A protein phosphatase 2A-associated protein, regulates ABA-regulated gene expression in Arabidopsis.
Abstract
TAP46 is a protein phosphatase2A (PP2A)-associated protein that regulates PP2A activity in Arabidopsis (Arabidopsis thaliana). To study how PP2A is involved in abscisic acid (ABA) signaling in plants, we studied the function of TAP46 in ABA-regulated seed maturation and seedling development. Expression of TAP46 coincides with the action of ABA in developing seeds and during seed germination, and the TAP46 transcript reaches to the highest level in mature seeds. Real-time polymerase chain reaction analysis indicates that external ABA can increase TAP46 transcript level transiently during seed germination. Overexpression of TAP46 increases plant sensitivity to ABA, while tap46 knockdown mutants are less sensitive to ABA during seed germination, suggesting that TAP46 functions positively in ABA signaling. Overexpression of TAP46 also leads to lower PP2A activity, while tap46-1 knockdown mutant displays higher PP2A activity, suggesting that TAP46 negatively regulates PP2A activity in Arabidopsis. Both TAP46 and PP2A interact with the ABA-regulated transcription factor ABA INSENSITIVE5 (ABI5) in vivo, and TAP46’s binding to ABI5 can stabilize ABI5. Furthermore, TAP46’s binding to the phosphorylated ABI5 may prevent PP2A or PP2A-like protein phosphatases from removing the phosphate from ABI5, thereby maintaining ABI5 in its active form. Overexpression of TAP46 and inhibition of activities of PP2A or PP2A-like protein phosphatases can increase transcript levels of several ABI5-regulated genes, suggesting that TAP46 is a positive factor in the ABA-regulated gene expression in Arabidopsis.
The phytohormone abscisic acid (ABA) plays critical roles in many physiological processes that include seed maturation, dormancy, and germination and in plant response to abiotic and biotic stresses such as drought, salt, cold, and pathogen infections (Koornneef et al., 1998; Finkelstein et al., 2002, 2008; Ton et al., 2009).
The major ABA signal transduction pathway in seed development involves four basic components: ABA receptors (PYR/PYL/RCAR), phosphatases of the protein phosphatase 2C (PP2C) family, kinases of the SNF1-related kinase2 (SnRK2) family, and transcription factors (ABA-responsive element-binding factor [ABF2]; Cutler et al., 2010). The ABA/receptor complex inhibits the activity of PP2C, which allows three SnRK2s to phosphorylate transcription factors, thereby activating downstream genes (Cutler et al., 2010). However, the ABA signaling pathway in guard cell closure is slightly different, as the last step in this pathway involves channel proteins instead of transcription factors (Hua et al., 2012). For example, the S-type anion channel protein SLAC1 (for SLOW ANION CHANNEL-ASSOCIATED PROTEIN1) on the guard cell plasma membrane is phosphorylated and activated, upon ABA stimulation,by several kinases such as OPEN STOMATA1 (SnRK2.6), GUARD CELL HYDROGEN PEROXIDE-RESISTANT1, and the calcium-dependent protein kinases, which leads to Cl– ion efflux from guard cells, resulting in stomatal closure (Mori et al., 2006; Geiger et al., 2010; Brandt et al., 2012; Hua et al., 2012).
Early genetic studies identified several ABA insensitive loci, such as abi1, abi2, abi3, abi4, and abi5 (Koornneef et al., 1984; Finkelstein, 1994), and these genes have been assigned functions in the ABA signaling pathways. For example, ABI1 and ABI2 encode PP2Cs (Leung et al., 1994; Meyer et al., 1994) that negatively regulate activities of three SnRK2s in the absence of ABA and become inactive upon binding to ABA/ABA receptor complex (Cutler et al., 2010). The other three genes, ABI3, ABI4, and ABI5 encode transcription factors that function in the last step in this ABA signaling pathway, and all three genes play important roles in seed maturation and seedling development (Finkelstein et al., 2002, 2008). For example, mutations at the ABI3 locus result in reduced seed dormancy and decreased sensitivity to exogenous ABA (Giraudat et al., 1992). Loss-of-function mutants of ABI4 and ABI5 display insensitivity to ABA in seed germination and seedling development (Finkelstein et al., 1998; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000).
PP2A belongs to a major Ser/Thr phosphatase family in eukaryotic cells, and it plays crucial roles in numerous signaling pathways in animals and plants by regulating the dephosphorylation to affect the substrate conformation, cellular localization, activity, or stability (Janssens and Goris, 2001; País et al., 2009). The PP2A holoenzyme is composed of three subunits: the scaffolding subunit A that brings together the regulatory subunit B and the catalytic subunit C. There are three A genes, 17 B genes, and five C genes for PP2A subunits in Arabidopsis (Arabidopsis thaliana; Farkas et al., 2007). The five PP2A catalytic subunit genes are classified into two subfamily groups (Farkas et al., 2007; País et al., 2009): subfamily I, including PP2A-C1, PP2A-C2, and PP2A-C5, and subfamily II, including PP2A-C3 and PP2A-C4. Overexpression of PP2A-C2 decreases ABA sensitivity and alters plant response to drought, high salt, and sugar stresses, indicating that this subunit is negatively involved in ABA signaling (Pernas et al., 2007). Our early study indicated that ubiquitylation of the A subunits of PP2A by the E3 ligase AtCHIP (for Arabidopsis thaliana carboxy terminus of Hsc70-interacting protein) increases PP2A activity in Arabidopsis, which leads to increased ABA sensitivity, an indication that PP2A is positively involved in ABA signaling (Luo et al., 2006). The different roles of PP2A in ABA signaling might be due to the actions of different B or C subunits involved.
The TAP46 protein was initially identified as an interacting protein of the PP2A-C1 subunit in Arabidopsis (Harris et al., 1999). Earlier studies indicated that TAP46 homologs Tap42 and α4 were regulators of PP2A in yeast (Saccharomyces cerevisiae) and human, respectively (Di Como and Arndt, 1996; Murata et al., 1997; Nanahoshi et al., 1998). The yeast Tap42 is an essential regulator in the Target of Rapamycin (TOR) signaling pathway by directly associating with PP2A and SIT4, a type 2A-related phosphatase initially defined from characterizing a temperature-sensitive mutant4 [sit4] in yeast (Di Como and Arndt, 1996). Under conditions of nutrient deficiency or rapamycin treatment, dephosphorylated Tap42 releases PP2A-C and/or SIT4, which allows PP2A and SIT4 to dephosphorylate downstream substrates such as GLN3 and NPR1 (Crespo and Hall, 2002; Düvel and Broach, 2004). However, under conditions of sufficient nutrient or absence of rapamycin, Tap42 is phosphorylated by the TOR kinase, which enables Tap42 to bind the C subunit of PP2A and SIT4, thereby inhibiting the activities of PP2A or PP2A-like protein phosphatase (PP2A-l; Crespo and Hall, 2002; Düvel and Broach, 2004).
The α4 protein was initially identified as a B-cell receptor associated protein in human (Inui et al., 1995), and later it was found to interact with PP2A catalytic subunits and other PP2A-ls such as PP4 and PP6 (Murata et al., 1997; Chen et al., 1998). In the absence of α4, total cellular PP2A activity decreased by more than 70% (Kong et al., 2009). Furthermore, loss of function in the α4 gene led to degradation of PP2A catalytic subunits, PP4, and PP6, whereas overexpression of the α4 gene could prevent the degradation of phosphatases and stabilize these phosphatases, indicating that α4 plays a critical role in stabilizing PP2A, PP4, and PP6 in animal cells (Kong et al., 2009).
Recently, Ahn et al. (2011) provided important information on the functions of TAP46 in Arabidopsis. They demonstrated that TAP46 is essential for plant growth and development, because loss of function in TAP46 by using virus-induced gene silencing or RNA interference (RNAi) could lead to programmed cell death in tobacco (Nicotiana tabacum) and Arabidopsis. More importantly, they demonstrated that TAP46 could be phosphorylated by the TOR kinase, and TAP46 is a critical regulator in the TOR signaling pathway in Arabidopsis, similar to what was found in the yeast system (Di Como and Arndt, 1996). Their work indicated that the TAP46-involved TOR signaling pathway is conserved in plants.
In this study, the function of TAP46 in ABA-regulated developmental processes was analyzed. We found that TAP46 is highly expressed in seeds and is induced by ABA. Overexpression of TAP46 decreases PP2A activity and increases ABA sensitivity in seed germination, whereas down-regulation of TAP46 leads to increased PP2A activity and decreased ABA sensitivity in seed germination. Furthermore, TAP46 interacts with ABI5 in vivo, and overexpression of TAP46 leads to higher abundance of ABI5 in both free form and phosphorylated form, with the concomitant increase in the transcript levels of many downstream genes of ABI5. Our data indicate that TAP46 is a positive regulator in ABA-regulated gene expression. More specifically, TAP46 might function antagonistically with PP2A or PP2A-ls on ABI5 and is required for ABI5 stability in vivo. So, in addition to what was known about TAP46’s important role in the TOR signaling pathway in plants (Ahn et al., 2011), TAP46 also plays important roles in the ABA signaling pathway.
RESULTS
TAP46 Is a Negative Regulator in Seed Germination
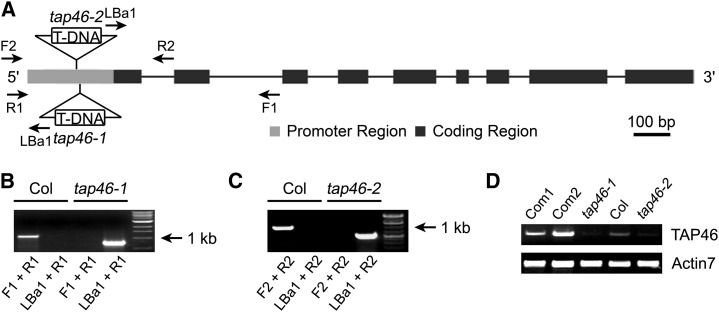
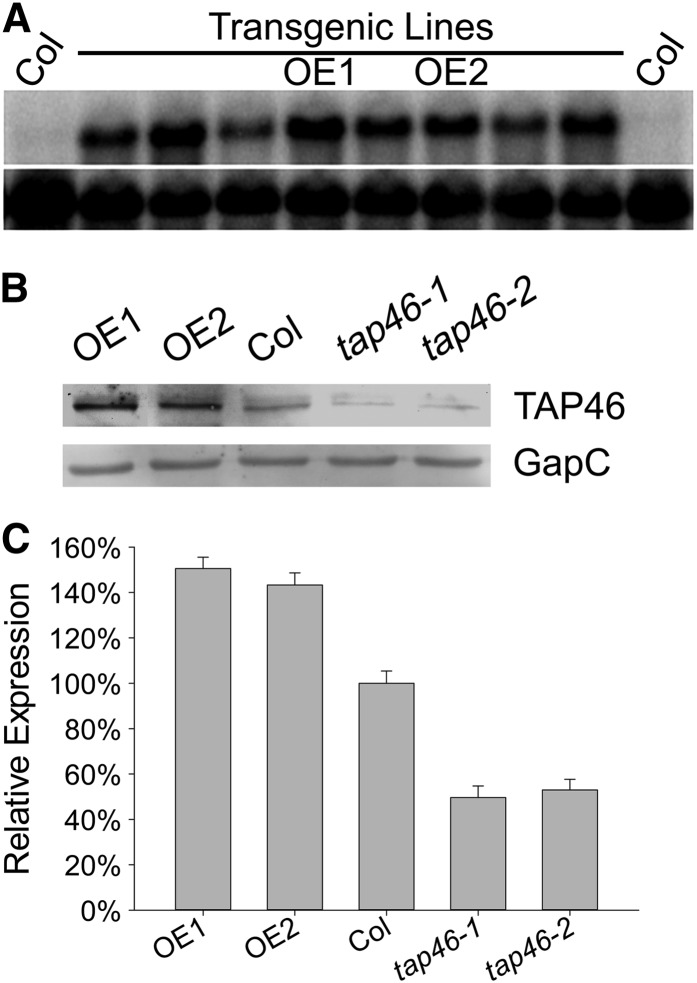
To further study the function of TAP46 (At5g53000) in Arabidopsis, we analyzed two transfer DNA (T-DNA) insertion mutants: tap46-1 and tap46-2. Genotyping PCR experiments confirmed that the T-DNA was inserted into the promoter region of the TAP46 gene (Fig. 1, A to C), and these T-DNA insertions led to decreased transcript level in both mutants according to the result of reverse transcription (RT)-PCR analysis (Fig. 1D). To analyze the consequence of gain of function, we overexpressed TAP46 by placing TAP46 under the control of a strong promoter, i.e. the 35S promoter from Cauliflower mosaic virus. A total of 18 independent TAP46-overexpressing lines were created and analyzed. One RNA blot data with eight independent transgenic lines is shown in Figure 2A, which indicates that the TAP46 transcript was highly expressed in all transgenic plants (>10-fold increase compared with the level in wild-type plants). We then used two TAP46-overexpressing lines, OE1 and OE2 (first homozygous lines obtained), for further molecular and physiological analyses. In western-blot analysis, we found that the steady-state levels of TAP46 were about 1.4 times higher in the 1-d-old germinating seeds of OE1 and OE2 than that in wild-type plants (Fig. 2B). The discrepancy between the RNA level and the protein level suggests that posttranscriptional regulation might play a role in the final steady-state level of TAP46 in TAP46-overexpressing plants. Because the T-DNA insertion sites are in the promoter region of TAP46, the two tap46 mutants are not knock-out mutants, but knock-down mutants, leading to a reduction at the protein level by almost 50% (Fig. 2, B and C). Despite this, under normal growth conditions, tap46-1 and tap46-2 mutants looked similar to wild-type plants (Supplemental Fig. S1), whereas the TAP46 RNAi plants created by Ahn et al. (2011) looked very small and could not produce progenies.
Figure 1.
Molecular analysis of tap46 mutants. A, The TAP46 genomic structure and T-DNA insertion sites within the promoter sequence of the TAP46 gene. Dark boxes are exons, and the lines are introns. Primers that were used for PCR confirmation of the tap46-1 and tap46-2 mutants are marked by arrows. The primers tap46-F1 and tap46-R1 (F1 and R1) and tap46-F2 and tap46-R2 (F2 and R2) are specific for TAP46 DNA, and LBa1 (left border primer) is specific for the T-DNA sequence. B and C, PCR experiments demonstrate that the tap46-1 and tap46-2 mutants are homozygous. The primer combination for PCR reaction is shown below each lane. D, RT-PCR experiments demonstrate that the TAP46 transcript is reduced in the tap46-1 and tap46-2 mutants and elevated in complemented lines of the tap46-1 mutant. Com1 and Com2 indicate two complemented lines of tap46-1, respectively. Actin7 was used as the internal control.
Figure 2.
RNA-blot analysis of TAP46-overerexpressing plants and western-blot analysis of TAP46-overerexpressing and tap46 mutant plants. A, RNA-blot analysis of TAP46-overerexpressing plants. A cDNA fragment of TAP46 was used as the probe in the blot (top) and the 18S rRNA was used as the RNA loading control (bottom). Lanes 1 and 10 are wild-type Arabidopsis (Col-0) and lanes 2 to 9 are independent TAP46-overerexpressing transgenic lines. OE1 and OE2 indicates two TAP46-overerexpressing transgenic lines selected for further analysis. B, Western-blot analysis of TAP46-overerexpressing and tap46 mutant plants. GapC was used as the loading control. C, Relative levels of TAP46 in OE1, OE2, Col-0, tap46-1, and tap46-2 plants with respect to GapC (obtained by analyzing the western blot shown in B using the densitometry method).
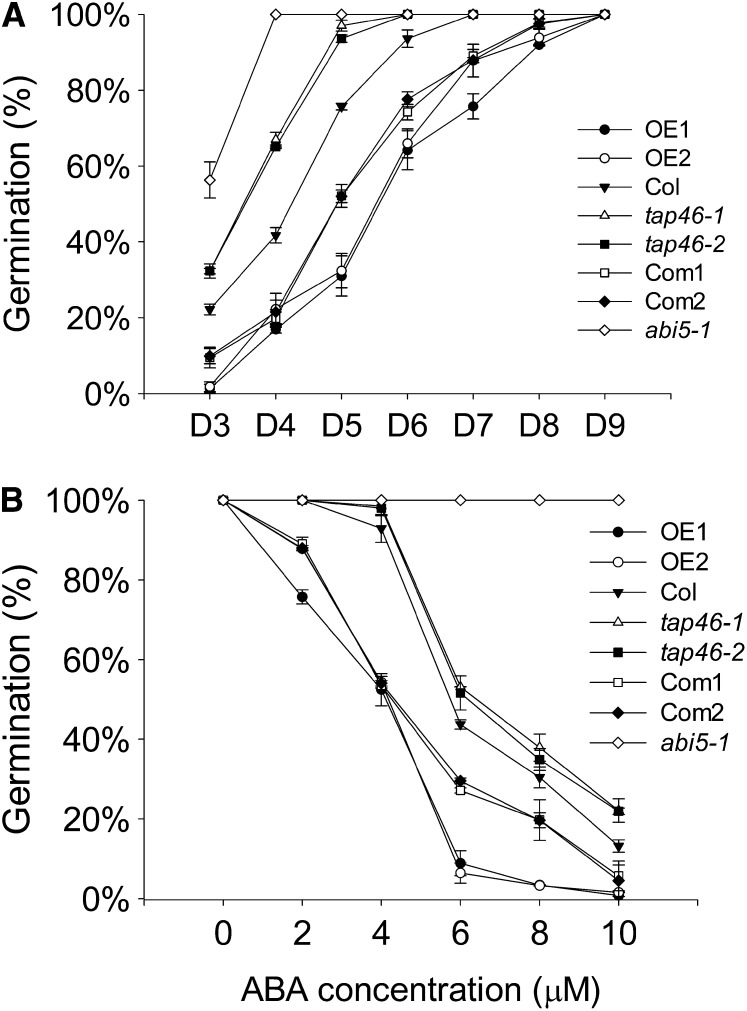
With the loss-of-function and gain-of-function mutants in hand, we analyzed how these plants would respond to ABA treatment during seed germination. Under normal condition, all plants showed similar growth at germination and postgermination stages (Supplemental Fig. S2). In the presence of 2 µm of ABA, the loss-of-function mutants tap46-1 and tap46-2 were less sensitive to ABA treatment in germination compared with wild-type ecotype Columbia (Col-0; the progenitor line of tap46-1 and tap46-2), whereas the gain-of-function mutants OE1 and OE2 were more sensitive to ABA compared with wild-type plants in the early stages of germination (Fig. 3A). For example, at day 4, close to 65% of tap46-1 and tap46-2 plants already germinated, whereas only about 40% of wild-type plants germinated and about 17% of TAP46-overexpressing plants germinated (Fig. 3A). As a control, the ABA-insensitive mutant abi5-1 demonstrated 100% germination by day 4 (Fig. 3A). The two TAP46-overexpressing plants also displayed lower germination rates at higher ABA concentrations compared with the wild type and loss-of-function mutants tap46-1 and tap46-2 (Fig. 3B).
Figure 3.
Germination rates of wild-type plants, tap46 mutants, complemented lines, and TAP46-overexpressing plants in the presence of ABA. A, Germination rates of wild-type (Col-0), tap46 mutants (tap46-1 and tap46-2), complemented lines of tap46-1 (Com1 and Com2), TAP46-overexpressing plants (OE1 and OE2), and abi5-1 mutant in the presence of 2 μm of ABA from day 3 to day 9 (D3–D9). B, Germination rates of wild-type (Col-0), tap46 mutants (tap46-1 and tap46-2), complemented lines of tap46-1 (Com1 and Com2), TAP46-overexpressing plants (OE1 and OE2), and abi5-1 mutant in the absence and presence of various concentrations of ABA on day 7. Three independent experiments were performed.
To confirm that the ABA-insensitive phenotype of tap46-1 is due to the T-DNA insertion into TAP46, we rescued this mutant phenotype by introducing a wild-type TAP46 gene into this mutant with a different vector, pCambia1302 mGFP5 (Kim et al., 2007). After transformation and screening, two lines that expressed the transgene transcript at relatively high level, Com1 and Com2 (Fig. 1D), were chosen for complementation analysis. Our results indicated that the wild-type TAP46 gene driven by the 35S promoter could rescue the ABA-insensitive phenotype (Fig. 3A). Because the rescued lines used a strong constitutive promoter, these rescued lines were more sensitive to ABA than their wild-type progenitor line Col-0. However, they were not as sensitive as the two TAP46-overexpressing lines. Our data clearly indicate that TAP46 plays a positive role in the ABA-regulated inhibition of germination.
TAP46 Is Induced by ABA and Mainly Expressed in Seed
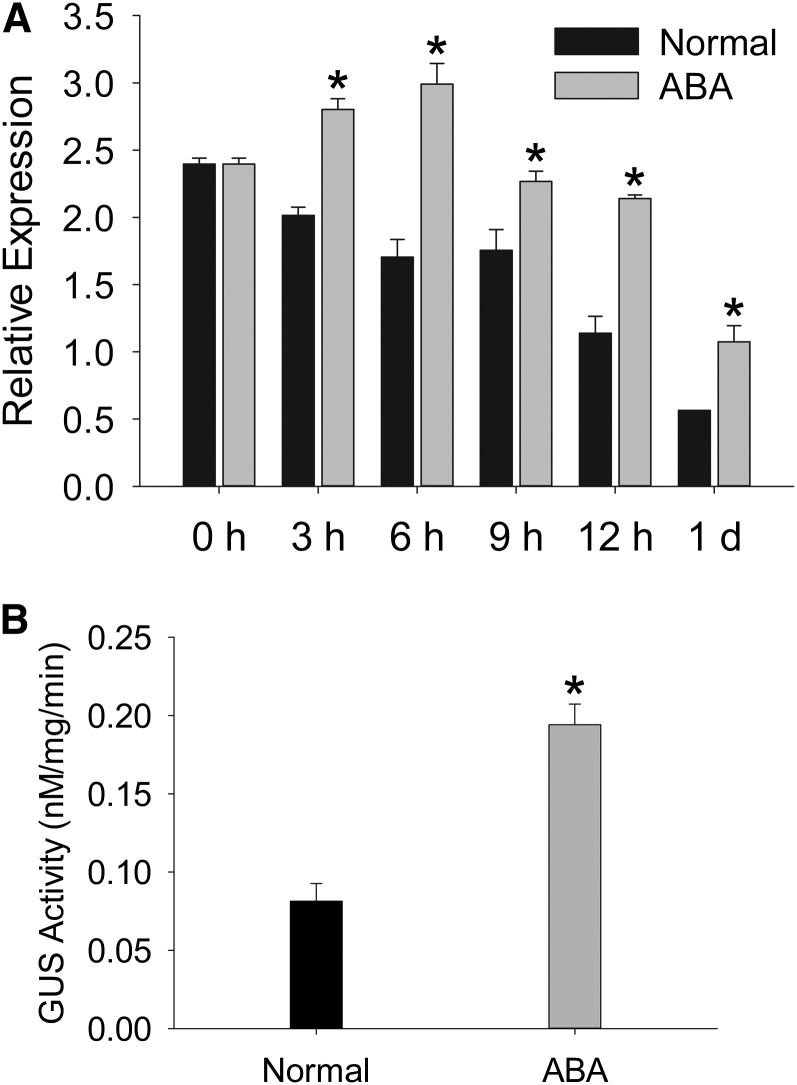
The positive role of TAP46 in ABA signaling during seed germination led us to investigate this gene’s expression pattern in Arabidopsis. We first tested whether ABA influences TAP46’s transcript during germination by using the real-time PCR technique. Under normal condition, the TAP46 transcript decreased quickly after stratification, and within 1 d, it decreased to 20% of the level found at 0 h (just before moving seeds from 4°C to room temperature of 22°C; Fig. 4A). However, if ABA was provided in the germination media, the TAP46 transcript was induced transiently (Fig. 4A), reaching the highest level at 6 h. Then, it decreased, but it maintained at higher levels than that under normal condition after 24 h (Fig. 4A).
Figure 4.
Expression of TAP46 is inducible by ABA. A, Real-time PCR analysis of TAP46 transcript in germinating seeds in the absence and presence of ABA. Three technical repeats and two biological repeats were performed. B, Expression of TAP46 is induced by ABA in the 1-d-old germinating seeds. The induction of TAP46 expression by ABA was analyzed by measuring the GUS activities of the TAP46 promoter::GUS fusion plants in the absence and presence of ABA. Three independent experiments were performed. ABA concentration used was 10 µm. Asterisk indicates statistically significant at 5% by Student’s t test.
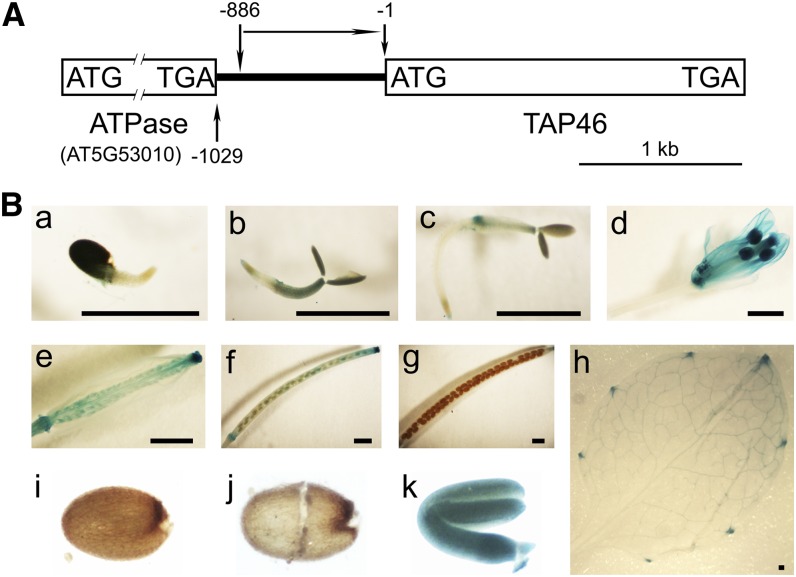
To study the temporal and spatial expression pattern of TAP46, we fused the promoter sequence of TAP46 (–1 to –886 bases 5′ to ATG; Fig. 5A) to the GUS gene. Two independent homozygous transgenic lines containing the PTAP46::GUS construct were used to determine the expression pattern of TAP46, and similar results were obtained with these two lines. We first analyzed transgenic seeds to see if GUS is induced by ABA treatment. These transgenic seeds were soaked on wet filter paper saturated with water or ABA solution for 1 d, and then we analyzed GUS activities. The GUS activity from the ABA-treated seeds was 2 times higher than that from water-treated seeds (Fig. 4B), which supports the real-time PCR data (Fig. 4A) that ABA increases TAP46 transcript.
Figure 5.
Spatial expression patterns of TAP46 in Arabidopsis. A, The 5′ sequence information of TAP46. The 5′ sequence –1 to –886 of TAP46 was used for the GUS fusion construct. B, GUS staining patterns of TAP46 promoter::GUS transgenic plants. a, A 1-d-old germinating seed. b, A 2-d-old seedling. c, A 3-d-old seedling. d, A flower. e, A developing silique 5 d after flowering. f, A developing silique 10 d after flowering. g, A mature silique 17 d after flowering. h, A mature rosette leaf. i, An intact mature seed. j, A seed coat plus endosperm. k, An embryo. Bar = 1 mm.
We then studied the spatial expression pattern of TAP46 using the PTAP46::GUS transgenic plants. The GUS activity was found in the cotyledon leaves of 1- to 2-d-old germinating seeds (Fig. 5B, a and b). At day 3, the GUS activity was limited to the junction area between shoot and root (Fig. 5Bc). The GUS activity was not detected in adult plants except in the hydrathodes of rosette leaves (Fig. 5Bh). In flowers, TAP46 was highly expressed in anthers and pistil tissues and at the joint tissues between carpel and flower stock (Fig. 5Bd). GUS activity was also found in developing seeds (Fig. 5B, e and f). However, in matured siliques and in dry seeds, no GUS staining was found (Fig. 5B, g and i), which could be due to the difficulty for the GUS staining solution to get into dry seeds. Therefore, we manually separated the embryo from the endosperm/seed coat and then stained them with the GUS staining solution. We still could not see GUS straining in the endosperm/seed coat (Fig. 5Bj), but we could see strong GUS straining in the embryo (Fig. 5Bk). This result suggests that TAP46 is specifically expressed in the embryo, not in the endosperm or seed coat.
To confirm these GUS straining patterns, we conducted real-time PCR experiments to quantify the transcript levels of TAP46 in different tissues (Supplemental Fig. S3). Compared with TAP46 expression in roots where the lowest level of TAP46 was found, the highest relative expression level, about 30 times that of roots, was found in 17-d siliques and dry seeds. With the silique developing, the accumulation of TAP46 transcript increased as well. In other tissues, including stems, flowers, and leaves, the relative expression levels were much lower than those in dry seeds.
TAP46 Is a Negative Regulator for PP2A Activity in Arabidopsis
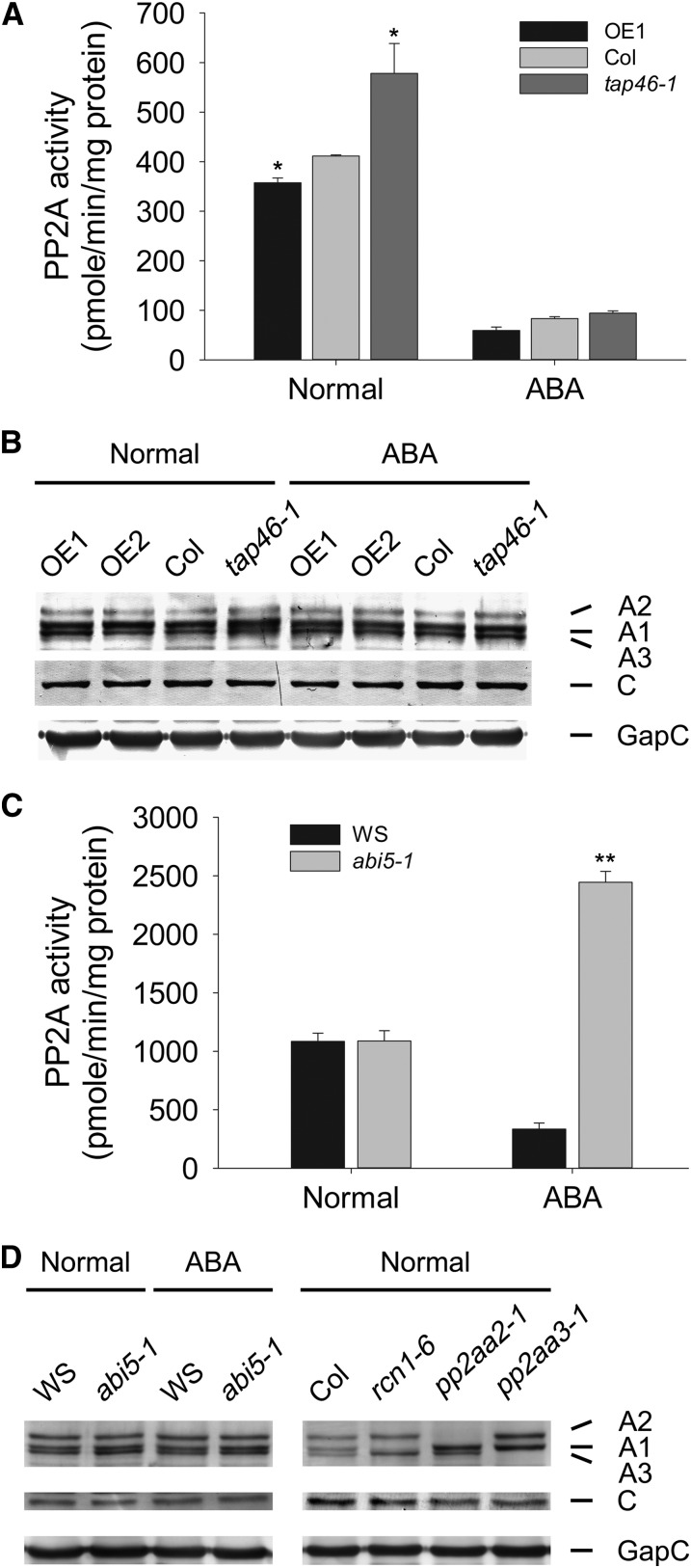
TAP46 was originally identified as a PP2A-associated protein that interacts with the C1 catalytic subunit of PP2A in a yeast two-hybrid screening (Harris et al., 1999). We confirmed this interaction between TAP46 and C1, and furthermore we found that TAP46 also interacts strongly with C2, the closest C subunit in Arabidopsis PP2A, by using the yeast two-hybrid assay (Supplemental Fig. S4). Because TAP46’s homologs in yeast and human, TAP42 and α4, were shown to be regulators of PP2A activities (Nanahoshi et al., 1998; Jiang, 2006), we tested whether TAP46 could affect the PP2A activity in germinating seeds. Under normal conditions, the loss-of-function mutant tap46-1 displayed 40% higher PP2A activity than wild-type plants, whereas the TAP46-overexpressing plant OE1 displayed lower PP2A activity in 1-d-old germinating seeds (Fig. 6A). These data indicate that TAP46 is a negative regulator of PP2A activity in Arabidopsis. In the presence of 10 µm of ABA, PP2A activities were depressed to very low levels in all three plants, yet the tap46-1 mutant still had the highest level of PP2A activity and OE1 plant had the lowest PP2A activity (Fig. 6A). The drastic impact of ABA in inhibiting PP2A activity in all three plants suggests that in addition to TAP46, other factors activated by ABA are also responsible for inhibiting PP2A activity and these factors are more potent in inhibiting PP2A activity.
Figure 6.
PP2A activities in TAP46-overexpressing, wild-type, tap46-1, and abi5-1 plants in the absence and presence of ABA. A, PP2A activities of 1-d-old germinating seeds of TAP46-overexpressing (OE1), wild-type (Col-0), and tap46-1 plants in the absence and presence of ABA. The ABA concentration used was 10 µm, and three independent experiments were performed. Asterisk indicates statistically significant at 5% by Student’s t test. B, Western-blot analyses of the A and C subunits of PP2A in the absence and presence of ABA in TAP46-overexpressing (OE1 and OE2), wild-type (Col-0), and tap46-1 plants. A1, A2, and A3 indicate the three A subunits of PP2A, and C indicates the C subunit(s) of PP2A. C, PP2A activities of 1-d-old germinating seeds of the wild type (Ws) and abi5-1 mutant in the absence and presence of ABA. The ABA concentration used was 10 µm, and three independent experiments were performed. Two asterisks indicate statistically significant at 1% by Student’s t test. D, Western-blot analyses of the A and C subunits of PP2A in the absence and presence of ABA in the wild type and abi5-1 mutant. rcn1-6, pp2aa2-1, and pp2aa3-1 indicate three A subunit mutants of the Arabidopsis PP2A (used as reference lines for marking the A1, A2, and A3 subunit, respectively).
To rule out the possibility that the reduced PP2A activity was due to altered expression of PP2A subunit genes, we analyzed the steady-state levels of A and C subunits of PP2A in the presence and absence of ABA. Our western-blot data indicate that the three A subunits were not reduced by the ABA treatment and the C subunits were also not affected by the ABA treatment (Fig. 6B). Our data show that the ABA-induced suppression of PP2A activity is likely due to posttranslational events.
Interestingly, when we analyzed PP2A activity in the ABA-insensitive mutant abi5-1, we found that the PP2A activity was significantly higher in abi5-1 mutant in the presence of ABA, but not in the absence of ABA (Fig. 6C). Although it is not known why the basal level of PP2A activity is much higher in ecotype Wassilewskija (Ws) than in Col-0, it is clear that ABA inhibits PP2A activity in both ecotypes of Arabidopsis. Because the abi5-1 mutant was isolated from the Ws background, we used wild-type Ws as the reference line. The increased PP2A activity in abi5-1 in the presence of ABA was not due to increased expression of PP2A subunit genes, as the steady-state levels of A and C subunits were about the same in the presence or absence of ABA (Fig. 6D). Because ABI5 is a transcription factor that functions downstream of TAP46 (see below), the increased PP2A activity in the abi5-1 mutant indicate that some genes downstream of ABI5 might be negative regulators of PP2A.
TAP46 Interacts with ABI5 in Vivo
Previous studies reported that the transcriptional factor ABI5 plays important roles in ABA-regulated seed maturation and seed germination (Carles et al., 2002; Finkelstein et al., 2008). Furthermore, the expression pattern of TAP46 mimics that of ABI5 (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000). We therefore studied the relationship between TAP46 and ABI5. In yeast two-hybrid assay, TAP46 interacted with ABI5 (Fig. 7A). We then performed coimmunoprecipitation experiments to further test these two proteins’ interaction in vivo. Firstly, anti-ABI5 antibodies were used to pull down the ABI5 complex from cellular extracts of TAP46-GFP fusion plant. Then, anti-GFP antibodies were used to analyze the complex in the western-blot experiments. Our data indicate that only the cellular extracts from TAP46-GFP transgenic plants contain a protein of 72 kD that could be recognized by GFP antibodies (Fig. 7B). This Mr matches the expected Mr of TAP46-GFP fusion protein, and no similar protein could be found in the cellular extracts from abi5-1 mutant or wild-type Arabidopsis plants (Fig. 7B). These data indicate that TAP46 does interact with ABI5 in vivo.
Figure 7.
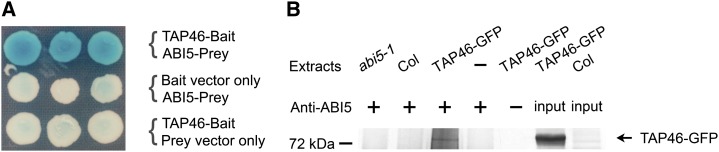
TAP46 interacts with ABI5. A, TAP46-ABI5 interaction in the yeast two-hybrid system. B, TAP46-ABI5 interaction as demonstrated by coimmunoprecipitation experiments. Cellular extracts were made from abi5-1 mutant, wild-type (Col-0), and TAP46-GFP transgenic plants. Plus or minus indicates with or without the pulling ABI5 antibodies, and input indicates purified TAP46-GFP and cellular proteins of wild-type Arabidopsis directly loaded in the western-blot experiments. The fusion protein, marked on the right, is recognized by anti-GFP antibodies.
TAP46 Stabilizes ABI5 and Prevents Dephosphorylation of ABI5 in Vivo
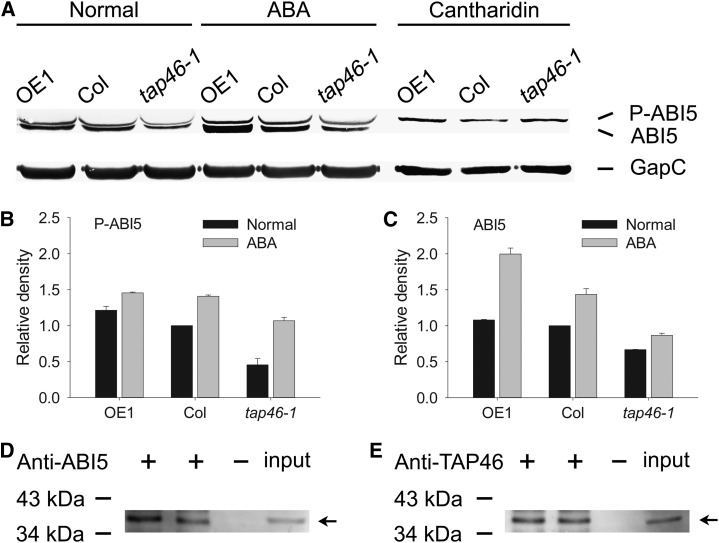
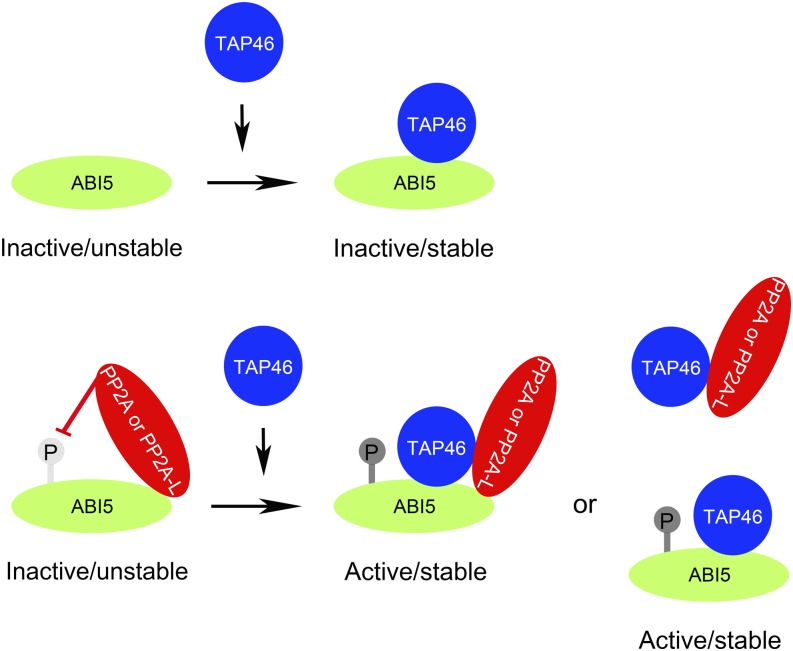
Lopez-Molina et al. (2001) reported that protein extracts from germinating seeds contained two forms of ABI5 that could be separated as 52- and 50-kD proteins in western-blot analysis, which were identified as phosphorylated ABI5 and free ABI5. Because ABA treatment leads to phosphorylation and stabilization of ABI5 (Lopez-Molina et al., 2001), it was suggested that phosphorylated ABI5 is the active and stable form, whereas the free ABI5 is the inactive and unstable form (Fujii et al., 2007). Because TAP46 could bind to ABI5 in vivo, then this TAP46-ABI5 interaction might stabilize ABI5 and protect ABI5 from phosphatase’s action in removing the phosphate from ABI5. If this hypothesis was correct, we would expect to see higher levels of free and phosphorylated ABI5 in the TAP46-overexpressing plants than in the wild type or tap46-1 mutant.
Under normal conditions, more free ABI5 and phosphorylated ABI5 were found in the 5-d-old seedlings of TAP46-overexpressing plants than in wild-type plants, and the tap46-1 mutants produced the smallest amount of both forms of ABI5 (Fig. 8A, left). Our quantitative analysis of the band intensity in the western blots indicates that TAP46-overexpressing plants produced 12% more phosphorylated ABI5 and 8% more free ABI5 than wild-type plants, and wild-type plants in turn produced 120% more phosphorylated ABI5 and 50% more free ABI5 than tap46-1 plants (Fig. 8, B and C). After ABA treatment for 1 d, both phosphorylated ABI5 and free ABI5 were increased in all plants (Fig. 8A, middle), which was consistent with the early discoveries that ABA induces ABI5 expression at seedling stages (Lopez-Molina et al., 2001; Brocard et al., 2002). Importantly, the biggest increase was found in the TAP46-overexpressing plants, and the smallest increase was found in the tap46-1 mutant. Again, our quantitative analysis of the band intensity in the western blots indicates that TAP46-overexpressing plants produced 3% more phosphorylated ABI5 and 39% more free ABI5 than wild-type plants, and wild-type plants in turn produced 32% more phosphorylated ABI5 and 66% more free ABI5 than tap46-1 plants after ABA treatment (Fig. 8, B and C). Our data clearly indicate that TAP46 overexpression is able to stabilize both forms of ABI5 and maintains more ABI5 in active form after ABA treatment.
Figure 8.
ABI5 is stabilized by TAP46 and dephosphorylated by PP2A or PP2A-ls, and both ABI5 and TAP46 can interact with the C subunit(s) of PP2A in vivo. A, TAP46 stabilizes ABI5 and protects ABI5 from being dephosphorylated by phosphatases. OE1 indicates TAP46-overexpressing plants, tap46-1 indicates mutant, P-ABI5 indicates phosphorylated ABI5, and ABI5 indicates free ABI5. The concentration used for ABA and cantharidin was 10 µm. B, The relative abundance of the phosphorylated ABI5 band in TAP46-overexpressing, the wild type, and tap46-1 mutant in A. C, The relative abundance of the free ABI5 band in TAP46-overexpressing, the wild type, and tap46-1 mutant in A. The error bar was calculated based on four independent experiments performed. D Protein-protein interaction between ABI5 and C subunit(s) of PP2A as demonstrated by coimmunoprecipitation experiments. Plus or minus indicates with or without ABI5 antibodies in pulling down cellular proteins, input indicates cellular proteins directly loaded in the western-blot experiments, and the arrow indicates the C subunit(s) of PP2A. E, Protein-protein interaction between TAP46 and C subunit(s) of PP2A as demonstrated by coimmunoprecipitation experiments.
To test what type of phosphatase is responsible for removing the phosphate from ABI5, we treated 4-d-old seedlings with cantharidin, a phosphatase inhibitor that is specific for PP2A or PP2A-ls such as PP4 and PP6. We then detected one ABI5 band in the western-blot experiment, and this band migrated at the 52-kD position (Fig. 8A, right), indicating that all of the ABI5 was in phosphorylated form after cantharidin treatment. Furthermore, the impact of TAP46 on ABI5’s stability became unimportant after inhibition of phosphatases, as the steady-state levels of phosphorylated ABI5 were roughly the same in all three plants, indicating that the phosphorylation state of ABI5 is more important in the stability of ABI5. To prove that cantharidin treatment could have kept ABI5 in active form (i.e. phosphorylated form) in Arabidopsis seedlings, we performed real-time PCR experiments to analyze the expression of three ABI5 downstream genes, Desiccation-Responsive29A (RD29A), RD29B, and 9-cis-epoxycarotenoid dioxygenase3 (NCED3). The transcripts of these three genes were significantly increased after cantharidin treatment (Supplemental Fig. S5).
ABI5 Directly Interacts with PP2A in Vivo
Because TAP46 is a regulator of PP2A, TAP46’s impact on ABA response might involve PP2A. More importantly, a study by Dai et al. (2013) showed that PP6 could dephosphorylate ABI5 in vivo, which prompted us to investigate the possible role of PP2A in regulating the ABI5 phosphorylation. We tested if PP2A could interact with ABI5 in vivo by conducting coimmunoprecipitation experiments. We used anti-ABI5 antibodies to immunoprecipitate ABI5 complex from cellular extracts and followed with anti-PP2A-C antibodies to detect the signal in western-blot experiment. The result shows that ABI5 could interact with the C subunits of PP2A in vivo (Fig. 8D). Using the same approach, we detected interaction between TAP46 and C subunits of PP2A as well (Fig. 8E), which supports the yeast two-hybrid data on TAP46/PP2A-C subunit interactions (Harris et al., 1999; Ahn et al., 2011; Supplemental Fig. S4). Our data indicate that in addition to PP6, PP2A is also a protein phosphatase that could dephosphorylate ABI5 in vivo.
TAP46 Regulates Downstream Genes of ABI5
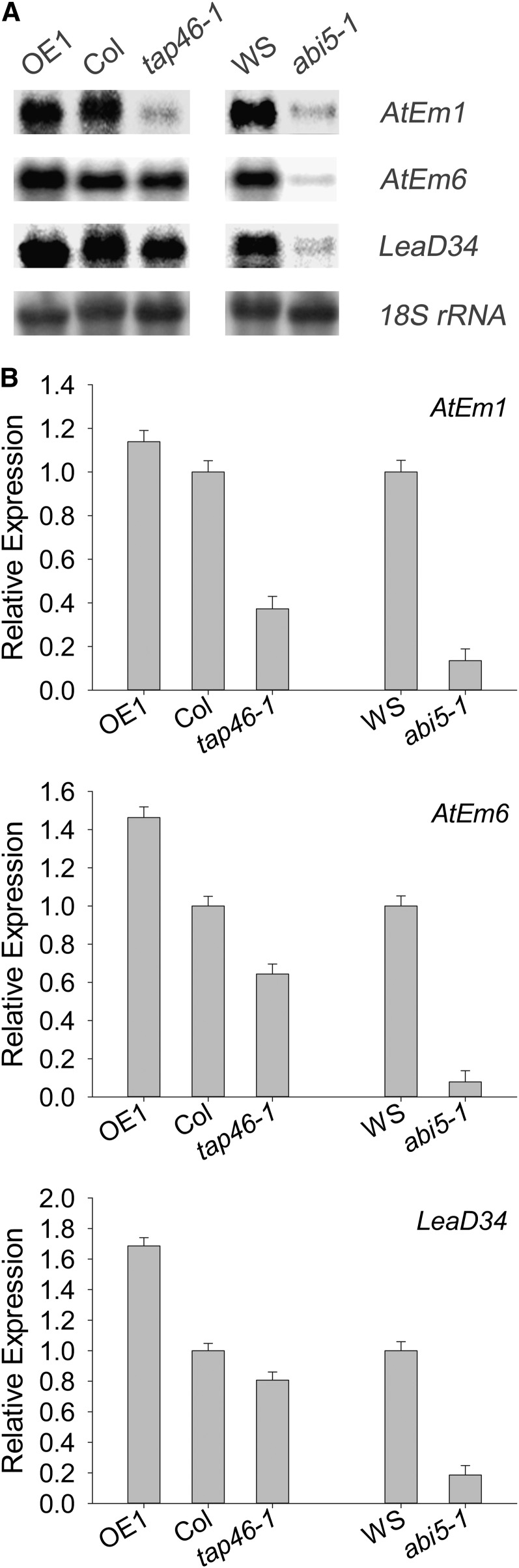
Because TAP46 is highly expressed during seed maturation (Fig. 5; Supplemental Fig. 3), which mimics that of ABI5 (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000), it is likely that downexpression of TAP46 and overexpression of TAP46 would affect transcript levels of genes downstream of ABI5. To test this possibility, we conducted RNA-blot analysis with a few ABI5-regulated genes in seeds. Early studies indicated that many late embryogenesis abundant (LEA) genes such as Arabidopsis thaliana late embryogenesis abundant gene1 (AtEm1) and AtEm6 were ABI5-regulated genes, as their transcripts were significantly reduced in abi5 mutants (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Carles et al., 2002). We analyzed the transcript levels of three LEA genes, AtEm1, AtEm6, and LeaD34. We isolated total RNAs from dry seeds of TAP46-overexpressing, wild-type, and tap46-1 mutant plants and compared the transcript levels of these genes. As expected, transcripts of these genes were reduced in abi5-1 mutant (Fig. 9). When TAP46 was overexpressed, transcripts of these genes were up-regulated, approximately by 20% to 70% higher than that of wild-type plants, whereas in tap46-1 mutant, the transcript levels of these genes were down-regulated, approximately by 20% to 60% lower than that of wild-type plants (Fig. 9). The gene expression pattern in the tap46-1 mutant is similar to that in abi5-1 mutant.
Figure 9.
RNA-blot analysis of three genes downstream of ABI5. A, RNA-blot analysis of dry Arabidopsis seeds. OE1 indicates TAP46-overexpressing line 1, and tap46-1 and abi5-1 indicate mutants. B, Relative levels of AtEm1, AtEm6, and LeaD34 with respect to the 18S rRNA in A. Three independent experiments were performed.
Because TAP46 expression is induced by ABA during seedling development (Fig. 4) and overexpression of TAP46 leads to an increase in both phosphorylated ABI5 and free ABI5 after ABA treatment (Fig. 8A), it is likely that some stress-inducible genes with promoters containing ABA response elements would also be regulated by TAP46. We therefore analyzed transcripts of three genes, RD29A, RD29B, and NCED3, that are representatives of such stress-inducible genes (Shinozaki and Yamaguchi-Shinozaki, 2007). The expressions of these three genes are very low in the 5-d-old Arabidopsis seedlings under normal condition (Fig. 10). However, with the addition of ABA in the germination media, the transcripts of these three genes were all induced (Fig. 10). However, these genes were all expressed at higher levels in the TAP46-overexpressing plants and at lower levels in the tap46-1 mutant (RD29A and RD29B) or about the same (NCED3) compared with wild-type plants (Fig. 10). These data indicate that other ABA-inducible genes are also likely regulated by TAP46.
Figure 10.
Real-time PCR analysis of three genes downstream of ABI5 in 5-d-old seedlings. OE1 indicates TAP46-overexpressing line 1, and tap46-1 indicates mutant. Actin8 was used as the reference. The ABA concentration used was 10 µm, and three independent experiments were performed.
DISCUSSION
In this report, we provide strong evidence that TAP46 is positively involved in the ABA signaling pathway, as overexpression of TAP46 in Arabidopsis made transgenic plants more sensitive to ABA in germination (Fig. 3). The TAP46 transcript was up-regulated by ABA treatment during germination (Fig. 4), the expression of TAP46 was found mainly during seed maturation (Fig. 5; Supplemental Fig. 3), and the highest transcript level was found in dry seeds (Supplemental Fig. 3), all suggesting that TAP46 plays positive roles in ABA-regulated seed maturation and inhibition of germination. Because TAP46 was initially found as a PP2A-associated protein in Arabidopsis and TAP46’s homologs in yeast (Tap42) and mammals (α4) regulate PP2A activities (Nanahoshi et al., 1998; Jiang, 2006), PP2A likely plays an important role in ABA-regulated processes in plants. Our early work with PP2A suggests that PP2A is positively involved in ABA signaling (Luo et al., 2006), and here we find evidence that PP2A is negatively involved in ABA signaling. TAP46 inhibits PP2A activity, as PP2A activity is higher in the tap46-1 mutant (Fig. 6A), and treatment of ABA in Arabidopsis seedlings reduces PP2A activity substantially (Fig. 6A), indicating an opposite role of PP2A in the ABA-regulated seeding development.
The molecular mechanism of TAP46 and PP2A’s involvement in ABA signaling during seed maturation and seedling development is likely centered on the regulation of ABI5 activity. It was previously shown that ABI5 was stabilized by ABA-induced phosphorylation, and upon removal of ABA, ABI5 was quickly degraded by the 26S proteasome (Lopez-Molina et al., 2001). Our data here show that TAP46 interacts with ABI5 in vivo (Fig. 8C), and in particular, overexpression of TAP46 increases the steady-state levels of phosphorylated ABI5 and free ABI5 (Fig. 8A), suggesting that TAP46-ABI5 interaction stabilizes both forms of ABI5 and likely prevents the action of phosphatases that remove the phosphate group from ABI5. The phosphatases involved are PP2A or its related protein phosphatases, because by using a protein phosphatase inhibitor, cantharidin, that is specific for PP2A, PP4, and PP6, we could keep most ABI5 in its stable form, i.e. phosphorylated form (Fig. 8A). Furthermore, we demonstrated that overexpression of TAP46 increases the transcript levels of ABI5-regulated genes during seed development and seed germination (Figs. 9 and 10). Based on our data, we propose that a major function of TAP46 is to stabilize ABI5, and another function of TAP46 is to interact with PP2A and PP2A-ls such as PP4 and PP6 to prevent PP2A or PP2A-l from binding to ABI5 and dephosphorylating ABI5, thereby keeping ABI5 in its active form (Fig. 11). This mechanism explains how TAP46 effectively stabilizes ABI5 and protects ABI5 from phosphatases’ action. However, TAP46’s binding to and protection of ABI5 should be a regulated event, as ABA-triggered stabilization of ABI5 is clearly mediated by TAP46, and TAP46 itself is also likely regulated by other factors, as expression of TAP46 is induced by ABA treatment (Fig. 4) and activity of TAP46 might be regulated by phosphorylation and dephosphorylation (Ahn et al., 2011).
Figure 11.
Working model showing how TAP46 stabilizes ABI5 and protects ABI5 from the action of PP2A or PP2A-ls in plant cells. ABI5 is phosphorylated upon ABA treatment and becomes active in triggering gene expression. Binding of TAP46 to ABI5 stabilizes ABI5 and prevents PP2A or PP2A-ls (e.g. PP6 or PP4) from removing the phosphate on ABI5, binding of TAP46 to PP2A or PP2A-l may prevent PP2A or PP2A-l from binding to ABI5, or the binding of TAP46 to ABI5 may prevent PP2A or PP2A-l from binding to ABI5. Whatever the mechanisms, the TAP46’s involvement is a positive factor in stabilizing ABI5 and keeping ABI5 in phosphorylated form, thereby staying active.
PP2A is one of the most abundant phosphatase families in plants (MacKintosh and Cohen, 1989), and in particular, the highest transcript levels of A subunit genes of PP2A were found in developing rape seeds (Brassica napus; Slabas et al., 1994). Gonzalez-Garcia et al. (2006) discovered that a C subunit of PP2A was found in nucleus and cytoplasm, and this C subunit gene was expressed in developing seeds and induced by gibberellin acid and repressed by ABA. This evidence indicates that PP2A is likely a candidate that TAP46 regulates in ABA-regulated seed maturation and inhibition of germination. It appears that the ABI5-controlled gene expression plays a major role in repressing PP2A activity in seed germination, as abi5-1 mutants contains much higher PP2A activity than wild-type plants in the presence of ABA (Fig. 6C). This positive feedback regulation might be very important in ABA-regulated inhibition of germination, which allows ABI5 staying active, then ABI5-regulated gene activities further strengthen ABA’s role in repressing PP2A. The other two transcription factors, ABI3 and ABI4, also play important roles in ABA-regulated seed maturation and inhibition of germination (Giraudat et al.,1992; Finkelstein et al., 1998), and these two transcription factors may be subject to similar regulation by other proteins like TAP46 and PP2A. We did not detect interaction between ABI3 and TAP46, but we found strong interaction between TAP46 and ABI4 in the yeast two-hybrid assay (Y. Zhu, R. Hu, J. Chen, G. Shen, and H. Zhang, unpublished data). It is clear that further study of the relationships between TAP46 and ABI4 and between PP2A and ABI4 will likely reveal more insight into the molecular mechanisms of ABA-regulated seed maturation and seedling development.
TAP46 plays other important roles in plant cells in addition to its roles in the ABA signaling pathway, as Ahn et al. (2011) demonstrated that reducing TAP46 expression either by virus-induced gene silencing or RNAi could lead to programmed cell death, reduction of global translation activities, induction of autophage, and anomaly in mitosis. We were lucky in a sense that both tap46-1 and tap46-2 mutants were weak allele mutants, which allowed us to maintain these mutants and study these mutants for many generations. Our discovery that TAP46 directly regulates activity of transcription factor ABI5 is not surprising, as it has been shown that α4 regulates the dephosphorylation of the transcriptional factors c-Jun and p53 in mice (Kong et al., 2004). Our finding suggests that the TAP46 regulation of transcriptional factors might also be a conserved mechanism, and plants apparently tapped this mechanism in regulating ABI5 activity in the ABA signaling pathway.
Ahn et al. (2011) showed that TAP46 interacts with PP4 and PP6, indicating a possibility that TAP46’s role in ABI5 regulation might involve PP4 and PP6 as well. We confirmed that TAP46 could interact with one isoform of PP4 (i.e. Protein Phosphatase X1) and two isoforms of PP6 (i.e. Arabidopsis FLOWER-SPECIFIC PHYTOCHROME-ASSOCIATED PROTEIN PHOSPHATASE1 and Arabidopsis FLOWER-SPECIFIC PHYTOCHROME-ASSOCIATED PROTEIN PHOSPHATASE3) in the yeast two-hybrid system (Supplemental Fig. S6). Dai et al. (2013) provided direct evidence that PP6 could dephosphorylate ABI5 in vivo and down-regulate the activity of ABI5. Their model regarding PP6’s role in regulating ABI5 is similar to ours. Therefore, we incorporated their discovery into our model (Fig. 11), and this model illustrates how TAP46 regulates PP2A and PP2A-related phosphatases such as PP4 and PP6 in the ABA-regulated gene expression. The catalytic subunit of PP6 is very similar to the catalytic subunits of PP2A (Farkas et al., 2007), and PP6 also physically interacts with the A subunits of PP2A (Dai et al., 2012). All of this evidence suggests that TAP46 could regulate PP2A and its related protein phosphatases in plants.
Harris et al. (1999) isolated TAP46 as an interacting protein for the C1 subunit of PP2A, and we found that TAP46 also interacts with the C2 subunit as strongly as with C1 in the yeast two-hybrid system (Supplemental Fig. S4). This is not surprising because C1 and C2 are highly homologous and they share greater than 90% identity in amino acid sequences (Ariño et al., 1993). TAP46 also interacts with C3 (Supplemental Fig. S4), which is similar to what was reported by Ahn et al. (2011). Interestingly, the C5 subunit (At1G69960), although slightly closer to C1 and C2 based on the phylogenetic analysis of amino acid sequences (Farkas et al., 2007), does not interact with TAP46 (Supplemental Fig. S4). The C4 subunit (At3G58500) also does not interact with TAP46 (Supplemental Fig. S4).
There are at least 17 B subunits of PP2A in Arabidopsis, but we do not know which B or how many B subunits are involved in recognizing ABI5. Consequently, we do not know what combinations of these PP2A subunits are involved in regulating ABI5’s activity in the ABA-regulated seed development and germination. Nevertheless, having identified TAP46, PP2A, and PP6 as new regulatory factors for ABI5, we are one step further in filling the gaps in our understanding of the ABA signaling pathways. Identifying the specific B subunit(s) involved in recognizing ABI5 will help explain the fine-tuning of the transcriptional regulation of ABI5-regulated gene expression. It will also provide information on PP2A’s functional allocations in ABA-regulated cellular processes. Some PP2As are negatively involved in ABA action, such as the case given here and by Pernas et al. (2007), and others are positively involved in ABA functions as demonstrated previously (Kwak et al., 2002; Luo et al., 2006). Only until we identify each specific B subunit for a specific ABA-regulated process can we understand why PP2A can perform functions that are in opposite directions in the ABA signaling pathways. The PP2A-associated protein TAP46 clearly plays a critical role in the ABA-regulated seed development and germination.
MATERIALS AND METHODS
Confirmation of tap46 Mutants
Two T-DNA insertion mutants, tap46-1 (Salk_091669) and tap46-2 (Salk_067216C), were obtained from the Arabidopsis Biological Resources Center at Ohio State University. The T-DNA insertion sites of these two mutants were confirmed by conducting PCR experiments using gene-specific primers tap46-F1 and tap46-R1 and T-DNA-specific primer LBa1 for tap46-1 and tap46-F2, tap46-R2, and T-DNA-specific primer LBa1 for tap46-2. The PCR reaction conditions for confirming these two mutants are 95°C for 2 min first, then 35 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s, followed by an extension of 5 min at 72°C. To extract genomic DNAs for PCR analysis, two fresh flowers were ground in DNA extraction buffer, which contains 50 mm Tris, pH 8.0, 10 mm EDTA, pH 8.0, 100 mm NaCl, 1.0% (w/v) SDS, and 10 mm β-mercaptoethanol, and centrifuged 5 min at room temperature at 13,000g. DNAs were precipitated with equal volume of isopropanol and then resuspended into Tris-EDTA buffer (pH 8.0). Information on PCR primers is listed in Supplemental Table S1.
Creation of TAP46-Overexpressing Plants
To overexpress TAP46 in Arabidopsis (Arabidopsis thaliana), the coding region of TAP46 was amplified from a complementary DNA (cDNA) library of 6- to 8-d seedlings using TAP46 primers TAP46-OF1 and TAP46-OR2 and cloned into the pBI121 vector (Jefferson et al., 1987) by replacing the GUS gene. The PCR reaction conditions for amplifying TAP46 are 95°C for 30 s, 56°C for 30 s, and 72°C for 60 s, 35 cycles. The Agrobacterium tumefaciens strain GV3101 containing the TAP46-overexpression construct was used to transform wild-type Arabidopsis (Col-0) using the floral dip method of Clough and Bent (1998). Kanamycin-resistant plants were selected from T1 and T2 seeds. Transgenic plants with 3 to 1 ratio of kanamycin resistance versus sensitivity were used for obtaining homozygous lines for molecular characterization.
Creation of TAP46 Promoter-GUS Fusion Plants
The promoter sequence of TAP46 was amplified from the genomic DNA of wild-type Arabidopsis (Col-0) using the primers TAP46-Pro-F and TAP46-Pro-R by PCR. The PCR reaction conditions are as follows: 95°C for 30 s, 56°C for 30 s, and 72°C for 60 s, 35 cycles. The PCR product was inserted into the pBI121 vector by replacing the Cauliflower mosaic virus 35S promoter with HindIII and XbaI restriction enzymes. Then the construct was introduced into A. tumefaciens strain GV3101, which was used for Arabidopsis transformation. The homozygous transgenic plants for this PTAP46::GUS construct were obtained and used for histochemical staining and GUS activity assays.
Complementation of the tap46-1 Mutant
The oligonucleotide primers TAP46-Com-F and TAP46-Com-R were used to amplify the wild-type TAP46 cDNA from an Arabidopsis cDNA library in a PCR experiment (95°C for 30 s, 56°C for 30 s, and 72°C for 60 s, 35 cycles). The PCR-amplified DNA fragment was cut by using the restriction enzymes NcoI and SpeI, and the DNA fragment was inserted into the binary transforming vector pCambia1302 mGFP5 vector (Kim et al., 2007). The resulting recombinant binary vector was then introduced into the A. tumefaciens GV3101, which was then used to transform the tap46-1 mutant using the floral dip method of Clough and Bent (1998).
Plant Growth and Germination Assay
Arabidopsis seeds (>100 seeds per line) were surface sterilized in 15% bleach for 10 min, followed by three to five times of wash in sterile water (5 min each time). The seeds were then incubated at 4°C for 4 d in darkness and transferred to a growth chamber with a photoperiod of 16-h light and 8-h dark at 22°C. For ABA treatments, more than 100 sterilized seeds were placed on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962), supplemented with different concentrations of ABA. After stratification at 4°C, seeds were transferred to a growth chamber for germination assay at indicated times. The germination was scored as radical tip emerged from seed coat. The data were based on at least three experiments.
Histochemical Staining and GUS Activity Assay
When analyzing the GUS activity in seed, dry seeds were dissected into embryo and endosperm/seed coat by soaking the seed in water for 4 to 6 h at 4°C as described by Penfield et al. (2006), and then the dissected embryo and endosperm/seed coat were put directly into a staining solution that is made of 50 mm NaPO4, pH 7.0, 10 mm Na2EDTA, pH 8.0, 0.1% (v/v) Triton X-100, and 0.5 mg mL–1 5-bromo-4-chloro-3-indolyl β-d-glucuronide and incubated at 37°C in the dark overnight. The GUS activity (blue color) was recorded with a camera-enabled dissecting microscope after washing the tissues in 70% ethanol two to three times.
For quantitative GUS activity assay, total proteins were extracted with the extraction buffer (50 mm NaPO4, pH 7.0, 10 mm Na2EDTA, pH 8.0, 0.1% [v/v] Triton X-100, 0.1% sodium lauryl sarcosine, and 10 mm β-mercaptoethanol). Then the chemical 4-methylumbelliferyl-β-d-glucuronide was used to measure the GUS activity using a spectrofluorometer as described in Jefferson et al. (1987).
RNA-Blot Analysis
Ten-day-old Arabidopsis plants or dry seeds were ground in liquid nitrogen, and total RNAs were extracted by using the PureLink Plant RNA Reagent (Invitrogen) according to the manufacturer’s instructions. The extracted RNAs were run in a 1.2% (w/v) denaturing agarose gel and then transferred onto a BioTrans(+) nylon membrane (ICN Biochemicals). The 32P-labeled probes were then used to hybridize with the nylon membrane. After hybridization, the membrane was washed extensively as described by Church and Gilbert (1984) and then exposed to a PhosphorImager screen for 4 h before data were collected. The 18S ribosomal RNA (rRNA) was used as the RNA loading control. The oligonucleotide primers used for amplifying cDNAs for making hybridization probes were AtEm1-F1 and AtEm1-R1for AtEm1 (AT3G51810), AtEm6-F1 and AtEm6-R1 for AtEm6 (AT2G40170), and LeaD34-F1 and LeaD34-R1 for LeaD34 (AT3G22490).
RT-PCR and Real-Time Quantitative PCR Analyses
For RT, the first-strand cDNA was synthesized from 1 µg of total RNAs using the superscript reverse transcriptase from Invitrogen with oligo(dT)18 as the primer according to the manufacturer’s instructions. The PCR reaction was 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s, 28 cycles, followed by an extension of 5 min at 72°C. The oligonucleotide primers were TAP46-RT-F1 and TAP46-RT-R1 for TAP46 and Actin7-F and Actin7-R for Actin7. The final PCR products were analyzed by gel electrophoresis. Real-time quantitative PCR was performed with a PCR machine (7500 Sequence Detection System, Applied Biosystems) using the SYBR Green Supermix (Bio-Rad Laboratories). One microliter of the cDNA product (from RT reaction above) was used as the template in a 25-µL reaction. PCR was conducted after a preincubation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 15 s and extension at 55°C for 40 s. Two biological and three technical replicates were used for each experiment. The oligonucleotide primers were TAP46-qPCR-F1 and TAP46-qPCR-R1 for TAP46, Actin8-F and Actin8-R for Actin8, RD29A-F1 and RD29A-R1 for RD29A (At5G52310), RD29B-F1 and RD29B-R1 for RD29B (At5G52300), and NCED3-F1 and NCED3-R1 for NCED3 (At3G14440).
PP2A Activity Assay
The protein Ser/Thr phosphatase assay system (New England Biolabs) was used to analyze the PP2A activity directly from seed protein extracts according to manufacturer’s instructions. The protein substrate used in this assay was myelin basic protein (MyBP), and EDTA was added to inhibit the activities of PP2C. A specific protein phosphatase inhibitor2 (I-2; New England Biolabs) was used to inhibit the activity of PP1. The PP2A activity is defined as the release of phosphate from MyBP. Different ecotypes of Arabidopsis have different baseline PP2A activities. Therefore, mutant was compared to its parental ecotype when PP2A activity was measured (Fig. 6).
TAP46-ABI5 Interaction in Yeast Two-Hybrid System
The full-length cDNAs of TAP46 and ABI5 genes were amplified from a cDNA library using oligonucleotide primers TAP46-YF1 and TAP46-YR1 and ABI5-YF1 and ABI5-YR1, respectively, digested with restriction enzymes EcoRI and XhoI, and then inserted into the prey vector (ABI5) and the bait vector (TAP46), respectively. The information on the prey vector pJG4-5 and bait vector pEG202 and the procedure for analyzing protein-protein interaction in this yeast (Saccharomyces cerevisiae) two-hybrid system was described by Golemis et al. (1996).
Protein Coimmunoprecipitation Assay (Pull-Down Experiment)
The coding sequence of TAP46 was amplified from a cDNA library using oligonucleotide primers TAP46-GFP-F1 and TAP46-GFP-R1 and cloned into the pBI121-GFP vector (Shen et al., 2010) with restriction enzymes XbaI and BamHI. The resulting vector was then transformed into the A. tumefaciens strain GV3101, which was used for Arabidopsis transformation to create TAP46-GFP fusion transgenic plants. When homozygous lines of TAP46-GFP fusion transgenic plants were obtained, they were used for pull-down experiment.
Prestratified Arabidopsis seeds (4°C for 3 d) of abi5-1, the wild type (Col-0), and TAP46-GFP fusion transgenic lines were put into MS solution for 1 d, followed by adding 10 μm ABA for another day. After grinding seeds in liquid nitrogen, the protein extraction buffer (10 mm EDTA, 0.1% [v/v] Triton X-100, 0.1% [w/v] sodium lauryl sarcosine, 40 mm sodium phosphate buffer, pH 7.0, 10 mm β-mercaptoethanol, 1 μg mL–1 leupeptin, 1 μg mL–1 aprotonin, and 1 mm phenylmethylsulfonyl fluoride) was added to extract total soluble proteins and then centrifuged at 13,000g for 10 min at 4°C to collect supernatant. The Bio-Rad protein assay system (Bradford, 1976) was used to determine protein concentration. Approximately 800 μg protein extracts were incubated with 10 μL ABI5 antibodies (see Supplemental Materials and Methods S1 for how antibodies were made) for 2 h at 4°C, followed by adding 50 μL Protein A-agarose slurry to bind antibodies for 2 h. Agarose-immune complexes were washed in Nonidet P-40 buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 1% Nonidet P-40) five times and then mixed with 100 μL 1× SDS protein loading buffer (50 mm Tris-HCl, pH 6.8, 2% SDS [w/v], 10% glycerol [v/v], 100 mm dithiothreitol, and 0.01% bromphenol blue). After boiling for 5 min and centrifugation at 13,000g for 5 min at room temperature, the supernatants were subject to SDS-PAGE and western-blot analysis using GFP antibodies (purchased from Invitrogen).
Western-Blot Analysis
The antibody against the C subunits of PP2A was purchased from Millipore, and this antibody could recognize all five catalytic subunits of PP2A in Arabidopsis, as it was raised against the most conserved part of all PP2A catalytic subunits. This antibody was used by many PP2A researchers in the plant science community, including Pernas et al. (2007) and Wu et al. (2011). Proteins from wild-type, tap46-1, tap46-2, and various transgenic plants were subjected to electrophoresis in a 8% SDS polyacrylamide gel. The polyclonal antibodies against cytosolic glyceraldehyde-3-P-dehydrogenase (GapC) were used as the loading control in the western-blot experiments. The conditions for blotting and color development were the same as described previously by Shen et al. (2010). The band intensity in the western blot was obtained by using a densitometry ImageQuant TL software (Amersham Biosciences), and each band was calibrated with the loading control band (i.e. GapC) and then compared to the band in wild-type plants that was set as 100%.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At5G53000 (TAP46).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes of the wild type, TAP46-overexpressing, and tap46 mutants under normal growth conditions for 16 d.
Supplemental Figure S2. Phenotypes of wild-type, TAP46-overexpressing, and tap46-1 mutant plants on MS plate for 14 d.
Supplemental Figure S3. Transcript analysis of TAP46 in various tissues of Arabidopsis.
Supplemental Figure S4. Protein-protein interactions between TAP46 and the C subunits of PP2A in the yeast two-hybrid system.
Supplemental Figure S5. Expression of the three ABI5 downstream genes RD29A, RD29B, and NCED3 is activated by the PP2A inhibitor cantharidin in 5-d-old seedlings.
Supplemental Figure S6. TAP46 interacts with PP4 and PP6, in addition to ABI5, in the yeast two-hybrid system.
Supplemental Table S1. List of oligonucleotide primers used for PCR and cloning experiments.
Supplemental Materials and Methods S1. Materials and Methods for supplemental materials.
Acknowledgments
We thank Ruth Finkelstein for providing the abi5-1 mutant and for advice throughout our research and Alison Delong for providing PP2A-A subunit mutants a1, a2, and a3 and sharing information on PP2A.
Glossary
- ABA
abscisic acid
- RNAi
RNA interference
- T-DNA
transfer DNA
- Col-0
ecotype Columbia
- Ws
ecotype Wassilewskija
- cDNA
complementary DNA
- MS
Murashige and Skoog
- rRNA
ribosomal RNA
- RT
reverse transcription
- GapC
glyceraldehyde-3-P-dehydrogenase
References
- Ahn CS, Han JA, Lee HS, Lee S, Pai HS. (2011) The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 23: 185–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariño J, Pérez-Callejón E, Cunillera N, Camps M, Posas F, Ferrer A. (1993) Protein phosphatases in higher plants: multiplicity of type 2A phosphatases in Arabidopsis thaliana. Plant Mol Biol 21: 475–485 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brandt B, Brodsky DE, Xue S, Negi J, Iba K, Kangasjärvi J, Ghassemian M, Stephan AB, Hu H, Schroeder JI. (2012) Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc Natl Acad Sci USA 109: 10593–10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR. (2002) Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M. (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373–383 [DOI] [PubMed] [Google Scholar]
- Chen J, Peterson RT, Schreiber SL. (1998) α4 associates with protein phosphatases 2A, 4, and 6. Biochem Biophys Res Commun 247: 827–832 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crespo JL, Hall MN. (2002) Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol Mol Biol Rev 66: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dai M, Xue Q, Mccray T, Margavage K, Chen F, Lee JH, Nezames CD, Guo L, Terzaghi W, Wan J, et al. (2013) The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25: 517–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Zhang C, Kania U, Chen F, Xue Q, McCray T, Li G, Qin G, Wakeley M, Terzaghi W, et al. (2012) A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell 24: 2497–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Arndt KT. (1996) Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev 10: 1904–1916 [DOI] [PubMed] [Google Scholar]
- Düvel K, Broach JR. (2004) The role of phosphatases in TOR signaling in yeast. Curr Top Microbiol Immunol 279: 19–38 [DOI] [PubMed] [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C. (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 12: 169–176 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR. (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5: 765–771 [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu J. (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, et al. (2010) Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA 107: 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis EA, Gyuris J, Brent R (1996) Interaction trap/two-hybrid system to identify interacting proteins. In FM Ausubel, R Brent, RE Kingston, DD Moore, JG Seidman, JA Smith, K Struhl, eds, Current Protocols in Molecular Biology. John Wiley and Sons, New York, pp 20.1.1–20.1.27 [Google Scholar]
- Gonzalez-Garcia MP, Rodriguez D, Nicolas C, Nicolas G, Lorenzo O. (2006) A protein phosphatase 2A from Fagus sylvatica is regulated by GA3 and okadaic acid in seeds and related to the transition from dormancy to germination. Physiol Plant 128: 153–162 [Google Scholar]
- Harris DM, Myrick TL, Rundle SJ. (1999) The Arabidopsis homolog of yeast TAP42 and mammalian α4 binds to the catalytic subunit of protein phosphatase 2A and is induced by chilling. Plant Physiol 121: 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua D, Wang C, He J, Liao H, Duan Y, Zhu Z, Guo Y, Chen Z, Gong Z. (2012) A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 24: 2546–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui S, Kuwahara K, Mizutani J, Maeda K, Kawai T, Nakayasu H, Sakaguchi N. (1995) Molecular cloning of a cDNA clone encoding a phosphoprotein component related to the Ig receptor-mediated signal transduction. J Immunol 154: 2714–2723 [PubMed] [Google Scholar]
- Janssens V, Goris J. (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 353: 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. (2006) Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70: 440–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim OT, Bang KH, Shin YS, Lee MJ, Jung SJ, Hyun DY, Kim YC, Seong NS, Cha SW, Hwang B. (2007) Enhanced production of asiaticoside from hairy root cultures of Centella asiatica (L.) Urban elicited by methyl jasmonate. Plant Cell Rep 26: 1941–1949 [DOI] [PubMed] [Google Scholar]
- Kong M, Ditsworth D, Lindsten T, Thompson CB. (2009) α4 is an essential regulator of PP2A phosphatase activity. Mol Cell 36: 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong M, Fox CJ, Mu J, Solt L, Xu A, Cinalli RM, Birnbaum MJ, Lindsten T, Thompson CB. (2004) The PP2A-associated protein α4 is an essential inhibitor of apoptosis. Science 306: 695–698 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Leon-Kloosterziel KM, Schwartz SH, Zeevaart JAD. (1998) The genetic and molecular dissection of abscisic acid biosynthesis and signal transduction in Arabidopsis. Plant Physiol Biochem 36: 83–89 [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI. (2002) Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH. (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41: 541–547 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand SB, Chua NH. (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Shen G, Yan J, He C, Zhang H. (2006) AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J 46: 649–657 [DOI] [PubMed] [Google Scholar]
- MacKintosh C, Cohen P. (1989) Identification of high levels of type 1 and type 2A protein phosphatases in higher plants. Biochem J 262: 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al. (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiol 15: 473–497 [Google Scholar]
- Murata K, Wu J, Brautigan DL. (1997) B cell receptor-associated protein α4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc Natl Acad Sci USA 94: 10624–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanahoshi M, Nishiuma T, Tsujishita Y, Hara K, Inui S, Sakaguchi N, Yonezawa K. (1998) Regulation of protein phosphatase 2A catalytic activity by α4 protein and its yeast homolog Tap42. Biochem Biophys Res Commun 251: 520–526 [DOI] [PubMed] [Google Scholar]
- País SM, Téllez-Iñón MT, Capiati DA. (2009) Serine/threonine protein phosphatases type 2A and their roles in stress signaling. Plant Signal Behav 4: 1013–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas M, García-Casado G, Rojo E, Solano R, Sánchez-Serrano JJ. (2007) A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J 51: 763–778 [DOI] [PubMed] [Google Scholar]
- Shen G, Kuppu S, Venkataramani S, Wang J, Yan J, Qiu X, Zhang H, Zhang H. (2010) ANKYRIN REPEAT-CONTAINING PROTEIN 2A is an essential molecular chaperone for peroxisomal membrane-bound ASCORBATE PEROXIDASE3 in Arabidopsis. Plant Cell 22: 811–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Slabas AR, Fordham-Skelton AP, Fletcher D, Martinez-Rivas JM, Swinhoe R, Croy RRD, Evans IM. (1994) Characterisation of cDNA and genomic clones encoding homologues of the 65 kDa regulatory subunit of protein phosphatase 2A in Arabidopsis thaliana. Plant Mol Biol 26: 1125–1138 [DOI] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci 14: 310–317 [DOI] [PubMed] [Google Scholar]
- Wu G, Wang X, Li X, Kamiya Y, Otegui MS, Chory J. (2011) Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal 4: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]