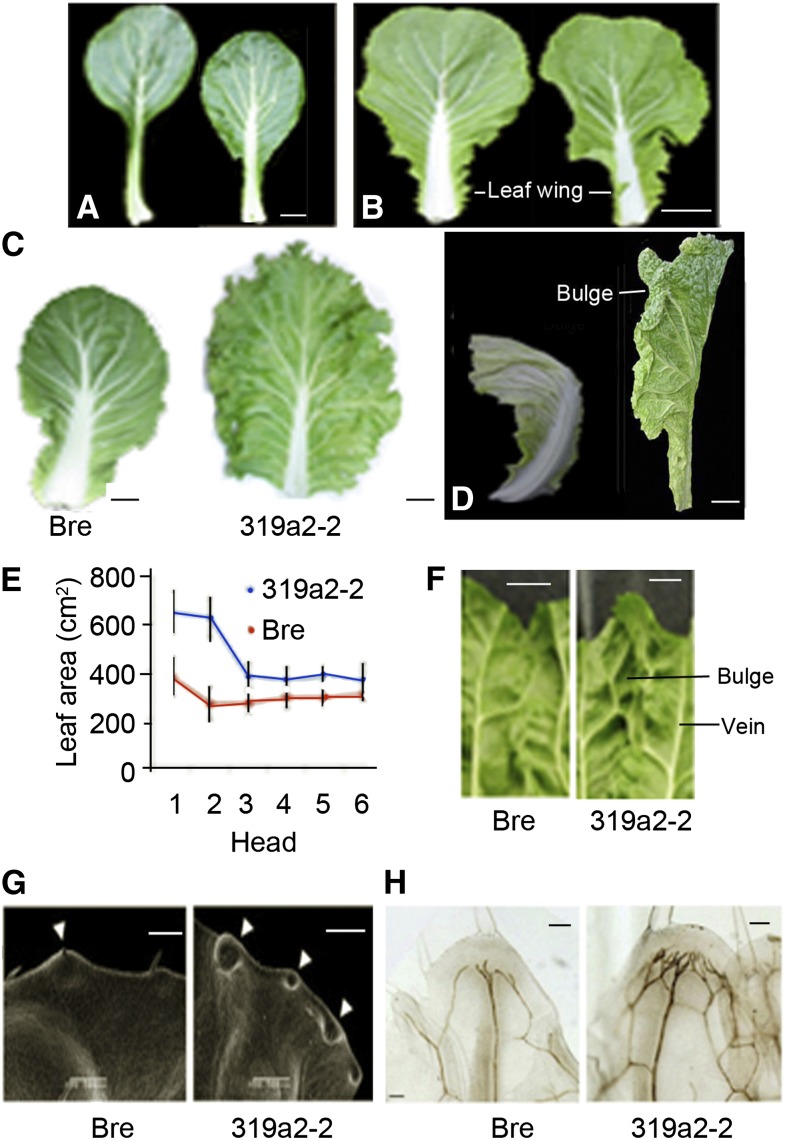
The leafy heads of cv Bre (the wild type) were round (Fig. 1, I and J), whereas those of 319a2-1, 319a2-2, and 319a2-3 lines were cylindrical (Fig. 1, K–N). The tops of 319a2-1, 319a2-2, and 319a2-3 heads were open (Fig. 1L), in contrast to the closed top of the wild type. Comparison of the three transgenic lines indicated that 319a2-1 and 319a2-3 leaves showed smaller bulges than did 319a2-2 leaves and that 319a2-1 heads were lower in height than 319a2-2 heads (Fig. 1M) and 319a2-3 heads were higher than 319a2-2 heads (Fig. 1N). Cross sections of shoot tips at the heading stage showed that the young head leaves of 319a2-2 had more wavy leaf surfaces than those of the wild type, and the stalk was higher than that of the wild type (Fig. 1, O and P). The diameter of a 319a2-2 head was 1.6 cm longer than that of the wild type (Table I). The head shape index (height-to-diameter ratio) of 319a2-2 heads was 2.62, much higher than that of the wild-type. Although the diameter and height of the 319a2-2 head increased (compared with the wild type), the height of leafy heads increased more than the diameter. Overall, the leafy heads of 319a2-2 were enlarged. The increase in fresh weight of a 319a2-2 heads was consistent with the change in height and diameter.
Table I. Parameters of leafy heads (mature) in Chinese cabbage plants (319a2-2) transgenic for the BrpMIR319a2 gene.
| Leafy Heads | Wild Type | 319a2-1 | 319a2-2 | 319a2-3 |
|---|---|---|---|---|
| Diameter (cm) | 16.10 ± 1.4 | 16.92 ± 1.5 | 17.70 ± 1.6 | 17.98 ± 1.8 |
| Height (cm) | 17.23 ± 1.6 | 44.42 ± 6.0 | 46.37 ± 6.3 | 48.37 ± 6.9 |
| Weight (kg) | 1.12 ± 0.09 | 1.28 ± 1.2 | 1.31 ± 1.3 | 1.36 ± 1.5 |
| Height-to-diameter ratio | 1.07 ± 0.12 | 2.53 ± 0.25 | 2.62 ± 0.29 | 2.82 ± 0.28 |
Excess Growth in the Apical Regions
Vegetative growth of Chinese cabbage is divided into four stages: seedling, rosette, folding, and heading. The seedling and rosette leaves grow progressively, but they are not fully expanded until they reach the final size at the heading stage. To examine whether and how miR319 might regulate excess growth, we observed and measured young and mature leaves of the transgenic plants at different stages. In 319a2-2 seedlings, one of the cotyledons was much wider than the wild type, downwardly curved, and wavy in the marginal region, while the other looked normal (Fig. 4E); the seedling leaves were similar in size to the wild type but possessed shorter petioles (Fig. 5A); the rosette leaves were more crinkly than the wild type (Fig. 5B), with more leaf lobes (leaf wings); and the rosette leaves matured later than did the wild type at the heading stage, and their final size was much larger than the wild type (Fig. 5C; Table II), as they grew longer than the wild type. The fully expanded 319a2-2 leaves had a lower length-to-width ratio than did the wild type, indicating increased lateral growth within the 319a2-2 rosette leaves. These fully expanded leaves were much more crinkly and wavy, especially in the tip and lateral regions (Figs. 4E and 5C); however, the incurvature level of rosette leaves was not changed compared with the wild type.
Figure 5.
Altered leaf phenotypes of the plants transgenic for BrpMIR319a2. A, The fifth seedling leaves of the wild type (left) and transgenic 319a2-2 (right) at the seedling stage (eight leaves in total). B, The fifth rosette leaves of the wild type (left) and 319a2-2 (right) at the rosette stage (21 leaves in total). C, The last rosette leaves of the wild type (left) and transgenic 319a2-2 (right) at the heading stage (43 leaves in total). D, The third head leaves (counted from outside to inside) of the wild type (left) and transgenic 319a2-2 (right) at the heading stage (43 leaves in total). E, Leaf areas of the first six rosette leaves of wild-type and 319a2-2 plants at the heading stage (43 leaves in total). The number of leaves for each measurement is more than 20. Error bars indicate sd. F, Local bulges in head leaf of cv Bre and 319a2-2. G, Scanning electron microscopy results showing the hydathodes on leaf margins of the fifth heading leaves of the wild type (left) and 319a2-2 (right) at the heading stage. Arrowheads indicate the hydathodes. H, Vein patterns in the local leaf margins of the wild type (left) and 319a2-2 (right) at the heading stage (43 leaves in total). Bars = 1 cm in A, 5 cm in B to D, and 0.5 cm in F to H. [See online article for color version of this figure.]
Table II. Parameters of head leaves in Chinese cabbage plants (319a2-2) transgenic for the BrpMIR319a2 gene.
The fifth rosette leaves at the rosette stage and the first head leaves at the heading stage were measured. The number of leaves for each treatment is more than 20.
| Leaf Parameters | Wild Type | 319a2-2 |
|---|---|---|
| Rosette leaves | ||
| Length (cm) | 19 ± 1.3 | 21 ± 1.8 |
| Width (cm) | 12 ± 1.1 | 14 ± 1.2 |
| Area (cm2) | 198 ± 11.2 | 229 ± 12.7 |
| Length-to-width ratio | 1.6 ± 0.1 | 1.5 ± 0.1 |
| Head leaves | ||
| Length (cm) | 26 ± 2.4 | 28 ± 2.6 |
| Width (cm) | 20 ± 1.7 | 25 ± 2.1 |
| Area (cm2) | 395 ± 27.5 | 610 ± 32.6 |
| Length-to-width ratio | 1.3 ± 0.1 | 1.1 ± 0.1 |
The wild-type head leaves were extremely incurved in both transverse and longitudinal directions at the heading stage (Fig. 5D). By contrast, the apical regions of the 319a2-2 head leaves were straightened along the longitudinal axis and remained incurved in the transverse direction. In addition, the head leaves of 319a2-2 were much larger than the wild type, and their length-to-width ratio was much less than the wild type (Table II). The first two head leaves of 319a2-2 showed excess growth relative to the other head leaves (Fig. 5E). The straightening of the apical regions and the increase in size of 319a2-2 head leaves was likely due to excess growth in these regions, especially when the growth in the central regions was not changed or, if so, only slowed.
Excess Growth of the Interveinal Region
The presence of local bulges in leaves is a result of excess growth in the interveinal regions (Nath et al., 2003). On the adaxial side of the wild-type head leaves, there were a few local bulges between the leaf veins (Fig. 5D). However, the bulges on head leaves of 319a2-2 plants were much greater in number and larger in appearance. An adaxial view of the bulges between the two mature veins in the uppermost portion of the first head leaf showed vigorous excess growth in the interveinal regions (Fig. 5F). In addition, the developing veins in the interveinal region of the 319a2-2 head leaves were much larger than in the wild type. Likely due to the excess growth of interveinal regions and developing veins, 319a2-2 head leaves were larger than the wild type. Excess growth of veins and interveinal regions occurred not only in the marginal regions of 319a2-2 head leaves but also in the central regions, albeit to a lesser extent. In the mature rosette and head leaves of 319a2-2 plants, there was a gradual increase of waviness from the center to the margins (Fig. 5, C and D). These data indicate that excess growth of the interveinal regions contributed to the extra expansion of the whole head leaves.
In leaf margins, the hydathodes of 319a2-2 leaves were denser than those of the wild type (Fig. 5G). The veins inside a local marginal region of the wild-type plants were simple, with few veins (Fig. 5H), whereas those of 319a2-2 plants were more complex, with many secondary veins. These results revealed that overaccumulation of miR319 promotes excess growth in interveinal regions and even enhances the differentiation of hydathodes and veins in leaf marginal regions.
Distribution of BrpTCP Transcripts in Leaf Regions
To better understand the molecular basis for the observed changes in head shape and leaf shape of 319a2-2, we examined the spatial expressional pattern of miR319 in the tip, central region, and base of leaves via northern blotting (Supplemental Fig. S3). In the wild-type rosette leaves, miR319a accumulated more in the tips than in the central region (Fig. 6A). In 319a2-2 leaves, miR319a expression increased and was almost equally high in all three regions. In situ hybridization also indicated that 319a2-2 and wild-type plants were distinct with regard to the temporal and spatial expression patterns of miR319a. At the rosette stage, miR319a accumulation in the wild type occurred in the shoot apical meristem and leaf primordia (Fig. 6B). In developing leaves, miR319 accumulation was mainly observed in vascular bundles and the epidermis (especially in early vascular tissues). By contrast, miR319 accumulation in 319a2-2 and was not localized either temporally or spatially.
Figure 6.
Temporal and spatial expression of BrpMIR319a2 and BrpTCP4 genes in three leaf regions. A, Northern blotting showing the expression levels of BrpMIR319a2 in the tip, middle (mid), and base of the fifth rosette leaves and the third heading leaves of the wild type (left) and 319a2-2 (right) at the heading stage. B, In situ hybridization showing the localized expression of BrpMIR319a and BrpTCP4 genes in the fifth rosette leaves and the third heading leaves of the wild type (left) and 319a2-2 (right) at the heading stage. Bars = 100 μm. C, Real-time PCR showing the relative expression of BrpTCP4-1 in the fifth rosette leaves and the third heading leaves of the wild type (left) and 319a2-2 (right) at the heading stage. ACTIN was used as the loading control. The error bars indicate three biological replicates. D, Longitudinal sections through the middle of the first head leaf at the heading stage showing H4 expression (only the top half of the section is shown). Bars = 100 μm.
BrpTCP4 expression in developing rosette leaves of the wild type was greatest in the apical and lateral regions (relative to the central region) and weakest in veins (relative to the interveinal regions). In the leaves, expression domains of BrpTCP4-1 were restricted to the adaxial side (Fig. 6B). At the heading stage, the expression domains of BrpTCP4 were smaller and weaker in the apical and lateral regions. In 319a2-2 plants, BrpTCP4 expression in whole rosette leaves decreased and was almost equally high in the apical and central regions. As expected, expression levels of BrpTCP genes were negatively controlled by miR319. However, overaccumulation of miR319 reduced the difference in expression levels of BrpTCP between the apical and lateral regions of leaves.
To further examine the temporal and spatial expressional patterns of BrpTCP genes, we performed real-time PCR using the RNA samples from apical, lateral, central, and basal regions of rosette leaves (outer leaves) and head leaves (inner leaves) at the heading stage. In wild-type rosette leaves, the expression of BrpTCP4 in apical regions was higher than in the central regions (Fig. 6C). In wild-type head leaves, the expression of BrpTCP4 in marginal regions was higher than in the central regions. In 319a2-2 plants, however, differences in expression levels of BrpTCP4 between apical and central regions were not observed.
We also compared the expression patterns of miR319 with those of BrpTCP4. In wild-type plants, miR319 accumulated in vascular bundles and epidermis in developing rosette leaves and head leaves, whereas BrpTCP4-1 expression was not observed, and both were stronger in the apical than in the central regions. In the apical regions, BrpTCP4 expression was preferential on the adaxial side, whereas miR319 accumulation was not. These data indicate that BrpTCP4 expression was excluded from the regions where miR319 accumulated. In 319a2-2 plants, miR319 was overaccumulated, whereas BrpTCP4 expression was reduced, suggesting that miR319 silenced BrpTCP4 genes. This is consistent with the findings reported in Arabidopsis (Palatnik et al., 2003; Liu et al., 2011).
Cell Division Arrest Directed by BrpTCP4 Genes
To better understand how miR319 accumulation affected cell division, we examined the expression pattern of HISTONE4 (H4) in the apical and central regions of 319a2-2 head leaves. In the wild type, H4 was sporadically distributed in the tips of the young head leaves, but this expression sharply declined in the central regions (Fig. 6D). In 319a2-2 plants, the sporadic signals of H4 in the apical and central regions were much greater than in the wild type, and the expression domains of H4 were greater than in the wild type. These data indicate that the cell division in 319a2-2 head leaves was much greater and occurred in a wider range of tissues than in the wild type.
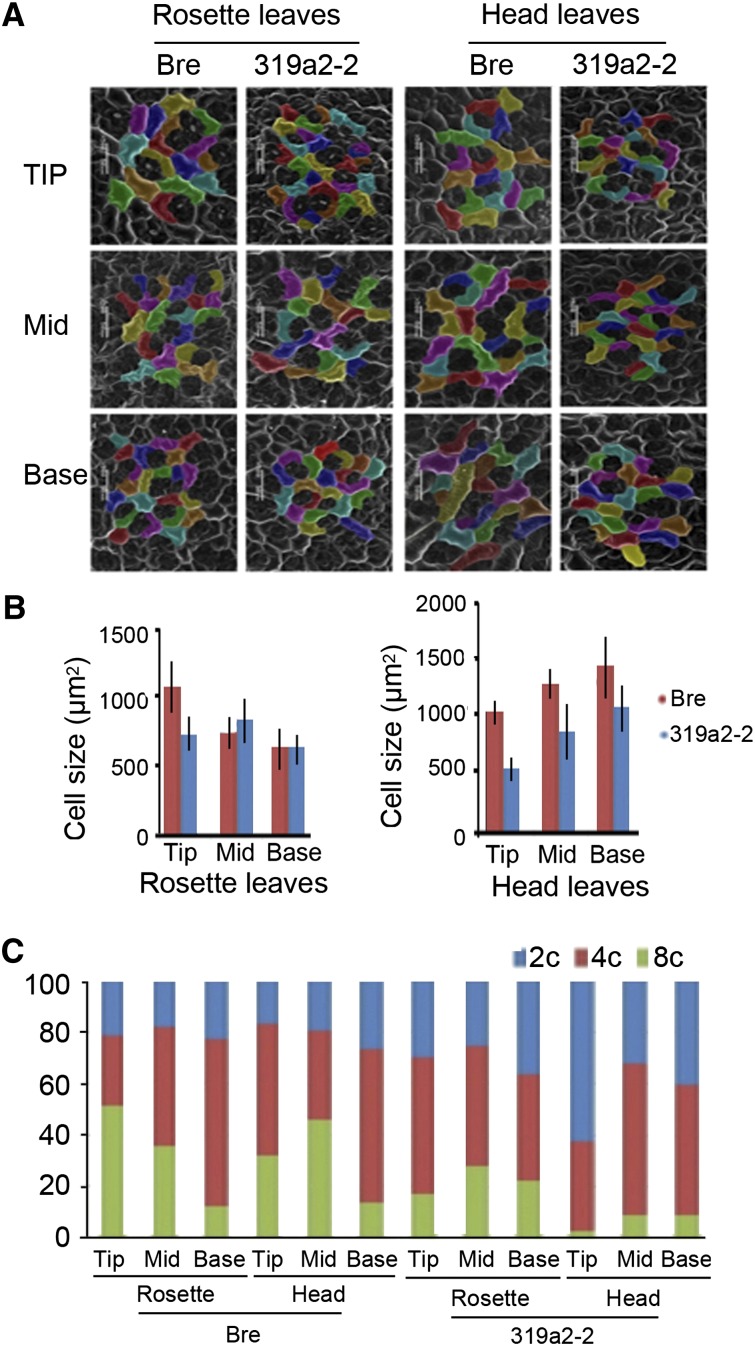
Normally, leaf cells remain small during proliferation but subsequently increase in size following the arrest of division, after which further growth occurs primarily via cell expansion (Nath et al., 2003; Aggarwal et al., 2011). We observed a remarkable difference in cell division between 319a2-2 and wild-type leaves. The 319a2-2 cells in the tips of both rosette leaves and head leaves were much smaller than those in the wild type (Fig. 7, A and B), revealing that cell division of 319a2-2 in the tip was accelerated. To further identify the role of miR319 in cell proliferation, we performed flow cytometry assays using the fifth rosette leaf and 15th head leaf at the heading stage (Fig. 7B). A positive relationship between DNA quantity and mature cell size (Melaragno et al., 1993) and a negative relationship between DNA quantity and cell division (Lyons and Doherty, 2004) have been reported. In a tip of 319a2-2 rosette leaves, the ratio of 8C to 2C cells was higher than that in the wild type, whereas the ratio in the middle leaf was lower than that in the wild type, suggesting that cell division was more active in the leaf tip (Fig. 7C). In the tip and middle of 319a2-2 head leaves, the ratio of 8C to 2C cells was much higher than that in the wild type. In the base of 319a2-2 head leaves, the ratio of 8C to 2C cells was almost the same as that in the wild type. These results confirmed that miR319 promoted cell division in the apical regions of rosette and head leaves.
Figure 7.
Size of the epidermal cells in three leaf regions of rosette and head leaves. A, Scanning electron microscopy images showing the relative cell size in the tip, middle (Mid), and base of the fifth rosette leaves and third head leaves of the wild type and 319a2-2 at the heading stage. B, Epidermal cell sizes of rosette and head leaves of the fifth rosette leaves and third head leaves of the wild type and 319a2-2. Bars = 50 μm. C, Flow cytometry assay showing the distributions of mean leaf cell ploidy (n = 5). The different patterns represent the percentage of cells with nuclei of 2C, 4C, and 8C (from bottom to top). [See online article for color version of this figure.]
DISCUSSION
The Cylindrical Shape of a Leafy Head Is Associated with the Decreased Expression of BrpTCP4
The leafy head is a three-dimensional structure. Round, cylindrical, oblong, and cone-like heads are the four basic shapes of leafy heads. In Arabidopsis, several miRNAs and their target genes have been identified as related to mutant phenotypes deficient in leaf size, shape, and curvature (Liu et al., 2011). However, it has proven quite difficult to define how the miRNA target genes contribute to the variation of leaf form in many crops. As such, box plots demonstrate that the difference in the medians of BrpTCP4 expression between the “cylindrical” and “round” subpopulations is highly significant. Importantly, the median of BrpTCP4 expression in the cylindrical subpopulation is much lower than BrpTCP4 expression in the parental line of the RIL population with the round head. This reflects the relationship between the cylindrical shape of the leafy head and the low level of BrpTCP4-1 expression. By contrast, the median of BrpTCP4 expression in the round subpopulation is higher than BrpTCP4 expression in cv Bre.
The association between the cylindrical shape of a leafy head and a relatively high level of BrpTCP4-1 expression is verified via the examination of transgenic plants with p35S::BrpMIR319a2. Silencing of BrpTCP genes causes the transition of a leafy head from round to cylindrical shape.
The differences in the medians of BrpTCP4-1 expression between cone-like, oblong, and round subpopulations were not significant. This suggests that the cone-like and oblong shapes of leafy heads are related to the high expression of BrpTCP4 genes, as the round shape is. However, some additional element is involved in the formation of cone-like and oblong shapes.
Slower Growth of the Apical Regions Is Relevant to the Differential Expression of BrpTCP4 Genes in Leaf Regions
In Antirrhinum majus leaves, a change in the shape and the progression of a cell division arrest front moves from the leaf tip toward the leaf base (Nath et al., 2003). In the head leaves of Chinese cabbage, BrpTCP4 expression in the apical regions is stronger than in the central regions, and BrpTCP4 expression in the interveinal regions is stronger than in veins. In the head leaves of the transgenic plants (319a2-2) with p35S::BrpMIR319a2, straightening of the apical regions and the appearance of dense bulges are concomitant with excess growth in the apical regions and interveinal regions. When the expression of BrpTCP4 genes decreases in the apical regions, the cell division arrest is relieved, resulting in excess growth in the apical regions. Among the three leaf regions of the transgenic plant, the apical region is where BrpTCP4 expression decreases the most. Therefore, the extent of excess growth in the apical region is greatest. Nevertheless, this type of excess growth does not change the original tendency of lateral incurvature. The lateral regions of head leaves are different from the apical regions in excess growth. Excess growth in the apical regions and interveinal regions contributes to the leaf size of 319a2-2 plants. The central regions display excess growth, albeit to a lesser extent. From the central to the marginal region, excess growth is progressively increased.
In the wild-type head leaves, the relatively strong expression of the BrpTCP4 gene in the apical region is essential for strong cell division arrest. This maintains slower growth in the apical regions compared with the central regions. We suggest that the relatively high expression of BrpTCP4 genes in the apical regions causes slow growth in these regions. The difference in expression levels of the BrpTCP4 gene between the apical and central regions is necessary for the overall incurvature of head leaves.
The young leaves of 319a2-2 and wild-type plants at the seedling and rosette stages are basically flat and similar in shape, size, and color. Although miR319a is overaccumulated in rosette and head leaves, the rosette leaves are never incurved, as head leaves were. Therefore, it may be possible that rosette leaves are not sensitive to a cell division arrest signal.
BrpTCP4 Genes Modulate Head Shape by Differential Cell Division Arrest in Leaf Regions
Leaf curvature is a process essential for the formation of leafy heads during the vegetative development of Chinese cabbage. According to Gaussian curvature theory, the leaves whose marginal regions grow more slowly than the central regions adopt a cup-like shape with positive Gaussian curvature (Nath et al., 2003). The head leaves of cv Bre plants are curved inward and are cup-like in appearance, possibly a result of slower growth in the apical, lateral, and basal regions than in the central region. It remains unknown what genetic elements control this incurvature. In Arabidopsis and Antirrhinum majus, a few mutants show leaf curvature. PEAPOD (PPD) regulates lamina size and curvature (White 2006). In a ppd leaf, downward curvature reflects the difference between excess growth of the central region and a limitation to the extension capacity of its perimeter. Leaves of cin display an excess growth in marginal regions (Nath et al., 2003), resulting in a wavy surface and the downward curvature of apical regions. Constitutive expression of miR319 has been found to alter plant development in transgenic creeping bentgrass (Agrostis stolonifera; Zhou et al., 2013). Down-regulation of several LANCEOLATE-like (transcription factor) genes using ectopic expression of miR319 resulted in larger leaflets and continuous growth of leaf margins (Ori et al., 2007). TCP4 was shown to induce miR396 and to repress GROWTH-REGULATING FACTOR activity (Rodriguez et al., 2010). In this regard, the data presented in this study confirm these previous results, but with the novelty of investigating Chinese cabbage. It is probable that a head leaf of Chinese cabbage and a ppd leaf of Arabidopsis may share relevant developmental information related to leaf incurvature.
According to the expression patterns and function of miR319, we propose that miR319 controls cell division in the apical regions, veins, and interveinal regions of head leaves in Chinese cabbage via differential cell division in leaf regions. Uneven accumulation of miR319a and BrpTCP4 genes in the apical and central regions of head leaves is required for the development of a round head shape, as the relatively high expression of BrpTCP4 genes in the apical region maintains stronger cell division arrest in this region in favor of leaf incurvature. The incurved leaves inside a round head reflect the difference between the relatively rapid growth of the central region and a limitation to the extension capacity of its apical and lateral regions. For the transgenic plants overexpressing BrpMIR319a2, the decreased expression and the even dispersion of BrpTCP4 transcripts in different leaf regions cause the transition of head shape from round to cylindrical. This suggests that the changes in the spatial expression patterns of BrpTCP4 affect the transition of head shape. Therefore, miR319a and BrpTCP4 genes are potentially important for the genetic improvement of head shape desirable for high commercial quality and high yield of Chinese cabbage.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of cv Bre, an inbred line of heading Chinese cabbage (Brassica rapa), and cv Wut, a inbred line of nonheading Chinese cabbage, were germinated on moisture-absorbent papers in a plant growth chamber at 25°C for 3 d and then transferred to the greenhouse with 80% ± 5% humidity, a 16-h-light/8-h-dark cycle, and 150 mmol m−2 s−1 light intensity at 22°C for 20 d. The seedlings were transplanted to the field at the Songjiang Farm Station of Shanghai Institute of Plant Physiology and Ecology in early September 2012. To induce reproductive growth for use in genetic transformation, the germinated seeds were grown in a chamber of 4°C for an additional period of vernalization for 25 d. The seedlings were then transferred to the greenhouse and grown for 1 month at 22°C for 20 d.
Gene Cloning and Genetic Transformation
The 834-bp genomic fragment of BrpMIR319a2 was cloned from cv Bre plants using specific primers (Supplemental Table S3) and constructed in pCAMBIA3301 binary vectors under the control of the CaMV 35S promoter. The procedures of in planta transformation of cv Bre via the vernalization-infiltration method were as described by Bai et al. (2013). Briefly, plants with small flower buds at the early bolting stage were placed upside down in a vacuum desiccator that contained infiltration medium and the engineered Agrobacterium tumefaciens (Bechtold and Pelletier, 1998) for vacuum infiltration. The Agrobacterium tumefaciens-infected plants were transferred to a dark growth room, incubated for 2 d at 22°C/18°C, and then transferred to a growth room. These plants were pollinated by hand with the pollen from the cv Bre plants without vacuum infiltration. The pots were placed back in the growth room. The resulting seeds were then harvested.
For the selection of transgenic plants, seeds were sterilized and germinated on agar medium containing 30 mg L−1 phosphinothricin. The seedlings resistant to the herbicide Basta were transplanted to the greenhouse and self-pollinated for three generations to obtain homozygous lines. The transgenic lines were identified using two pairs of the primers: one specific for the CaMV 35S promoter to detect its existence, and the other specific for the T-DNA fragment containing polylinker and BrpMIR319a2 to detect the correct connection of BrpMIR319a2 with the CaMV 35S promoter in the transgenic plants. The PCR products were sequenced. The homologous transgenic lines as identified by Southern hybridization were used for the analysis of phenotypic and genotypic expression.
Southern Hybridization
Genomic DNA of Brassica rapa in leaf samples was extracted using a modified cetyl-trimethyl-ammonium bromide method (Aldrich and Cullis, 1993; Ausubel et al., 1994). DNA samples were treated with phenol/chloroform extraction to remove polysacchride contamination. Twenty micrograms of genomic DNA was used for EcoRI or HindIII digestion. The positive control of plasmid pCAMBIA3301-35S:Brp319a2 was digested with EcoRI and XbaI. Digested DNA was separated on a 1% agarose gel at 80 V for 3 h, transferred to a Hybond membrane (Amersham Biosciences, GE Healthcare), and incubated in 1× Tris-acetate-EDTA overnight at 250 mA. The UV cross-linked membrane was hybridized in DIG EASY Hyb buffer (Roche) using probes amplified from CaMV 35S or a GUS reporter gene sequence by PCR DIG probe synthesis mix (Roche). The hybridization signals were detected using CDP-Star (Roche) and imaged with a FLA-5000 phosphorimager (FujiFilm).
miRNA Northern Blotting and Quantitative Real-Time PCR
RNA samples were extracted from leaves using TRIzol (Invitrogen). For northern blotting of small RNA, 50 μg of total RNAs was resolved by 19% PAGE in 1× Tris-borate-EDTA at 80 V for 4 to 6 h, transferred to a Hybond membrane (Amersham Biosciences, GE Healthcare), and incubated in 0.5× Tris-borate-EDTA overnight at 28 mA. The UV cross-linked membrane was hybridized in ULTRAhyb Ultrasensitive Hybridization buffer (Ambion) using the probes of 3′ biotin-labeled DNA oligo (TaKaRa) antisense to the mature miR319a or U6 transcript (Supplemental Table S3). Hybridization signals were detected using the LightShift EMSA kit (Thermo Scientific) and imaged with a FLA-5000 phosphorimager (FujiFilm).
For quantitative real-time PCR, total RNA was treated with DNase I (TaKaRa) followed by phenol/chloroform extraction to remove DNA contamination. Approximately 4 μg of purified RNAs was used for first-strand complementary DNA synthesis using PrimeScript Reverse Transcriptase (TaKaRa) with oligo(dT) primers. Real-time PCR was performed using specific primer pairs (Supplemental Table S3) in the MyiQ2 Two-Color Real-Time PCR Detection System (Bio-Rad). Quantitative PCR for each gene was performed on at least three biological replicates. Relative transcript levels were determined for each sample by normalizing them to BrpACTIN complementary DNA levels.
Sequence Analysis
The pre-miR319a (176 nucleotides) and miR319-targeted TCPs from Arabidopsis (Arabidopsis thaliana) were compared with the genomic sequences of Chinese cabbage (var Chiifu-401-42) in the Brassica Database (http://brassicadb.org/brad/index.php). Multiple sequence alignment was performed, and phylogenetic trees were constructed using the maximum likelihood method based on the Tamura-Nei model by MEGA5.
Measurement of Head Parameters and Association Analysis
To characterize the head shapes between the RILs and between the wild-type and the transgenic plants, mature heads were harvested from the field, and the outer leaves (wrapper) were removed. The heads were classified into four shapes: round, oblong, cylindrical, and cone like. Head height was the distance between the bottom and top of a head; head diameter was measured in the middle of a head; head weight was the fresh weight per head; and the head shape index was equal to the height divided by the diameter. The leaf area, length (including the petiole), and width of the outer leaves at the widest points orthogonal to the midrib were measured. For the observation of head structure, the head was sectioned at the shoot apex along the longitudinal axis.
The box plot was drawn according to the method of Hoffmann (1995). The data of BrpTCP4-1 expression in the RILs were gathered, and the median and quartiles were found using R Statistical Software. The significance of any differences between the subpopulations was detected by the Kruskal-Wallis test (Kruskal and Wallis, 1952).
Histology and Scanning Electron Microscopy
For vasculature observation, the head leaves were cleared of chlorophyll in 95% alcohol overnight and treated with distilled, deionized water:glycerol:phenol:lactic acid (1:1:1:1) for 20 min at 90°C. For leaf surface scanning, leaves were fixed in 50% (v/v) ethanol, 5% (v/v) acetic acid, and 3.7% (v/v) formaldehyde, dried, and then dissected with a stereomicroscope and mounted on scanning electron microscopy stubs. Mounted leaves were coated with palladium-gold and then examined using a JSM-6360LV scanning electron microscope (JEOL) with an acceleration voltage of 7 to 15 kV. The fifth rosette leaf and 15th head leaf of cv Bre and 319a2-2 were taken for cell size calculation by Image J.
Flow Cytometry
The fifth rosette leaf and the 15th head leaf of cv Bre and 319a2-2 were removed. Fresh leaf tissues (less than 0.2 g) were chopped with a razor blade by hand for 5 min in 1 mL of FASC buffer (40 mm MgCl2, 30 mm sodium citrate, 20 mm MOPS, and 1% Triton). The released nuclei were passed through a 40-μm nylon mesh to remove cellular debris. 4′,6-Diamino-phenylindole (2 μL of 1 mg mL−1) was added to the filtered extracts for nuclear staining. After 20 to 60 min of incubation on ice, the suspensions were loaded onto a FACS Calibur flow cytometer (Becton Dickinson), and 10,000 flow cytometric events were recorded. The output was gated to eliminate signal from chloroplasts and debris. For a comparative study of developed leaves, the ratio of octaploids to diploids was designated as the mature index.
In Situ Hybridization
Sections (7 μm thick) of shoot apices from both wild-type and transgenic plants were prepared following pretreatment and the hybridization methods described previously (Jackson, 1991). Hybridization probes corresponding to coding sequences were defined as follows: coding sequence of BrpTCP4-1. Digoxigenin-labeled probes were prepared by in vitro transcription (Roche) according to the manufacturer’s protocol. Locked nucleic acid-modified probes of miR319a were synthesized, labeled with digoxigenin at the 3′ end by TaKaRa, and used for the in situ hybridization of miR319.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number KJ130320.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Parental plants from which the RIL lines were derived.
Supplemental Figure S2. Identification of the transgenic lines with p35S::BrpMIR319a2 by PCR.
Supplemental Figure S3. Apical, lateral, central, and basal regions of a rosette leaf of heading Chinese cabbage.
Supplemental Table S1. Numbers of the RILs with different forms of rosette leaves.
Supplemental Table S2. Relative expression levels of BrpTCP4-1 across 150 RILs and parents.
Supplemental Table S3. Primers and probes used in this study.
Acknowledgments
We thank an anonymous reviewer for advice for statistical analysis.
Glossary
- miRNA
microRNA
- QTL
quantitative trait locus
- RIL
recombinant inbred line
- CaMV
cauliflower mosaic virus
- T-DNA
transfer DNA
References
- Aggarwal P, Padmanabhan B, Bhat A, Sarvepalli K, Sadhale PP, Nath U. (2011) The TCP4 transcription factor of Arabidopsis blocks cell division in yeast at G1→S transition. Biochem Biophys Res Commun 410: 276–281 [DOI] [PubMed] [Google Scholar]
- Aldrich J, Cullis CA. (1993) RAPD analysis in flax: optimization of yield and reproducibility using KlenTaq 1 DNA polymerase, Chelex 100, and gel purification of genomic DNA. Plant Mol Biol Rep 11: 128–141 [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GT, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Ausubel FH, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. (1994) Current Protocols in Molecular Biology. John Wiley & Sons, Hoboken, NJ [Google Scholar]
- Bai J, Wu F, Mao Y, He Y (2013) In planta transformation of Brassica rapa and B. napus via vernalization-infiltration methods. Protocol Exchange, http://www.nature.com/protocolexchange/protocols/2769 (August 7, 2013)
- Bechtold N, Pelletier G. (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y. (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, Goud S, Song L, Fedoroff NV. (2004) The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA 101: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R. (1995) The Same and Not the Same. Columbia University Press, New York [Google Scholar]
- Horiguchi G, Ferjani A, Fujikura U, Tsukaya H. (2006) Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J Plant Res 119: 37–42 [DOI] [PubMed] [Google Scholar]
- Jackson D (1991) In situ hybridization in plants. In SJ Gurr, M McPherson, DJ Bowles, eds, Molecular Plant Pathology: A Practical Approach. Oxford University Press, Oxford, pp 163–174 [Google Scholar]
- Kruskal WH, Wallis WA. (1952) Use of ranks in one-criterion variance analysis. J Am Stat Assoc 47: 583–621 [Google Scholar]
- Liu Z, Jia L, Wang H, He Y. (2011) HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J Exp Bot 62: 4367–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons AB, Blake SJ, Doherty KV (2004) Flow cytometric analysis of cell division by dilution of CFSE and related dyes. Curr Protoc Cytom, Unit 9.11 http://www.currentprotocols.com/WileyCDA/CPUnit/refId-cy0911.html (April, 2013) [DOI] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW. (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E. (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, et al. (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39: 787–791 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP. (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- White DW. (2006) PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci USA 103: 13238–13243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Yu L, Cao W, Mao Y, Liu Z, He Y. (2007) The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. Plant Cell 19: 914–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wang H, Zhong W, Bai J, Liu P, He Y. (2013) QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PLoS ONE 8: e76059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Li D, Li Z, Hu Q, Yang C, Zhu L, Luo H. (2013) Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol 161: 1375–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]