A single gene mutation yields novel morphologies and uncovers a divergent pattern of paralogous genes within the Solanaceae.
Abstract
Physalis spp. develop the “Chinese lantern” trait, also known as inflated calyx syndrome, that is a morphological novelty. Here, we identified the double-layered-lantern1 (doll1) mutant, a recessive and monofactorial mutation, in Physalis floridana; its corolla and androecium were transformed into the calyx and gynoecium, respectively. Two GLOBOSA-like MADS-box paralogous genes PFGLO1 and PFGLO2 were found in Physalis floridana, while the mutated phenotype was cosegregated with a large deletion harboring PFGLO1 and was complemented by the PFGLO1 genomic locus in transgenic plants, and severe PFGLO1 knockdowns phenocopied doll1. Thus, DOLL1 encodes the PFGLO1 protein and plays a primary role in determining corolla and androecium identity. However, specific PFGLO2 silencing showed no homeotic variation but rather affected pollen maturation. The two genes featured identical floral expression domains, but the encoding proteins shared 67% identity in sequences. PFGLO1 was localized in the nucleus when expressed in combination with a DEFICIENS homolog from Physalis floridana, whereas PFGLO2 was imported to the nucleus on its own. The two proteins were further found to have evolved different interacting partners and regulatory patterns, supporting the hypothesis that PFGLO2 is functionally separated from organ identity. Such a divergent pattern of duplicated GLO genes is unusual within the Solanaceae. Moreover, the phenotypes of the PFGLO1PFGLO2 double silencing mutants suggested that PFGLO2, through genetically interacting with PFGLO1, also exerts a role in the control of organ number and tip development of the second floral whorl. Our results, therefore, shed new light on the functional evolution of the duplicated GLO genes.
Morphological diversification is usually accompanied by an increase in the complexity of genetic material. The process is often achieved through gene duplication events that can provide the raw material for diversification and enable a sort of playground from which novelties of gene structures, protein function, and orgamismal morphology can emerge during evolution (Irish and Litt, 2005; Freeling and Thomas, 2006). However, the genetic architectures of the morphological diversification and the emergence of morphological novelty remain largely unknown. Nonetheless, in these evolutionary processes, it is assumed that processes such as gene expression, protein-protein interactions, and regulation targets of the orthologous genes or the duplicated genes can be altered. These dynamic processes lead to subfunctionalization, neofunctionalization, or loss of genes, thus bringing about morphological innovation (Irish and Litt, 2005; Zhao et al., 2013). The “Chinese lantern,” also called inflated calyx syndrome (ICS) in Physalis spp., is a morphological novelty within the Solanaceae (Fig. 1A; He and Saedler, 2005). The trait is triggered by fertilization and is controlled by action of hormones (He and Saedler, 2005, 2007). Furthermore, the molecular genetic basis underlying the origin of this novel trait has started to be empirically deciphered (He and Saedler, 2005, 2007; He et al., 2007; Zhao et al., 2013). The MADS-box gene2 from Physalis floridana (MPF2) belongs to a small subfamily of the MADS-box genes (He and Saedler, 2005). Heterotopic expression of MPF2 is key to the origin of ICS; the gene mainly controls ICS size (He and Saedler, 2005, 2007). The second functionally characterized MADS-box protein associated with the development of ICS was MPF3 (encoded by MPF3, the MADS-box gene3 from Physalis floridana), the euAP1 ortholog of APETALA1 (AP1) in Arabidopsis (Arabidopsis thaliana), and SQUAMOSA (SQUA) in Antirrhinum majus (Huijser et al., 1992; Mandel et al., 1992; Zhao et al., 2013). MPF3 exerts its role in the development of ICS through the MPF3-MPF2/MPF2 interaction and regulatory circuit (He et al., 2007; Zhao et al., 2013). Both MPF2 and MPF3 secure their new roles in male fertility through the evolution of novel genetic and physical interactions with the key components for male functionality, for example, the GLOBOSA (GLO) proteins (He and Saedler, 2005; He et al., 2007; Zhao et al., 2013). Thus, the ICS origin is assumed to be associated with fertility, an assertion that is corroborated by the observation that successful fertilization is required for ICS development (He and Saedler, 2005).
Figure 1.
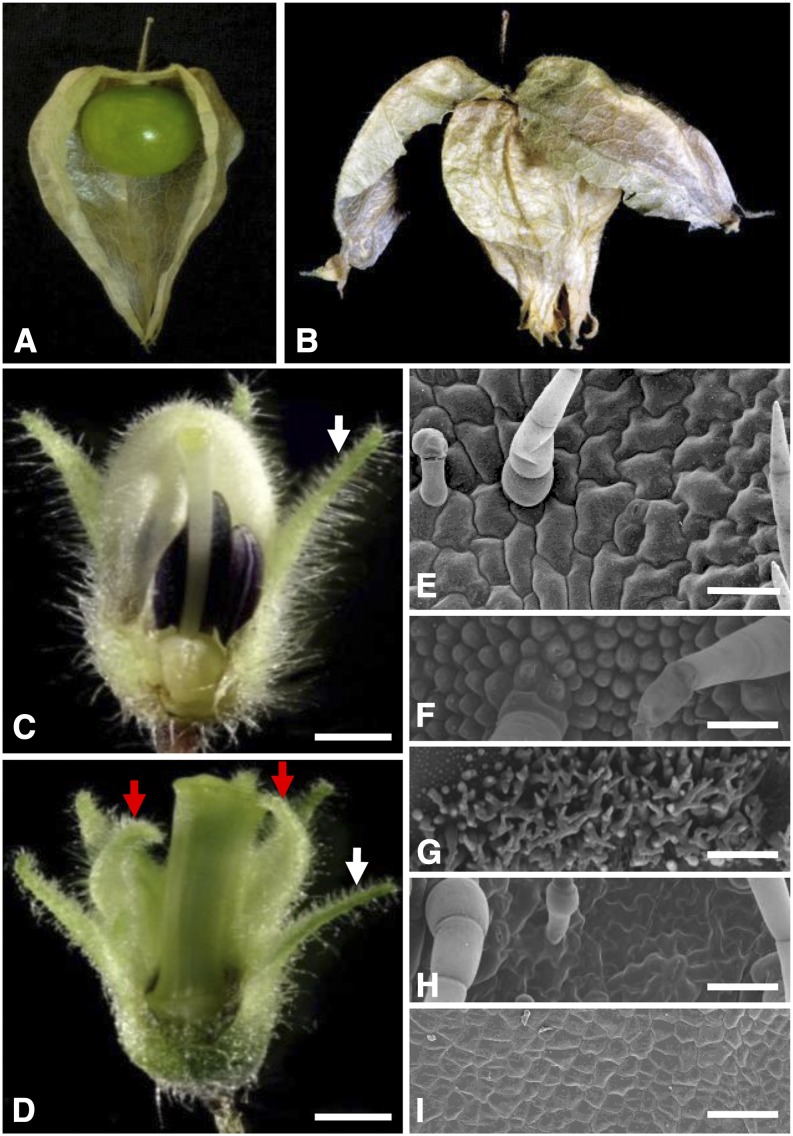
Floral phenotypic analyses of the doll1 mutant. A, A wild-type fruit. Part of the ICS was removed to show the berry inside. B, A fruit from the doll1 mutant. The first layer of the lantern was broken to show the second layered lantern. C, A wild-type flower. D, A mutated flower. Parts of the vegetative organs of the flower were removed to show the reproductive structures. White arrows indicate the tip of the calyx, while the red arrows indicate the tip of the transformed calyx in the mutated flower. Bars = 1 mm. E, Epidermal cells on the abaxial zone proximal to the tip of the calyx. F, Epidermal cells on the abaxial tip zone of the corolla from a wild-type flower. G, Epidermal cells on the adaxial base of the corolla from a wild-type flower. H, Epidermal cells on abaxial tip zone of the second whorl of the mutated flower. I, Epidermal cells on adaxial base zone of the second whorl of the mutated flower. Bars = 100 um.
Although apparently simple, ICS is actually a complex trait. To delve deep into the molecular origin of this morphological novelty, mutagenesis of Physalis floridana via γ-ray radiation was initiated (Lönnig, 2010). One line of an M2 family (M1750) segregating with floral homeotic transformation was obtained. The mutant featured two homeotic alterations related to the corolla and androecium. Unlike wild-type plants (Fig. 1A), once the berry was set, the mutant featured two whorls of the “Chinese lantern” (Fig. 1B), thus being named the double-layered-lantern1 (doll1) mutant. According to the ABC model of flower development, A functions alone specifies sepal identity, A and B function together to control petal identity, B and C function together to control stamen identity, and C functions alone to specify carpel identity (Coen and Meyerowitz, 1991). We hypothesized that the doll1 mutant might result from a typical and strong mutation of the B-function MADS-box genes, perhaps like orthologs of the GLO/PISTILLATA (PI) and DEFICIENS (DEF)/AP3 genes in Antirrhinum majus and Arabidopsis, respectively (Sommer et al., 1990; Jack et al., 1992; Tröbner et al., 1992; Goto and Meyerowitz, 1994; Egea-Cortines et al., 1999; Lamb and Irish, 2003). GLO and DEF are closely related paralogs, and both gene lineages were duplicated before the ancestor of the core asterids (Hernández-Hernández et al., 2007; Viaene et al., 2009), thus resulting in the GLO1 and GLO2 clades in the GLO lineage and the DEF and TOMATO MADS6 (TM6) clades in the DEF lineage. The two duplicates for each lineage redundantly partitioned the role of the B function with a variable subfunctionalization process in Petunia hybrida, Nicotiana benthamiana, and Solanum lycopersicum. This supposition is supported by the observations that a single mutation in each lineage in a species shows a partially mutated B-type phenotype in these Solanaceous species (Vandenbussche et al., 2004; de Martino et al., 2006; Rijpkema et al., 2006; Geuten and Irish, 2010).
In Physalis spp., GLO genes are also duplicated, and PFGLO1 and PFGLO2 were isolated from Physalis floridana (Zhao et al., 2013). As male fertility, the primary function of the androecium, is associated with ICS development (He and Saedler, 2005; He et al., 2007; Zhao et al., 2013), we further assumed that the B-functional genes might have a potential association with the development of the novelty ICS in Physalis spp. To empirically address this hypothesis, we here analyzed the doll1 mutants and characterized B-class MADS-box genes with an emphasis on the functional divergence of the two GLO duplicates. We demonstrated that PFGLO1 and PFGLO2 have diverged in terms of their molecular interactions and developmental roles, thus representing a new divergent pattern of the GLO duplicates within the Solanaceae. The functional evolution of the GLO genes and their potential roles in the development and evolution of the “Chinese lantern” are discussed.
RESULTS
The doll1 Mutant Features Double-Layered Calyces
The four-whorl floral organs of the wild-type Physalis floridana are, from the outer side to the inner side, the calyx, corolla, androecium, and gynoecium (Fig. 1C). However, in the doll1 mutant, the corolla was replaced by the calyx and the androecium was replaced by the gynoecium (Fig. 1D). The tip of the outer floral calyx in the wild-type flower and in the mutated flower stretched outward (Fig. 1, C and D, highlighted in white arrows), while the transformed floral calyx in the mutant bent inward (highlighted by red arrows in Fig. 1D), like the tip of the corolla (Fig. 1C). Scanning electronic microscopy (SEM) analyses revealed that in both wild-type and doll1 mutant plants, the epidermal cells in the zones proximal to the tip of the calyx began to enlarge and became lobate at anthesis. Moreover, the trichomes and stomatal apparatus were developed (Fig. 1E). In wild-type flowers, epidermal cells on the second floral whorl were under differentiated statuses in different zones. In the abaxial tip zone, they were regular dome cells, and trichomes developed occasionally (Fig. 1F), while the adaxial zone proximal to base was covered by extensive and tiny trichomes (Fig. 1G). In contrast to this, the cells on the second floral whorl of the mutant became calyx like, i.e. they were lobate and imbedded with trichomes and stomatal cells (Fig. 1H), while adaxial cells at the base zone, rather than developing the extensive trichomes as on the corolla (Fig. 1G), remained small and undifferentiated (Fig. 1I), like the cells on calyx base (Hu and Saedler, 2007). The epidermal cells on the transformed calyx were significantly deviated from the epidermal cells on the corolla, while actually resembling the cells on the calyx, further supporting that the corolla was transformed into the calyx in the doll1 mutant. The transformed gynoecium from the androecium was often fused with the original gynoecium in the mutated flower (Fig. 1D). Lacking an androecium, the mutant was male sterile. Once fertilized with wild-type pollen grains, two layers of the “Chinese lantern” were formed in the mutant (Fig. 1B). However, the fruiting rate was less than 2%, hinting that female fertility was also affected in this mutant. We thus assumed that mutations in the B-class MADS-box genes might cause the mutated phenotype, and hence these genes were further investigated in Physalis floridana.
Molecular Characterization of Physalis floridana B-Class MADS-Box Genes
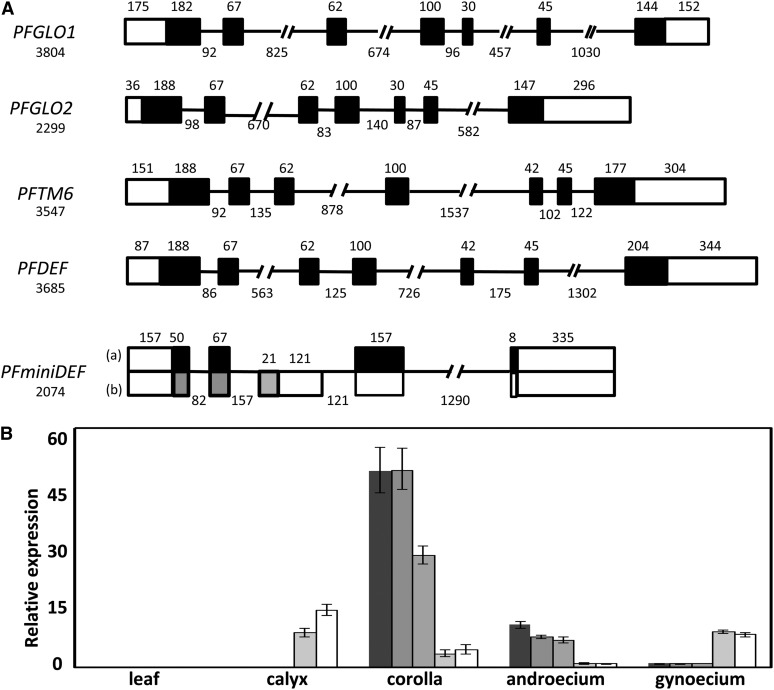
Six B-class MADS-box gene transcripts were isolated in Physalis floridana and named PFGLO1, PFGLO2, PFDEF, PFTM6, PFminiDEFa, and PFminiDEFb. Phylogenetic analyses with multiple methods revealed that these Physalis floridana genes fell into one of two groups: the GLO or DEF lineages (Supplemental Fig. S1; Supplemental Data Set S1). In the Solanaceae, each of these two groups was duplicated during evolution, yielding the GLO1- and GLO2-like variations in the GLO lineage and the DEF- and TM6-like variations in the DEF lineage (Supplemental Fig. S1). Sequencing these genes revealed that PFGLO1 (3,804 bp), PFGLO2 (2,299 bp), PFDEF (3,685 bp), and PFTM6 (3,547 bp) had identical structures with seven exons for each gene (Fig. 2A). PFminDEF (2,074 bp), which resulted from a duplication of PFDEF, encoded the two transcripts PFminiDEFa and PFminiDEFb, a result of alternative splicing (Supplemental Figs. S1 and S2). Moreover, the structure of PFminiDEF was altered dramatically and resulted in an unusual MADS-domain protein (Fig. 2A; Supplemental Fig. S2), indicating that it was very likely a pseudogene.
Figure 2.
Molecular characterizations of the B-class MADS-box Genes in Physalis floridana. A, The exon-intron structure of the B-class MADS-box genes. Five B-class MADS-box genes were obtained. Boxes stand for the exons. Black boxes represent the coding regions, while the empty boxes represent untranslated regions. The lines represent introns. The length of each gene and the size of each intragenic region are given in base pairs. The digit above the box indicates the length of the corresponding exon, while the digits under the line indicate the length of the corresponding intron. For PFminDEF, two alternative splicing transcripts, a and b, are shown. B, Relative expression of the B-class MADS-box genes. Expression of PFGLO1, PFGLO2, PFDEF, PFTM6, and PFminDEF from left to right was investigated via qRT-PCR. PFACTIN was used as an internal control. The experiments were repeated three times using three independent biological samples. Mean expression values and sd are presented.
We also investigated the expression domains of these Physalis floridana genes. The results of quantitative reverse transcription (qRT)-PCR analyses demonstrated that transcripts of none of these genes could be detected leaves. PFGLO1, PFGLO2, and PFDEF were predominantly expressed in the corolla and androecium; PFTM6 was abundantly expressed in the calyx, corolla, and gynoecium (Fig. 2B). Surprisingly, PFminDEF, a putative pseudogene, had a similar expression pattern to PFTM6 (Fig. 2B).
In the doll1 mutant, PFGLO1 seemed not to be expressed in flowers, and no PFGLO1 complementary DNA (cDNA) was obtained, while other B-class MADS-box genes were transcribed as in the wild type (Supplemental Fig. S3A), and the cDNA sequences were identical to these of the wild type. Moreover, genomic fragment of PFGLO1 was not amplified in the doll1 mutant (Supplemental Fig. S3B). These analyses suggest that PFGLO1 might have been deleted from the doll1 genome, resulting in the nonexpression of this gene.
Floral Expressions of MADS-Box Genes in the Wild Type and the doll1 Mutants
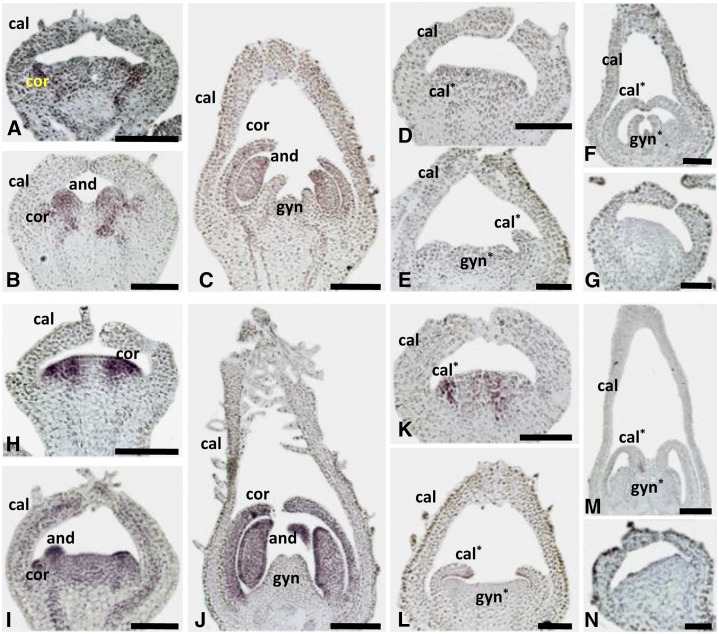
To further confirm the above notion, the spatial expression of PFGLO1 and PFGLO2 during the early floral development was investigated using mRNA in situ hybridization. In the wild type, PFGLO1 was expressed in the primordium of the corolla and androecium (Fig. 3, A–C). In the doll1 mutant background, no PFGLO1 expression signal was detected (Fig. 3, D–F). PFGLO2 shared an identical expression domain with PFGLO1 in the wild type (Fig. 3, H–J). However, in the doll1 mutant, expression of PFGLO2 was attenuated (Fig. 3, K–M); it was only observed in the primordium of the second whorl (Fig. 3K) and, later on, was restricted to the tip of the transformed calyces (Fig. 3, L and M). Hybridizations with sense probes of PFGLO1 (Fig. 3G) or PFGLO2 (Fig. 3N), respectively, were used as the controls. In further qRT-PCR analyses, no PFGLO1 was detected at all, while PFGLO2, PFDEF, and MPF2 were significantly down-regulated in the mutated flowers, MPF3, PFAG (for an AGAMOUS homolog from Physalis floridana), and PFTM6 were significantly up-regulated, and PFSEP1 (for a SEPALLATA 1 homolog from Physalis floridana) and PFSEP3 were slightly up-regulated compared with the wild type (Supplemental Fig. S4). These findings confirmed that PFGLO1 was not expressed in the doll1 mutant and suggested that these altered genes might be potential targets of PFGLO1.
Figure 3.
mRNA in situ expression of PFGLO1 and PFGLO2. A to F, Expression of PFGLO1. Antisense probe in the wild type (A–C) and in the doll1 mutant (d–F). G, Sense probe of PFGLO1. H to M, Expression of PFGLO2. Antisense probe in the wild type (H–J) and in the doll1 mutant (K–M). N, Sense probe of PFGLO2. The primordium of the calyx (cal), corolla (cor), transformed calyx (cal*), androecium (and), gynoecium (gyn), and transformed gynoecium (gyn*) are shown. Asterisk indicates the transformed organ primordium in the doll1 mutant. Bars = 100 μm.
The PFGLO1 Locus, Genetic Segregation, and Genomic Complementation
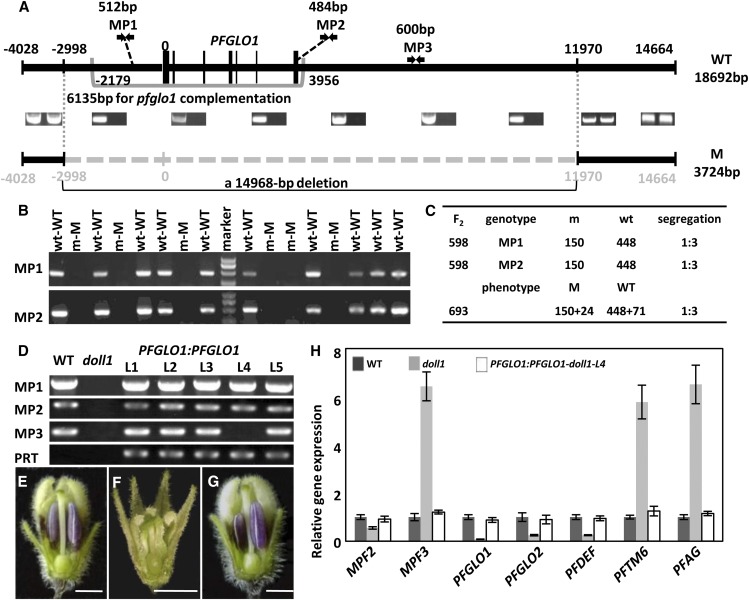
To understand the nature of PFGLO1 silencing in the doll1 mutant, we analyzed the genomic structure of the PFGLO1 locus. An 18,692-bp-long fragment was assembled in the wild type, including 4,028 bp of upstream and 14,664 bp of downstream sequence (Fig. 4A), if the first nucleotide of PFGLO1’s open reading frame (ORF) was defined as position 1. Any fragment between positions –2,998 and 11,970 was not detected in the doll1 mutant genome, thus the mutant resulted from a large genomic deletion (14,968 bp) harboring PFGLO1 (Fig. 4A). Gene annotation of the wild-type sequences indicated that no other ORFs linked with PFGLO1 were found in the large deleted fragment, suggesting that possibly a single gene (PFGLO1) was deleted in the mutant.
Figure 4.
Reconstruction of the PFGLO1 locus, genetic segregation analysis, and genomic complementation of the doll1 mutant. A, Reconstruction of the PFGLO1 locus. Black lines represent the genomic sequences obtained; the dashed line represents the lost sequences in the mutant. The intron-exon structure of PFGLO1 is shown for the wild type (WT). MP1 and MP2 are two PFGLO1-based markers used to screen the homozygous doll1 mutant. The bracketed region was used for functional complementation in the doll1 mutant. B, Linkage and cosegregation analyses between the two PFGLO1-based markers and the mutated phenotype in the F2 population from a cross of the wild type and the doll1 mutant. W, Wild type; M, mutant; w, sample with amplified band; m, sample without amplified band. C, Segregation rate of the PFGLO1-based markers and mutated phenotypes in the F2 population. D, Generation of transgenic plants harboring PFGLO1:PFGLO1 in the doll1 mutant. MP1, MP2, and MP3 were amplified in the genomes of the wild type, the doll1 mutant, and the five transgenic lines (L1, L2, L3, L4, and L5), indicating that L4 is PFGLO1:PFGLO1-doll1 plant. The PRT (primers for the pBAR-vector test) marker from the binary vector was amplified to show the identity of the transgenic plants. E, A wild-type flower. F, A doll1 flower. G, A doll1 flower with expression of PFGLO1 driven by its native promoter (PFGLO1:PFGLO1-doll1-L4). Part of the calyx and corolla was removed to show the organs inside. Bars = 2.5 mm. H, Relative expression of PFGLO1 and other MADS-box genes in flowers from wild-type, doll1, and PFGLO1:PFGLO1-doll1-L4 plants. PFACTIN was used as an internal control. The expression experiments were repeated three times using independent biological samples. Mean expression values and sd are presented. [See online article for color version of this figure.]
To genetically link the deletion and the mutated phenotypes, we conducted genetic analyses. The F1 hybrids resulting from pollination of the mutant gynoecium with wild-type pollen grains had the wild-type Physalis floridana floral phenotype. The inheritance of the mutated phenotype was evaluated in a total of 693 F2 progeny. The mutated phenotype segregated in a manner consistent with Mendel’s law (wild type [519]: mutant [174] = 3:1; χ2 = 0.004 for 3:1, P = 0.95; Fig. 4C). Genomic DNA from 598 lines of the segregating population was analyzed for genetic-phenotype linkage analysis using PFGLO1-linked molecular markers Primers for Marker1 (PM1) and MP2 (Fig. 4A). The genomic DNA fragments of the expected size could be only amplified from the individuals with wild-type phenotypes (Fig. 4B). These markers cosegregated well with the mutated phenotype; the segregation of the wild type (448) and the mutant (150) was 3:1 (χ2 = 0.002 for 3:1, P = 0.96; Fig. 4, B and C), indicating that the doll1 mutant resulted from a monofactorial and recessive mutation with a high likelihood of occurring in the PFGLO1 locus.
We next transformed a 6,135-bp genomic fragment covering the complete PFGLO1 locus from the wild type (Fig. 4A) into the F2 progenies and obtained five transgenic plants. Then, we used MP1, MP2 (two markers on the transformed fragment), and MP3 (a marker outside of the transformed fragment) to characterize these transgenic plants. One transgenic plant in the homozygous doll1 background (PFGLO1:PFGLO1-doll1-L4) was found, as indicated by the lack of amplification of MP3 (Fig. 4D). Compared with the wild-type flower (Fig. 4E) and the doll1 flower (Fig. 4F), the mutated phenotype was completely rescued in the PFGLO1:PFGLO1-doll1-L4 flowers (Fig. 4G). Moreover, the expression of the significantly altered MADS-box genes in the doll1 mutants (Supplemental Fig. S4) was restored to an equivalent level to that of the wild type by the PFGLO1:PFGLO1 expression (Fig. 4H). Thus, we conclude that the DOLL1 encodes the PFGLO1 protein. These results also hinted at a functional divergence of PFGLO1 from PFGLO2.
Extensive Variations at Protein Sequences Predict Functional Divergence
Because the expression domains of PFGLO1 and PFGLO2 were kept unaltered, the alteration at protein level might likely account for the functional divergence. Sequence comparison revealed that the paralogous protein pair shared 67% identity. The diverged sites (33%) had occurred conservative (amino acid changes with similar physicochemical property) or radical (amino acid changes with different physicochemical property) substitutions during evolution. In total, 70 substituted residues between PFGLO1 and PFGLO2 and 34 radical substitutions were found in the four domains of these MADS-domain proteins (Supplemental Fig. S5). Compared with other Solanaceous GLO orthologous pairs, we found that 25 mutational sites that had radically substituted were clade specific to separate GLO1- and GLO2-like. Moreover, substantial species-specific mutated sites were observed between each paralogous pair (Supplemental Fig. S5). Particularly, L18 and I163 in the M and K domains of PFGLO2 and P61 in the I domain of PFGLO1 were found to be the Physalis floridana-specific mutated residues. These domains are essential in both protein-protein interactions and protein-DNA interactions of the MADS-domain regulatory proteins (Immink et al., 2010). Therefore, the spectrum and specificity of the molecular interaction targets of PFGLO1 and PFGLO2 could be changed, thus leading to different developmental roles. We next investigated their divergence at these various levels.
Virus-Induced Gene Silencing Analyses Reveal Diverged Developmental Roles
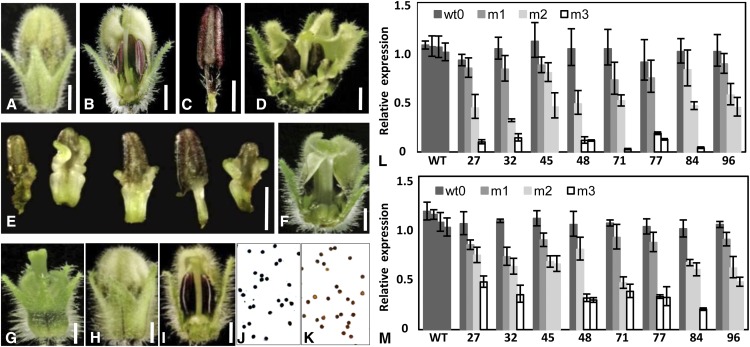
We exploited virus-induced gene silencing (VIGS) to dissect the functional divergence of PFGLO1 and PFGLO2. Eight-four of 115 PFGLO1-VIGS plants produced similar and visible floral variation. Compared with the wild type (Fig. 5, A–C), three categories of mutated phenotypes related to homeotic transformation of both corolla and androecium into the calyx and gynoecium (m1, m2, and m3) were observed in these lines (Fig. 5, D–G). PFGLO1 was knocked down to differing extents in the m1, m2, and m3 flowers (Fig. 5L). The extent of the phenotypic transformation apparently depended on the dose of PFGLO1 mRNA. In the m1 grade, the androecium was only mutated partially (Fig. 5, D and E), and in the m2 grade, the androecium was completely transformed into a gynoecium, while the corolla was not changed at all (Fig. 5F). The phenotype in the most severe category m3 (Fig. 5G), in which PFGLO1 expression was only 10% of that observed in the wild type (Fig. 5L), resembled the doll1 mutant (Fig. 1D). We also found that the expression of PFGLO2 was attenuated in these PFGLO1 down-regulated flowers (Fig. 5M), as was observed in the doll1 mutant.
Figure 5.
Down-regulating PFGLO1 in VIGS analyses phenocopies the doll1 mutant. A, An intact wild-type (WT) flower. B, A dissected wild-type flower to show the androecium and gynoecium. C, A stamen from a wild-type flower. D, A grade m1 flower from the PFGLO1-VIGS plants. E, Partially carpel-like androecium from a grade m1 flower. F, A grade m2 flower from the PFGLO1-VIGS plants. G, A grade m3 flower from the PFGLO1-VIGS plants. H, An intact pfglo2-flower from PFGLO2-VIGS plants. I, A dissected pfglo2 flower to show the androecium and gynoecium. Bars = 2 mm. J, I2-KI-stained pollen from a wild-type flower. K, I2-KI stained pollen of a pfglo2 flower from PFGLO2-VIGS plants. Active pollen is blue, while sterile pollen is tawny. L, Expression of PFGLO1 in PFGLO1-VIGS flowers. M, Expression of PFGLO2 in PFGLO1-VIGS flowers. Flowers that featured three grades of mutated phenotype (m1, m2, and m3) in eight VIGS-infected lines (27, 32, 45, 48, 71, 77, 84, and 96) were investigated. The expression of MADS-box genes in the wild-type flowers and the wild-type flowers from the VIGS-infected plants (Wt0) were also checked. PFACTIN was used as an internal control. The qRT-PCR analyses were repeated three times using independent biological samples. Mean expression values and sd are presented.
PFGLO2 was specifically expressed in floral whorl 2 when PFGLO1 was absent, hinting that PFGLO2 might have a particular role in the development of the second floral whorl. To reveal this, PFGLO2-VIGS lines were produced. The results of qRT-PCR analyses showed that the expression of PFGLO2 was specifically and efficiently knocked down, as evidenced by the fact that PFGLO2 expression in some flowers was no more than 10% of that seen in the wild type (Supplemental Fig. S6A), while the expression of PFGLO1 was not affected in these PFGLO2-VIGS flowers (Supplemental Fig. S6B), suggesting that PFGLO1 is not regulated by PFGLO2. Even the most severe down-regulation of PFGLO2 did not reveal any visible phenotypic variation (Fig. 5, H and I). Pollination with wild-type pollen grains led to normal fruit development, indicating that the functionality of the gynoecium is not impaired. In I2-KI staining assays, 99.8% of wild-type pollen grains were deeply stained blue (Fig. 5J), while less than 50% of the pollen grains from the PFGLO2-VIGS flowers were stained blue. In one extreme case, only 10% of the pollen grains assayed were mature (Fig. 5K). As a negative control, the phytoene desaturase gene from Physalis floridana-VIGS flowers developed normal pollens as the wild type (Zhao et al., 2013). Therefore, down-regulation of PFGLO2 alone did not affect organ identity but may conceivably directly or indirectly block sugar translocation to the pollen, thereby specifically preventing maturation of pollen grains.
Divergent Subcellular Localization and Protein-Protein Interactions
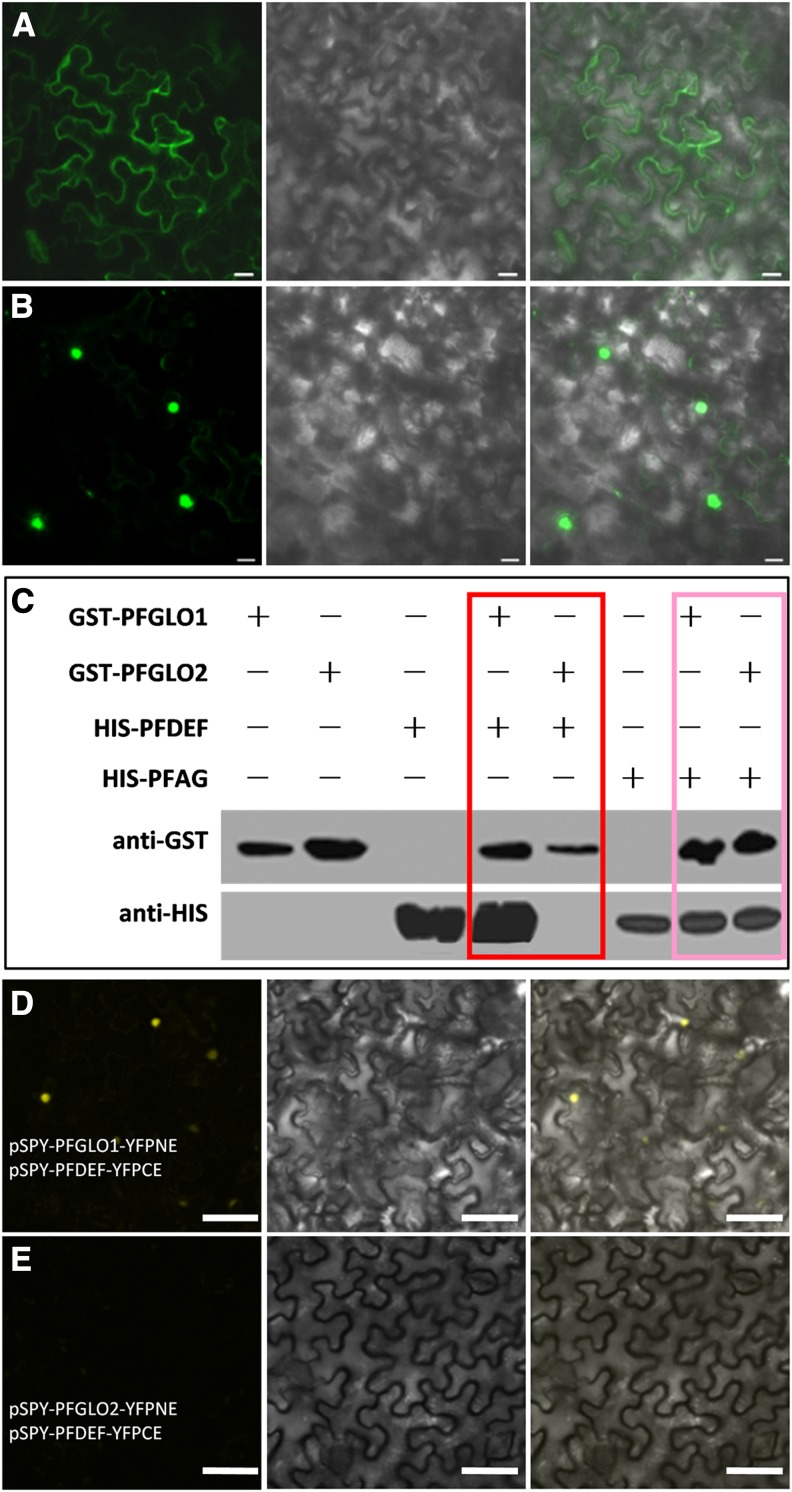
Import into the cell nucleus is critically important for transcription factors to exert their roles. To reveal the subcellular localization of PFGLO1 and PFGLO2, the 35S promoter-driven fusion proteins PFGLO1-GFP and PFGLO2-GFP were produced in N. benthamiana leaves. PFGLO1-GFP was mainly observed in the cytoplasm, and if there was any present in the nucleus, it was not highly accumulated (Fig. 6A), while PFGLO2-GFP was mainly detected in the nucleus (Fig. 6B), suggesting different subcellular localizations of the two proteins.
Figure 6.
Subcellular localization of PFGLO1 and PFGLO2 and characterizations of their interacting proteins. A and B, Subcellular localization of PFGLO1 and PFGLO2, respectively. From right to left in both A and B are images with green fluorescence, light images, and the merged images. Bars = 20 µm. C, Pull-down assay to confirm that PFGLO1 and PFGLO2 interact with PFDEF and PFAG, respectively. Their interactions with PFDEF are highlighted in the red box, and their interactions with PFAG are highlighted in the pink box. Other lanes are controls. D, PFGLO1 interacts with PFDEF in BiFC. E, Coexpression of PFGLO2-YFPNE and PFDEF-YFPCE. Bars = 50 μm.
The nuclear import of MADS-domain proteins is often facilitated by formation of homodimers or heterodimers with other MADS-domain proteins (McGonigle et al., 1996; Theissen and Saedler, 2001; Hernández-Hernández et al., 2007). We therefore characterized the interaction of various MADS-domain proteins with both PFGLO1 and PFGLO2. In yeast (Saccharomyces cerevisiae), both PFGLO1 and PFGLO2 could not self-activate and formed homodimers, and they were found to have common interacting partners such as MPF3, PFAG, and PFSEP3. In addition, we found that PFDEF interacted with MPF3, PFGLO1, PFAG, and PFSEP3 and that PFTM6 dimerized with MPF3, PFGLO2, PFSEP1, and PFSEP3 (Supplemental Fig. S7). Heterodimerizations of PFGLO1 and PFGLO2 with PFDEF and PFAG were particularly verified in a pull-down assay (Fig. 6C). Glutathione transferase (GST) was fused with both PFGLO1 and PFGLO2 to produce the fusion proteins GST-PFGLO1 and GST-PFGLO2. PFDEF and PFAG, respectively, were labeled with a HIS tag. Western hybridizations using antibodies of HIS (anti-HIS) or GST (anti-GST) confirmed that both PFGLO1 and PFGLO2 interacted with PFAG, while only PFGLO1 maintained the ability to interact with PFDEF.
Heterodimerization of GLO and DEF proteins is essential for their nuclear import and subsequent functions in flower development (Jack et al., 1992; Schwarz-Sommer et al., 1992). The differences in affinity between both PFGLO1/PFGLO2 proteins with PFDEF were further verified with a bimolecular fluorescence complementation (BiFC) assay. In a combination of PFGLO1-YFPNE (for the N-terminal end of YFP) and PFDEF-YFPCE (for the C-terminal end of YFP), the yellow fluorescence protein (YFP) signal was exclusively detected in the nucleus (Fig. 6D), indicating that PFGLO1 and PFDEF interact in the nucleus, further solidifying the notion that the heterodimerization of GLO-DEF is required for their nuclear import, while no YFP signal was observed in the combination of PFGLO2-YFPNE and PFDEF-YFPCE (Fig. 6E), verifying that PFGLO2 and PFDEF do not form heterodimers.
Different Consequences of PFGLO1 and PFGLO2 Overexpression in Mutants
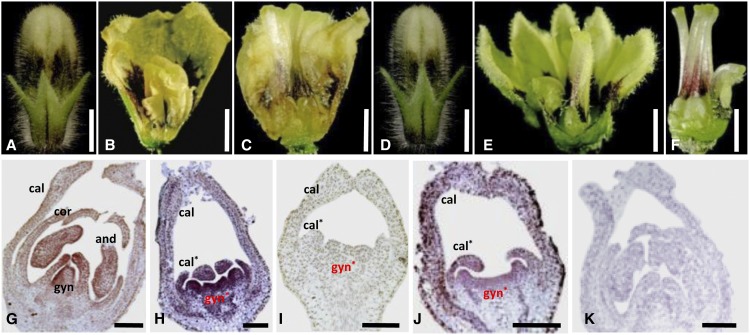
To further investigate the divergence, we expressed full-length, 35S promoter-driven cDNAs of PFGLO1 and PFGLO2, respectively, into the doll1 mutant backgrounds. In total, four lines of 35S:PFGLO1-pfglo1 (1L1–1L4) and six lines of 35S:PFGLO2-pfglo1 (2L1–2L6) were generated (Supplemental Fig. S8). The calyx organ identity of the first floral whorl in all of these transgenic lines was not altered (Fig. 7, A and D). Nonetheless, in the 35S:PFGLO1-pfglo1 lines, the mutated corolla phenotypes were restored, while the inner whorls were further transformed into the corolla-like structure (Fig. 7, B and C), indicating that C function may be strongly repressed. The second floral whorl of the 35S:PFGLO2-pfglo1 lines also had a wild-type-like corolla, but the transformed gynoecia were not altered. Only brown coloration, which is characteristic of the corolla, was seen at the base of the gynoecium (Fig. 7, E and F), indicating that PFGLO2 completely restores the corolla phenotype and does not repress C function.
Figure 7.
Overexpression further reveals functional divergence. A, A flower bud from a 35S:PFGLO1 overexpression transgenic plant in the pfglo1 mutant (1L1). B, A flower from a 35S:PFGLO1 overexpression transgenic plant in the pfglo1 mutant (1L1). The calyx and corolla were partially removed to show the petaloid structure transformed from the reproductive organs. C, The corolla transformed from the gynoecium in 35S:PFGLO1 overexpressed transgenic plants in the pfglo1 mutant (1L1). D, A flower bud from 35S:PFGLO2 in the pfglo1 mutant (2L6). E, A flower from 35S:PFGLO2 in the pfglo1 mutant (2L6). The calyx and corolla were partially removed to show the inner floral organs. F, The transformed fused gynoecium in 35S:PFGLO2 in the pfglo1 mutant (2L6). Bars = 3 mm. G to J, Expression of PFAG revealed by in situ hybridization. Antisense probe of PFAG in wild type (G), in the pfglo1 mutant (H), in 35S:PFGLO1 in pfglo1 (I), and in 35S:PFGLO2 in pfglo1 (J). K, Sense probe of PFAG. The primordium of the calyx (cal), corolla (cor), transformed calyx (cal*), androecium (and), gynoecium (gyn), and transformed gynoecium (gyn*) are shown. Asterisk indicates the transformed organ primordia in the doll1 mutant. Bars = 100 μm.
The repression of C function, putatively exerted by PFAG (He et al., 2007; Zhao et al., 2013), was further confirmed via RNA in situ hybridization. In both the wild type and in the doll1 mutant backgrounds, PFAG expression in floral organ primordia was restricted to the inner three whorls (Fig. 7, G and H). However, PFAG expression was completely abolished when PFGLO1 was overexpressed in the pfglo1 background (Fig. 7I). In the 35S:PFGLO2-pfglo1 transgenic lines, the expression of PFAG appeared to be normal (Fig. 7J). No visible signal was seen when the sense PFAG probe was used (Fig. 7K).
PFGLO1 and PFGLO2 Evolved Distinct Regulatory Patterns
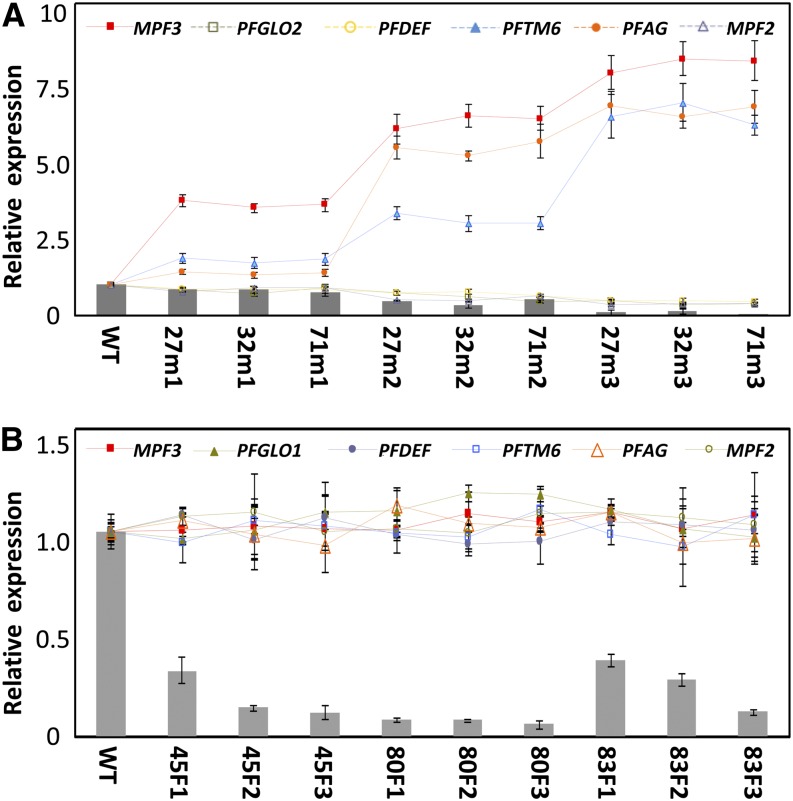
To reveal more differences in regulatory targets of PFGLO1 and PFGLO2, the expression of the significantly altered MADS-box genes in doll1 mutant (Supplemental Fig. S5) was investigated in the flowers of both PFGLO1-VIGS and PFGLO2-VIGS flowers. Three categories of flowers (m1, m2, and m3) from the three PFGLO1-VIGS plants (27, 32, and 71) were randomly selected and used to monitor gene expression. The expression tendency of these genes was similar to that in the doll1 mutant, and the extent of alteration (either up- or down-regulated) apparently depended on the residual PFGLO1 mRNA levels (Fig. 8A). Similar experiments were performed in the PFGLO2-VIGS plants. However, in these PFGLO2-down-regulated flowers, the expression of these MADS-box genes was not affected (Fig. 8B). These results suggest that PFAG, PFGLO2, PFDEF, PFTM6, MPF3, and MPF2 are direct or indirect target genes of PFGLO1 but that PFGLO2 does not regulate any of these genes. Thus, both genes have evolved distinct regulatory targets and patterns. Because PFGLO2 became a downstream gene of PFGLO1, the two genes may interact genetically to regulate floral development.
Figure 8.
Evolution of distinct regulatory targets and patterns. A, Floral expression of the MADS-box genes in PFGLO1-VIGS lines. Flowers that featured three grades of phenotypic variations (m1, m2, and m3) in three VIGS-infected lines (27, 32, and 71) were subjected to qRT-PCR analysis. The experiments were repeated three times using independent biological samples. Mean expression values and sd are presented. B, Floral expression of the MADS-box genes in the PFGLO2-VIGS lines. Three flowers (F1, F2, and F3) in three VIGS-infected lines (45, 80, and 83) were subjected to qRT-PCR analysis. The experiments were repeated three times. Mean expression values and sd are presented. In both A and B, the expression of MPF3, PFGLO1, PFGLO2, PFDEF, PFTM6, PFAG, and MPF2 were compared between wild-type (WT) and mutated flowers. PFACTIN was used as an internal control. The columns in A and B represent expression of PFGLO1 and PFGLO2, respectively, while the lines indicate the expression of other genes.
Genetic Interaction of PFGLO1 and PFGLO2 in Corolla Development
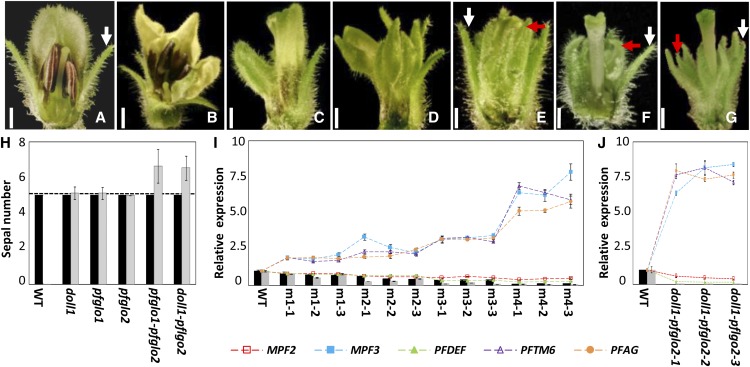
To reveal genetic interactions, we used a VIGS strategy to produce PFGLO1PFGLO2 double mutants: PFGLO1-PFGLO2-VIGS lines and PFGLO2-VIGS-doll1 (pfglo1) lines. Compared with the wild-type flower (Fig. 9A), four categories of mutated phenotypes were seen in the co-VIGS mutated plants (Fig. 9, B–E). Compared with the single mutants (the doll1 and the most severe PFGLO1-VIGS mutated flowers, like in Fig. 9F), there were two different floral phenotypic variations observed in the mutated flowers from the double mutants. The tip of the transformed calyx (the second whorl) in the single pfglo1 mutant bent inward (highlighted by a red arrow in Fig. 9F), while in the double mutants, i.e. the most severe lines of PFGLO1-PFGLO2-VIGS (Fig. 9E) and PFGLO2-VIGS-pfglo1 (Fig. 9G), the tip of the second whorl stretched outward or was erected as the calyx developed in the wild type (highlighted by white arrows in Fig. 9A; Fig. 9, E and G). In addition, the organ number in the first and second floral whorls of the wild type and the single mutants was five, while the number of the second floral whorl (the transformed calyx) was increased in the double mutants (Fig. 9H). These phenotypic variations apparently correlated with the degree of cosilencing PFGLO1 and PFGLO2 (Fig. 9, I and J). The variation in the expression of the MADS-box genes MPF3, PFAG, PFTM6, and MPF2 in these double mutants was similar to that observed in the PFGLO1 mutants (Figs. 8B and 9, I and J). These findings indicate that PFGLO1 is epistatic to PFGLO2 and that PFGLO2 interacts with PFGLO1 genetically to regulate the organ number and the tip development of the second whorl organs.
Figure 9.
Genetic interaction between PFGLO1 and PFGLO2. A, A wild-type (WT) flower. B to E, Different grades of floral variations m1 (B), m2 (C), m3 (D), and m4 (E) occurred in the double knockdowns of PFGLO1 and PFGLO2 via VIGS. F, A flower from the doll1 mutant. G, A flower from a PFGLO2-silencing plant in the doll1 mutant background. Part of the calyx or the second whorl is removed to show the inside organs. Bars = 2 mm. The white arrows indicate the normal calyx, and the red arrows indicate the transformed calyx. H, The sepal number of the Physalis floridana. calyx in various mutant lines, as indicated. Five sepals are typical of the wild-type flowers, as indicated by the dashed line. The black column stands for the original whorl calyx, and the gray column stands for the transformed calyx in the second floral whorl of the various mutants. I, Expression of the MADS-box genes in different grades (m1, m2, m3, and m4) of the double-gene VIGS mutated flowers, as indicated. For each grade, three flowers were collected. J, Expression of MADS-box genes in the double doll1-pfglo2 mutants. Three mutated flowers were harvested for gene expression. In both I and J, the expression of MPF3, PFGLO1, PFGLO2, PFDEF, PFTM6, PFAG, and MPF2 were monitored and compared with the wild-type flowers. PFACTIN was used as an internal control. Mean expression values and sd are presented.
DISCUSSION
The calyx of Physalis spp. inflates to a “Chinese lantern” structure as the berry develops following fertilization. MADS-box genes, such as MPF2 and MPF3, are associated with the evolution and development of this novel structure (He and Saedler, 2005; Zhao et al., 2013). Here, we investigated the doll1 mutant with the two-layered “Chinese lantern” that resulted from a lack of expression of the GLO gene PFGLO1 in Physalis floridana. We found that PFGLO1, instead of PFGLO2, had its primary role in complete B function, while PFGLO2 alone exerted a role in male fertility, but controlled the organ number and tip development of the second floral whorl in a PFGLO1-dependent manner. Such a diverging pattern of the GLO duplicates is distinct from the purely redundant patterns observed in Petunia hybrida, S. lycopersicum, or N. benthamiana (Vandenbussche et al., 2004; Geuten and Irish, 2010).
Functions of the Duplicated GLO Genes in Physalis spp. Floral Organ Development
The doll1 mutant resembled the typical mutations in B-function genes from various model plant species. We therefore characterized the closely related B-class homologous genes from Physalis floridana and designated as PFGLO1, PFGLO2, PFDEF, PFTM6, and PFminiDEF. However, the mutant was recessively inherited and resulted from a mutation in one locus. Concomitantly, both expression and locus of PFGLO1 were not detected. Local genome comparison revealed that a large fragment (about 15 kb) containing PFGLO1 was deleted from the genome in this mutant. Genetic linkage analysis further showed that the mutated phenotype closely cosegregated with the loss of PFGLO1. Failures in full complementation by PFGLO1 cDNA indicated that genomic elements like introns might be required for proper gene expression, as described for ERECTA (Karve et al., 2011), or overexpressed PFGLO1 could damage the normal MADS-complex formation. Nonetheless, an approximate 6-kb genomic sequence harboring PFGLO1 fully complemented the mutated phenotypes, while knocking down PFGLO1 expression in the wild-type background phenocopied the doll1 mutant. Thus, we conclude that DOLL1 encodes the PFGLO1 protein and is dominantly involved in specifying organ identity of the corolla and the androecium in Physalis floridana.
These results also imply that the paralogous genes PFGLO1 and PFGLO2 might have dramatically diverged in function. In the doll1 mutant, overexpression of PFGLO2 cDNA (35S:PFGLO2-pfglo1) rescued only the mutated corolla phenotype, while knocking PFGLO2 down in the wild type did not reveal any visible homeotic variation but inhibited pollen maturation, indicating that PFGLO2 is not necessary in corolla development but is specifically involved in the regulation of male fertility. Nonetheless, PFGLO2 might have retained a potential role in corolla identity but was masked by PFGLO1. Such a complete functional separation of GLO-like genes from stamen organ identity was not observed in Petunia hybrida, S. lycopersicum, or N. benthamiana (Vandenbussche et al., 2004; Geuten and Irish, 2010). However, a similar functional separation was observed for the C-class paralogous genes in Antirrhinum majus, in which FARINELLI (FAR) is involved in male fertility, while PLENA exerts roles in organ identity specification of the stamen and carpel (Davies et al., 1999).
PFGLO2 had an identical expression domain with PFGLO1 in the development of the wild-type floral organs. However, in the doll1 mutant, expression of PFGLO1 was completely abolished, while PFGLO2 was expressed in the primordium of the second floral whorl and subsequently restricted to the tip of the transformed organs. When PFGLO2 was down-regulated, the expression of PFGLO1 was not affected. These observations suggest that PFGLO1 defines the expression region of and controls the expression level of PFGLO2 but that PFGLO2 does not regulate the expression of PFGLO1. The particular expression domain of PFGLO2 in the doll1 suggests that PFGLO2 has a specific role in the development of the second floral whorl and that this role is likely independent of organ identity. The observed physical interactions of PFGLO2 with PFTM6 may be preventing the second floral whorl organs from taking on full calyx morphology because PFGLO2 was not expressed in the calyx but always in the second floral whorl organs, and PFTM6 was expressed in both calyx and corolla. PFGLO1PFGLO2 cosilenced mutants gave rise to new phenotypic variations, which were not observed in either the pfglo1 (doll1), PFGLO1 down-regulated, or PFGLO2 down-regulated mutants. Thus, PFGLO2, genetically interacting with PFGLO1, has been restricted to function in the tip development of the second floral whorl prior to anthesis and also functions in controlling the organ number of this floral whorl. The role of PFGLO2 in the tip development might be a residual of corolla organ identity. However, such functional divergence and genetic interaction of two GLO genes for the phenotypic determination were not observed in Petunia hybrida, S. lycopersicum, or N. benthamiana (Vandenbussche et al., 2004; Geuten and Irish, 2010).
In pfglo1, MPF3 was significantly up-regulated, thus the new whorl of the calyx developed in place of the corolla. Nonetheless, MPF3 is expressed in the calyx and the corolla (Zhao et al., 2013), while PFGLO1 and PFGLO2 were mainly expressed in the corolla and the androecium. Thus, a direct role of GLO proteins in the development of ICS is unlikely. However, the development of ICS is closely associated with fertility (He and Saedler, 2005, 2007; Zhao et al., 2013). The structural and molecular basis affecting male and female fertility in both PFGLO2 down-regulated and doll1 (pfglo1) mutants remains elusive. However, fertility seems to be an integral factor for the development of ICS in Physalis spp. Maintaining the proper fertility might serve as a strong selective pressure for the evolution of the “Chinese lantern” (He et al., 2007).
Molecular Bases of the Diverged Functions of the Physalis floridana GLO Paralogs
MADS-domain regulatory proteins usually form complexes to function in plant development (Theissen and Saedler, 2001; Immink et al., 2010). We discovered that both PFGLO1 and PFGLO2 shared some interacting partners, including MPF3, PFAG, and PFSEP3. MPF3, as an ortholog of AP1 and SQUA, is essential to the calyx development and male fertility (Zhao et al., 2013). PFAG, an ortholog of FAR in Antirrhinum majus and AGAMOUS (AG) in Arabidopsis (Bowman et al., 1991; Davies et al., 1999; He et al., 2007), controls male fertility and organ identities of the stamen and the carpel. PFSEP3, a SEPALLATA-like protein, is required for normal floral organ development (Pelaz et al., 2000; He et al., 2007; Immink et al., 2009). However, the direct interactions between B-class proteins and E-class (PFSEP3) proteins were observed in tomato (Solanum lycopersicum) but not in Arabidopsis, Antirrhinum majus, Petunia hybrida, or Eschscholzia californica (Davies et al., 1996; Vandenbussche et al., 2004; de Folter et al., 2005; de Martino et al., 2006; Leseberg et al., 2008; Lange et al., 2013), indicating that the dimerization pattern of the homeotic orthologous proteins is variable among angiosperms. However, our findings on protein-protein interactions provide a common basis underlying the role of PFGLO1 and PFGLO2 in floral development in Physalis floridana. PFGLO1 interacted with PFAG, PFSEP3, and MPF3 to specify androecium identity, while through these interactions, PFGLO2 was mainly involved in male fertility. Moreover, TM6-like genes play roles in carpels, particularly in ovules (de Martino et al., 2006; Geuten and Irish, 2010). We found that PFTM6, negatively controlled by PFGLO1, was also expressed in the gynoecium and that its encoding proteins interacted with PFGLO2, MPF3, PFAG, and PFSEP3. These could explain the poor female fertility in the doll1 mutant. In addition to the roles in fertility, PFGLO1 exerted its full role in the specification of organ identity according to the proposed floral quartets organized by PFGLO1, PFDEF, MPF3, and PFSEP3 in the corolla and those organized by PFGLO1, PFDEF, PFAG, and PFSEP3 in the androecium (He et al., 2007; Theissen and Saedler, 2001). PFGLO2 may fulfill its role in the corolla via a sequestered complex consisting of PFGLO2, MPF3, PFSEP3, PFTM6, and PFDEF. Overexpression of PFGLO1 or PFGLO2 restored the mutation in the second floral whorl in the doll1 mutant, basically confirming the assumptions on their role in corolla. As PFDEF was not expressed in the calyx, overexpression of neither PFGLO1 nor PFGLO2 produced a double flower, as has been observed in Tulipa gesneriana and Clermontia parviflora. In later cases, B-class MADS-box genes are heterotopically expressed in the first floral whorl (Kanno et al., 2003; Hofer et al., 2012).
Corroborated with sequence variations, molecular interactions associated with PFGLO1 and PFGLO2 were remarkably diverged. In line with the previous report that simultaneous expression of PI (GLO) and AP3 (DEF) in Arabidopsis is required for their nuclear localization (McGonigle et al., 1996), heterodimerization with PFDEF facilitated the import of PFGLO1 into the nucleus, while PFGLO2 lost the ability to heterodimerize with PFDEF and was likely localized to the nucleus via homodimerization. Furthermore, the petaloid structure was seen when AG was mutated in Arabidopsis (Yanofsky et al., 1990; Bowman et al., 1991). This was observed in plants overexpressing 35S:PFGLO1 in the pfglo1 background, thus suggesting that a C function is repressed by the overexpressed PFGLO1. Our expression studies revealed that PFGLO1 repressed PFAG, the putative ortholog of AG in Physalis floridana. A presence of PFGLO1 genomic fragment in the doll1 background completely restored both the wild-type floral phenotypes and the expression of all detected MADS-box genes, including PFAG. However, our findings are contrary to studies that showed activation of AG by AP3 and/or PI in Arabidopsis (Bowman et al., 1991; Wuest et al., 2012) and that demonstrated that the C-function gene AG2 is reduced in the seirena1 mutant, a GLO mutant in California poppy (Eschscholzia californica; Lange et al., 2013). Therefore, transcriptional regulation of AG homologs by GLO proteins should be considered to be variable among angiosperms. Moreover, in addition to PFAG, we found that PFGLO1 regulates PFGLO2, PFDEF, MPF2, MPF3, and PFTM6, while PFGLO2 does not regulate any of these genes.
The proposed transcriptional regulations and protein-protein interaction complexes remain to be substantiated functionally. Nonetheless, these findings support their roles of PFGLO1 and PFGLO2 in flower development, while variations in sequences and molecular interactions may largely explain the observed functional divergence.
Divergent Patterns of the Duplicated GLO Genes within the Solanaceae
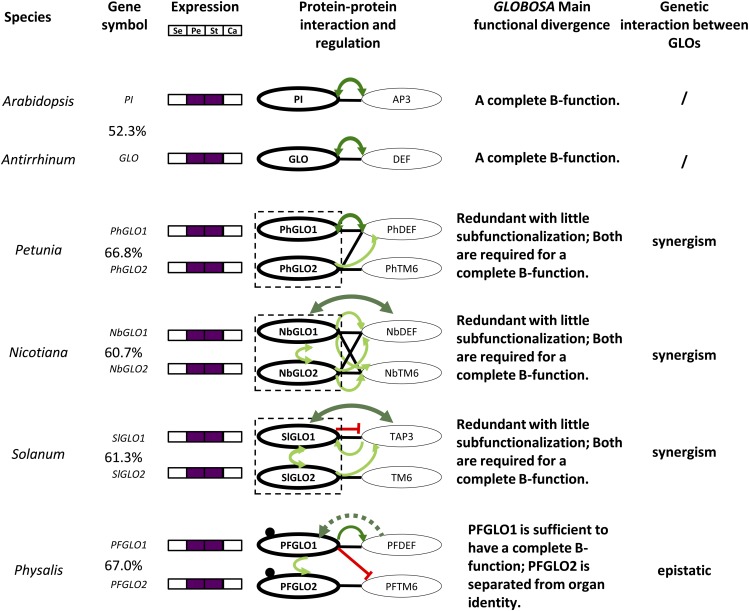
The auto- and cross-regulatory circuit of obligate heterodimers, such as PI-AP3 in Arabidopsis and GLO-DEF in Antirrhinum majus, is required for the specification of petal and stamen identity in angiosperms (Zahn et al., 2005). In the Solanaceae, the circuit is duplicated (Fig. 10); a more complicated but adaptive evolution in the interaction and regulatory networks thus occurred for their functional diversification in flower development (Vandenbussche et al., 2004; Hernández-Hernández et al., 2007; Geuten and Irish, 2010). Divergence is an important strategy in maintaining the duplicated genes (Moore and Purugganan, 2005; Innan and Kondrashov, 2010). GLO genes are expressed in the corolla and the androecium (Fig. 10), but the coding sequences of these paralogous regulatory factors dramatically diverged in a species (Supplemental Fig. S5). Hence, new protein-protein interactions and novel regulatory roles could be established. Comparisons of these molecular interactions among species could show the evolutionary trajectory of the duplicated paralogous genes (VanderSluis et al., 2010).
Figure 10.
Divergence of the various GLO duplicate pairs within the Solanaceae. The protein sequence identity was given between each paralogous pair. In the column of the protein dimerization and regulation, a black line indicates heterodimerization, and a dot near the proteins indicates homodimerization. The curved line with arrows in light green or dark green indicates activation or autoregulation, respectively. The blocked line in red indicates gene repression. The dashed arrow indicated a proposed role needing verification. A dashed box indicates that the two duplicates function as a unit. The functional evolutionary fate and the genetic interaction of each paralogous pair are given. A complete B function is defined as the mutation(s) that could lead to a complete homeotic transformation of the petal and stamen. A slash indicates “no” for this item. Calyx and corolla are homologous to sepals and petals, respectively. For details, please see text. Se, Sepal; Pe, petal; St, stamen; Ca, carpel.
While homodimerization was only detected for the GLO proteins in Physalis floridana, heterodimer pairs GLO1-DEF and GLO2-TM6 were found to form stably in the Solanaceae (Vandenbussche et al., 2004; de Martino et al., 2006; Rijpkema et al., 2006; Geuten and Irish, 2010); however, maximal cross dimerizations were observed in N. benthamiana (Fig. 10). The origin of the specificity of GLO1-DEF and GLO2-TM6 interactions seemed to start from Petunia hybrida, and the heterodimer formation became restricted in S. lycopersicum and Physalis floridana (Fig. 10). Transcriptional regulation of DEF-like genes by GLO1-like proteins occurred in all species reported, while the regulation by GLO2-like proteins was observed in Petunia hybrida, N. benthamiana, and S. lycopersicum. The reciprocal autoregulation of the GLO duplicates occurs in N. benthamiana and S. lycopersicum, while in Physalis floridana, interestingly, PFGLO1 is epistatic to PFGLO2 and activates PFGLO2 (Fig. 10).
According to the functional consequences of a mutation in either of the GLO (PI)-DEF (AP3) heterodimer pairs, we proposed that both GLO1- and GLO2-like proteins exert their role as a functional unit to bear the complete B function in these species (Fig. 10, highlighted in the dashed box), as the GLO proteins in Antirrhinum majus and PI in Arabidopsis do. Moreover, the two GLO genes may have a synergistic role in a Solanaceous species. Such subfunctionalization with variability represents an oft-seen diverging pattern of duplicated GLO genes, supporting the robustness and evolvability of the B function of the duplicated GLO genes (Geuten et al., 2011). However, if one duplicate retains the full B function, another one might deviate from the original developmental program, either becoming lost, as occurred in the basal asterids (Viaene et al., 2009) and in the euasterids I (Lee and Irish, 2011), evolving a new role, or being diverged as happened in Physalis floridana.
However, the observed variations in sequences and molecular interactions did not completely explain functional divergence patterns. Extensive sequence alterations between the orthologous pair GLO/PI with a low identity (52.3%) did not change their functions (Fig. 10), hinting a potential role of the coevolution of GLO/PI and DEF/AP3 in the complete B function. In Solanaceous species, the clade-specific mutated sites that originated prespeciation could discriminate the GLO1 and GLO2 clade, while species-specific mutations between the duplicate that occurred after speciation might contribute to the specificity of certain interacting targets, including proteins and DNAs, thus leading to the different developmental consequences. However, similar functional differentiation pattern evolved in Petunia hybrida, N. benthamiana, and S. lycopersicum, where their GLO paralogs respectively shared 66.8%, 60.7%, and 61.3%, while Physalis floridana. GLO paralogs (sharing 67% identity) functionally diverged to an extreme extent (Fig. 10). These observations hinted that each GLO paralogous pair might have been under different forces after speciation. The selected formation of GLO1-DEF and GLO2-TM6 during evolution might be the key for functional separation in Physalis floridana; however, it did not lead to a significant divergence between the two paralogous pairs in S. lycopersicum (Fig. 10). Thus, these findings represent a new functionally divergent pattern of the duplicated GLO genes within the Solanaceae. Comparisons in higher order complex formation and complete regulatory networks associated with GLO paralogs among these Solanaceous species will provide further insights into the evolution of these various functional divergence patterns.
MATERIALS AND METHODS
Plant Materials
Physalis floridana P106 (He and Saedler, 2005), the doll1 (Lönnig, 2010), and their F1 and F2 progenies and transgenic plants were grown in a growth chamber under long-day conditions (16-h/8-h light/dark cycle) with a constant temperature of 22°C. Leaves and floral organs were harvested for genomic DNA or total RNA isolation.
Genome Walking and Gene Annotation
Genome sequences flanking the PFGLO1 locus in Physalis floridana were isolated by rapid amplification of genomic DNA ends (RAGE), using the Universal Genome Walker kit (Clontech). Multiple PCR reactions using genomic DNA from either the doll1 mutant or wild-type plants as a template were performed to further clarify the obtained sequences. Amplification bands are always expected to appear in the wild type; however, once the amplification bands appeared in both the wild type and mutant, the deletion region in the mutant is thus defined. We used 3,500 bp of upstream sequences and 8,500 bp of downstream sequences of the PFGLO1 locus (1ATG-TAA3956) for gene annotation and used the gene prediction program FGENESH (http://linux1.softberry.com/) to search for ORFs. This and similar gene prediction algorithms are typically thought to have rather low accuracy and to yield many false positive results. To preclude these known potential deficiencies from influencing our final conclusions regarding the structure of the deletion, we used the predicted ORFs from FGENESH to search against the sequence database of the Tomato Genome Sequencing Project (http://mips.helmholtz-muenchen.de/plant/tomato/index.jsp), and no homologous sequences were detected. Thus, our gene annotation analysis indicated that no other ORFs were detected in the deleted fragment.
Genetic Linkage Analyses
Homozygous doll1 plants were pollinated with wild-type pollen to produce F1 seeds, and then F2 seeds were produced through self-fertilization of F1 plants. The phenotype of the F1 plants was identical to that of the wild type. Six hundred ninety-three progeny from F1 plants were grown for segregation analyses. The phenotypes were recorded for each individual in the F2 population. Genomic DNA was isolated from wild-type, doll1 mutant, and F1 plants and each individual of the F2 population for use in segregation analyses with the PFGLO1-linked markers MP1, MP2, and MP3.
RNA in Situ Hybridization
A cDNA fragment covering a portion of the C domain and part of the 3′ untranslated region from each of PFGLO1 (279 bp), PFGLO2 (291 bp), or PFAG (299 bp) was used as a probe template. Probes were synthesized using the T7 RNA polymerase driven by a T7 promoter and labeled with digoxigenin using the DIG RNA Labeling Kit (Roche). Hybridization was performed as described in Carr and Irish (1997), with the alteration that we washed the slides at a temperature of 50°C.
qRT-PCR Analyses
Total RNA was extracted from young floral buds using Plant RNA Reagent (Invitrogen). First-strand cDNA was synthesized with SuperScript III Reverse Transcriptase (Invitrogen) using the Poly (T) primer 5′-CCGGATCCTCTAGAGCGGCCGC (T)17–30. qRT-PCR was performed using the SYBR Premix Ex Taq (Perfect Real Time) kit (TaKaRa) according to the manufacturer’s manuals of the Mx3000p Real Time System (Stratagene). The amplification conditions were 30 s at 95°C, one cycle, followed by 40 cycles of 5 s at 95°C, and 20 s at 60°C. Data were monitored to detect dissociation curves. PFACTIN was used as the housekeeping gene, and relative quantification was performed as previously described (Livak and Schmittgen, 2001).
Yeast Two-Hybrid Assays
ORFs of all MADS-box genes in this study were cloned in frame into the pGADT7 and pGBKT7 vectors (Clontech). The bait and prey construct combinations were cotransformed into the AH109 yeast (Saccharomyces cerevisiae) strain and plated on the synthetic dextrose/-Leu-His plates. Then the survived cells were spotted on selective medium (synthetic dextrose/-Leu-His-Trp) supplemented with 3.0 mm 3-amino-1, 2, 4-triazole. The plates were incubated at 28°C for 2 to 5 d.
Pull-Down Assays
ORFs of PFDEF and PFAG were cloned into the pET-30a (+) vector (containing 6× His tag), expressed in Escherichia coli strain BL21 (DE3), and then purified on Ni Sepharose columns (GE Healthcare). ORFs of PFGLO1 and PFGLO2 were cloned into the pGEX-4T-1 vector (containing GST tag), expressed in E. coli strain BL21, and then infused onto Glutathione Sepharose columns (GE Healthcare). The purified recombinant proteins HIS-PFDEF and HIS-PFAG were mixed into the Glutathione Sepharose columns containing the GST-PFGLO1 or GST-PFGLO2 recombinant proteins, respectively, the columns were washed four times with binding buffer, and the bound proteins were eluted with washing buffer. The eluted proteins were heated and then separated on 12% (w/v) SDS polyacrylamide gels. Proteins were blotted onto nitrocellulose filter membranes (Cwbiotech) using the semidry electrophoretic transfer method, and then the membranes were blocked with 5% (w/v) bovine serum albumin-Tris-buffered saline plus Tween 20 buffer for 1 h. The membranes were initially incubated with anti-GST or anti-HIS antibodies for 2 h, and then anti-HRP antibodies were used as the secondary antibodies for a 1-h incubation. Following each round of incubation, the membranes were washed three times with Tris-buffered saline plus Tween 20 buffer. Radioautographs of the membranes were obtained using Kodak film at room temperature for 1 min and fixed according to standard procedures.
Transient Protein Expression Assays
For subcellular localization studies, the ORFs of PFGLO1 and PFGLO2 were cloned into the Super1300 expression vector (Chen et al., 2009) using the XbaI/KpnI (Roche) restriction sites and fused to GFP. For the BiFC assays, the ORFs of PFGLO1, PFGLO2, and PFDEF were cloned into the pSPYNE-35S and pSPYCE-35S vector pair (Walter et al., 2004) using the XbaI/BamHI (Roche) restriction sites. These paired vectors were designed to express either the N- or C-terminal halves of YFP. The recombinant constructs of PFGLO1-GFP and PFGLO2-GFP and the construct combination of two proteins fused with the N- or C-terminal halves of YFP were transformed into Agrobacterium tumefaciens and were then injected into leaf epidermal cells of Nicotiana benthamiana (Walter et al., 2004). Forty-eight hours after injection, the fluorescence signal of the GFP or YFP was detected using a confocal laser scanning microscope (Olympus FV1000MPE).
VIGS, Plant Transformation, and Genotyping
VIGS procedures were performed according to previously described methods (Zhang et al., 2013). A 479-bp cDNA fragment of PFGLO1, a 416-bp cDNA fragment of PFGLO2, and their respective chimerical fragments were transformed into the tobacco rattle virus vector2 binary vector for the creation of VIGS constructs for both single and double gene silencing. Full-length cDNAs of PFGLO1 and PFGLO2 were cloned into the pBAR plant binary vector for overexpression analysis. A 6-kb genomic DNA sequence consisting of the PFGLO1 locus was also cloned into pBAR for complementation analysis. As no seed from the homozygous mutant was produced, each construct was transformed into F2 progenies from a heterozygous mutant line. The LBA4404 strain of Agrobacterium tumefaciens was used for the Physalis floridana transformation. The media and transformation procedures used here were identical to previously described methods (He and Saedler, 2005). The transgenic plants were genotyped via various analyses of the PCR using genomic DNA as the template and routine reverse transcription-PCR. The gene silencing in VIGS-infected flowers was confirmed using qRT-PCR analysis.
Morphological Analyses
For SEM analyses, fresh materials were fixed in formalin-acetic-alcohol solution (95% ethyl alcohol:glacial acetic acid:formaldehyde [8:1:1]), sputter coated with gold, and examined with a digital scanning microscope (Hitachi S–4800). Pollen maturation was detected using I2-KI staining. The floral buds, mature flowers, androecium, and pollen grains were photographed using a Zeiss microscope.
Phylogenetic Reconstructions
BLAST searches were performed in the National Center for Biotechnology Information Blast Suite (http://www.ncbi.nlm.nih.gov/) using the Physalis floridana B-class MADS-box genes isolated as queries. Information for all of the sequences downloaded from the National Center for Biotechnology Information database is available in Supplemental Table S1. Coding sequences were aligned as Supplemental Data Set S1 using MEGA5 (Tamura et al., 2011) and manually adjusted in BioEdit version 7.0.9 (Hall, 1999). The Bayesian inference, maximum likelihood, and neighbor-joining phylogeny trees were constructed using protein sequences. The maximum likelihood tree was constructed using the PhyML v3.0 program (Guindon and Gascuel, 2003) under the Jones, Taylor, and Thornton model (bootstrap, 100; γ distribution parameter, estimated). The neighbor-joining tree was constructed with MEGA5 (Kimura two-parameter model; bootstrap, 100; Tamura et al., 2011), and the Bayesian inference tree was generated with MrBayes version 3.1.2 (Huelsenbeck and Ronquist, 2001; prset aamodelpr = mixed; ngen = 1,000,000; samplefreq = 100).
Sequencing Analyses
The 3′ and 5′ cDNA ends of the B-class MADS-box genes from Physalis floridana. were amplified according to the standard manual of the 3′/5′ RACE kit (Roche). All sequences were amplified using the TaKaRa LA Taq Polymerase. Amplified fragments were then cloned into the pGEM T-Easy Vector (Promega). All constructs made were commercially sequenced by Beijing Genomics Institute. Primer information used is available in Supplemental Table S2. The sequences reported in this study were deposited in the National Center for Biotechnology Information database under the following accession numbers: JX467691 (PFGLO1), KC174706 (PFGLO2), KC174703 (PFDEF), KC174704 (PFTM6), and KC174705 (PFminDEF).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic analyses of B-class MADS-box genes.
Supplemental Figure S2. Genomic loci and their gene products differ between PFDEF and PFminiDEF.
Supplemental Figure S3. Existence of B-class MADS-box genes in the doll1 mutant.
Supplemental Figure S4. Floral expression of some MADS-box genes in the wild type and the doll1 mutant.
Supplemental Figure S5. Pairwise comparison of the GLO paralogous MADS-domain proteins.
Supplemental Figure S6. Expression of PFGLO genes in PFGLO2-tobacco rattle virus vector2-infected flowers.
Supplemental Figure S7. Protein-protein interactions of Physalis floridana MADS-domain proteins in yeast.
Supplemental Figure S8. Genotyping analyses of PFGLO genes’ overexpressors in the doll1 mutant.
Supplemental Table S1. B-class MADS-box genes for phylogenetic analyses.
Supplemental Table S2. Primer information used in this work.
Supplemental Data Set S1. Alignment of B-class MADS-domain proteins for phylogenetic analysis.
Acknowledgments
We thank Dr. Wolf-Ekkehard Lönnig for generously offering the doll1 mutant seeds and the double-lantern picture (Fig. 1B), Yinhou Xiao for SEM analyses, and Dr. Jingquan Li for operation of the confocal laser scanning microscopic analyses.
Glossary
- ICS
inflated calyx syndrome
- SEM
scanning electronic microscopy
- qRT
quantitative reverse transcription
- cDNA
complementary DNA
- ORF
open reading frame
- VIGS
virus-induced gene silencing
- GST
glutathione transferase
- YFP
yellow fluorescence protein
- BiFC
bimolecular fluorescence complementation
References
- Bowman JL, Drews GN, Meyerowitz EM. (1991) Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3: 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr SM, Irish VF. (1997) Floral homeotic gene expression defines developmental arrest stages in Brassica oleracea L. vars. botrytis and italica. Planta 201: 179–188 [DOI] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH. (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Davies B, Egea-Cortines M, de Andrade Silva E, Saedler H, Sommer H. (1996) Multiple interactions amongst floral homeotic MADS box proteins. EMBO J 15: 4330–4343 [PMC free article] [PubMed] [Google Scholar]
- Davies B, Motte P, Keck E, Saedler H, Sommer H, Schwarz-Sommer Z. (1999) PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J 18: 4023–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martino G, Pan I, Emmanuel E, Levy A, Irish VF. (2006) Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18: 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H. (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. (2006) Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res 16: 805–814 [DOI] [PubMed] [Google Scholar]
- Geuten K, Irish V. (2010) Hidden variability of floral homeotic B genes in Solanaceae provides a molecular basis for the evolution of novel functions. Plant Cell 22: 2562–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuten K, Viaene T, Irish VF. (2011) Robustness and evolvability in the B-system of flower development. Ann Bot (Lond) 107: 1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 8: 1548–1560 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- He CY, Saedler H. (2005) Heterotopic expression of MPF2 is the key to the evolution of the Chinese lantern of Physalis, a morphological novelty in Solanaceae. Proc Natl Acad Sci USA 102: 5779–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CY, Saedler H. (2007) Hormonal control of the inflated calyx syndrome, a morphological novelty, in Physalis. Plant J 49: 935–946 [DOI] [PubMed] [Google Scholar]
- He CY, Sommer H, Grosardt B, Huijser P, Saedler H. (2007) PFMAGO, a MAGO NASHI-like factor, interacts with the MADS-domain protein MPF2 from Physalis floridana. Mol Biol Evol 24: 1229–1241 [DOI] [PubMed] [Google Scholar]
- Hernández-Hernández T, Martínez-Castilla LP, Alvarez-Buylla ER. (2007) Functional diversification of B MADS-box homeotic regulators of flower development: adaptive evolution in protein-protein interaction domains after major gene duplication events. Mol Biol Evol 24: 465–481 [DOI] [PubMed] [Google Scholar]
- Hofer KA, Ruonala R, Albert VA. (2012) The double-corolla phenotype in the Hawaiian lobelioid genus Clermontia involves ectopic expression of PISTILLATA B-function MADS box gene homologs. Evodevo 3: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JY, Saedler H. (2007) Evolution of the inflated calyx syndrome in Solanaceae. Mol Biol Evol 24: 2443–2453 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Huijser P, Klein J, Lönnig WE, Meijer H, Saedler H, Sommer H. (1992) Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J 11: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RG, Tonaco IA, de Folter S, Shchennikova A, van Dijk AD, Busscher-Lange J, Borst JW, Angenent GC. (2009) SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RG, Kaufmann K, Angenent GC. (2010) The ‘ABC’ of MADS domain protein behaviour and interactions. Semin Cell Dev Biol 21: 87–93 [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Irish VF, Litt A. (2005) Flower development and evolution: gene duplication, diversification and redeployment. Curr Opin Genet Dev 15: 454–460 [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697 [DOI] [PubMed] [Google Scholar]
- Kanno A, Saeki H, Kameya T, Saedler H, Theissen G. (2003) Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol Biol 52: 831–841 [DOI] [PubMed] [Google Scholar]
- Karve R, Liu W, Willet SG, Torii KU, Shpak ED. (2011) The presence of multiple introns is essential for ERECTA expression in Arabidopsis. RNA 17: 1907–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RS, Irish VF. (2003) Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc Natl Acad Sci USA 100: 6558–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange M, Orashakova S, Lange S, Melzer R, Theißen G, Smyth DR, Becker A. (2013) The seirena B class floral homeotic mutant of California poppy (Eschscholzia californica) reveals a function of the enigmatic PI motif in the formation of specific multimeric MADS domain protein complexes. Plant Cell 25: 438–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HL, Irish VF. (2011) Gene duplication and loss in a MADS box gene transcription factor circuit. Mol Biol Evol 28: 3367–3380 [DOI] [PubMed] [Google Scholar]
- Leseberg CH, Eissler CL, Wang X, Johns MA, Duvall MR, Mao L. (2008) Interaction study of MADS-domain proteins in tomato. J Exp Bot 59: 2253–2265 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lönnig WE. (2010) Mutagenesis in Physalis pubescens L. ssp. floridana: some further research on Dollo’s law and the law of recurrent variation. Floriculture Ornamental Biotech 4: 1–21 [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- McGonigle B, Bouhidel K, Irish VF. (1996) Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev 10: 1812–1821 [DOI] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD. (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8: 122–128 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Rijpkema AS, Royaert S, Zethof J, van der Weerden G, Gerats T, Vandenbussche M. (2006) Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell 18: 1819–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönnig WE, Saedler H, Sommer H. (1992) Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J 11: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H, Beltrán JP, Huijser P, Pape H, Lönnig WE, Saedler H, Schwarz-Sommer Z. (1990) Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J 9: 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Saedler H. (2001) Plant biology. Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lönnig WE, Saedler H, Sommer H, Schwarz-Sommer Z. (1992) GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J 11: 4693–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M, Zethof J, Royaert S, Weterings K, Gerats T. (2004) The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell 16: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderSluis B, Bellay J, Musso G, Costanzo M, Papp B, Vizeacoumar FJ, Baryshnikova A, Andrews B, Boone C, Myers CL. (2010) Genetic interactions reveal the evolutionary trajectories of duplicate genes. Mol Syst Biol 6: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaene T, Vekemans D, Irish VF, Geeraerts A, Huysmans S, Janssens S, Smets E, Geuten K. (2009) Pistillata—duplications as a mode for floral diversification in (Basal) asterids. Mol Biol Evol 26: 2627–2645 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wuest SE, O’Maoileidigh DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F. (2012) Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc Natl Acad Sci USA 109: 13452–13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]
- Zahn LM, Leebens-Mack J, DePamphilis CW, Ma H, Theissen G. (2005) To B or Not to B a flower: the role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. J Hered 96: 225–240 [DOI] [PubMed] [Google Scholar]
- Zhang JS, Zhao J, Zhang SH, He CY. (2014) Efficient gene silencing mediated by tobacco rattle virus in an emerging model plant Physalis. PLoS ONE 9: e85534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Tian Y, Zhang JS, Zhao M, Gong PC, Riss S, Saedler R, He CY. (2013) The euAP1 protein MPF3 represses MPF2 to specify floral calyx identity and displays crucial roles in Chinese lantern development in Physalis. Plant Cell 25: 2002–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]