A flavodiiron protein operon encodes for an alternative electron transfer mechanism at the PSII acceptor side and is dependent on functional phycobilisomes.
Abstract
Oxygenic photosynthesis evolved with cyanobacteria, the ancestors of plant chloroplasts. The highly oxidizing chemistry of water splitting required concomitant evolution of efficient photoprotection mechanisms to safeguard the photosynthetic machinery. The role of flavodiiron proteins (FDPs), originally called A-type flavoproteins or Flvs, in this context has only recently been appreciated. Cyanobacterial FDPs constitute a specific protein group that evolved to protect oxygenic photosynthesis. There are four FDPs in Synechocystis sp. PCC 6803 (Flv1 to Flv4). Two of them, Flv2 and Flv4, are encoded by an operon together with a Sll0218 protein. Their expression, tightly regulated by CO2 levels, is also influenced by changes in light intensity. Here we describe the overexpression of the flv4-2 operon in Synechocystis sp. PCC 6803 and demonstrate that it results in improved photochemistry of PSII. The flv4-2/OE mutant is more resistant to photoinhibition of PSII and exhibits a more oxidized state of the plastoquinone pool and reduced production of singlet oxygen compared with control strains. Results of biophysical measurements indicate that the flv4-2 operon functions in an alternative electron transfer pathway from PSII, and thus alleviates PSII excitation pressure by channeling up to 30% of PSII-originated electrons. Furthermore, intact phycobilisomes are required for stable expression of the flv4-2 operon genes and for the Flv2/Flv4 heterodimer-mediated electron transfer mechanism. The latter operates in photoprotection in a complementary way with the orange carotenoid protein-related nonphotochemical quenching. Expression of the flv4-2 operon and exchange of the D1 forms in PSII centers upon light stress, on the contrary, are mutually exclusive photoprotection strategies among cyanobacteria.
Photosynthetic light reactions are evolutionarily highly conserved among oxygenic photosynthetic organisms from cyanobacteria to higher plants. Because of dangerous chemistry of the water splitting reactions, oxygenic photosynthesis produces reactive oxygen species (ROS) and other radicals that potentially could destroy the photosynthetic machinery. To avoid permanent damage, all oxygenic photosynthetic organisms are equipped with an array of various photoprotective and regulatory mechanisms. Accumulating evidence on these regulatory mechanisms has revealed vast evolutionary differences between organisms performing oxygenic photosynthesis.
Photosynthetic organisms have a capacity to adjust to different light intensities and to changes in the availability of electron sinks, which depends largely on metabolic cues. When light or metabolic conditions change, photosystems can dissipate excess energy as heat in nonphotochemical energy dissipation processes in the light-harvesting antenna systems (for review, see Horton et al., 1996; Müller et al., 2001). Cyanobacteria have phycobilisomes (PBs) as light-harvesting antenna, which also participate in state transitions (for review, see van Thor et al., 1998; Mullineaux and Emlyn-Jones, 2005) and nonphotochemical quenching (NPQ) of excitation energy (for review, see Bailey and Grossman, 2008). Both of these processes are involved in short-term regulation of light-harvesting processes and concomitantly function as photoprotective mechanisms. These nonphotochemical energy quenching mechanisms, however, have only limited capacity, and it often occurs that more electrons are excited than can be safely used in photochemistry for reduction of natural metabolic electron acceptors, particularly under stress conditions. In such situations, electrons can be directed, for example, to molecular oxygen resulting in production of ROS. To avoid harmful reactions by ROS that threaten the cell viability, a repertoire of different photoprotection mechanisms have evolved in cyanobacteria as well as in all other oxygenic photosynthetic organisms.
Flavodiiron proteins (FDPs), originally called A-type flavoproteins or Flvs (Wasserfallen et al., 1998), were recently demonstrated to have an important role in photoprotection of the photosynthetic machinery (Zhang et al., 2009, 2012; Allahverdiyeva et al., 2011, 2013; Ermakova et al., 2013). FDPs in general are most widespread among strict and facultative anaerobic bacteria. Many of their FDPs have been characterized structurally and functionally, showing homodimeric or homotetrameric forms (Vicente et al., 2008b, 2009). A typical FDP consists of a core composed of a metallo-β-lactamase-like domain and a C-terminal flavodoxin domain. The former domain contains a nonheme diiron center, whereas the latter harbors a FMN moiety. It has been shown that FDPs in anaerobic bacteria are involved in O2 and/or NO detoxification (Vicente et al., 2008a). Completely unique FDPs are, however, found in specific groups of oxygenic photosynthetic organisms. FDPs found in cyanobacteria and some photosynthetic eukaryotes possess an extra C-terminal flavin reductase domain (Zhang et al., 2009). This particular domain composition theoretically allows NAD(P)H oxidation to be coupled with O2 reduction in the same enzyme.
Synechocystis sp. PCC 6803 (hereafter Synechocystis), a widely used model organism among cyanobacteria in photosynthesis research, contains four FDPs encoded by the sll1521 (flv1), sll0219 (flv2), sll0550 (flv3), and sll0217 (flv4) genes. In vivo, Flv1 and Flv3 acquire electrons after PSI and deliver them further to molecular oxygen, reducing it to water. We have denominated this process as a Mehler-like reaction (Allahverdiyeva et al., 2013) because the excess of electrons is donated to O2, similarly to the genuine plant-type Mehler reaction, but there is no production of ROS in the FDP-mediated reaction (Helman et al., 2003). Up to 60% of the electrons produced by the oxygen splitting activity of PSII are redirected to Flv1- and Flv3-mediated Mehler-like reactions in severe inorganic carbon starvation conditions (Allahverdiyeva et al., 2011). Flv1 and Flv3 proteins form a very important electron sink that protects PSI against oxidative damage under fluctuating light conditions (Allahverdiyeva et al., 2013).
Flv2 and Flv4 can be found only in cyanobacteria and they have been assigned a role in photoprotection of PSII (Zhang et al., 2009, 2012). PSII is historically known to be extremely vulnerable to oxidative damage upon illumination, with the severity of damage being dependent on light intensity and on the availability of electron acceptors. At air-level CO2 concentrations (low CO2 or LC) and/or high light (HL) irradiances, terminal acceptors are consistently limiting the electron flow, making PSII particularly sensitive to these conditions, widely exceeding the repair capacity of damaged PSII centers (Aro et al., 1993).
Flv2 and Flv4 proteins are encoded in an operon including a small Sll0218 protein. Importantly, the flv4-2 operon is strongly induced in LC and HL conditions (Zhang et al., 2009). Flv4 and Flv2 proteins form a heterodimer that localizes in cytoplasm but also has a high affinity to membrane in the presence of cations (Zhang et al., 2012). Sll0218, the 19-kD protein encoded by the flv4-2 operon, locates in the thylakoid membrane and forms a high molecular mass complex in association with unknown partners. In the model proposed by Zhang et al. (2012), Sll0218 stabilizes PSII dimers and facilitates the opening of a novel electron transfer pathway through the Flv2/Flv4 heterodimer, which associates with the thylakoid membrane in light. The Flv2/Flv4 complex is also important for proper energy transfer from PBs to PSII as evidenced by a high emission peak at 685 nm in the 77K fluorescence spectra. This effect is caused by uncoupled PB terminal emitters, as deduced from detailed examination of the deconvoluted emission spectra (Zhang et al., 2012). This strongly suggested a distorted energy transfer from PB terminal emitters to the PSII reaction centers in flv4-2 operon deletion mutants. However, the photoprotection mechanism induced by the flv4-2 operon is not yet clearly understood. Here, with an overexpression approach, we provide evidence that Flv2/Flv4 acts as an important electron sink at the PSII acceptor side, allowing the maintenance of the plastoquinone (PQ) pool in an oxidized state and preventing the production of singlet oxygen in PSII. Furthermore, regular PBs are required for the Flv2/Flv4-related mechanism to be expressed. Genome mining of sequenced cyanobacteria strains provided evidence for the loss of the flv4-2 operon in the genomes of cyanobacteria that have acquired a stress-inducible D1 copy.
RESULTS
Overexpression of the flv4-2 Operon in Synechocystis
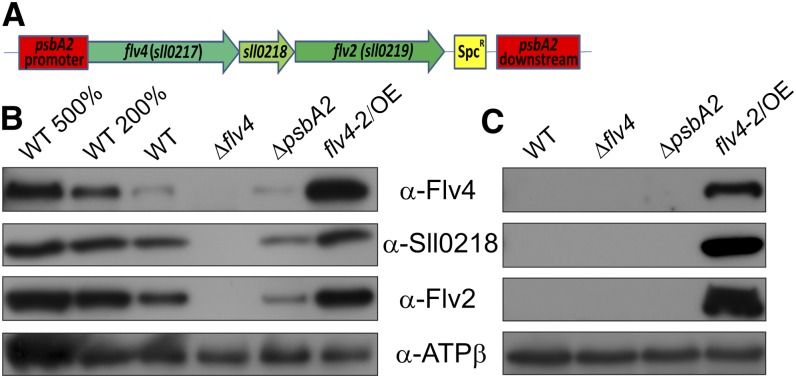
The sll0217-sll0219 (flv4-flv2) operon was overexpressed in Synechocystis in order to investigate and clarify its role in photoprotection. The flv4-2 operon was placed under the control of the psbA2 promoter and integrated into the chromosome by replacement of the psbA2 gene (Fig. 1A). The obtained flv4-2 overexpression mutant (flv4-2/OE) contained the flv4-2 operon under control of the psbA2 promoter in addition to the native operon, and was devoid of the psbA2 gene. Therefore, the ΔpsbA2 mutant containing the spectinomycin resistance cassette instead of the psbA2 gene (Zhang et al., 2012) was used as the control strain together with the wild type in functional studies of the overexpression mutant. In addition, the experiments were also performed with the Δflv4 mutant, which does not express the Flv4, Sll0218, and Flv2 proteins (Fig. 1B; Zhang et al., 2012). The latter mutant contained the kanamycin cassette in sll0217 (flv4), the first gene of the operon (Zhang et al., 2012).
Figure 1.
Construction and characterization of the flv4-2/OE mutant. A, Schematic representation of the insertion of flv4-2 operon into the Synechocystis chromosome under the control of the psbA2 gene promoter. B and C, Western blots were performed with protein samples isolated from cultures grown at air-level CO2 (LC; B) and at 3% CO2 (HC; C). Twenty micrograms of total protein from the cell extracts were loaded into each lane (= 100%), if not otherwise indicated. WT, wild type. [See online article for color version of this figure.]
Western-blot analyses revealed that the level of Flv2, Sll0218, and Flv4 proteins in the flv4-2/OE mutant increased more than 2-fold compared with the wild type when cells were grown in LC conditions (Fig. 1B), whereas the ΔpsbA2 mutant showed the lowest content of flv4-2 operon-encoded proteins in LC conditions (Fig. 1B). Importantly, the flv4-2/OE mutant also expressed the Flv4, Sll0218, and Flv2 proteins under the high CO2 (HC) condition (Fig. 1C), whereas these proteins were strongly down-regulated by the excess of CO2 in the wild type and ΔpsbA2. The expression of the Flv4, Sll0218, and Flv2 proteins in the flv4-2/OE mutant in HC conditions was related to the light-inducible psbA2 promoter.
Growth Phenotype of the flv4-2/OE Mutant in Different Light Intensity Conditions
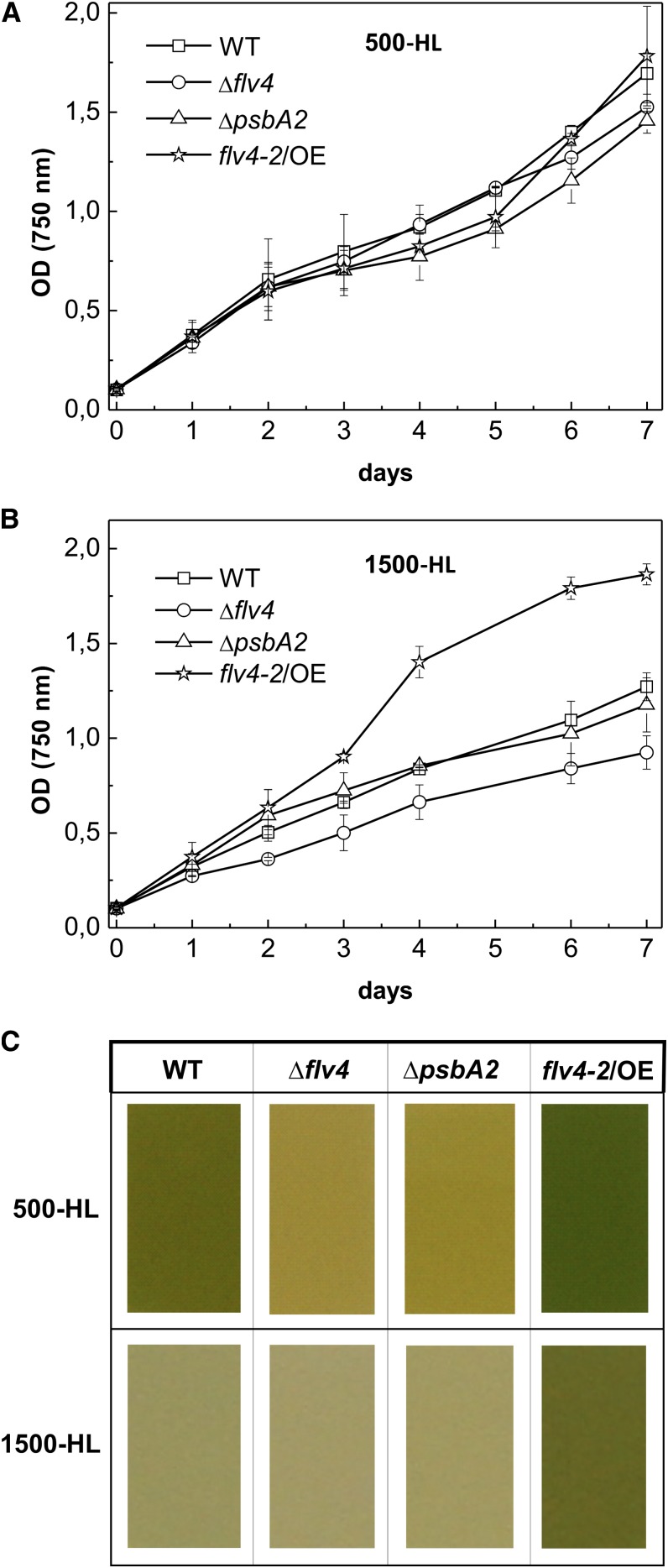
The above-described strains were tested for sensitivity to strong light irradiances in LC conditions. When cells were grown at 500 µmol photons m−2 s−1 (500-HL), no significant differences were observed in growth curves (Fig. 2A). Increase of the light intensity to 1,500 µmol photons m−2 s−1 (1,500-HL) caused slower growth of the wild type, ΔpsbA2, and Δflv4 (Fig. 2B), whereas the growth of the flv4-2/OE cells was not hampered (Fig. 2B). Because alterations in thylakoid membrane content might cause inconsistencies in the correlation of OD750 and cell number (Fuhrmann et al., 2009), we calculated cells of all four strains grown in the different tested conditions using a cell counting chamber. Only minor differences in OD750 per cell number between the strains were observed (data not shown), confirming the results presented in growth curves (Fig. 2, A and B).
Figure 2.
Growth phenotype of wild-type and mutant strains. A and B, Growth curves were obtained with cells cultivated at 500-HL (A) or at 1,500-HL (B). C, The difference in color of cells grown for 7 d at corresponding light intensities. WT, wild type.
After 7 d of growth, clear differences in the color of the cell cultures were observed. At 500-HL, the overexpression mutant showed a darker green color than the wild-type strain, whereas the ΔpsbA2 and Δflv4 strains had acquired a clear yellow color (Fig. 2C). The dark green color of the flv4-2/OE culture suggested that the increased expression of the flv4-2 operon-encoded proteins counteracted the negative effect caused by the absence of psbA2. Moreover, the flv4-2/OE mutant cells remained unbleached after 7 d of growth at 1,500-HL in contrast with the other three strains, including the wild type. Growth experiments clearly showed that the overexpression mutant tolerated HL stress better than the wild-type, ΔpsbA2, and Δflv4 strains.
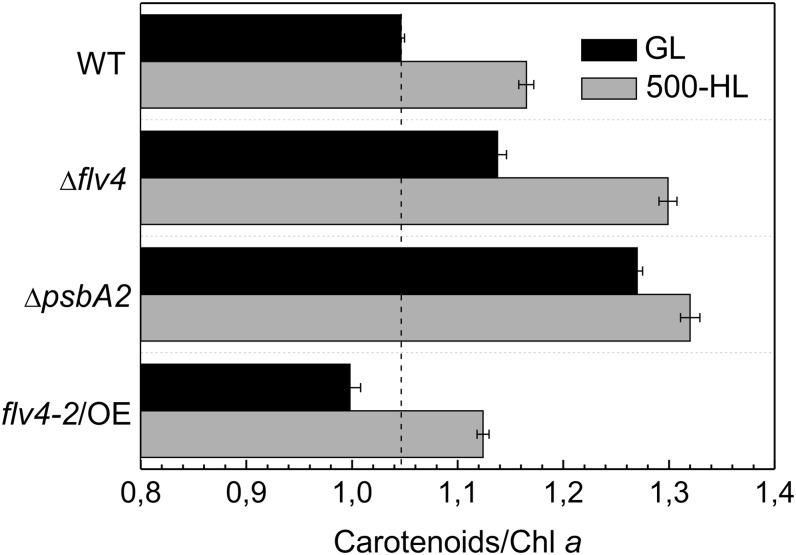
Whole-cell absorption spectra (Supplemental Fig. S1) of cells grown in standard growth light (GL) and 500-HL were recorded to analyze the content of carotenoids that typically accumulate in photosynthetic organisms under stressful conditions. When the strains were grown in GL and 500-HL, the ΔpsbA2 and Δflv4 strains showed the highest carotenoid/chlorophyll a ratio (Car/Chl a), whereas the flv4-2/OE mutant had the lowest ratio among all of the strains. The overexpression mutant in particular had an approximately 22% lower Car/Chl a, whereas Δflv4 had an approximately 10% higher Car/Chl a compared with the respective control strains (Fig. 3). These findings clearly indicated that the photoprotection phenotype of the flv4-2/OE mutant is based on a specific function of the flv4-2 operon but not on accumulation of carotenoids.
Figure 3.
Car/Chl a ratio measured from cultures cultivated at standard GL conditions (black bars) and at high light intensity of 500-HL (gray bars). The cultures were adjusted to an OD750 = 0.6 before the measurements. Values are means ± sd of three independent experiments. WT, wild type.
Production of Singlet Oxygen in flv Mutants
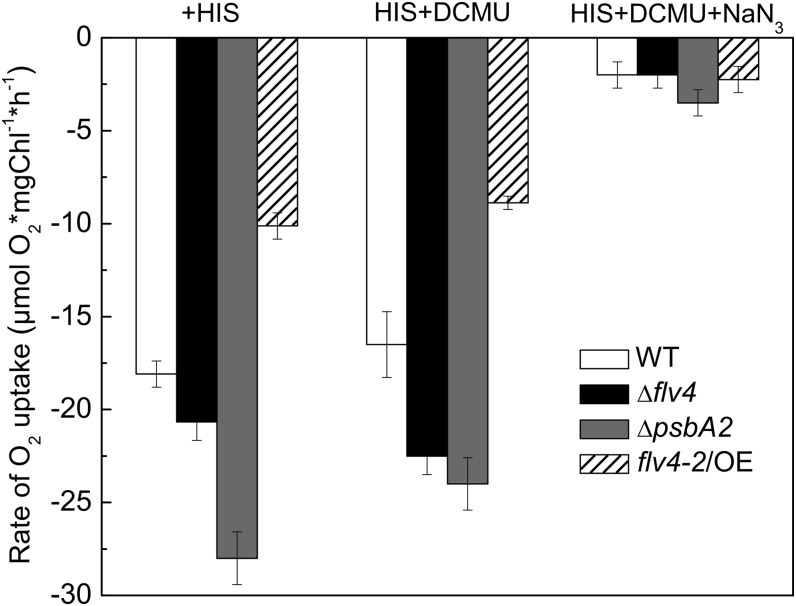
One important role of carotenoids in photosynthetic organisms is the protection that they offer against singlet oxygen (1O2; for review, see Frank and Cogdell, 1993), which is particularly harmful to the photosynthetic apparatus. To test whether the production of singlet oxygen is altered in the flv4-2/OE mutant, we assessed singlet oxygen production by measuring a His-mediated oxygen uptake in all four strains. The assay is based on the interaction of the generated 1O2 with the aromatic side chain of His, leading to the formation of an oxidation product of His that decreases the concentration of dissolved oxygen in the suspension. Thus, the extent of O2 uptake in the presence of His is proportional to the amount of singlet oxygen (Rehman et al., 2013). To demonstrate that the O2 uptake signal indeed arises from 1O2, we also applied 10 mm of sodium azide, which is a specific quencher of 1O2. Because sodium azide inhibits O2 evolution (Rehman et al., 2013), its 1O2 quenching effect was tested in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) in which the artifact that would arise from the decrease in the O2 evolution rate was avoided.
In the absence of the Flv4, Sll0218, and Flv2 proteins (Δflv4 strain), the extent of 1O2 production increased compared with the wild type, despite the somewhat increased level of carotenoids, which confirmed that the products of flv4-2 operon avoid generation of ROS (Fig. 4). This idea was further corroborated by a large decrease in 1O2 production in the flv4-2/OE mutant compared with all other strains, even though it contains fewer carotenoids than the wild type. The lack of psbA2 significantly increased the production of 1O2 compared with the wild type (Fig. 4). When 1O2 production was compared between the flv4-2/OE strain and the ΔpsbA2 control strain, we found that overexpression of the flv4-2 operon decreased singlet oxygen production by approximately 60%. These data are consistent with relative light sensitivities of the strains and provide strong support for the idea that the flv4-2 operon has an important role in photoprotection via decreasing the production of 1O2, which is a key factor of PSII photodamage (Vass et al., 1992).
Figure 4.
Production of singlet oxygen in wild-type, ΔpsbA2, flv4-2/OE, and Δflv4 strains in the presence of 5 mm His upon illumination of the cells at 2,300 μmol photon m−2 s−1. The cultures were adjusted to a chlorophyll concentration of 5 µg mL−1. To demonstrate that the O2 uptake signal indeed arises from 1O2, 10 mm sodium azide, which is specific quencher of 1O2, was also applied. Because sodium azide inhibits O2 evolution, its 1O2 quenching effect was tested in the presence of DCMU in which the artifact that would arise from the decreased of O2 evolution rate could be avoided. The results are a mean ± sd of three independent experiments. NaN3, sodium azide; WT, wild type.
Photochemistry of PSII in the flv4-2/OE Mutant
The performance of PSII in the flv4-2/OE, Δflv4, and control strains grown in LC conditions was characterized with various biochemical and biophysical methods. When cells were grown under GL conditions, the flv4-2/OE mutant cells demonstrated 10% to 20% higher maximum photochemical efficiency of PSII in the dark-adapted state (Fv/Fm) and 50% higher effective quantum yield of PSII [Y(II)] compared with the wild-type and ΔpsbA2 strains (Supplemental Table S1). These results indicated an improved photochemistry of PSII when the flv4-2 operon was overexpressed. To note, both the Y(II) and the Fv/Fm values were especially low in the Δflv4 mutant (Supplemental Table S1). The Y(II) of cells grown in the 500-HL condition (Supplemental Table S1) showed trends that were similar to the Y(II) of cells grown in the GL condition, but the differences between the strains were larger. In the 500-HL condition, the ΔpsbA2 strain also exhibited a very low Y(II) (Supplemental Table S1). This is in line with earlier findings showing that under GL conditions, the deletion of the psbA2 gene copy is fully compensated by expression of the psbA3 gene, whereas the expression of both genes is essential in HL conditions (Mohamed et al., 1993).
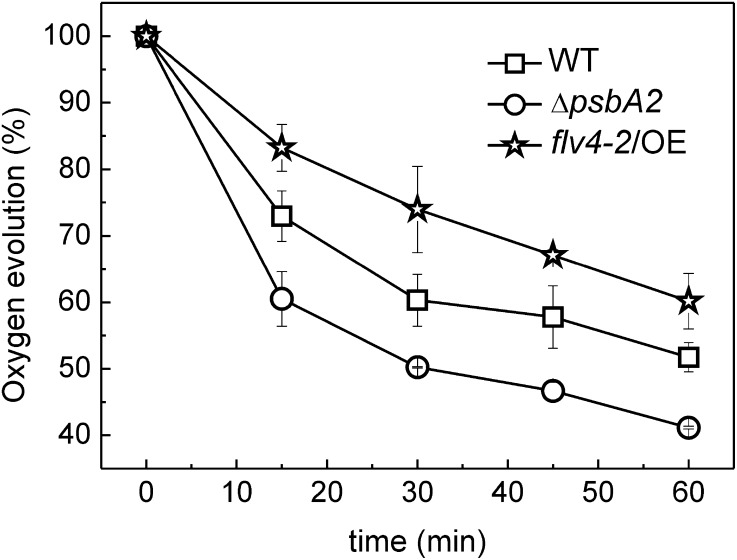
To reveal the sensitivity of PSII to photoinhibition in the flv4-2/OE mutant, the cells were exposed to HL in the presence of lincomycin, an inhibitor of de novo protein synthesis and thus the repair of damaged PSII. The cells grown in the GL and LC conditions were illuminated by 500-HL for up to 60 min. The PSII activity was measured during the course of illumination by monitoring the oxygen-evolving activity of the cells using 2 mm 2,6-dimethyl-p-benzoquinone (DMBQ) as an artificial electron acceptor. The flv4-2/OE mutant presented significantly higher PSII activity, showing 20% to 25% higher oxygen evolution rates compared with the control ΔpsbA2 strain and approximately 10% higher oxygen evolution rates compared with the wild type (Fig. 5). These measures indicate a decisive role of the flv4-2 operon in photoprotection of PSII.
Figure 5.
PSII photoinhibition kinetics in wild-type, ΔpsbA2, and flv4-2/OE strains. The strains were illuminated with white light intensity of 500 µmol photons m−2 s-1in the presence of lincomycin (300 µg mL−1). Oxygen evolution rates were measured in the presence of 2 mm DMBQ as an artificial electron acceptor. The cultures were adjusted to a chlorophyll concentration of 5 µg mL−1. Values are means ± sd of three independent experiments. WT, wild type.
PQ Pool Redox State in the flv4-2/OE Mutant
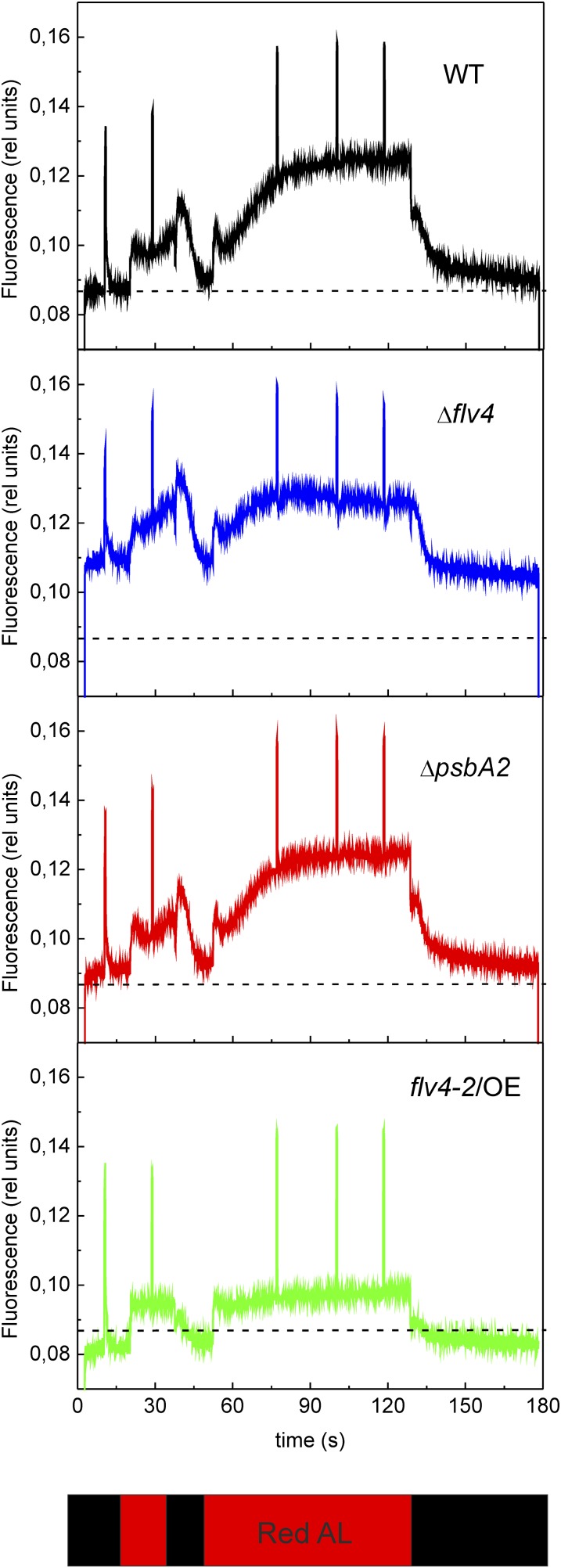
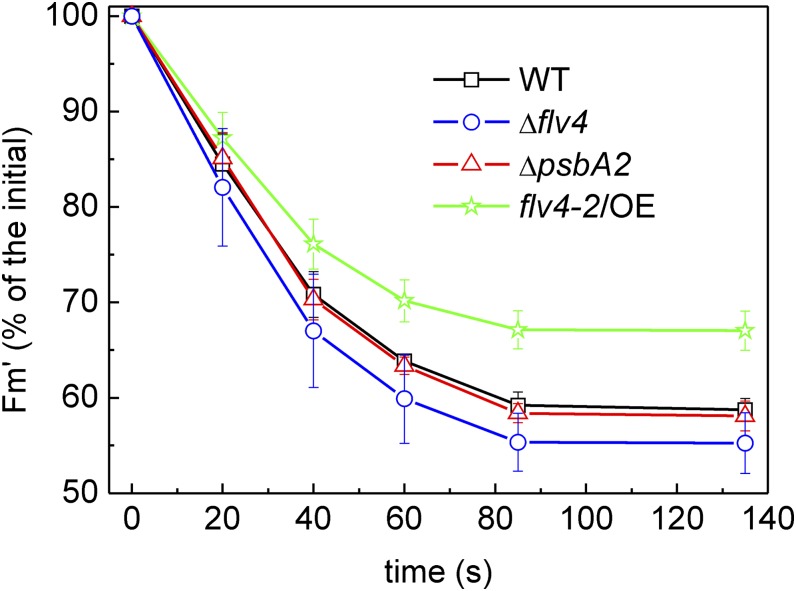
Fluorescence induction kinetics, using red actinic light of 120 µmol photons m−2 s−1, was recorded for all four strains to gain information about the redox state of the PQ pool (Fig. 6) in LC conditions. Comparison of the maximal fluorescence in the dark-adapted state (FmD) with the maximum PSII fluorescence in the light-adapted state (Fm′) obtained during illumination with actinic light (Fig. 6; Supplemental Fig. S2) revealed a much smaller difference in these fluorescence parameters in the flv4-2/OE mutant than in the other strains. This finding suggested stronger PSII contribution to the fluorescence level after darkness in the flv4-2/OE mutant, indicating a more oxidized state of the PQ pool during dark incubation of the cells. The transient increase of fluorescence after termination of actinic light (F0 rise) is considered to arise from reduction of the PQ pool by NAD(P)H or other reducing substances that accumulated in the light (Mi et al., 1995). As demonstrated in Supplemental Figure S3, the F0 rise was lower in flv4-2/OE and much higher in Δflv4 compared with respective control strains, implying the more oxidized PQ pool in darkness in the flv4-2/OE mutant and the more reduced state in the Δflv4 mutant.
Figure 6.
Fluorescence induction curves from wild-type, ΔpsbA2, flv4-2/OE, and Δflv4 strains. Cells were dark adapted and then exposed to red actinic light of 120 µmol photons m−2 s−1. The cultures were adjusted to a chlorophyll concentration of 10 µg mL−1. AL, actinic light; WT, wild type. [See online article for color version of this figure.]
The flv4-2/OE mutant showed higher variable fluorescence (Fv) in the dark-adapted state as well as a higher Fq value (Fm′ − Fs) in the light-adapted state (Fig. 6), indicating a higher fraction of open PSII centers compared with the other strains. This is in line with the Y(II) values (Supplemental Table S1). The lower F0 and steady state fluorescence level in the light-adapted state (Fs), in turn, indicated that the primary electron-accepting plastoquinone of PSII (QA) was less reduced in flv4-2/OE cells than in other strains. In contrast with flv4-2/OE, the Δflv4 mutant had higher F0 and Fs levels, but no significant difference in Fq compared with the wild type. Because of altered energy transfer from PBs to PSII reaction centers (Zhang et al., 2012), a strong contribution of PBs to fluorescence cannot be directly excluded in the Δflv4 mutant. To exclude the contribution of PBs to the fluorescence, an actinic blue light of 44 µmol photons m−2 s−1 was used to record the fluorescence kinetics of the Δflv4 mutant (Supplemental Fig. S4). The results clearly showed that the F0 level was still higher in the Δflv4 mutant compared with the wild type, whereas the Fq level was lower. This indicates that the mutant has fewer PSII centers open during the illumination compared with the control cells.
Although flv4-2/OE showed an oxidized state of the PQ pool in darkness as well as in the light-adapted state, the state transition kinetics of flv4-2/OE, recorded during illumination with PSII and PSI lights, were not modified compared with the control strains (Supplemental Fig. S2).
Orange Carotenoid Protein-Related NPQ in the flv4-2/OE Mutant
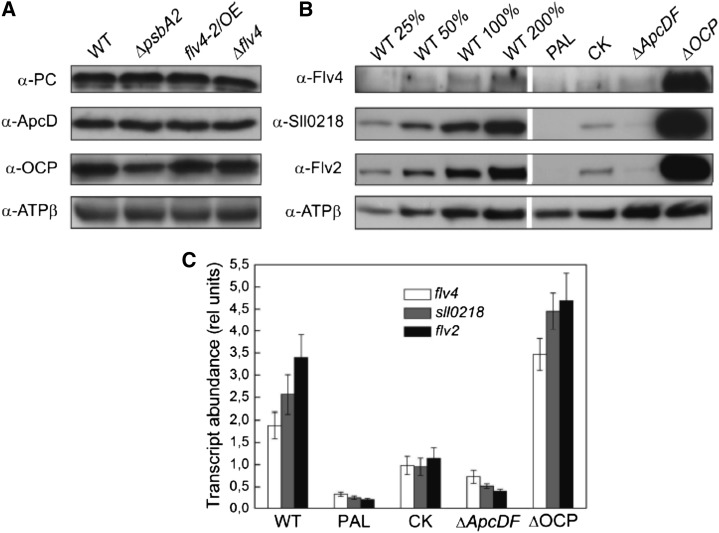
Because the energy absorbed by chlorophyll in the photosynthetic apparatus decays via photochemistry, NPQ, and fluorescence, which are in equilibrium with each other (for review, see Bailey and Grossman 2008), we tested how the stronger photochemistry of PSII observed in the flv4-2/OE mutant compared with other strains is reflected to the NPQ of the strains. To this end, the orange carotenoid protein (OCP)-related NPQ (Wilson et al., 2006) was measured, exposing the cells to high intensities of blue-green light. The results revealed approximately 20% lower amplitude of quenching of maximal fluorescence in flv4-2/OE than the control strains (Fig. 7). Immunoblotting of the OCP protein did not, however, reveal significant differences between the wild type and the flv4-2/OE mutant (Fig. 8A). Interestingly, the flv4-2 operon transcripts and encoded proteins were more abundant in the ΔOCP strain compared with the wild type (Fig. 8, B and C).
Figure 7.
Decrease of maximal fluorescence induced by OCP-mediated NPQ upon illumination of cells with strong blue light intensity (750 µmol photons m−2 s−1). The cultures were adjusted to a chlorophyll concentration of 10 µg mL−1. The values shown are the mean ± sd of three independent experiments. WT, wild type. [See online article for color version of this figure.]
Figure 8.
A, Protein immunoblots showing the content of ApcD, PC, and OCP in wild-type, ΔpsbA2, flv4-2/OE, and Δflv4 strains. Ten micrograms of total protein from the cell extract were loaded in each lane. B, Protein immunoblots showing the content of Flv4, Sll0218, and Flv2 in wild-type, PAL, CK, ΔApcDF, and ΔOCP mutants. Twenty micrograms of total protein from the cell extract were loaded in each lane (= 100%), if not otherwise stated. This is a representative picture of three independent immunoblots. C, Transcript accumulation of flv4-2 operon in the wild-type, PAL, CK, ΔApcDF, and ΔOCP mutants analyzed by real-time quantitative RT-PCR. Transcript abundance is shown as relative units. The transcript level of the rnpB gene is used as a reference. The results are the mean from three independent experiments ± sd. ApcD allophycocyanin D; PC, phycocyanin; rel unit, relative unit; WT, wild type.
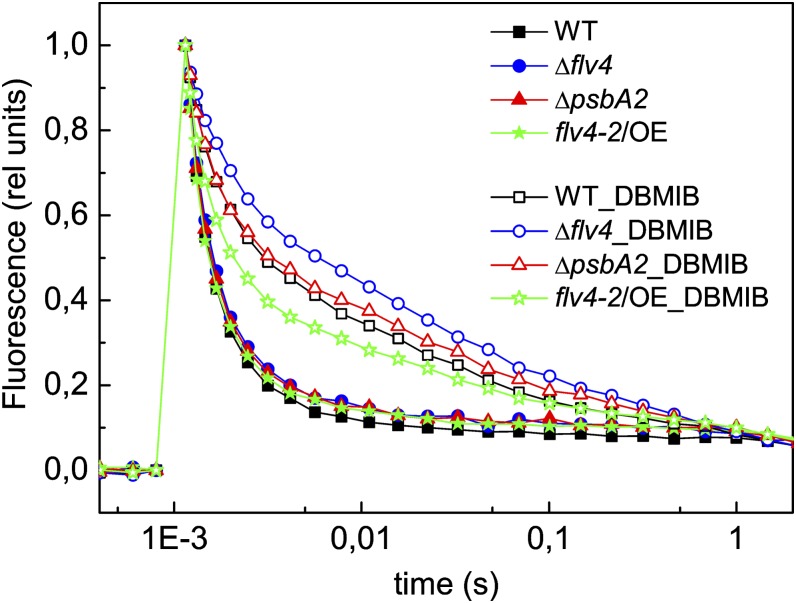
PSII Properties in flv Mutants
The flash-induced increase in fluorescence yield and its subsequent decay in darkness provide important information about functional modifications in PSII. The method was applied here for comparison of the flv4-2/OE strain, Δflv4 mutant, and control strains, all grown in LC conditions. Measurements were performed in the presence of 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB), which blocks the Q0 site of the cytochrome b6f complex and thus also the main reoxidation route of plastoquinol (Trebst, 1980). The results revealed that the absence of Flv2 and Flv4 causes slower QA− oxidation compared with the wild type (Fig. 9; Zhang et al., 2012). In line with these results, QA− reoxidation was shown to be significantly faster in the flv4-2/OE mutant compared with the wild type, especially compared with ΔpsbA2 (Fig. 9). These data provide strong support for the role of the Flv2/Flv4 heterodimer in electron transfer by facilitating the oxidation of QA− and maintaining the oxidized PQ pool.
Figure 9.
Flash-induced chlorophyll fluorescence relaxation curves in darkness from wild-type, ΔpsbA2, flv4-2/OE, and Δflv4 strains, grown in LC conditions. The curves were recorded in the absence (closed symbols) and in the presence of 20 µM DBMIB (open symbols). The cultures were adjusted to a chlorophyll concentration of 5 µg mL−1. To facilitate the comparison of the curves, F0 and FmD values were normalized to 0 and 1, respectively. The fluorescence traces are the mean of three independent experiments. rel unit, relative unit; WT, wild type. [See online article for color version of this figure.]
To characterize this novel electron transfer route in more detail, the oxygen evolution measurements were performed using different electron acceptors (Table I). In the absence of Flv2 and Flv4 proteins (the Δflv4 mutant), the rate of oxygen evolution from water to the 2,6-dichloro-p-benzoquinone (DCBQ) electron acceptor was higher than the rates measured with DMBQ, whereas in the presence of Flv2 and Flv4 (the wild type), the oxygen evolution rates with DMBQ were higher than those with DCBQ (Table I), in accordance with the results of Zhang et al. (2012). The flv4-2/OE mutant revealed increased DMBQ-mediated rates of O2 evolution compared with the wild type, whereas the rates with DCBQ were lower. These results indicated that the presence of DCBQ inhibits the electron transfer to the Flv2/Flv4 heterodimer, whereas DMBQ allows this electron transfer mechanism to take place. To further characterize the electron transfer rates to the Flv2/Flv4 heterodimer, the oxygen evolution measurements were performed in the presence of DBMIB at low concentrations (2 µm). When electron transfer to cytochrome b6f was partially inhibited with DBMIB, the Δflv4 mutant showed approximately 30% smaller oxygen evolution rates than the wild type, whereas the flv4-2/OE mutant had approximately 10% higher rates than the control ΔpsbA2 strain and the wild type.
Table I. Oxygen evolution rates in wild-type, Δflv4, ΔpsbA2, and flv4-2/OE strains with different electron acceptors and inhibitors.
Steady state oxygen evolution (in µmol O2 mg−1 chlorophyll h−1) was measured with an oxygen electrode under saturating light conditions. The cultures were adjusted to a chlorophyll concentration of 5 µg mL−1. Each value is an average of at least three independent experiments ± sd.
Relation of flv4-2 Operon with PBs
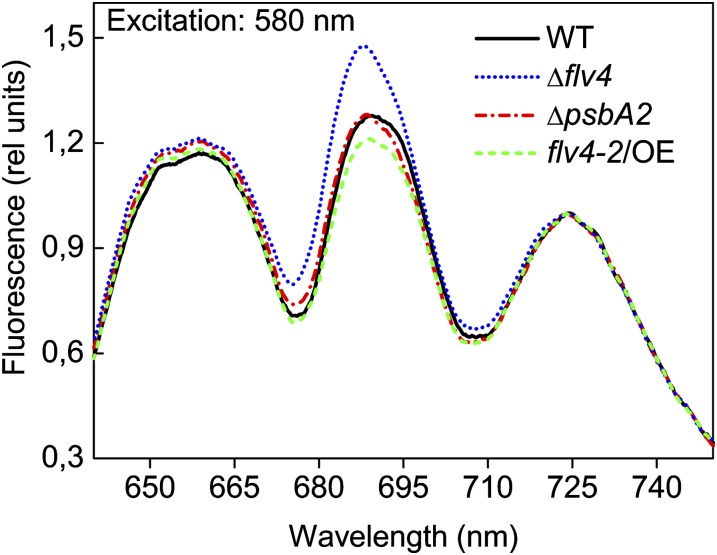
We next addressed the question of whether flv4-2/OE has an influence on energy transfer from PBs to the PSII reaction centers, which in the absence of the flv4-2 operon was previously shown to be distorted in LC-grown cells (Zhang et al., 2012). The 77K fluorescence spectra, using excitation light of 580 nm, demonstrated a high peak at 685 nm in the Δflv4 mutant. This result was also shown by Zhang et al. (2012), who concluded that because the increase of the F685-nm band was not accompanied by enhancement of the F695-nm band, the intensification of the F685-nm band results from disturbed energy transfer from terminal emitters of PBs to the PSII core. The flv4-2/OE mutant revealed completely different 77K fluorescence spectra, demonstrating a lower peak at 685 nm compared with the control strains or Δflv4 (Fig. 10), which is a signal of enhanced efficiency of the energy transfer from PBs to the PSII reaction centers. Importantly, no significant differences were observed in the 77K fluorescence spectra of the different strains when chlorophyll was excited with 440 nm of light (Supplemental Fig. S5). Furthermore, western blots made for evaluation of the phycocyanin and allophycocyanin D contents in the Δflv4, flv4-2/OE, and control strains revealed no difference in the protein amounts (Fig. 8A). Likewise, the evaluation of the phycobilin/chlorophyll a ratios from whole-cell absorption spectra did not reveal any significant differences between the strains (Supplemental Fig. S6). The ΔpsbA2 strain, when grown in 500-HL conditions, was the only exception showing a relevant increase in this ratio. Therefore, changes in phycobilin amount cannot explain the increased F0 levels detected in the Δflv4 mutant (Fig. 6). Instead, the high F0 is an indication of reduced energy transfer from PBs to PSII reaction centers (Campbell et al., 1998a) in Δflv4, again confirming the results deduced from the 77K fluorescence spectra.
Figure 10.
The 77K fluorescence emission spectra of wild-type, ΔpsbA2, flv4-2/OE, and Δflv4 strains excited at 580 nm. The spectra were averaged from three independent experiments. Each spectrum was normalized to PSI fluorescence peak at 723 nm (set as 1). The cultures were adjusted to a chlorophyll concentration of 5 µg mL−1. WT, wild type. [See online article for color version of this figure.]
To understand the relevance of PBs to the mechanism of the flv4-2 operon-mediated photoprotection, we investigated different PB mutants with respect to the expression of the Flv4, Sll0218, and Flv2 proteins in LC conditions (Fig. 8B). In the PAL mutant in which PBs are completely absent (Ajlani and Vernotte, 1998), no flv4-2 operon-encoded proteins were detected. In the CK mutant (ΔcpcC; constructed by Ughy and Ajlani and described in Thomas et al., 2006) containing PBs deprived of phycocyanin rods and composed of only the allophycocyanin core, the amounts of Flv4, Sll0218, and Flv2 proteins were reduced by approximately 75% compared with those in the wild type. In the ΔApcDF mutant (Jallet et al., 2012) that lacks the allophycocyanin D and allophycocyanin F terminal emitters (emitting at 680 nm), a severe reduction in energy transfer from PBs to the PSII reaction centers has been demonstrated (Ashby and Mullineaux, 1999; Jallet et al., 2012). The ΔApcDF mutant was also nearly depleted of the Flv4, Flv2, and Sll0218 proteins compared with the high amounts present in the wild type (Fig. 8B). On the contrary, the ΔOCP mutant that is incapable of quenching the excitation energy absorbed by the PBs and funneled to PSII reaction centers, demonstrated significant up-regulation of the Flv2, Flv4, and Sll0218 proteins compared with the control cells.
Finally, the strong correlation between the content and composition of PBs with the accumulation of the flv4-2 operon proteins prompted us to solve whether the down-regulation of the flv4-2 operon expression in PB mutants occurred at the transcript or protein level. To this end, reverse transcription PCR (RT-PCR) experiments were performed with RNA extracted from wild-type, PAL, CK, ΔApcDF, and ΔOCP strains grown in LC conditions for 24 h (after a shift from the HC condition in which no transcripts were present). The abundance of the flv4-2 operon transcripts was significantly lower in the CK, ΔApcDF, and PAL mutants compared with the wild type (more than 2-, 5-, and 10-fold lower in the mutants, respectively; Figure 8C). On the contrary, the ΔOCP strain had an almost 2-fold higher amount of flv4-2 operon transcripts than the wild type (Fig. 8C). The RT-PCR results are in line with the western-blot experiments (Fig. 8B). Thus, a down-regulation in the transcript level of the flv4-2 operon occurred when dysfunctional PBs were present. In the absence of the OCP photoprotective mechanism, an up-regulation of flv4-2 operon transcripts was observed.
DISCUSSION
Overexpression of the flv4-2 Operon Maintains the PQ Pool Oxidized Even in HL
The experiments conducted with the flv4-2/OE mutants of Synechocystis clearly demonstrate that overexpression of the flv4-2 operon results in a more photoprotected phenotype in LC conditions. Cultures of flv4-2/OE mutant grown under strong light were less photosensitive and continued growth even at 1,500-HL, whereas the other strains showed slower growth (Fig. 2). Strong photoprotection was reflected in higher maximal photochemistry of PSII and Y(II) values in GL- and HL-grown cells (Supplemental Table S1). Importantly, the flv4-2/OE mutants kept the PQ pool more oxidized, allowing PSII to function properly with a decreased risk of photoinhibition. In fact, the PSII oxygen-evolving activity was higher in the flv4-2/OE exposed to 500-HL than in the control strains, showing a phenotype resistant to photoinhibition (Fig. 5). By contrast, in the Δflv4 strain, the PQ pool was more reduced (Fig. 6; Supplemental Fig. S3) and the cells were previously shown to be sensitive to photoinhibition of PSII (Zhang et al., 2009). Generally, the mutations causing a change in the redox level of the PQ pool might also modify the state transitions (Schreiber et al., 1995). Therefore, it is noteworthy that in the case of the flv4-2/OE mutant, the higher oxidation level of the PQ pool is not connected with modification in the state transitions (Supplemental Fig. S2).
Resistance of the flv4-2/OE Mutant to Photodamage Correlates with Reduced Production of Singlet Oxygen
Singlet oxygen is the main ROS produced in the reaction center of PSII (Durrant et al., 1990; Macpherson et al., 1993; Hideg et al., 1994; for review, see Krieger-Liszkay, 2005; Krieger-Liszkay et al., 2008), in which the chlorophyll triplet state can react with 3O2 to produce the very reactive 1O2, as a result of a charge recombination reaction of the primary pair, P680+Pheo−. This occurs especially under stress conditions such as exposure of the cells to HL intensities or drought (Hideg et al., 2001, 2002; Trebst et al., 2002). In these conditions, the PQ pool may become highly reduced, and the light-induced loss of PSII activity occurs (for review, see Adir et al., 2003). 1O2 is a very reactive oxygen species that can attack proteins, pigments, nucleic acids, and lipids, and is thought to be the most harmful ROS responsible for light-induced loss of PSII activity (Vass et al., 1992), degradation of the D1 protein (one of the PSII reaction center proteins), and pigment bleaching (for review, see Prasil et al., 1992; Aro et al., 1993; Nixon et al., 2005; Vass and Aro, 2007; Tyystjärvi, 2008).
Clearly, the more oxidized state of the PQ pool in the flv4-2/OE mutant correlates with decreased production of singlet oxygen (Fig. 4) and the lowered extent of photodamage to the PSII centers (Fig. 5), both being indicative of better tolerance to photoinhibition by overexpression of the flv4-2 operon. This protection was not related to the enhanced carotenoid production. On the contrary, the light-sensitive Δflv4 and especially the ΔpsbA2 strains showed an increased level of carotenoid production, whereas the carotenoid level was lower in the phototolerant flv4-2/OE strain (Fig. 3). This indicates that the Flv2 and Flv4 proteins and carotenoids participate in protection against 1O2-mediated damage in a complementary fashion; therefore, when the Flv2-4 proteins provide sufficient photoprotection, the content of carotenoids remains at a low level.
The Flv2/Flv4 Heterodimer Functions as an Important Electron Sink at the PSII Acceptor Side
As stated above, the flv4-2 operon clearly modulates the PQ pool redox state in LC conditions. In the presence of DBMIB, which blocks the main reoxidation route of plastoquinol, the decay of the flash-induced fluorescence in darkness slowed down in Δflv4 but became faster in flv4-2/OE compared with respective control strains (Fig. 9). These results imply that QA− reoxidation is faster when the flv4-2 operon is overexpressed and slower when the operon proteins are absent, indicating that Flv2 and Flv4 are dissipating excess electrons and thereby preventing over-reduction of the PQ pool. Similar conclusions can be drawn from oxygen evolution measurements, which in the presence of DMBQ that accepts electrons from the PQ pool, revealed low activity in the Δflv4 mutant and the highest activity in the flv4-2/OE mutant (Table I). Furthermore, the measurements performed with DCBQ, which mainly accepts the electron at the secondary electron-accepting PQ (QB) pocket of PSII (Graan and Ort, 1986), showed results opposite of those with DMBQ. The oxygen evolution rate in the presence of DCBQ is strikingly lower in the flv4-2/OE strain in particular compared with that with DMBQ (Table I). These data provide strong evidence for the involvement of Flv2 and Flv4 in an alternative electron transfer process that intercepts electrons at the QB pocket of PSII, and in this way maintains the PQ pool oxidized in LC conditions. The expression of the flv4-2 operon is, in fact, induced only at LC and/or HL conditions, when cyanobacterial cells show a chronically sensitive phenotype to photoinhibition. In addition, the operon is induced in the absence of oxygen (Summerfield et al., 2011), a condition in which the function of terminal oxidases and the Flv1/Flv3-mediated Mehler-like reactions are inhibited, leading to an increased reduction level of the PQ pool. This effect can be compensated by the induction of Flv2/Flv4 that helps to relieve the over-reduction of the PQ pool.
In the HC condition, which actually relates to the original atmospheric CO2 condition at the time of evolution of oxygenic photosynthesis, the photosynthetic processes in Synechocystis are working optimally because of the high amount of CO2 that provides an unlimited sink for electrons originating from the water splitting PSII. In those conditions, the function of the flv4-2 operon does not provide benefit for the cells and indeed the fluorescence phenotype of the flv4-2/OE mutant does not differ from that of the control strains in HC growth conditions, as can be deduced from the flash fluorescence decay curve in Supplemental Figure S7. Interestingly, the antisense RNA As1_flv4 prevents unfavorable synthesis of the proteins from the flv4-2 operon in the early phase after a change from HC to LC conditions (Eisenhut et al., 2012). Thus, the tight control on the expression of the flv4-2 operon is necessary to optimize the efficiency of photosynthesis in unstressed cells in HC conditions, whereas it ensures photoprotection upon stressful conditions in LC conditions.
In the present-day LC condition, the terminal electron acceptor CO2 is present in a limited amount and carbon concentration mechanisms are highly expressed in cyanobacteria in order to concentrate as much CO2 as possible around Rubisco and at the same time induce cyclic electron flow to generate more ATP (for review, see Kaplan and Reinhold, 1999; Badger and Price, 2003; Giordano et al., 2005; Price et al., 2008). In this way, photosynthesis can be functional but the acceptors are present in a more reduced form than in HC conditions, causing a higher risk of photoinhibition. The expression of the flv4-2 operon provides PSII with a higher amount of electron acceptors (having a Flv2/Flv4 heterodimer and a more oxidized state of the PQ pool), thus preventing the over-reduction of the PQ pool. The addition of a low concentration of DBMIB to the cells mimics the conditions of high excitation pressure on PSII (e.g. occurring in LC and HL conditions). Oxygen evolution measurements performed in these conditions revealed that the Flv2/Flv4-mediated electron transfer mechanism can absorb up to 30% of the electrons originating from PSII, making it an important electron sink at the PSII acceptor side.
PBs Are Necessary for Flv2/Flv4-Mediated Photoprotection Mechanisms
The 77K fluorescence emission spectra suggested a distorted energy transfer from PB terminal emitters to the PSII reaction centers in Δflv4, whereas the lower 685-nm peak of the flv4-2/OE mutant indicated an improved energy transfer to PSII reaction centers compared with the control strains (Fig. 10). We discovered that the expression of the flv4-2 operon is dependent on the presence of PBs. In their absence (PAL mutant), none of the flv4-2 operon-encoded proteins are found in the cell. Moreover, the amounts of Flv4, Sll0218, and Flv2 are low in mutants containing truncated PBs such as the CK and ΔApcDF mutants (Fig. 8B). The reduced amount of Flv4, Sll0218, and Flv2 proteins in the different PB mutants is a consequence of the down-regulation of the flv4-2 operon at transcript level (Fig. 8C). On the contrary, an increased accumulation of Flv4, Sll0218, and Flv2 proteins is observed in the ΔOCP mutant, which lacks the ability to dissipate the excess of energy collected by the PBs. These results provide evidence for the crucial importance of Flv2, Flv4, and Sll0218 in efficient allocation of the energy collected by the PB antenna to PSII in LC conditions.
Furthermore, these data support the idea that a certain level of excitation pressure on PSII, reflected in the redox state of the PQ pool, is essential for efficient transcription of the flv4-2 operon. In fact, the operon is most actively transcribed under the combined HL and LC conditions when the PQ pool is over-reduced (Zhang et al., 2009), whereas the transcription of the flv4-2 operon is down-regulated when the PQ pool remains more oxidized (e.g. in the presence of DCMU; Hihara et al., 2003). Indeed, the regulation of the flv4-2 operon has turned out to be extremely complicated and is, in addition to a putative redox regulation by the PQ pool, under strict control of the NdhR transcription factor and several noncoding RNAs (Eisenhut et al., 2012).
Flv2/Flv4-Mediated Electron Transfer Mechanism Competes with OCP-Mediated NPQ for Excess Energy Dissipation
The overexpression of the flv4-2 operon resulted in an enhanced photochemistry of PSII. Because photochemistry competes with NPQ processes that dissipate the absorbed light energy as heat, we found that the flv4-2/OE mutant, exposed to high blue light levels, has approximately 20% lower amplitude of quenching of maximal fluorescence than the control strains (Fig. 7). The fact that the Flv2/Flv4-mediated electron transfer mechanism is part of the photochemical processes, whereas OCP-mediated fluorescence quenching represents the nonphotochemical processes, puts these photoprotective mechanisms theoretically in competition for energy dissipation. However, in natural environmental conditions, they are likely to be active in different situations. OCP-mediated NPQ is more active in HL conditions (El Bissati et al., 2000), whereas we have shown that the Flv2/Flv4-mediated electron transfer mechanism is active in LC conditions already at GL. Moreover, upon exposition of cells to 350 µmol photons m−2 s−1 of blue light for 1 h, only about 29% of the PBs were quenched by the OCP mechanism in vivo (Tian et al., 2011). This observation implies that the Flv2/Flv4 heterodimer might be important at the acceptor side of those PSII complexes that received a correct energy transfer from PBs. Furthermore, we showed that a very high amount of Flv4, Sll0218, and Flv2 proteins were produced in the ΔOCP mutant compared with the wild type (Fig. 8B), again confirming the important role of the flv4-2 operon proteins in photoprotection, especially when the OCP photoprotective mechanism is absent.
Evolutionary Trends in PSII Photoprotective Mechanisms in Cyanobacteria
We have demonstrated that the flv4-2 operon is important for PSII photoprotection in Synechocystis. Considering the evolution of the flv4-2 operon (Zhang et al., 2012), it is remarkable to note that it is present in the genomes of nearly all β-cyanobacteria (containing β-carboxysomes and the form 1B of Rubisco), with only a few exceptions (Synechococcus sp. PCC 7002, Synechococcus elongatus PCC 7942, Thermosynechococcus elongatus BP-1, and Trychodesmium IMS 101). These flv4-2 operon-less cyanobacteria, which nevertheless use a PB antenna system, are distinct from the other cyanobacteria with regard to the diversity of the psbA gene family products, encoding the D1 protein of the PSII reaction center (for review, see Mulo et al., 2009). In fact, they possess a further D1 copy, called D1:2, in addition to the standard D1:1 copy. The main difference of D1:2 from the other D1 proteins is a substitution of Gln to Glu at position 130 (Q130E). D1:2 expression is induced by HL conditions, where it reaches 70% of the total D1 proteins in T. elongatus BP-1 (Kós et al., 2008; Sander et al., 2010). Deletion mutants of the D1:2 copy show impaired growth in HL (Kulkarni and Golden, 1995; Sander et al., 2010). In fact, the exchange of D1 protein forms with a preference for D1:2 in HL provides increased phototolerance to the cells (Krupa et al., 1991; Clarke et al., 1993), which is accompanied by accelerated nonradiative charge recombination, with a consequent reduced production of singlet oxygen (Tichý et al., 2003; Cser and Vass, 2007; Sander et al., 2010).
It is conceivable that the flv4-2 operon has played a pivotal role in the maintenance of oxygenic photosynthesis at its early emergence during evolution; however, it was subsequently lost because of its apparently high metabolic cost for the cell, compared with other effective and less expensive photoprotection mechanisms. The appearance of the D1:2 copy might have contributed to the elimination of the flv4-2 operon relatively early during evolution (Table II). The D1 protein from algae and plants, which is encoded by a single psbA gene, shares more common residues with the D1:2 cyanobacterial D1 copy, including the 130E residue (Svensson et al., 1991; Winhauer et al., 1991; Campbell et al., 1998b; Takishita and Uchida, 1999). These observations suggest that the Q130E D1 form was a successful strategy for the photosynthetic organisms, becoming ubiquitous among them.
Table II. Occurrence of different photoprotection mechanism in the genomes of representative strains of cyanobacteria and photosynthetic eukaryotes.
The D1:2 copy homology analysis was performed with BLASTP (http://blast.ncbi.nlm.nih.gov). Part of the data were imported from Zhang et al. (2012). LHCII/I, Light-harvesting complex II/I; Pcb, chlorophyll a/b binding light-harvesting protein; LL, low light ecotype; HL, high light ecotype.
| Organism | Type | Ecological Niche | Antenna System | flv4-2 Operon | D1:2 Copy (Q130E) |
|---|---|---|---|---|---|
| Cyanobacteria | |||||
| Synechocystis sp. PCC 6803 | β | Freshwater | PBs | + | − |
| Microcystis aeruginosa NIES-843 | β | Marine | PBs | + | − |
| Microcystis aeruginosa sp. PCC 7806 | β | Freshwater | PBs | + | − |
| Cyanothece sp. PCC 8801 | β | Freshwater | PBs | + | − |
| Cyanothece sp. ATCC 51142 | β | Marine | PBs | + | + |
| Anabaena sp. PCC 7120 | β | Freshwater | PBs | + | + |
| Trychodesmium IMS 101 | β | Marine | PBs | − | + |
| Synechococcus sp. PCC 7002 | β | Marine | PBs | − | + |
| Synechococcus elongatus PCC 7942 | β | Freshwater | PBs | − | + |
| Thermosynechococcus elongatus BP-1 | β | Freshwater-thermophile | PBs | − | + |
| Synechococcus sp. WH7803 | α | Marine | PBs | − | + |
| Synechococcus sp. WH8102 | α | Marine | PBs | − | + |
| Prochlorococcus marinus str. MIT9211 | α | Marine (LL) | Pcb | − | − |
| Prochlorococcus marinus MED4 | α | Marine (HL) | Pcb | − | − |
| Photosynthetic eukaryote | |||||
| Chlamydomonas reinhardtii | Freshwater | LHCII/I | − | + | |
| Arabidopsis | Land | LHCII/I | − | + |
In conclusion, the flv4-2 operon encodes for an electron sink mechanism at the PSII acceptor side, which allows the PQ pool to be in oxidized state, prevents PSII from photodamage, and concomitantly reduces the production of singlet oxygen. Flv2, Sll0218, and Flv4 protein accumulation is related to the presence of functional PBs. These results provide evidence that the Flv2/Flv4 electron transfer mechanism requires an efficient energy transfer from PBs to PSII. It is also demonstrated that the OCP-mediated NPQ functions in competition for energy dissipation with the photochemical protection mechanism provided by the Flv2/Flv4 heterodimer. The flv4-2 operon has, however, disappeared in the course of evolution of higher plants and is also missing from many cyanobacteria strains. Protection of PSII by the Flv2/Flv4 heterodimer is energetically expensive and was, therefore, replaced by evolution of a more efficient and stress tolerant form of the D1 protein.
MATERIALS AND METHODS
Synechocystis Strains and Growth Conditions
The Synechocystis sp. PCC 6803 Glc-tolerant strain (Williams, 1988) was used as the wild type. The Δflv4 strain was described by Zhang et al. (2012). To obtain flv4-2/OE, the operon fragment was amplified by PCR with the 1Fw primer (5′-AATACCATATGGTTACCCTAATTGATTCTCC-3′) and the 2Rv primer (5′-AGTTCGCTAGCCTAATATTGTCCCCCCGATTTG-3′), and then inserted in the NdeI-NheI sites of the modified pPSBA2KS plasmid (Lagarde et al., 2000) containing both spectinomycin- and kanamycin-resistance cassettes. Insertion of the flv4-2 operon at the NdeI-NheI sites resulted in placing of the operon under control of the psbA2 promoter and deletion of the kanamycin-resistance cassette. The operon was integrated into the chromosome by replacement of the native psbA2 gene of Synechocystis (Fig. 1A). Positive transformants were selected with spectinomycin (25 µg mL−1). Segregation of the strain was confirmed by PCR using the 3Fw primer (5′-GCCCCAAAATACATCCCCCTAA-3′) and the 4Rv primer (5′-AACCTGCCTCCAATCCACACTG-3′).
Wild-type and mutant strains were grown at 30°C in BG-11 (Allen, 1968) buffered with 20 mm HEPES-NaOH (pH 7.5). White light was used for illumination, with intensity of 50 µmol photons m−2 s−1 (GL); in some cases higher light intensities were used (500, 1,500 µmol photons m−2 s−1). The cultures were grown in flasks shaking at 120 rpm. For HC conditions, air was enriched with 3% CO2. For LC conditions, normal air was used, and sodium carbonate was omitted from the culture ingredients.
For physiological experiments, the cells were harvested at the logarithmic phase (OD750 between 0.6 and 1.1), resuspended in fresh BG-11 medium, and adjusted to chlorophyll concentrations of 5 to 10 µg mL−1.
Protein Isolation, Electrophoresis, and Immunodetection
Total cell extracts and the soluble fractions of Synechocystis cells were isolated as described by Zhang et al. (2009). Proteins were separated by 12% (w/v) SDS/PAGE containing 6 m urea. The proteins were transferred to a polyvinylidene fluoride membrane (Immobilon-P; Millipore) and analyzed with protein-specific antibodies.
Oxygen Evolution Measurements
Steady state oxygen evolution was measured with a Clark-type oxygen electrode (Hansatech) at 30°C under saturating white light with intensity of 1,000 µmol photons m−2 s−1. Before measurements, the cells were harvested and resuspended in fresh growth medium at a chlorophyll concentration of 5 μg mL−1. The PSII electron transfer rates (water to quinone) were measured in the presence of the artificial electron acceptor, either 0.5 mm DCBQ or 2 mm DMBQ. These measurements were performed also in the presence of 10 mm sodium bicarbonate. To estimate PSII electron transfer rates when the PQ pool was over-reduced, oxygen evolution measurements were performed in the presence of 2 µM DBMIB.
Fluorescence
A pulse amplitude modulated fluorometer Dual-PAM-100 (Walz) was used to monitor chlorophyll a fluorescence in intact cells adjusted to a chlorophyll concentration of 10 μg mL−1. Measurements were performed in stirred 1 cm × 1 cm cuvettes at 30°C. Red (620 nm) or blue (460 nm) actinic lights were used depending on the experiment. Saturating pulses (5,000 µmol photons m−2 s−1, 300 ms) were applied to transiently close all PSII centers in order to measure FmD and Fm′. The maximal photochemical efficiency of PSII (Fv/Fm) was measured in the presence of DCMU. Y(II) was measured in cells exposed to an actinic light of 120 µmol photons m−2 s−1 for 2 min.
Low-temperature fluorescence emission spectra at 77K of whole cells were measured by a USB4000-FL-450 (Ocean Optics) spectrofluorometer. The cells were harvested at OD750 = 0.6 to 0.8 and resuspended in fresh BG-11 (sodium carbonate-free) medium at a chlorophyll concentration of 5 µg mL−1. The resuspended cultures were acclimated under the same growth conditions for 1 h. The cells were frozen in liquid nitrogen directly from the GL. The cells were excited by 580 nm or 440 nm light obtained via interference filters 10 nm in width.
The flash-induced increase and the subsequent decay of chlorophyll fluorescence yield was measured using a fluorometer FL 3500 (PSI Instruments) according to the method of Vass et al. (1999). The cells were resuspended in fresh BG-11 medium at a chlorophyll concentration of 5 μg mL−1, and dark adapted for 5 min before the excitation with a single 10-μs saturation flash in the absence and presence of 20 µm DBMIB.
In Vivo Absorption Spectra
Absorption spectra were measured in vivo with a UV-3000 spectrophotometer (Shimadzu) from 400 to 800 nm. Carotenoid content (in relation to chlorophyll a content, 678 nm peak) was determined measuring the absorption spectra at room temperature at 485 nm.
Singlet Oxygen Production
His-mediated oxygen uptake measurements were applied, which were based on the oxidation of His by 1O2, for quantification of singlet oxygen production. This process leads to removal of dissolved oxygen in aqueous media that can be detected by a standard oxygen electrode. This method, which was originally applied for isolated PSII reaction center complexes (Telfer et al., 1994), has recently been extended for intact cyanobacterial cells (Rehman et al., 2013). The rate of singlet oxygen-induced oxygen uptake was measured by using a Hansatech DW2 O2 electrode. Measurements were performed in the presence of 5 mm His, at 2,300 μmol photon m−2 s−1, at a chlorophyll concentration of 5 µg mL−1, and in the absence of artificial electron acceptors (Rehman et al., 2013). Before the O2 uptake measurements, Synechocystis cells were centrifuged and resuspended in fresh BG-11 medium.
Bioinformatics
A BLAST search was performed with the Synechococcus elongatus PCC 7942 psbA3 gene sequence using a genome database of cyanobacteria and other photosynthetic organisms listed in the National Center for Biotechnology Information database. The presence of a conserved 130E residue was used to discriminate the D1:2 form among other D1 protein sequences.
RNA Isolation and Real-Time Quantitative RT-PCR Analysis
Total RNA was extracted using TRIsure (Bioline) reagent at 65°C. The RNA was further purified by extraction with phenol:chloroform:isoamylalcohol (25:24:1) and precipitated by isopropanol. Traces of genomic DNA were removed with 1 unit of DNase (Ambion Turbo DNase kit). The first-strand complementary DNA was synthesized from 1 µg of purified RNA. Reverse transcription was performed with random hexamer primers and SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s protocol. Synthesized complementary DNA was diluted 5-fold and used as a template for quantitative RT-PCR.
To perform the flv4, sll0218, and flv2 gene transcript analysis, the following primers were designed with Primer3 Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi): 5′-GATCGCCGTTTTTACTTTGA-3′ and 5′-GGTTGCTGTATTGCCATAGG-3′ for flv2; 5′-CTGGCAGCTTGTTCATTTCT-3′ and 5′-ATGGGGACTCCTTACCAGAC-3′ for sll0218; and 5′-GCGATACTTTCGTGCTCAAT-3′and 5′-GCGACTCGTCCAGTTTGTAT-3′ for flv4. The primers were designed for generating a similar length (approximately 200 bp) of amplicons. The primers for the reference gene rnpB were the same as in Zhang et al. (2009). The real-time quantitative RT-PCR was carried out on an IQ5 system (Bio-Rad) at the optimized annealing temperature. The melting curve analysis was performed to ensure the specificity of the products. The efficiency of these reactions was estimated for each triplicate using the LineReg program (Ramakers et al., 2003), and an average of the three was used in the subsequent estimation of the expression levels. Relative changes in gene expression were calculated using the qbaseplus (Biogazelle) program. For all genes, expression was normalized to rnpB.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Absorption spectra of wild-type, ΔpsbA2, flv4-2/OE, and Δflv4 strains grown at standard GL conditions and at HL intensity of 500 µmol photons m−2 s−1.
Supplemental Figure S2. State transitions kinetics in wild-type, ΔpsbA2, flv4-2/OE, and Δflv4 strains.
Supplemental Figure S3. Transient increase of fluorescence after termination of illumination (F0 rise) in wild-type, ΔpsbA2, flv4-2/OE, and Δflv4 strains.
Supplemental Figure S4. Fluorescence induction curves from wild-type and Δflv4 strains.
Supplemental Figure S5. The 77K fluorescence emission spectra of wild-type, ΔpsbA2, flv4-2/OE, and Δflv4 strains excited at 440 nm.
Supplemental Figure S6. Phycobilin/chlorophyll a ratios from cultures cultivated at standard GL conditions and at HL intensity of 500 µmol photons m−2 s−1.
Supplemental Figure S7. Flash-induced chlorophyll fluorescence relaxation curves in darkness from wild-type, ΔpsbA2, flv4-2/OE, and Δflv4 strains, grown in HC conditions.
Supplemental Table S1. Fv/Fm and Y(II) measured from dark-acclimated wild-type, ΔpsbA2, flv4-2/OE, and Δflv4 mutants.
Acknowledgments
We thank Dr. Ghada Ajlani for the gift of PAL and CK mutants; Dr. Diana Kirilovsky for providing ΔOCP, ΔApcDF mutants, and OCP antibodies; Dr. Taina Tyystjärvi for the gift of PC antibodies; and Essi Ruohisto for providing excellent technical assistance.
Glossary
- FDP
flavodiiron protein
- ROS
reactive oxygen species
- PB
phycobilisome
- NPQ
nonphotochemical quenching
- LC
low CO2
- HL
high light
- PQ
plastoquinone
- HC
high CO2
- 500-HL
500 µmol photons m−2 s−1
- 1,500-HL
1,500 µmol photons m−2 s−1
- GL
growth light
- Car/Chl a
carotenoid/chlorophyll a ratio
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- Fv/Fm
maximum photochemical efficiency of PSII in the dark-adapted state
- Y(II)
quantum yield of PSII
- DMBQ
2,6-dimethyl-p-benzoquinone
- FmD
maximal fluorescence after initial dark incubation
- Fm′
maximum PSII fluorescence in the light-adapted state
- F0 rise
the transient increase of fluorescence after termination of actinic light
- F0
fluorescence after termination of actinic light
- Fv
variable PSII fluorescence in the dark-adapted state
- Fq
Fm′ − Fs
- Fs
steady state fluorescence level in the light-adapted state
- QA−
negatively charged primary electron-accepting plastoquinone of PSII
- OCP
orange carotenoid protein
- DBMIB
2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone
- DCBQ
2,6-dichloro-p-benzoquinone
- RT-PCR
reverse transcription PCR
References
- Adir N, Zer H, Shochat S, Ohad I. (2003) Photoinhibition - a historical perspective. Photosynth Res 76: 343–370 [DOI] [PubMed] [Google Scholar]
- Ajlani G, Vernotte C. (1998) Construction and characterization of a phycobiliprotein-less mutant of Synechocystis sp. PCC 6803. Plant Mol Biol 37: 577–580 [DOI] [PubMed] [Google Scholar]
- Allahverdiyeva Y, Ermakova M, Eisenhut M, Zhang P, Richaud P, Hagemann M, Cournac L, Aro EM. (2011) Interplay between flavodiiron proteins and photorespiration in Synechocystis sp. PCC 6803. J Biol Chem 286: 24007–24014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdiyeva Y, Mustila H, Ermakova M, Bersanini L, Richaud P, Ajlani G, Battchikova N, Cournac L, Aro EM. (2013) Flavodiiron proteins Flv1 and Flv3 enable cyanobacterial growth and photosynthesis under fluctuating light. Proc Natl Acad Sci USA 110: 4111–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MM. (1968) Simple conditions for growth of unicellular blue-green algae on plates. J Phycol 4: 1–4 [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B. (1993) Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Ashby MK, Mullineaux CW. (1999) The role of ApcD and ApcF in energy transfer from phycobilisomes to PSI and PSII in a cyanobacterium. Photosynth Res 61: 169–179 [Google Scholar]
- Badger MR, Price GD. (2003) CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot 54: 609–622 [DOI] [PubMed] [Google Scholar]
- Bailey S, Grossman A. (2008) Photoprotection in cyanobacteria: regulation of light harvesting. Photochem Photobiol 84: 1410–1420 [DOI] [PubMed] [Google Scholar]
- Campbell D, Hurry V, Clarke AK, Gustafsson P, Oquist G. (1998a) Chlorophyll fluorescence analysis of cyanobacterial photosynthesis and acclimation. Microbiol Mol Biol Rev 62: 667–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D, Eriksson MJ, Oquist G, Gustafsson P, Clarke AK. (1998b) The cyanobacterium Synechococcus resists UV-B by exchanging photosystem II reaction-center D1 proteins. Proc Natl Acad Sci USA 95: 364–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AK, Hurry VM, Gustafsson P, Oquist G. (1993) Two functionally distinct forms of the photosystem II reaction-center protein D1 in the cyanobacterium Synechococcus sp. PCC 7942. Proc Natl Acad Sci USA 90: 11985–11989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cser K, Vass I. (2007) Radiative and non-radiative charge recombination pathways in Photosystem II studied by thermoluminescence and chlorophyll fluorescence in the cyanobacterium Synechocystis 6803. Biochim Biophys Acta 1767: 233–243 [DOI] [PubMed] [Google Scholar]
- Durrant JR, Giorgi LB, Barber J, Klug DR, Porter G. (1990) Characterization of triplet-states in isolated photosystem II reaction centres—oxygen quenching as a mechanism for photodamage. Biochim Biophys Acta Bioenerg 1017: 167–175 [Google Scholar]
- Eisenhut M, Georg J, Klähn S, Sakurai I, Mustila H, Zhang P, Hess WR, Aro EM. (2012) The antisense RNA As1_flv4 in the cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply. J Biol Chem 287: 33153–33162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bissati K, Delphin E, Murata N, Etienne AL, Kirilovsky D. (2000) Photosystem II fluorescence quenching in the cyanobacterium Synechocystis PCC 6803: involvement of two different mechanisms. Biochim Biophys Acta 1457: 229–242 [DOI] [PubMed] [Google Scholar]
- Ermakova M, Battchikova N, Allahverdiyeva Y, Aro EM. (2013) Novel heterocyst-specific flavodiiron proteins in Anabaena sp. PCC 7120. FEBS Lett 587: 82–87 [DOI] [PubMed] [Google Scholar]
- Frank HA, Cogdell RJ (1993) Photochemistry and function of carotenoids in photosynthesis. In A Young, G Britton, eds, Carotenoids in Photosynthesis, Chapman and Hall, London, pp 252–326 [Google Scholar]
- Fuhrmann E, Gathmann S, Rupprecht E, Golecki J, Schneider D. (2009) Thylakoid membrane reduction affects the photosystem stoichiometry in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 149: 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M, Beardall J, Raven JA. (2005) CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol 56: 99–131 [DOI] [PubMed] [Google Scholar]
- Graan T, Ort DR. (1986) Detection of oxygen-evolving photosystem II centers inactive in plastoquinone reduction. Biochim Biophys Acta Bioenerg 852: 320–330 [Google Scholar]
- Helman Y, Tchernov D, Reinhold L, Shibata M, Ogawa T, Schwarz R, Ohad I, Kaplan A. (2003) Genes encoding A-type flavoproteins are essential for photoreduction of O2 in cyanobacteria. Curr Biol 13: 230–235 [DOI] [PubMed] [Google Scholar]
- Hideg E, Barta C, Kálai T, Vass I, Hideg K, Asada K. (2002) Detection of singlet oxygen and superoxide with fluorescent sensors in leaves under stress by photoinhibition or UV radiation. Plant Cell Physiol 43: 1154–1164 [DOI] [PubMed] [Google Scholar]
- Hideg E, Spetea C, Vass I. (1994) Singlet oxygen production in thylakoid membranes during photoinhibition as detected by EPR spectroscopy. Photosynth Res 39: 191–199 [DOI] [PubMed] [Google Scholar]
- Hideg É, Ogawa K, Kálai T, Hideg K. (2001) Singlet oxygen imaging in Arabidopsis thaliana leaves under photoinhibition by excess photosynthetically active radiation. Physiol Plant 112: 10–14 [DOI] [PubMed] [Google Scholar]
- Hihara Y, Sonoike K, Kanehisa M, Ikeuchi M. (2003) DNA microarray analysis of redox-responsive genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 185: 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Ruban AV, Walters RG. (1996) Regulation of light harvesting in green plants. Annu Rev Plant Physiol Plant Mol Biol 47: 655–684 [DOI] [PubMed] [Google Scholar]
- Jallet D, Gwizdala M, Kirilovsky D. (2012) ApcD, ApcF and ApcE are not required for the orange carotenoid protein related phycobilisome fluorescence quenching in the cyanobacterium Synechocystis PCC 6803. Biochim Biophys Acta 1817: 1418–1427 [DOI] [PubMed] [Google Scholar]
- Kaplan A, Reinhold L. (1999) CO2-concentrating mechanisms in photosynthetic microorganisms. Annu Rev Plant Physiol Plant Mol Biol 50: 539–570 [DOI] [PubMed] [Google Scholar]
- Kós PB, Deák Z, Cheregi O, Vass I. (2008) Differential regulation of psbA and psbD gene expression, and the role of the different D1 protein copies in the cyanobacterium Thermosynechococcus elongatus BP-1. Biochim Biophys Acta 1777: 74–83 [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A. (2005) Singlet oxygen production in photosynthesis. J Exp Bot 56: 337–346 [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A, Fufezan C, Trebst A. (2008) Singlet oxygen production in photosystem II and related protection mechanism. Photosynth Res 98: 551–564 [DOI] [PubMed] [Google Scholar]
- Krupa Z, Oquist G, Gustafsson P. (1991) Photoinhibition of photosynthesis and growth responses at different light levels in psbA gene mutants of the cyanobacterium Synechococcus Plant Physiol 82: 1–8 [Google Scholar]
- Kulkarni RD, Golden SS. (1995) Form II of D1 is important during transition from standard to high light intensity in Synechococcus sp. strain PCC 7942. Photosynth Res 46: 435–443 [DOI] [PubMed] [Google Scholar]
- Lagarde D, Beuf L, Vermaas W. (2000) Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl Environ Microbiol 66: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AN, Telfer A, Truscott TG, Barber J. (1993) Direct detection of singlet oxygen from isolated photosystem II reaction centres. Biochim Biophys Acta Bioenerg 1143: 301–309 [Google Scholar]
- Mi H, Endo T, Ogawa T, Asada K. (1995) Thylakoid membrane-bound, NADPH-specific pyridine-nucleotide dehydrogenase complex mediates cyclic electron-transport in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 36: 661–668 [Google Scholar]
- Mohamed A, Eriksson J, Osiewacz HD, Jansson C. (1993) Differential expression of the psbA genes in the cyanobacterium Synechocystis 6803. Mol Gen Genet 238: 161–168 [DOI] [PubMed] [Google Scholar]
- Müller P, Li XP, Niyogi KK. (2001) Non-photochemical quenching. A response to excess light energy. Plant Physiol 125: 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux CW, Emlyn-Jones D. (2005) State transitions: an example of acclimation to low-light stress. J Exp Bot 56: 389–393 [DOI] [PubMed] [Google Scholar]
- Mulo P, Sicora C, Aro EM. (2009) Cyanobacterial psbA gene family: optimization of oxygenic photosynthesis. Cell Mol Life Sci 66: 3697–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon PJ, Barker M, Boehm M, de Vries R, Komenda J. (2005) FtsH-mediated repair of the photosystem II complex in response to light stress. J Exp Bot 56: 357–363 [DOI] [PubMed] [Google Scholar]
- Prasil O, Adir N, Ohad I (1992) Dynamics of photosystem II: mechanism of photoinhibition and recovery process. In J Barber , ed, Topics in Photosynthesis, The Photosystems: Structure, Function and Molecular Biology, Elsevier, Amsterdam, pp 220–250 [Google Scholar]
- Price GD, Badger MR, Woodger FJ, Long BM. (2008) Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59: 1441–1461 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rehman AU, Cser K, Sass L, Vass I. (2013) Characterization of singlet oxygen production and its involvement in photodamage of photosystem II in the cyanobacterium Synechocystis PCC 6803 by histidine-mediated chemical trapping. Biochim Biophys Acta 1827: 689–698 [DOI] [PubMed] [Google Scholar]
- Sander J, Nowaczyk M, Buchta J, Dau H, Vass I, Deák Z, Dorogi M, Iwai M, Rögner M. (2010) Functional characterization and quantification of the alternative PsbA copies in Thermosynechococcus elongatus and their role in photoprotection. J Biol Chem 285: 29851–29856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber U, Endo T, Mi H, Asada K. (1995) Quenching analysis of chlorophyll fluorescence by the saturation pulse method: particular aspects relating to the study of eukaryotic algae and cyanobacteria. Plant Cell Physiol 36: 873–882 [Google Scholar]
- Summerfield TC, Nagarajan S, Sherman LA. (2011) Gene expression under low-oxygen conditions in the cyanobacterium Synechocystis sp. PCC 6803 demonstrates Hik31-dependent and -independent responses. Microbiology 157: 301–312 [DOI] [PubMed] [Google Scholar]
- Svensson B, Vass I, Styring S. (1991) Sequence analysis of the D1 and D2 reaction center proteins of photosystem II. Z Naturforsch C 46: 765–776 [DOI] [PubMed] [Google Scholar]
- Takishita K, Uchida A. (1999) Molecular cloning and nucleotide sequence analysis of psbA from the dinoflagellates: origin of the dinoflagellate plastid. Phycol Res. 47: 207–216 [Google Scholar]
- Telfer A, Bishop SM, Phillips D, Barber J. (1994) Isolated photosynthetic reaction center of photosystem II as a sensitizer for the formation of singlet oxygen. Detection and quantum yield determination using a chemical trapping technique. J Biol Chem 269: 13244–13253 [PubMed] [Google Scholar]
- Thomas JC, Ughy B, Lagoutte B, Ajlani G. (2006) A second isoform of the ferredoxin:NADP oxidoreductase generated by an in-frame initiation of translation. Proc Natl Acad Sci USA 103: 18368–18373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, van Stokkum IH, Koehorst RB, Jongerius A, Kirilovsky D, van Amerongen H. (2011) Site, rate, and mechanism of photoprotective quenching in cyanobacteria. J Am Chem Soc 133: 18304–18311 [DOI] [PubMed] [Google Scholar]
- Tichý M, Lupínková L, Sicora C, Vass I, Kuviková S, Prásil O, Komenda J. (2003) Synechocystis 6803 mutants expressing distinct forms of the photosystem II D1 protein from Synechococcus 7942: relationship between the psbA coding region and sensitivity to visible and UV-B radiation. Biochim Biophys Acta 1605: 55–66 [DOI] [PubMed] [Google Scholar]
- Trebst A. (1980) Inhibitors in electron flow: tools for the functional and structural localization of carriers and energy conservation sites. Methods Enzymol 69: 675–715 [Google Scholar]
- Trebst A, Depka B, Holländer-Czytko H. (2002) A specific role for tocopherol and of chemical singlet oxygen quenchers in the maintenance of photosystem II structure and function in Chlamydomonas reinhardtii. FEBS Lett 516: 156–160 [DOI] [PubMed] [Google Scholar]
- Tyystjärvi E. (2008) Photoinhibition of photosystem II and photodamage of the oxygen evolving manganese cluster. Coord Chem Rev 252: 361–376 [Google Scholar]
- van Thor JJ, Mullineaux CW, Matthijs HCP, Hellingwerf KJ. (1998) Light-harvesting and state transitions in cyanobacteria. Bot Acta 111: 430–443 [Google Scholar]
- Vass I, Aro EM (2007) Photoinhibition of photosynthetic electron transport. In G Renger , ed, Primary Processes in Photosynthesis, Comprehensive Series in Photochemical and Photobiological Sciences, RSC Publishing, The Royal Society of Chemistry, Cambridge, UK, pp 393–425 part 1 [Google Scholar]
- Vass I, Kirilovsky D, Etienne A-L. (1999) UV-B radiation-induced donor- and acceptor-side modifications of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry 38: 12786–12794 [DOI] [PubMed] [Google Scholar]
- Vass I, Styring S, Hundal T, Koivuniemi A, Aro EM, Andersson B. (1992) Reversible and irreversible intermediates during photoinhibition of photosystem II: Stable reduced QA species promote chlorophyll triplet formation. Proc Natl Acad Sci USA 89: 1408–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente JB, Carrondo MA, Teixeira M, Frazão C. (2008a) Structural studies on flavodiiron proteins. Methods Enzymol 437: 3–19 [DOI] [PubMed] [Google Scholar]
- Vicente JB, Justino MC, Gonçalves VL, Saraiva LM, Teixeira M. (2008b) Biochemical, spectroscopic, and thermodynamic properties of flavodiiron proteins. Methods Enzymol 437: 21–45 [DOI] [PubMed] [Google Scholar]
- Vicente JB, Testa F, Mastronicola D, Forte E, Sarti P, Teixeira M, Giuffrè A. (2009) Redox properties of the oxygen-detoxifying flavodiiron protein from the human parasite Giardia intestinalis. Arch Biochem Biophys 488: 9–13 [DOI] [PubMed] [Google Scholar]
- Wasserfallen A, Ragettli S, Jouanneau Y, Leisinger T. (1998) A family of flavoproteins in the domains Archaea and Bacteria. Eur J Biochem 254: 325–332 [DOI] [PubMed] [Google Scholar]
- Williams JKG. (1988) Construction of specific mutations in PSII photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol 167: 766–778 [Google Scholar]
- Wilson A, Ajlani G, Verbavatz JM, Vass I, Kerfeld CA, Kirilovsky D. (2006) A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell 18: 992–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winhauer T, Jäger S, Valentin K, Zetsche K. (1991) Structural similarities between psbA genes from red and brown algae. Curr Genet 20: 177–180 [DOI] [PubMed] [Google Scholar]
- Zhang P, Allahverdiyeva Y, Eisenhut M, Aro EM. (2009) Flavodiiron proteins in oxygenic photosynthetic organisms: photoprotection of photosystem II by Flv2 and Flv4 in Synechocystis sp. PCC 6803. PLoS ONE 4: e5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Eisenhut M, Brandt AM, Carmel D, Silén HM, Vass I, Allahverdiyeva Y, Salminen TA, Aro EM. (2012) Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell 24: 1952–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]