A carbon-nitrogen lyase from the tree legume Leucaena leucocephala degrades the toxin mimosine.
Abstract
The tree legume Leucaena leucocephala contains a large amount of a toxic nonprotein aromatic amino acid, mimosine, and also an enzyme, mimosinase, for mimosine degradation. In this study, we isolated a 1,520-bp complementary DNA (cDNA) for mimosinase from L. leucocephala and characterized the encoded enzyme for mimosine-degrading activity. The deduced amino acid sequence of the coding region of the cDNA was predicted to have a chloroplast transit peptide. The nucleotide sequence, excluding the sequence for the chloroplast transit peptide, was codon optimized and expressed in Escherichia coli. The purified recombinant enzyme was used in mimosine degradation assays, and the chromatogram of the major product was found to be identical to that of 3-hydroxy-4-pyridone (3H4P), which was further verified by electrospray ionization-tandem mass spectrometry. The enzyme activity requires pyridoxal 5′-phosphate but not α-keto acid; therefore, the enzyme is not an aminotransferase. In addition to 3H4P, we also identified pyruvate and ammonia as other degradation products. The dependence of the enzyme on pyridoxal 5′-phosphate and the production of 3H4P with the release of ammonia indicate that it is a carbon-nitrogen lyase. It was found to be highly efficient and specific in catalyzing mimosine degradation, with apparent Km and Vmax values of 1.16 × 10−4 m and 5.05 × 10−5 mol s−1 mg−1, respectively. The presence of other aromatic amino acids, including l-tyrosine, l-phenylalanine, and l-tryptophan, in the reaction did not show any competitive inhibition. The isolation of the mimosinase cDNA and the biochemical characterization of the recombinant enzyme will be useful in developing transgenic L. leucocephala with reduced mimosine content in the future.
Leucaena leucocephala is an important agroforestry tree legume of the tropics, and its foliage can be used as a protein-rich fodder (Garcia et al., 1996; Soedarjo and Borthakur, 1998). L. leucocephala is highly tolerant to drought (Shelton and Brewbaker, 1994) and resistant to many pests and diseases. The protein-rich foliage and tolerance to various abiotic and biotic stresses make L. leucocephala a promising legume for use as a fodder. In spite of these desirable attributes, the use of L. leucocephala as a fodder is rather limited because its foliage also contains an N-heterocyclic nonprotein amino acid, known as mimosine, which is toxic to both prokaryotic cells (Soedarjo et al., 1994) and eukaryotic cells (Lalande, 1990). Mimosine inactivates a variety of enzymes either by chelating bivalent metallic ions and thereby limiting their availability for use as cofactors by several metallic ion-dependent enzymes, such as ribonucleotide reductase, alkaline phosphatase, and dopamine β-hydroxylase (Chang, 1960; Hashiguchi and Takahashi, 1977; Dai et al., 1994), or by forming a stable complex with pyridoxal-5′-phosphate (PLP), leading to the inactivation of PLP-dependent enzymes, such as cystathionine synthetase, cystathionase, Asp-Glu transaminase, Tyr decarboxylase, tyrosinase, and l-dopa decarboxylase (Crounse et al., 1962; Lin et al., 1962, 1963; Hylin, 1969). The inactivation of important enzymes by mimosine causes various physiological abnormalities, including enlarged thyroid glands, infertility, birth defects, and loss of hairs (Crounse et al., 1962; Hamilton et al., 1968; Joshi, 1968; Dewreede and Wayman, 1970; Reis et al., 1975; Jones et al., 1976).
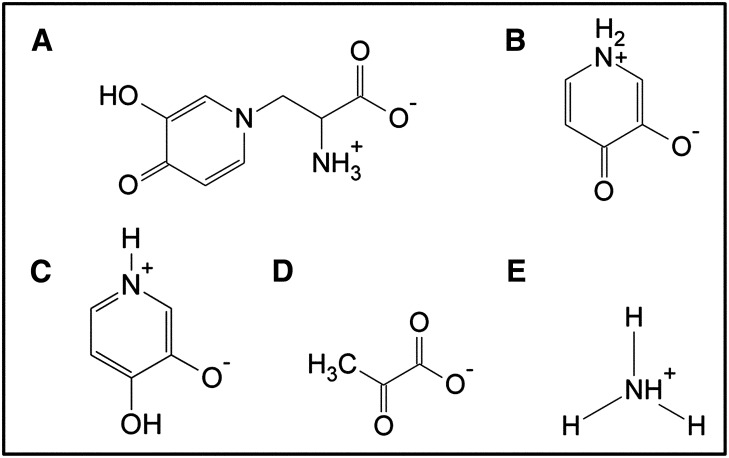
Mimosine is abundant in all parts of L. leucocephala, and on a dry weight basis, L. leucocephala leaves contain approximately 5% mimosine (Soedarjo and Borthakur, 1998). Such high mimosine content in the foliage indicates that mimosine may have some functional role in the plant. Previously, mimosine has been shown to inhibit DNA synthesis in many DNA viruses by chelating iron required by ribonucleotide reductase (Dai et al., 1994), suggesting its role in defense against virus attacks. Besides this, other possible roles of mimosine in L. leucocephala are not well established. Considering its biochemical properties of inactivating various enzymes that require either bivalent metallic ions or PLP as cofactors, mimosine may have a role in plant defense, and based on its chemical composition, it may serve as a reservoir of carbon and nitrogen for survival and growth under nutrient-limiting conditions. But the utilization of mimosine as a source of carbon and nitrogen is possible only if the plant has specific enzymes to catabolize it. Interestingly, the presence of such mimosine-degrading enzymes has been reported from seedling extracts of L. leucocephala and Mimosa pudica, another mimosine-containing plant (Suda, 1960; Smith and Fowden, 1966). Smith and Fowden (1966) identified the mimosine-degrading enzyme from L. leucocephala seedling extracts as a carbon-nitrogen (C-N) lyase that converted mimosine into 3,4-dihydroxypyridine (3,4DHP), pyruvic acid, and ammonia (Fig. 1). Additionally, a mimosine-degrading enzyme, mimosinase, was purified from L. leucocephala leaves (Tangendjaja et al., 1986) and was found to degrade mimosine into 3-hydroxy-4-pyridone (3H4P; Fig. 1). However, the genes encoding the mimosine-degrading enzymes from L. leucocephala have not been isolated and characterized.
Figure 1.
Chemical structures of mimosine (A), 3H4P (B), 3,4DHP (C), pyruvate (D), and ammonium (E).
The goals of this study were to isolate complementary DNA (cDNA) for a mimosine-degrading enzyme from L. leucocephala and to determine the biochemical and kinetic properties of the encoded enzyme. This will help us to understand roles of mimosine and mimosine-degrading enzymes in L. leucocephala. Additionally, it may be useful in developing transgenic L. leucocephala with reduced mimosine content, which will make this tree legume suitable for use as a nutritious fodder for animals in the future.
RESULTS
Isolation of cDNA for a Mimosine-Degrading Enzyme from L. leucocephala
Previously, we isolated a set of 406 clones from the cDNA library of L. leucocephala, made from young shoots from approximately 8-week-old seedlings, through interspecies suppression subtractive hybridization (iSSH) using cDNAs from L. leucocephala and a related tree legume, Acacia confusa, as the tester and driver, respectively (Negi et al., 2011). These cDNA clones represent either L. leucocephala-specific genes that are missing in A. confusa or genes that are highly expressed in L. leucocephala. Since mimosine-degrading enzyme activity is specific to L. leucocephala, we expected that one of these 406 cDNA clones from the iSSH library might encode the mimosine-degrading enzyme. We also expected the mimosine-degrading enzyme from L. leucocephala to be a C-N lyase for two reasons: (1) Smith and Fowden (1966) showed that a C-N lyase from L. leucocephala had mimosine-degrading activity; and (2) recently, we have found that the mimosine degradation geneD (midD) of the L. leucocephala symbiont Rhizobium sp. strain TAL1145 encodes a C-N lyase that degrades mimosine into 3H4P, pyruvate, and ammonia (Negi et al., 2013). Therefore, we analyzed the L. leucocephala iSSH library of 406 cDNA clone sequences to identify any sequence that shows homology to lyases. BLASTP analysis of the deduced amino acid sequences of the L. leucocephala iSSH cDNA library against reference protein sequence databases identified one 764-bp cDNA clone, named seq3, which showed homology with cystathionine β-lyases (CBLs) from soybean (Glycine max; NP_001242026; 78% similarity), Ricinus communis (XP_002512818; 76% similarity), grape (Vitis vinifera; XP_002274313; 76% similarity), and Arabidopsis (Arabidopsis thaliana; NP_850712; 75% similarity). CBL catalyzes cystathionine degradation into homocysteine, ammonia, and pyruvate. Two of the products of this reaction, pyruvate and ammonia, are the same as the products of mimosine degradation catalyzed by the enzymes from L. leucocephala (Smith and Fowden, 1966) and Rhizobium sp. strain TAL1145 (Negi et al., 2013), in which the third product is either 3,4DHP or its isomer 3H4P. Considering the similarity in the products from the degradation reactions catalyzed by CBL and the mimosine-degrading C-N lyases from L. leucocephala and Rhizobium sp. strain TAL1145, as well as the homology of seq3 with CBL, we decided to obtain the full-length cDNA for this partial cDNA fragment, expecting that it might be the cDNA for the mimosine-degrading enzyme from L. leucocephala. RNA ligase-mediated (RLM)-RACE of the seq3 cDNA fragment resulted in the isolation of a 1,520-bp cDNA fragment that consists of a 1,332-bp-long open reading frame (ORF). The BLASTP analysis of the deduced amino acid sequence of the full-length ORF of seq3 using a reference protein database of the National Center for Biotechnology Information (NCBI) showed its homology with various proteins that belong to the aspartate aminotransferase (AAT) superfamily. The highest sequence similarity of the seq3 sequence was observed to be 71% with the chloroplast-like CBL of soybean. Interestingly, the BLASTP analysis of the deduced amino acid sequence of the seq3 ORF against the nonredundant protein database showed that seq3 had 100% sequence similarity with a sequence for mimosinase from L. leucocephala (accession no. AB298597.1). This mimosinase sequence in the nonredundant protein database (direct submission by Masakazu Fukuta) has not been experimentally established to be the sequence for mimosinase, and the enzyme activity for the encoded protein has not been demonstrated. Therefore, we decided to test the enzymatic activity of the protein encoded by the 1,332-bp seq3 ORF.
Codon Optimization of the seq3 ORF for Expression in Escherichia coli
In order to establish if seq3 encodes mimosinase, it was necessary to express the 1,332-bp ORF in E. coli, purify the recombinant protein, and determine its enzyme activity. Hydrophobic regions, if present, usually make protein purification difficult by forming inclusion bodies (Fink, 1998). Signal peptides of proteins are hydrophobic and are not essential for in vitro protein activity. Based on the homology of seq3 with chloroplast-like CBL, we expected that seq3 might also have a chloroplast signal peptide. Therefore, for efficient expression of the seq3-encoded protein in E. coli, we decided to eliminate any possible signal peptide sequence and optimize the sequence based on the E. coli codon preferences. The deduced amino acid sequence of the seq3 ORF was subjected to the TargetP 1.1 server using plant networks. The TargetP 1.1 server predicted a 43-amino acid chloroplast transit peptide with a reliability class value of 2 at the N terminus of the 443-amino acid sequence (Supplemental Table S1). The low reliability class value indicates strong prediction of the transit peptide, suggesting that the encoded protein may be localized in the chloroplast. The 126-bp sequence for the predicted chloroplast transit peptide from the 5′ end of the ORF, excluding the start codon, was eliminated, and the remaining 1,206-bp sequence was analyzed for the presence of rare codons of E. coli, and a synthetic derivative of the ORF was obtained by replacing the rare codons with commonly used codons of E. coli (Supplemental Fig. S1). In the 1,206-bp synthetic ORF, a total of 258 out of 402 codons were changed by replacing 301 nucleotides to obtain synthetic seq3 (syn-seq3).
syn-seq3 Expression in E. coli and Purification of the Encoded Enzyme
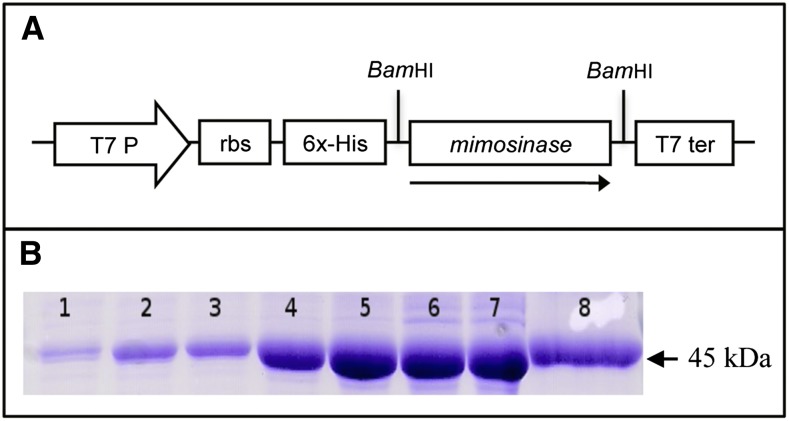
Recombinant protein encoded by syn-seq3 was obtained by expressing it in E. coli under the control of an inducible T7 promoter. An expression plasmid, pET-seq3, was constructed by inserting syn-seq3 into the pET14-b vector downstream from the poly-His tag and T7 promoter (Fig. 2A) and expressed in E. coli. SDS-PAGE analysis of total soluble proteins from the isopropyl β-d-1-thiogalactopyranoside-induced culture showed a major band of approximately 45 kD, which was absent from the uninduced culture. The poly-His-tagged protein purified from the induced culture appeared as a single band with a molecular mass of approximately 45 kD in SDS-PAGE (Fig. 2B). The induction experiment showed that the optimum induction time for expression of the 45-kD protein in E. coli was 6 h. Therefore, for subsequent purification of this protein, the E. coli cultures were induced for 6 h. The purified recombinant protein from E. coli was used for enzymatic assays.
Figure 2.
Cloning and expression of a seq3-encoding protein in E. coli. A, The plasmid for seq3 expression was constructed by cloning the codon-optimized synthetic seq3 ORF lacking the coding region for chloroplast signal peptide at the BamHI restriction site of the pET-14b vector in the sense orientation. B, SDS-PAGE of recombinant protein expressed in E. coli. Lanes 1 to 7, total soluble protein from E. coli containing pET-seq3 induced for 0, 1, 2, 4, 6, 8 and 10 h, respectively; lane 8, purified recombinant protein from 6-h induced E. coli containing pET-seq3. RBS, ribosome binding site. [See online article for color version of this figure.]
Mimosine Degradation Assay Using the Recombinant Protein
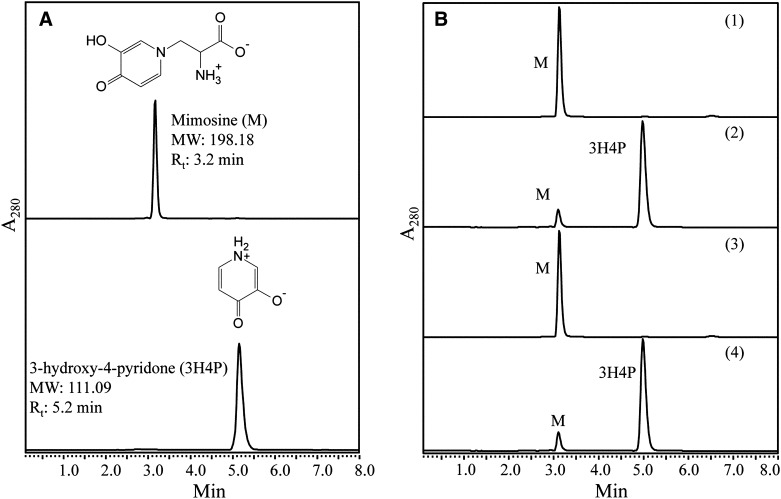
To determine if the purified 45-kD recombinant protein has mimosine-degrading (mimosinase) activity, it was used in an in vitro enzyme assay with mimosine as the substrate. In the HPLC analysis, mimosine and synthetic 3H4P, used as standards, showed retention times of 3.2 and 5.2 min, respectively (Fig. 3A). HPLC results from control reactions that contain heat-inactivated enzyme did not show any decrease in the amount of mimosine. However, the reactions with purified recombinant protein exhibited a sharp decrease in the amount of mimosine, and the product formed had an identical retention time as that of 3H4P (Fig. 3B). This result establishes that the seq3-encoded enzyme has mimosine-degrading activity, and the major degradation product may be 3H4P, an isomer of 3,4DHP, which was previously identified as a product of mimosine degradation catalyzed by a C-N lyase from L. leucocephala (Smith and Fowden, 1966). However, it remains to be established if seq3 encodes the C-N lyase found in L. leucocephala by Smith and Fowden (1966) or if it encodes another mimosine-degrading enzyme.
Figure 3.
HPLC results of standards and test samples in mimosine degradation assays. A, The chromatograms represent mimosine and 3H4P that were used as standards. The chromatograms of mimosine and 3H4P had retention times (Rt) of 3.2 and 5.2 min, respectively. B, HPLC results of mimosine degradation assays: 1, a reaction in which heat-inactivated recombinant enzyme was added exhibited a single peak of unused substrate, mimosine (M); 2, a reaction catalyzed by functionally active recombinant enzyme in the absence of exogenously added α-KG and PLP exhibited a large peak of mimosine degradation product with the same retention time as that of 3H4P; 3, a reaction catalyzed by functionally active recombinant enzyme in the presence of 50 μm hydroxylamine had only one peak of unutilized substrate; 4, a functionally active recombinant enzyme-catalyzed reaction in the presence of inhibitor, 50 μm hydroxylamine, restored the enzyme activity when supplemented with 0.1 μm PLP and showed a large peak of mimosine degradation product with the same retention time as that of 3H4P.
In the previous section, it was shown that the amino acid sequence of seq3 belongs to the AAT superfamily. However, this does not mean that it is an aminotransferase, because all enzymes in the AAT superfamily are not aminotransferases. The seq3-encoded enzyme is homologous to CBL, which is a lyase but not an aminotransferase, although it belongs to the AAT superfamily. To determine if the purified enzyme was a lyase or an aminotransferase, we performed the reactions in the presence and absence of α-ketoglutarate (α-KG) in the reaction buffer, which is used as a cosubstrate in the reactions catalyzed by aminotransferases. Since aminotransferases and lyases are both PLP-dependent enzymes, we conducted the experiments in the presence and absence of PLP in the reaction buffer. The reactions with the purified protein resulted in significant decreases in mimosine irrespective of the presence or absence α-KG in the reaction buffer (Fig. 3B). Since α-KG is not essential as the cosubstrate for the reaction, we concluded that the enzyme is not an aminotransferase. Therefore, it is likely that the seq3-encoded enzyme is a lyase. However, the seq3-encoded enzyme degraded mimosine even in the absence of exogenously added PLP; we did not observe any detectable difference in the products formed in reaction mixtures with or without PLP, which is an essential cofactor for lyases. This suggests that the recombinant enzyme from E. coli does not require exogenously added PLP. However, it may also be possible that the recombinant enzyme carried PLP from the E. coli host cell. To test this possibility, we performed the mimosine degradation reaction in the presence and absence of hydroxylamine, which is a potent inhibitor of PLP-dependent enzymes. Hydroxylamine is known to react with PLP in the enzyme’s active site, in a reversible single-step reaction forming spectrally distinct oxime adducts, which result in loss of the enzyme activity due to the removal of the bound PLP. Hydroxylamine was added to the reaction at a final concentration of 0.01 to 50 mm, which at a final concentration of 50 µm completely inhibited the catalytic activity of the recombinant enzyme. However, supplementation of the hydroxylamine-added reaction with 0.1 µm PLP successfully overcame the inhibitory effects of hydroxylamine by restoring the catalytic activity of the enzyme (Fig. 3B). This confirms that seq3 encodes for a PLP-dependent mimosine-degrading enzyme.
Biochemical Properties of the seq3-Encoded Mimosine-Degrading Enzyme
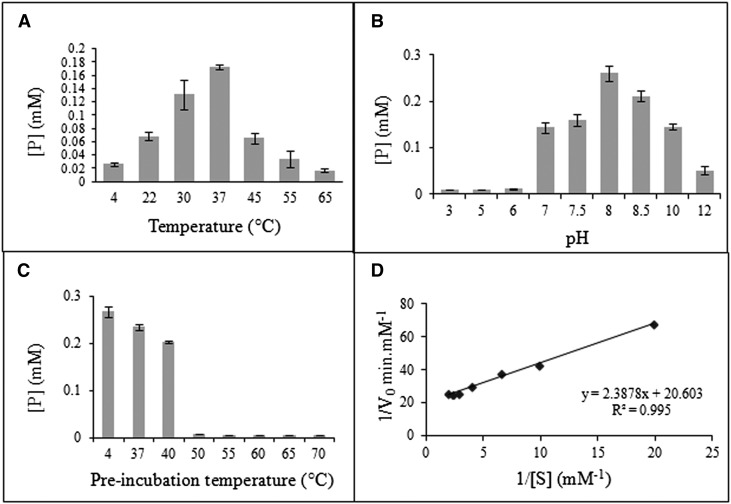
The catalytic activity of the purified mimosine-degrading enzyme was calculated as the concentration of product formed in 1 h. The highest catalytic activity of the enzyme was observed at 37°C, suggesting that the optimum temperature for the enzyme is 37°C (Fig. 4A). The enzyme catalytic activity sharply decreased at 45°C or higher and below 22°C. The enzyme had high catalytic activities in a pH range from 7.5 to 10, with maximum activity at pH 8.5 (Fig. 4B). To determine the thermal stability of the enzyme, the percentage enzyme activity for different preincubation temperatures was measured with reference to the enzyme activity at the preincubation temperature of 4°C. The enzyme activity at a 37°C preincubation temperature was found to be 86% of that of the reference, and it was reduced to 74.5% when the enzyme was preincubated at 40°C (Fig. 4C). However, at a preincubation temperature of 50°C or higher, a sharp reduction in the enzyme activity was observed. This indicates that the enzyme is fairly stable up to 40°C.
Figure 4.
Biochemical and kinetic properties of a mimosine-degrading enzyme. A to C, Catalytic activity of the enzyme was determined as the product formed [P] at different temperatures (A), different pH levels (B), and different preincubation temperatures (C). D, Estimation of kinetic parameters of the enzyme obtained by plotting a Lineweaver-Burk plot of the initial velocities at different substrate concentrations.
Kinetic properties of the seq3-encoded enzyme were determined at the optimum temperature (37°C) and pH (8). The rate of reaction for different substrate concentrations ranging from 0.05 to 0.5 mm was found to be linear from 0 to 2 min (data not shown). Initial velocities of the enzyme for each substrate concentration were calculated as the slope of product formed at 0 and 2 min. The purified recombinant enzyme followed a typical Michaelis-Menten kinetic model of single substrate reaction. The experimental determination of the apparent Km and Vmax values for the recombinant enzyme was calculated from the linear regression of the Lineweaver-Burk plot (Fig. 4D). The Km and Vmax for the enzyme were found to be 1.16 × 10−4 m and 5.05 × 10−5 mol s−1 mg−1, respectively. Assuming one active site per enzyme molecule, the total enzyme concentration was calculated to be 0.22 × 10−7 mol mg−1. The turnover number of the seq3-encoded enzyme was estimated to be 2,300 s−1.
The aromatic amino acids l-Tyr, l-Trp, and l-Phe, which are the structural analogs of mimosine, were tested as possible competitive inhibitors of mimosine. The amount of product formed in a reaction where only mimosine was added as the substrate (control reaction) was found to be similar to that of the test reactions, where the mimosine was supplemented with the 1-, 2-, or 3-fold concentrations of structural analogs in separate reactions (Supplemental Fig. S2), indicating that these aromatic amino acids are not competitive inhibitors of mimosine catalysis by the seq3-encoded enzyme.
Characterization of the seq3-Catalyzed Mimosine Degradation Product
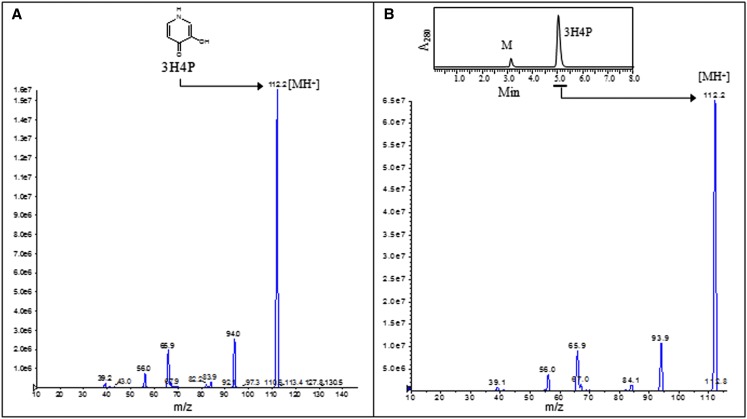
The HPLC results for synthetic 3H4P and for one of the mimosine degradation products formed in the reaction catalyzed by the recombinant enzyme were identical, indicating that 3H4P is the major degradation product of mimosine in the reaction. To verify this, synthetic 3H4P and the HPLC-purified product of mimosine degradation were subjected to fragmentation through tandem mass spectrometry (MS/MS) of the parent ion (mass-to-charge ratio [m/z] 112.2). The resulting fragmentation patterns of synthetic 3H4P (reference sample) and the mimosine degradation product (test sample) were found to be identical, generating common internal structural fragments of m/z 94.0 ([M-H2O]+), 65.9 ([M-H2O-CO]+), 56.0, and 39.1 (Fig. 5). This confirms that 3H4P is the major product of the mimosine degradation reaction catalyzed by the recombinant enzyme.
Figure 5.
MS/MS spectra of the synthetic 3H4P (A; reference sample) and the HPLC-purified mimosine degradation product formed in a mimosinase-catalyzed reaction (B). [See online article for color version of this figure.]
The seq3-Encoded Enzyme Is a C-N Lyase
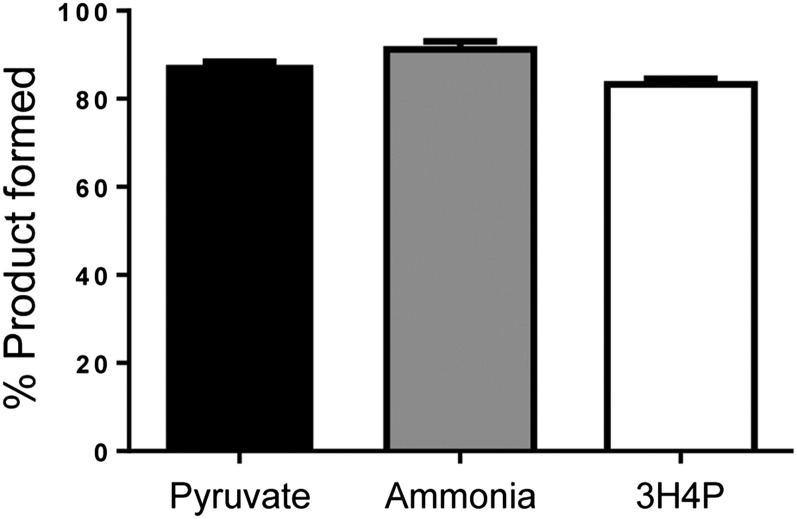
3H4P as the major degradation product of mimosine accounts for only eight carbons and two nitrogens of mimosine among 11 carbons and three nitrogens in mimosine. This suggests that there must be other products formed in the degradation reaction that could account for the remaining three carbons and one nitrogen. Previously, an enzyme from M. pudica was reported to degrade mimosine into Ser and pyruvate (Suda, 1960), whereas an enzyme from L. leucocephala seedling extract was found to degrade mimosine into 3,4DHP, pyruvate, and ammonia (Smith and Fowden, 1966). Therefore, we tested the reaction products catalyzed by the seq3-encoded recombinant enzyme for the presence of Ser, pyruvate, and ammonia. Considering that Ser is structurally similar to Ala, we also tested the reaction products for the presence of Ala. Neither Ser nor Ala appeared in the HPLC scans of reactions catalyzed by the recombinant enzyme. However pyruvate and ammonia were detected spectrophotometrically in the reaction products. The amount of pyruvate, ammonia, and 3H4P produced in the mimosine degradation reaction were found to be 87.7%, 91.2%, and 83.2%, respectively, of mimosine, indicating that these products are formed in equimolar quantities as that of the substrate (Fig. 6). Based on these observations, the completely balanced reaction of mimosine degradation catalyzed by the seq3-encoded enzyme can be as follows:
Figure 6.
The mass balance of the substrate and products in the mimosinase-catalyzed reaction. Bars represent the percentage of products formed with respect to the substrate used. Error bars represent the sd of three replicates.
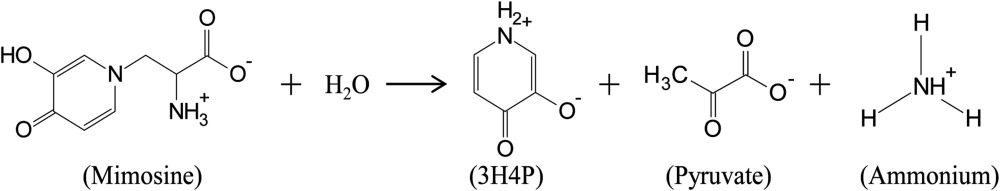 |
Thus, like C-N lyases, the seq3-encoded enzyme is PLP dependent and the reaction products contain pyruvate and ammonia. Therefore, we conclude that the seq3-encoded enzyme is a C-N lyase, and under treaties and by convention, as described by the International Union of Biochemistry and Molecular Biology, mimosinase can be classified in the enzyme category with the EC number 4.3.1.
The seq3-Encoded C-N Lyase Has Conserved Active Site Residues of Arabidopsis CBL
Mimosinase in BLASTP analysis with the Protein Data Bank reference database showed 69% similarity with CBL from Arabidopsis (Supplemental Fig. S3). A previous study of the three-dimensional structure of Arabidopsis CBL identified the key active site residues of the enzyme (Breitinger et al., 2001). Most of the known active site residues of CBL are also present in mimosinase (Supplemental Fig. S3). The high degree of sequence similarity between Arabidopsis CBL and L. leucocephala mimosinase and their conserved active site residues suggest that mimosinase may have a similar catalytic mode of action as described by Breitinger et al. (2001) for CBL.
High Temperature Results in Increased Mimosine Degradation without Affecting the Level of Mimosinase
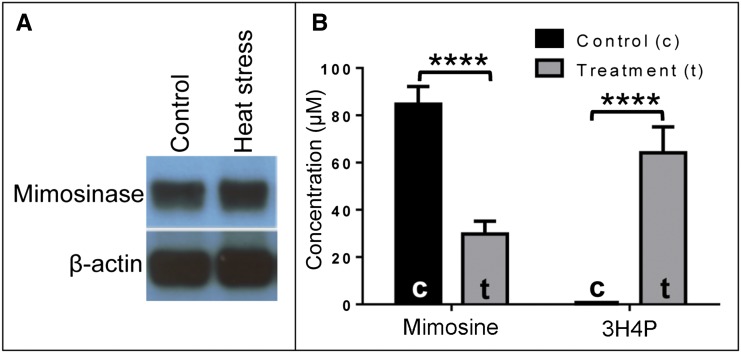
Previously, Tangendjaja et al. (1984) isolated mimosinase from L. leucocephala leaves and determined the mimosine contents of intact leaves at different temperatures. They observed that mimosine content decreased with increasing temperature above 45°C and recorded the lowest amounts of mimosine after exposure of the leaves to 70°C for 5 to 30 min. These authors did not consider mimosinase to be a chloroplast-localized enzyme, although they mentioned that some tissue damage is required for mimosine to be in contact with the enzyme. The decrease in mimosine content in the L. leucocephala leaves with increasing temperature may happen due to two possibilities: (1) increased expression of mimosinase at higher temperature; or (2) partial breakdown of chloroplast at higher temperatures, resulting in the release of mimosinase and consequent decrease in mimosine content. We tested these ideas by conducting two experiments. In the first experiment, we did a western-blot analysis of mimosinase in intact young leaves from L. leucocephala seedlings grown at 25°C and exposed to 70°C for 10 min. As seen in Figure 7A, the mimosinase content appeared to be similar at both temperatures, suggesting that the expression of mimosinase did not increase at high temperature. Therefore, we considered the second possibility, which was based on a previous report, that lesions occurred in the chloroplast envelope membrane of different plant species at temperatures between 53°C and 57°C (McCain et al., 1989). We hypothesized that, due to lesions occurring in chloroplast membranes at high temperature, mimosinase would leak out of the chloroplast and degrade mimosine in the cytosol. As a result, the mimosine content should decrease and the content of its degradation product 3H4P should increase. As expected, we found that the mimosine content in the heat-exposed leaves decreased but the 3H4P content increased. The mimosine contents in control and heat-treated leaf samples were 85 ± 7.5 and 30 ± 5.36 µm, respectively. No 3H4P was detected in control samples compared with 64 ± 10.93 µm 3H4P detected in heat-treated leaf (Fig. 7B).
Figure 7.
Effect of heat stress on mimosine content and its degradation product and on the expression level of mimosinase. A, Western-blot analysis of mimosinase (45 kD) expression in leaf tissue under controlled temperature (25°C) and heat treatment (70°C). Rabbit anti-mimosinase polyclonal antibody and mouse anti-actin (plant) monoclonal antibody were used as primary antibodies for mimosinase and β-actin, respectively. B, The concentrations of mimosine and 3H4P were measured using HPLC in 0.3 g of intact leaf tissues from 8-week-old L. leucocephala. For heat treatment (t), the 8-week-old plants were incubated at 70°C for 10 min before collecting the intact leaf tissues. For each control (c) and heat treatment group, three biological replicates were used (n = 3). The bars show means of mimosine and 3H4P contents in control and treatment groups, and error bars represents sd. [See online article for color version of this figure.]
DISCUSSION
In this study, we isolated a cDNA from L. leucocephala encoding a mimosine-degrading enzyme and expressed the coding region of the cDNA in E. coli as a recombinant enzyme. We also demonstrated that the purified enzyme converts mimosine into 3H4P and produces pyruvate and ammonia as by-products in the reaction. Previous studies reported either 3,4DHP (Smith and Fowden, 1966) or 3H4P (Tangendjaja et al., 1986) as the major degradation product of mimosine by a mimosine-degrading enzyme from L. leucocephala seedlings and leaves, respectively. Both 3,4DHP and 3H4P are isomers of each other, and based on the experimental conditions of the mimosine degradation reaction, different researchers might have found different isomers of the same compound. Since the above experiments were done with enzymes isolated from different L. leucocephala tissues, it is not clear if the enzymes were the same or different in the two experiments. It is possible that L. leucocephala has more than one isoform of mimosinase that may express at different developmental stages. The low Km, high Vmax, and estimated turnover number of the recombinant enzyme show that it is highly efficient in cleaving the alanyl side chain of mimosine and thereby converting it to 3H4P.
The optimum pH of the recombinant enzyme was 8, which is the same as that found for the mimosine-degrading enzyme from L. leucocephala seedling extract (Smith and Fowden, 1966) and from L. leucocephala leaves (Tangendjaja et al., 1986). The optimum temperature for the catalytic activity of the recombinant enzyme was determined to be 37°C, which is significantly different from the reported optimum temperature of 45°C for the mimosine-degrading enzyme from L. leucocephala leaves (Tangendjaja et al., 1984, 1986). The mimosinase enzyme in our study may be different from the enzyme described by Tangendjaja et al. (1986). It is likely that L. leucocephala has more than one isoform of mimosinase that may express in different tissues or under different physiological conditions. Like other enzymes catalyzing single-substrate reactions, mimosinase also followed typical Michaelis-Menten kinetics. Our results show that the recombinant mimosine-degrading enzyme is a C-N lyase that breaks the C-N bond of mimosine to produce 3H4P. This finding supports the report of Smith and Fowden (1966), in which they identified the mimosine-degrading enzyme from L. leucocephala seedling extracts as a mimosine C-N lyase. We have shown that, like other lyases, the recombinant mimosinase is PLP dependent and is inhibited by the presence of hydroxylamine in the reaction, and the enzyme activity in the hydroxylamine-containing reaction can be restored by exogenously added PLP to the reaction.
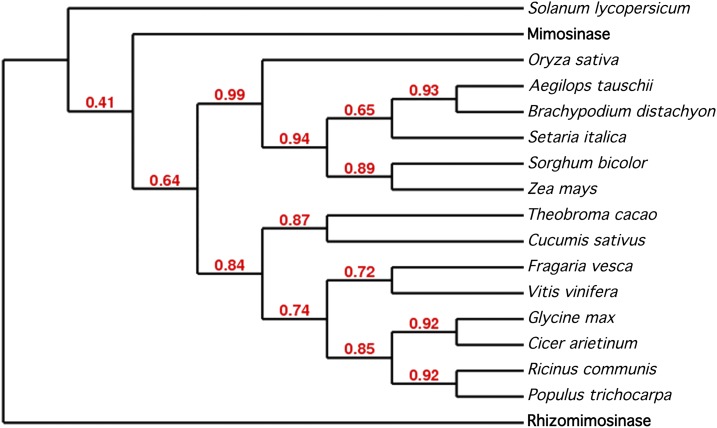
Recently, we have characterized another mimosine-degrading enzyme, rhizomimosinase (Negi et al., 2013), from Rhizobium sp. strain TAL1145, an L. leucocephala root-nodulating bacterial strain. The L. leucocephala enzyme mimosinase and the Rhizobium sp. strain TAL1145 enzyme rhizomimosinase both are C-N lyases and have similar catalytic functions, biochemical properties, and kinetic properties (Table I). However, the deduced amino acid sequences of mimosinase (BAF80449) and rhizomimosinase (AAG47972) in pairwise global alignment using the EMBOSS needle Web server exhibited only 26.9% similarity and 16.7% identity (Supplemental Fig. S4). Additionally, in the phylogenetic analysis, mimosinase and rhizomimosinase appeared in different clades (Fig. 8). Therefore, mimosinase and rhizomimosinase represent a good example of enzymes from two different systems that have similar catalytic roles but show little sequence similarity.
Table I. Comparison of rhizobial rhizomimosinase and L. leucocephala mimosinase.
| Features | Rhizomimosinase | Mimosinase |
|---|---|---|
| Amino acid residues | 406 | 400 (excluding 43 amino acid residues for predicted chloroplast signal peptide) |
| Size | Approximately 45 kD | Approximately 45 kD |
| Optimum temperature | 37°C | 37°C |
| Optimum pH | 8.5 | 8.0 |
| Km | 1.27 × 10−4 m | 1.16 × 10−4 m |
| Vmax | 4.96 × 10−5 mol s−1 mg−1 | 5.05 × 10−5 mol s−1 mg−1 |
| Turnover number | 2,250 s−1 | 2,300 s−1 |
| PLP dependence | Yes | Yes |
| Degrades mimosine into | 3H4P, ammonia, and pyruvate | 3H4P, ammonia, and pyruvate |
| Homology shown in BLASTP analysis | 58% positive residues with CBL from Bacillus bataviensis (WP_007083900) in BLAST | 70% positive residues with CBL from chloroplast-like tomato (XP_004249212) in BLASTP analysis |
Figure 8.
Cladogram of mimosinase and rhizomimosinase with CBLs from various plants. The Web-based phylogeny server www.phylogeny.fr/version2_cgi/advanced.cgi was used to construct the phylogenetic tree, and the statistical analysis for branch support was performed using the approximate likelihood-ratio test. The accession numbers of the sequences used are given in “Materials and Methods.” [See online article for color version of this figure.]
The recombinant enzyme was predicted to have a chloroplast signal peptide, indicating that it may be a chloroplast-localized protein. Tangendjaja et al. (1984) reported a decrease in mimosine content with increasing temperature, with the lowest amount of mimosine recorded at 70°C. However, it was not established if the decrease in mimosine content was because of overexpression of the enzyme at higher temperature. Our results show that the decrease in mimosine content and the increase in its major degradation product 3H4P at 70°C were not associated with the overexpression of mimosinase; therefore, it is possible that exposure of intact leaf to 70°C might have caused damage to the chloroplast membrane, resulting in the release of mimosinase to the cytoplasm, where it degraded mimosine into 3H4P. L. leucocephala leaves contain ample amounts of mimosine, comprising 3% to 5% of plant dry weight (Soedarjo and Borthakur, 1998).
The production of such a high amount of mimosine is a significant investment of carbon and nitrogen resources by the plant. We estimate that if the energy resources invested for the production of mimosine were diverted for growth, the L. leucocephala plant would have grown at least 20% larger. The forage yield of L. leucocephala is reported to be 6 to 18 tons dry matter ha−1 (National Academy of Sciences, 1984). For our calculations, we assumed the forage yield of L. leucocephala as 10 tons ha−1 and the mimosine content as 3%. Therefore, 10 tons of dry L. leucocephala forage should contain approximately 42 kg of nitrogen due to mimosine (the molecular weight of mimosine is 198 and it has two nitrogens). Assuming that most plants can utilize up to 200 kg nitrogen ha−1 for maximum growth, and considering that nitrogen is the only limiting factor for growth and that other nutrients, water, and environmental conditions are optimal, the 42 kg of nitrogen from mimosine, if diverted to growth, should result in 21% (42/200 × 100) additional growth.
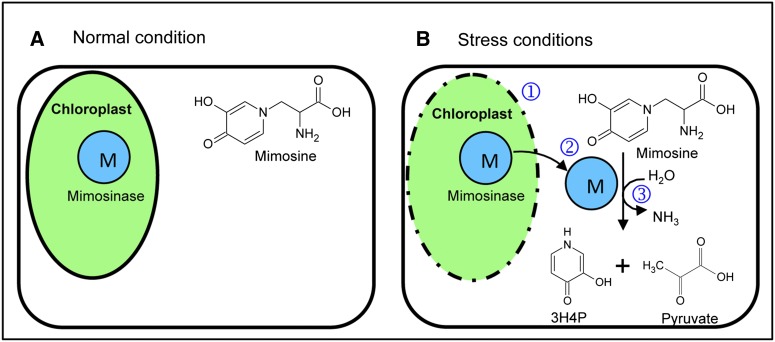
The role of mimosine in L. leucocephala is not well established. It is likely that as a nitrogen-rich compound, mimosine plays a role in maintaining homeostasis in the growth of L. leucocephala under stress conditions, such as high temperature and drought. Under such stress conditions, when nutrient availability becomes limited, the stored mimosine may be used as a source of carbon and nitrogen to produce amino acids and other secondary metabolites. Thus, mimosinase may function as the first enzyme in channeling mimosine into other biochemical pathways, including amino acid synthesis during stress conditions. The ammonia released in the reaction may be used to convert Glu to Gln by Gln synthase; pyruvate may enter the tricarboxylic acid cycle; and 3H4P may be degraded further by other plant enzymes. Although plant enzymes involved in the conversion of 3H4P to smaller compounds have not been identified, a dioxygenase and a hydrolase, encoded by the pyridone degradation genes A and B (pydA and pydB), respectively, have been isolated and characterized from Rhizobium sp. strain TAL1145 that forms nitrogen-fixing nodules on L. leucocephala (Awaya et al., 2005). It was shown that these enzymes convert 3H4P into ammonia, pyruvate, and formate. It is likely that L. leucocephala leaves may also have similar enzymes for the conversion of 3H4P to useable compounds during stress conditions. To explain the possible role of mimosine and mimosinase in L. leucocephala, we have proposed a compartmentalization model according to which, under favorable growth conditions, L. leucocephala synthesizes mimsoine and stores it in the cytoplasm. To protect the stored mimosine from untimely degradation by mimosinase, L. leucocephala might have adapted the strategy of keeping mimosine and mimosinase apart from each other by compartmentalizing mimosinase in the chloroplast. Based on this hypothesis, we propose that mimosinase and other downstream enzymes provide a mechanism for the utilization of mimosine under stress conditions (Fig. 9). This hypothesis also explains one possible way that L. leucocephala manages to grow well under drought stress conditions when the availability of carbon and nitrogen is scarce.
Figure 9.
Compartmentalization model showing the possible roles of mimosine and mimosinase in dealing with nutrient-limiting stress conditions such as drought. A, Under normal conditions of growth, mimosine is synthesized and stored in the cell cytoplasm. The enzyme mimosinase is localized in the chloroplast, and thus mimosine is inaccessible to mimosinase. B, However, stress conditions may cause damage to the chloroplast membrane (1), resulting in the release of mimosinase from the chloroplast to the cytoplasm (2), where the substrate mimosine is accessible to mimosinase, which catalyzes the first step of mimosine catabolism into 3H4P, pyruvate, and ammonia (3). These degradation products of mimosine are rich in carbon and nitrogen, and hence mimosine may serve as a source of stored carbon and nitrogen at the time of need. [See online article for color version of this figure.]
MATERIALS AND METHODS
Identification and Isolation of a cDNA That Encodes a Mimosine-Degrading Enzyme
For the identification of a Leucaena leucocephala cDNA that encodes a mimosine-degrading enzyme, a set of 406 cDNA sequences obtained from the iSSH library of L. leucocephala (Negi et al., 2011) were screened for their homology with C-N lyase using the BLASTX tool and a reference protein database of the NCBI. The cDNA fragment of the iSSH clone that exhibited similarity with C-N lyase was amplified to full-length cDNA by using the FirstChoice RLM-RACE kit (Ambion) according to the manufacturer’s instructions. The primers used for RLM-RACE, seq3-F and seq3-R, are listed in Supplemental Table S2. The resultant full-length cDNA sequence was cloned in pGEMT-easy vector, and the sequence was verified.
Sequence Analyses for the Prediction of Signal Peptide and the Presence of Rare Codons
The deduced amino acid sequence from the ORF of the full-length cDNA was analyzed for the presence of any N-terminal signal peptide including chloroplast transit peptide, mitochondrial targeting peptide, or secretory pathway signal peptide using the TargetP 1.1 server (Emanuelsson et al., 2000). For enhanced expression of the plant gene in Escherichia coli, the full-length ORF was evaluated for the presence of E. coli rare codons based on its codon usage bias using the GenScript Rare Codon Analysis Tool (GenScript), and a synthetic ORF, codon optimized for enhanced expression in E. coli, was obtained from GenScript. The BamHI restriction site was introduced at both ends of the synthetic gene to facilitate its cloning in the expression vector.
Cloning and Construction of the seq3 Expression Plasmid
The codon-optimized synthetic ORF and the expression vector pET-14b (Novagen) were digested with BamHI (Promega) followed by dephosphorylation of 5′ phosphate from the vector using shrimp alkaline phosphatase (Promega). BamHI-digested synthetic ORF was cloned into the BamHI site of the expression vector pET-14b behind the T7 promoter and introduced into E. coli JM109 (Promega). The resultant plasmid was verified by sequencing and termed pET-seq3 (Supplemental Table S2).
Synthetic ORF Expression in E. coli and Purification of the Encoded Protein
The expression construct, pET-seq3, was transformed into E. coli strain BL21 (DE3) pLysS (Promega). The transformed cells were grown in Luria-Bertani medium, and at midlog phase (optical density at 600 nm = 0.6), isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 1 mm to induce the expression of the recombinant protein. The optimum induction time for protein expression was determined by inducing the cells for 0 to 10 h. Samples (200 µL) from the induced cells were collected at 0, 1, 2, 4, 6, 8, and 10 h of induction. The sample collected at 0 h served as the negative control. The collected samples were centrifuged at 4,000 rpm for 10 min at 4°C, and the pellet was resuspended in SDS-PAGE loading buffer followed by heating at 95°C for 5 min to extract total protein. The recombinant protein was purified from the induced culture by using the MagneHis Protein Purification System (Promega) according to the manufacturer’s instructions. Total soluble proteins of samples and purified recombinant protein were analyzed through SDS-PAGE. The purified recombinant protein was sent to GenScript for the development of polyclonal antibody in rabbit.
Enzymatic Assay
The purified recombinant enzyme was used in an in vitro mimosine degradation assay using mimosine as the substrate. For each 1,000-µL reaction, 0.016 mg of the purified enzyme and 1 mm mimosine were used in 0.1 m Tris-HCl buffer with a final pH of 7.5, and the reaction was incubated at 37°C for 1 h unless otherwise stated. To determine whether the recombinant enzyme is an aminotransferase, the mimosine degradation assay was performed in both the presence and absence of 1 mm α-KG and 20 µm PLP in the reaction buffer. Each reaction was carried out in three replications, and the reaction was assayed by HPLC using a C18 column (4.6 × 250 mm; Dionex Acclaim 120). For quantitative estimation of the product formed, different concentrations of chemically synthesized 3H4P (synthetic 3H4P) were used as standards. Synthetic 3H4P was obtained from Dr. Edward J. Behrman (Ohio State University), who previously described an improved method for the synthesis of 3H4P (Behrman, 2009). The peak area of known concentrations of synthetic 3H4P was used to plot the standard curve, which was then used to quantify the amount of product formed from the peak area of test samples. An isocratic solvent system of 0.02 m o-phosphoric acid with a flow rate of 1 mL min−1 and a UV detection photodiode array (200–400 nm) was used for HPLC analysis. The enzyme was also tested for PLP dependence in enzymatic assays performed by supplementing 0.01 to 50 mm hydroxylamine in the reaction buffer containing enzyme but no substrate. The reaction mixture was incubated for 5 min before adding the substrate. In a separate assay, the reaction mixture that contained the least concentration of hydroxylamine required to inhibit the enzyme was supplemented with 0.1 to 20 µm PLP after 5 min of incubation. The reaction mixture was again incubated for 5 min followed by the addition of the substrate. Each reaction was analyzed using HPLC as described above.
Characterization of the Mimosine-Degrading Recombinant Enzyme
The optimum temperature for the catalytic activity of the recombinant enzyme was determined by performing mimosine degradation assays at different temperatures, 4°C, 22°C, 30°C, 37°C, 45°C, 55°C, and 65°C, but the pH of the reaction buffer in all the reactions was constant, pH 7.5. Similarly, for determining the pH optima, the reactions were set at different pH levels ranging from pH 3 to pH 12 and incubated at a constant temperature of 37°C. Enzymatic reactions were terminated by heating the reaction mixtures at 100°C for 3 min. The subsequent assays were conducted at optimum temperature (37°C) and optimum pH (8). The thermal stability of the recombinant enzyme was studied by preincubating the enzyme with reaction buffer lacking substrate at different temperatures (4°C, 37°C, 40°C, 50°C, 55°C, 60°C, 65°C, and 70°C) for 30 min, followed by adding the substrate and incubating at 37°C for 1 h. The mimosine degradation product in each enzymatic reaction was quantified by HPLC as mentioned in the previous section.
The enzyme was also tested for any possible competitive inhibition with aromatic amino acids, l-Tyr, l-Trp, and l-Phe, which are structural analogs of mimosine. For this, the mixture of each amino acid with mimosine in ratios of 1:1, 2:1, and 3:1 was used as the substrate in the mimosine degradation assay. The reaction containing only mimosine as the substrate was used as the control. The product formed in each reaction was quantitated using HPLC.
Kinetic Studies of the Mimosine-Degrading Recombinant Enzyme
The enzymatic assays to study the kinetic properties of mimosinase were performed at optimum temperature and pH. To determine initial velocity, the rates of reaction for 0.05, 0.1, 0.15, 0.25, 0.35, 0.5 mm mimosine were calculated at time points of 0, 2, 3, 5, 10, 20, and 30 min. The rate of reaction was linear between 0 and 2 min; therefore, the initial velocities of the recombinant enzyme-catalyzed reaction was calculated for different substrate concentrations as the slopes formed by the amount of product formed at 0 and 2 min of reaction time. A Lineweaver-Burk plot was plotted using the calculated values of the initial velocities and substrate concentrations, and the kinetic parameters including Vmax and Km for the encoded enzyme were determined from linear regression of the plot. The total enzyme concentration (mol of enzyme active sites per mg of the enzyme) was calculated assuming that there is only one active site per enzyme molecule, and the turnover number of the enzyme was estimated using Vmax and total enzyme concentration.
Characterization of the Mimosine Degradation Products
The primary product of the mimosine degradation reaction catalyzed by the recombinant enzyme was characterized by MS/MS of the purified degradation product, and the reference compound used was synthetic 3H4P. The product was purified via HPLC from the enzyme-catalyzed reaction. The HPLC-purified sample and synthetic 3H4P were freeze dried and individually dissolved in carrier solvent (50%/50% [v/v] of a 0.09% [v/v] aqueous formic acid and 90%/10% [v/v] CH3CN/0.09% [v/v] aqueous formic acid). The samples were delivered to the atmospheric pressure ionization source of the AB/MDS-Sciex ESI-MS API 3000 triple quadrupole mass spectrometer using a continuous flow of 5 to 10 µL min−1 provided by a microsyringe infusion pump (Harvard Apparatus). The electrospray ionization-mass spectrometry system was calibrated manually in positive mode with PPG 3000 (AB/MDS-Sciex) to achieve less than 5 ppm mass accuracy, as per the manufacturer’s protocol before delivering the samples.
MS/MS of test and reference samples was performed with N2 bombardment confined to quadrupole-2 with a collision cell gas thickness of 3 × 1014 atoms cm−2 and a collision energy (quadrupole-0 to quadrupole-2 rod offset voltage) typically set at approximately 20 to 40 eV. The resulting MS/MS (daughter ion) spectra were obtained by scanning quadrupole-3 from m/z 10 to 150 in 0.6 s with a step size of 0.1 D. Electrospray ionization-mass spectrometry data analysis was assisted with the use of Mac BioSpec version 1.01 (PE Sciex).
To identify the degradation products other than 3H4P in the mimosine degradation reaction so that all carbons and nitrogens of mimosine are accounted for, the reaction products were tested for the presence of Ala, Ser, pyruvate, and ammonia. For Ala and Ser detection, the reaction products were derivatized using o-phthalaldehyde (OPA) and β-mercaptoethanol. In a basic environment (pH 9–11) and in the presence of β-mercaptoethanol, OPA reacts with primary amines to form a fluorescent isoindole derivative. The reaction buffer used in previous mimosine degradation assays contained Tris, which is a primary amine that may also react with OPA. Therefore, we used 40 mm sodium phosphate as the buffer for the enzymatic assay in this case. The reaction was set at optimum temperature and optimum pH for 1 h using 1 mm mimosine as the substrate. The reaction products in 40 mm sodium phosphate buffer were first analyzed for the presence of 3H4P using a C18 column as described in the previous section to verify if the sodium phosphate buffer does not interfere with the mimosine degradation reaction. Ala and Ser (1 mm each) were prepared in the 40 mm sodium phosphate buffer for use as controls. The pH of the reactions and controls was adjusted to 10 before performing the OPA derivatization. The reactions and controls were applied to precolumn derivatization for 5 min at room temperature using OPA at a final concentration of 5 mm. The derivatized samples were then analyzed through HPLC using a Kinetex C18 column (2.6 μ, 100 × 4.6 mm) in a gradient system in which the mobile phase consists of 40 mm potassium phosphate, pH 7.8 (buffer A), and 50:50 methanol:acetonitrile (buffer B). The flow rate was 1.5 mL min−1, and the run time was 20 min with 3% to 60% buffer B. The sample detection was performed at 338 nm using a UV detection photodiode array (200–400 nm). Pyruvate and ammonia in the reaction were detected by using a pyruvate assay kit (Biovision) and an ammonia assay kit (Sigma-Aldrich), respectively, according to each manufacturer’s instructions. In order to analyze mass balance between products and reactants, the reaction was performed to achieve the catalysis of the substrate to near completion by using excess of the enzyme (0.2 mg) and lower substrate concentration (500 µm mimosine) in 40 mm phosphate buffer. The reaction was performed at optimum temperature and optimum pH with a reaction time of 1 h, and pyruvate, ammonia, and 3H4P were quantified using the line equation from the respective standard curves.
Prediction of Conserved Residues of the Recombinant Enzyme
The deduced amino acid sequence of the full-length seq3 ORF was used in a BLASTP search against the protein databank database in NCBI. The resulting sequence with highest similarity was aligned with the deduced amino acid sequence of the seq3-encoded protein sequence using the ClustalW multiple sequence alignment program, and the conserved residues were analyzed.
Phylogenetic Analysis
The CBL sequences from various plant species, which showed the highest similarities with the mimosinase sequence (without signal peptide) in the BLASTP analysis, were used for phylogenetic analysis. The top 15 sequences (excluding hypothetical proteins) and rhizomimosinase (AAG47972), which showed 26.9% similarities and 16.7% identities with mimosinase (BAF80449), were included for the phylogenetic tree construction. The accession numbers of CBLs from various plant species are as follows: tomato (Solanum lycopersicum; XP_004249212), Theobroma cacao (EOY09022), cucumber (Cucumis sativus; XP_004166746), grape (Vitis vinifera; XP_002274313), poplar (Populus trichocarpa; ERP51507), Ricinus communis (XP_002512818), soybean (Glycine max; XP_003554134), Setaria italica (XP_004964713), chickpea (Cicer arietinum; XP_004514725), sorghum (Sorghum bicolor; XP_002436582), maize (Zea mays; NP_001148100), Brachypodium distachyon (XP_003561024), strawberry (Fragaria vesca; XP_004294046), rice (Oryza sativa; BAA95830), and Aegilops tauschii (EMT05183). The signal peptides from these sequences were predicted using the TargetP 1.1 server and removed prior to analysis. The Web-based phylogeny server http://www.phylogeny.fr/version2_cgi/advanced.cgi was used to construct a phylogenetic tree, and the statistical analysis for branch support was performed using the “approximate likelihood-ratio test.” The Clustal format of multiple sequence alignment that was used to create phylogenetic tree was made using MUSCLE 3.7 (Supplemental Fig. S5).
Plant Growth and Heat-Stress Treatment
L. leucocephala cv K-636 seeds were obtained from the University of Hawaii research station Waimanalo in Honolulu. Surface sterilization followed by scarification of mature seeds were done as described previously (Pal et al., 2012a). The seeds were germinated on one-half-strength Murashige and Skoog medium for 2 to 3 d under sterile conditions at 28°C in the dark, and the resulting germinated seedlings were transferred to vermiculite. The seedlings were watered with Hoagland solution twice per week and grown for 8 weeks. The plants were maintained at 25°C ± 2°C with a 16/8-h light/dark photoperiod with an irradiance of 30 µmol s−1 m−2. For heat stress treatment, the 8-week-old plants were incubated at 70°C for 10 min before collecting the intact leaf tissues, whereas for controls, the intact leaf tissues were directly collected. To avoid any enzyme-catalyzed degradation of mimosine after collecting the intact leaf samples, the samples (0.3 g) from control and treatment groups were immediately homogenized in 3 mL of 0.1 n HCl with a mortar and pestle. The homogenized samples were centrifuged at 10,000 rpm for 5 min at 4°C. The supernatants were filtered using a 0.45-μm pore size filter (Corning), and the filtrates were used for the determination of mimosine concentration by HPLC as described above.
Western-Blot Analysis
The effect of heat (70°C) condition on the expression of mimosinase compared with controls was studied using western-blot analysis using a rabbit polyclonal antibody developed against purified recombinant mimosinase (rabbit anti-mimosinase). The rabbit anti-mimosinase antibody was used in a 1:5,000 dilution, and the western blotting was performed as described previously (Pal et al., 2012b). For a loading control, the polyvinylidene difluoride blots were stripped off using ReBlot plus strong antibody-stripping solution (Millipore) and were blotted using mouse monoclonal anti-actin (plant) antibody (Sigma) as the primary antibody at a 1:1,000 dilution and horseradish peroxidase-conjugated goat anti-mouse IgG antibody (Bio-Rad) as the secondary antibody at a 1:3,000 dilution to detect and quantify actin.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Codon comparison of synthetic mimosinase with native open reading frame.
Supplemental Figure S2. HPLC chromatograms of mimosine degradation assays performed in presence of structural analogs of mimosine.
Supplemental Figure S3. Pairwise alignment of mimosinase with cystathionine β-lyase.
Supplemental Figure S4. Pairwise alignment of mimosinase with rhizomimosinase.
Supplemental Figure S5. Multiple sequence alignment in Clustal that was used to create phylogenetic tree.
Supplemental Table S1. Prediction of sub-cellular localization of mimosinase.
Supplemental Table S2. Plasmids and primers used in this study.
Acknowledgments
We thank Edward J. Behrman (Department of Chemistry and Biochemistry, Ohio State University) for generously providing us the synthetic 3H4P, and Nguyen Hue and Paul Singleton (Department of Tropical Plant and Soil Sciences, University of Hawaii at Manoa) for useful discussions.
Glossary
- PLP
pyridoxal-5′-phosphate
- C-N
carbon-nitrogen
- 3,4DHP
3,4-dihydroxypyridine
- 3H4P
3-hydroxy-4-pyridone
- cDNA
complementary DNA
- iSSH
interspecies suppression subtractive hybridization
- CBL
cystathionine β-lyase
- ORF
open reading frame
- NCBI
National Center for Biotechnology Information
- AAT
aspartate aminotransferase
- α-KG
α-ketoglutarate
- MS/MS
tandem mass spectrometry
- m/z
mass-to-charge ratio
- OPA
o-phthalaldehyde
References
- Awaya JD, Fox PM, Borthakur D. (2005) pyd genes of Rhizobium sp. strain TAL1145 are required for degradation of 3-hydroxy-4-pyridone, an aromatic intermediate in mimosine metabolism. J Bacteriol 187: 4480–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman EJ. (2009) Synthesis of 4-pyridone-3-sulfate and an improved synthesis of 3-hydroxy-4-pyridone. Chem Cent J 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitinger U, Clausen T, Ehlert S, Huber R, Laber B, Schmidt F, Pohl E, Messerschmidt A. (2001) The three-dimensional structure of cystathionine β-lyase from Arabidopsis and its substrate specificity. Plant Physiol 126: 631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LT. (1960) The effect of mimosine on alkaline phosphatase of mouse kidney. J Formos Med Assoc 59: 108–114 [Google Scholar]
- Crounse RG, Maxwell JD, Blank H. (1962) Inhibition of growth of hair by mimosine. Nature 194: 694–695 [DOI] [PubMed] [Google Scholar]
- Dai Y, Gold B, Vishwanatha JK, Rhode SL. (1994) Mimosine inhibits viral DNA synthesis through ribonucleotide reductase. Virology 205: 210–216 [DOI] [PubMed] [Google Scholar]
- Dewreede S, Wayman O. (1970) Effect of mimosine on the rat fetus. Teratology 3: 21–27 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fink AL. (1998) Protein aggregation: folding aggregates, inclusion bodies and amyloid. Fold Des 3: R9–R23 [DOI] [PubMed] [Google Scholar]
- Garcia GW, Ferguson TU, Neckles FA, Archibald KAE. (1996) The nutritive value and forage productivity of Leucaena leucocephala. Anim Feed Sci Technol 60: 29–41 [Google Scholar]
- Hamilton RI, Donaldson LE, Lambourne LJ. (1968) Enlarged thyroid glands in calves born to heifers fed a sole diet of Leucaena leucocephala. Aust Vet J 44: 484 [Google Scholar]
- Hashiguchi H, Takahashi H. (1977) Inhibition of two copper-containing enzymes, tyrosinase and dopamine β-hydroxylase, by L-mimosine. Mol Pharmacol 13: 362–367 [PubMed] [Google Scholar]
- Hylin JW. (1969) Toxic peptides and amino acids in foods and feeds. J Agric Food Chem 17: 492–496 [Google Scholar]
- Jones RJ, Blunt CG, Holmes JHG. (1976) Enlarged thyroid glands in cattle grazing Leucaena pastures. Trop Grassl 10: 113–116 [Google Scholar]
- Joshi HS. (1968) The effect of feeding on Leucaena leucocephala (Lam) de Wit. on reproduction in rats. Aust J Agric Res 19: 341–352 [Google Scholar]
- Lalande M. (1990) A reversible arrest point in the late G1 phase of the mammalian cell cycle. Exp Cell Res 186: 332–339 [DOI] [PubMed] [Google Scholar]
- Lin JY, Lin KT, Ling KH. (1963) Studies on the mechanism of toxicity of mimosine (β-(N-[3-hydroxy pyridone])-α-amino propionic acid). 3. The effect of mimosine on the activity of L-dopa decarboxylase, in vitro. J Formos Med Assoc 62: 587–592 [Google Scholar]
- Lin JY, Shih YM, Ling KH. (1962) Studies on the mechanism of toxicity of mimosine (β-(N-[3-hydroxypyridone])-α-aminopropionic acid). 1. Studies of the reactions of mimosine and pyridoxal 5-phosphate using the spectrophotometric method. J Formos Med Assoc 61: 997–1003 [Google Scholar]
- McCain DC, Croxdale J, Markley JL. (1989) Thermal damage to chloroplast envelope membranes. Plant Physiol 90: 606–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academy of Sciences (1984) Leucaena: Promising Forage and Tree Crop for the Tropics, Ed 2. National Academy Press, Washington, DC [Google Scholar]
- Negi VS, Bingham JP, Li QX, Borthakur D. (2013) midD-encoded ‘rhizomimosinase’ from Rhizobium sp. strain TAL1145 is a C-N lyase that catabolizes L-mimosine into 3-hydroxy-4-pyridone, pyruvate and ammonia. Amino Acids 44: 1537–1547 [DOI] [PubMed] [Google Scholar]
- Negi VS, Pal A, Singh R, Borthakur D. (2011) Identification of species-specific genes from Leucaena leucocephala using interspecies suppression subtractive hybridisation. Ann Appl Biol 159: 387–398 [Google Scholar]
- Pal A, Negi VS, Borthakur D. (2012a) Efficient in vitro regeneration of Leucaena leucocephala using immature zygotic embryos as explants. Agrofor Syst 84: 131–140 [Google Scholar]
- Pal A, Negi VS, Khanal S, Borthakur D. (2012b) Immunodetection of curcin in seed meal of Jatropha curcas using polyclonal antibody developed against curcin-L. Current Nutrition and Food Science 8: 213–219 [Google Scholar]
- Reis PJ, Tunks DA, Hegarty MP. (1975) Fate of mimosine administered orally to sheep and its effectiveness as a defleecing agent. Aust J Biol Sci 28: 495–501 [DOI] [PubMed] [Google Scholar]
- Shelton HM, Brewbaker JL (1994) Leucaena leucocephala: the most widely used forage tree legume. In RC Gutteridge, HM Shelton, eds, Forage Tree Legumes in Tropical Agriculture. CAB International, Wallingford, UK, pp 1–13 [Google Scholar]
- Smith IK, Fowden L. (1966) A study of mimosine toxicity in plants. J Exp Bot 17: 750–761 [Google Scholar]
- Soedarjo M, Borthakur D. (1998) Mimosine, a toxin produced by the tree-legume Leucaena provides a nodulation competition advantage to mimosine-degrading Rhizobium strains. Soil Biol Biochem 30: 1605–1613 [Google Scholar]
- Soedarjo M, Hemscheidt TK, Borthakur D. (1994) Mimosine, a toxin present in leguminous trees (Leucaena spp.), induces a mimosine-degrading enzyme activity in some Rhizobium strains. Appl Environ Microbiol 60: 4268–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda S. (1960) On the physiological properties of mimosine. Bot Mag 73: 142–147 [Google Scholar]
- Tangendjaja B, Lowry JB, Wills RBH. (1984) Optimisation of conditions for the degradation of mimosine in Leucaena leucocephala leaf. J Sci Food Agric 35: 613–616 [Google Scholar]
- Tangendjaja B, Lowry JB, Wills RBH. (1986) Isolation of a mimosine degrading enzyme from leucaena leaf. J Sci Food Agric 37: 523–526 [Google Scholar]