The splicing of nuclear pre-mRNAs is a fundamental process required for the expression of most metazoan genes.1 The majority of the approximately 25,000 genes encoded by the human genome2 has been shown to produce more than one kind of transcripts through alternative splicing.3 Alternative splicing of pre-mRNAs can lead to the production of multiple protein isoforms from a single gene, significantly enriching the proteomic diversity of higher eukaryotic organisms.3–7 Because regulation of this process determines the timing and location that a particular protein isoform is produced, changes of alternative splicing patterns have the potential to modulate many cellular activities. Consequently, pre-mRNA splicing must occur with a high degree of specificity and fidelity to ensure the appropriate expression of functional mRNAs. Here we review recent progress made in understanding the extent of alternative splicing within the human genome with particular emphasis on splicing fidelity.
Alternative splicing
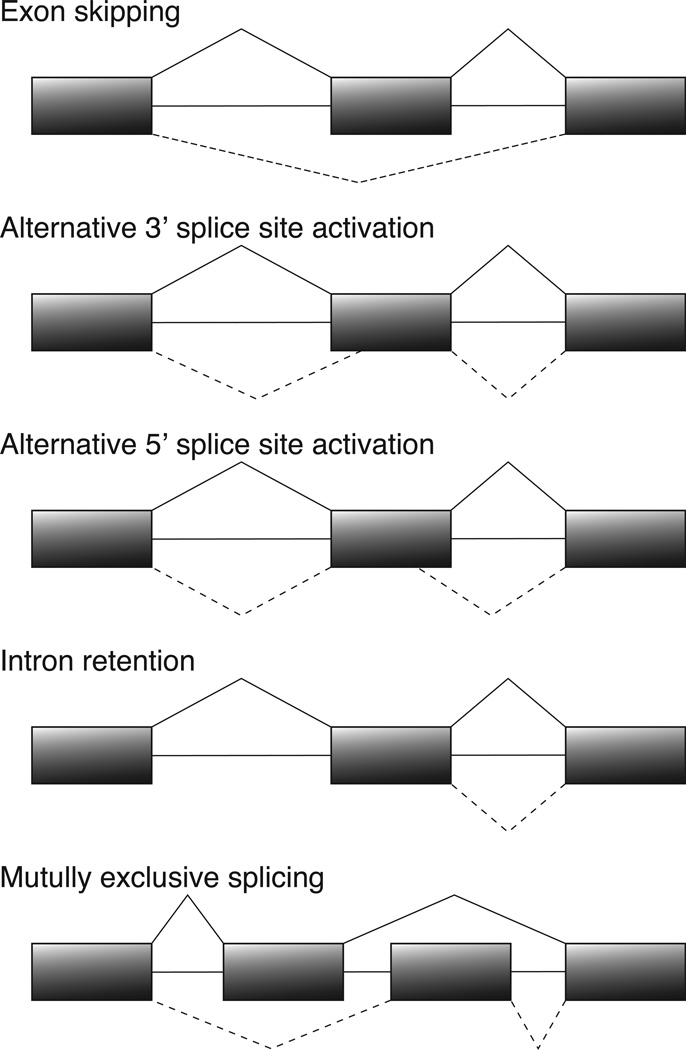
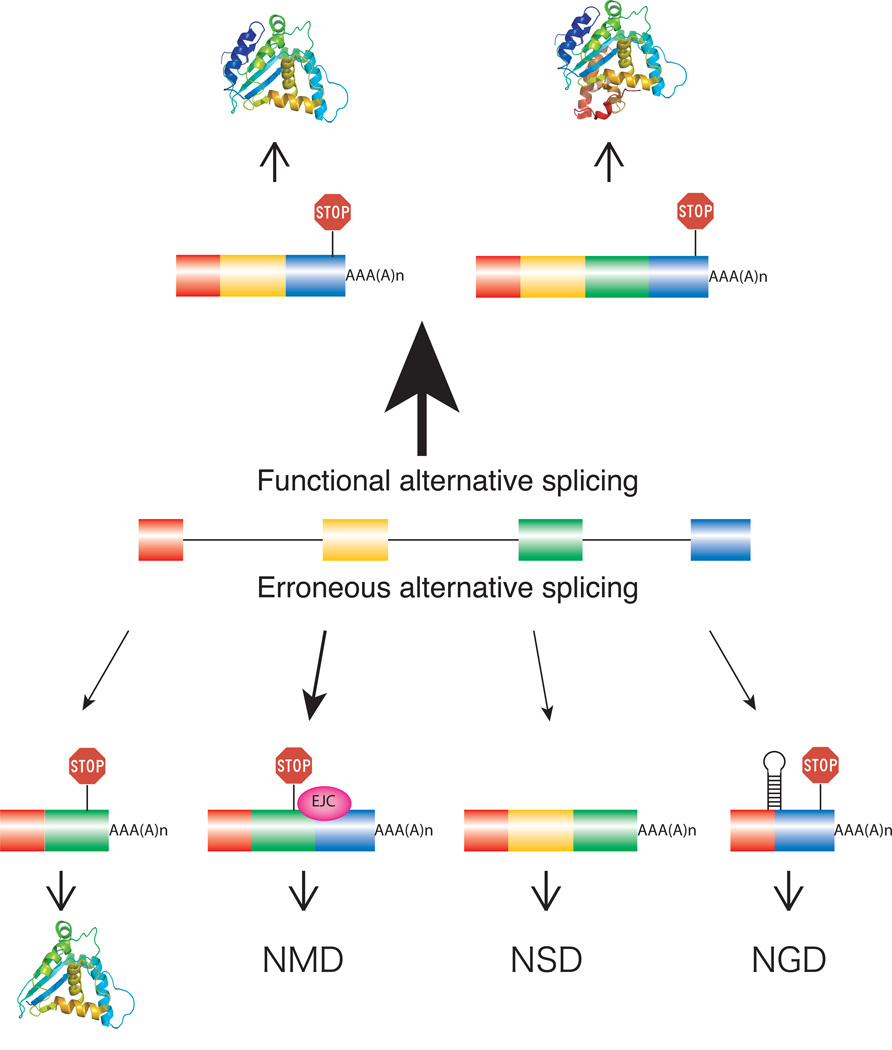
A critical step in pre-mRNA splicing is the recognition and correct pairing of 5' and 3' splice sites. While the 5' splice site junction is defined by a single element of 9 conserved nucleotides, the 3' splice site is defined by three sequence elements usually located within 40 nts upstream of the 3' intron/exon junction.8 Given the complexity of higher eukaryotic genes and the relatively low level of splice-site conservation,9 it is not surprising that many alternative splicing events accompany the processing of pre-mRNAs, regardless of their function.10 Predominant types of alternative splicing include exon skipping, alternative splice-site activation, intron retention, and mutually exclusive splicing (Fig. 1).11–14 In addition, it is known that minor perturbations of the template sequence such as single base mutations can frequently lead to alternative splicing.15–18 To ensure the quality of mRNA isoforms and their resulting protein products, several RNA quality control steps are implemented either pre- or co-translationally to degrade mRNA molecules or prevent them from engaging in protein synthesis (Fig. 2). These include nonsense-mediated decay (NMD),19 non-stop decay (NSD),20 no-go decay (NGD),21 or the rapid mammalian deadenylation-dependent decay pathway.22
Figure 1. Major types of alternative splicing.
Boxes denote exons and horizontal lines introns. Solid lines above and dashed lines below the gene structure represent different alternative splicing events.
Figure 2. RNA surveillance mechanisms controlling the expression of mRNA isoforms generated through alternative splicing.
The majority of pre-mRNA molecules are processed to generate protein isoforms serving diverse cellular functions (functional alternative splicing). Point mutations within the template or mis-regulation of splice site pairing can lead to erroneously spliced mRNA products (erroneous alternative splicing). A small fraction of erroneously spliced mRNAs is translated into a different protein product (bottom left) with the potential to negatively or positively influence cellular functions. The majority of erroneously spliced mRNAs will be the target of RNA surveillance mechanisms such as NMD (triggered by premature stop codons), NSD (lack of stop codon), NGD (ribosome stalling), or the rapid mammalian deadenylation-dependent decay pathway, resulting in selective degradation of erroneously spliced mRNA isoforms. The thickness of the arrows above and below the pre-mRNA reflects the frequency of these events.
The widespread generation of alternative mRNA isoforms has evoked a debate whether the majority of mRNA isoforms are generated by mistake or with a biological purpose.5, 23 Biological functions for mRNA isoforms have been demonstrated in a number of cases studied.4, 6 Yet, it is also likely that a significant fraction of mRNA isoforms is translated without an obvious biological function. Thus, increased proteomic output through alternative splicing may come at the cost of periodically generating isoforms that initially do not have biological functions in the cell.24 However, reducing the fidelity of the spliceosome to increase proteome diversification could be problematic, as there are housekeeping processes that cannot afford decreased levels of certain gene products. These considerations raise the question how the splicing machinery has evolved to balance high fidelity and an apparent promiscuity during intron removal.
Defining erroneous splicing
When evaluating the fidelity of the splicing reaction one is initially faced with the challenge of defining what constitutes a splicing error. Unlike the processes of RNA transcription or translation where incorporation of nucleotides or tRNAs is template-directed, intron removal depends on binding affinities between the pre-mRNA and components of the splicing machinery and the catalytic steps following initial splice site pairing. Once the spliceosome has assembled around a pair of splice sites, multiple structural rearrangements and quality control steps take place to ensure that the correct nucleotides defining the exon/intron boundary are selected for the ensuing phosphodiester transfer reactions. Recent experiments have demonstrated that spliceosomal rearrangement steps that incorporate proofreading mechanisms guarantee high fidelity of spliceosomal catalysis.25, 26 However, no such mechanisms are known as of yet for splice site pairing. Therefore, it is difficult to categorize an alternatively spliced mRNA isoform as a consequence of functional alternative splicing or erroneous alternative splicing. Such a classification may be more straightforward if the biological consequence of alternative splicing is taken into account. Here we define functional alternative splicing as alternative splicing events that generate products with biological functions and erroneous splicing events as those that do not (Fig. 2).
In several documented cases, erroneous alternative splicing causes disease.15–18 One striking example is the alternatively spliced exon 10 of the microtubule-associated protein tau (MAPT) gene. In normal cells, the ratio of exon 10-included and exon 10–excluded mRNAs is close to 1:1. Mutations within or downstream of exon 10 disrupt splicing regulatory elements and perturb the well-maintained balance between these two isoforms, resulting in the neuropathological disorder Frontotemporal Dementia.27 Thus, erroneous alternative splicing events include those caused by mutations in the pre-mRNA templates. Because functional alternative splicing events are often characterized by high relative abundance, tissue specificity and conservation, we will also refer to erroneous alternative splicing as events that are of low abundance, that do not exhibit tissue- or developmental-specific regulation, and that are not conserved among closely related species.
The frequency of erroneous splice site pairing
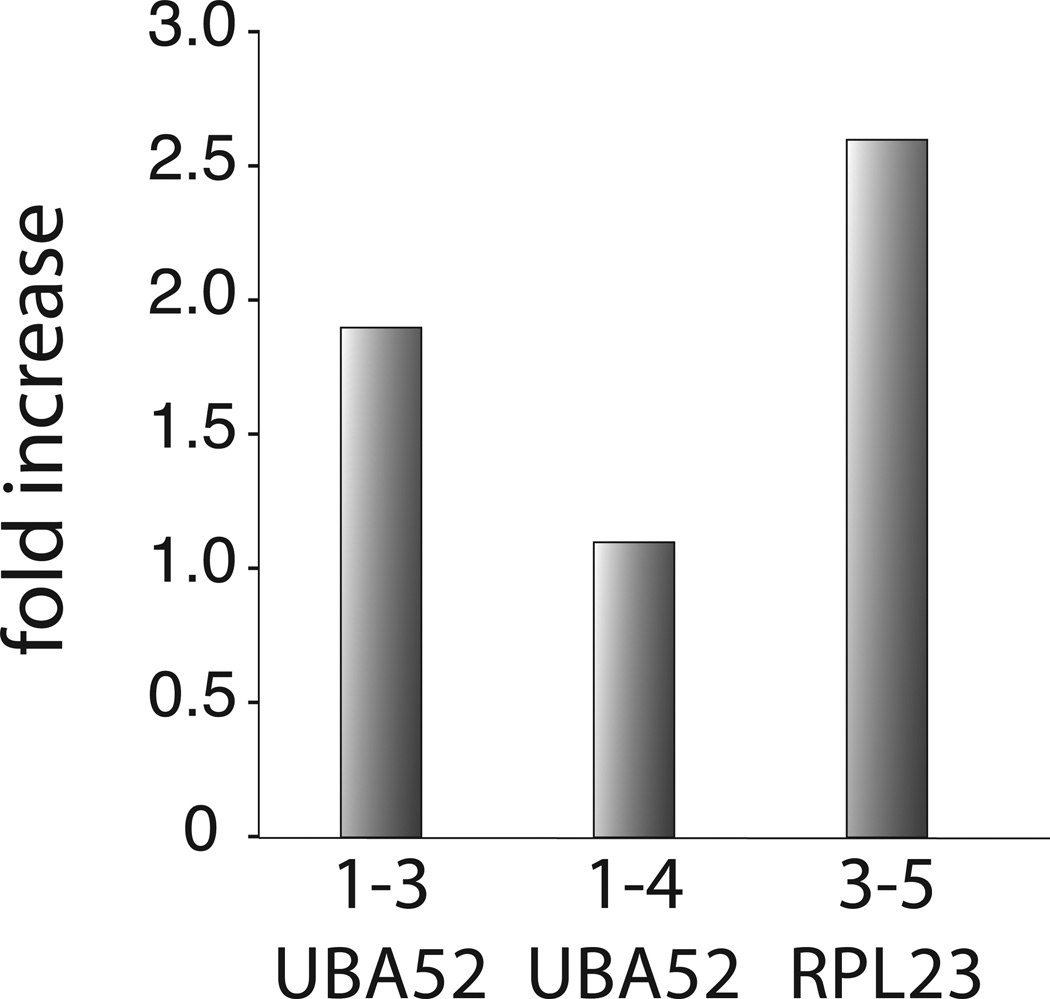
Based on the above criteria, we recently selected two highly conserved human genes (UBA52 and RPL23) as a gold standard for constitutive splicing.28 Any splice pattern that deviated from the only annotated, constitutively spliced mRNA sequence was considered an outcome of erroneous splice site pairing. Using quantitative real-time PCR, we showed that signals for all possible exon skipping mRNA isoforms could be detected, however, at significantly reduced levels. The lowest error rate of splice site pairing measured was around one per 105 splicing events. These observations provided the foundation for several conclusions and suggestions. First, they showed that the process of splice-site pairing is an extremely high fidelity process. This conclusion implies that the high levels of alternative splicing detected in higher eukaryotes are the result of splice sites that have evolved to offer a weak binding potential for components of the spliceosome. Second, the results suggested that the accuracy of splice-site pairing is limited by the fidelity of transcription. It is known that the transcription machinery makes a mistake once in every 103–105 nucleotide insertion step, even with documented proofreading.29–31 With such a high fidelity, transcription of constitutively spliced genes such as UBA52 or RPL23 generates nucleotide mis-incorporations in a very small fraction of pre-mRNA copies. Analogous to disease-causing mutations, the resulting pre-mRNA then contains at least one nucleotide difference that can trigger erroneous pre-mRNA splicing such as exon skipping. Further support for the idea that the error rate of splice site pairing is limited by the fidelity of transcription comes from recent experiments designed to knock down TFIIS, an elongation factor known to be involved in proofreading nucleotide insertion during transcription.30 As expected from the idea that transcription and splicing errors are directly connected, reduction of cellular TFIIS levels through siRNA increased the error rate of splice site pairing (Fig. 3). Irrespective of the frequency, some mistakes will occur during the synthesis of pre-mRNAs, once in a while causing alternative splicing of the transcript. These considerations argue strongly that every multi-intron containing gene, including constitutively spliced genes such as UBA52 and RPL23, will exhibit some level of alternative splicing, albeit with drastically different efficiencies.
Figure 3. The effect of TFIIS knockdown on the error rate of splice site pairing.
The graph depicts the increase in erroneous splice site pairing of the constitutively spliced genes UBA52 and RPL23 as a consequence of siRNA-mediated knockdown of TFIIS, a Pol II-associated transcription factor that ensures high fidelity nucleotide insertion during RNA transcription. The y-axis shows the relative increase of splice site pairing errors in TFIIS knockdown experiments over the error rate observed in control siRNA experiments. As confirmed by quantitative real-time PCR and western blot, siRNA treatment reduced TFIIS levels by ~60% (data not shown).
The extent of alternative splicing measured through high-throughput sequencing of the human transcriptome
Throughout the years new experimental methodologies have offered solutions to advance our knowledge of the extent of alternative splicing. For example, direct cloning and sequencing in combination with genomic sequence analysis of Drosophila Dscam32, 33 and N-Cadherin34, 35 molecules identified and confirmed the expression of many previous unknown mRNA isoforms. Nevertheless, this approach is labor-intensive and could not be applied to the genome-wide scale. Alternative high throughput methodologies such as microarrays designed to detect genome-wide alternative splicing suffer from incomplete lists of all plausible splice junctions, non-specific hybridization of probes with the cDNA, and the inability to provide absolute quantitative information.36 Lately, the rapid development of high-throughput sequencing technologies is coming to fruition and permits re-examining the human transcriptome with deep coverage at a reasonable price. The advantages of the high-throughput sequencing technologies include the ability to generate an enormous set of sequencing data within a short period of time, with a high read coverage, with quantifiable expression levels, and with no requirement for prior knowledge of potential splicing junctions.
The extent of alternative splicing in the human genome has recently been analyzed in several different laboratories using these high-throughput sequencing approaches.37–39 After applying appropriate computational tools, the high-throughput sequencing analyses demonstrated that alternative splicing occurs in as high as ~98% of human multi-exon genes, which account for 94% of all human genes. Pan et al showed that the median number of alternative splicing events per exon is between 0.5 and 0.75 for genes with intermediate to high levels of sequence coverage.38 Based on the average number of exons per multi-exon human gene,11 this result suggests that there are multiple alternative splicing events per gene, indicating that alternative splicing of human genes is a universal phenomenon. However, it is still unclear whether these mRNA isoforms serve physiological functions in the cell or whether they are just noise caused by erroneous alternative splicing.
What is apparent from these results is that the splicing error and high-throughput sequencing analyses agree in their main conclusion that alternative splicing of multi-exon genes is the rule rather than the exception. Yet, the two approaches differ considerably in their sensitivity of detection and in the type of alternative splicing observed. While the real-time PCR permitted detection of very low abundant transcript isoforms, the high-throughput sequencing reactions identified mRNA isoforms of more significant representation. The important challenge now is to determine at which point alternatively spliced mRNA isoforms become biologically important and how the cell deals with the existence of many very low abundance transcripts. Some insights into these questions can be derived from the high-throughput sequencing analysis. Even when considering only alternative splicing events that exceed a frequency of 15%, more than 85% of human genes were estimated to produce significant levels of alternatively spliced isoforms. Furthermore, depending on the type of alternative splicing, up to 74% of events showed variations between tissues, implying tissue-specificity and regulation of alternative splicing.39 Because these alternative splicing events are reasonably abundant and tissue-specific, it is very likely that they are biologically significant and exert some impact on the cell. The coverage of EST databases is usually lower than what can be derived from high-throughput sequencing reactions. This suggests that mRNA isoforms listed in EST databases are expressed at an appreciate level. If functional alternative splicing events are those of high abundance and with tissue-specific expression patterns, it is therefore likely that many alternative splicing events in EST databases are biologically significant as well.
By contrast, the very low abundance mRNA isoforms, such as those generated through errors during splice-site pairing, may not interfere significantly with cellular processes. In most cases it is anticipated that the activities of mRNA quality control steps, such as NMD, NSD, NGD, and rapid deadenylation-dependent decay, limit the translation of potentially harmful mRNA isoforms.19, 22 A second layer of control lies in the relative abundance of the mRNA isoforms. Even if some of erroneously spliced mRNA escaped the various quality control surveillance mechanisms, their infrequent occurrence renders them biologically irrelevant.
What do these results suggest for day-to-day RNA processing?
When facing exon/intron boundary sequence elements of high affinity, the splicing machinery is exceedingly accurate in selecting splice junctions, exhibiting error frequencies as low as one per 105 splicing events.28 However, when taking into account the significant sequence variation of splice site signals and the high frequency of splicing in cells, it is anticipated that many erroneously spliced mRNAs are produced. The vast majority of these mis-spliced mRNA will be removed from cells through NMD or similar RNA quality control steps (Fig 2). Given the high energetic cost involved in RNA synthesis, pre-mRNA processing, and mRNA surveillance, one is left wondering why cells permit this seemingly wasteful way of doing business? As is documented by many examples of functional alternative splicing with proven biological impact, the most likely answer to this question is that the low levels of erroneous alternative splicing tolerated provide an evolutionary advantage. Even if the gene product of a lowly expressed erroneous alternative splicing event is harmful to the cell, it is not expected to disrupt the normal biological process because it is masked by highly abundant correctly spliced mRNAs. However, if the gene product of an alternative isoform benefits the survival of an organism, its expression level could be selected for through up-regulation of the alternative splicing event. Through these selection mechanisms, very low abundance transcripts can transition to become minor or even major spliced isoforms with appreciable levels.40 As such, transcriptional and spliceosomal imperfections could permit diversification of protein functions and adaptation to environmental changes throughout time.
Acknowledgements
We are grateful to the Hertel laboratory for helpful comments on the manuscript. This work was supported by NIH grant GM 62287 (K.J.H.).
References
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.INTERNATIONAL HUMAN GENOME SEQUENCING CONSORTIUM. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 4.Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 5.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 6.Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 8.Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 9.Burge CB, Tuschl T, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, CTR, Atkins JF, editors. The RNA World. Cold Spring Harbor, New York: CSHL Press; 1999. pp. 525–560. [Google Scholar]
- 10.Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 11.Lee C, Atanelov L, Modrek B, Xing Y. ASAP: the Alternative Splicing Annotation Project. Nucleic Acids Res. 2003;31:101–105. doi: 10.1093/nar/gkg029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thanaraj TA, Stamm S, Clark F, Riethoven JJ, Le Texier V, Muilu J. ASD: the Alternative Splicing Database. Nucleic Acids Res. 2004;32:D64–D69. doi: 10.1093/nar/gkh030. Database issue: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leipzig J, Pevzner P, Heber S. The Alternative Splicing Gallery (ASG): bridging the gap between genome and transcriptome. Nucleic Acids Res. 2004;32:3977–3983. doi: 10.1093/nar/gkh731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 16.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 17.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 19.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 20.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 22.Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell. 2006;24:943–953. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 23.Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Boue S, Letunic I, Bork P. Alternative splicing and evolution. Bioessays. 2003;25:1031–1034. doi: 10.1002/bies.10371. [DOI] [PubMed] [Google Scholar]
- 25.Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Mol Neurodegener. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox-Walsh KL, Hertel KJ. Splice-site pairing is an intrinsically high fidelity process. Proc Natl Acad Sci U S A. 2009;106:1766–1771. doi: 10.1073/pnas.0813128106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cochella L, Green R. Fidelity in protein synthesis. Curr Biol. 2005;15:R536–R540. doi: 10.1016/j.cub.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 30.Jeon C, Agarwal K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc Natl Acad Sci U S A. 1996;93:13677–13682. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nesser NK, Peterson DO, Hawley DK. RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proc Natl Acad Sci U S A. 2006;103:3268–3273. doi: 10.1073/pnas.0511330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 33.Celotto AM, Graveley BR. Alternative splicing of the Drosophila Dscam pre-mRNA is both temporally and spatially regulated. Genetics. 2001;159:599–608. doi: 10.1093/genetics/159.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ting CY, Yonekura S, Chung P, Hsu SN, Robertson HM, Chiba A, et al. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development. 2005;132:953–963. doi: 10.1242/dev.01661. [DOI] [PubMed] [Google Scholar]
- 35.Hsu SN, Yonekura S, Ting CY, Robertson HM, Iwai Y, Uemura T, et al. Conserved alternative splicing and expression patterns of arthropod N-cadherin. PLoS Genet. 2009;5:e1000441. doi: 10.1371/journal.pgen.1000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calarco JA, Saltzman AL, Ip JY, Blencowe BJ. Technologies for the global discovery and analysis of alternative splicing. Adv Exp Med Biol. 2007;623:64–84. doi: 10.1007/978-0-387-77374-2_5. [DOI] [PubMed] [Google Scholar]
- 37.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 38.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 39.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang XH, Chasin LA. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc Natl Acad Sci U S A. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]