Abstract
We have adapted our existing compression-induced fracture technology to cell culture studies, by generating linear patterns on a complex cell culture well structure, rather than on simple solid constructs. We present a simple method to create 1D, submicron, linear patterns of extracellular matrix on a multilayer silicone material. We identified critical design parameters necessary to optimize compression-induced fracture patterning on the wells, and applied stresses using compression Hoffman clamps. Finite-element analyses show that the incorporation of the well improves stress homogeneity (stress variation = 25%), and, thus, crack uniformity over the patterned region. Notably, a shallow well with a thick base (vs. deeper wells with thinner bases) reduces out-of-plane deflections by greater than a sixth in the cell culture region, improving clarity for optical imaging. The comparison of cellular and nuclear shape indices of a neuroblast line cultured on patterned 1D lines and unpatterned 2D surfaces reveals significant differences in cellular morphology, which could impact many cellular functions. Since 1D cell cultures recapitulate many important phenotypical traits of 3D cell cultures, our culture system offers a simple means to further study the relationship between 1D and 3D cell culture environments, without demanding expensive engineering techniques and expertise.
Keywords: fracture, patterning, biomaterial design, polydimethyl siloxane, compression
1. Introduction
Culturing cells in three-dimensional (3D) environments has emerged as a crucial in-vitro cell culture paradigm to simulate native cell function1,2 and regulate cell response to therapeutics3. 3D microenvironments mediate significant effects on cellular morphology,4,5 cell-cell interactions, viability, proliferation,6 motility,7,8 adhesion,9,10 signaling11 and differentiation,12 and, hence, 3D culture systems are growing in popularity. However, biologists culturing cells in 3D must control a host of changes in other environmental parameters13 including differences in transport phenomena14, ligand presentation13, and transmission of intrinsic15 and applied mechanical forces16,17 to the cells. The need to control carefully these parameters, in conjunction with the associated technical challenges inherent in 3D culture and imaging, drives the need for simpler, ‘minimally 3D’ systems.
Yamada and co-workers recently developed a promising approach to simulate 3D-like functionality while addressing these challenges by culturing cells along “one-dimensional” (1D) micropatterns. They demonstrated that 1D adhesive patterns on a flat surface prompts cells to change morphology and adopt behavior similar to that in 3D.7 They demonstrated that phenotypes of fibroblasts cultured on micropatterned lines were similar to fibroblasts cultured in 3D, and distinct from those cultured in 2D environments. These phenotypes included cell motility, spreading rates, polarization, uniaxial shape, expression of focal adhesion proteins, microtubule stability, and posterior centrosome orientation.7 As these phenotypes are related to cytoskeletal arrangement, these environmental conditions may also significantly influence higher order cell functions.18 The micropatterning approach is compatible with conventional biological imaging and assay tools, and suggests that 1D culture may be used as a simplified and precisely controlled substitute for 3D culture for many assays of biological function.
However, creating 1D adhesive patterns generally requires considerable expertise in micro/nano-fabrication and patterning techniques. For a pattern to be considered ‘1D’ based on cell function, Doyle et al. suggest that lines need to be < 1.5 μm in width7, a size which is challenging to fabricate reliably and consistently even for those with experience in the field. While both manual and automated techniques exist to address this problem19, they each require complex and specialized equipment unavailable to most biologists. To enable a broad spectrum of biologists to study the relationship between 1- and 3-dimensional environments, a simpler patterning and fabrication methodology is required. To address this need, we present a simple, versatile and inexpensive method to generate arrays of nanometer-to-micron scale linear cell-adhesive patterns in silicone cell culture wells via compression-induced fracture of the material.
Previous work in our lab20 demonstrated the use of fracture in multilayered materials for patterning cells along linear adhesive structures. Polydimethylsiloxane (PDMS), a biocompatible and easily processed elastomer commonly used in soft lithography21 forms a thin, brittle silica-like surface layer when treated with a plasma oxidizer. When strain is applied, the toughness mismatch between this brittle film and the underlying elastomer causes the formation of an array of parallel cracks within the film.22 If the uncracked surface is coated with a non-adhesive layer, cells will adhere to the relatively adhesive crack walls20. This simple approach provides the added capability of dynamically altering pattern dimensions by varying the applied strain.
While previous use of this method for cell culture purposes was based on applying tensile strain to the substrate, this set-up requires a specialized and expensive rig to clamp either end of the material and apply the required deformation. Unless custom-machined for the purpose, commercially available stretching rigs are generally large, and rarely compatible with cell culture incubator conditions. We recently demonstrated that compressive strains may also be used to generate cracks, via a lateral tensile stress resulting from constraint at the fixed edge. In this case, resulting cracks align along the direction of compression.23 Compressive stresses can be applied with simple clamps that are inexpensive, readily available, and have small footprints compatible with standard cell culture dishes. In this work we extend our previous study in compressing geometrically simple structures (spheres and cubes), towards more complex well-shaped geometries necessary for cell culture studies. We identify critical design parameters in using silicone wells for compression-induced fracture patterning, and apply compressive stresses with Hoffman clamps (used generally to clamp and control flow in flexible tubing) to rapidly and easily create 1D adhesive patterns. The entire set-up can be enclosed in a standard petri dish, enabling easy handling and minimizing contamination and evaporation. We then utilize this system to pattern immortalized dopaminergic neurons, a model cell line, on the 1D protein adhesions, and demonstrate that such patterns influence cell and nuclear morphology, similar to changes demonstrated by others in 3D cell culture environments.
2. Materials and Methods
2.1 Finite-Element Analysis
Compression of cell culture wells was simulated using commercially-available finite-element analysis software (ANSYS; Canonsburg, PA, USA). Quarter-symmetry models of the structures (cuboid, 19 mm × 19 mm × 10 mm, with a square well, 5 mm × 5 mm, of varying depth) were meshed with tetrahedral mesh elements (SOLID186, Young’s modulus E = 3 MPa, Poisson ratio ν = 0.49). The oxidized PDMS layer was assumed to have a thickness of 200 nm, and was modeled with triangular shell elements (SHELL41, E = 37 MPa, ν = 0.2).24 Clamped surfaces were constrained against movement in all dimensions, and compressive stresses were applied. Well depths were varied such that the substrate thickness was 0.5 mm (deep wells), 5 mm (medium wells), or 9.5 mm (shallow wells).
2.2 PDMS Substrate Preparation
A molding template was prepared by using glass adhesive to affix eight evenly spaced 5 × 5 × 0.55 mm glass pieces (UQG Optics Ltd, Cambridge, England) on a 75 × 38 mm glass microscope slide (Corning). A PDMS (Sylgard 184, Dow Corning) mixture was prepared from 10 parts elastomer and 1 part crosslinking agent, and cast against the molding template to a final thickness of 10 mm. The dishes were stored overnight at room temperature to allow the prepolymer mixture to de-gas and partially cure. The PDMS replica was cured at 60° C for 3 hours and 150° C for 24 hours to fully crosslink the PDMS. The PDMS slab was cut into 19 × 19 × 10 mm substrates centered on the wells.
2.3 Crack Generation
The silicone wells were exposed to oxygen plasma (Covance, Femto Science, Hwaseong, Korea) at 100 W for 5 min. The well surfaces of the PDMS substrates were positioned approximately 2 cm over a 1:1 mixture of (Tridecafluoro-1,1,2,2-Tetrahydrooctyl)-1-Trichlorosilane (T2492, United Chemical Technologies, Bristol, PA) and mineral oil (Sigma) inside a vacuum chamber. The samples were subjected to vacuum at 100 mm Hg for 15 minutes. This vapor deposition process results in the formation of a thin silane film on the interior well surface, and this coating serves as a binding agent for subsequent Pluronic treatment. Wells were filled with 0.1% Pluronic F127 in distilled water, a blocking agent, for 1.5 hours; gently rinsed and then submerged in water. Wells were filled with 0.1 mg/mL of either tetramethylrhodamine-isothiocyanate-labeled bovine serum albumin (TRITC-BSA) for crack visualization, or candidate extracellular matrix proteins for cell culture. The silicone well was then centered under the screw between the stationary base and mobile top bar of the closed jaw Hoffman clamp. A standard glass microscope slide was placed under a laterally positioned Hoffman clamp to maintain levelness and steady application of pressure. The screw was gently tightened just enough to restrict movement, and then further advanced until the desired displacement was achieved, fracturing the well surface. Protein adsorbed to the surfaces of the resulting cracks (Figure 1). The crack spacing and width were determined by analysis of optical images with ImageJ software (NIH).
Figure 1.
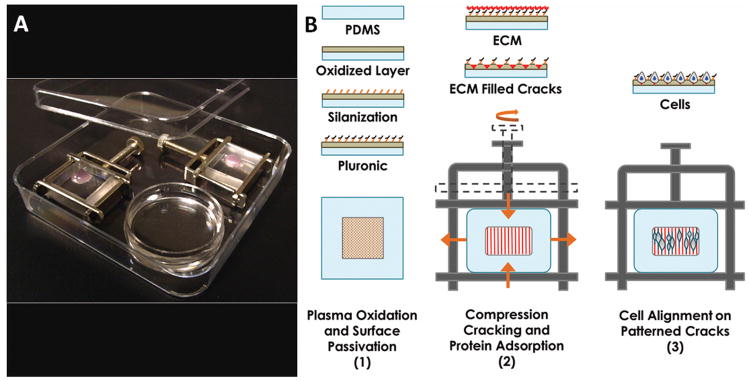
Device set-up and cell alignment scheme. (A) Wells mounted in Hoffman clamps are housed in square petri dishes, containing an open petri dish (round) of water for humidification. (B) Plasma oxidized wells are coated with silane that serves as a binding site for Pluronic blocking agents. The passivated well is filled with extracellular matrix (ECM) protein solution and mounted in a Hoffman clamp. Cracks form in the oxidized surface layer when pressure is applied by tightening the screw clamp, and protein fills the cracks, presenting an adhesive surface for cells.
2.4 Cell Cultivation in Cracks
Hoffman clamps were sterilized in an autoclave prior to cell culture. Each oxidized and surface-treated PDMS well was filled with 75 μL fibronectin (0.1 mg/mL, Sigma F1141) in phosphate buffered saline (PBS). Cracks were formed as described above. The entire set-up was exposed to germicidal ultraviolet light in a cell culture hood for 30 minutes. Fibronectin solution was removed from the wells. The wells were rinsed twice with PBS, filled with medium, and placed in a 37°C (5% CO2, 20% O2) incubator until the cells were loaded.
Rat dopaminergic neuroblasts (1RB3AN27 or N27) were propagated in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 1% antibiotic-antimycotic solution, and 2 mM GlutaMax (Invitrogen). N27s were harvested and resuspended in a 2.5 × 105 cells/mL cell suspension. The medium in wells was replaced with 100 μL of the cell suspension. The cells were allowed to settle into the cracks of the wells at room temperature for 20 minutes. Two Hoffman clamps were placed in a square petri dish (4 × 4 in.), along with a small round petri dish (1.5 cm diameter) containing sterile water to humidify the chamber and minimize evaporation. To compare the morphology of cells in patterned 1D culture with unpatterned 2D cultures, cells were plated at a density of 2 × 104 cells/cm2 in 24-well tissue culture plates (Becton, Dickinson & Co.).
2.5 Cell Fixation and Staining
Cells were rinsed twice with warm PBS, fixed with 4% paraformaldehyde (Sigma) for 15 minutes, and rinsed thrice (5 minutes between rinses) with cold PBS. Fixed cells were permeabilized with Triton X-100 (0.1% in PBS) for 3 minutes, blocked with BSA (1% in PBS) for 30 minutes, stained for F-actin with Phalloidin 594 (1:40 dilution in blocking solution) for 20 minutes, and stained for nucleic acids with 4′,6-diamidino-2-phenylindole, DAPI (1:1000 dilution in PBS), for 5 minutes in the dark. All steps of the staining procedure were performed at room temperature. Cells were rinsed twice with PBS between each step of the procedure, after cell fixation, and thrice following the final step.
The wells were imaged either with substrates mounted or not mounted on the Hoffman clamps. The PBS-filled wells were sealed with 9 × 9 mm square cover glasses (Bellco) when mounted, or with 48 × 65 mm rectangular cover glasses (Thermo Fisher Scientific, Gold Seal) when not mounted. Kimwipes were used to absorb excess PBS that created a seal between the glass and PDMS.
2.6 Statistical Analysis
Statistical analysis was conducted with commercially-available software (SigmaStat 3.5, Systat Software Inc., San Jose, CA), using a standard ANOVA test to compare population means. Post-hoc comparisons were conducted using the Tukey method.
3. Results
3.1 Stress uniformity in well-based culture systems
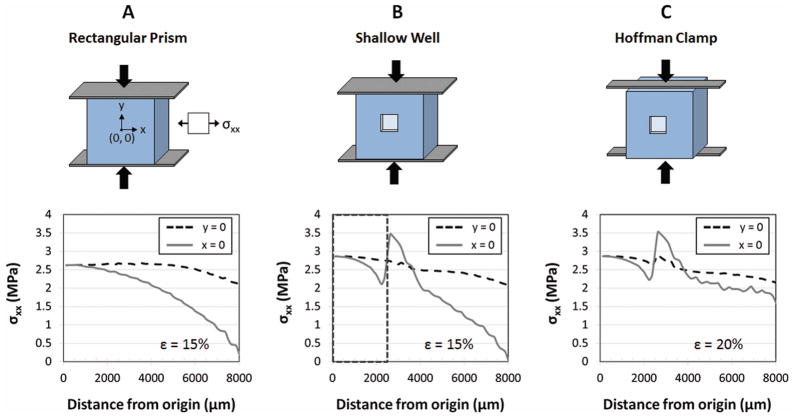
Finite-element analysis reveals that stresses generated as a result of compressive strains vary greatly across the surface of a simple rectangular prism (Figure 2). Under compressive strains of 15%, lateral stresses vary by as much as 212% (from 2.7 to −3.0 MPa), which would result in significant variation in crack dimensions across the entire culture surface.24 However, inside the shallow well within which cultured cells are confined, the stresses varied by only 25%. Hence, including a cell culture well is not only a requirement to maintain a sufficient quantity of media for cell culture, but also the well feature improves stress uniformity (Figure 2B) and, therefore, crack uniformity.
Figure 2.
Comparison of lateral surface stresses (σxx) generated perpendicular to applied compressive stresses in various culture systems. (A) Compression of a simple rectangular prism, under ideal no-slip clamping conditions, yields stresses ranging from 0 to 2.6 MPa across the strained surface for an applied compression of 15%. (B) Incorporating a shallow well, with ideal clamping conditions, limits cells to an area (dotted box) in which stresses range from 2.1 to 2.7 MPa for similar applied strains. (C) Utilizing a Hoffman clamp, which allows some freedom of the clamped ends, can yield stresses, within the cell culture region, that are similar in magnitude and uniformity to those in (B) when the applied strain is increased to 20%. Lateral stress (σxx) is shown along line y = 0 and line x = 0 at a distance from the origin (0, 0). Cell culture region spans 2500 μm from the origin in all directions along x and y.
Standard Hoffman clamps have compressive plates 4 mm thinner than the 10 mm thick PDMS slabs. Simulations reveal that this difference does not influence stress uniformity within the cell culture well, but does reduce the stress magnitude (data not shown). By increasing the applied compressive strains from 15% to 20%, stress magnitudes and uniformities with Hoffman clamp conditions were comparable to ‘ideal’ clamping conditions (Figure 2C).
3.2 Design principles for stress uniformity in wells
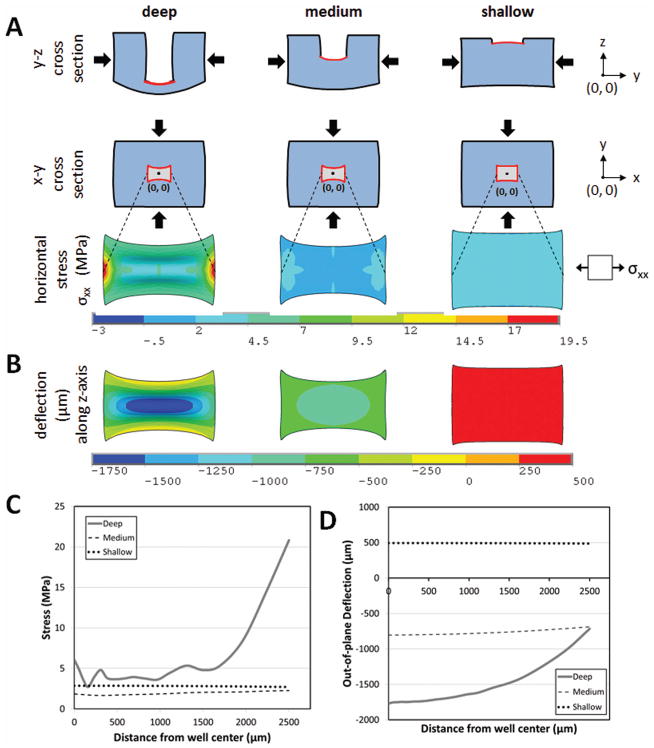
Finite-element simulations for a rectangular prism (dimensions 19 × 19 × 10mm) with a deep, medium or shallow well (0.5mm × 0.5mm culture surface) (Figure 3), demonstrate that the magnitude of the stresses and their uniformity along the well surface are strongly dependent on the well depth (Figure 3A). For a deep well, stresses vary by as much as 87% across the available culture surface, which would yield wider cracks at the edges than at the center of the well. In contrast, stresses vary by only 26% in the case of medium and shallow wells (Figure 3C), which would result in more uniform crack widths across the culture surface. However, another important parameter for microscopy and imaging in cell culture is uniformity of the out-of-plane deflections across the surface. The out-of-plane deflection (Figure 3B, D) varied most for deep wells (1.75 mm), less (166 μm) for medium wells and least (25 μm) for shallow wells. While greater well depths would allow greater cell culture volumes and hence, longer-term cultures, all further experiments were conducted with the shallowest well depths for optimal stress uniformity, and to facilitate microscopy.
Figure 3.
Effect of geometry on mechanical behavior of culture wells under compression. (A) Lateral stresses generated perpendicular to applied compressive strains of 15% in deep (0.5 mm thick base), medium (5 mm thick base) and shallow (9.5 mm thick base) wells. (B) Out-of-plane deflection of the wells resulting from applied compression. (C) Lateral stresses and (D) out-of-plane deflection across the center of the wells (centered at origin x = 0, y = 0) demonstrate an increase in stress uniformity and decrease in magnitude of out-of-plane displacement with decreasing well depth. Compressive stress, tensile stress, and deflection are reported along the y-axis, x-axis, and z-axis respectively.
3.3 Crack Formation
After the PDMS well surfaces were oxidized, they were uniaxially compressed in the clamps to 10% or 20%. As described previously, transverse deformation induced by the Poisson effect in materials undergoing compression results in surface tensile stresses when the loading surfaces do not slip against the clamp face. In this multilayered material system, the toughness mismatch between the PDMS and the oxidized PDMS film results in the formation of an array of cracks (V-shaped in cross-section) perpendicular to the resultant tensile stresses.23
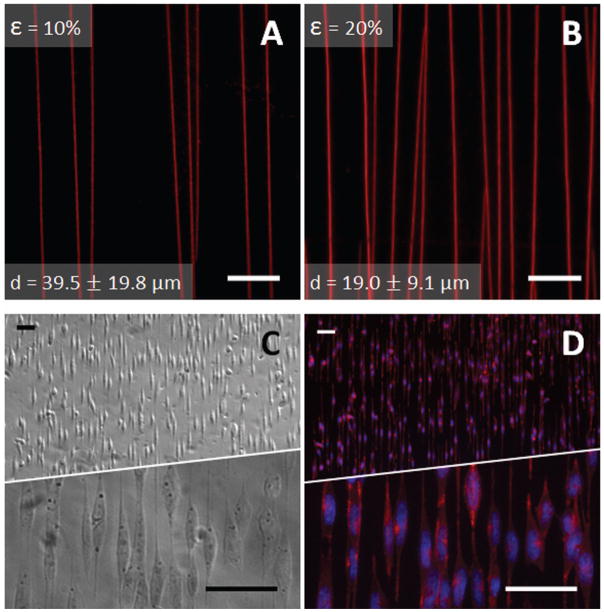
Crack features were fluorescently labeled with adsorbed TRITC-BSA, and the average spacing between the cracks was found to be 39.5 ± 19.8 μm and 19.0 ± 9.1 μm for 10% and 20% compressive loads respectively (Figure 4A and 4B). As expected based on our previous findings, the average crack spacing and spacing uniformity decreased with an increased compressive load.23 Previous work also suggests that the depth of the cracks is limited by the thickness of the oxidized PDMS layer, a few hundred nm for PDMS substrates subjected to several minutes of oxidation at high power.24 Crack widths were visually confirmed to be 1.4 ± 0.17 μm for 10% applied compression, similar to previous structures prepared with these oxidized PDMS multilayer material systems.20
Figure 4.
Crack spacing, protein adherence, and staining of aligned cells. Cracks were visualized with TRITC-BSA (red), and the average spacing resulting from (A) 10% and (B) 20% compressive loads was respectively 39.5 ± 19.8 μm and 19.0 ± 9.1 μm (mean ± S.D.). Data was collected from five different fields in the well region of each of three cuboids for each compressive load. Scale bar = 25 μm (C) Phase-contrast image of N27 cells aligned on cracks. (D) Aligned N27 cells were counterstained with FITC-labeled Phalloidin (red) and DAPI (blue, to label nuclei) revealing aligned actin filaments and elongated nuclei. Scale Bar = 50 μm.
3.4 Demonstration of 1D Patterning and Comparison to 2D Cell Culture
N27 cells seeded in the crack-patterned wells spread cellular extensions to align along the direction of the cracks (Figure 4C). Staining of N27 cells, with phalloidin and DAPI (Figure 4D), reveals aligned elongation of the cell extensions along the cracks, and a consequent elongation of nuclei on the 1D patterns.
A comparison between N27 cells on our 1D substrates with unpatterned N27 cells cultured on conventional tissue culture plastic (TCP) reveals significant differences in cell and nuclear morphology (Figure 5 A–F). To quantify the degree to which cells are influenced by the 1D patterns, the cell size index (CSI) and nuclear size index (NSI) were measured (Figure 5G, H). The cell and nuclear size indices are defined as the aspect ratio between the length of the longest region of the cell or nucleus and the perpendicular width of the widest region of the cell or nucleus. The CSI on 1D patterns (Figure 5G) was significantly increased when compared to the cells cultured in 2D, with an increased spread of CSI values centered on an aspect ratio of 10. In contrast, cells in 2D culture were significantly less elongated in morphology, the highest frequency of which occurs at an aspect ratio of 2, confirming that 1D culture drives significant changes in cell morphology. Importantly, this difference in aspect ratio is also reflected in the nuclear shape. NSI measurements reveal that while similar spreads in nuclear shape exist, these distributions are centered on an NSI of 2 for TCP, and 4 for 1D patterns (Figure 5H). Statistical analysis reveals a significant difference between mean CSI and NSI values (p < 0.001) when comparing cells cultured on 1D patterned and 2D unpatterned cell culture substrates.
Figure 5.
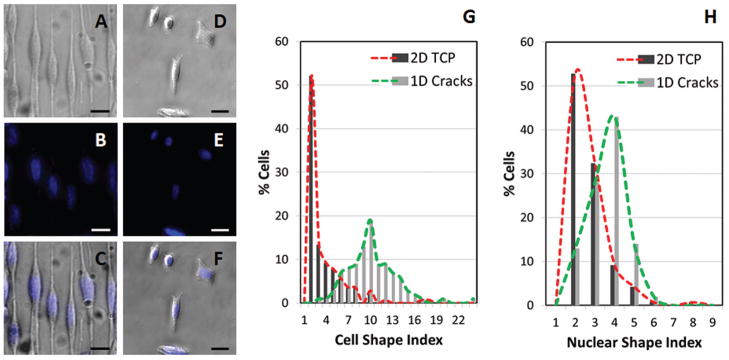
Comparison of patterned 1D and unpatterned 2D N27 cells quantified by Cell Shape Index (CSI) and Nuclear Shape Index (NSI). Images of cells (phase-contrast) and nuclei (stained with DAPI, blue) and overlay of cells and nuclei for cells aligned on 1D crack substrates (A, B, C) and on 2D tissue culture plastic (TCP) substrates (D, E, F). Scale bar = 25 μm. Histogram comparisons of patterned and nonpatterned cells for CSI (G) and NSI (H). Total cells for 1D crack = 100. Total cells for 2D TCP = 142. Cell culture duration = 8 hrs. Significant differences exist between both the CSI and NSI mean values when comparing morphologies on the two culture surfaces (p < 0.001).
4. Discussion
Mimicking native cell culture environments is an important approach in next-generation drug screening applications,25 as well as in disease modeling in a culture dish. Hence, 3D culture is emerging as an important paradigm in in-vitro cell culture models, but such systems present unique technical and control challenges that cannot be easily addressed using conventional approaches.13 We present a culture system that is applicable to a wide variety of cell types for studying cell behavior in a well-defined 1D system, as a useful alternative to 3D cultures.
Recent findings that 1D protein patterns may simulate 3D functionality, particularly for migration studies,7 presents an interesting opportunity to study cell function in precisely defined contexts that recapitulate much of the 3D culture models; but forming 1D culture substrates has been challenging. 1D adhesive patterns, with micron-scale features, have been generated by an assortment of techniques involving one or more of the following processes: photolithography, reactive ion etching electron beam lithography, wet etching,26 and soft lithography transfer molding, contact printing and capillary molding.27 Doyle, et al. created 1D linear patterns by combining confocal microscopy with photolytic ablation to precisely etch linear adhesive regions on an antifouling polyvinyl alcohol coated surface.7 These patterning methods require steep initial fabrication costs, expensive infrastructure, and considerable expertise; often do not reliably transfer patterns due to collapsing molds;28 and are prone to cutting errors.26 Such technological challenges can prevent researchers from studying this phenomenon. While 1D culture has been demonstrated as a potential substitute for 3D systems for applications in cell morphology and migration, further research is necessary to determine how broadly applicable 1D culture is in simulating 3D functionality. Differences in ligand presentation for example, may drive differential functionality. Hence technologies that are reliable, easily accessible and inexpensive are necessary to thoroughly explore the relationship between 1D and 3D culture. In this work, we demonstrate a simple, versatile system for 1D culture of cells, based on compression-induced formation of protein-patterned cracks.
Previously, we developed a custom apparatus to compress oxidized PDMS cubes and spheres, and this method served as the foundation for creating the current crack fabrication system23. Here, we made the compression system compatible to cell culture by introducing a well reservoir to contain cells. Based on FEA simulations, reservoir features contribute to the homogeneity of stress distribution; and since crack dimensions are directly related to stress magnitudes,24 this feature improves the uniformity of generated cracks. Design parameters for the well include well depth and thickness of the underlying culture substrate. Deeper wells (and thinner culture substrates) allow the system to hold more medium, and thus enable longer term cultures that are less prone to evaporation and the need for medium replenishment. However, the thickness of the culture substrate influences ease of microscopy, with thinner substrates improving image clarity. Even though we achieved crack formation with deep wells and 2 mm thin culture surfaces (data not shown), the substrate was prone to buckling, which made imaging through the bottom of the well difficult. FEA simulations reveal that deep wells (thin bases) give rise to heterogeneous stress distributions with a high degree of out-of-plane deflection. As a practical compromise, we opted for shallow wells with thick bases, where the well height was adjusted to give sufficient medium volume capacity and optimal visual clarity (shallow wells were inverted to view cells from the well top rather than through the base). Minimizing cell culture well depths reduced out-of-plane deflection. Hence, a shallow well reservoir is an important design feature essential to utilizing this system for cell culture applications.
To demonstrate the applicability of this technique to culturing and probing cell function in 1D systems, we successfully cultured a neuroblast cell line on the protein-patterned cracks and further demonstrated distinct differences in cellular and nuclear morphology, compared to cells grown on 2D tissue culture treated polystyrene surfaces (Figure 5), a broadly applicable tool for biologists. Our results indicate that cell morphology is significantly influenced by the underlying pattern, and, similar to studies performed in conventional 3D systems8, nuclear shape is also mediated by the features of the culture surface. Since nuclear shape is a critical influence on cell function29, these results demonstrate that our 1D patterning system successfully organizes the cell structure to recapitulate characteristic features demonstrated by others in more complex culture systems.8
Control of cellular shape has broad applications and significance for adherent cells.30,31,32,33 Individual cells from a supposedly homogenous source, such as cell lines from a single clone, when grown on a uniform 2D substrate, display widely heterogeneous cell morphologies and possibly atypical cellular functions31. Hence, control of cell shape is a critical parameter in predictive control over cell function, as well as in simulating normal (in-vivo) microenvironmental niches.
In addition to regulating cellular shape,34, 35 topographical landscape governs cellular behaviors such as adhesion,34 proliferation,34, 36 migration, and differentiation.36 These behaviors have significant implications in biological processes including cancer metastasis, wound healing,36 and organogenesis36,37 where cells can actually express unique topography-dependent migration patterns.36 Grooved substrates, the most commonly exploited type, 38 in particular can alter shape, polarity and migration of certain cell types. A variety of cells types can respond to a unique set of groove dimensions34, 39 in one way by aligning parallel to the grooves, and such orientation is most pronounced with the groove features that are less than or equal to the cell width.39 Cytoskeletal components align along the grooves36, 38, 40 evidenced by not only cell shape or polarity, but stimulating signal transduction cascades,34, 35, 36, 39 and altered gene and protein expression patterns.34, 35 Our previous and current work indicate that v-shaped grooved features with 1–1.5 μm widths and no more than a few hundred nanometers of depth are sufficient to coerce alignment of N27 rat dopaminergic neuroblasts.20 The alteration of signal transduction pathways of N27 cells due to alignment, response of other cells to the groove features, and generation of other groove dimensions remains to be explored.
In order to better simulate the 3D topography of native microenvironments, in vitro systems, such as those described here, can be easily engineered with built-in microscale and nanoscale features similar to native extracellular topography.34, 38 We believe that the 1D environments produced in our cell culture set-up will be used by biologists to further study the connection between environmental cues, including topography, and cell phenotype2.
While utilizing 1D patterns to simulate 3D behaviour might not necessarily translate to multiple cell types, 41, 42, 43 novel methods are needed to generate these patterns to enable a variety of researchers to investigate the potential of such an approach to reduce the complexity of cell culture models in chemical library screens.
5. Conclusion
Our simple cell culture system utilizes an easily fabricated PDMS well, and a ubiquitously available Hoffman clamp to generate a 1D pattern of nano- to micron-scale adhesive surfaces on which cells can be predictably aligned. An evaluation of the design parameters for the wells suggests that shallow wells serve to enhance stress uniformity over the patterned region, thereby improving uniformity of crack features. We also present a simple analysis of differences in cell morphologies on 1D crack versus 2D tissue culture surfaces, demonstrating the compatibility of this approach with cell culture techniques, and demonstrating the utility of this approach in manipulating cell and nuclear shape. This approach is significantly more accessible than conventional engineering technologies used to create linear micropatterns of adhesive structures. We anticipate that this inexpensive and user-friendly system has a broad range of research applications in studying the implications of cellular alignment, such as in neuron alignment, 44 guided stem cell differentiation, 45, 46 and topographical effects on cell cycling.47 Potential similarities between cells cultured in 1D and 3D environments suggests that this technology may be broadly applicable in creating simplified cell culture models.
Acknowledgments
We acknowledge contributions and helpful discussions with Tejash Patel, Dr. Xiaoyue Zhu and Dr. Tomoyuki Uchida. The authors gratefully acknowledge support from the University of Michigan National Center for Institutional Diversity postdoctoral fellowship program to ARD, and the Natural Sciences and Engineering Research Council of Canada / Banting postdoctoral fellowship programs to CM. This research was funded by the NIH (EB 003793 and HG 004653) and NSF (CMMI 0700232).
References
- 1.Smalley K, Lioni M, Herlyn M. Life isn’t flat: taking cancer biology to the next dimension. In Vitro Cell Dev Biol Anim. 2006;42:242–247. doi: 10.1290/0604027.1. [DOI] [PubMed] [Google Scholar]
- 2.Pampaloni F, Reynaud E, Stelzer E. The third dimension bridges the gap between cell culture and live tissue. Nature Rev Mol Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 3.Justice B, Badr N, Felder R. 3D cell culture opens new dimensions in cell-based assays. Drug Discov Today. 2009;14:102–107. doi: 10.1016/j.drudis.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Yamada K, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Kraning-Rush C, Carey S, Califano J, Smith B, Reinhart-King C. The role of the cytoskeleton in cellular force generation in 2D and 3D environments. Phys Biol. 2011;8(1):015009. doi: 10.1088/1478-3975/8/1/015009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cukierman E, Pankov R, Yamada KM. Cell interactions with three-dimensional matrices. Curr Opin Cell Biol. 2002;14:633–640. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 7.Doyle A, Wang F, Matsumoto K, Yamada K. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nathan A, Baker B, Nerurkar N, Mauck R. Mechano-topographic modulation of stem cell nuclear shape on nanofibrous scaffolds. Acta Biomater. 2011;7:57–66. doi: 10.1016/j.actbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cukierman E, Pankov R, Stevens D, Yamada K. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 10.Harunaga J, Yamada K. Cell-matrix adhesions in 3D. Matrix Biol. 2011;30:363–368. doi: 10.1016/j.matbio.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green A, Yamada K. Three-dimensional microenvironments modulate fibroblast signaling responses. Adv Drug Deliv Rev. 2007;59:1293–1298. doi: 10.1016/j.addr.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraehenbuehl T, Langer R, Ferreira L. Three-dimensional biomaterials for the study of human pluripotent stem cells. Nat Methods. 2011;8:731–736. doi: 10.1038/nmeth.1671. [DOI] [PubMed] [Google Scholar]
- 13.Baker B, Chen C. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghavan S, Shen CJ, Desai RA, Sniadecki NJ, Nelson CM, Chen CS. Decoupling diffusional from dimensional control of signaling in 3D culture reveals a role for myosin in tubulogenesis. J Cell Sci. 2010;123:2877–2883. doi: 10.1242/jcs.055079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huebsch N, Arany P, Mao A, Shvartsman D, Ali O, Bencherif S, Rivera-Feliciano J, Mooney D. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moraes C, Wang G, Sun Y, Simmons C. A microfabricated platform for high-throughput unconfined compression of micropatterned biomaterial arrays. Biomaterials. 2010;31:577–584. doi: 10.1016/j.biomaterials.2009.09.068. [DOI] [PubMed] [Google Scholar]
- 17.Moraes C, Sun Y, Simmons C. (Micro)managing the mechanical microenvironment. Integr Biol. 2011;3:959–971. doi: 10.1039/c1ib00056j. [DOI] [PubMed] [Google Scholar]
- 18.Treiser M, Yang E, Gordonov S, Cohen D, Androulakis I, Kohn J, Chen C, Moghe P. Cytoskeleton-based forecasting of stem cell lineage fates. Proc Natl Acad Sci U S A. 2010;107:610–615. doi: 10.1073/pnas.0909597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Théry M. Micropatterning as a tool to decipher cell morphogenesis and functions. J Cell Sci. 2010;123:4201–4213. doi: 10.1242/jcs.075150. [DOI] [PubMed] [Google Scholar]
- 20.Zhu X, Mills K, Peters P, Bahng J, Liu E, Shim J, Naruse K, Csete M, Thouless MD, Takayama S. Fabrication of reconfigurable protein matrices by cracking. Nat Mater. 2005;4:403–406. doi: 10.1038/nmat1365. [DOI] [PubMed] [Google Scholar]
- 21.Xia Y, Whitesides GM. Soft ithography. Annu Rev Mater Sci. 1998;28:153–184. [Google Scholar]
- 22.Thouless MD, Li Z, Douville NJ, Takayama S. Periodic cracking of films supported on compliant substrates. J Mech Phys Solids. 2011;59:1927–1937. doi: 10.1016/j.jmps.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchida T, Mills KL, Kuo C-H, Roh W, Tung Y-C, Garner A, Koide K, Thouless MD, Takayama S. External compression-induced fracture patterning on the surface of poly(dimethylsiloxane) cubes and microspheres. Langmuir. 2009;25:3102–3107. doi: 10.1021/la801986k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mills KL, Zhu X, Takayama S, Thouless MD. The mechanical properties of a surface-modified layer on poly(dimethylsiloxane) J Mater Res. 2008;23:37–48. doi: 10.1557/JMR.2008.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moraes C, Mehta G, Lesher-Perez SC, Takayama S. Organs-on-a-chip: a focus on compartmentalized microdevices. Ann Biomed Eng. 2012;40:1211–1227. doi: 10.1007/s10439-011-0455-6. [DOI] [PubMed] [Google Scholar]
- 26.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF. Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomaterials. 1999;20:573–588. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 27.ten Elshof JE, Khan SU, Göbel OF. Micrometer and nanometer-scale parallel patterning of ceramic and organic–inorganic hybrid materials. J Eur Ceramic Soc. 2010;30:1555–1577. [Google Scholar]
- 28.Zhou W, Huang Y, Menard E, Aluru NR, Rogers JA, Alleyne AG. Mechanism for stamp collapse in soft lithography. Appl Phys Lett. 2005;87:251925. doi: 10.1021/la0502185. [DOI] [PubMed] [Google Scholar]
- 29.Dahl K, Ribeiro A, Lammerding J. Nuclear Shape, Mechanics, and Mechanotransduction. Circ Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Micropatterned surfaces for control of cell shape, position, and function. Biotechnol Prog. 1998;14:356–363. doi: 10.1021/bp980031m. [DOI] [PubMed] [Google Scholar]
- 31.Li F, Li B, Wang Q-M, Wang JHC. Cell shape regulates collagen type I expression in human tendon fibroblasts. Cell Motil Cytoskeleton. 2008;65:332–341. doi: 10.1002/cm.20263. [DOI] [PubMed] [Google Scholar]
- 32.McBeath R, Pirone D, Nelson C, Bhadriraju K, Chen C. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 33.Kilian K, Bugarija B, Lahn B, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natil Acad Sci U S A. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kriparamanan R, Aswath P, Zhou A, Tang L, Nguyen KT. Nanotopography: cellular responses to nanostructured materials. J Nanosci Nanotechno. 2006;6(7):1905–1919. doi: 10.1166/jnn.2006.330. [DOI] [PubMed] [Google Scholar]
- 35.Kulangara K, Leong KW. Substrate topography shapes cell function. Soft Matter. 2009;5(21):4072–4076. [Google Scholar]
- 36.Kim D-H, Provenzano PP, Smith CL, Levchenko A. Matrix nanotopography as a regulator of cell function. J Cell Biol. 2012;197(3):351–360. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D-H, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh K-Y, Tung L, Levchenko A. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proc Natl Acad Sci USA. 2010;107(2):565–570. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF. Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomaterials. 1999;20(6):573–588. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 39.Curtis A, Wilkinson C. Topographical control of cells. Biomaterials. 1997;18(24):1573–1583. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 40.Martínez E, Lagunas A, Mills CA, Rodríguez-Seguí S, Estévez M, Oberhansl S, Comelles J, Samitier J. Stem cell differentiation by functionalized micro- and nanostructured surfaces. Nanomedicine-UK. 2008;4(1):65–82. doi: 10.2217/17435889.4.1.65. [DOI] [PubMed] [Google Scholar]
- 41.Picone R, Ren X, Ivanovitch K, Clarke J, McKendry R, Baum B. A polarised population of dynamic microtubules mediates homeostatic length control in animal cells. PLoS Biol. 2010;8(11):e1000542. doi: 10.1371/journal.pbio.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levina E, Kharitonova M, Rovensky Y, Vasiliev J. Cytoskeletal control of fibroblast length: experiments with linear strips of substrate. J Cell Sci. 2001;114:4335–4341. doi: 10.1242/jcs.114.23.4335. [DOI] [PubMed] [Google Scholar]
- 43.Vignaud T, Blanchoin L, Théry M. Directed cytoskeleton self-organization. Trends Cell Biol. 2012;22:671–682. doi: 10.1016/j.tcb.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 44.Hoffman-Kim D, Mitchel JA, Bellamkonda RV. Topography, cell response, and nerve Regeneration. Annu Rev Biomed Eng. 2010;12:203–231. doi: 10.1146/annurev-bioeng-070909-105351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee MR, Kwon KW, Jung H, Kim HN, Suh KY, Kim K, Kim K-S. Direct differentiation of human embryonic stem cells into selective neurons on nanoscale ridge/groove pattern arrays. Biomaterials. 2010;31:4360–4366. doi: 10.1016/j.biomaterials.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 46.Yim EKF, Pang SW, Leong KW. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp Cell Res. 2007;313(9):1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Kooten TG, Whitesides JF, von Recum AF. Influence of silicone (PDMS) surface texture on human skin fibroblast proliferation as determined by cell cycle analysis. J Biomedi Mater Res. 1998;43:1–14. doi: 10.1002/(sici)1097-4636(199821)43:1<1::aid-jbm1>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]