A transcription factor increases gibberellin biosynthesis and improves seed longevity.
Abstract
Seed longevity is crucial for agriculture and plant genetic diversity, but it is limited by cellular damage during storage. Seeds are protected against aging by cellular defenses and by structures such as the seed coat. We have screened an activation-tagging mutant collection of Arabidopsis (Arabidopsis thaliana) and selected four dominant mutants with improved seed longevity (isl1-1D to isl4-1D) under both natural and accelerated aging conditions. In the isl1-1D mutant, characterized in this work, overexpression of the transcription factor ARABIDOPSIS THALIANA HOMEOBOX25 (ATHB25; At5g65410) increases the expression of GIBBERELLIC ACID3-OXIDASE2, encoding a gibberellin (GA) biosynthetic enzyme, and the levels of GA1 and GA4 are higher (3.2- and 1.4-fold, respectively) in the mutant than in the wild type. The morphological and seed longevity phenotypes of the athb25-1D mutant were recapitulated in transgenic plants with moderate (4- to 6-fold) overexpression of ATHB25. Simultaneous knockdown of ATHB25, ATHB22, and ATHB31 expression decreases seed longevity, as does loss of ATHB25 and ATHB22 function in a double mutant line. Seeds from wild-type plants treated with GA and from a quintuple DELLA mutant (with constitutive GA signaling) are more tolerant to aging, providing additional evidence for a role of GA in seed longevity. A correlation was observed in several genotypes between seed longevity and mucilage formation at the seed surface, suggesting that GA may act by reinforcing the seed coat. This mechanism was supported by the observation of a maternal effect in reciprocal crosses between the wild type and the athb25-1D mutant.
Seed longevity is an important trait for maintaining plant genetic diversity in nature and seed banks and for providing reliable crop seeds to farmers. Seeds gradually lose their viability during storage, and the rate of aging is strongly influenced by environmental and genetic factors (Rajjou and Debeaujon, 2008). The accumulation of reactive oxygen species and free radicals has often been considered as one of the most important factors influencing seed aging (Bailly, 2004). Accordingly, ferric-chelate reductase1, a mutant in mitochondrial NADH dehydrogenase showing constitutive reactive oxygen species accumulation, produces seeds very sensitive to aging (Clerkx et al., 2004). Reactive oxygen species induce lipid oxidation, and Arabidopsis (Arabidopsis thaliana) mutants (vitamin E deficient1 and vitamin E deficient2) affected in the biosynthesis of vitamin E (tocopherol, a lipophilic antioxidant) produce seeds with reduced longevity (Sattler et al., 2004). Repair of oxidized proteins by Met sulfoxide reductase (Châtelain et al., 2013), of oxidized DNA by DNA glycosylase/Apurinic/Apyrimidinic lyase (Chen et al., 2012), and of chemically degraded proteins by l-isoaspartyl methyltransferase (Ogé et al., 2008) enhances seed longevity. Other important factors are chaperones or heat shock proteins. Transgenic tobacco (Nicotiana tabacum) plants overexpressing the seed-specific transcription factor Heat Shock FactorA9 (Kotak et al., 2007) produce seeds with increased amounts of heat shock proteins that have improved longevity (Prieto-Dapena et al., 2006). On the other hand, a double mutant in two redundant peptidyl-prolyl cis-trans-isomerases, rotamase fkbp1 rotamase fkbp2, has seeds more sensitive to aging than the wild type (Bissoli et al., 2012). In addition to cellular stress defenses and repair systems, the protective effects of the seed coat or testa are very important for seed longevity (Rajjou and Debeaujon, 2008). Seeds from both structural and pigmentation mutants in the seed coat lose viability during aging faster than wild-type seeds (Debeaujon et al., 2000; Clerkx et al., 2004).
The differentiation of the seed coat, including mucilage production, occurs during the morphogenesis phase of seed development, while the accumulation of storage compounds and stress defenses and the acquisition of dormancy and desiccation tolerance occur during the second maturation phase (Holdsworth et al., 1999; Braybrook and Harada, 2008; Western, 2012). These processes are orchestrated by the LEAFY COTYLEDON (LEC) transcription factors (LEC1, LEC2, and FUSCA3 in Arabidopsis) and mediated by the growth hormones auxin and gibberellin (GA), the stress hormone abscisic acid (ABA), and the seed-specific, ABA-activated transcription factor ABSCISIC ACID INSENSITIVE3 (ABI3) (Holdsworth et al., 1999; Gutierrez et al., 2007; Braybrook and Harada, 2008; Suzuki and McCarty, 2008). ABI3 activates the previously mentioned transcription factor HSFA9 (Kotak et al., 2007). Mutants affected in seed development, such as lec1, lec2, fus3, and abi3, have seeds of poor desiccation tolerance and longevity (Holdsworth et al., 1999; Clerkx et al., 2003, 2004).
In the maturation phase of seed development, a progressive increase in ABA levels prepares the seeds to resist desiccation and aging in a dormant state. The levels of GAs are reduced during the latest stages of maturation, but an increase in GA is required to break the ABA-induced dormancy at the time of germination (Holdsworth et al., 1999; Braybrook and Harada, 2008). No connection between GA and seed longevity has been found in previous genetic analyses (Clerkx et al., 2004).
Blind genetic screens to identify crucial genes for seed longevity have never been made. A genetic analysis to map quantitative trait loci for seed longevity has been performed (Nguyen et al., 2012), but no genes were identified so far. Also, all mutants previously described were obtained from known genes of stress defenses or involved in seed development. We have screened an “activation-tagging” mutant collection of Arabidopsis (Weigel et al., 2000), selecting for tolerance of seeds to natural and accelerated aging (Tesnier et al., 2002; Tejedor-Cano et al., 2010). As a first result, we report that the transcription factor ARABIDOPSIS THALIANA HOMEOBOX25/ZINC FINGER HOMEODOMAIN2/ZINC FINGER HOMEODOMAIN1 (ATHB25/ZFHD2/ZHD1; At5g65410) is an important determinant of seed longevity by positively regulating the GIBBERELLIC ACID3-OXIDASE2 (GA3OX2) gene and, therefore, GA biosynthesis. Thus, GA has a novel and unexpected role during seed development, increasing longevity by a mechanism unrelated to ABA and stress defenses. This may involve reinforcement of the seed coat, because the athb25-1D mutation (overexpressing ATHB25) exhibits a maternal effect.
RESULTS
Isolation of Arabidopsis Mutants with Increased Seed Longevity
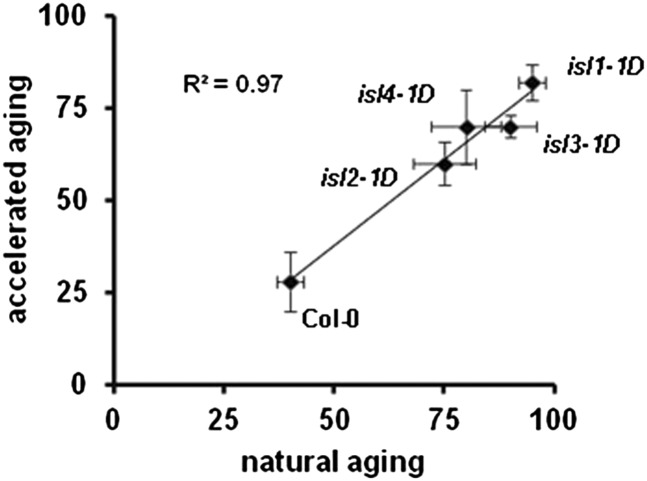
We screened 30,000 lines of an Arabidopsis activation-tagging mutant collection (approximately 150,000 seeds) for increased seed longevity as determined by an accelerated aging procedure of imbibed seeds (24 h at 42°C in water; see “Materials and Methods”). In order to discern if our aging procedure induces secondary dormancy, we tested if there is an increase of ABA (Holdsworth et al., 1999; Braybrook and Harada, 2008), because it has been reported that high temperatures induce ABA biosynthesis during germination (Toh et al., 2008). Under our experimental conditions (high temperature during accelerated aging but normal temperature during germination), ABA levels are not increased (Supplemental Fig. S1); therefore, our mutants are not related to the breaking of secondary dormancy. Four dominant mutants were identified and named isl1-1D (for increased seed longevity, allele 1, dominant) to isl4-1D, and we could demonstrate a good correlation between the germination of wild-type and mutant seeds after our accelerated aging procedure and after natural aging in the dry state during 30 months at 20°C to 22°C and 40% to 50% humidity (Fig. 1). The characterization of the isl1-1D mutant is the subject of this work. The other mutants are not allelic to isl1-1D and will be described elsewhere.
Figure 1.
Correlation between germination after accelerated aging and after natural aging in the mutants with increased seed longevity. Seeds from wild-type Columbia (Col-0) and from four isl mutants were subjected to natural aging (dry seeds incubated during 30 months at room temperature and humidity) or to accelerated aging (seeds imbibed in water and incubated for 24 h at 42°C). The percentage of germination (mean of three experiments with 100 seeds each with se in both coordinates) is shown for the five genotypes as well as the correlation coefficient.
Overexpression of ATHB25 Increases Seed Longevity
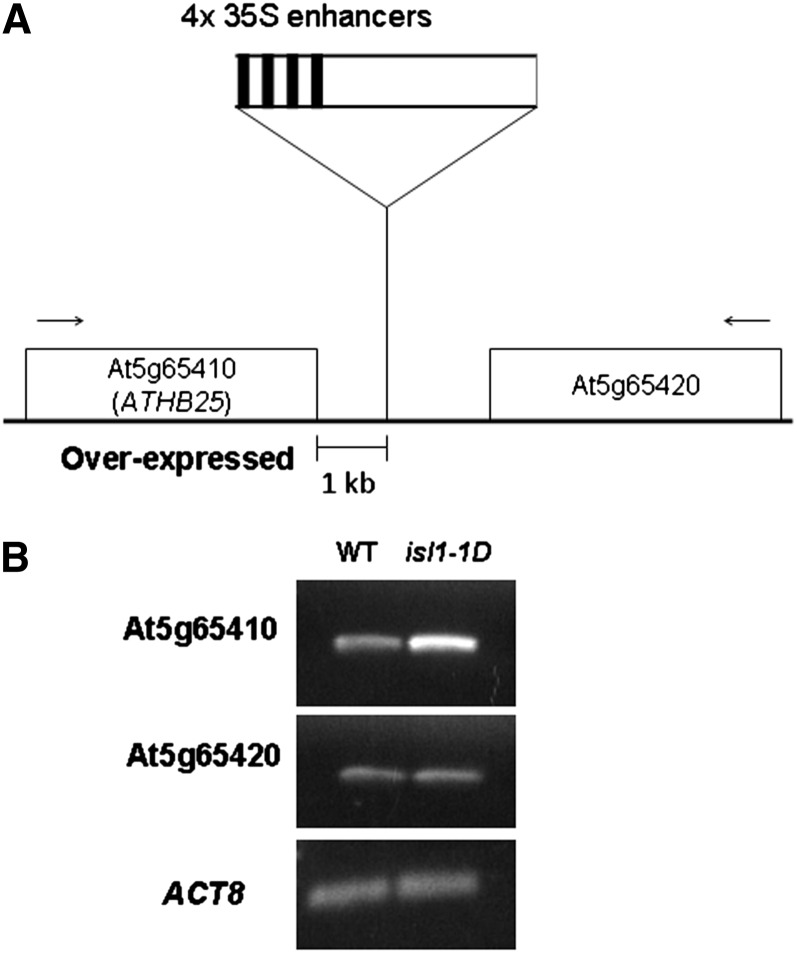
We detected in the isl1-1D mutant the presence of only one transfer DNA (T-DNA) insertion by Southern blot (Supplemental Fig. S2). The insertion was located by plasmid rescue and sequencing to a region between the At5g65410 and At5g65420 genes (Fig. 2A). The 35S transcriptional enhancer was close to the promoter of the At5g65410 gene, encoding transcription factor ATHB25/ZFHD2/ZHD1. This gene, but not At5g65420, was overexpressed in the mutant (Fig. 2B). In order to test whether the tolerance to aging of this mutant was linked to the T-DNA insertion, homozygous mutant plants were crossed to Columbia wild-type plants. From the segregating F2 generation, 30 plants from seeds tolerant to accelerated aging were selected, and the T-DNA was detected by PCR in all of them. Accordingly, we conclude that the phenotype of this mutant is linked to the T-DNA (χ2 = 10 and P < 0.0025 for the hypothesis that it follows a 3:1 segregation, which must be rejected).
Figure 2.
Molecular characterization of the isl1-1D mutant. A, Location of the T-DNA in the intergenic region between At5g65410 and At5g65420 as determined by plasmid rescue. B, RT-PCR semiquantitative analysis of At5g65410, At5g65420, and ACTIN8 (ACT8) mRNAs from the wild type (WT) and the isl1-1D mutant.
As indicated above, the At5g65410 gene has received three different names: ATHB25 (Tan and Irish, 2006), ZFHD2 (Tran et al., 2007), and ZHD1 (Hu et al., 2008). We respect the first naming (ATHB25), and the overexpression mutant isl1-1D will be renamed athb25-1D.
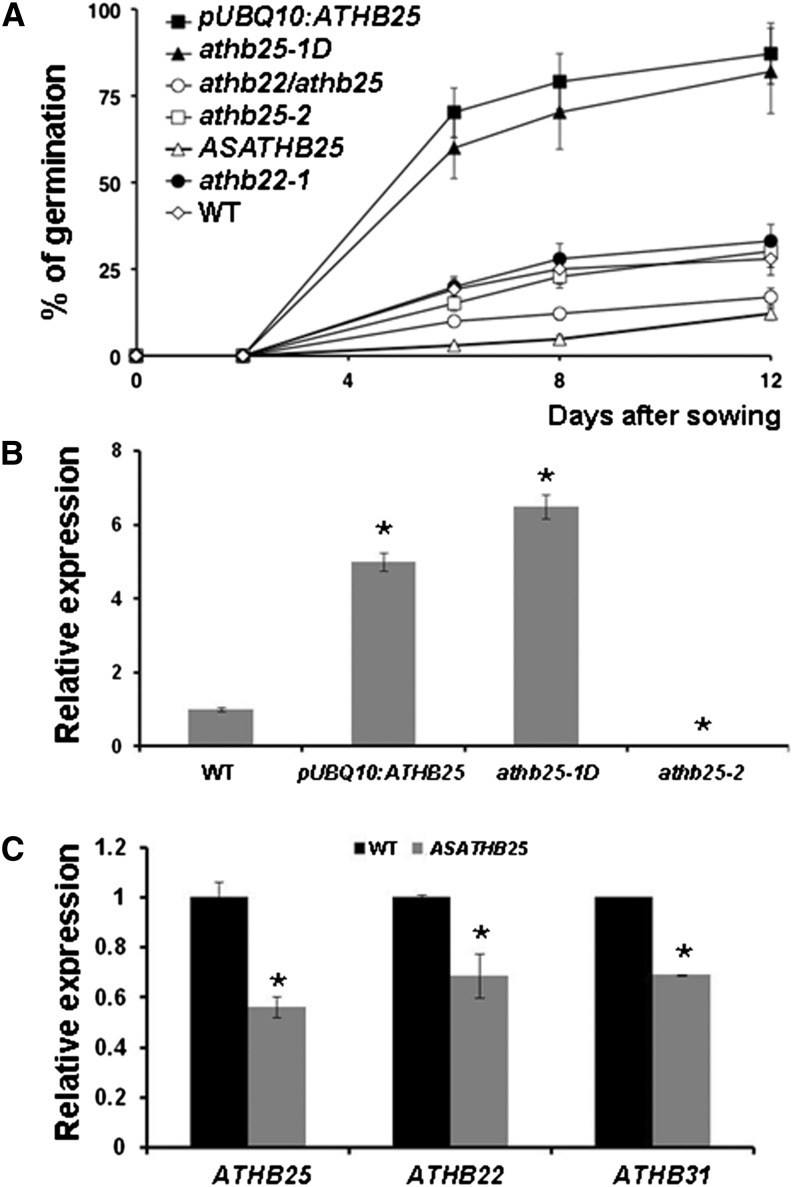
To confirm the relationship between ATHB25 and seed longevity, we generated transgenic plants in which the complementary DNA (cDNA) of the gene was cloned in sense and antisense orientations under the control of two promoters expressed in nearly all tissues: the strong 35S promoter (Benfey and Chua, 1990) and the moderate UBIQUITIN10 promoter (Grefen et al., 2010). Transgenic lines overexpressing ATHB25 from the UBQ10 promoter expressed ATHB25 at levels similar to those of the athb25-1D mutant (5- to 6-fold wild-type levels; Fig. 3B) and exhibited improved germination after accelerated aging (Fig. 3A). It must be remarked that all lines overexpressing ATHB25 exhibited normal germination in the absence of the aging treatment (data not shown), and the expression of ATHB25 was not significantly affected by the accelerated aging treatment (Supplemental Fig. S3). Five tested transgenic lines containing the 35S promoter had very high expression levels of ATHB25 (40- to 50-fold wild-type levels) and did not show any improvement in seed longevity. These 35S promoter lines did not exhibit the GA-dependent phenotypes of the original mutant and the UBQ10 promoter lines (see below). These results suggest that the increased expression of ATHB25 should be tightly regulated to improve seed longevity.
Figure 3.
ATHB25 (At5g65410) plays an important role in the seed longevity of Arabidopsis. A, Seeds from pUBQ10:ATHB25, activation-tagging mutant isl1-1D (referred to as athb25-1D), double null mutant athb22-1 athb25-2, single null mutant athb25-2, ASATHB25, single null mutant athb22-1, and the wild type (WT) were subjected to accelerated aging for 24 h and sown on MS plates. The percentage of germination was recorded at the indicated times. The results are averages of six experiments with 100 seeds per line, and the se is indicated by error bars. Untreated lines germinated more than 99% after 3 d. B, Expression levels of ATHB25 in the wild type and mutants with gain (pUBQ10:ATHB25 and athb25-1D) and loss (athb25-2) of function of ATHB25 determined by quantitative RT-PCR. C, Expression levels of ATHB25 (At5g65410) and related genes ATHB22 (At4g24660) and ATHB31 (At1g14440) in the wild type and ASATHB25 (three independent lines were tested with similar results). In B and C, results are averages of three determinations, and error bars correspond to se. Asterisks indicate significant differences with controls (the wild type) at P < 0.05. In the case of pUBQ10:ATHB25 and ASATHB25, three independent transgenic lines were tested with similar results. In B and C, plants (10-d-old seedlings grown in solid MS medium) were grown under long-day conditions before RNA extraction.
Further evidence supporting the role of ATHB25 in seed longevity is the poor seed viability observed after accelerated aging treatment of the lines expressing an antisense construct of this gene with the 35S promoter (Fig. 3A), where the expression level of ATHB25 is compromised (Fig. 3C). However, a loss-of-function mutant (athb25-2) did not show sensitivity to aging despite greatly reduced expression of this gene (Fig. 3B). This can be explained by the high sequence similarity (more than 75% sequence identity at the nucleotide level) and genetic redundancy of the Arabidopsis zinc finger-homeobox gene family, consisting of 14 members with overlapping regulatory roles in Arabidopsis floral development (Tan and Irish, 2006). As indicated in Figure 3C, the antisense construct of ATHB25 inhibits the expression of two other members of this family, the closely related genes At4g24660 (ATHB22) and At1g14440 (ATHB31), and this may explain the observed phenotype. This cross reaction of antisense constructs has been observed in other gene families with high sequence identity (Tieman et al., 2001).
ATHB22 is the zinc finger homeodomain most similar to ATHB25 (Tan and Irish, 2006). Additional evidence for a redundant role of the corresponding genes (ATHB25 and ATHB22) in seed longevity has been provided by determining the tolerance to accelerated aging of a double mutant in both genes. As indicated in Figure 3A, the double mutant exhibits a reduced tolerance to accelerated aging, a phenotype similar to that of the antisense line.
GA-Dependent Phenotypes in ATHB25-Overexpressing Lines
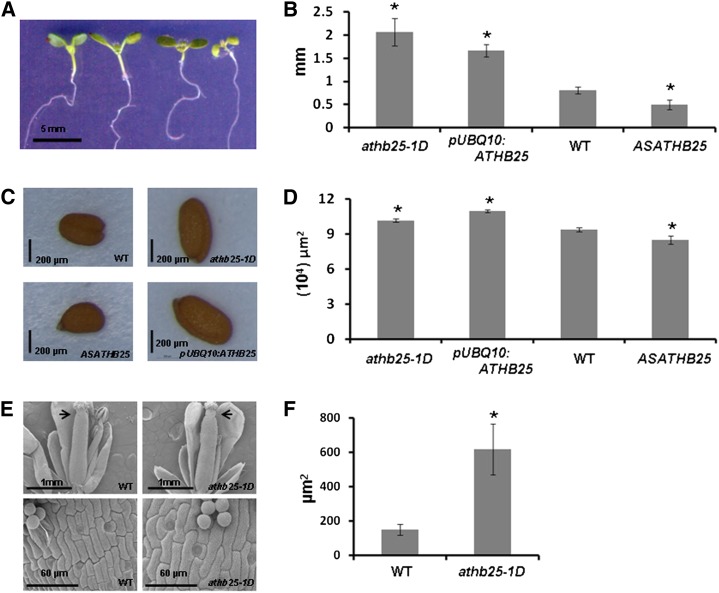
ATHB25 is significantly expressed in the shoot apex, vegetative rosette, flowers, and seed embryo (Arabidopsis electronic Fluorescent Pictograph browser; http://bbc.botany.utoronto.ca); therefore, we carried out a phenotypic characterization in shoots of the athb25-1D mutant and transgenic plants overexpressing ATHB25 from the UBQ10 promoter. At first glance, both lines displayed pleiotropic phenotypes that may be associated with typical responses to growth-promoting hormones such as GAs and auxins. These include elongated petioles and hypocotyls (Fig. 4, A and B), earlier flowering, larger seeds (Fig. 4, C and D), wider siliques, and increased cell size (Fig. 4, E and F). Plants expressing an antisense construct of ATHB25 from the 35S promoter have shorter petioles and hypocotyls (Fig. 4, A and B) and smaller seeds (Fig. 4, C and D).
Figure 4.
ATHB25 regulates GA-dependent phenotypes. A, Hypocotyl length of representative seedlings of athb25-1D, pUBQ10:ATHB25, the wild type, and ASATHB25 (from left to right, with one representative plant for every line) on MS medium under long-day conditions for 7 d. B, Means from three independent experiments as in A with 10 plants from each line. C, Representative dry seeds of athb25-1D, pUBQ10:ATHB25, the wild type (WT), and ASATHB25. D, Means from three independent experiments of the projected area of 50 dry seeds from each line as in C. E, Scanning electron micrographs of flowers (top) and cells of the style (bottom) of the wild type and athb25-1D. F, Means from 10 cells of the style areas from the lines shown in E. Arrows indicate the locations of the cells measured in the flower; error bars indicate se, and asterisks indicate significant differences from controls (the wild type) at P < 0.05. In the case of pUBQ10:ATHB25 and ASATHB25, three independent transgenic lines were tested with similar results. [See online article for color version of this figure.]
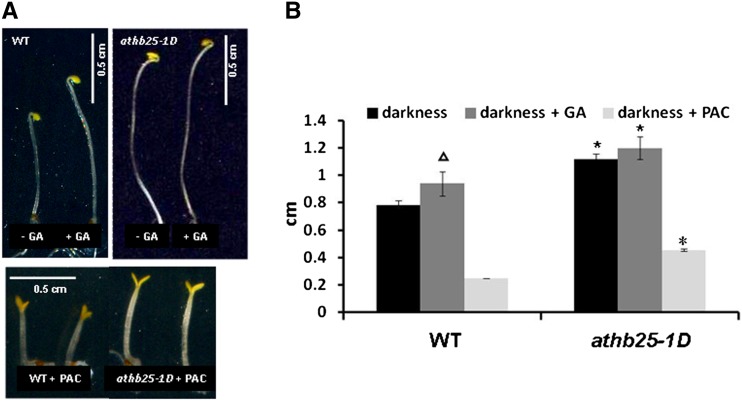
In order to determine which growth-promoting hormone(s) could explain these phenotypes, other auxin-related phenotypes, such as apical dominance or epinastic cotyledons (Kim et al., 2007), were analyzed, but no changes with respect to the wild type were observed (data not shown). Next, we tested whether GA signaling is activated in athb25-1D by monitoring hypocotyl length in the presence of paclobutrazol, an inhibitor of GA biosynthesis (Cowling and Harberd, 1999). In darkness and after paclobutrazol treatment, the reduction in the hypocotyl elongation of athb25-1D (58%) was less effective than in the wild type (70%; Fig. 5). This result and the failure of the already elongated hypocotyls of athb25-1D to respond to exogenous GA for elongation (Fig. 5A) reveal that ATHB25 is somehow related to the synthesis or the signal transduction of this important phytohormone.
Figure 5.
Effects of GA and paclobutrazol (PAC) in the athb25-1D mutant. A, Hypocotyls of representative 5-d-old seedlings of the wild type (WT) and athb25-1D grown in darkness with or without 50 µm GA (top) and with 0.5 µm PAC (bottom). B, Means from three independent experiments quantifying the hypocotyl length in 10 plants of lines as in A. Error bars indicate se, and asterisks indicate significant differences from controls (the wild type) at P < 0.05; the triangle indicates a significant difference (P < 0.05) between darkness and darkness + GA in the wild type. [See online article for color version of this figure.]
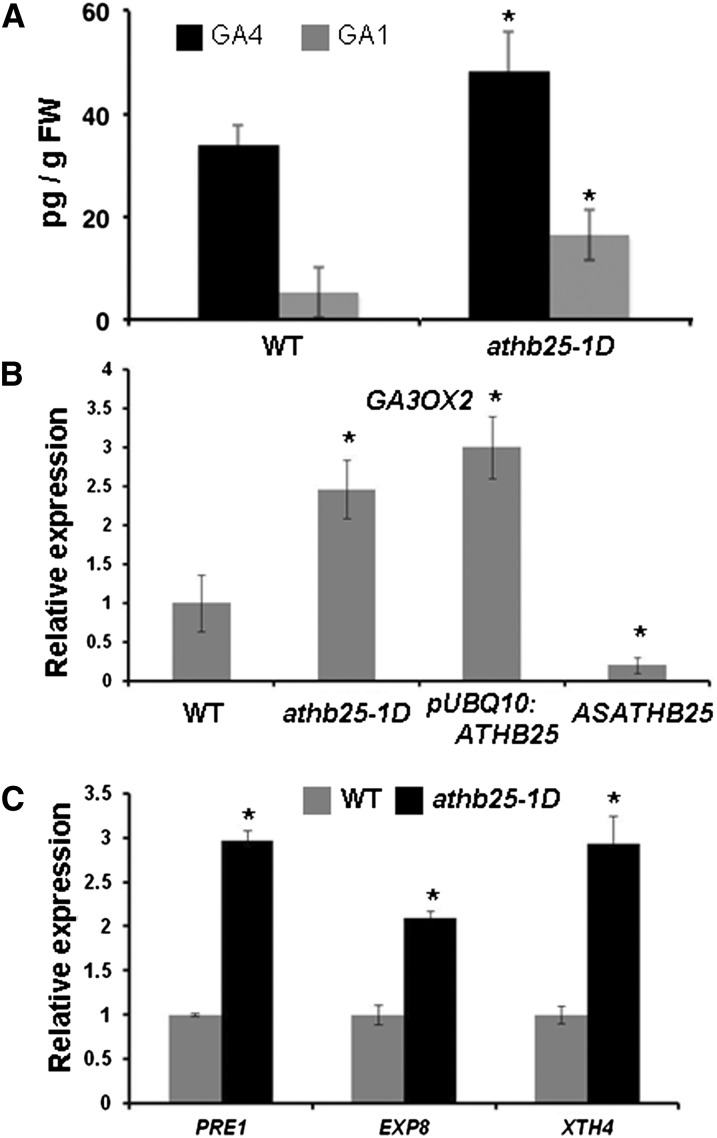
In order to determine whether GA-dependent phenotypes exhibited by ATHB25-overexpressing lines are due to the overproduction of this hormone, as observed in lines overexpressing GA20OX1 (Huang et al., 1998), or due to the constitutive activation of the GA signaling pathway, as in spindly mutants (Jacobsen and Olszewski, 1993), the level of the endogenous bioactive GA was measured in 10-d-old seedlings. Wild-type seedlings accumulated the expected levels of both GA4 and GA1 (Huang et al., 1998). However, athb25-1D seedlings accumulated 1.4 times more GA4 and showed a 3.2-fold increase in the level of GA1 (Fig. 6A). Then, we used quantitative reverse transcription (RT)-PCR to measure the expression levels of the four genes of GA 3-oxidases and the five genes of GA 20-oxidases in Arabidopsis, whose induction could explain the high level of GA accumulated in the athb25-1D mutant (Hedden and Thomas, 2012). The expression levels of GA3OX1 and GA3OX3 were below detection, but we found a 2.5-fold induction of GA3OX2 in this mutant (Fig. 6B). As indicated in Supplemental Figure S4, GA3OX4 and some GA 20-oxidase genes (GA20OX1 and GA20OX3) were only slightly induced (less than 1.5-fold) in the athb25-1D mutant, while GA20OX4 and GA20OX5 were repressed in this mutant. GA3OX2 is a key gene responsible for GA biosynthesis (Curaba et al., 2004) and is mainly expressed in germinating seeds (Yamaguchi et al., 2001), predominantly in the hypocotyls (Mitchum et al., 2006). The regulation of the expression of this GA 3-oxidase by ATHB25 was confirmed by the induction of GA3OX2 in the pUBQ10:ATHB25 lines and by the repression in the antisense (ASATHB25) lines (Fig. 6B). In agreement with the described results, we observed that several GA response genes implicated in the control of cell elongation, such as PACLOBUTRAZOL RESISTANCE1 (PRE1), EXPANSIN8 (EXP8), XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE4 (XTH4) (Ogawa et al., 2003; Bai et al., 2012), have increased expression in the athb25-1D mutant with respect to the wild type (Fig. 6C). This may explain why the athb25-1D mutant presents larger cells in the style of the flower (Fig. 4, E and F).
Figure 6.
ATHB25 increases GA levels and the expression of GA3OX2. A, Endogenous GA4 and GA1 levels in 10-d-old athb25-1D and wild-type (WT) seedlings. FW, Fresh weight. B, Quantitative RT-PCR analyses were performed in triplicate on RNA samples from 10-d-old seedlings. Relative values refer to the expression levels of the GA3OX2 gene in the athb25-1D mutant, pUBQ10:ATHB25, and ASATHB25 with respect to the wild type, which was taken as 1. Error bars indicate se, and asterisks indicate significant differences from controls (the wild-type) at P < 0.05. C, Quantitative RT-PCR analyses were performed in triplicate on RNA samples from 10-d-old seedlings. Relative values refer to the expression levels of PRE1, EXP8, and XTH4 genes in the athb25-1D mutant with respect to the wild type, which was taken as 1. Error bars indicate se, and asterisks indicate significant differences from controls (the wild type) at P < 0.05.
Global Gene Expression of the ATHB25 Overexpression Mutant Reveals an Induction of GA-Responsive Genes
In order to further characterize the athb25-1D mutant, we studied its differential gene expression with respect to the wild type by microarray experiments. A 2-fold change threshold was used to identify 598 differentially expressed genes; 208 genes were up-regulated and 390 genes were down-regulated in athb25-1D as compared with wild-type plants (Table I; Supplemental Table S1). We failed to identify any overrepresented functional categories among the up-regulated genes using Gene Ontology. However, by comparing this group of genes with those induced after GA3 treatment (Nottingham Arabidopsis Stock Centre [NASC] array-177), we found a significant overlap (P = 0.037) between these two sets of genes: 24 genes (11.5%) induced in athb25-1D were also induced after GA3 treatment (Tables I and II). Analyzing down-regulated genes, we found several overrepresented Gene Ontology categories involved in “response to stimulus.” The dominant categories for repressed genes were “defense response” (P = 1.06e−10), including “response to water deprivation” (P = 3.94e−5), “response to biotic stimulus” (P = 2.41e−3), and “response to ABA” (P = 3.7e−3; Table I).
Table I. Summary of microarray data of genes differentially expressed in the athb25-1D mutant with respect to the wild type.
| Regulation | No. of Genes | Functional Category Overrepresented | Motifs Overrepresented |
|||
|---|---|---|---|---|---|---|
| Sequence | Window | Percentage in Microarray | Percentage in Arabidopsis | |||
| % | ||||||
| Up-regulated | 208 | GA-responsive genes (24) | TATAWA | (−44, −18) | 38.97 | 25.24 |
| AATAAA | (−106, −85) | 15.44 | 9.44 | |||
| AAAG | (−140, −90) | 65.44 | 58.47 | |||
| TGAC | (−93, −68) | 22.79 | 16.85 | |||
| AMCWAMC | (−141, −107) | 11.76 | 5.26 | |||
| TGTCTC | (−29, 52) | 11.76 | 6.85 | |||
| TAATTA | (−250, −150) | 17.5 | 14 | |||
| Down-regulated | 390 | Response to water deprivation (15) Response to biotic stimulus (22) Response to ABA (15) Programmed cell death (15) Transmembrane receptor activity (13) | TATAWA | (−38, −23) | 34.74 | 22.67 |
| ACGT | (−241, −12) | 53.78 | 46.1 | |||
| ACGTG | (−227, −2) | 35.95 | 25.9 | |||
| CACGTG | (−222, −8) | 12.69 | 8.68 | |||
| ACGTGKC | (−188, −11) | 12.08 | 8.28 | |||
| BACGTGKM | (−244, −11) | 16.92 | 11.97 | |||
| GATA | (−148, −142) | 12.08 | 6.21 | |||
Table II. Genes induced in the athb25-1D mutant with respect to the wild type and that are responsive to GAs according to NASC array-177 (2- to 9-fold induction).
| GAs Response Genes | Fold Change | Description |
|---|---|---|
| At3g46480 | 6.1 | 2-Oxoglutarate and Fe(II)-dependent oxygenase superfamily protein |
| At2g44570 | 5.5 | GLYCOSYL HYDROLASE9B12 |
| At5g15360 | 5.4 | Unknown protein |
| At1g18220 | 4.6 | PURINE PERMEASE9 |
| At4g29200 | 4.1 | β-Galactosidase-related protein |
| At1g43880 | 3.9 | Transposable element gene |
| At5g49250 | 3.8 | β-Galactosidase-related protein |
| At4g08840 | 3.7 | PUMILIO11 |
| At1g29090 | 3.6 | Cys proteinase superfamily protein |
| At1g10420 | 3.6 | Protein coding |
| At5g48850 | 3.4 | SULFUR DEFICIENCY INDUCED1 |
| At2g01200 | 3.4 | INDOLE-3-ACETIC ACID INDUCIBLE32 |
| At3g56890 | 3.4 | F-box-associated ubiquitination effector family protein |
| At2g17430 | 3.0 | MILDEW RESISTANCE LOCUS O7 |
| At1g26530 | 2.8 | PIN domain-like family protein |
| At1g59640 | 2.8 | Basic helix-loop-helix-encoding gene (BIGPETAL) |
| At3g15130 | 2.8 | Tetratricopeptide repeat-like superfamily protein |
| At1g14970 | 2.4 | O-Fucosyltransferase family protein |
| At5g28680 | 2.2 | Receptor-like kinase (ANXUR2) |
| At1g64290 | 2.2 | F-box protein related |
| At1g62940 | 2.2 | ACYL-COENZYME A SYNTHETASE5 |
| At4g37770 | 2.1 | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE8 |
| At5g38070 | 2.1 | RING/FYVE/PHD zinc finger superfamily protein |
| At1g04645 | 2.0 | Plant self-incompatibility protein S1 family |
Determining the expression of several genes by quantitative RT-PCR validated the microarray results. Among induced genes, three of the GA-responsive genes, PRE1, EXP8, and XTH4, were already analyzed (Fig. 6C). In addition, the expression of the gene encoding the epithiospecifier protein was tested. In the group of repressed genes, LEUCINE RICH REPEAT PROTEIN1 (Choi et al., 2012) and MITOGEN ACTIVATED PROTEIN KINASE/EXTRACELLULAR SIGNAL REGULATED KINASE KINASE KINASE1 (Suarez-Rodriguez et al., 2007) were analyzed as representative kinases involved in defense signaling. All of them confirmed the microarray results (Supplemental Fig. S5).
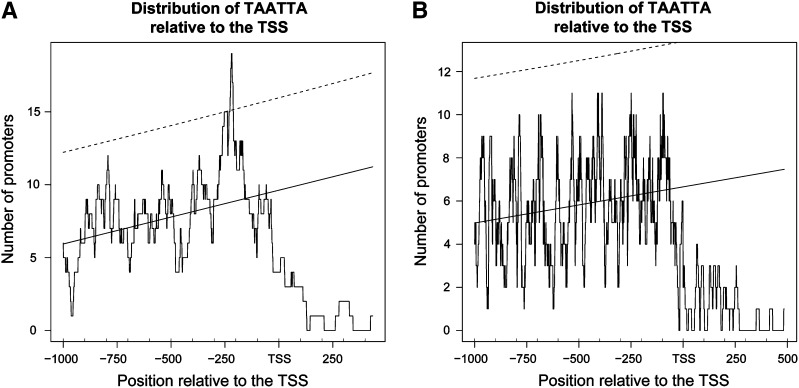
We carried out a “preferentially located motif” study (Bernard et al., 2010) in the two sets of differentially regulated genes (Table I). This approach allows the identification of overrepresented motifs in a specific location relative to the transcription start site. Among the genes up-regulated in the athb25-1D samples, we observed, at positions −150 to −250 from the transcription start site, the TAATTA motif (Fig. 7). This is directly bound by Arabidopsis ATHB33 and includes the sequence ATTA, the canonical binding site for homeodomain proteins (Tan and Irish, 2006). Also enriched in the promoters of up-regulated genes were AAAG sites (the core site required for the binding of DNA binding with one finger [Dof] transcription factors and for the induction of some genes by GA; Sun and Gubler, 2004) and TGTCTC sites (the binding site for auxin response transcription factors; Guilfoyle and Hagen, 2007). The motif most represented for down-regulated genes is ABRE (YACGTGKC; International Union of Biochemistry coding: Y = T or C; K = G or T; Yamaguchi-Shinozaki and Shinozaki, 2005), a motif for ABA-induced genes included in the promoters of many GA down-regulated genes (Ogawa et al., 2003).
Figure 7.
Distribution of the TAATTA motif in promoters of genes modulated by overexpression of ATHB25. A, The set of 136 promoters for induced genes, where a preferential position of this motif has been detected in the region [−250, −150] before the transcription start site (TSS). B, The set of 331 promoters for repressed genes, where no significant peak was detected. In both graphs, the solid line is the linear mean and the dashed line is the 95% confidence interval.
Seed Longevity in Arabidopsis Is Regulated by GAs
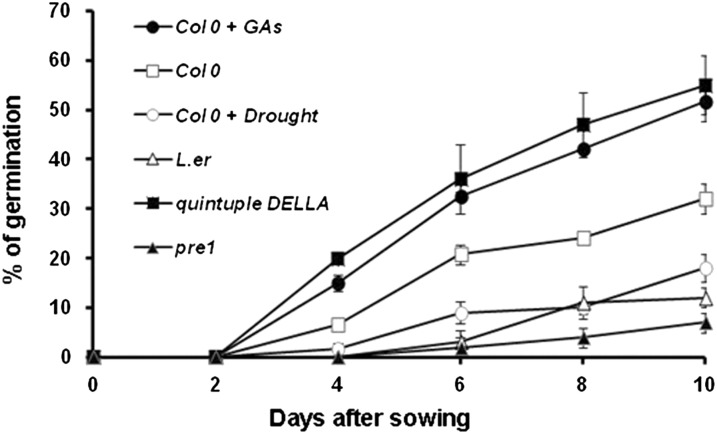
GAs have been recognized as plant growth regulators that act as endogenous hormones controlling various aspects of growth and development, such as seed germination, stem elongation, fruit growth, leaf shape, and flower development (Swain and Singh, 2005). However, we are unaware of any published study linking GA and seed longevity. Therefore, the key question suggested by our results is whether the ability of ATHB25-overexpressing lines to resist seed aging is due to the overaccumulation of bioactive GA. We carried out an accelerated aging experiment of seeds collected from plants either treated with GA3 or subjected to drought stress. We also included the quintuple DELLA mutant (gibberellic acid insensitive-t6 [gai-t6] repressor of gibberellic acid-t2 [rga-t2] repressor of gibberellic acid like1-1 [rgl1-1] rgl2-1 rgl3-1) and pre1 mutant lines in this analysis. While all of these lines exhibited normal germination in the absence of the aging treatment (data not shown), all demonstrated a correlation between GA and seed longevity. GA3-treated plants and the quintuple DELLA mutant, which shows constitutive activation of the GA signaling pathway, produced seeds that are more resistant to accelerated aging, as compared with their respective controls. However, plants subjected to drought stress, with an expected decrease in GA content (Wang et al., 2008), and the pre1 mutant, with a compromised response to GA (Bai et al., 2012), produced seeds less resistant to accelerated aging, as compared with their respective controls (Fig. 8). Finally, the addition of GA3 to the germination plates did not rescue aged seeds to germinate (data not shown), as expected if these seeds were irreversibly damaged and not just dormant.
Figure 8.
Seed longevity in Arabidopsis is regulated by GA. Seeds from control Columbia (Col 0), from Columbia treated with 50 µm GA (Col 0 + GA) or subjected to drought stress (Col 0 + drought), the pre1 mutant (in the Columbia background), control Landsberg erecta (L.er), and the quintuple DELLA mutant (in the Landsberg erecta background) were subjected to accelerated aging treatment for 24 h and sown on MS plates. The percentage of germination was recorded after 4, 6, 8, and 10 d. The results are means of six experiments with 100 seeds per line, and error bars indicate se. Untreated lines germinated 100% after 3 d.
Seed Longevity Dependent on GA Correlates with Mucilage Formation and Shows a Maternal Effect
The mechanism of GA action on seed longevity cannot be related to cellular stress defenses because these hormones do not induce them as ABA does and, in addition, GAs repress ABA-induced genes (Ogawa et al., 2003). As GAs regulate growth and development (Swain and Singh, 2005), the alternative mechanism could be that GAs promote the formation of some parts of the seed coat, an important structure for seed longevity (Debeaujon et al., 2000; Clerkx et al., 2004; Rajjou and Debeaujon, 2008).
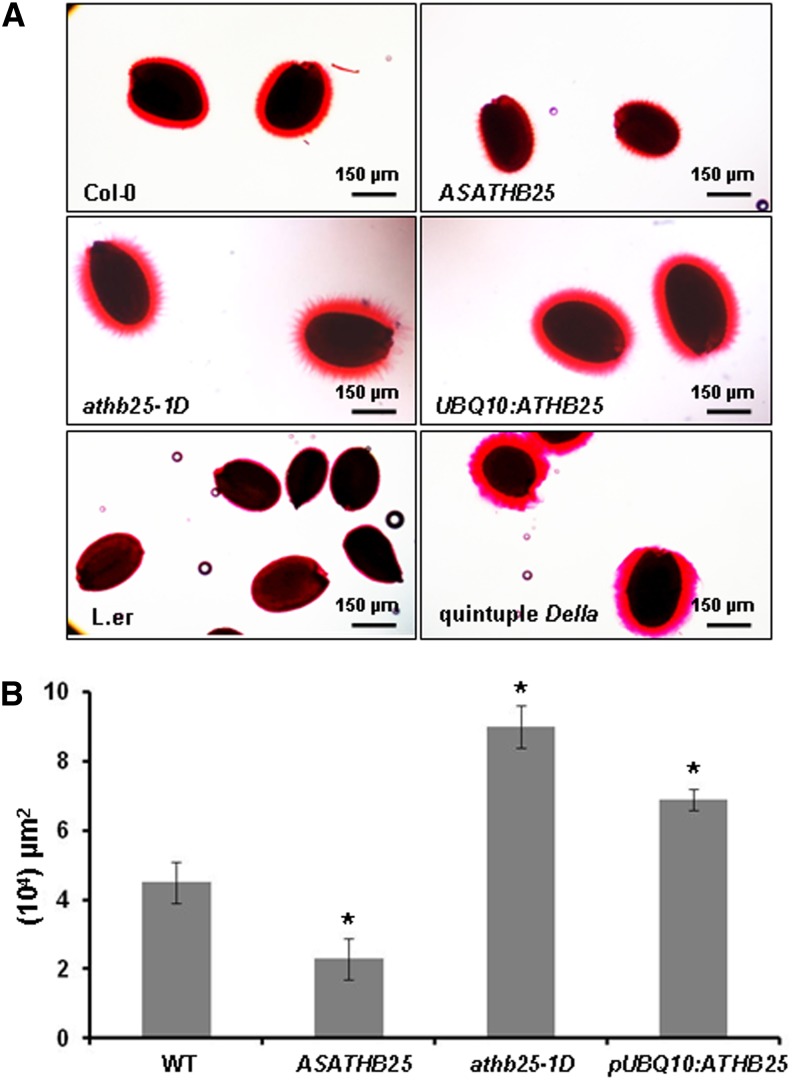
Kim et al. (2005) have reported that the seed coat of a knockout mutant in the GA3OX4 gene (encoding a seed-specific enzyme of GA biosynthesis) is distorted, with abnormal cell shape and reduced mucilage. The latter is a pectin complex secreted by specialized epidermal cells of the seed coat (Arsovski et al., 2010). We have investigated the mucilage of ATHB25 and DELLA mutants and, as indicated in Figure 9, the increased expression of ATHB25 in athb25-1D and pUBQ10:ATHB25 mutants is accompanied by increased mucilage, while the reduced expression in the antisense mutant (ASATHB25) correlates with decreased mucilage. Increased GA signaling in the DELLA mutant is also accompanied by increased mucilage. Therefore, one feature of the seed coat, mucilage formation, correlates with seed longevity in the GA mutants investigated in this work.
Figure 9.
Mucilage staining in seeds from genotypes with differences in seed longevity. A, Seeds stained with ruthenium red after first shaking in water. B, Measure of the surrounded mucilage area. Each value is the mean of 20 seeds chosen at random from two different batches, 10 each. Error bars indicate se, and asterisks indicate significant differences from the control (the wild type [WT]) at P < 0.05. [See online article for color version of this figure.]
Any seed coat mechanism should have a maternal effect, because this structure is of maternal origin (Haughn and Chaudhury, 2005; Rajjou and Debeaujon, 2008). We made reciprocal crosses between the wild type and the athb25-1D mutant and found that the F1 seeds are tolerant to accelerated aging if the mother plant is a mutant but not when the mother plant is the wild type (Supplemental Fig. S6). These results are in agreement with a role of the seed coat in the improved seed longevity of the ATHB25-overexpressing mutants.
DISCUSSION
Many protocols for accelerated aging of plant seeds (sometimes referred to as “controlled deterioration tests”) have been developed to speed up studies in seed longevity. Seeds are partially or completely rehydrated and exposed to high temperatures, leading to rapid loss of seed viability (e.g. days instead of years, as in natural aging; Woodstock and Taylorson, 1981; Tesnier et al., 2002; Prieto-Dapena et al., 2006; Tejedor-Cano et al., 2010). Accelerated aging mimics natural aging (Tesnier et al., 2002; Rajjou et al., 2008), and similar results are obtained with controlled hydration or with seeds imbibed in water (Tejedor-Cano et al., 2010). Our aging procedure with imbibed seeds incubated at 42°C for 1 to 2 d has been validated by the observed correlation between tolerance to natural aging and to accelerated aging in four different mutants obtained in our screening (Fig. 1).
Most mutants in seed longevity are of two types: recessive mutants, with poor longevity affected in genes required for seed development (such as lec1, lec2, fus3, and abi3; Holdsworth et al., 1999; Clerkx et al., 2003, 2004), and dominant mutants, with improved longevity obtained by reverse genetics overexpressing stress defenses and repair systems such as small heat shock proteins (Prieto-Dapena et al., 2006), Met sulfoxide reductase (Châtelain et al., 2013), DNA glycosylase/AP lyase (Chen et al., 2012), and l-isoaspartyl methyltransferase (Ogé et al., 2008). To our knowledge, our athb25-1D mutant is the first mutant obtained by forward genetics and the first dominant mutant not related to stress defenses or repair systems. Gain-of-function mutants such as those obtained by overexpression obviate genetic redundancy and can identify bottlenecks in biological pathways (Weigel et al., 2000; Alejandro et al., 2007).
Analyses of mutants with gain and loss of function of the ATHB25 gene, encoding a zinc finger homeobox transcription factor, suggest a novel mechanism of seed longevity based on the action of GA during seed development. That ATHB25 acts by increasing GA biosynthesis is demonstrated by the observations that the expression of ATHB25 correlates with hypocotyl length, seed and ovary cell size (Fig. 4), sensitivity to paclobutrazol (Fig. 5), concentrations of the active GAs GA4 and GA1, and expression of GA3OX2, encoding a crucial enzyme in GA synthesis (Fig. 6). Further evidence is provided by transcriptomic analysis of the athb25-1D mutant, which reveals increased expression of many GA-induced genes, decreased expression of many GA-repressed genes, enrichment of the AAAG motif (present in many GA-induced genes) in the promoter of induced genes, and enrichment of the ABRE motif (present in many ABA-induced genes, typically repressed by GA) in repressed genes (Tables I and II). Finally, treatment of plants with GA produces seeds with improved longevity, and mutants with increased (quintuple DELLA mutant) or decreased (pre1 mutant) response to GA have seeds with increased or decreased longevity, respectively.
Our results suggest that, in addition to the accumulation of cellular stress defenses by the action of ABA, the promotion of some aspects of seed development by GA during seed maturation improves longevity. A plausible mechanism for the latter effect may consist of the formation of the seed coat or testa. GAs are required for the normal formation of this structure through starch degradation (Kim et al., 2005), and the seed coat is very important to protect the embryo from stresses during storage (Clerkx et al., 2004; Rajjou and Debeaujon, 2008). The maternal effect observed during reciprocal crosses between wild-type and ATHB25-overexpressing plants suggests that a plausible mechanism for this transcription factor to increase seed longevity is that, by increasing GA synthesis, it reinforces the seed coat. The observed correlation between expression levels of ATHB25 and seed mucilage and the increased mucilage in the quintuple DELLA mutant (Fig. 8) supports this hypothesis. Several transcription factors (AP2, TRANSPARENT TESTA GLABRA1 [TTG1], TTG2, ENHANCER OF GLABRA3, TRANSPARENT TESTA2 [TT2], TT8, MYB DOMAIN PROTEIN5 [MYB5], MYB61, and GLABRA2) are required for the differentiation of the outer layers of the seed coat and the secretion of mucilage (Arsovski et al., 2010), but the expression of none of them is altered in the athb25-1D mutant (Supplemental Table S1).
It must be noted that the observed correlation between GA levels, seed longevity, and the amount of mucilage does not imply that the increased mucilage is specifically or solely responsible for the observed phenotype of seed longevity. As a hormone, GA could be acting on many aspects of the seed coat differentiation. Other portions of the seed coat that could play a role in seed coat reinforcement and seed longevity are the secondary cell walls of the epidermal mucilage cells (columellae) and the subepidermal palisade cells (Debeaujon et al., 2000; Rajjou and Debeaujon, 2008). It is plausible that during starch degradation promoted by GA, these walls are also made thicker, in addition to the increased mucilage production (Kim et al., 2005). It is also possible that altered GA production could increase proanthocyanidin synthesis in the endothelial layer of the seed coat, which has also been implicated in seed longevity (Rajjou and Debeaujon, 2008).
We do not expect ATHB25 to directly regulate GA3OX2, because it was not possible to detect binding of this transcription factor to the promoter of this gene in chromatin immunoprecipitation experiments (data not shown). The transcription factors that directly regulate the expression of GA3OX genes in Arabidopsis are only partially known (Hedden and Thomas, 2012). Mutation of genes encoding some transcription factors, such as AT-HOOK PROTEIN OF GA FEEDBACK1 (Matsushita et al., 2007), SCARECROW-LIKE13 (Zhang et al., 2011), and SOMNUS (Kim et al., 2008), affects GA3OX1 and GA3OX2 expression, but there is no evidence for their binding to the promoter of the genes. Only FUS3 (Curaba et al., 2004) and DOF AFFECTING GERMINATION1 (Gabriele et al., 2010) have been shown to repress GA3OX1 and GA3OX2, respectively, by binding to the promoter of the regulated gene. None of the genes encoding all these transcription factors are regulated by ATHB25, according to our microarray experiments (Supplemental Table S1).
CONCLUSION
The identification of the ATHB25 gene in a screening of activation-tagging mutants with selection for increased seed longevity has uncovered a novel role for GAs in seed longevity. The ATHB25 transcription factor induces the expression of the GA biosynthetic enzyme GA3OX2 and increases the levels of bioactive GAs. The mechanism of GA-dependent longevity may be based on reinforcement of the seed coat, and these findings may be of relevance to improve seed longevity, with important benefits for biodiversity and agriculture.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown and germinated on Murashige and Skoog (MS) plates with Suc as described (Alejandro et al., 2007). Plants treated with GA3 were sprayed twice per week with a 50 µm solution. Plants subjected to drought were grown with irrigation for 3 weeks, and then watering was stopped. We used the following Arabidopsis strains: wild-type Columbia, wild-type Landsberg erecta, activation-tagging mutant overexpressing ATHB25 (athb25-1D), transgenic lines overexpressing ATHB25 under the control of the UBQ10 promoter (pUBQ10:ATHB25), transgenic lines overexpressing ATHB25 under the control of the 35S promoter (35S:ATHB25), lines overexpressing antisense At5g65410 (ASATHB25), loss of function of ATHB25 (NASC line N665321; athb25-2), loss of function of ATHB22 (NASC line N667869), quintuple DELLA mutant (NASC line N16298; gai-t6 rga-t2 rgl1-1 rgl2-1 rgl3-1), and paclobutrazol-resistant mutant pre1 (Bai et al., 2012).
Accelerated Aging
This procedure was a modification of the “basal thermotolerance assay” of Tejedor-Cano et al. (2010). Seeds were imbibed in water, stratified for 2 d at 4°C, and incubated at 42°C for 1 to 2 d (relative humidity of 100%). In the initial screening for mutants, the seeds were exposed to 42°C for 48 h because, after this time, the germination of wild-type seeds was completely inhibited. In other experiments, incubation at 42°C was for 24 h.
Molecular Techniques
Plasmid rescue from mutant plants, RNA extraction, RT-PCR (qualitative and quantitative or real time) to measure gene expression, PCR to detect T-DNA, and Southern-blot analysis to determine the number of T-DNA insertions were performed as described (Alejandro et al., 2007). Plants (10-d-old seedlings grown in solid MS medium) were grown under long-day conditions. The sequences of the primers are shown in Supplemental Table S2. The constructs for overexpression of the ATHB25 (At5g65410) gene in plants under the control of the cauliflower mosaic virus 35S promoter were made as follows. The cDNA containing the complete open reading frame was obtained in plasmid pUNI51 from the Genomics Science Center at RIKEN. The cDNA was released by digestion with SfiI, blunted, subcloned into plasmid pBluescript SK+ (Stratagene), digested with XbaI and NotI, and blunted. Recombinant plasmids with both orientations of the cDNA were obtained. For expression from the cauliflower mosaic virus 35S promoter, a SacI-BamHI fragment including the cDNA was subcloned into pBI121 by replacing the GUS coding region between the SacI and BamHI sites. For expression from the UBQ10 promoter, the pBluescript SK+ recombinant plasmid was digested with SacI, blunted, and then digested with ApaI. The fragment including the cDNA was subcloned into plasmid pUBQ10:MCS (Grefen et al., 2010), digested with SpeI, blunted, and then digested with ApaI.
Global Gene Expression Using Long Oligonucleotide Microarrays
Plants (10-d-old seedlings grown in solid MS medium) were grown under long-day conditions, RNA extraction, amplification, labeling, and hybridization were performed as described previously (Bueso et al., 2007), and the analysis of microarray data was as described by Bissoli et al. (2012). Long oligonucleotide microarrays were provided by Dr. David Galbraith (University of Arizona; http://www.ag.arizona.edu/microarray/). Oligonucleotides were from the Operon Arabidopsis Genome Oligo Set version 3.0 (https://www.operon.com/), which contains 29,110 70-mer probes, representing 26,173 protein-coding genes, 28,964 protein-coding gene transcripts, and 87 microRNAs. The design is based on ATH1 release 5.0 of The Institute for Genomic Research Arabidopsis genome annotation database (http://www.tigr.org/tdb/e2k1/ath1/) and release 4.0 of the microRNA registry at the Sanger Institute (http://www.sanger.ac.uk/Software/Rfam/mirna/index.shtml).
The microarray data from this article have been submitted to the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) and assigned the identifier E-MEXP-3857.
Measure of Hypocotyl Length and Seed Area
Photographs were taken with the manual fluorescence stereomicroscope Leica MZ16 F, and hypocotyl length and seed area were measured with the Java-based image-processing program ImageJ (http://rsb.info.nih.gov/ij/).
Measure of Cell Area
For scanning electron microscopy, samples were fixed in 50% ethanol, 5% acetic acid, and 3.7% formaldehyde for 16 h at 4°C. Samples were dehydrated in an ethanol series and critical point dried in liquid CO2 (Polaron E300 apparatus). Dried samples were mounted on slides with activated carbon. Then, samples were coated with gold palladium (4:1) in a Sputter Coater SCD005 (BALTEC). Scanning electron microscopy was performed with a JEOL JSM-5410 microscope (10 kV).
GA Extraction and Quantification
Aliquots of 100 mg fresh weight from 10-d-old seedlings were homogenized in cold 80% (v/v) methanol-water, stirred overnight at 4°C, and reextracted twice for 30 min with one-half volume of methanol using [17,17-2H]GA1, [17,17-2H]GA4, [17,17-2H]GA8, [17,17-2H]GA9, [17,17-2H]GA19, [17,17-2H]GA20, [17,17-2H]GA29, [17,17-2H]GA34, and [17,17-2H]GA44 as internal standards. The methanolic extracts were combined, taken to dryness in vacuo, and dissolved in 0.5 mL of 10% methanol solution. This extract was then applied to a preequilibrated 500-mg SAX column (BondElut SS-SAX; Varian Scharlau). The column was washed with water, pH 8.0, before being eluted with 4 mL of 0.2 m formic acid solution. The formic acid eluate was run directly onto a preequilibrated 500-mg C18 column (BondElut C18; Varian Scharlau) that was then washed with water, pH 3.0, and finally eluted with 5 mL of 80% methanol. The methanol eluate was dried, dissolved in 0.2 mL of 2-propanol, and then methylated with trimethylsilyl-diazomethane in hexane. After methylation, the residue was dried, and the samples were trimethylsilylated using 7.5 µL of pyridine and 7.5 µL of N,O-bis(trimethylsilyl)trifluoroacetamide solution containing 1% trimethyl chlorosilane. For gas chromatography-mass spectrometry analysis, the samples were injected dissolved in the mix of pyridine/N,O-bis(trimethylsilyl)trifluoroacetamide into a gas chromatograph (7890A; Agilent Technologies) coupled to a mass spectrometer (7000 Triple Quad; Agilent Technologies). Samples were injected in splitless mode into an HP 5890 gas chromatograph (Hewlett-Packard), fitted with a fused silica glass capillary column (30 m long, 0.25 mm i.d.) with a chemically bonded 0.25-µm DB-5MS stationary phase (J&W Scientific). The injector temperature was 270°C. The column temperature program varied depending on which GA was being analyzed. The column effluent was introduced into the ion source of a JMS-SX/SX102A mass spectrometer (JEOL). The interface temperature was 280°C, and the ion source temperature was 250°C. The amount of GA in the samples was quantified by isotopic dilution, and we used the MassHunter Quantitative software provided by Agilent.
Identification of Preferentially Located Motifs
We studied a set of 199 known motifs coming from the AGRIS database (Yilmaz et al., 2011), and for each motif, we extracted all occurrences in a promoter set. The promoter sequences were then divided into two regions. First, the [−1,000, −300] region was used to learn the distribution model using a simple linear regression and determine a 95% confidence interval. Second, within the [−300, untranslated region (UTR)] region, we searched for nonevenly distributed motifs (i.e. those exhibiting a peak above the confidence interval). When a motif is considered, like an overrepresented motif in a set of promoters, we performed a binomial probability to compare percentages between this considered set and the whole promoter set of Arabidopsis. Each motif with a probability of less than 5% was considered significant. We then applied the same methodology to study all the four- and six-base motif lengths. This methodology to detect if a motif had a preferential location has been described in detail (Bernard et al., 2010). We used FLAGdb++ (Dèrozier et al., 2011), an integrative database of plant model genomes, to define the transcriptional units by aligning all the available transcripts to gene models (gene models, ESTs, and full-length cDNAs are defined in The Arabidopsis Information Resource [http://www.arabidopsis.org/]). The promoter was defined by 1 kb upstream of the predicted transcription start site and contained the whole 5′ UTR. We excluded promoters with a 5′ UTR smaller than 10 bases in order to avoid false promoter regions. Thus, we defined promoters for 19,921 genes of Arabidopsis as well as 136 and 331 promoters, respectively, for the induced and repressed gene sets studied.
Staining of the Mucilage with Ruthenium Red
Seeds were imbibed in water for 3 min before the application of 0.2% (w/v) aqueous ruthenium red solution for 15 min. Photographs were taken with Nikon’s Eclipse E600 research microscope, and mucilage area was measured with the Java-based image-processing program ImageJ.
Extraction and Quantification of ABA
ABA was extracted overnight at 4°C from 50 mg of seeds in 2.5 mL of extraction buffer containing 80% (v/v) acetone, 100 mg L−1 butylated hydroxytoluene, and 500 mg L−1 citric acid and quantified with the Phytodetek-ABA kit (Agdia).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers BT002385.1 GI: 27311558 (At5g65410).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Endogenous ABA content of Arabidopsis seeds of the wild type and the isl1-1D mutant after accelerated aging.
Supplemental Figure S2. The isl1-1D mutant has a single insertion of T-DNA.
Supplemental Figure S3. ATHB25 expression during accelerated aging treatment.
Supplemental Figure S4. Expression levels of GA20OX and GA3OX genes in the wild type and the athb25-1D mutant.
Supplemental Figure S5. Microarray validation.
Supplemental Figure S6. Maternal effect of increased seed longevity conferred by the athb25-1D mutation.
Supplemental Table S1. Genes induced more than 2-fold and genes repressed more than 2-fold in the athb25-1D mutant with respect to the wild type.
Supplemental Table S2. Primers used in this work.
Acknowledgments
We thank Thomas Moritz (Swedish Metabolomics Centre) for the facilities to analyze GAs, David Alabadí (Instituto de Biología Molecular y Celular de Plantas) for the quintuple DELLA mutant, and Jörg Kudla (Westfälische Wilhelms-Universität Münster) for the pUBQ10:MCS plasmid.
Glossary
- ABA
abscisic acid
- T-DNA
transfer DNA
- cDNA
complementary DNA
- RT
reverse transcription
- MS
Murashige and Skoog
- NASC
Nottingham Arabidopsis Stock Centre
- UTR
untranslated region
References
- Alejandro S, Rodríguez PL, Bellés JM, Yenush L, García-Sanchez MJ, Fernández JA, Serrano R. (2007) An Arabidopsis quiescin-sulfhydryl oxidase regulates cation homeostasis at the root symplast-xylem interface. EMBO J 26: 3203–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Haughn GW, Western TL. (2010) Seed coat mucilage cells of Arabidopsis thaliana as a model for plant cell wall research. Plant Signal Behav 5: 796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Fan M, Oh E, Wang ZY. (2012) A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C. (2004) Active oxygen species and antioxidants in seed biology. Seed Sci Res 14: 93–107 [Google Scholar]
- Benfey PN, Chua NH. (1990) The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250: 959–966 [DOI] [PubMed] [Google Scholar]
- Bernard V, Lecharny A, Brunaud V. (2010) Improved detection of motifs with preferential location in promoters. Genome 53: 739–752 [DOI] [PubMed] [Google Scholar]
- Bissoli G, Niñoles R, Fresquet S, Palombieri S, Bueso E, Rubio L, García-Sánchez MJ, Fernández JA, Mulet JM, Serrano R. (2012) Peptidyl-prolyl cis-trans isomerase ROF2 modulates intracellular pH homeostasis in Arabidopsis. Plant J 70: 704–716 [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Harada JJ. (2008) LECs go crazy in embryo development. Trends Plant Sci 13: 624–630 [DOI] [PubMed] [Google Scholar]
- Bueso E, Alejandro S, Carbonell P, Perez-Amador MA, Fayos J, Bellés JM, Rodriguez PL, Serrano R. (2007) The lithium tolerance of the Arabidopsis cat2 mutant reveals a cross-talk between oxidative stress and ethylene. Plant J 52: 1052–1065 [DOI] [PubMed] [Google Scholar]
- Châtelain E, Satour P, Laugier E, Ly Vu B, Payet N, Rey P, Montrichard F. (2013) Evidence for participation of the methionine sulfoxide reductase repair system in plant seed longevity. Proc Natl Acad Sci USA 110: 3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chu P, Zhou Y, Li Y, Liu J, Ding Y, Tsang EW, Jiang L, Wu K, Huang S. (2012) Overexpression of AtOGG1, a DNA glycosylase/AP lyase, enhances seed longevity and abiotic stress tolerance in Arabidopsis. J Exp Bot 63: 4107–4121 [DOI] [PubMed] [Google Scholar]
- Choi DS, Hwang IS, Hwang BK. (2012) Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkx EJ, Blankestijn-De Vries H, Ruys GJ, Groot SPC, Koornneef M. (2004) Genetic differences in seed longevity of various Arabidopsis mutants. Physiol Plant 121: 448–461 [Google Scholar]
- Clerkx EJ, Vries HB, Ruys GJ, Groot SPC, Koornneef M. (2003) Characterization of green seed, an enhancer of abi3-1 in Arabidopsis that affects seed longevity. Plant Physiol 132: 1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling RJ, Harberd NP. (1999) Gibberellins control Arabidopsis hypocotyl growth via regulation of cellular elongation. J Exp Bot 50: 1351–1357 [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G. (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136: 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dèrozier S, Samson F, Tamby JP, Guichard C, Brunaud V, Grevet P, Gagnot S, Label P, Leplé JC, Lecharny A, et al. (2011) Exploration of plant genomes in the FLAGdb++ environment. Plant Methods 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P. (2010) The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J 61: 312–323 [DOI] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR. (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. (2007) Combined networks regulating seed maturation. Trends Plant Sci 12: 294–300 [DOI] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A. (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Hedden P, Thomas SG. (2012) Gibberellin biosynthesis and its regulation. Biochem J 444: 11–25 [DOI] [PubMed] [Google Scholar]
- Holdsworth M, Kurup S, McKibbin R. (1999) Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Plant Sci 4: 275–280 [Google Scholar]
- Hu W, dePamphilis CW, Ma H. (2008) Phylogenetic analysis of the plant-specific zinc finger-homeobox and mini zinc finger gene families. J Integr Plant Biol 50: 1031–1045 [DOI] [PubMed] [Google Scholar]
- Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM. (1998) Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol 118: 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G. (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, Baek D, Lee SY, Blakeslee JJ, Murphy AS, Bohnert HJ, et al. (2007) yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol 145: 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Nakajima M, Nakayama A, Yamaguchi I. (2005) Contribution of gibberellins to the formation of Arabidopsis seed coat through starch degradation. Plant Cell Physiol 46: 1317–1325 [DOI] [PubMed] [Google Scholar]
- Kotak S, Vierling E, Bäumlein H, von Koskull-Döring P. (2007) A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita A, Furumoto T, Ishida S, Takahashi Y. (2007) AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol 143: 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP. (2006) Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J 45: 804–818 [DOI] [PubMed] [Google Scholar]
- Nguyen TP, Keizer P, van Eeuwijk F, Smeekens S, Bentsink L. (2012) Natural variation for seed longevity and seed dormancy are negatively correlated in Arabidopsis. Plant Physiol 160: 2083–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogé L, Bourdais G, Bove J, Collet B, Godin B, Granier F, Boutin JP, Job D, Jullien M, Grappin P. (2008) Protein repair L-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell 20: 3022–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Dapena P, Castaño R, Almoguera C, Jordano J. (2006) Improved resistance to controlled deterioration in transgenic seeds. Plant Physiol 142: 1102–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Debeaujon I. (2008) Seed longevity: survival and maintenance of high germination ability of dry seeds. C R Biol 331: 796–805 [DOI] [PubMed] [Google Scholar]
- Rajjou L, Lovigny Y, Groot SP, Belghazi M, Job C, Job D. (2008) Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiol 148: 620–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. (2004) Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16: 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ. (2007) MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol 143: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F. (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Suzuki M, McCarty DR. (2008) Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol 11: 548–553 [DOI] [PubMed] [Google Scholar]
- Swain SM, Singh DP. (2005) Tall tales from sly dwarves: novel functions of gibberellins in plant development. Trends Plant Sci 10: 123–129 [DOI] [PubMed] [Google Scholar]
- Tan QK, Irish VF. (2006) The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol 140: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejedor-Cano J, Prieto-Dapena P, Almoguera C, Carranco R, Hiratsu K, Ohme-Takagi M, Jordano J. (2010) Loss of function of the HSFA9 seed longevity program. Plant Cell Environ 33: 1408–1417 [DOI] [PubMed] [Google Scholar]
- Tesnier K, Strookman-Donkers HM, van Pijlen JG, van der Geest AH, Bino RJ, Groot SP. (2002) A controlled deterioration test for Arabidopsis thaliana reveals genetic variation in seed quality. Seed Sci Technol 30: 149–165 [Google Scholar]
- Tieman DM, Ciardi JA, Taylor MG, Klee HJ. (2001) Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J 26: 47–58 [DOI] [PubMed] [Google Scholar]
- Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, Jikumaru Y, Hanada A, Aso Y, Ishiyama K, Tamura N, et al. (2008) High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol 146: 1368–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LSP, Nakashima K, Sakuma Y, Osakabe Y, Qin F, Simpson SD, Maruyama K, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J 49: 46–63 [DOI] [PubMed] [Google Scholar]
- Wang C, Yang A, Yin H, Zhang J. (2008) Influence of water stress on endogenous hormone contents and cell damage of maize seedlings. J Integr Plant Biol 50: 427–434 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL. (2012) The sticky tale of seed coat mucilages: production, genetics and role in seed germination and dispersal. Seed Sci Res 22: 1–25 [Google Scholar]
- Woodstock LW, Taylorson RB. (1981) Ethanol and acetaldehyde in imbibing soybean seeds in relation to deterioration. Plant Physiol 67: 424–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y, Sun T. (2001) Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J 28: 443–453 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10: 88–94 [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E. (2011) AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res 39: D1118–D1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Ogawa M, Fleet CM, Zentella R, Hu J, Heo JO, Lim J, Kamiya Y, Yamaguchi S, Sun T. (2011) SCAREROW-LIKE 3 promotes gibberellin signalling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci USA 108: 2160–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]