The plasma membrane-localized carbon/nitrogen nutrient regulator ATL31 positively controls papilla formation in response to powdery mildew fungi by interacting with a vesicle-trafficking protein.
Abstract
The carbon/nitrogen (C/N) balance of plants is not only required for growth and development but also plays an important role in basal immunity. However, the mechanisms that link C/N regulation and basal immunity are poorly understood. We previously demonstrated that the Arabidopsis (Arabidopsis thaliana) Arabidopsis Tóxicos en Levadura31 (ATL31) ubiquitin ligase, a regulator of the C/N response, positively regulates the defense response against bacterial pathogens. In this study, we identified the plasma membrane-localized soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor SYNTAXIN OF PLANTS121 (SYP121) as a novel ATL31 interactor. The syp121-1 loss-of-function mutant showed similar hypersensitivity to C/N stress conditions as the atl31 atl6 double mutant. SYP121 is essential for resistance to penetration by powdery mildew fungus and positively regulates the formation of cell wall appositions (papillae) at fungal entry sites. Microscopic analysis demonstrated that ATL31 was specifically localized around papillae. In addition, ATL31 overexpressors showed accelerated papilla formation, enhancing their resistance to penetration by powdery mildew fungus. Together, these data indicate that ATL31 plays an important role in connecting the C/N response with basal immunity by promoting papilla formation through its association with SYP121.
Carbon (or carbohydrates) and nitrogen nutrients are crucial for plant cellular functions, including plant immunity (Bolton, 2009; Massad et al., 2012). In addition to being a source of carbohydrates, carbon skeletons, and energy, soluble sugars contribute to the immune response as priming molecules (Bolouri Moghaddam and Van den Ende, 2012). For example, Suc treatment transcriptionally induces pathogenesis-related genes (Thibaud et al., 2004). In contrast to sugars, plant nitrogen supply, as nitrate and/or ammonium ions, generally decreases defense responses. For example, the high availability of nitrogen sources significantly increased the susceptibility of potato (Solanum tuberosum) plants to the oomycete Phytophthora infestans (Ros et al., 2008). In addition to the individual importance of carbon and nitrogen, the carbon/nitrogen (C/N) ratio is critical for plant adaptation to environmental conditions (Coruzzi and Bush, 2001). However, the mechanisms by which C/N contribute to plant immune responses have not been determined.
The Arabidopsis Tóxicos en Levadura (ATL) gene family encodes 91 plant-specific putative RING-type ubiquitin ligases with a transmembrane domain (Aguilar-Hernández et al., 2011). ATL31 and its closest homolog ATL6 are membrane-associated ubiquitin ligases that are involved in the C/N response by regulating the stability of 14-3-3 proteins through ubiquitination (Sato et al., 2009, 2011). Plants overexpressing full-length ATL31 or ATL6 (35S-ATL31 and 35S-ATL6), under the control of the constitutive cauliflower mosaic virus 35S promoter, were found to be insensitive to high C/N stress conditions. In contrast, single knockout mutants of atl31-1 and atl6-1, as well as an atl31-1 atl6-1 double knockout, showed increased sensitivity to high C/N stress (Sato et al., 2009). We recently found that both of these C/N response regulators are involved in the plant immune response (Maekawa et al., 2012). Overexpression of 35S-ATL31 and 35S-ATL6 showed enhanced callose deposition in response to Flg22, the main component of bacterial flagella (Felix et al., 1999), as well as enhanced resistance to the bacterial pathogen Pseudomonas syringae pv tomato (Pst) DC3000 (Katagiri et al., 2002). In contrast, only the double mutant, and not the single mutants, caused increased susceptibility to Pst DC3000 (Maekawa et al., 2012).
Powdery mildews are among the most important diseases of food and ornamental plants, with a high annual global economic impact (for review, see Glawe, 2008; Micali et al., 2008). These fungal species rely on living host plant tissues for survival, with many of these fungal species infecting a very narrow range of plant species. For example, the barley (Hordeum vulgare) powdery mildew fungus Blumeria graminis f. sp. hordei (Bgh) only infects barley and its close relatives. In contrast to barley, Arabidopsis (Arabidopsis thaliana) can resist Bgh penetration by the activation of basal immunity (Tucker and Talbot, 2001).
Powdery mildew fungus infection is initiated by the germination of conidiospores on the plant leaf surface, followed by the formation of structures called appressoria, from which develop infection hyphae called penetration pegs. The hyphae penetrate host epidermal cell walls, giving rise to infection-induced dome-shaped extensions of the inner surface of the wall, called papillae. The tips of the infecting hyphae then expand to form feeding structures called haustoria, which invaginate into but do not penetrate the host plasma membrane (Ellis, 2006; Glawe, 2008). A common response by plants to fungal attack is the formation of papillae, which include callose, phenolics, reactive oxygen species, and antimicrobial compounds, all of which act as physical and chemical barriers to slow pathogen invasion (Ellis, 2006). This early defense response provides the host plant time to initiate subsequent defense reactions, including the production of reactive oxygen species and antibacterial substances, such as phenolic compounds and phytoalexins, the activation of defense-related genes, and the export of pathogenesis-related proteins (Senthil-Kumar and Mysore, 2013).
Genetic screening for Arabidopsis mutants with increased penetration by Bgh resulted in the identification of the PENETRATION1 (PEN1) gene (Collins et al., 2003). A pen1-1 loss-of-function mutant caused delayed formation of papillae, resulting in increased Bgh infection (Assaad et al., 2004). PEN1 encodes the plasma membrane-localized SNARE SYNTAXIN OF PLANTS121 (SYP121; Collins et al., 2003). The fusion of secretory vesicles to the plasma membrane is mediated by specific binding between donor membrane-associated R-SNAREs and plasma membrane-associated Q-SNAREs. The Q-SNAREs are divided into three subgroups, Qa-, Qb-, and Qc-SNAREs, based on sequence similarities. A SNARE fusion complex consists of an R-SNARE motif and a Qa-, Qb-, and Qc-ternary complex or a Qa and Qb+Qc binary complex (Saito and Ueda, 2009). SYP121 (Qa-SNARE), SOLIBLE N-ETHYLMALEIMIDE-SENSITIVE FACTOR ADAPTOR PROTEIN33 (SNAP33; Qb+Qc), and VESICLE-ASSOCIATED MEMBRANE PROTEIN721 (VAMP721)/VAMP722 (R-SNARE) form the plasma membrane-localized SNARE complex, which is required for timely papilla formation at the fungal entry site (Assaad et al., 2004; Kwon et al., 2008).
Based on their physiological and transcriptional relationship, we show that ATL31 and SYP121 interact in vivo. We also demonstrate that the syp121 loss-of-function mutant shows hypersensitivity to C/N stress conditions. In addition, our analyses of responses to powdery mildew fungus revealed that ATL31 accumulates at fungal penetration sites and regulates papilla formation, thus contributing to resistance to fungal penetration. Taken together, these findings indicate that the C/N regulatory factor ATL31 positively controls basal immunity by accelerating callose deposition via its association with SYP121.
RESULTS
ATL31 Interacts with SYP121 in Vivo
We previously reported that ATL31 regulates the plant C/N response and plant immunity (Sato et al., 2009; Maekawa et al., 2012). To gain further insight into the function of ATL31, we performed coexpression analysis using Arabidopsis Coexpression Data Mining Tools (Manfield et al., 2006), with ATL31 and ATL6, the closest homolog of ATL31 (Sato et al., 2009), as query genes (Supplemental Fig. S1). Assessment of the SNARE complex consisting of SYP121, SNAP33, and VAMP721/VAMP722 showed that transcription of the genes encoding all of these proteins, except for VAMP721, was highly positively correlated with the transcription of ATL31 and ATL6. The SYP121-SNAP33-VAMP721/VAMP722 complex is involved in resistance to penetration by the nonadapted powdery mildew fungus Bgh, and loss of plasma membrane-localized SYP121 function significantly increases the Bgh penetration rate (Collins et al., 2003; Kwon et al., 2008). Since we previously found that plasma membrane-localized ATL31 played a role in basal immunity against bacterial pathogens (Maekawa et al., 2012), we examined the effects of the relationship between ATL31 and SYP121 on basal immunity against fungal pathogens. The physical interaction between ATL31 and SYP121 was examined by coimmunoprecipitation. We took advantage of a point mutation (C143S) in the RING domain of ATL31, a mutation that results in a loss of ubiquitination activity (Sato et al., 2009), as this mutant is presumably more stable than intact ATL31. FLAG-tagged ATL31C143S and GFP-tagged SYP121 or its close homolog SYP122 were transiently expressed in Nicotiana benthamiana leaves by the syringe agroinfiltration method (Kapila et al., 1997; D’Aoust et al., 2009). The crude extracts were subjected to immunoprecipitation using the anti-FLAG M2 affinity gel. Immunoblot analysis with an anti-GFP antibody demonstrated that SYP121, but not SYP122, coimmunoprecipitated with ATL31C143S (Fig. 1A). Furthermore, the interaction between ATL31 and SYP121 was confirmed using anti-SYP121 to immunoprecipitate the complex from stable 35S-ATL31C143S-FLAG transgenic MM2d cells (Fig. 1B). These findings indicated that ATL31 specifically interacts with SYP121 in vivo. The interaction between ATL6 and SYP121 was also confirmed by immunoprecipitating the complex from stable 35S-ATL6-FLAG transgenic MM2d cells (Fig. 1C).
Figure 1.
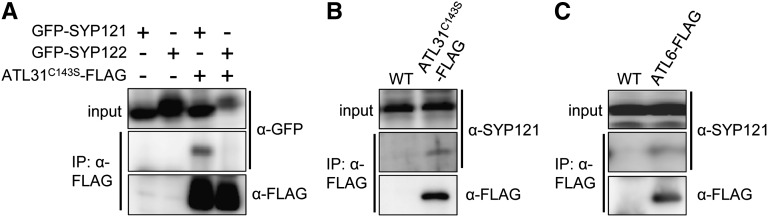
In vivo interaction of ATL31 and ATL6 with SYP121 or SYP122 by coimmunoprecipitation analyses. A, ATL31C143S-FLAG was transiently expressed with GFP-SYP121 or GFP-SYP122 in N. benthamiana leaves by the syringe agroinfiltration method. Crude extracts were immunoprecipitated (IP) with anti-FLAG M2 affinity gel, followed by immunoblot analysis with anti-FLAG and anti-GFP antibodies. B and C, Crude extracts from MM2d cells for wild-type (WT) and 35S-ATL31C143S-FLAG (B) and 35S-ATL6-FLAG (C) were immunoprecipitated on anti-FLAG M2 affinity gel, followed by immunoblot analysis with anti-SYP121 and anti-FLAG antibodies.
SYP121 Is Essential for the C/N Stress Response
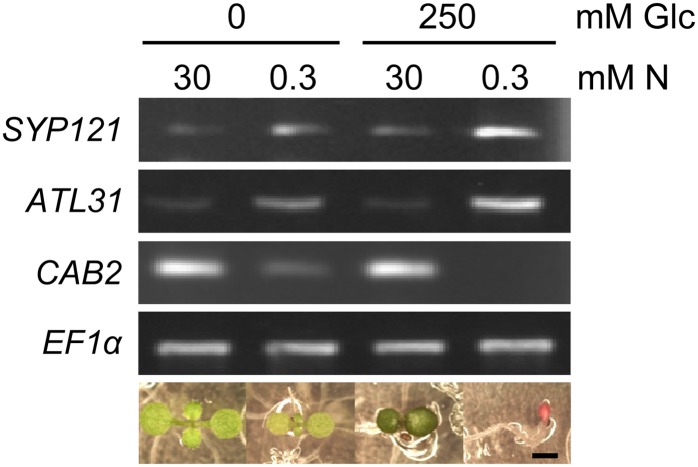
The identification of SYP121 as interacting with the C/N regulatory factor ATL31 prompted us to analyze the role of SYP121 in the C/N response. First, we examined the expression of SYP121 under various C/N stress conditions. RNA was extracted from wild-type seedlings grown in C/N-modified medium, and SYP121 expression was assessed by reverse transcription (RT)-PCR. SYP121 transcripts were abundant in plants grown under high-carbon (250 mm Glc) and low-nitrogen (0.3 mm nitrogen) conditions but were down-regulated in plants grown under high-nitrogen (30 mm nitrogen) conditions (Fig. 2). The pattern of expression of ATL31 was similar to that of SYP121, consistent with results showing an expressional correlation (Supplemental Fig. S1).
Figure 2.
Expression pattern of SYP121 under various C/N conditions. Total RNA was extracted from whole 7-d-old wild-type plants grown in various modified C/N conditions as indicated. Standard RT-PCR was performed. Expression of the CHLOROPHYLL a/b BINDING PROTEIN2 (CAB2), which was down-regulated by the high C/N stress condition (Martin et al., 2002), was assayed, with TRANSLATION ELONGATION FACTOR1α (EF1α) mRNA used as an internal control. The bottom gels show representative 7-d-old wild-type phenotypes under each C/N condition. N, Nitrogen. Bar = 1 mm.
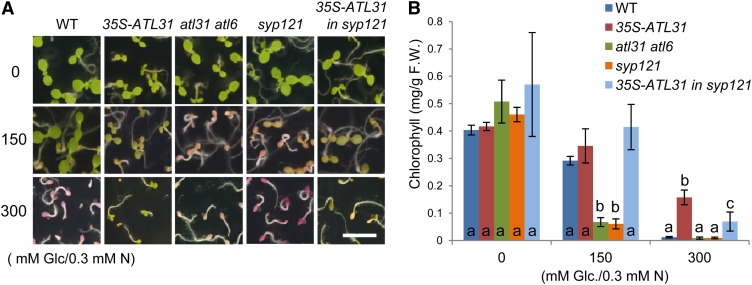
Next, we evaluated SYP121 function in the C/N stress response by growing seedlings in medium containing no Glc, 150 mm Glc (moderate C/N stress), or 300 mm Glc (high C/N stress) with low amounts (0.3 mm) of nitrogen. In the absence of exogenous Glc, all plants showed green expanded cotyledons (Fig. 3A) and contained similar amounts of total chlorophyll (Fig. 3B). As reported previously (Sato et al., 2009), the atl31-1 atl6-1 double mutant exhibited a hypersensitive phenotype under moderate C/N stress conditions (150 mm Glc and 0.3 mm nitrogen), such as growth arrest with red pigment accumulation (Fig. 3A) and low chlorophyll amounts (Fig. 3B). Interestingly, the syp121-1 mutant also had a hypersensitive phenotype, similar to that of the atl31-1 atl6-1 double mutant, under moderate C/N conditions (150 mm Glc and 0.3 mm nitrogen; Fig. 3). Plants expressing 35S-ATL31 were insensitive to high C/N stress (300 mm Glc and 0.3 mm nitrogen; Fig. 3), as reported previously (Sato et al., 2009). Introduction of 35S-ATL31 to the syp121-1 mutant reduced the hypersensitivity of the latter to high C/N stress, although not to the level of resistance observed in 35S-ATL31 alone (Fig. 3). When the C/N stress assay was repeated using medium with moderate concentrations of Glc (150 mm) and various concentrations of nitrogen (0.1, 0.3, and 1 mm), transgenic plants and mutants showed response patterns similar to those observed with various Glc concentrations and low-nitrogen conditions (Supplemental Fig. S2). These results indicate that SYP121 is an essential component of the C/N response, as are ATL31 and ATL6.
Figure 3.
Postgerminative growth phenotype under different C/N conditions. Postgerminative growth phenotypes are shown for the wild type (WT), 35S-ATL31, atl31-1 atl6-1, syp121-1, and 35S-ATL31 in syp121-1 germinated on medium containing different concentrations of Glc (0, 150, and 300 mm) with 0.3 mm nitrogen (N). A, Photographs taken 7 d after germination. Bar = 0.5 cm. B, Total chlorophyll amounts in plants 7 d after germination. Error bars represent sd (n = 3). Statistically significant differences were determined by ANOVA, followed by a post hoc Tukey test. Means that differed significantly (P < 0.05) are indicated by different letters. F.W., Fresh weight.
ATL31 Accumulates at Bgh Entry Sites
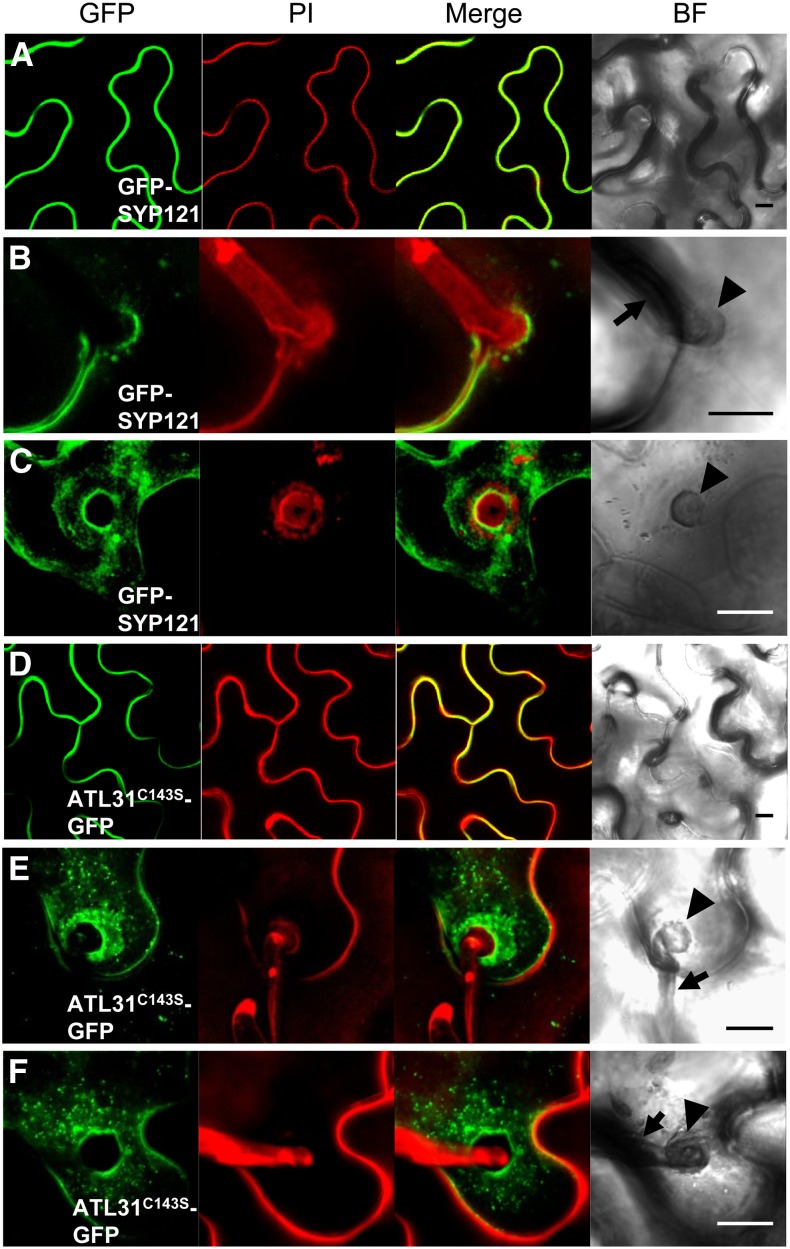
Since SYP121 plays a crucial role in resistance to penetration by nonadapted powdery mildew fungi (Collins et al., 2003; Assaad et al., 2004) and ATL31 is transcriptionally up-regulated by treatment with chitin (Maekawa et al., 2012), a carbohydrate present in the cell walls of fungi (Shibuya and Minami, 2001), we analyzed whether ATL31 is involved in the response to infection by nonadapted powdery mildew fungi. SYP121 has been shown to accumulate in fungi-induced papilla (Assaad et al., 2004; Meyer et al., 2009). To test whether ATL31 also accumulates at fungal entry sites, we analyzed its subcellular location. We were unable to detect ATL31-GFP fluorescence in Arabidopsis leaves, either in stable transgenic plants or plants with transient expression, even when ATL31 expression was driven by the 35S promoter. We then used the syringe agroinfiltration method to transiently express ATL31C143S in N. benthamiana leaves. To verify that these leaves were a suitable model system for testing changes in Bgh-induced localization, we first assessed GFP-SYP121 fluorescence in these leaves in the absence of Bgh infection, finding GFP-SYP121 fluorescence 3 d after agroinfiltration at the plasma membrane (Fig. 4A), as described previously (Uemura et al., 2004; Enami et al., 2009). N. benthamiana leaves were subsequently infected with Bgh 24 h after agroinfiltration; 40 to 50 h later, fluorescence resembling a cloud and apparently associated with papillae was observed (Fig. 4, B and C). This GFP-SYP121 localization pattern was in agreement with results from a previous study of Arabidopsis leaves (Assaad et al., 2004). However, in our system, fluorescence was restricted to the margin of the papillae (Fig. 4, B and C), whereas in Arabidopsis, GFP-SYP121 accumulated inside the papillae (Assaad et al., 2004; Meyer et al., 2009; Nielsen et al., 2012). This difference may be due to the differences in exocytosis components between N. benthamiana and Arabidopsis; however, SYP121 accumulation at Bgh entry sites was apparent in N. benthamiana.
Figure 4.
ATL31C143S localization at Bgh infection sites. ATL31C143S-GFP and GFP-SYP121 were transiently expressed in N. benthamiana leaves by the syringe agroinfiltration method, followed 24 h later by infection with Bgh. ATL31C143S-GFP (A–C) and GFP-SYP121 (D–F) fluorescence is shown 40 to 50 h after infection without (A and D) and with (B, C, E, and F) Bgh. To show the fungal structure and plant cell walls, detached leaves were stained with propidium iodide (PI). Three-dimensional reconstructions of image stacks are shown. Arrowheads and arrows indicate Bgh-induced papillae and appressorial germ tubes, respectively, in the bright field (BF). Bars = 10 μm.
Our ability to successfully detect GFP-SYP121 in N. benthamiana leaves upon agroinfiltration led us to assess ATL31C143S-GFP localization. In the absence of Bgh infection, ATL31C143S-GFP was observed in the cell periphery of leaf epidermal cells (Fig. 4D). After Bgh infection, ATL31C143S-GFP accumulated around papillae and in vesicle-like structures near the papillae, similar to the localization of GFP-SYP121 (Fig. 4, E and F).
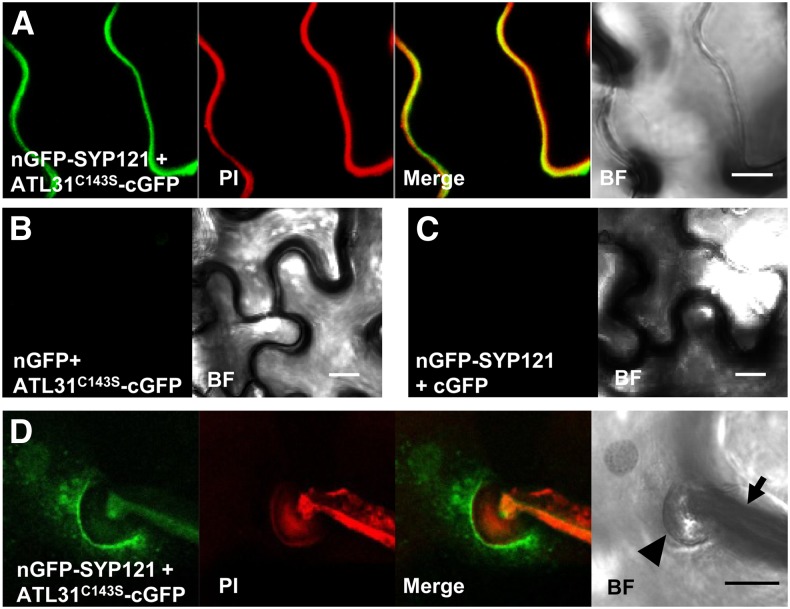
We analyzed the in vivo interaction between ATL31 and SYP121 using the bimolecular fluorescence complementation (BiFC) method (Walter et al., 2004). ATL31C143S and SYP121 fused to the C- and N-terminal halves of GFP (cGFP and nGFP), respectively, were transiently expressed in N. benthamiana leaves using the syringe agroinfiltration method. We observed a GFP signal at the cell periphery upon dual expression of ATL31C143S-cGFP and nGFP-SYP121 (Fig. 5A) but not upon the expression of ATL31C143S-cGFP and nGFP (Fig. 5B) or nGFP-SYP121 and cGFP (Fig. 5C). Although BiFC fluorescence intensity was relatively low in Bgh-infected cells, and we could not completely exclude the autofluorescence of the Bgh structure in the GFP field, GFP fluorescence was observed in the plasma membrane around the papillae with vesicle-like structures (Fig. 5D). Taken together, these results suggest that ATL31 and SYP121 are recruited to and interact at the site of Bgh entry.
Figure 5.
BiFC analysis of ATL31 with SYP121 at the Bgh infection sites. A, Fluorescence was observed in the cell periphery of N. benthamiana leaf epidermal cells, resulting from complementation of the N-terminal part of GFP fused with SYP121 (nGFP-SYP121) and the C-terminal part of GFP fused with ATL31C143S (ATL31C143S-cGFP). B and C, No fluorescence was observed when ATL31C143S-cGFP was coexpressed with nGFP (B) or when nGFP-SYP121 was coexpressed with cGFP (C). D, N. benthamiana leaves were infected with Bgh 24 h after agroinfiltration of ATL31C143S-cGFP and nGFP-SYP121, with BiFC fluorescence assessed 50 h after Bgh infection. To show fungal structures and plant cell walls, detached leaves were stained with propidium iodide (PI). Three-dimensional reconstructions of image stacks are shown. The arrowhead and arrow indicate Bgh-induced papillae and appressorial germ tubes, respectively, in the bright field (BF). Bars = 10 μm.
ATL31 Positively Regulates Resistance to Penetration by the Nonadapted Powdery Mildew Fungus
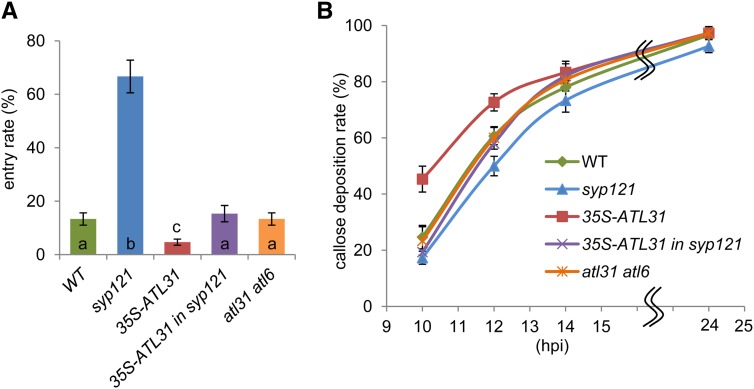
To further investigate the function of ATL31 in response to fungal penetration, we analyzed the Bgh entry rate at 48 h post inoculation (hpi) in wild-type Arabidopsis, plants overexpressing ATL31 (35S-ATL31), syp121-1, syp121-1 mutant overexpressing ATL31 (35S-ATL31 in syp121), and the atl31-1 atl6-1 double mutant. The Bgh entry rate was higher in syp121-1 than in wild-type plants, as reported previously (Collins et al., 2003), but was significantly lower in 35S-ATL31 than in wild-type plants (Fig. 6A). The fungal entry rates into 35S-ATL31 in syp121 and the atl31-1 atl6-1 double mutant were similar to that of the wild type (Fig. 6A).
Figure 6.
Resistance to penetration by Bgh. A, Frequency of Bgh entry. Percentage Bgh penetration was analyzed at 48 hpi in wild-type (WT), syp121-1, 35S-ATL31, 35S-ATL31 in syp121-1, and atl31-1 atl6-1 plants. For each leaf, a minimum of 50 germinated spores were scored. Error bars represent sd (n = 3). Statistical significance was determined by ANOVA, followed by a post hoc Tukey test. Means that differed significantly (P < 0.05) are indicated by different letters. B, Frequency of callose accumulation at Bgh attack sites. Percentage callose accumulation at Bgh attack sites in wild-type, syp121-1, 35S-ATL31, 35S-ATL31 in syp121-1, and atl31-1 atl6-1 plants was analyzed at the indicated time points. For each leaf, a minimum of 50 germinated spores were scored. Error bars represent sd (n = 5).
Since the increased Bgh entry rate in syp121 correlates with delayed papilla formation in this mutant (Assaad et al., 2004), we investigated whether the timing of papilla formation at the Bgh entry site was altered in ATL-related transgenic plants and mutants. We determined the percentage of germinated Bgh conidia with papillary callose focal deposition at entry sites. At 24 hpi, all mutants and transgenic plants showed similar rates of callose deposition (Fig. 6B). However, a more detailed time-course analysis revealed a delay in callose deposition in the syp121-1 mutant at approximately 12 hpi (Fig. 6B), as reported previously (Assaad et al., 2004). In contrast, and consistent with Bgh entry rate results (Fig. 6A), ATL31 overexpressors showed accelerated callose deposition compared with wild-type plants at 10 to 12 hpi (Fig. 6B). The callose accumulation rates of plants expressing the atl31-1 atl6-1 double mutant or the syp121-1 mutant overexpressing ATL31 were similar to that of wild-type plants at all time points (Fig. 6B). Together, these data show that ATL31 positively regulates papilla formation at the fungal entry site, thereby enhancing resistance to penetration by nonadapted fungi.
DISCUSSION
In this study, SYP121 was identified as a novel ATL31 interactor. The syp121-1 mutant showed hypersensitivity to C/N stress conditions, similar to that of the atl31-1 atl6-1 double mutant. After fungal infection, ATL31 accumulated at Bgh penetration sites and interacted with SYP121. Plants overexpressing ATL31 showed enhanced resistance to penetration, with accelerated papilla formation, in response to Bgh invasion. These results strongly indicate that ATL31, together with SYP121, regulates fungal penetration resistance and the C/N response.
It is important to understand the significance of the interaction between ATL31 and SYP121. For example, ATL31 localization may be regulated by SYP121. Indeed, the post-Golgi traffic of two plasma membrane-localized proteins, potassium channel KAT1 (Eisenach et al., 2012) and maize (Zea mays) PLASMA MEMBRANE INTRINSIC PROTEIN2;5 (Besserer et al., 2012), is regulated by SYP121. However, the expression of ATL31C143S-GFP in syp121-1 mesophyll protoplasts resulted in a level of GFP fluorescence in the plasma membrane similar to that in wild-type protoplasts (Supplemental Fig. S3). Although we could not exclude the possibility of SNARE function redundancy, these results suggest that SYP121 does not control the plasma membrane localization of ATL31. Another possibility is that SYP121 is the target of ATL31 ubiquitin ligase. ATL family proteins may bind to their targets at the C-terminal domain (Serrano et al., 2006), with 14-3-3χ, a ubiquitination target of ATL31, interacting with the C-terminal region of ATL31 (Sato et al., 2011). However, we found that C-terminal truncated ATL31 interacted with SYP121 (Supplemental Fig. S4). These results, together with the results of C/N stress and Bgh infection assays, suggest that ATL31 does not directly polyubiquitinate SYP121 for proteasomal degradation. However, ATL31 may ubiquitinate SYP121 to regulate membrane trafficking. Indeed, the plasma membrane-localized iron-regulated transporter1, which localizes to the trans-Golgi network compartment as a consequence of endocytosis, was found to mediate ubiquitination by ATL14/IRON-REGULATED TRANSPORTER1 DEGREDATION FACTOR1 (Barberon et al., 2011; Shin et al., 2013).
SNAREs other than SYP121 can associate with ATL31 and function in basal immunity and the C/N response. Since secretion into the papilla matrix can occur in syp121-1 with only a slight delay (Assaad et al., 2004), other syntaxins may mediate this secretion (Nielsen et al., 2012). Interestingly, the syp121-1 mutant overexpressing ATL31 showed an intermediate response between the ATL31 overexpressor and the syp121-1 mutant on C/N stress and Bgh infection assays. These results not only suggest that the interaction between ATL31 and SYP121 plays definitive roles in resistance to penetration and the C/N response but also that ATL31 interacts with other syntaxins. Indeed, SYP132, a plasma membrane-localized Qa-SNARE, was shown to partially complement the fungal entry rate of syp121-1 (Reichardt et al., 2011). Thus, ATL31 may interact with other plasma membrane-localized SNAREs and function in basal immunity and the C/N response.
ATL members other than ATL31 and ATL6 can also interact with SYP121 and function in basal immunity. Whereas the atl31-1 atl6-1 double knockout mutant became hypersensitive to C/N stress, it did not show any alterations in papilla formation, suggesting that SYP121 interacts with other ATL family members to function in the acceleration of papilla formation. The ATL family can be classified into 14 groups (groups A–N) based on a protein distance matrix (Serrano et al., 2006). Of the group G proteins, which include ATL31 and ATL6, endoplasmic reticulum-localized ATL9 has an important role in powdery mildew resistance. ATL9 expression was induced by chitin treatment, and an atl9 loss-of-function mutant showed enhanced susceptibility to Arabidopsis-adapted powdery mildew fungi (Ramonell et al., 2005; Berrocal-Lobo et al., 2010). Additionally, other ATL members have been reported to be defense-related genes (Guzmán, 2012) as well as being putative membrane fusion proteins (Serrano et al., 2006), and these ATL proteins may be involved in the defense response associated with SNARE proteins.
ATL31 may function in callose accumulation during early stages of the plant defense response. We previously reported that ATL31 and ATL6 positively regulate resistance to the bacterial pathogen Pst DC3000 (Maekawa et al., 2012). This study showed that ATL31 overexpressors displayed higher amounts of callose deposition than the wild type upon Flg22 treatment as well as accelerated callose deposition in response to Bgh. These findings, together with results showing an early induction of ATL31 transcripts by Flg22 and chitin (Maekawa et al., 2012), indicate that ATL31 may function in basal immunity by positively regulating callose deposition during early stages of pathogen infection.
ATL31 may play an important role in resistance to general powdery mildew fungi by regulating callose deposition. SYP121 has been shown to accumulate not only at entry sites of the barley powdery mildew fungus Bgh (Assaad et al., 2004) but at entry sites of the Arabidopsis-adapted fungi Golovinomyces cichoracearum and Golovinomyces orontii (Bhat et al., 2005; Meyer et al., 2009). SYP121 also contributes to resistance to penetration by powdery mildew fungi, regardless of whether these fungi are adapted or nonadapted to Arabidopsis (Collins et al., 2003; Zhang et al., 2007). Taken together with findings showing that early elevation of callose deposition enhances resistance to penetration by both Arabidopsis-adapted and nonadapted powdery mildew fungi (Ellinger et al., 2013), these findings suggest that ATL31, together with SYP121, may have a definitive role in resistance against general powdery mildew fungi.
SYP121 may be involved in the nutrient response by regulating plasma membrane-localized proteins. The finding that syp121-1 showed hypersensitivity to the C/N stress condition suggests that SYP121 may be involved in the nutrient response. SYP121 has been shown to regulate potassium uptake by regulating both the activity and localization of the plasma membrane-associated potassium channel KAT1 (Eisenach et al., 2012). Furthermore, the membrane trafficking system has been reported to contribute to the regulation of nutrient uptake in mammals. The GLUT4 Glc transporter, which plays a central role in whole-body Glc homeostasis, is regulated by the membrane trafficking system. Following nutrient uptake and the secretion of insulin, GLUT4 translocates from intracellular vesicles to the plasma membrane through exocytosis (Stöckli et al., 2011). Thus, SYP121 may regulate the localization of plasma membrane-localized C/N regulatory factors.
Carbon and nitrogen metabolites are regulated through the defense response to powdery mildew fungus. Barley leaves infected with the B. graminis isolate A6 exhibited increased levels of Suc, decreased starch levels, and increased intermediates of Suc biosynthesis compared with control plants (Molitor et al., 2011). Furthermore, glycolysis and the tricarboxylic acid cycle were diminished upon B. graminis infection, indicating altered flux into the anaplerotic tricarboxylic acid cycle that provides carbon skeletons for nitrogen assimilation into Gln. Consequently, the Gln-Glu ratio and the content of almost all amino acids, except Glu, were highly increased in leaves infected with B. graminis (Molitor et al., 2011). Although C/N metabolites were apparently altered by pathogen infection, it remains unclear whether these changes resulted from the plant defense against pathogens or the pathogen manipulates plant metabolism to retrieve more nutrients from the cell. Our finding, that the C/N regulatory factor ATL31 is involved in basal immunity, suggests that metabolomic analyses of ATL31 mutants in response to pathogens will provide insight into the relationship between C/N metabolism and defense responses.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type Columbia-0 and all other Arabidopsis (Arabidopsis thaliana) materials used in this study were grown as described (Morita-Yamamuro et al., 2005). Nicotiana benthamiana, which was used for transient expression analyses, was prepared as described (Igawa et al., 2009). Routine subculture and transformation of cultured suspensions of Arabidopsis MM2d cells were performed as described (Menges and Murray, 2002; Hirano et al., 2008). All Arabidopsis seeds and cultured cells related to ATL31 and ATL6 were obtained as described (Sato et al., 2009; Maekawa et al., 2012).
Molecular Cloning
Plasmids containing complementary DNA (cDNA) encoding mutated ATL31 (pENTRATL31C143S) and intact ATL6 (pENTRATL6) were prepared as described (Sato et al., 2009; Maekawa et al., 2012). cDNAs encoding truncated ATL31C143S, SYP121, and SYP122 were amplified by PCR using pENTRATL31C143S cDNA encoding SYP121 and SYP122 (Uemura et al., 2004; Sato et al., 2009) as templates and introduced into the pENTR/D-TOPO vector (Life Technologies) to generate the plasmids pENTRATL31C143S1-168, pENTRATL31C143S1-267, pENTRSYP1211-177, and pENTRSYP122. The primers used for PCR are listed in Supplemental Table S1. ATL31C143S was cloned into the pB4GWcG and pUGW5 transfer DNA (T-DNA) binary vectors; SYP121 was cloned into the pB4nGGW binary vector; ATL6, ATL31C143S1-168, and ATL31C143S1-267 were cloned into the pGWB11 T-DNA binary vector (Nakagawa et al., 2007); SYP121 and SYP122 were cloned into the pGWB6 T-DNA binary vector (Nakagawa et al., 2007); and SYP1211-177 was cloned into the pDEST-his T-DNA binary vector (Tsunoda et al., 2005), as described by the manufacturer (Life Technologies). All PCR products and inserts were verified by DNA sequencing.
Agrobacterium tumefaciens-Mediated Transient Gene Expression Method
Constructs were used to transform the A. tumefaciens strain GV3101::pMP90 by electroporation. A. tumefaciens cells were cultured, harvested by centrifugation, suspended in water and 150 μm acetosyringone to an A600 of 0.8, and infiltrated into leaves of N. benthamiana (4–5 weeks old) using a syringe without a needle. Leaves were harvested 3 d after inoculation.
Immunoprecipitation
MM2d cells were transformed with a construct containing ATL6-FLAG under the control of the cauliflower mosaic virus 35S promoter. Transformed MM2d cells or transiently expressed N. benthamiana were used for immunoprecipitation experiments as described (Sato et al., 2011) with minor modifications. Proteins precipitated with anti-FLAG M2 affinity gel were eluted with SDS sample buffer (125 mm Tris-HCl, pH 6.8, 20% [v/v] glycerol, 4% [w/v] SDS, and 10% [v/v] β-mercaptoethanol) and resolved by SDS-PAGE. Following transfer to polyvinylidene difluoride membranes, the latter were immunoblotted with anti-FLAG (Wako; 1:5,000), anti-GFP (Clontech; 1:10,000), and anti-SYP121 (1:1,000) antibodies.
Anti-SYP121 Antibody
The protein 6xHis-SYP1211-177, expressed and purified as described (Maekawa et al., 2012), was injected into a rabbit. The resultant anti-SYP121 antibody was confirmed as specifically recognizing Arabidopsis SYP121 protein (Supplemental Fig. S5).
Gene Expression Analysis
RNA was isolated, reverse transcribed, and PCR amplified as described (Morita-Yamamuro et al., 2005). The primers used are described in Supplemental Table S1. RT-PCR was performed using normalized cDNA samples. PCR products were electrophoresed on an agarose gel and visualized by ethidium bromide staining.
C/N Response Assay
Surface-sterilized seeds were sown on Murashige and Skoog medium (Murashige and Skoog, 1962) containing various concentrations of sugars and total nitrogen. The ratio of potassium nitrate to ammonium nitrate was maintained in each experiment. Potassium chloride was added to the medium to compensate for the lower potassium ion concentration in medium containing reduced potassium nitrate concentrations. Chlorophyll concentration was quantified as described (Porra et al., 1989).
Fluorescence Microscopic Analysis
Propidium iodide staining was performed as described (Bhat et al., 2005) with minor modifications. N. benthamiana leaves inoculated with Blumeria graminis f. sp. hordei spores were mounted in a solution of 2.5% mannitol, 0.01% Silwet, and 0.5% propidium iodide to stain fungal structures and plant cell walls for visualization by fluorescence microscopy. GFP and propidium iodide fluorescence were analyzed by confocal laser scanning microscopy (LSM510; Zeiss; http://microscopy.zeiss.com/microscopy), using a Plan Apochromat 40×/0.95 objective. The 488-nm argon ion laser line was used to cross excite GFP with propidium iodide. Images were acquired and processed using LSM510 software. Each image was a z-series of images; the obtained stack was “flattened” into a single image by maximum projection using ImageJ software (National Institutes of Health).
Infection with Bgh
The barley (Hordeum vulgare)-adapted powdery mildew fungus Bgh (a Japanese isolate Race I [Hiura, 1960] maintained on barley) was inoculated as described (Zimmerli et al., 2004) with minor modifications. Arabidopsis plants (28 to 32 d old) were placed in a settling tower 0.65 m in height, coated with a 0.94-μm nylon mesh, and inoculated with Bgh by dusting the conidia from heavily infected barley seedlings at 10 dpi into the top of the settling tower. After 10 min, the plants were returned to the growth chamber. Penetration frequencies were counted as described (Assaad et al., 2004) with lactophenol trypan blue staining (Koch and Slusarenko, 1990) for visualizing haustoria. Infected leaves were cleared in methanol for more than 24 h and incubated at 65°C in lactophenol trypan blue staining solution (10 mL of water, 10 mL of lactic acid, 10 mL of glycerol, 10 g of phenol, and 10 mg of trypan blue) for 15 min. The stained leaves were subsequently washed with water and viewed with a microscope (DMR; Leica; http://en.leica-camera.com/). Callose depositions were stained with aniline blue as described previously (Degrave et al., 2008) with minor modifications. Infected leaves were fixed in ethanol for more than 24 h and incubated overnight at room temperature in aniline blue staining solution (0.05% aniline blue in 70 mm NaHPO4, pH 9.0). Callose spots were analyzed by microscopy (DMR; Leica) excited by light in the UV spectrum.
Protoplast Transfection Assay
Protoplasts, isolated using the “tape-Arabidopsis sandwich method” (Wu et al., 2009), were transfected by a modified “transient expression of recombinant genes using Arabidopsis mesophyll protoplasts” method (Yoo et al., 2007). Approximately 5 × 104 protoplasts in 0.2 mL of MMg solution were mixed with approximately 20 μg of plasmid DNA at room temperature. An equal volume of a freshly prepared solution of 40% (w/v) polyethylene glycol (molecular weight 8,000) with 0.1 m CaCl2 and 0.2 m mannitol was added, and the mixture was incubated at room temperature for 5 min. Three milliliters of W5 solution was added slowly, the solution was mixed, and protoplasts were pelleted by centrifugation at 100g for 1 min. This W5 wash step was repeated twice. The protoplasts were resuspended gently in 1 mL of W5 and incubated on six-well plates at room temperature for 16 h. Protoplasts were observed as described above. The 488-nm argon ion laser line was used to cross excite GFP with chloroplast autofluorescence.
Sequence data for this study were retrieved from The Arabidopsis Information Resource (http://www.arabidopsis.org/index.jsp) under the following accession numbers: At5g27420 (ATL31), At3g05200 (ATL6), At3g11820 (SYP121), and At3g52400 (SYP122).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Correlation scatterplots of ATL31 and ATL6.
Supplemental Figure S2. Postgerminative growth phenotypes under different C/N conditions.
Supplemental Figure S3. ATL31C143S localization in mesophyll protoplast cells of wild-type and syp121-1 cells.
Supplemental Figure S4. Interaction of truncated ATL31 with SYP121 in vivo.
Supplemental Figure S5. Anti-SYP121 antibody specifically reacts with SYP121.
Supplemental Table S1. Primer sequences for vector constructs and gene expression assays.
Acknowledgments
We thank Dr. Tsuyoshi Nakagawa (Shimane University) and Dr. Shoji Mano (National Institute for Basic Biology) for the vector pB4GWcG/pB4nGGW, Dr. Tomohiro Uemura (University of Tokyo) for plasmids containing SYP121/SYP122 cDNAs, and Dr. Paul Schulze-Lefert (Max Planck Institute) for the syp121-1 seeds. We also thank Shitomi Nakagawa (Nara Institute of Science and Technology) for excellent technical support.
Glossary
- C/N
carbon/nitrogen
- Pst
Pseudomonas syringae pv tomato
- Bgh
Blumeria graminis f. sp. Hordei
- RT
reverse transcription
- BiFC
bimolecular fluorescence complementation
- hpi
hours post inoculation
- cDNA
complementary DNA
- T-DNA
transfer DNA
References
- Aguilar-Hernández V, Aguilar-Henonin L, Guzmán P. (2011) Diversity in the architecture of ATLs, a family of plant ubiquitin-ligases, leads to recognition and targeting of substrates in different cellular environments. PLoS ONE 6: e23934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad FF, Qiu JL, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, Wanner G, Peck SC, Edwards H, Ramonell K, et al. (2004) The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell 15: 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G. (2011) Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Stone S, Yang X, Antico J, Callis J, Ramonell KM, Somerville S. (2010) ATL9, a RING zinc finger protein with E3 ubiquitin ligase activity implicated in chitin- and NADPH oxidase-mediated defense responses. PLoS ONE 5: e14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A, Burnotte E, Bienert GP, Chevalier AS, Errachid A, Grefen C, Blatt MR, Chaumont F. (2012) Selective regulation of maize plasma membrane aquaporin trafficking and activity by the SNARE SYP121. Plant Cell 24: 3463–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R. (2005) Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA 102: 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri Moghaddam MR, Van den Ende W. (2012) Sugars and plant innate immunity. J Exp Bot 63: 3989–3998 [DOI] [PubMed] [Google Scholar]
- Bolton MD. (2009) Primary metabolism and plant defense: fuel for the fire. Mol Plant Microbe Interact 22: 487–497 [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Coruzzi G, Bush DR. (2001) Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol 125: 61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aoust MA, Lavoie PO, Belles-Isles J, Bechtold N, Martel M, Vézina LP. (2009) Transient expression of antibodies in plants using syringe agroinfiltration. Methods Mol Biol 483: 41–50 [DOI] [PubMed] [Google Scholar]
- Degrave A, Fagard M, Perino C, Brisset MN, Gaubert S, Laroche S, Patrit O, Barny MA. (2008) Erwinia amylovora type three-secreted proteins trigger cell death and defense responses in Arabidopsis thaliana. Mol Plant Microbe Interact 21: 1076–1086 [DOI] [PubMed] [Google Scholar]
- Eisenach C, Chen ZH, Grefen C, Blatt MR. (2012) The trafficking protein SYP121 of Arabidopsis connects programmed stomatal closure and K+ channel activity with vegetative growth. Plant J 69: 241–251 [DOI] [PubMed] [Google Scholar]
- Ellinger D, Naumann M, Falter C, Zwikowics C, Jamrow T, Manisseri C, Somerville SC, Voigt CA. (2013) Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol 161: 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. (2006) Insights into nonhost disease resistance: can they assist disease control in agriculture? Plant Cell 18: 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami K, Ichikawa M, Uemura T, Kutsuna N, Hasezawa S, Nakagawa T, Nakano A, Sato MH. (2009) Differential expression control and polarized distribution of plasma membrane-resident SYP1 SNAREs in Arabidopsis thaliana. Plant Cell Physiol 50: 280–289 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Glawe DA. (2008) The powdery mildews: a review of the world’s most familiar (yet poorly known) plant pathogens. Annu Rev Phytopathol 46: 27–51 [DOI] [PubMed] [Google Scholar]
- Guzmán P. (2012) The prolific ATL family of RING-H2 ubiquitin ligases. Plant Signal Behav 7: 1014–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano H, Harashima H, Shinmyo A, Sekine M. (2008) Arabidopsis RETINOBLASTOMA-RELATED PROTEIN 1 is involved in G1 phase cell cycle arrest caused by sucrose starvation. Plant Mol Biol 66: 259–275 [DOI] [PubMed] [Google Scholar]
- Hiura U. (1960) Studies on the disease-resistance in barley. IV. Genetics of the resistance to powdery mildew. Berichte des Ohara Instituts fur Landwirtschaftliche Biologie 11: 235–300 [Google Scholar]
- Igawa T, Fujiwara M, Takahashi H, Sawasaki T, Endo Y, Seki M, Shinozaki K, Fukao Y, Yanagawa Y. (2009) Isolation and identification of ubiquitin-related proteins from Arabidopsis seedlings. J Exp Bot 60: 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapila J, DeRycke R, VanMontagu M, Angenon G. (1997) An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci 122: 101–108 [Google Scholar]
- Katagiri F, Thilmony R, He SY. (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis Book 1: e0039, doi/10.1199/tab.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch E, Slusarenko A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al. (2008) Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840 [DOI] [PubMed] [Google Scholar]
- Maekawa S, Sato T, Asada Y, Yasuda S, Yoshida M, Chiba Y, Yamaguchi J. (2012) The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Mol Biol 79: 217–227 [DOI] [PubMed] [Google Scholar]
- Manfield IW, Jen CH, Pinney JW, Michalopoulos I, Bradford JR, Gilmartin PM, Westhead DR. (2006) Arabidopsis Co-expression Tool (ACT): Web server tools for microarray-based gene expression analysis. Nucleic Acids Res 34: W504–W509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Oswald O, Graham IA. (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massad TJ, Dyer LA, Vega CG. (2012) Costs of defense and a test of the carbon-nutrient balance and growth-differentiation balance hypotheses for two co-occurring classes of plant defense. PLoS ONE 7: e47554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Murray JA. (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Meyer D, Pajonk S, Micali C, O’Connell R, Schulze-Lefert P. (2009) Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J 57: 986–999 [DOI] [PubMed] [Google Scholar]
- Micali C, Göllner K, Humphry M, Consonni C, Panstruga R. (2008) The powdery mildew disease of Arabidopsis: a paradigm for the interaction between plants and biotrophic fungi. The Arabidopsis Book 6: e0115, doi/10.1199/tab.0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor A, Zajic D, Voll LM, Pons-Kühnemann J, Samans B, Kogel KH, Waller F. (2011) Barley leaf transcriptome and metabolite analysis reveals new aspects of compatibility and Piriformospora indica-mediated systemic induced resistance to powdery mildew. Mol Plant Microbe Interact 24: 1427–1439 [DOI] [PubMed] [Google Scholar]
- Morita-Yamamuro C, Tsutsui T, Sato M, Yoshioka H, Tamaoki M, Ogawa D, Matsuura H, Yoshihara T, Ikeda A, Uyeda I, et al. (2005) The Arabidopsis gene CAD1 controls programmed cell death in the plant immune system and encodes a protein containing a MACPF domain. Plant Cell Physiol 46: 902–912 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nielsen ME, Feechan A, Böhlenius H, Ueda T, Thordal-Christensen H. (2012) Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc Natl Acad Sci USA 109: 11443–11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S. (2005) Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol 138: 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt I, Slane D, El Kasmi F, Knöll C, Fuchs R, Mayer U, Lipka V, Jürgens G. (2011) Mechanisms of functional specificity among plasma-membrane syntaxins in Arabidopsis. Traffic 12: 1269–1280 [DOI] [PubMed] [Google Scholar]
- Ros B, Mohler V, Wenzel G, Thümmler F. (2008) Phytophthora infestans-triggered response of growth- and defense-related genes in potato cultivars with different levels of resistance under the influence of nitrogen availability. Physiol Plant 133: 386–396 [DOI] [PubMed] [Google Scholar]
- Saito C, Ueda T (2009) Functions of RAB and SNARE proteins in plant life. Int Rev Cell Mol Biol 274: 183–233 [DOI] [PubMed] [Google Scholar]
- Sato T, Maekawa S, Yasuda S, Domeki Y, Sueyoshi K, Fujiwara M, Fukao Y, Goto DB, Yamaguchi J. (2011) Identification of 14-3-3 proteins as a target of ATL31 ubiquitin ligase, a regulator of the C/N response in Arabidopsis. Plant J 68: 137–146 [DOI] [PubMed] [Google Scholar]
- Sato T, Maekawa S, Yasuda S, Sonoda Y, Katoh E, Ichikawa T, Nakazawa M, Seki M, Shinozaki K, Matsui M, et al. (2009) CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J 60: 852–864 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS. (2013) Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu Rev Phytopathol 51: 407–427 [DOI] [PubMed] [Google Scholar]
- Serrano M, Parra S, Alcaraz LD, Guzmán P. (2006) The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. J Mol Evol 62: 434–445 [DOI] [PubMed] [Google Scholar]
- Shibuya N, Minami E. (2001) Oligosaccharide signaling for defence responses in plant. Pathology 59: 223–233 [Google Scholar]
- Shin LJ, Lo JC, Chen GH, Callis J, Fu H, Yeh KC. (2013) IRT1 degradation factor1, a ring E3 ubiquitin ligase, regulates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell 25: 3039–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckli J, Fazakerley DJ, James DE. (2011) GLUT4 exocytosis. J Cell Sci 124: 4147–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud MC, Gineste S, Nussaume L, Robaglia C. (2004) Sucrose increases pathogenesis-related PR-2 gene expression in Arabidopsis thaliana through an SA-dependent but NPR1-independent signaling pathway. Plant Physiol Biochem 42: 81–88 [DOI] [PubMed] [Google Scholar]
- Tsunoda Y, Sakai N, Kikuchi K, Katoh S, Akagi K, Miura-Ohnuma J, Tashiro Y, Murata K, Shibuya N, Katoh E. (2005) Improving expression and solubility of rice proteins produced as fusion proteins in Escherichia coli. Protein Expr Purif 42: 268–277 [DOI] [PubMed] [Google Scholar]
- Tucker SL, Talbot NJ. (2001) Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu Rev Phytopathol 39: 385–417 [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. (2004) Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. (2009) Tape-Arabidopsis sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Feechan A, Pedersen C, Newman MA, Qiu JL, Olesen KL, Thordal-Christensen H. (2007) A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J 49: 302–312 [DOI] [PubMed] [Google Scholar]
- Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Somerville S. (2004) Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J 40: 633–646 [DOI] [PubMed] [Google Scholar]