Opposite changes in the cutin polyester component in tomato fruit cuticle mutants lead to increased fruit glossiness.
Abstract
The cuticle is a protective layer synthesized by epidermal cells of the plants and consisting of cutin covered and filled by waxes. In tomato (Solanum lycopersicum) fruit, the thick cuticle embedding epidermal cells has crucial roles in the control of pathogens, water loss, cracking, postharvest shelf-life, and brightness. To identify tomato mutants with modified cuticle composition and architecture and to further decipher the relationships between fruit brightness and cuticle in tomato, we screened an ethyl methanesulfonate mutant collection in the miniature tomato cultivar Micro-Tom for mutants with altered fruit brightness. Our screen resulted in the isolation of 16 glossy and 8 dull mutants displaying changes in the amount and/or composition of wax and cutin, cuticle thickness, and surface aspect of the fruit as characterized by optical and environmental scanning electron microscopy. The main conclusions on the relationships between fruit brightness and cuticle features were as follows: (1) screening for fruit brightness is an effective way to identify tomato cuticle mutants; (2) fruit brightness is independent from wax load variations; (3) glossy mutants show either reduced or increased cutin load; and (4) dull mutants display alterations in epidermal cell number and shape. Cuticle composition analyses further allowed the identification of groups of mutants displaying remarkable cuticle changes, such as mutants with increased dicarboxylic acids in cutin. Using genetic mapping of a strong cutin-deficient mutation, we discovered a novel hypomorphic allele of GDSL lipase carrying a splice junction mutation, thus highlighting the potential of tomato brightness mutants for advancing our understanding of cuticle formation in plants.
The epidermis of all aerial plant organs is covered with an extracellular layer, the cuticle, which is synthesized by the epidermal cells. The cuticle is localized on the outer face of primary cell walls and is largely composed of cutin embedded with polysaccharides, filled with intracuticular waxes, and covered with a thin layer of epicuticular waxes (Nawrath, 2006). Cutin is a polyester of glycerol, hydroxy, and epoxy fatty acids; in most species, the main cutin monomers are C16 and C18 ω-hydroxy fatty acids (Pollard et al., 2008). Besides cutin, another lipid polyester named suberin typically also contains α,ω-dicarboxylic acids, hydroxycinnamic acids, and fatty alcohols. Suberin forms a hydrophobic layer in cell walls of specific plant organs (e.g. roots and seeds) or is synthesized in response to stress (Pollard et al., 2008). Waxes are a mixture of very long chain fatty acids (VLCFAs) (C24 to C34) and their derivatives (e.g. alkanes, aldehydes, primary and secondary alcohols, ketones, or esters) and occasionally include triterpenoids and phenylpropanoids (Kunst and Samuels, 2009).
In recent years, availability of the Arabidopsis (Arabidopsis thaliana) genome sequence, high-throughput gene expression analysis tools, and mutant collections enabled deciphering the biosynthetic pathways and transport networks involved in cutin, suberin, and wax biosynthesis (Pollard et al., 2008; Li-Beisson et al., 2009; Beisson et al., 2012; Yeats and Rose, 2013). The synthesis of the cutin monomer starts with the synthesis of long chain fatty acids in the plastids. Fatty acids are then transported to the cytoplasm where they undergo a series of modifications, including the activation to CoA thioesters by long chain acyl-CoA synthetases, oxidation by cytochrome P450 (CYP)–dependent fatty acid oxidases, and esterification to glycerol-based acceptors by glycerol-3-phosphate acyl transferases to produce acyl-glycerols (Pollard et al., 2008; Li-Beisson et al., 2009). Although the sequential order of the reactions remains to be determined, the implication of several long chain acyl-CoA synthetases, CYP86A, CYP77A, and glycerol-3-phosphate acyl transferases in cutin biosynthesis has been confirmed in Arabidopsis. Mechanisms of transport of the cutin monomers and their assembly into the cuticle remain largely unknown. The plasma membrane ATP-binding cassette transporters have been implicated in the transport of both wax and cutin to the apoplast, whereas lipid transfer proteins very likely contribute to the transport of cutin monomers through the cell wall to the cutin layer (Yeats and Rose, 2013). The implication of enzymes of the GDSL lipase family in cutin assembly, which is long suspected (Reina et al., 2007; Mintz-Oron et al., 2008), was recently demonstrated in tomato (Solanum lycopersicum; Girard et al., 2012; Yeats et al., 2012a).
Molecular genetic studies were also of considerable help in the recent identification of key proteins in suberin and wax biosynthesis. The same gene families as for cutin likely contribute to suberin formation (Franke et al., 2012), which also includes specific steps such as fatty acid elongation involving β-keto acyl-CoA synthases and primary alcohol synthesis implicating fatty acyl reductases (Domergue et al., 2010). Likewise, several major enzymes of wax biosynthesis were only recently identified (Bernard and Joubès, 2013). Wax biosynthesis involves several steps including the synthesis of VLCFAs through a multi-enzyme fatty acid elongase complex and the synthesis of VLCFA derivatives through either the alcohol-forming pathway, which gives rise to primary alcohols and wax esters, or the alkane-forming pathway producing aldehydes, alkanes, secondary alcohols, and ketones. Export of wax through plasma membrane occurs via ATP-binding cassette transporters and glycosylphosphotidylinositol-anchored lipid transfer proteins (Kunst and Samuels, 2009; Yeats and Rose, 2013).
Coordinated regulation of metabolic pathways controlling cuticle formation has been demonstrated for transcription factors of the SHINE/WAX-INDUCER family (Aharoni et al., 2004; Broun et al., 2004) and recently for MIXTA-like myeloblastosis (MYB) transcription factors (Oshima et al., 2013). Several of their downstream targets involved in cutin synthesis, modification, and assembly, as well as in cell wall formation and structure and in epidermal cell patterning (Shi et al., 2013), have been identified. Other transcription factors of the MYB protein family, Homeodomain-leucine-zipper (HD-ZIP) IV family, and WW domain protein were shown to control cuticle formation, some of which and were suggested to take part in a regulatory network controlling epidermal cell patterning (Yeats and Rose, 2013).
Despite these advances and technological developments, much remains to be determined regarding the regulation, synthesis, assembly, and structure of the cuticle and their influence on cuticle properties. The thick and easy-to-isolate peel from tomato has long been used for studying the biomechanical properties of plant cuticle (Domínguez et al., 2011). In recent years, the increased availability of tomato genomic resources, including saturated genetic linkage maps, markers, and very recently the tomato genomic sequence (Tomato Genome Consortium, 2012), offered unprecedented possibilities for exploiting tomato as a model for studying plant cuticle (Hovav et al., 2007; Adato et al., 2009; Isaacson et al., 2009; Girard et al., 2012; Yeats et al., 2012b; Shi et al., 2013). To fully explore the relationships between cuticle composition and cuticle properties and to discover new genes and functions, new sources of genetic variability affecting tomato genes involved in wax, cutin, and suberin synthesis and regulation are required.
In this study, we focused on tomato cuticle mutants identified through screening for fruit brightness (glossy or dull fruits) in a tomato ethyl methanesulfonate (EMS) mutant collection in the miniature cultivar Micro-Tom. Up to 24 tomato fruit cuticle mutants with modified fruit brightness but no otherwise major plant or fruit phenotypic changes were selected. Cytological and biochemical analyses revealed that mutant fruit with altered brightness displayed wide variations in cutin load and composition and in epidermal patterning. Only few changes in wax composition and increases in wax load were observed. Fruit brightness modifications could not be attributed to a single cause but rather to combinations of various and sometimes opposite alterations of fruit cuticle. We further identified the mutation underlying a glossy cutin-deficient mutant by genetic mapping of the corresponding cuticle-associated traits and discovered a novel allele of the tomato GDSL lipase involved in cutin polymerization (Girard et al., 2012; Yeats et al., 2012b). Thus, our study highlights how the exploitation of artificially induced genetic diversity and of genomic tools currently available in tomato can efficiently contribute to the study of cuticle biosynthesis and properties in plants.
RESULTS
Screening a ‘Micro-Tom’ EMS Mutant Collection for Plants with Altered Fruit Brightness
Analysis of cuticle formation and properties using reverse genetic approaches may be hampered by other modifications of plant and fruit physiology often induced in strong cuticle mutants. In this study, we chose to focus on mild tomato fruit cuticle mutants displaying no obvious phenotypic alterations such as dwarfism, wilting, or organ fusion in plants or large cracks, multiple microcracks (russeting), and strong water loss in fruit. Toward this end, the criterion we used for selecting tomato mutants was fruit brightness, the variation of which results from milder modification of fruit surface. In addition, thin cuticle and glossy fruit appearance associated with adequate postharvest shelf-life and absence of fruit defects are among the desirable breeding traits for several types of fresh-market tomatoes.
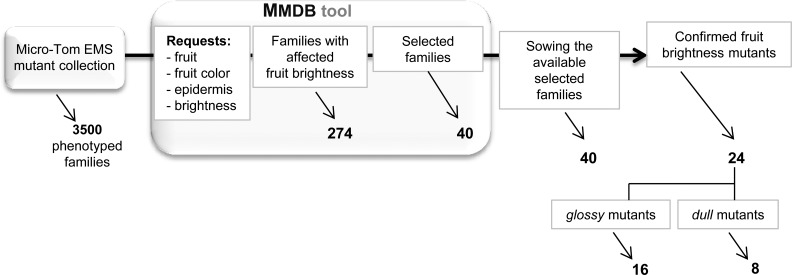
The tomato mutant collection used was an EMS mutant population generated in the miniature cultivar Micro-Tom by our group (Rothan and Causse, 2007; Just et al., 2013). The collection comprises approximately 3,500 highly mutagenized mutant families thoroughly phenotyped (12 plants per M2 families) for approximately 150 plant and fruit phenotypic criteria ranged in categories and subcategories. All of the phenotypic data are compiled and stored in a dedicated web-searchable database called the MicroTom Mutant Database. Selection of fruit brightness mutants was done using requests centered on the fruit (category: fruit; subcategories: color, epidermis, and brightness [i.e. fruit glossier or duller than the wild-type control]). Strong fruit cuticle/color mutants, such as the slcyp86a9 cuticle synthesis mutant recently identified from the same ‘Micro-Tom’ mutant collection (Shi et al., 2013), were excluded from the analysis. As shown in Figure 1, the first query using these criteria resulted in the identification of 274 mutant families. Taking into account additional criteria such as the presence/absence of other gross plant and fruit phenotypic alterations and the previous confirmation (or not) of the observed fruit brightness phenotypes in independent cultures of the retained M2 families allowed the final selection of 40 mutant families.
Figure 1.
Strategy used for the selection of the fruit brightness tomato mutants through the MicroTom Mutant Database (MMDB) tool. Visual selection of fruit brightness mutants was done in comparison with wild-type fruits at the RR stage.
For each selected family, 24 plants were grown in a greenhouse. Visual evaluation of fruit brightness was done for each plant of each family on fruits at Red Ripe (RR) stage compared with wild-type fruits (Supplemental Table S1). In the M2 families studied, which segregates for the EMS-induced mutations, a plant was considered as a fruit brightness mutant when all of the fruits on that plant displayed the glossy or dull trait. Previous fruit brightness phenotypic annotation (dull or glossy fruit) was confirmed for 20 families (i.e. at least one plant in the family showing uniformly dull or glossy fruits), whereas 7 families displayed fruit brightness phenotypes opposite to those previously observed. In addition, two displayed both dull/glossy fruits on the same plants and 11 did not show any visual difference with the wild-type fruits. We finally selected the 20 confirmed mutant families plus 4 families displaying fruit brightness phenotypes opposite to those previously observed (i.e. one plant with dull or glossy fruits for each of the selected families). Among these 24 fruit brightness mutant plants, 16 were glossy mutants and 8 were dull mutants (Supplemental Figure S1).
Cuticle Is Altered in Fruit Brightness Mutants That Display Remarkable Changes in Cutin Load and Composition
To investigate whether variations in fruit brightness could be associated with quantitative and/or qualitative differences of cuticle, we analyzed wax and cutin load and composition of RR fruits (approximately 45 d post-anthesis [DPA]) collected on the 24 selected mutant lines.
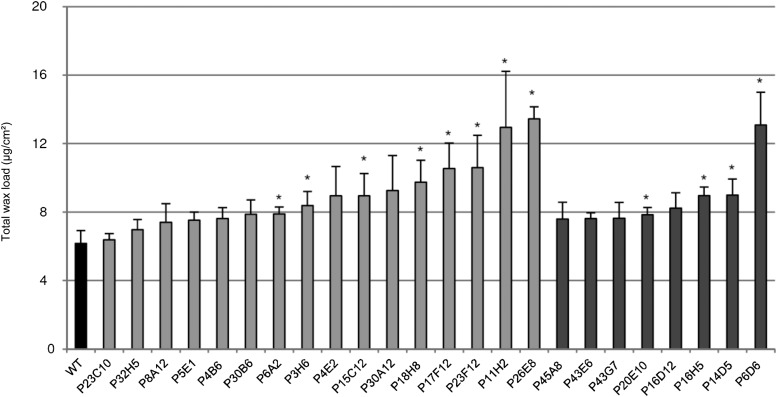
Both the glossy and dull mutants exhibited a wide range of variation in wax (identified compounds) load (Fig. 2; Table I). For up to 50% of the mutants, no significant differences in cuticular wax load were observed relative to the wild type (Fig. 2). Remarkably, the glossy and dull mutants displaying wax load variation all showed an increase in wax load compared with the wild type. However, whereas the glossy mutants showed a gradual variation in wax load from approximately 6 µg/cm2 (i.e. close to wild-type values) to 13.5 µg/cm2, most of the dull mutants had similar wax loads (7.6 to 9 µg/cm2) with the exception of one (P6D6 mutant; 13.1 µg/cm2). When we took into account the unidentified wax compounds, which represent 17% of the total wax in the wild type and up to 40% in the P4E2 mutant, similar variations were observed (data not shown).
Figure 2.
Total wax load in fruit cuticle from the wild type (WT) and selected tomato mutants. Mean values (in micrograms per square centimeter) are given with sd (n = 4). Unidentified compounds are not included. Black bar, Wild type; light gray bar, glossy mutants; dark gray bar, dull mutants. Asterisk indicates significant difference from wild-type fruit (t test, P < 0.01).
Table I. Wax composition of fruit cuticle from the wild type and selected tomato mutants.
Mean values (in micrograms per square centimeter × 10) of total wax load and of individual compounds (n = 4). The percentage of total wax load is indicated for individual compounds. Unidentified compounds are not included in the table.
| Plant | Total Identified Load | n-Alkanes | Isoalkanes | Alcohols | Amyrins | Fatty Acids | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± sd | Mean ± sd | % | Mean ± sd | % | Mean ± sd | % | Mean ± sd | % | Mean ± sd | % | |
| Wild type | 61.7 ± 7.5 | 42.0 ± 4.7 | 68.0 | 4.7 ± 0.9 | 7.6 | 2.1 ± 0.5 | 3.4 | 11.0 ± 2.4 | 17.8 | 2.0 ± 0.5 | 3.3 |
| Glossy | |||||||||||
| P23C10 | 63.8 ± 3.6 | 41.4 ± 1.0 | 65.0 | 3.4 ± 0.1 | 5.4 | 1.5 ± 0.3 | 2.3 | 15.4 ± 3.5 | 24.2 | 2.0 ± 0.3 | 3.2 |
| P32H5 | 69.8 ± 5.9 | 45.1 ± 4.0 | 64.6 | 5.4 ± 1.1 | 7.8 | 2.5 ± 0.2 | 3.6 | 13.3 ± 0.9 | 19.0 | 3.5 ± 1.1 | 5.0 |
| P8A12 | 74.1 ± 10.9 | 44.6 ± 6.5 | 60.2 | 5.2 ± 0.7 | 7.0 | 2.9 ± 0.2 | 3.9 | 18.6 ± 4.1 | 25.1 | 2.8 ± 0.6 | 3.8 |
| P5E1 | 75.3 ± 4.7 | 52.9 ± 3.2 | 70.2 | 5.4 ± 0.3 | 7.2 | 2.5 ± 0.2 | 3.3 | 10.5 ± 0.9 | 13.9 | 4.1 ± 0.6 | 5.4 |
| P4B6 | 76.3 ± 6.3 | 49.9 ± 3.8 | 65.4 | 5.1 ± 1.0 | 6.7 | 2.5 ± 0.7 | 3.3 | 15.3 ± 0.6 | 20.1 | 3.4 ± 1.2 | 4.5 |
| P30B6 | 78.8 ± 8.4 | 51.7 ± 5.5 | 65.6 | 4.3 ± 0.2 | 5.5 | 2.5 ± 0.6 | 3.2 | 18.1 ± 4.0 | 23.0 | 2.2 ± 0.2 | 2.7 |
| P6A2 | 78.9 ± 4.2 | 47.4 ± 3.0 | 60.1 | 6.8 ± 0.2 | 8.6 | 3.6 ± 0.7 | 4.6 | 18.9 ± 4.7 | 23.9 | 2.2 ± 0.5 | 2.8 |
| P3H6 | 83.9 ± 8.1 | 54.0 ± 6.0 | 64.4 | 5.6 ± 0.4 | 6.7 | 3.3 ± 1.0 | 3.9 | 19.0 ± 1.7 | 22.6 | 2.0 ± 0.7 | 2.4 |
| P4E2 | 89.5 ± 17.1 | 58.7 ± 11.6 | 65.6 | 5.1 ± 1.3 | 5.7 | 10.6 ± 3.5 | 11.8 | 13.1 ± 2.6 | 14.6 | 2.1 ± 0.5 | 2.3 |
| P15C12 | 89.6 ± 12.9 | 53.8 ± 8.8 | 60.1 | 5.8 ± 1.0 | 6.5 | 3.0 ± 1.1 | 3.4 | 25.9 ± 2.5 | 28.9 | 1.1 ± 0.4 | 1.2 |
| P30A12 | 92.5 ± 20.5 | 61.0 ± 9.6 | 66.0 | 5.0 ± 1.0 | 5.4 | 3.8 ± 1.1 | 4.1 | 20.1 ± 9.0 | 21.7 | 2.7 ± 0.4 | 2.9 |
| P18H8 | 97.6 ± 12.7 | 67.2 ± 8.4 | 68.8 | 4.8 ± 0.4 | 4.9 | 3.2 ± 0.0 | 3.3 | 20.4 ± 4.7 | 20.9 | 2.0 ± 0.2 | 2.1 |
| P17F12 | 105.4 ± 14.9 | 62.2 ± 3.9 | 59.0 | 5.4 ± 0.9 | 5.1 | 4.4 ± 0.9 | 4.2 | 31.0 ± 10.5 | 29.4 | 2.4 ± 0.9 | 2.3 |
| P23F12 | 106.0 ± 18.9 | 61.7 ± 11.3 | 58.2 | 5.7 ± 0.6 | 5.3 | 2.5 ± 0.6 | 2.4 | 34.6 ± 7.1 | 32.6 | 1.5 ± 0.4 | 1.4 |
| P11H2 | 129.5 ± 32.8 | 84.0 ± 23.8 | 64.9 | 9.1 ± 1.4 | 7.0 | 9.3 ± 3.8 | 7.2 | 22.7 ± 5.4 | 17.5 | 4.4 ± 1.4 | 3.4 |
| P26E8 | 134.5 ± 7.0 | 91.4 ± 5.0 | 68.0 | 5.4 ± 1.4 | 4.0 | 7.4 ± 2.0 | 5.5 | 27.0 ± 2.9 | 20.1 | 3.2 ± 0.5 | 2.4 |
| Dull | |||||||||||
| P45A8 | 76.0 ± 9.8 | 44.0 ± 2.0 | 57.9 | 7.3 ± 1.4 | 9.6 | 2.2 ± 0.1 | 2.9 | 20.5 ± 6.5 | 26.9 | 2.1 ± 0.5 | 2.7 |
| P43E6 | 76.3 ± 3.3 | 51.9 ± 2.9 | 68.0 | 7.3 ± 0.7 | 9.5 | 3.0 ± 0.2 | 3.9 | 11.6 ± 0.5 | 15.1 | 2.7 ± 0.3 | 3.5 |
| P43G7 | 76.4 ± 9.2 | 47.4 ± 5.2 | 62.1 | 8.4 ± 2.5 | 10.9 | 2.5 ± 1.1 | 3.2 | 14.8 ± 2.1 | 19.4 | 3.4 ± 0.5 | 4.4 |
| P20E10 | 78.5 ± 4.2 | 56.7 ± 4.7 | 72.2 | 6.4 ± 1.6 | 8.2 | 1.4 ± 0.1 | 1.8 | 12.0 ± 1.1 | 15.2 | 2.0 ± 0.2 | 2.5 |
| P16D12 | 82.4 ± 8.9 | 55.0 ± 10.0 | 66.7 | 9.0 ± 2.1 | 10.9 | 2.2 ± 0.2 | 2.7 | 13.5 ± 1.4 | 16.4 | 2.6 ± 0.3 | 3.2 |
| P16H5 | 89.6 ± 5.0 | 56.2 ± 2.3 | 62.7 | 7.0 ± 0.6 | 7.8 | 2.8 ± 0.1 | 3.1 | 21.8 ± 2.4 | 24.4 | 1.8 ± 0.2 | 2.0 |
| P14D5 | 89.9 ± 9.4 | 57.4 ± 5.3 | 63.8 | 8.4 ± 1.9 | 9.3 | 2.9 ± 0.4 | 3.3 | 19.1 ± 1.8 | 21.2 | 2.1 ± 0.4 | 2.4 |
| P6D6 | 130.8 ± 19.2 | 75.6 ± 8.5 | 57.8 | 10.1 ± 3.6 | 7.7 | 3.3 ± 1.2 | 2.5 | 39.5 ± 6.0 | 30.2 | 2.2 ± 0.5 | 1.7 |
As shown in Table I, the compounds identified in the chloroform-soluble wax from the Micro-Tom cultivar are mainly made up of alkanes, which represent more than 60% of the wax, triterpenoids (α-, β-, and δ-type amyrin; approximately 18%), isoalkanes (7% to 8%), and alcohols (approximately 3% to 4%). Although changes in alkane and in wax load showed the same trend in most mutants, several mutants showed remarkable modifications of wax composition. Amyrins increased to 29% to 33% of total wax load in the P17F12 and P23F12 glossy mutants and to almost 30% in the P6D6 dull mutant. Alcohols increased to 7% to 12% in the P11H2 and P4E2 glossy mutants, respectively. In conclusion, most fruit brightness mutants displayed no or slight variations in wax load and glossy mutants could not be distinguished from dull mutants based only on single wax analysis.
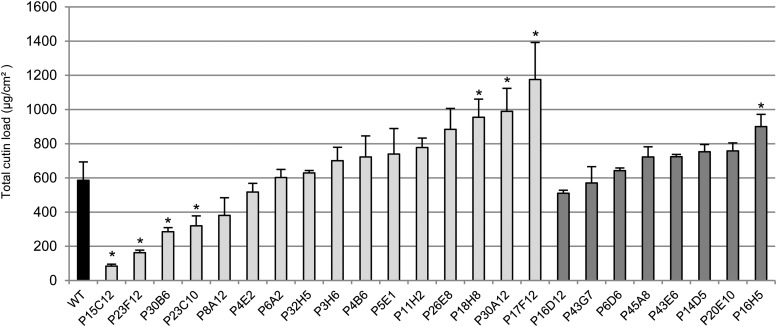
A wider range of variation was observed for cutin (identified compounds) load among the mutants (Fig. 3; Table II). Again, as for wax load, the dull mutants displayed fewer variations than the glossy mutants, with the larger variations in cutin load in these mutants being a 60% increase (P16H5) relative to the wild type. By contrast, glossy mutants exhibited large disparities between the most extreme phenotypes (Supplemental Fig. S2). Both strong reductions (down to 15% of the wild type) and increases (up to 220% of the wild type) in cutin load were observed, unlike wax load. The residual cutin loads of the three most affected glossy mutants were 84.1 µg/cm2 (P15C12), 162.6 µg/cm2 (P23F12), and 285 µg/cm2 (P30B6), which is below the wild type load of 586.1 µg/cm2. By contrast, up to five glossy mutants displayed significant increases in cutin load, which ranged from approximately 722.9 µg/cm2 (P4B6) to 1175.5 µg/cm2 (P17F12). As in other tomato cultivars described to date (Mintz-Oron et al., 2008; Girard et al., 2012; Nadakuduti et al., 2012), cutin monomers are mostly made up of polyhydroxy fatty acids (65%, with 9,16-dihydroxy-palmitic acid representing 95% of the total) and ω-hydroxy fatty acids (11%), and in lesser amounts by fatty acids (approximately6%) and dicarboxylic acids (approximately 5%) (Table II). Whereas changes in cutin load were usually paralleled by similar changes in the various classes of cutin monomers, several glossy mutants displayed remarkable and specific patterns of accumulation of cutin monomers. Indeed, further exploration of the cutin composition data by principal component analysis (PCA) and hierarchical clustering analysis (HCA) revealed groups of mutants displaying similar cutin composition changes (Fig. 4). The first principal component (PC1) explaining 53% of the total variability clearly separated three clusters on the positive side and one cluster on the negative side. The second principal component (PC2) explaining 23% of the total variability separated two clusters on the positive side and two clusters on the negative side. A bulk of eight mutants, either dull or glossy, was not separated by the principal components PC1 and PC2 and were aggregated within the wild-type cluster.
Figure 3.
Total cutin load in fruit cuticle from the wild type (WT) and selected tomato mutants. Mean values (in micrograms per square centimeter) are given with sd (n = 4). Unidentified compounds are not included. Black bar, Wild type; light gray bar, glossy mutants; dark gray bar, dull mutants. Asterisk indicates significant difference from wild-type fruit (t test, P < 0.01).
Table II. Cutin monomer composition of fruit cuticle from the wild type and selected tomato mutants.
Mean values (in micrograms per square centimeter) of total cutin load and of individual compounds (n = 4). Unidentified compounds are not included in the table. The percentage of total cutin load is indicated for individual compounds. ND, not detected.
| Plant | Total Load | Fatty Acids | Dicarboxylic Acids | ω-Hydroxy Acids | Cinnamic Acid | Polyhydroxy Acids | 2-Hydroxy Fatty Acids | Fatty Alcohols | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± sd | Mean ± sd | % | Mean ± sd | % | Mean ± sd | % | Mean ± sd | % | Mean ± sd | % | Mean ± sd | % | Mean ± sd | % | |
| Wild type | 586.1 ± 107.4 | 35.2 ± 8.6 | 6.0 | 31.9 ± 8.5 | 5.4 | 65.8 ± 26.2 | 11.2 | 17.9 ± 3.8 | 3.1 | 385.3 ± 85.7 | 65.7 | 6.6 ± 0.9 | 1.1 | 43.5 ± 12.4 | 7.4 |
| Glossy | |||||||||||||||
| P15C12 | 84.1 ± 11.3 | 2.6 ± 0.5 | 3.1 | 3.3 ± 0.1 | 3.9 | 6.2 ± 1.9 | 7.4 | 10.0 ± 2.3 | 11.9 | 57.1 ± 2.2 | 67.8 | 1.5 ± 0.2 | 1.8 | 3.4 ± 0.6 | 4.1 |
| P23F12 | 162.6 ± 14.8 | 11.8 ± 1.9 | 7.2 | 7.2 ± 1.1 | 4.4 | 19.1 ± 0.9 | 11.7 | 6.3 ± 0.9 | 3.9 | 102.2 ± 9.8 | 62.8 | ND | 0 | 16.1 ± 2 | 9.9 |
| P30B6 | 285.0 ± 24.0 | 17.6 ± 2.1 | 6.2 | 10.6 ± 3.9 | 3.7 | 19.9 ± 2 | 7 | 8.8 ± 0.5 | 3.1 | 198.7 ± 23.3 | 69.7 | ND | 0 | 29.5 ± 5.7 | 10.3 |
| P23C10 | 320.0 ± 58.0 | 13.3 ± 2.5 | 4.1 | 20.0 ± 6.2 | 6.2 | 28.9 ± 6.3 | 9 | 8.8 ± 0.7 | 2.7 | 218.6 ± 34.4 | 68.3 | 3.2 ± 0.8 | 1 | 27.3 ± 8.3 | 8.5 |
| P8A12 | 380.8 ± 103.6 | 18.2 ± 6.6 | 4.8 | 34.3 ± 8.6 | 9 | 28.9 ± 9.4 | 7.6 | 10.1 ± 5.3 | 2.6 | 244.2 ± 53.4 | 64.1 | 2.8 ± 0. 9 | 0.7 | 42.3 ± 12.5 | 11.1 |
| P4E2 | 517.2 ± 51.1 | 16.4 ± 5.6 | 3.2 | 14.3 ± 5.6 | 2.8 | 83.3 ± 10.5 | 16.1 | 23.9 ± 3.2 | 4.6 | 349.7 ± 31.4 | 67.6 | 3.4 ± 0.1 | 0.7 | 26.1 ± 6.1 | 5.1 |
| P6A2 | 602.4 ± 47.6 | 40.1 ± 5.4 | 6.7 | 28.5 ± 3.7 | 4.7 | 50.1 ± 3.1 | 8.3 | 16.8 ± 1.1 | 2.8 | 413.4 ± 27.6 | 68.6 | 4.0 ± 0.6 | 0.7 | 49.7 ± 9 | 8.2 |
| P32H5 | 629.1 ± 14.0 | 23.6 ± 3 | 3.7 | 36.8 ± 2.7 | 5.9 | 66.8 ± 10.3 | 10.6 | 28.9 ± 6 | 4.6 | 399.3 ± 30.2 | 63.5 | 6.7 ± 0.9 | 1.1 | 67.0 ± 10.7 | 10.6 |
| P3H6 | 701.0 ± 78.5 | 57.5 ± 12.2 | 8.2 | 36.0 ± 13.1 | 5.1 | 54.3 ± 9.3 | 7.7 | 15.2 ± 3.8 | 2.2 | 485.3 ± 57.1 | 69.2 | 2.1 ± 1.1 | 0.3 | 50.6 ± 6.6 | 7.2 |
| P4B6 | 722.9 ± 122.9 | 32.5 ± 7.5 | 4.5 | 65.7 ± 19.3 | 9.1 | 65.5 ± 10.8 | 9.1 | 24.9 ± 6.3 | 3.4 | 427.0 ± 55.4 | 59.1 | 7.7 ± 1.7 | 1.1 | 99.6 ± 2.6 | 13.8 |
| P5E1 | 739.4 ± 149.8 | 36.6 ± 6.9 | 5.0 | 134.2 ± 27.6 | 18.2 | 58.9 ± 18.1 | 8 | 21.1 ± 7 | 2.9 | 360.4 ± 49.2 | 48.7 | 12.4 ± 2 | 1.7 | 115.8 ± 25.6 | 15.7 |
| P11H2 | 778.1 ± 54.9 | 30.5 ± 10.8 | 3.9 | 28.6 ± 6.8 | 3.7 | 140.7 ± 17.3 | 18.1 | 27.2 ± 1.6 | 3.5 | 502.4 ± 23.4 | 64.6 | 5.0 ± 1.3 | 0.6 | 48.8 ± 8.2 | 6.3 |
| P26E8 | 883.9 ± 122.8 | 35.6 ± 8 | 4.0 | 138.9 ± 40.1 | 15.7 | 77.5 ± 9.3 | 8.8 | 30.8 ± 4.1 | 3.5 | 449.6 ± 40.7 | 50.9 | 14.2 ± 0.4 | 0.6 | 137.4 ± 36.1 | 6.3 |
| P18H8 | 954.6 ± 106.0 | 82.4 ± 14 | 8.6 | 41.8 ± 14.7 | 4.4 | 142.0 ± 34.1 | 14.9 | 34.6 ± 1.8 | 3.6 | 564.6 ± 47.6 | 59.1 | 7.4 ± 2.1 | 0.8 | 81.8 ± 13 | 8.6 |
| P30A12 | 989.5 ± 134.7 | 66.3 ± 23.5 | 6.7 | 49.5 ± 0.6 | 5 | 112.8 ± 17.9 | 11.4 | 45.0 ± 6.6 | 4.6 | 631.8 ± 74.1 | 63.9 | 7.6 ± 0.8 | 0.8 | 76.5 ± 29.8 | 7.7 |
| P17F12 | 1175.5 ± 216.6 | 39.6 ± 8 | 3.4 | 106.2 ± 14 | 9 | 81.7 ± 16.8 | 6.9 | 36.8 ± 9.2 | 3.1 | 541.2 ± 113.9 | 46 | 201.4 ± 46.7 | 17.1 | 168.6 ± 49.7 | 14.3 |
| Dull | |||||||||||||||
| P16D12 | 510.6 ± 17.2 | 21.2 ± 0.5 | 4.1 | 32.1 ± 2.9 | 6.3 | 56.3 ± 5.3 | 11 | 18.9 ± 0.9 | 3.7 | 321.1 ± 8.1 | 62.9 | 5.9 ± 0.7 | 1.2 | 55.1 ± 5.4 | 10.8 |
| P43G7 | 570.1 ± 95.7 | 21.9 ± 3.7 | 3.8 | 57.3 ± 10.9 | 10.1 | 53.8 ± 26.9 | 9.4 | 18.1 ± 7.2 | 3.2 | 344.3 ± 59.1 | 60.4 | 6.0 ± 2.2 | 1.1 | 68.8 ± 9.8 | 12.1 |
| P6D6 | 642.8 ± 15.3 | 15.6 ± 3.1 | 2.4 | 15.2 ± 4.6 | 2.4 | 104.4 ± 8.6 | 16.2 | 21.7 ± 1.1 | 3.4 | 454.0 ± 11 | 70.6 | ND | 0 | 32.0 ± 2.5 | 5 |
| P45A8 | 722.7 ± 59.0 | 17.9 ± 3.2 | 2.5 | 24.3 ± 2 | 3.4 | 103.4 ± 7 | 14.3 | 38.8 ± 4 | 5.4 | 501.7 ± 44 | 69.4 | 1.7 ± 0.5 | 0.2 | 34.9 ± 3.5 | 4.8 |
| P43E6 | 724.0 ± 13.1 | 32.8 ± 9 | 4.5 | 39.4 ± 19 | 5.4 | 61.7 ± 8.3 | 8.5 | 16.5 ± 0.8 | 2.3 | 533.9 ± 26.2 | 73.7 | 5.1 ± 0.8 | 0.7 | 34.8 ± 8.5 | 4.8 |
| P14D5 | 752.8 ± 42.4 | 32.5 ± 0.2 | 4.3 | 40.8 ± 1.5 | 5.4 | 42.0 ± 4.7 | 5.6 | 15.0 ± 2 | 2 | 564.8 ± 7.7 | 75 | 3.7 ± 0.2 | 0.5 | 53.9 ± 0.8 | 7.2 |
| P20E10 | 757.9 ± 46.9 | 18.8 ± 4.4 | 2.5 | 27.9 ± 3.6 | 3.7 | 108.8 ± 8.1 | 14.4 | 33.5 ± 1.5 | 4.4 | 531.8 ± 32.6 | 70.2 | 3.7 ± 0.7 | 0.5 | 33.4 ± 4.8 | 4.4 |
| P16H5 | 900.2 ± 71.2 | 23.1 ± 2.5 | 2.6 | 41.3 ± 4.6 | 4.6 | 78.2 ± 2.9 | 8.7 | 42.9 ± 0.9 | 4.8 | 667.5 ± 56.1 | 74.1 | 4.4 ± 0.5 | 0.5 | 42.9 ± 5.4 | 4.8 |
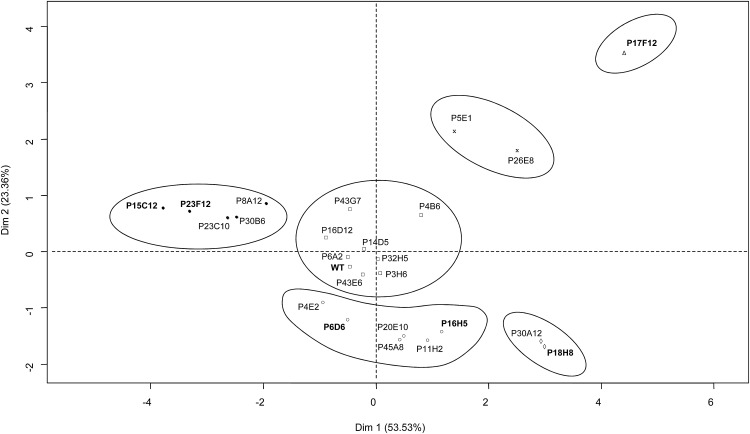
Figure 4.
HCA plot of the two first principal components showing the 24 mutants and the wild type (WT) analyzed for cutin composition of fruit cuticle. Data were centered and scaled for the PCA. The Euclidian distance and Ward aggregation method were used for the HCA. The x axis and y axis are the two principal components with the percentage of contribution to the two dimensions indicated between brackets. Cluster 1 is represented with black circles, cluster 2 with white triangles, cluster 3 with black crosses, cluster 4 with white diamonds, cluster 5 with white squares, and cluster 6 with white circles. Bold type indicates wild type and fruit brightness mutants further analyzed for fruit surface and cuticle.
Examination of PC1 and PC2 loadings (Supplemental Fig. S3) allowed the identification of cutin monomers responsible for cutin modifications in the various mutants. On the positive side of PC1 and PC2, the primary alcohols and 2-hydroxy fatty acids and the dicarboxylic acids discriminated the P17F12 glossy mutant and the P5E1/P26E8 cluster of glossy mutants. The P17F12 mutant, in which the total cutin load is increased by more than 2-fold, exhibited a 30-fold enhancement of 2-hydroxy fatty acids, which increased from trace amounts in the wild type (6.6 µg/cm2) to approximately 16% of the total cutin monomers in the mutant (approximately 201 µg/cm2). The dicarboxylic acid contents were strikingly similar between the P5E1 and P26E8 mutants and increased from approximately 32 µg/cm2 in the wild type to 134 to 139 µg/cm2 in the mutants. Another remarkable cluster groups the strong cutin-deficient mutants that are similarly affected for all cutin monomers (e.g. the P15C12 and the P23F12 mutants clustered on the negative side of PC1). On the positive side of PC1 and negative side of PC2, the polyhydroxy fatty acids, ω-hydroxy fatty acids, fatty acids, and cinnamic acid compounds were discriminating the P30A12/P18H8 cluster of glossy mutants. These mutants also accumulate large amounts of cutin monomers (X 1.6). However, in contrast to P17F12, their 2-hydroxy fatty acids content is similar to that of the wild type (Table II). Finally, a cluster discriminated on the negative side of PC2 groups several dull and glossy mutants with moderate to large changes in cutin load.
Thus, although cutin is altered in most dull and glossy mutants, no obvious link could be made between fruit cutin load and/or composition and fruit brightness, as already observed for waxes. Moreover, glossy mutants can show either strong deficiencies (e.g. P15C12) or increases (e.g. P17F12) in cutin load (Fig. 3). To further explore the possible effect of wax and cutin covariations on fruit brightness, we combined the biochemical (wax and cutin composition and load) and phenotypic data and analyzed them by HCA (Supplemental Figs. S4 and S5). However, as for cutin alone, no obvious relationships could be found between cutin and wax variations and the dull or glossy aspect of the fruit.
Epidermal and Cuticle Architecture Affects Fruit Brightness
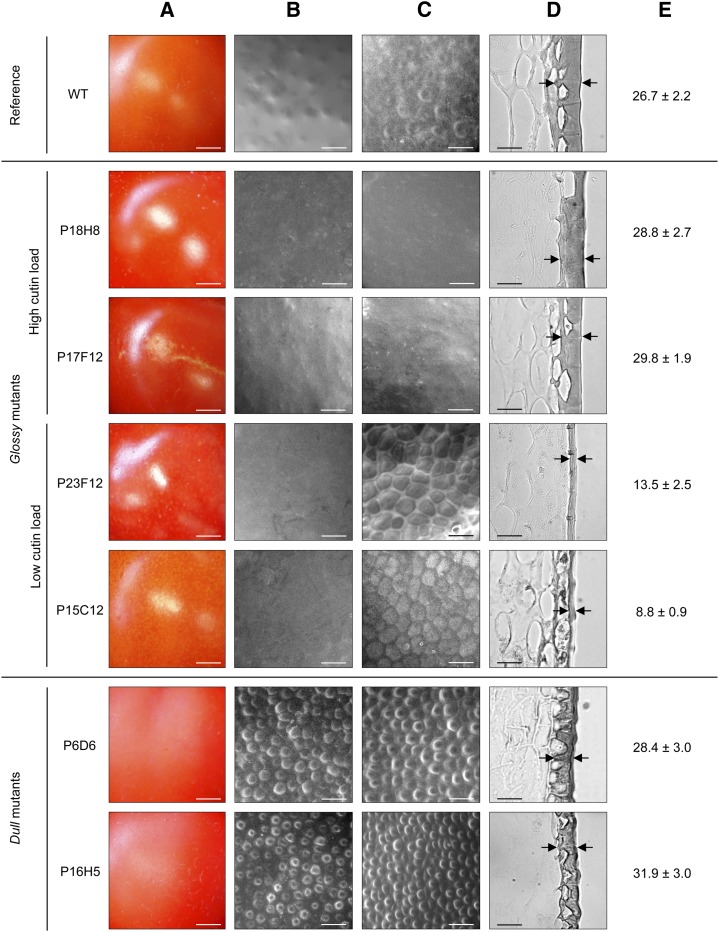
To obtain additional insight into the relationships between fruit brightness and cuticle, we selected three groups of mutants according to cutin monomer and/or wax load of the fruits and further characterized them by optical and scanning electron microscopy. Among these were glossy mutants with high cutin load (P18H8 and P17F12) and low cutin load (P23F12 and P15C12) and dull mutants with either high cutin load (P16H5) or high wax load (P6D6) (Fig. 5A). As expected, glossy mutants with low cutin load exhibited very thin cuticles compared with the wild type, with cuticle thickness reduced by approximately 49% and 67% for P23F12 and P15C12, respectively (Fig. 5, D and E). Surprisingly, cuticle thickness was very similar between wild-type fruits and glossy mutants with high cutin loads, despite the strong difference in cutin load between these mutants and the wild type (60% to 120% more cutin in P18H8 and P17F12, respectively). Examination of exocarp sections indicated that neither tissue structure nor epidermal cell size and shape were affected in these glossy mutants (data not shown). Dull mutants did not show any significant difference in cuticle thickness relative to the wild type but both high cutin and high wax dull mutants displayed distinctive morphological alterations of the cuticle-encased epidermal cells, which appeared less elongated and more conical-shaped than in the wild type and other mutants.
Figure 5.
The surface and cuticle from the wild type (WT) and selected glossy and dull mutants fruits at RR stage. A, Light macroscopy images showing cuticle brightness. B, ESEM of native fruit surface. C, ESEM of de-waxed fruit surface. D, Light microscopy of fruit exocarp sections stained with Sudan IV. Black arrows show examples of cuticle thickness measurement points. E, Cuticle thickness measurements of pericarp sections stained with Sudan IV. Mean values (in micrometers) of 40 measures on 3 different sections are given with sd. Scale bar = 5 mm in A; 40 µm in B and C; 30 µm in D. [See online article for color version of this figure.]
When examined under environmental scanning electron microscopy (ESEM), native fruit surface from glossy mutants with low cutin load mutants looked much smoother than that of the wild-type fruits, which showed more irregular surface with small domes (Fig. 5B). By contrast, fruit surface from dull mutants was rough and entirely covered with circular-shaped, dome-like structures, in agreement with optical microscopy observations (Fig. 5D). These differences appeared even more clearly when epicuticular waxes were removed from cuticle by treatment with chloroform (Fig. 5C). Cutinized epidermal cell walls were clearly visible in the wild type, whereas the surface of high cutin load glossy mutants remained remarkably smooth with no (P18H8) or very few (P17F12) surface irregularities. In contrast, de-waxing low cutin load glossy mutants revealed very thin-walled epidermal cells underneath the cuticle proper, with no cutin deposit on top of these cells unlike the wild type. De-waxing did not change the surface aspect of the dull fruits, although the dome-like structures that correspond to epidermal cells (Fig. 5D) were even more apparent. Their quantification clearly indicates that the fruit epidermal cell number is increased by 66% to 122% in the dull mutants (Supplemental Table S2). Together, these observations suggest that both the cuticle and the development of the epidermis are altered in dull fruit mutants. By contrast, only the cuticle is affected in glossy fruit mutants and increased fruit brightness can be provoked by both deficiency and increased accumulation of cutin.
Cutin-Composition Mutants P5E1 and P26E8 Show Constitutive Alteration of Suberin Biosynthesis in the Fruit
To investigate the mutations underlying the variations in fruit brightness, we focused on the glossy mutants because dull mutations are likely pleiotropic and affect the epidermis. We further focused on cutin mutants because cutin load is possibly the major factor controlling fruit brightness in tomato (Girard et al., 2012; Nadakuduti et al., 2012; Yeats et al., 2012b), and the possible effects of wax changes on glossiness have already been well described in Arabidopsis (Chen et al., 2003; Aharoni et al., 2004; Bourdenx et al., 2011). One way to identify the possible origin of the mutation is to combine the wealth of information now available on cuticle biosynthesis and regulation with the data on fruit surface chemistry. Two of the cutin-accumulation mutants that clustered together in PCA-HCA (P5E1 and P26E8 lines; Fig. 4) showed unusual high dicarboxylic acid content (15% to 18% of total monomers). Cutin compositions of these mutants are strikingly similar (Table II; Supplemental Table S3), suggesting that they are allelic. In addition, close examination of the dicarboxylic acid components of the P5E1 and P26E8 mutants (Supplemental Table S3) indicates that both display a 2.5-fold increase in C16:0 dicarboxylic acid and a 5-fold increase in C18:1 dicarboxylic acid compared with the wild type. In most species, except Arabidopsis and other Brassicaceae, this composition is considered as indicative of suberin (Pollard et al., 2008). Remarkably, no russeting or any other visible mark of suberin accumulation had been observed in the fruit sample analyzed, which presented a uniform glossy appearance. After cuticle composition analysis, we therefore planted one of these mutants (P26E8) in order to observe its cuticle phenotype. Close examination of the fruits revealed the obvious accumulation of suberin-like material at the distal end of the fruit (Supplemental Fig. S6), a trait not observed in the fruit sample previously analyzed. This result reinforces the hypothesis that the biosynthesis of both cutin and suberin are altered in these mutants. Because growth period and position of the fruit on the plant apparently influence the extent of accumulation of this suberin-like material, this is likely under the control of the environmental conditions.
The P15C12 Cuticle Mutant Displays Constitutive Cutin Deficiency in the Fruit
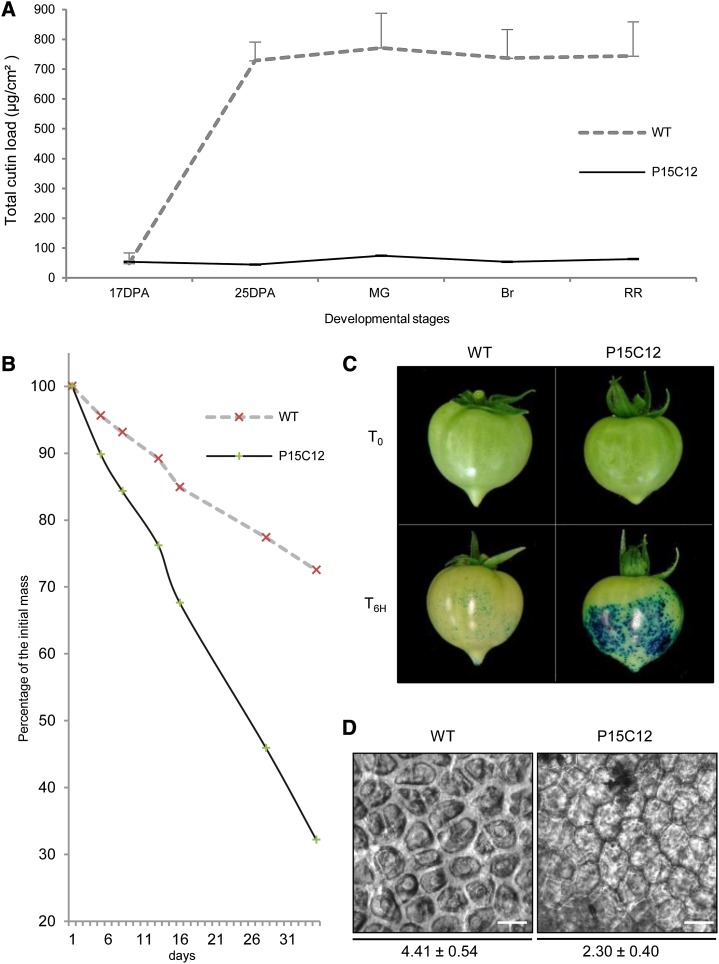
Two other obvious targets are the P15C12 and P17F12 cutin mutants, which are strong cutin-deficient (cutin load: 84.1 µg/cm2) and strong cutin-accumulating (cutin load: 1175.5 µg/cm2) mutants, respectively. Fruits from the P17F12 mutant display obligatory parthenocarpy and plants carrying the cuticle mutation did not set seeded fruits, despite attempts of pollination with wild-type pollen. This mutant was therefore not further studied. By contrast, the P15C12 mutant, in which plant and fruit were not affected by the mutation except for cuticle alteration, was further characterized along fruit development for cutin load and water loss (Fig. 6, A to C). At 17 DPA, fruits from the wild type and P15C12 mutant exhibited low cutin loads (between 30 and 50 µg/cm2). From then on, a striking accumulation of cutin occurred in the wild-type fruit during the next 8 d (i.e. during the cell expansion phase of fruit growth). Cutin load reached approximately 615 µg/cm2 at 25 DPA in the wild type and slightly increased thereafter until 695 µg/cm2 at the RR stage. By contrast, cutin load of the P15C12 mutant continuously stayed at the same low level throughout fruit development until the RR stage. The effect of the mutation on cutin accumulation in the cell walls from epidermal cells could be scored by analyzing the de-waxed fruit epidermis and measuring the thickness of the cutinized cell walls between two adjacent epidermal cells, hereafter called cutin width (Supplemental Figure S7). The cutin width was almost twice as thin in the P15C12 mutant than in the wild type (Fig. 6D).
Figure 6.
Cuticle properties of the glossy P15C12 cutin-deficient mutant. A, Total cutin load of the wild type (WT) and P15C12 fruits at 17 DPA, 25 DPA, MG, Breaker (Br), and RR stages. Mean values (in micrograms per square centimeter) of total cutin load are given with sd (n = 4). B, Water loss of RR fruit during storage. Fruits were stored at 28°C under pulsed air, and masses were measured at time zero (T0) and after 4, 7, 12, 15, 27, and 34 d of storage. C, Permeability to toluidine blue of MG fruits incubated with the dye during 6 h (T6H). D, Cutin width measurements on fresh pealed outer epidermis from RR fruits are expressed in micrometers. Twelve independent measures of the width of the cutinized cell walls were done after picture acquisition. Scale bar = 30 µm. [See online article for color version of this figure.]
Alterations of cutin biosynthesis in the P15C12 mutant were also accompanied by large modifications of the cuticle permeability to water, although mutant wax load was 40% higher than that of the wild type. The water loss kinetics of representative fruits from the wild type and P15C12 mutant (Fig. 6B) clearly show the impaired ability of P15C12 detached fruits to avoid water loss, with the mutant fruit retaining only 32.2% of its original weight at the end of the experiment versus 72.5% in the wild type. Consistently, the tests of cuticle permeability performed on Mature Green (MG) fruit demonstrated that mutant fruit was very permeable to the toluidine blue dye, unlike the wild-type fruit (Fig. 6C). Thus, besides fruit brightness, the mutation affecting fruit cutin load in the P15C12 mutant has dramatic consequences on the integrity and properties of its cuticle. Because these various phenotypic traits (fruit brightness, cutin width, water loss, permeability to toluidine blue) describe various aspects of the same mutation affecting cutin load, they were therefore used for characterizing tomato genotypes carrying the cuticle-deficiency allele found in P15C12.
Mapping of the P15C12 Locus and Identification of the gdsl2-b Mutation through the Candidate Gene Approach
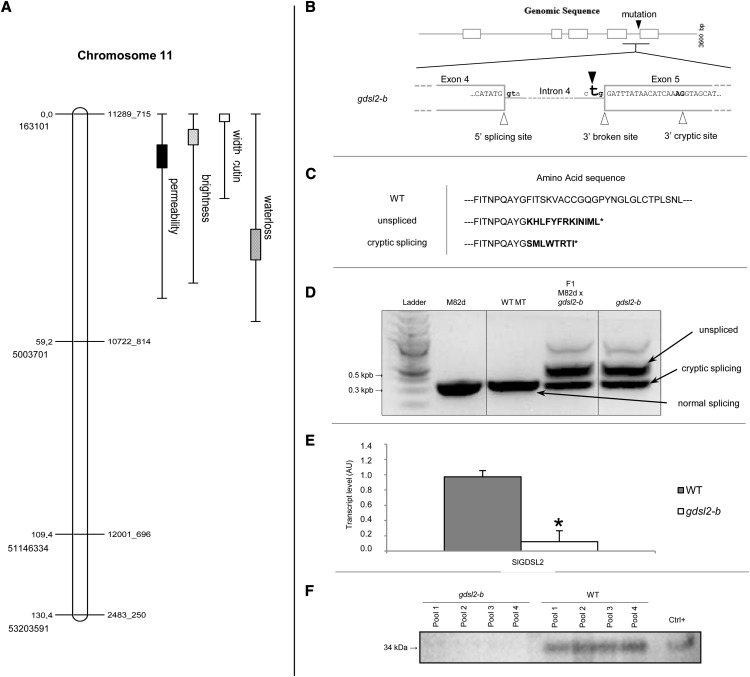
A F2 mapping population segregating for the cuticle-deficiency mutation found in P15C12 was generated through crossing the Micro-Tom P15C12 homozygous mutant with a M82 dwarf mutant, previously selected among EMS mutants generated in the widely used tomato cultivar M82 (Menda et al., 2004). There are multiple rationale for this strategy (Just et al., 2013). First, by crossing ‘Micro-Tom’ with a cultivar from the same tomato species (S. lycopersicum) and not with a wild tomato such as a Solanum pimpinellifolium accession, we avoid the segregation of multiple polymorphic traits, among which those affecting the cuticle (Yeats et al., 2012a). Second, the F2 mapping population of 110 plants can be grown in the greenhouse on a limited space (6 m2 instead of 25 m2 for S. pimpinellifolium). Finally, a whole set of single nucleotide polymorphism (SNP) markers between Micro-Tom and M82 was recently developed (Shirasawa et al., 2010), allowing us to select 48 SNP markers evenly distributed along the 12 tomato chromosomes (Tomato Genome Consortium, 2012). The mapping population was scored as follows for cuticle-associated traits previously characterized in the P15C12 mutant: fruit brightness (three classes; Supplemental Fig. S8, A and B), cuticle permeability to toluidine blue dye (six classes; Supplemental Fig. S8, C and D), water loss (five classes; Supplemental Fig. S8, E and F), and cutin width (four classes; Supplemental Fig. S8, G and H). Frequency distributions show complex inheritance patterns for the various traits, probably revealing both the imprecisions in scoring the traits (fruit brightness, cuticle permeability) and the effects on cuticle properties of the interactions between cutin load and other cuticular traits that could segregate in the F2 population (e.g. epidermal patterning or fruit surface chemistry). Nevertheless, the major locus controlling each trait was located in a 4.84-Mb region on chromosome 11, between markers 11289_715 and 10722_814 (Fig. 7A). The log of the odds (LOD) score, R2 percentage and effect of quantitative trait loci (QTL) are presented in Table III.
Figure 7.
Mapping of the P15C12 glossy mutant and characterization of the mutation. A, The four cuticle-related traits (brightness, water loss, permeability, cutin width) were mapped to chromosome 11 between SNP markers 11289_715 and 10722_814. Genetic distance (in centimorgans) and physical position (ITAG2.40) are indicated on the left. Markers are indicated on the right. 1-LOD and 2-LOD support intervals of each QTL are marked by boxes and thin bars, respectively. B, SlGDSL2 gene splicing model. Exons, introns, and splicing sites (in bold) are represented by white boxes, gray lines, and white triangles, respectively. The black triangle shows the A to T mutation in the 3′ consensus splice site of intron 4, which results in a broken splicing site in the gdsl2-b mutant. The 3′ cryptic splicing site is shown on the right. C, Amino acid sequences deduced from native SlGDSL2 transcript (wild-type plant, SlGDSL2 gene), unspliced transcript (in-frame reading through intron 4 in the gdsl2-b mutant), and alternative cryptic site transcript (splicing of intron 4 at 3′ cryptic site in exon 5 in the gdsl2-b mutant). Incorrect amino acids are indicated in bold. D, Electrophoretic analysis of the SlGDSL2 transcripts in M82dwarf (M82d), wild-type ‘Micro-Tom’ (WT MT), gdsl2-b mutant, and F1 hybrid between M82d and gdsl2-b. E, Real-time reverse-transcription PCR analysis of SlGDSL2 expression in 20 DPA fruits in wild-type ‘Micro-Tom’ (gray box) and gdsl2-b mutant (white box). Mean values of three biological replicates are shown with sd. Asterisk indicates significant difference from wild-type fruits (t test, P < 0.01). F, Immunoblot analysis of GDSL2 protein in four independent pools of gdsl2-b and wild-type epidermis. Positive control is a recombinant GDSL extract (Girard et al., 2012). AU, Arbitrary units; Ctrl, control; LOD, log of the odds; QTL, quantitative trait loci.
Table III. Summary of the major locus identified for fruit brightness mutant P15C12 on chromosome 11 between markers 11289_715 and 10722_814.
LOD score is maximum logarithm of odds for the presence of a locus controlling each analyzed trait by simple interval mapping (MapMaker/QTL). R2 is the percentage of variance explained by the locus. Additive allele effect was determined by QTL Cartographer.
| Trait | LOD Score | R2 | Additive Allele Effect |
|---|---|---|---|
| % | |||
| Permeability | 15.90 | 75.3 | 1.2807 |
| Brightness | 13.64 | 61.1 | 0.6287 |
| Cutin width | 4.76 | 24 | −3.715 |
| Water loss | 8.13 | 65.6 | −25.885 |
We next screened the chromosomal region of interest for candidate genes with known implication in cutin synthesis or regulation. Among these were genes identified as specifically expressed in the outer epidermis of tomato by laser microdissection of various tomato pericarp tissues followed by RNA sequencing analysis (Matas et al., 2011). Two genes (Solyc11g007540 and Solyc11g006250; Sol Genomics Network [SGN] database, http://solgenomics.net/) were localized between the markers of interest on chromosome 11. The first presents a strong homology with an Arabidopsis gene (AT5GO4660.1) encoding a CYP77A4 cytochrome P450 oxidase catalyzing the epoxidation of free fatty acids (Sauveplane et al., 2009). No mutation in this gene was detected upon sequencing. The second encodes an acyl-transferase of the GDSL esterase/lipase protein family recently shown to be involved in the extracellular deposition of cutin in tomato fruit (Girard et al., 2012; Yeats et al., 2012b). The current locus name of Solyc11g006250 (1,004,368 to 1,006,899 nucleotides on chromosome 11) in the Solanaceae SGN database is GDSL2, whereas its given names were previously SlGDSL1 (Girard et al., 2012) and cutin synthase CD1 (Yeats et al., 2012b). In agreement with SGN, the gene was herein named SlGDSL2.
Sequencing the SlGDSL2 (Solyc11g006250) gene from the wild type and P15C12 plants revealed an A to T mutation disrupting the 3′ splice site of intron 4 (Fig. 7B). Because a mutated allele of SlGDSL2 was already described in tomato (Yeats et al., 2012b), the P15C12 mutant is thereafter named gdsl2-b. As in other introns belonging to the major class of introns processed by the U2 spliceosome, the splice site sequences of intron 4 from SlGDSL2 gene fit the canonical GT-AG consensus borders, where the nearly invariant GT dinucleotide is at the 5′ end and the AG dinucleotide at the 3′ end of the intron. Point mutations in this 3′ AG dinucleotide, which is essential to the definition of the 3′ splice junction, may lead to the production of mRNA with unspliced intron 4. Actually, missplicing of intron 4 of SlGDSL2 leads to the accumulation of several species of SlGDSL2 mRNA in both the P15C12 mutant and its F1 hybrid (Fig. 7D). The larger mRNA effectively corresponds to unspliced mRNA, as confirmed by sequencing. In-frame reading of intron 4 produces 13 additional incorrect residues after exon 4 (Fig. 7C), thus leading to the production of a truncated protein in which the 64 residue C-terminal region is missing. Interestingly, a second mRNA is produced whose size is consistent with that of correctly spliced SlGDSL2 mRNA. However, close examination of the mutated sequence reveals the presence of a cryptic 3′ splicing site in exon 5, 17 nucleotides downstream of the canonical 3′ consensus splice sequence of intron 4, which is predicted to be favored in the mutant (Human Splice Finder, http://www.umd.be/HSF/HSF.html). Indeed, sequencing the smaller-sized mRNA confirmed the alternative splicing at this site. The new open reading frame in exon 5 leads to the synthesis of eight new incorrect amino acids before truncation of the protein (Fig. 7C).
Real-time quantitative PCR analysis of SlGDSL2 expression in 20 DPA fruit revealed that accumulation of SlGDSL2 transcripts was reduced by approximately 96% in the P15C12 mutant compared with the wild type (Fig. 7E). Immunoblot analysis of SlGDSL2 protein in 20 DPA fruit epidermis failed to detect the protein in the epidermis from the gdsl2-b fruit, in contrast with the wild-type fruit (Fig. 7F), suggesting that the gdsl2-b mutant carries a strong hypomorphic or null allele of the SlGDSL2 gene, which is likely responsible for the cutin-deficiency phenotype observed in the P15C12 mutant line.
DISCUSSION
Considerable progress in research on cuticle synthesis and regulation has been made in recent years owing to the availability of Arabidopsis mutants and genomic tools. Main pathways for the synthesis of waxes and cuticle polymers have also been deciphered (Pollard et al., 2008; Bernard and Joubès, 2013) and increasing evidence on the mechanisms of transport of cuticular components and on the regulation of cuticle biosynthesis is now available (Yeats and Rose, 2013). Currently, the main challenges are to understand how cuticle properties are linked with cuticle composition and structure, how cuticle components interact with cell wall polymers, and how this will affect plant characteristics (e.g. plant growth or resistance to biotic and abiotic stresses) (Domínguez et al., 2011).
Although Arabidopsis remains the model of choice for plant functional genomics, this species is not well adapted for studying cuticle properties due to its very thin cuticle and some specificities (e.g. the high level of dicarboxylic acids in Arabidopsis cutin, unlike most other plants). By contrast, tomato fruit has a thick and easy-to-study cuticle synthesized along early fruit development (Yeats and Rose, 2013). Tomato has thus long been used for studying cuticle biomechanics and permeability (Schreiber, 2010; Domínguez et al., 2011) and has recently emerged as a new model for functional genomics of cuticle formation in plants. Because tomato is both a major crop species and a model for fleshy fruits, a wealth of information and genomic tools is now available for this species (Tomato Genome Consortium, 2012). In addition, several major agronomical traits in tomato and in other fleshy fruit species (e.g. fruit growth, visual aspect, cracking, water loss, resistance to pathogens, and postharvest shelf-life) are highly dependent on fruit cuticle (Bargel and Neinhuis, 2005; Saladié et al., 2007; Matas et al., 2009; Domínguez et al., 2011; Parsons et al., 2012). An increasing number of studies highlight the possibilities offered by tomato for analyzing cuticle architecture, mechanical properties, and permeability (López-Casado et al., 2007; Saladié et al., 2007; Mintz-Oron et al., 2008; Buda et al., 2009; Isaacson et al., 2009; Wang et al., 2011) and for discovering genes contributing to cuticle synthesis and regulation (Hovav et al., 2007; Mintz-Oron et al., 2008; Girard et al., 2012; Nadakuduti et al., 2012; Yeats et al., 2012b; Shi et al., 2013). Nevertheless, to further our understanding of the relationships between cuticle composition and architecture and cuticle properties and performance in plants, new tomato cuticle mutants are highly needed (Domínguez et al., 2011).
Tomato EMS Mutants for Studying Cuticle Composition and Properties
Collections of artificially induced genetic diversity resulting from fast-neutron or EMS mutagenesis are increasingly being used in tomato because generating saturated T-DNA collections remains out of reach in this species (Emmanuel and Levy, 2002). Although wild tomato species probably display larger variability in cuticle composition and architecture than cultivated tomato (Hovav et al., 2007; Yeats et al., 2012a), EMS mutants provide several interesting features. The genetic background is almost identical for all mutant lines generated in a given cultivar, except for the induced point mutations, therefore allowing direct comparison between lines. Because point mutations induced by EMS are distributed randomly over the whole genome, allelic series that include both loss-of-function and weak and strong hypomorphic alleles can be found for each target gene (Just et al., 2013). Tomato EMS mutant collections can be further exploited by the targeting-induced local lesions in genomes (TILLING) approach, which allows the discovery of unknown point mutations in known candidate genes (Okabe et al., 2011), or through forward genetic approaches aiming at identifying the mutation underlying the mutant trait. Less straightforward than TILLING, this approach relies first on the phenotypic characterization of the mutant trait and then on the identification of the mutation through map-based cloning, combined (or not) with the candidate gene approach or, more recently, through whole-genome sequencing (Abe et al., 2012; Just et al., 2013). Using this strategy, new cuticle-related genes involved in cuticle regulation (the cd2 HD-ZIP IV mutant; Isaacson et al., 2009), cutin monomer biosynthesis (the cyp86A9 mutant; Shi et al., 2013), and cutin polymerization (the cutin synthase mutant; Yeats et al., 2012b) were recently uncovered in tomato EMS mutant collections. However, when considering the large number of candidate genes involved in the regulation, synthesis, and transport processes necessary for cuticle formation (Matas et al., 2011; Yeats and Rose, 2013), very few tomato cuticle mutants have been described to date (Kimbara et al., 2012; Nadakuduti et al., 2012; Shi et al., 2013).
In this study, screening the highly mutagenized EMS mutant collection of 3,500 families generated in the miniature Micro-Tom cultivar (Just et al., 2013) revealed hundreds of mutants possibly affected in cuticle. Fruits from the selected mutants showed either strong fruit surface defects such as microcracks (russeting), cracks, strong shriveling, or peel browning, or less severe alterations such as increased water loss and altered fruit color or brightness (data not shown). Because strong cuticle alterations are detrimental to fruit quality, we preferred to focus in a first step on the phenotypic alterations responsible only for variations in fruit brightness (glossy or dull fruits), which likely arise from more subtle changes in cuticle properties. Pleiotropic mutations were also excluded because the origin of the cuticular defect is not always easy to trace (e.g. in fruit developmental mutants; Czerednik et al., 2012). The disadvantage of using fruit brightness as a screen for detecting cuticle mutations is that this trait can be very sensitive to environmental conditions such as the growing season (data not shown). However, of the 40 emutants examined (from the 274 originally found in the mutant database), more than 20 were further confirmed as being fruit brightness mutants. Among them, eight mutants displayed the glossy/dull trait for both fruit and leaf (Supplemental Table S1), indicating that fruit can be used as an attractive model for studying alterations of leaf cuticle.
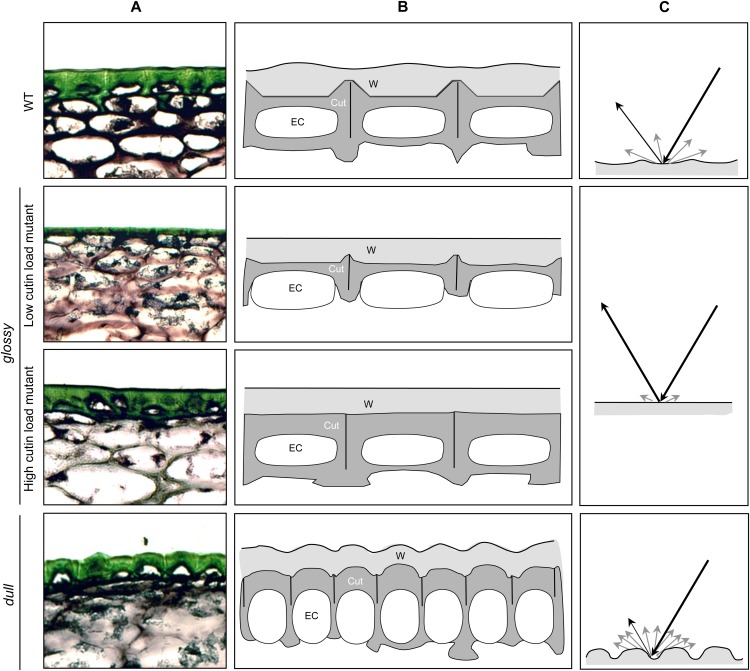
Our study clearly shows that although all selected brightness mutants displayed cuticle alterations, the glossy/dull fruit trait is not due to one single alteration. Recent studies of tomato mutants and transgenic lines established that increased fruit glossiness was associated with cutin deficiency (Isaacson et al., 2009; Girard et al., 2012; Nadakuduti et al., 2012; Shi et al., 2013). We indeed confirmed this relationship for several mutants (Fig. 4) but further showed that more complex cuticle architecture can be responsible for this trait. Most of the glossy mutants showed significant variations in either total cutin load or cutin composition (Table II). As shown in Supplemental Figure S2 and summarized in Figure 8, both low and high cutin load mutants may exhibit increased fruit glossiness. Incident light will be either reflected in a mirror-like manner (specular reflection) or reflected in a broad range of directions (diffuse reflection) due to surface irregularities and light scattering (e.g. by wax crystals; Pfündel et al., 2006). In wild-type tomato, the cutin layer is thicker at the junction of the epidermal pavement cells (anticlinal pegs), thinner elsewhere (Fig. 4D; Bargel and Neinhuis, 2005; Girard et al., 2012; Yeats et al., 2012a), and therefore presents surface irregularities (Fig. 4B). The cutin layer is further covered by epicuticular waxes mostly made up of aliphatic compounds, which form a film at the surface of the fruit (Vogg et al., 2004; Buschhaus and Jetter, 2011). Because of these characteristics, wild-type fruit is moderately glossy. In the high cutin load mutant, a thick cutin layer covers all cell surface and thus likely levels most irregularities resulting in a smooth surface, which can be seen in both native and de-waxed fruit (Fig. 4). This likely increases the specular reflection, hence giving a glossy aspect to the fruit. By contrast, the de-waxed surface of low cutin load mutants displays a very irregular aspect because of the presence of a thin to very thin cutin layer. The epicuticular wax film covering these small surface irregularities likely polishes the surface of the fruit, which therefore displays increased glossiness. By contrast, dull mutants present a highly irregular surface due to an increased number of small epidermal cells of different morphological aspects, as shown by ESEM (Fig. 4). The rugged aspect of the fruit surface, seen for both native and de-waxed fruit, is probably responsible for increased diffuse light reflection, hence giving the fruit a matte aspect (Fig. 8). The higher wax load of some mutants may also contribute to increased light scattering.
Figure 8.
Model of the influence of cuticle architecture on tomato fruit brightness. A, Light microscopy of fruit exocarp sections stained with toluidine blue. B, Schematic representation of the architecture of fruit epidermis in the wild type (WT), high and low cutin load glossy mutants, and dull mutants. Epidermal cells (EC) are encased in cutinized cell walls (Cut, in dark gray) covered by waxes (W, light gray). C, Light reflection on tomato fruit surface. Black lines indicate incident light and specular reflection. Light gray lines indicate diffuse reflection. [See online article for color version of this figure.]
No glossy tomato fruit mutants with reduced total wax load were observed in this study. In Arabidopsis, stem glossiness is generally indicative of altered epicuticular wax crystallization and is either due to general wax load reduction or to alteration of specific wax compounds (Jenks et al., 2002). Similar observations have been made in maize (Zea mays) and in rice (Oryza sativa) (Jenks et al., 2002; Islam et al., 2009). In addition, no organ fusion was observed among all the glossy/dull fruit mutants studied, in contrast with the glossy mutants described in Arabidopsis or the SlCER6 loss-of-function wax tomato mutant (Smirnova et al., 2013). Although the participation of wax in the glossy aspect of the fruit cannot be excluded (several glossy mutants display significant increases in wax load), our results strengthen the conclusion that the glossy/dull fruit appearance in our tomato mutants is not due to wax deficiency. However, the most striking result is that the dull fruit aspect of tomato cuticle mutants appears primarily due to different epidermal cell differentiation. The relationship between epidermal cell differentiation and cuticle development is now well established and genes controlling both cell morphogenesis and cuticle synthesis have been identified (Kurdyukov et al., 2006; Javelle et al., 2011; Oshima et al., 2013). More indirect effects may also arise from perturbations in fruit cell division and development that affect cuticle formation in tomato fruit (Czerednik et al., 2012).
In addition to fruit brightness, the tomato cuticle mutants identified should be instrumental for deciphering the bases of structural, mechanical, or water barrier properties of the cuticle (Schreiber, 2010; Domínguez et al., 2011) and their influence on fruit performance such as resistance to cracking, drought, and pathogens. For example, the water loss properties of tomato fruit cuticle are largely affected by strong cutin alterations, as in the gdsl2-b mutant, although the associated modifications in cuticle structure have not been studied in detail (Isaacson et al., 2009; Girard et al., 2012; Yeats et al., 2012b; Fig. 6). Likewise, the high proportion of cyclic triterpenoids found in the wax-rich glossy (P4E2) or dull (P6D6) mutants may modify the water loss properties of the fruit (Vogg et al., 2004; Buschhaus and Jetter, 2012).
Cuticle Composition Analysis of EMS Mutants as a Tool for Discovering Genes and Pathways
Insights into cuticle composition may provide some hints on the regulations or biochemical pathways affected in the various cuticle mutants. Before undertaking allelism tests by crossing the various mutants, as done for the wax glaucous mutants in Sorghum (Peters et al., 2009), we first analyzed mutants for their cutin and wax composition (Tables II and III). Indeed, several groups of mutants displayed remarkably similar cuticle characteristics suggesting the involvement of the mutated genes in related biochemical pathways (e.g. in the cutin-deficient mutants P15C12 and P23F12) or in their regulation (e.g. in the wax- and cutin-rich mutants P18H8 and P30A12). These groups may help to discover new genes and functions related to poorly known aspects of cuticle formation. In addition, as discussed below for the gdsl lipase gene, these mutants may help uncover new alleles of known candidate genes such as those involved in cutin formation or in the coordinated regulation of wax and cutin biosynthesis (Yeats and Rose, 2013). Finally, these groups may also reveal genes involved in suberin biosynthesis. In the P5E1 and P26E8 mutants, the accumulation in cutin fraction of dicarboxylic acids, which are considered as markers for suberin, and the associated visual changes (Supplemental Figure S6) may be indicative of the constitutive up-regulation of suberin biosynthetic pathway in these mutants. Indeed, the C18:1 unsaturated dicarboxylic acid content of P5E1 and P26E8 tomato fruit cuticle (approximately 99 µg cm−2; Supplemental Table S3) is close to that of potato (Solanum tuberosum) tuber periderm (approximately 80 to 100 µg cm−2; Serra et al., 2009). Because both species are closely related, these mutants may thus prove very interesting for investigating the relationships between cutin and suberin within the Solanaceae. Although both partially share a common biosynthetic pathway, suberin normally accumulates in the fruit only after wounding and in other stressful conditions.
The next step is the identification of the mutations underlying the mutant cuticle traits. Identification of unknown mutations in tomato classically proceeds through map-based cloning by crossing the S. lycopersicum mutant with a wild-related species such as S. pimpinellifolium (Isaacson et al., 2009; Yeats et al., 2012b). To reduce the inherent high genetic and phenotypic variability in the offspring of such a cross, we used as parental line a dwarf mutant of the cultivated M82 genotype, taking advantage of the development of SNP markers polymorphic between M82 and ‘Micro-Tom’ (Shirasawa et al., 2010). The combination of genetic mapping of the mutation and candidate gene approach resulted in the identification of a new hypomorphic or null allele of the SlGDSL2 gene responsible for a strong cutin-deficient mutation. Sequencing of the SlGDSL2 gene from the wild type and P15C12 plants revealed an A to T mutation disrupting the 3′ splice site of intron 4, which leads to the accumulation of unspliced mRNA and to mRNA resulting from the utilization of a cryptic 3′ splicing site in exon 5 and, in both cases, to truncated SlGDSL2 proteins. Thus, the evidence presented here indicates that SlGDSL2 mutant plants are not able to accumulate correctly spliced mRNA in the fruit epidermis. In both scenarios (no-splicing or missplicing of intron 4), the presence of premature translation-termination codons in the mRNAs would result in the generation of truncated GDSL lipases lacking the C-terminal region. This region has an essential role in SlGDSL2 activity because it harbors two residues of the GDSL lipase catalytic triad: the Asp (d-321) and the His (H-326; Akoh et al., 2004). Truncated proteins devoid of GDSL lipase enzymatic activity could therefore have dominant-negative effects on cutin formation (e.g. by interfering with a cutin-assembly machinery involving SlGDSL2). Actually, the very low SlGDSL2 transcript level and the absence of immuno-detected SlGDSL2 protein in gdsl2-b fruit epidermis (Fig. 7F) strongly suggest that the nonsense-mediated mRNA decay, a quality control mechanism present in plants and other eukaryotes that recognizes and degrades aberrant mRNA harboring premature translation-termination codons (Stalder and Mühlemann, 2008), prevents the production of truncated SlGDSL2 proteins that could possibly have detrimental effects on the plant.
Interestingly, one of the three fruit cutin-deficiency (cd1) mutants identified in the M82 tomato mutant collection (Menda et al., 2004; Isaacson et al., 2009) was recently identified as a SlGDSL2 truncation mutant (Yeats et al., 2012b). The discovery of SlGDSL2 mutations by two independent screenings of unrelated tomato mutant collections in M82 and ‘Micro-Tom’ is probably not a mere coincidence. SlGDSL2 was a likely candidate for cutin assembly in the cuticle due to its properties and its high and specific expression in tomato fruit epidermis (Lemaire-Chamley et al., 2005; Reina et al., 2007; Mintz-Oron et al., 2008; Matas et al., 2011). Reverse genetics approaches based on both screening of mutant collections in processing tomato and RNA interference–silencing strategy in cherry tomato (Girard et al., 2012; Yeats et al., 2012b) recently proved that SlGDSL2 plays a key role as an acyl-transferase for the assembly of monomers to form the cutin polyester in the cuticle (Yeats and Rose, 2013). The new gdsl2 allelic variant described in this study will aid in investigating the role of the GDSL lipase in planta by allowing comparison of GDSL function in cutin polymerization in different genetic backgrounds (cultivated tomato S. lycopersicum, M82, and ‘Micro-Tom’; cherry tomato S. lycopersicum var cerasiformae Wva 106).
In conclusion, tomato mutants characterized herein or present in tomato EMS mutant collections constitute rich sources of genetic variability for exploring cuticle properties and for identifying new genes and alleles involved in cuticle formation. For example, several other strong cutin deficiency mutants identified in this study do not map with other known candidate genes. The recessive P23F12 mutation maps to chromosome 9 unlike other mutations identified to date in tomato (Supplemental Figure S9). The P30B6 mutation, which maps in a different location to chromosome 9, displays a different inheritance pattern (overdominance; data not shown). These results open the way for the discovery of new genes involved in cutin formation in tomato. The continual technological and scientific advances in tomato genomics and in cuticle research now greatly facilitate the identification of the mutations underlying the mutant traits. In addition to traditional map-based cloning (Isaacson et al., 2009), the combination of genetic mapping and candidate gene approaches, which are already proven successful (this study and Shi et al., 2013), can be further extended to the identification of the most promising target genes by RNA sequencing analysis of cuticle fruit mutants. Another very promising approach is the identification of the mutation by whole-genome sequencing (Abe et al., 2012; Just et al., 2013), which only involves crosses between wild-type ‘Micro-Tom’ and mutant ‘Micro-Tom’ and therefore excludes all other undesirable variations of other cuticle compounds that could alter cuticle properties.
MATERIALS AND METHODS
Plant Materials
Fruit brightness mutants were isolated from an EMS mutant tomato (Solanum lycopersicum) collection generated in the miniature cv Micro-Tom at the Institut National de la Recherche Agronomique as previously described (Rothan and Causse 2007; Just et al., 2013). Plants were grown at a density of 200 plants per square meter in a greenhouse under a 14-h light/10-h dark cycle, 18°C to 28°C temperature, and 60% to 80% relative humidity. Plants were watered twice a week with solution 1 [pH 5.8, oligoelements plus 3.5 mm KNO3, 1 mm K2SO4, 2 mm KH2PO4, 6 mm Ca(NO3)2, 2 mm MgSO4] until first fruit set on the first truss, and then with solution 2 [pH 5.8, oligoelements plus 4 mm KNO3, 1.5 mm K2SO4, 1.5 mm KH2PO4, 4 mm Ca(NO3)2, 1.5 mm MgSO4] until fruits were ripe. Flowers were regularly vibrated to ensure optimal self-pollination and thus fruit development. Seeds were treated with 4% calcium hypochlorite for 15 min, rinsed for 30 min, air-dried for 2 d, and stored at 4°C with a desiccant until use.
Genetic Mapping
A mapping F2 population of 110 plants was created by crossing the P15C12 Micro-Tom glossy mutant with a dwarf mutant from the M82 cultivar EMS population (Menda et al., 2004). Plants from the mapping population were grown in greenhouse in the conditions indicated above, except that plant density was 21 plants per square meter and plants were watered 3 times a week. Cuticle-associated traits were measured at two developmental stages of the fruit, first at the MG stage (brightness and permeability) and then at the RR stage (brightness, water loss, width of the cutin layer surrounding fruit epidermal cells). Genomic DNA was extracted from young leaves of each plant by a cetyl-trimethyl-ammonium bromide (CTAB) method. Five-hundred microliters of CTAB extraction buffer (145 mM sorbitol, 850 mm NaCl, 125 mm Tris-HCl [pH 8.0], 25 mm EDTA, 0.8% [w/v] Sarcosyl [n-lauroyl-sarcosine], 0.8% [w/v] CTAB) was added with 2.2 mg of sodium metabisulfite per sample, incubated at 65°C for 90 min, and 450 µl of chloroform:isoamyl alcohol (24:1) was added to each sample, mixed by inversion and centrifuged for 10 min at 4500g. Subsequently, the supernatant was transferred to a fresh tube and treated with 500 µl of cold isopropanol. DNA was dried 15 min at 45°C under vacuum, suspended in 50 µl of distilled water and samples were treated with ribonuclease. Primer sets of 48 SNP markers well spread along the genome and displaying polymorphism between the ‘Micro-Tom’ and the ‘M82’ (Shirasawa et al., 2010) were chosen for genotyping each plant. Genotype and phenotype data were analyzed using MapMaker/Exp v3.0 (Lander et al., 1987), MapMaker/QTL v1.1 (Paterson et al., 1988), and QTL Cartographer (Basten et al., 2002) software.
Fruit Brightness Assessment
Fruit brightness of the mutant lines was visually estimated at the MG and RR (Breaker stage plus 7 d) stages by comparison with wild-type ‘Micro-Tom’ plants grown side-by-side and disseminated randomly among mutant lines. Mutants differing from wild-type ‘Micro-Tom’ were classified as glossy mutants (i.e. fruits with homogeneous glossy aspect) or as dull mutants (i.e. those with no light reflect on the fruit). An important criterion for classifying mutants as fruit brightness mutants was that all fruits on the plant displayed the same phenotype. However, several selected mutant lines carried both glossy and dull fruits, which were clearly different from the wild-type fruits. Analysis of fruit brightness in the segregating P15C12 Micro-Tom mutant × M82 dwarf population was done similarly except that fruit brightness was estimated by comparison of all genotypes to each other and that genotypes were further classified in three brightness groups.
Water Loss and Cuticle Permeability Measurements
For water loss measurements, one RR fruit was harvested from each plant of the F2 mapping population, including the parents. Fruit sealing wax was then applied on stalks and fruits were stored at room temperature. Fruit fresh weight was recorded at time zero and each week, until 6 weeks. Water loss was calculated as a percentage of weight loss. For measurements of cuticle permeability to stain, one MG fruit was collected from each plant of the F2 mapping population and from the parents, and dropped in 1% toluidine blue solution during 6 h as described in Tanaka et al. (2004).
Cutin Monomer and Wax Analysis
Cuticular waxes were extracted by fruit immersion during 30 s in 6 mL of chloroform containing 6 µg of docosane, as internal standard. Extracts were dried under moderate nitrogen flux and lipids were derivatized 15 min at 110°C in 100 µL BSTFA-TMCS (N,O-bis[trimethylsilyl trifluoroacetamide]: trimethyl chlorosilane [99:1]). The BSTFA-TMCS was dried under nitrogen and samples were suspended in 500 µL of hexane. Wax was then analyzed and quantified as previously described (Bourdenx et al., 2011).
For the cutin monomer analysis, a 1-cm diameter disc was cut off from a RR fruit epidermis, carefully scratched with a scalpel blade in order to remove exocarp cells, and incubated 30 min in isopropanol at 85°C. The disc was then delipidated by successive deeps in chloroform/methanol (C/M), at different ratios: C/M (2:1) for 24 h, C/M (1:1) for 24 h, C/M (1:2) for 24 h, and 100% methanol for 24 h. The delipidated epidermis disc was dried for 48 h under pulsed air and for 48 h in a desiccator. Cutin was then depolymerized, analyzed, and quantified as previously described (Domergue et al., 2010).
Light Microscopy
For cuticle thickness measurements, fruit exocarp (including cuticle) was obtained from two independent RR fruits of the wild type and mutant lines. Samples were fixed and embedded in paraffin as previously described (Mounet et al., 2009). Eight-micrometer slides of exocarp were stained using saturated and filtered Sudan Red solution in ethanol. Mean cutin thickness was assessed from 60 measurements.
For analyzing the thickness of the cutin layer between two adjacent fruit epidermal cells (thereafter called cutin width) in the mapping population, a 10-mm2 square tissue sample was peeled off from one RR fruit harvested from each plant of the F2 mapping population, including the parents. As many epidermal cells as possible were then removed from the peel by scratching the internal surface with a scalpel blade. The resulting cuticle-enriched fragment was immerged in water, placed flatten on a glass slide, and observed under optical microscope at ×20 magnification. Cutin width was then determined by measuring thickness of the cutinized cell wall between two adjacent epidermal cells (Supplemental Figure S6). Mean cutin width was assessed from 12 measurements.
ESEM
For the observation of wax on the surface of the fruit, 5-mm side cubes of exocarp including cuticle were excised from the equatorial part of RR fruits and placed directly into the observation chamber of a FEI Quanta 200 scanning electronic microscope, in environmental mode. For cutin observation, cubes were submerged 30 s into chloroform beforehand, with gentle agitation. Observations were performed with a plate at 4°C, a 6-Tor pressure in the chamber, and a 4.7-V voltage applied to the filament. Pictures were captured with ×600 and ×3000 magnifications.
PCR and Quantitative Reverse-Transcription PCR Analysis of SlGDSL2
Genomic DNA was extracted from 100 mg fresh weight of the wild type and P15C12 mutant leaves with Plant DNAzol reagent (Invitrogen), further amplified using four primer pairs (GDSL 1F/1R, GDSL 2F/2R, GDSL 3F/3R, GDSL 4F/4R) and sequenced for determination of allelic sequences of SlGDSL2 and gdsl2-b. Primer sequences are indicated in Supplemental Figure S10.
The cDNA sizing was performed after a total RNA extraction from 100 mg fresh weight and a reverse transcription, as previously described (Mounet et al., 2009; Girard et al., 2012). The amplification was performed using GDSL2-cDNA4F and GDSL2-cDNA5R primers. Primers sequences are indicated in Supplemental Figure S10.
SlGDSL2 expression was analyzed from bulks of plants segregating for both the fruit brightness and the gdsl2-b mutation in the F2 mapping population. The wild-type ‘Micro-Tom’, the ‘M82dwarf’, and the P15C12 (gdsl2-b) parental lines were included in the analysis. The glossy fruit bulk (homozygous for the mutant gdsl2-b allele) was made up of 21 RR fruits collected from 7 plants (3 fruits per plant). On each fruit, two discs of 1-cm diameter were collected and carefully scratched with a scalpel blade. Three pooled samples were then generated by distributing two discs per plant in each pool (i.e. 14 discs per pooled sample in total). The same strategy was used for the wild-type–like bulk (SlGDSL2 allele) except that the three pooled samples were prepared from 24 RR fruits collected on 4 plants (i.e. 16 discs per pooled sample in total). Samples were ground in liquid nitrogen and stored at −80°C until RNA extraction as previously described (Mounet et al., 2009). PCR analysis was performed on reverse-transcribed cDNAs as previously described (Girard et al., 2012), using primers GDSL2-53 and GDSL2-34. Primer sequences are indicated in Supplemental Figure S10.
SlGDSL2 Immunoblot Analysis
Four independent 15 DPA fruit pools were constituted for both the wild type and gdsl2-b mutant line P15C12. Each pool was composed of the outer epidermis from three fruits, ground, and lyophilized. Antibody and immunoblot analyses were as previously described (Girard et al., 2012).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of fruit brightness from selected glossy and dull mutants at RR stage.
Supplemental Figure S2. Comparison of the fruit brightness of mutants that showed the most extreme wax and cutin compositions.
Supplemental Figure S3. Variables factor map for cutin components, after a centered PCA analysis.
Supplemental Figure S4. Individual factor map after PCA of the wax and cutin compositions.
Supplemental Figure S5. Variables factor map for wax and cutin components, after a centered PCA analysis.
Supplemental Figure S6. Photograph of fruits from the P26E8 mutant showing suberin deposition at the distal end of the fruit.
Supplemental Figure S7. Microscopic observation of fresh pealed outer epidermis from RR fruit.
Supplemental Figure S8. Phenotyping the F2 progeny from the cross M82d × P15C12 .
Supplemental Figure S9. Mapping of the P23F12 glossy mutant.
Supplemental Figure S10. Nucleotide sequences of the primers used in this study.
Supplemental Table S1. Comparison of the plant and fruit brightness data obtained from the MicroTom Mutant Database and from the subsequent observation of the 40 selected families grown in the greenhouse.
Supplemental Table S2. Mean density of fruit epidermal cells in the indicated mutants.
Supplemental Table S3. Cutin and wax composition of fruit cuticle from the wild type and four selected tomato mutants.
Acknowledgments
The authors thank Michel Hernould for fruit cytology analyses, Aurélie Petit for her help in statistical analyses, Isabelle Atienza for taking care of the plants in the greenhouse, and Isabelle Svahn for her participation. The ESEM experiments were performed at the Electronic Imaging pole of the Bordeaux Imaging Center, and lipid analyses were performed at the Bordeaux Metabolome Facility.
Glossary
- VLCFA
very long chain fatty acid
- EMS
ethyl methanesulfonate
- RR
Red Ripe
- DPA
d post-anthesis
- PCA
principal component analysis
- HCA
hierarchical clustering analysis
- PC1
first principal component
- PC2
second principal component
- ESEM
environmental scanning electron microscopy
- MG
Mature Green
- SNP
single nucleotide polymorphism
- LOD
log of the odds
- QTL
quantitative trait loci
- CTAB
cetyl-trimethyl-ammonium bromide
- C/M
chloroform/methanol
References
- Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, et al. (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30: 174–178 [DOI] [PubMed] [Google Scholar]
- Adato A, Mandel T, Mintz-Oron S, Venger I, Levy D, Yativ M, Domínguez E, Wang Z, De Vos RC, Jetter R, et al. (2009) Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genet 5: e1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. (2004) GDSL family of serine esterases/lipases. Prog Lipid Res 43: 534–552 [DOI] [PubMed] [Google Scholar]
- Bargel H, Neinhuis C. (2005) Tomato (Lycopersicon esculentum Mill.) fruit growth and ripening as related to the biomechanical properties of fruit skin and isolated cuticle. J Exp Bot 56: 1049–1060 [DOI] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng Z-B (2002) QTL Cartographer, Version 1.16. Department of Statistics, North Carolina State University, Raleigh, NC. [Google Scholar]
- Beisson F, Li-Beisson Y, Pollard M. (2012) Solving the puzzles of cutin and suberin polymer biosynthesis. Curr Opin Plant Biol 15: 329–337 [DOI] [PubMed] [Google Scholar]
- Bernard A, Joubès J. (2013) Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog Lipid Res 52: 110–129 [DOI] [PubMed] [Google Scholar]
- Bourdenx B, Bernard A, Domergue F, Pascal S, Léger A, Roby D, Pervent M, Vile D, Haslam RP, Napier JA, et al. (2011) Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol 156: 29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 101: 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buda GJ, Isaacson T, Matas AJ, Paolillo DJ, Rose JKC. (2009) Three-dimensional imaging of plant cuticle architecture using confocal scanning laser microscopy. Plant J 60: 378–385 [DOI] [PubMed] [Google Scholar]
- Buschhaus C, Jetter R. (2011) Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J Exp Bot 62: 841–853 [DOI] [PubMed] [Google Scholar]
- Buschhaus C, Jetter R. (2012) Composition and physiological function of the wax layers coating Arabidopsis leaves: β-amyrin negatively affects the intracuticular water barrier. Plant Physiol 160: 1120–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XB, Goodwin SM, Boroff VL, Liu XL, Jenks MA. (2003) Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerednik A, Busscher M, Bielen BA, Wolters-Arts M, de Maagd RA, Angenent GC. (2012) Regulation of tomato fruit pericarp development by an interplay between CDKB and CDKA1 cell cycle genes. J Exp Bot 63: 2605–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue F, Vishwanath SJ, Joubès J, Ono J, Lee JA, Bourdon M, Alhattab R, Lowe C, Pascal S, Lessire R, et al. (2010) Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol 153: 1539–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez E, Cuartero J, Heredia A. (2011) An overview on plant cuticle biomechanics. Plant Sci 181: 77–84 [DOI] [PubMed] [Google Scholar]
- Emmanuel E, Levy AA. (2002) Tomato mutants as tools for functional genomics. Curr Opin Plant Biol 5: 112–117 [DOI] [PubMed] [Google Scholar]
- Franke RB, Dombrink I, Schreiber L. (2012) Suberin goes genomics: use of a short living plant to investigate a long lasting polymer. Front Plant Sci 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard AL, Mounet F, Lemaire-Chamley M, Gaillard C, Elmorjani K, Vivancos J, Runavot JL, Quemener B, Petit J, Germain V, et al. (2012) Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell 24: 3119–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav R, Chehanovsky N, Moy M, Jetter R, Schaffer AA. (2007) The identification of a gene (Cwp1), silenced during Solanum evolution, which causes cuticle microfissuring and dehydration when expressed in tomato fruit. Plant J 52: 627–639 [DOI] [PubMed] [Google Scholar]
- Isaacson T, Kosma DK, Matas AJ, Buda GJ, He Y, Yu B, Pravitasari A, Batteas JD, Stark RE, Jenks MA, et al. (2009) Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. Plant J 60: 363–377 [DOI] [PubMed] [Google Scholar]
- Islam MA, Du H, Ning J, Ye H, Xiong L. (2009) Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol Biol 70: 443–456 [DOI] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Rogowsky PM, Ingram GC. (2011) Epidermis: the formation and functions of a fundamental plant tissue. New Phytol 189: 17–39 [DOI] [PubMed] [Google Scholar]
- Jenks MA, Eigenbrode SD, Lemieux B. (2002) Cuticular waxes of Arabidopsis. The Arabidopsis Book 1: e0016, 10.1199/tab.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just D, Garcia V, Fernandez L, Bres C, Mauxion J, Petit J, Jorly J, Assali J, Bournonville C, Ferrand C, et al. (2013) Micro-Tom mutants for functional analysis of target genes and discovery of new alleles in tomato. Plant Biotechnol 30: 225–231 [Google Scholar]
- Kimbara J, Yoshida M, Ito H, Hosoi K, Kusano M, Kobayashi M, Ariizumi T, Asamizu E, Ezura H. (2012) A novel class of sticky peel and light green mutations causes cuticle deficiency in leaves and fruits of tomato (Solanum lycopersicum). Planta 236: 1559–1570 [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels L. (2009) Plant cuticles shine: advances in wax biosynthesis and export. Curr Opin Plant Biol 12: 721–727 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Nawrath C, Bär S, Voisin D, Efremova N, Franke R, Schreiber L, Saedler H, Métraux JP, et al. (2006) The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA. (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Lemaire-Chamley M, Petit J, Garcia V, Just D, Baldet P, Germain V, Fagard M, Mouassite M, Cheniclet C, Rothan C. (2005) Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiol 139: 750–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F. (2009) Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA 106: 22008–22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Casado G, Matas AJ, Domínguez E, Cuartero J, Heredia A. (2007) Biomechanics of isolated tomato (Solanum lycopersicum L.) fruit cuticles: the role of the cutin matrix and polysaccharides. J Exp Bot 58: 3875–3883 [DOI] [PubMed] [Google Scholar]
- Matas AJ, Gapper NE, Chung MY, Giovannoni JJ, Rose JK. (2009) Biology and genetic engineering of fruit maturation for enhanced quality and shelf-life. Curr Opin Biotechnol 20: 197–203 [DOI] [PubMed] [Google Scholar]
- Matas AJ, Yeats TH, Buda GJ, Zheng Y, Chatterjee S, Tohge T, Ponnala L, Adato A, Aharoni A, Stark R, et al. (2011) Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23: 3893–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D. (2004) In silico screening of a saturated mutation library of tomato. Plant J 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, et al. (2008) Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol 147: 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounet F, Moing A, Garcia V, Petit J, Maucourt M, Deborde C, Bernillon S, Le Gall G, Colquhoun I, Defernez M, et al. (2009) Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiol 149: 1505–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadakuduti SS, Pollard M, Kosma DK, Allen C, Jr, Ohlrogge JB, Barry CS. (2012) Pleiotropic phenotypes of the sticky peel mutant provide new insight into the role of CUTIN DEFICIENT2 in epidermal cell function in tomato. Plant Physiol 159: 945–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C. (2006) Unraveling the complex network of cuticular structure and function. Curr Opin Plant Biol 9: 281–287 [DOI] [PubMed] [Google Scholar]
- Okabe Y, Asamizu E, Saito T, Matsukura C, Ariizumi T, Brès C, Rothan C, Mizoguchi T, Ezura H. (2011) Tomato TILLING technology: development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol 52: 1994–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M. (2013) MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25: 1609–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons EP, Popopvsky S, Lohrey GT, Lü S, Alkalai-Tuvia S, Perzelan Y, Paran I, Fallik E, Jenks MA. (2012) Fruit cuticle lipid composition and fruit post-harvest water loss in an advanced backcross generation of pepper (Capsicum sp.). Physiol Plant 146: 15–25 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Lander ES, Hewitt JD, Peterson S, Lincoln SE, Tanksley SD. (1988) Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 335: 721–726 [DOI] [PubMed] [Google Scholar]
- Peters PJ, Jenks MA, Rich PJ, Axtell JD, Ejeta G. (2009) Mutagenesis, selection, and allelic analysis of epicuticular wax mutants in sorghum. Crop Sci 49: 1250–1258 [Google Scholar]
- Pfündel EE, Agati G, Cerovic ZG. (2006) Optical properties of plant surfaces. Annu Plant Rev 23: 219–249 [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Reina JJ, Guerrero C, Heredia A. (2007) Isolation, characterization, and localization of AgaSGNH cDNA: a new SGNH-motif plant hydrolase specific to Agave americana L. leaf epidermis. J Exp Bot 58: 2717–2731 [DOI] [PubMed] [Google Scholar]
- Rothan C, Causse M. (2007) Natural and artificially induced genetic variability in crop and model plant species for plant systems biology. EXS 97: 21–53 [DOI] [PubMed] [Google Scholar]
- Saladié M, Matas AJ, Isaacson T, Jenks MA, Goodwin SM, Niklas KJ, Xiaolin R, Labavitch JM, Shackel KA, Fernie AR, et al. (2007) A reevaluation of the key factors that influence tomato fruit softening and integrity. Plant Physiol 144: 1012–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauveplane V, Kandel S, Kastner PE, Ehlting J, Compagnon V, Werck-Reichhart D, Pinot F. (2009) Arabidopsis thaliana CYP77A4 is the first cytochrome P450 able to catalyze the epoxidation of free fatty acids in plants. FEBS J 276: 719–735 [DOI] [PubMed] [Google Scholar]
- Schreiber L. (2010) Transport barriers made of cutin, suberin and associated waxes. Trends Plant Sci 15: 546–553 [DOI] [PubMed] [Google Scholar]
- Serra O, Soler M, Hohn C, Sauveplane V, Pinot F, Franke R, Schreiber L, Prat S, Molinas M, Figueras M. (2009) CYP86A33-targeted gene silencing in potato tuber alters suberin composition, distorts suberin lamellae, and impairs the periderm’s water barrier function. Plant Physiol 149: 1050–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JX, Adato A, Alkan N, He Y, Lashbrooke J, Matas AJ, Meir S, Malitsky S, Isaacson T, Prusky D, et al. (2013) The tomato SlSHINE3 transcription factor regulates fruit cuticle formation and epidermal patterning. New Phytol 197: 468–480 [DOI] [PubMed] [Google Scholar]
- Shirasawa K, Isobe S, Hirakawa H, Asamizu E, Fukuoka H, Just D, Rothan C, Sasamoto S, Fujishiro T, Kishida Y, et al. (2010) SNP discovery and linkage map construction in cultivated tomato. DNA Res 17: 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova A, Leide J, Riederer M. (2013) Deficiency in a very-long-chain fatty acid β-ketoacyl-coenzyme a synthase of tomato impairs microgametogenesis and causes floral organ fusion. Plant Physiol 161: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Mühlemann O. (2008) The meaning of nonsense. Trends Cell Biol 18: 315–321 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y. (2004) A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J 37: 139–146 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogg G, Fischer S, Leide J, Emmanuel E, Jetter R, Levy AA, Riederer M. (2004) Tomato fruit cuticular waxes and their effects on transpiration barrier properties: functional characterization of a mutant deficient in a very-long-chain fatty acid beta-ketoacyl-CoA synthase. J Exp Bot 55: 1401–1410 [DOI] [PubMed] [Google Scholar]
- Wang Z, Guhling O, Yao R, Li F, Yeats TH, Rose JKC, Jetter R. (2011) Two oxidosqualene cyclases responsible for biosynthesis of tomato fruit cuticular triterpenoids. Plant Physiol 155: 540–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Buda GJ, Wang Z, Chehanovsky N, Moyle LC, Jetter R, Schaffer AA, Rose JK. (2012a) The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. Plant J 69: 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Martin LB, Viart HM, Isaacson T, He Y, Zhao L, Matas AJ, Buda GJ, Domozych DS, Clausen MH, et al. (2012b) The identification of cutin synthase: formation of the plant polyester cutin. Nat Chem Biol 8: 609–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JK. (2013) The formation and function of plant cuticles. Plant Physiol 163: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]