Abstract
Constitutive heterochromatin is important for maintaining chromosome stability but also delays the repair of DNA double strand breaks (DSB). DSB repair in complex mammalian genomes involves a fast phase (2–6 hrs) where most of the breaks are rapidly repaired, and a slow phase (up to 24 hrs) where the remaining damages in heterochromatin are repaired. We found that p53 deficiency delays the slow phase DNA repair after ionizing irradiation. P53 deficiency prevents down regulation of histone H3K9 trimethylation at pericentric heterochromatin after DNA damage. Moreover, p53 directly induces expression of the H3 K9 demethylase JMJD2b through promoter binding. P53 activation also indirectly down regulates expression of the H3 K9 methytransferase SUV39H1. Depletion of JMJD2b or sustained expression of SUV39H1 delays the repair of heterochromatin DNA and reduces clonogenic survival after ionizing irradiation. The results suggest that by regulating JMJD2b and SUV39H1 expression, p53 not only controls transcription but also promotes heterochromatin relaxation to accelerate a rate-limiting step in the repair of complex genomes.
Keywords: p53, JMJD2b, SUV39H1, heterochromatin, DNA damage, repair, histone methylation
Introduction
Different tumor types have variable response to ionizing radiation (IR). Long-term survival after IR has been shown to correlate with efficient DNA repair. P53 is highly inducible by IR through ATM-dependent inhibition of the MDM2 E3 ligase (1). P53 stabilization induces cell cycle arrest in most epithelial and mesenchymal cell types, and induces apoptosis in lymphocytes or proliferating cells of the intestine.
P53-null mice are sensitive to IR-induced tumorigenesis, suggesting that p53 is involved in DSB repair (2). Inactivation of p53 leads to chromosomal instability during tumor development in mouse models (3). P53-null MEFs become aneuploid after only a few passages in culture, indicating a significant defect in chromosome segregation (4). Fibroblasts from patients with Li-Fraumeni syndrome (germ line heterozygous p53 mutation) also have chromosome instability in culture (5). Bone marrow and spleen cells in p53-null mice show higher levels of chromosome abnormalities in the absence of any treatment, suggesting that p53 function is important for maintaining genomic stability under physiological conditions (6). P53 deficiency leads to increased homologous recombination in vivo and in vitro (7, 8). The potential of p53 in regulating multiple DNA repair pathways were generally explained by its ability to regulate the expression of genes directly involved in the DNA repair process, or by direct binding of p53 to DNA repair factors (9, 10).
The biological outcome of p53 activation (arrest, apoptosis, senescence) is dependent on cell type and stimuli. The p53 pathway is compromised to different degrees in all tumors, including partial to complete loss-of-function mutations, gain-of-function mutations, and functional inactivation by MDM2. While loss of wild type p53 activity per se may increase radio sensitivity, most tumors accumulate gain-of-function mutant p53 that increase resistance to DNA damage (11). The combination of these diverse and conflicting effects confounds earlier efforts to determine the prognostic value of p53 status in treatment response. The role of p53 in tumor sensitivity to radiation is controversial in clinical studies (12, 13). Wild type p53 has been reported to associate with favorable or unfavorable treatment response, or having no impact.
Numerous studies suggest that epigenetic marks on chromatin are important determinants of genomic stability and DNA repair efficiency. DNA double strand break repair involves a fast phase (2–6 hrs) when ~80% of the breaks are rapidly repaired, and a slow phase (up to 24 hrs) when the remaining damages are repaired. Completion of the slow phase repair specifically requires ATM signaling (14). Recent studies showed that DSB repaired during the slow phase are localized in constitutive pericentric heterochromatin (HC) regions of the genome (15). Although IR induces similar rate of DSB formation in heterochromatin and euchromatin (16), IR induces fewer γH2AX foci in heterochromatin shortly after IR (17), and these foci are cleared at a slower rate. Heterochromatin formation inhibits DSB repair by limiting access of repair factors to the breaks, and preventing chromatin remodeling near the break. It has been shown that mammalian cells with reduced linker histone H1 and more accessible chromatin are less sensitive to IR (18). ATM phosphorylation of the chromatin-modifying factor KAP1 promotes chromatin relaxation and efficient repair of heterochromatin DSB (19, 20). Study in Drosophila cells showed that heterochromatin domains become more mobile and expanded after IR, DSB foci in repetitive sequences relocate outside of the heterochromatin domain before being repaired by homologous recombination mechanism (21).
In mouse cells, H3 K9 trimethylation at pericentric heterochromatin is mainly mediated by the histone methyltransferase SUV39H1 and the testes-specific isoform SUV39H2 (22). Knockout of SUV39H1/2 in mice caused 3-fold reduction of pericentric H3 K9me3 level and 5-fold increase in the production of pericentric chromatin-derived non-coding RNA transcripts (23). SUV39H1/2 are also important for H3 K9me3 modification at telomeric heterochromatin. SUV39H1/2 deficiency leads to abnormal elongation of telomeres (24). MEFs derived from SUV39H1/2 double-null mice have significant chromosome mis-segregation and rapidly become aneuploid in culture (22). Drosophila embryos with Su(var)3-9 deficiency also have chromosome defects and spontaneous DNA damage in heterochromatin (25). Therefore, SUV39H1 is important for heterochromatin maintenance, and regulates the efficiency of DSB repair. The Jumonji domain 2 (JMJD2) family demethylase JMJD2b removes pericentric heterochromatin K9m3 marks, thus opposes the function of SUV39H1 (26). JMJD2b is overexpressed in breast tumors with chromosome instability. Overexpression of JMJD2b in cell lines also increases chromosome instability (27).
In this report, we show that p53 is required for down-regulation of H3 K9me3 level in pericentric heterochromatin after IR. P53 deficiency causes significant delay in the slow phase repair. To explore the mechanism, we found that p53 activates JMJD2b expression and represses SUV39H1. These results suggest that p53 induces relaxation of heterochromatin by regulating the enzymes involved in H3 K9 methylation. Therefore, regulation of JMJD2b and SUV39H1 level by p53 not only controls gene expression, but also facilitates the repair of damaged heterochromatin DNA, which is a rate-limiting step in global DNA repair of complex genomes.
Results
p53 inactivation delays heterochromatin DNA repair after IR
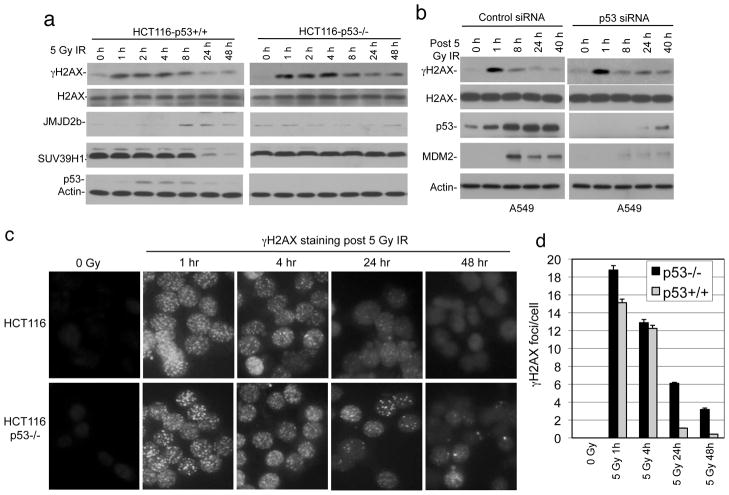
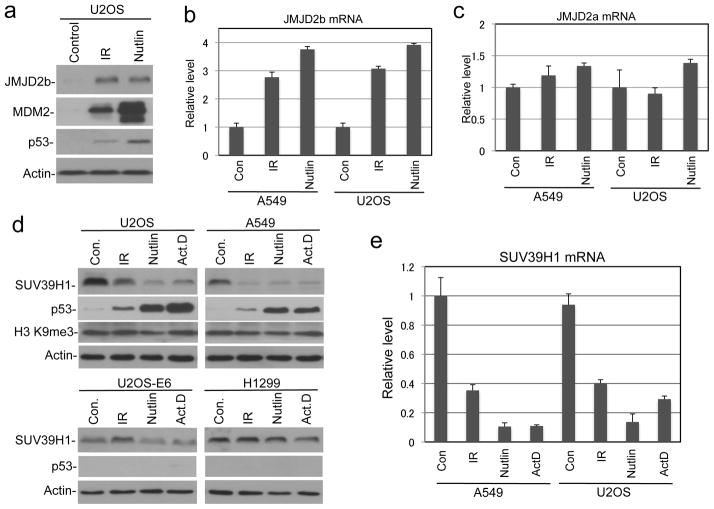
Previous study showed that p53 knockout in HCT116 reduces cell survival after IR (28). We also reported that p53 knockdown or overexpression of MDM2 in U2OS reduces colony formation after IR (29). However, the mechanisms of these observations were unclear. To test whether DNA repair plays a role, HCT116 and HCT116-p53−/− cells were analyzed for γH2AX level by western blot after IR, since γH2AX foci formation is a sensitive marker for the presence of DSB (30). IR treatment induced rapid increase of γH2AX in both cell lines. The γH2AX level returned to background after 24 hours in parental HCT116 cells, but remained significantly above background in HCT116-p53−/− at 24 and 48 hours after IR (Figure 1a). IR induced JMJD2b expression and suppressed SUV39H1 level in a p53-dependent fashion (Figure 1a, further addressed below). Transient knockdown of p53 in A549 cells also delayed the down regulation of γH2AX after IR, although the effect was moderate due to incomplete depletion of p53 (Figure 1b). Immunofluorescence staining of γH2AX showed significant number of DNA damage foci remaining in p53-null cells 48 hours after IR (Figure 1c, 1d). These results suggest that p53 deficiency prevents the timely repair of damaged DNA.
Figure 1. Loss of p53 delays DNA repair after IR.
(a) HCT116 with and without p53 were treated with 5 Gy IR and analyzed for γH2AX, SUV39H1, and JMJD2b levels at indicated time points by western blot. (b) A549 cells were treated with p53 siRNA for 48 hrs followed by 5 Gy IR treatment. Indicated markers were analyzed by western blot at different time points. (c) HCT116 cells with and without p53 were treated with IR and stained for γH2AX foci at indicated time points. (d) Quantitation of γH2AX foci density in the experiment described in (c).
p53 promotes the repair of heterochromatin DNA
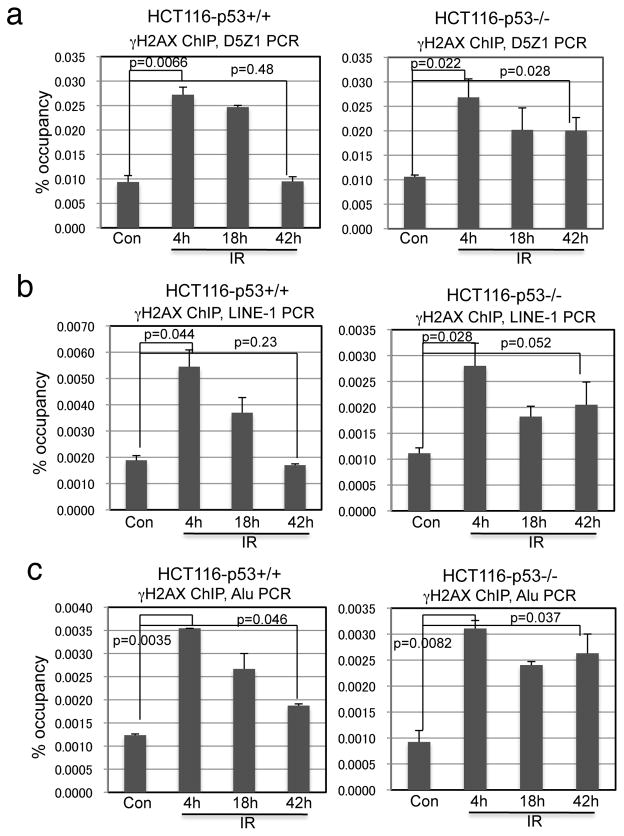
Since heterochromatin DNA is often repaired during the slow phase (15), we hypothesized that the γH2AX present in p53-null cells at late time points after IR may be associated with heterochromatin regions. We performed γH2AX ChIP analysis to compare the constitutive heterochromatin repair efficiency between HCT116 and HCT116-p53−/− cells. Cells were treated with 5 Gy IR and γH2AX ChIP was PCR amplified using D5Z1 primers specific for Chromosome 5 and 19 alpha satellite tandem repeats (27, 31), and primers that detect interspersed repetitive elements LINE-1 and Alu. As expected, the levels of alpha satellite, LINE-1, and Alu DNA pulled down by γH2AX ChIP remained elevated 42 hours after IR in HCT116-p53−/− cells, but largely returned to near-background levels in wild type HCT116 (Figure 2a, 2b, 2c). Attempts to analyze γH2AX ChIP signal on euchromatin were unsuccessful due to the extremely low probability of DNA strand breaks occurring in any single copy gene (~100 DSB/cell after 5 Gy IR). Overall, these results suggest that p53 is important for promoting the repair of damaged DNA in heterochromatin.
Figure 2. Loss of p53 delays heterochromatin repair.
HCT116 and HCT116-p53−/− were treated with 5 Gy IR and analyzed by γH2AX ChIP and qPCR using alpha satellite (a), LINE-1 (b), and Alu primers (c) at indicated time points.
p53 down regulates heterochromatin H3 K9 methylation after IR
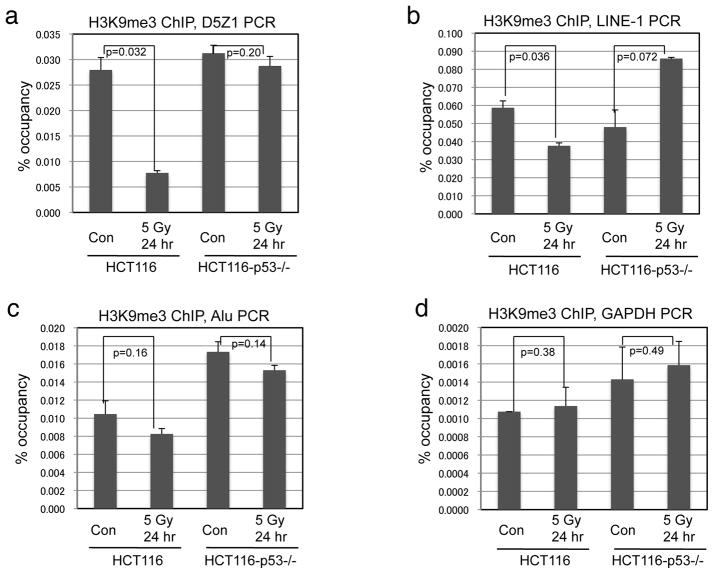
Recently, we found that the p53 transcription target MDM2 interacts with SUV39H1 (32), which is an enzyme important for H3 K9 trimethylation in pericentric heterochromatin. Furthermore, it was reported that MDM2 can ubiquitinate and degrade SUV39H1 (33). However, previous studies showed that DNA damage treatments do not reduce total H3 K9me3 level in human cell lines (19, 34). We also failed to detect robust decrease in bulk H3 K9me3 level after IR (Figure 6a). To test whether p53 activation specifically reduces H3 K9 methylation level in constitutive heterochromatin, HCT116 and HCT116-p53−/− cells were treated with IR and analyzed by H3 K9me3 and H3 K9me2 ChIP after 24 hours. IR treatment led to ~70% reduction of K9me3 level in pericentric alpha satellite DNA in HCT116 but not HCT116-p53−/− cells (Figure 3a). In addition to the pericentromeric tandem repeats, interspersed transposable elements (LINE, SINE, LTR, etc) are also mostly present in silenced states (35, 36). We found that LINE-1 and Alu repeats also have decreased H3 K9me3 level after IR (Figure 3b, 3c), although less significant than alpha satellite repeats presumably due to some presence near active genes. H3 K9me3 level at the GAPDH promoter did not show significant changes (Figure 3d). ChIP analysis of H3 K9me2 level also showed a similar trend as K9me3 (data not shown). Transient knock down of p53 in A549 cells also abrogated the down regulation of H3 K9me3 level at pericentric heterochromatin (Supplemental Figure 1). Therefore, p53 promotes the down-regulation of heterochromatin H3 K9 methylation after DNA damage.
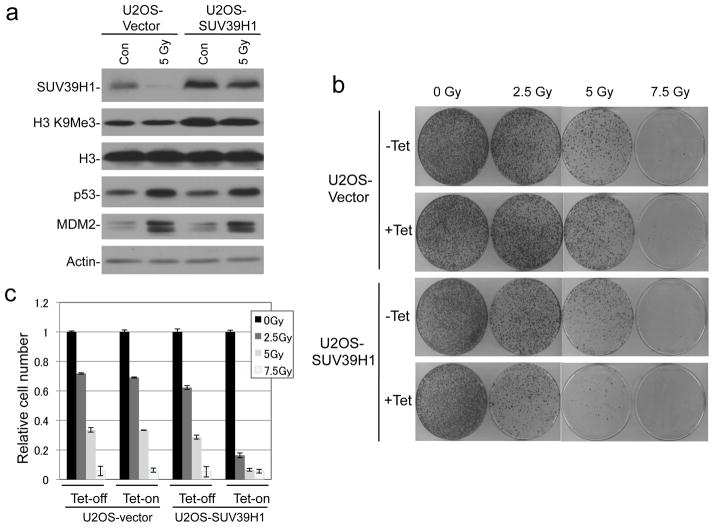
Figure 6. SUV39H1 expression increases radio sensitivity.
(a) U2OS cells with tetracycline-inducible SUV39H1 were induced for 24 hrs and treated with 5 Gy IR. SUV39H1 was detected by western blot. (b) U2OS-Tet-SUV39H1 cells were treated with IR, cultured for 14 days, and colonies were stained. (c) Quantification of stained cells in (b) by dye extraction and OD measurement.
Figure 3. p53 down regulates H3 K9me3 level in heterochromatin after IR.
HCT116 cells with and without p53 were treated with 5 Gy IR and analyzed by H3 K9me3 ChIP and qPCR using primers against pericentric alpha satellite (a), LINE-1 (b), Alu (c), and GAPDH promoter (e).
p53 activates JMJD2b expression and represses SUV39H1
To identify the mechanism of H3 K9me3 down regulation by p53, we tested whether the JMJD2 family of H3 K9 demethylases mediates the effect of p53 (26). We found that JMJD2b was induced by p53 at both protein and mRNA levels after IR and Nutlin treatments (Figure 1a, Figure 4a, 4b). In contrast, JMJD2a mRNA was not significantly induced by p53 (Figure 4c). A recent report showed that the p53-inducible E3 ligase MDM2 causes SUV39H1 degradation (33). Therefore, we tested whether p53 down regulates SUV39H1 through MDM2 induction. When U2OS and A549 cells were treated with IR, Nutlin or low dose actinomycin D, significant down regulation of SUV39H1 was observed (Figure 4d), correlating with p53 activation. In contrast, SUV39H1 level remained unchanged in p53-deficient H1299 and U2OS-E6 cells (Figure 4d, lower panels). Therefore, p53 activation regulates multiple enzymes that control H3 K9 methylation.
Figure 4. p53 regulates JMJD2b and SUV39H1 expression.
(a) p53 activation induces JMJD2b expression at the protein level. (b, c) p53 activation induces the mRNA level of JMJD2b but not JMJD2a as determined by qRT-PCR 24 hrs after treatments. (d) p53 activation down regulates SUV39H1. Cells were treated with 10 Gy IR, 8 μM Nutlin, 5 nM Act.D for 16 hrs and analyzed by western blot. (e) p53 represses SUV39H1 mRNA level as determined by qRT-PCR.
If SUV39H1 down regulation was due to degradation by MDM2, treatment with proteasome inhibitor should restore SUV39H1 level. However, we did not observe such rescue using MG132 (data not shown). Furthermore, we tested directly whether MDM2 overexpression can promote ubiquitination and degradation of SUV39H1 in cotransfection and failed to detect a reproducible effect (data not shown). RT-PCR analysis showed that SUV39H1 mRNA level was significantly reduced after activation of p53 (Figure 4e). The turnover rate of SUV39H1 mRNA after transcription block by high-dose actinomycin D was not affected by p53 activation (Supplemental Figure 2). Therefore, the loss of SUV39H1 was due to reduced transcription.
JMJD2b is a direct transcriptional target of p53
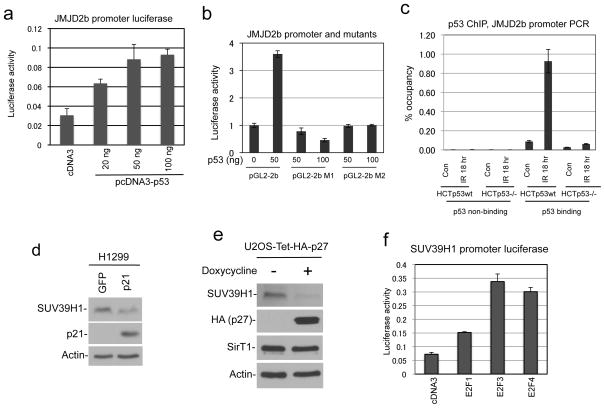
Since p53 induces JMJD2b expression at the mRNA level, we tested whether JMJD2b is a direct target of p53. Sequence analysis of the JMJD2b promoter revealed a putative p53 binding site located at −450 bp from the transcription start site, which contains two half sites without spacer [GCACCTGCCC CAGCCTGTCC. Underlined residues conform to p53 binding half site consensus: 5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′] (37). Therefore, we cloned a 1 kb fragment of the JMJD2b promoter and tested its response to p53 in luciferase reporter assay. The JMJD2b promoter was activated by p53 (~3 fold) in cotransfection assay as expected (Figure 5a). Furthermore, point mutations in the putative p53 binding site abrogated responsiveness to p53 (Figure 5b). P53 ChIP of HCT116 and HCT116-p53−/− cells confirmed that endogenous p53 binds to the JMJD2b promoter in vivo at −0.5 kb but not to a region at −1.8 kb from the transcription start site (Figure 5c), suggesting that JMJD2b is a bona fide target gene of p53.
Figure 5. p53 regulation of JMJD2b and SUV39H1 transcription.
(a) Luciferase reporter driven by a 1 kb fragment of JMJD2b promoter was cotransfected with p53 in U2OS cells. Promoter activation by p53 was determined by luciferase assay and normalized to cotransfected CMV promoter-driven beta galactosidase. (b) Wild type JMJD2b promoter and two mutants constructs (M1, M2) with base substitutions in the putative p53 bind site were tested for response to p53 in luciferase assay. (c) HCT116 cells with and without p53 were treated with 5 Gy IR and analyzed by p53 ChIP. The p53 ChIP samples were amplified using primers against a region 1.3 kb upstream of the putative p53 binding site, or flanking the putative p53 binding site. (d) H1299 cells were transfected with p21 for 48 hrs. SUV39H1 was detected by western blot. (e) U2OS with tetracycline-inducible p27 was induced for 24 hrs. SUV39H1 was detected by western blot. (f) Luciferase reporter driven by a 1 kb SUV39H1 promoter fragment was cotransfected with E2F expression plasmids into H1299 cells. Promoter activation by E2Fs was determined by luciferase assay.
P53 down regulates the transcription of many genes indirectly through inducing p21, which in turn activates the retinoblastoma protein and inhibits E2F transcription factors (38). When p21 was transiently transfected into H1299 cells, SUV39H1 expression was reduced (Figure 5d). Furthermore, when p27 was overexpressed in U2OS using tetracycline-inducible system, the cells undergoing G1 arrest also showed significant reduction of SUV39H1 (Figure 5e). We cloned the SUV39H1 promoter and found that it was activated by E2Fs in luciferase reporter assay (Figure 5f). These results suggest that p53 down regulates SUV39H1 through inducing p21 and repressing E2F activity.
Constitutive SUV39H1 expression delays DNA repair and reduces survival
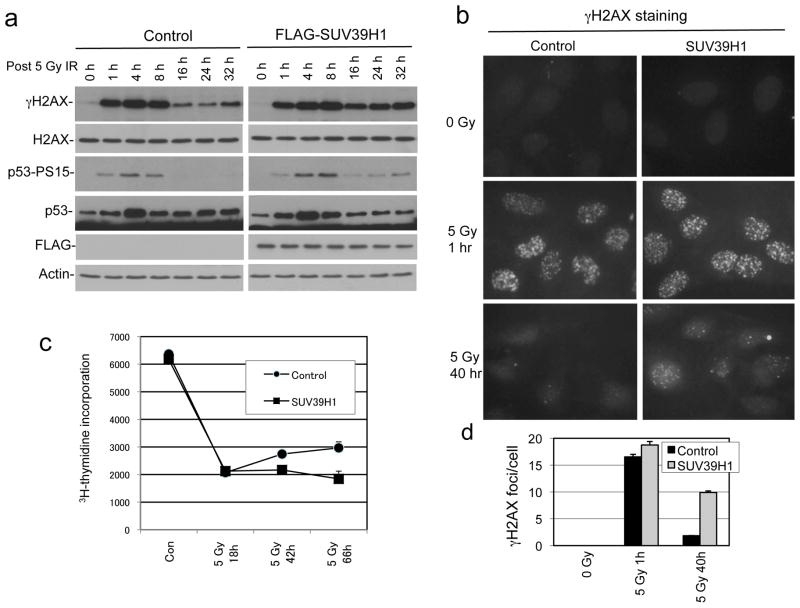
To test whether SUV39H1 down regulation by p53 is important for DNA repair and cell survival, we generated U2OS cells expressing Tet-inducible SUV39H1. Tetracycline treatment maintained total SUV39H1 at close to unstressed level even after DNA damage and p53 activation (Figure 6a). Sustained SUV39H1 expression led to reduction of colony formation efficiency after IR (Figure 6b, 6c), whereas the control U2OS-TR cells were not affected by tetracycline. Therefore, SUV39H1 down regulation after DNA damage facilitates long-term survival.
Western blot analysis showed that IR induced rapid γH2AX and p53 S15 phosphorylation irrespective of SUV39H1 induction. However, the down regulation of γH2AX and p53 phosphorylation at 16–32 hour time points after IR were significantly delayed in SUV39H1-expressing cells, indicating a repair deficiency and sustained ATM activation (Figure 7a). Immunofluoresence staining of γH2AX showed that SUV39H1 expression led to the presence of a significant number of γH2AX foci 40 hours after IR (Figure 7b, 7d). Therefore, SUV39H1 down regulation is important for the completion of DNA repair.
Figure 7. Sustained SUV39H1 expression inhibits DNA repair.
(a) U2OS-Tet-SUV39H1 cells were induced with tetracycline for 24 hrs and treated with IR. γH2AX level was determined by western blot at indicated time points after IR. (b) U2OS-Tet-SUV39H1 cells were induced with tetracycline and treated with IR. Cells were stained for γH2AX foci at indicated time points. (c) U2OS-Tet-SUV39H1 cells were induced with tetracycline and treated with IR. Cell proliferation was analyzed by 3H-thymidine incorporation. (d) Quantitation of γH2AX foci density in the experiment described in (b).
Sustained p53 phosphorylation in the presence of constitutive SUV39H1 is expected to prevent cell cycle re-entry after DNA damage, and possibly induce cellular senescence. SA-β-gal staining showed that SUV39H1 expression did not increase the number of cells displaying senescence phenotype before and after irradiation (Supplemental Figure 3). Following IR treatment, U2OS cells showed reduction of 3H-thymidine incorporation consistent with cell cycle arrest. At 48 and 66 hours after IR, control U2OS cells began to restore DNA synthesis, but SUV39H1 expressing cells remained arrested (Figure 7c). These results suggest that constitutive SUV39H1 expression prevents timely repair of DSB and causes sustained cell cycle arrest.
Constitutive SUV39H1 inhibits the repair of heterochromatin DNA
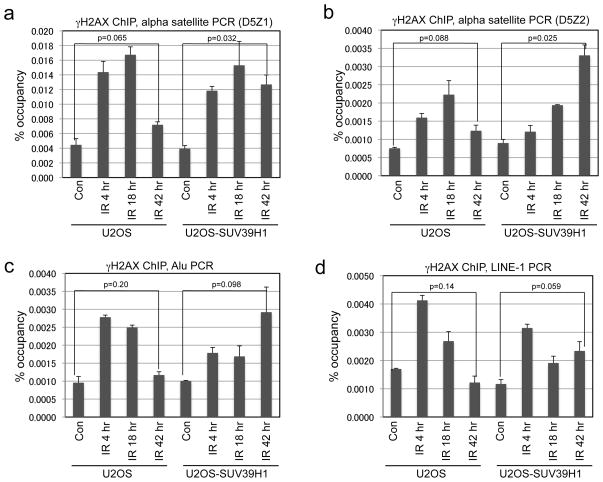
DSBs repaired during the slow phase are mostly localized in constitutive heterochromatin (15). SUV39H1 is involved in H3 K9 tri-methylation at pericentric heterochromatin (22). Therefore, SUV39H1 down regulation may be necessary for the efficient repair of heterochromatin DNA. To test this hypothesis, U2OS-tet-SUV39H1 cells were treated with 5 Gy IR and γH2AX ChIP was amplified using D5Z1 and D5Z2 primers (both are specific for Chromosome 5 and 19 alpha satellite) (27, 31). In control cells, the level of γH2AX associated with heterochromatin returned to near-basal level 42 hours after IR, consistent with completion of DSB repair. When SUV39H1 expression was sustained, heterochromatin-associated γH2AX level remained very high 42 hours after IR, suggesting the presence of persistent DNA damage (Figure 8a, 8b). PCR analysis using primers representing LINE-1 and Alu consensus sequence showed that the repair of DSBs at these repetitive elements were also delayed by constitutive SUV39H1 expression (Figure 8c, 8d). Therefore, down regulation of SUV39H1 expression is critical for the repair of heterochromatin DSB after IR.
Figure 8. Sustained SUV39H1 expression inhibits heterochromatin DNA repair.
U2OS-Tet-SUV39H1 cells were induced with tetracycline for 24 hrs and treated with 5 Gy IR. γH2AX ChIP was performed at indicated time points after IR and amplified using PCR primers specific for two variants of pericentric alpha satellites D5Z1 (a) and D5Z2 (b), interspersed repetitive element Alu (c) and LINE-1 (d).
JMJD2b expression facilitates repair of heterochromatin DNA
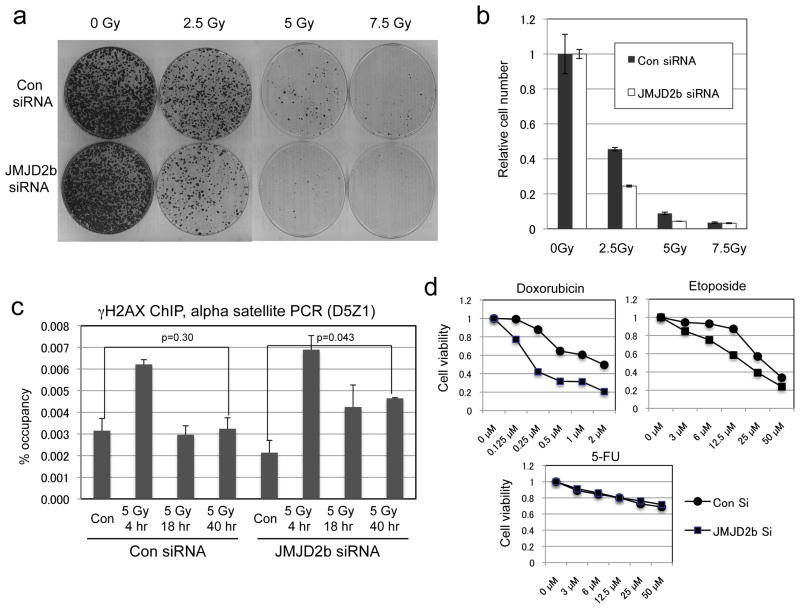
JMJD2b acts opposite to SUV39H1 in regulating H3 K9 methylation. Since its expression was induced by p53, we tested the effects of JMJD2b knockdown after IR treatment. The results showed that knockdown of JMJD2b by siRNA reduced cell survival after IR in colony formation assay (Figure 9a, 9b). Furthermore, γH2AX ChIP using alpha satellite primers showed that JMJD2b knockdown delayed the repair of heterochromatin DNA after IR (Figure 9c). These results are consistent with the interpretation that p53 down regulates SUV39H1 and induces JMJD2b to promote heterochromatin DSB repair and cell survival.
Figure 9. JMJD2b knockdown delays heterochromatin DNA repair.
(a) U2OS cells were transfected with JMJD2b siRNA for 48 hrs, treated with IR, and cultured for 14 days to determine colony formation efficiency. (b) Quantification of stained cells in (a) by dye extraction and OD measurement. (c) U2OS cells treated with JMJD2b siRNA and IR were analyzed by γH2AX ChIP using PCR primers specific for pericentric alpha satellite. (d) U2OS were treated with JMJD2b siRNA for 24 hrs, followed by addition of drugs and analysis of viability by MTT assay after 4 days.
To test whether inhibition of JMJD2b sensitize tumor cells to chemotherapy drugs, U2OS cells were treated with JMJD2b siRNA in combination with several DNA-damaging agents. JMJD2b knockdown by RNAi significantly reduced cell survival after DNA-damaging drug treatment (doxorubicin and etoposide), but had little effect on 5-FU (RNA-targeting drug at 50 μM) (Figure 9d). Therefore, targeting JMJD2b demethylase activity may be useful in combination with DNA-damaging drugs in cancer treatment.
Discussion
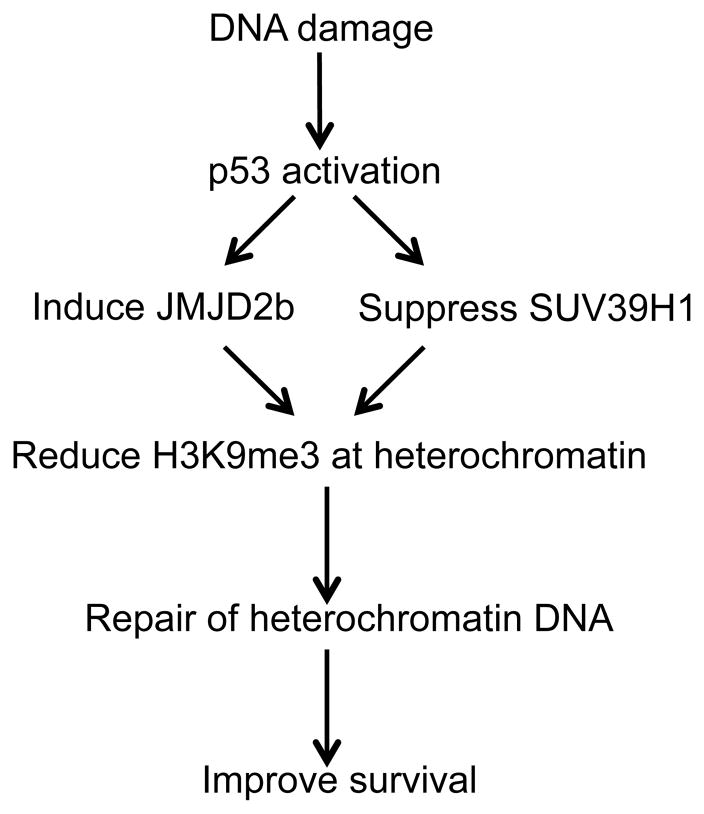
Although p53 mediates acute apoptotic response in rapidly proliferating cells such as thymocytes after irradiation, lose of p53 has been shown to paradoxically reduce long-term viability after IR in certain cell lines. Results described in this report show that p53 deficiency compromises the repair of DNA double strand breaks in constitutive heterochromatin. DNA double strand break triggers chromatin relaxation that involves ATM-dependent phosphorylation of KAP1 (39) (20). P53 has also been shown to facilitate chromatin decompaction after UV irradiation (40, 41), but the mechanism was unknown. Our results in this report suggest that p53 induction of JMJD2b and repression of SUV39H1 causes down regulation of H3 K9 trimethylation level at pericentric heterochromatin and interspersed repetitive elements after irradiation, thus promoting heterochromatin relaxation and increasing accessibility to DNA repair factors (Figure 10).
Figure 10.
A model of p53 promotion of heterochromatin DNA repair by regulating H3 K9 methylation level after damage.
SUV39H1 antagonizes p53 transcriptional function by interacting with MDM2, or directly modifying the chromatin of p53 target genes. Therefore, p53 repression of SUV39H1 should generate a positive feedback. Consistent with this model, results from our lab and a recent study by Mungamuri et al. showed that knockdown of SUV39H1 increase the expression of p53 target genes, whereas overexpression of SUV39H1 inhibits p53 activity (32, 42). Our current findings suggest that in addition to gene regulation, p53-mediated change in H3 K9 methylation has important functions in facilitating timely repair of DNA strand breaks. Proper condensation of pericentric heterochromatin is important for accurate segregation of sister chromosomes during mitosis. Inactivation of p53 causes abnormal regulation of SUV39H1 and JMJD2b expression, which may lead to chromosome instability in cancer. Telomeric repeats are also rich in H3 K9me3 modification (43). Mice deficient for SUV39H1/H2 have abnormal telomere length and H3 K9me3 level at telomeric heterochromatin (24). Although not analyzed in our study, it is likely that p53 also regulates the stability of telomeric heterochromatin. It has been shown that p53-deficient human tumors often have telomere length alterations (44, 45).
Cells with different radiation sensitivity often show similar level of DSB formation and similar ability to rapidly repair >70% of the DSBs a few hours after damage. Previous studies showed that radio sensitivity correlates with the number of DSB remaining after 24 hours (46). Radio-sensitive tumor cell lines and xenografts retain more γH2AX foci 24 hours after IR (47). Therefore, radio resistance correlates with the ability of the cells to down regulate γH2AX level 24 hours after IR. Recent studies indicate that DSBs that require up to 24 hours to repair are mostly localized in heterochromatin (15). IR induces fewer γH2AX foci in heterochromatin in human cells shortly after IR (17), despite similar rate of DSB in heterochromatin and euchromatin (16), suggesting that heterochromatin formation delays γH2AX foci assembly or disappearance. By regulating histone methylation at heterochromatin loci, p53 may help to overcome a rate-limiting step in global DNA repair. The DNA repair function of p53 may be particularly important in higher organisms because of increased complexity of their genomes. Up to 50% of human genomic DNA are composed of repetitive sequences, mostly present as silent chromatin with high levels of DNA and histone methylation (48).
Although p53 activation can cause apoptosis under some conditions, the pro-survival function observed in our experiment is consistent with several previous reports. P53 knockout in HCT116 increased sensitivity to IR and doxorubicin (28). Ectopic expression of a temperature-sensitive p53 in PC3 cells increased radio resistance in long-term survival assay (49). P53 wild type tumor cell lines generally show faster γH2AX down regulation after IR compared to p53 deficient cells (50). Therefore, it is likely that in many cell types, the physiological function of p53 is to promote DNA damage repair and cell survival, which is beneficial to organism fitness. Only severely damaged or rapidly cycling cells commit to apoptosis after p53 activation to reduce the chance of propagating mutations. Although there is no general consensus on the relationship between p53 mutation status and treatment response, in certain narrow setting such as breast cancer, p53 mutation is associated with favorable response to chemotherapy (51–53). A recent report showed that in a mouse model wild type p53 also reduced the efficacy of doxorubicin against mammary tumors (54).
Our results showed that p53-mediated cell cycle arrest indirectly represses SUV39H1 expression. In fact, overexpression of p21 or p27 also repressed SUV39H1. During the preparation of this manuscript, Mungamuri et al reported that p53 represses SUV39H1 transcription in a p21-dependent fashion (42), which is consistent with our finding. Many p53-repressed genes are transcriptional targets of E2F1 and are repressed through p21-mediated regulation of pRb (38). The SUV39H1 promoter is activated by E2Fs in reporter assay, suggesting that p53 suppresses SUV39H1 by inducing p21 and inactivating E2Fs. It has been reported that the Drosophila JMJD2b homolog is induced by the fly p53 after UV irradiation, leading to reduction in heterochromatin K9me3 level that is important for survival (55). Our results show that human JMJD2b is also inducible by p53, suggesting that regulation of heterochromatin H3K9 methylation is an important and evolutionarily conserved function of p53.
Activation of p53 has been shown to induce cellular senescence, accompanied by expression of the senescence marker SA-β-gal. The observation that p53 can induce senescence while repressing SUV39H1 important for heterochromatin formation appears self-contradictory. However, previous study showed that p53-dependent senescence is reversible after p53 inactivation (56). A recent study using Nutlin also showed that p53 only induces a senescence-like phenotype (SA-β-gal positive) in many epithelial tumor cell lines, which is reversible after removal of Nutlin (57). Therefore, the senescence phenotype induced by p53 is different from the pRb-mediated permanent arrest that involves irreversible silencing of E2F targets and formation of senescence-associated heterochromatin foci (SAHF) (58). The requirement of SUV39H1 in senescence is also cell type dependent. SUV39H1 is required for senescence by oncogenic ras in mouse splenocytes (59), but Narita et al. found that oncogenic ras can induce senescence in SUV39H1/2 double-null MEFs (58). Nutlin activation of p53 in epithelial cancer cells does not induce SAHF despite SA-β-gal positive staining (57). Therefore p53-induced senescence does not involve the level of heterochromatin formation that occurs in irreversible senescent cells. It is quite possible that repression of SUV39H1 and induction of JMJD2b by p53 contributes to the reversibility of senescence.
DNA damage induces rapid mobilization of HP1β on a global scale within minutes (60–62). P53-mediated changes of SUV39H1 and JMJD2b levels occur at a slower time scale (hours post damage) and may serve as a second wave response to reinforce the initial changes. P53 has been implicated in regulating multiple DNA repair pathways through regulation of genes involved in the repair or by direct binding to repair factors (9, 10). The results in this study suggest that p53 also facilitates DNA repair by regulating the levels of enzymes involved in H3 K9 methylation and modifying heterochromatin compaction on a global scale. Because perturbation of SUV39H1 and JMJD2b each has significant impact on DSB repair efficiency, these chromatin modifiers are likely to be important mediators of p53 function in maintaining the stability of highly repetitive DNA sequences.
Materials and methods
Cell lines and reagents
H1299 (non-small cell lung carcinoma, p53 null), U2OS (osteosarcoma, p53 wild-type), U2OS-E6 (U2OS stably infected with retrovirus vector expressing HPV E6), A549 (lung adenocarcinoma, p53 wild-type), and HCT116 (colon carcinoma, p53 wild-type) were maintained in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum. Transfection of H1299 cells was performed using standard calcium phosphate precipitation protocol. U2OS cells with stable expression of SUV39H1 were generated by infection with pLenti-FLAG-SUV39H1 virus followed by Zeocin selection (ViraPower T-REX lentiviral expression system, Invitrogen). Tetracycline-inducible U2OS-HA-p27 cells were generated previously (63). HCT116-p53−/− cells were provided by Dr. Bert Vogelstein. Racemic Nutlin-3a was purchased from Cayman Chemical Company (Ann Arbor, Michigan). Actinomycin D, 5-FU, doxorubicin and etoposide were purchased from Sigma. JMJD2b promoter-luciferase mutant constructs with altered p53 binding element were generated by site-directed mutagenesis (Wild type p53 binding site: GCACCTGCCCCAGCCTGTCC. M1: GaattCTGCCCCAGCCTGTCC. M2: GCACCaattCCAGCCTGTCC). SA-β-gal staining was performed using a kit from Sigma.
Western blot
Cells were lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride), centrifuged for 10 minutes at 14,000 × g and the insoluble debris was discarded. Cell lysate (10 to 50 μg of protein) was fractionated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to Immobilion P filters (Millipore). The filter was blocked for 1 h with phosphate-buffered saline (PBS) containing 5% nonfat dry milk and 0.1% Tween 20, incubated with primary and secondary antibodies, and the filter was developed using the Supersignal reagent (Thermo Scientific). MDM2 was detected using antibody 3G9. Anti-actin and HA tag antibodies were purchased from Santa Cruz Biotechnology. FLAG tag antibody was from Sigma-Aldrich. Anti-SUV39H1 antibody was from Millipore. Anti-JMJD2B antibody was from Bethyl. DO-1 for p53 was from Pharmingen and Phospho-p53 (Ser15) antibody was from Cell Signaling. Anti-histone H3 (trimethyl K9) and Anti-histone H3 (dimethyl K9) antibodies were from Abcam. Anti-histone H3 antibody, anti-γH2AX and H2AX antibodies were from Upstate.
Luciferase reporter assay
Cells (50,000 per well) were cultured in 24-well plates and transfected with a mixture containing 50 ng luciferase reporter plasmid, 5 ng CMV–lacZ plasmid, 50 ng E2Fs expression plasmids or p53 expression plasmid. Transfection was achieved using Lipofectamine PLUS reagents (Invitrogen). Cell lysate was analysed for luciferase and β-gal expression after 24 h. The ratio of luciferase/β-gal activity was used as an indicator of transcription activity.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was carried out using a published procedure (64). Protein-DNA complexes from cells lysate were immunoprecipitated with antibody and protein A agarose/salmon sperm DNA beads (Millipore). Coprecipitated chromatin was analyzed by quantitative PCR using the following primers: 5′-ACGAGGTCAGGAGATCGAGA and 5′-CTCAGCCTCCCAAGTAGCTG for Alu1 (65); 5′-CAGAATCTCTGGGACGCATT and 5′-ATTGTGATGTTCGGGTGTCA for LINE-1 (GenBank entry: X52235.1); 5′-TAGACAGAAATATTCTCACAATCGT and 5′-GCCCTCAAAGCGCTCCAAG for pericentric alpha satellite D5Z1; 5′-TTTTTGTGCAATTGGCAAATGGAG and 5′-AGACTGTTTCCTCACTGCTCT for pericentric alpha satellite D5Z2 (27). JMJD2b promoter −1.8 kb control primers: 5′-ACCAGTGTGCAGCTTTCCTTCTCT and 5′-AAATATCTCCCAACCGCTGAGCTAGG. JMJD2b −0.5 kb primers (bracketing the p53 binding site): 5′-GTCTCACGTACTCAGCCG and 5′-CACAGAGAGGAGCCAGTATTG. The readouts were normalized using 10% input chromatin for each sample. The experiments were repeated 3 or more times. The p-values were calculated using two-tailed Student’s t-Test and shown in the figures.
RNA isolation and quantitative PCR
Total RNA was extracted using the RNeasy Mini kit (Qiagen). cDNAs were prepared by reverse transcription of total RNA using the SuperScript™ III First-Strand Synthesis System (Invitrogen). The products were used for real-time PCR using the following primers: 5′-CTCAAGTGTGTGCGTATCCTCAA and 5′-CCGGAGCAGCTCCCTTTC for SUV39H1; 5′-GGAAGCCACGAGCATCCTATG and 5′-GGAACTCTCGAACAGTCATGG for JMJD2a; 5′-GGGGAGGAAGATGTGAGTGA and 5′-GACGGCTTTTGGAGGGTAAT for JMJD2b; 5′-GAGTCAACGGATTTGGTCGT and 5′-GACAAGCTTCCCGTTCTCAG for GAPDH.
RNA interference (RNAi)
Cells were transfected with 100 nM control siRNA or 100 nM Smartpool JMJD2b siRNAs (Dharmacon) using RNAiMAX (Invitrogen) according to instructions from the supplier. After 48 h of transfection, cells were treated with IR and analyzed for protein expression or chromatin immuno precipitation.
Immunofluorescence staining
Cells cultured on chamber slides were fixed with acetone-methanol (1:1) for 5 min at room temperature, blocked with PBS+10% normal goat serum (NGS) for 20 min, and incubated with anti-γH2AX in PBS with 10% NGS for 2 h. The slides were washed with PBS+0.1% Triton X-100, incubated with FITC-goat-anti-mouse IgG in PBS+10% NGS for 2 h, washed with PBS+0.1% Triton X-100 and mounted. The number of γH2AX foci per nucleus was determined using the Image Pro Plus (Media Cybernetics; Bethesda, MD) v6.2 software, ~20 cells were analyzed for each condition.
Cell death and colony formation assays
Cell death was measured by MTS assay using the Cell Titer kit (Promega). Five thousand cells were cultured in 24-well plates and treated with compounds for 72 h. Culture medium was replaced with fresh medium containing the MTS reagent and the cells were cultured for 15–30 min. Conversion of MTS reagent into color-absorbing product by metabolically active cells were measured by determining OD at 490 nm. For colony formation assay, five thousand cells were seeded in a 10-cm plate in triplicate, incubated for 8 h to allow attachment, treated with IR, and cultured for 7–10 days. After incubation, cells were washed with PBS, stained with 0.5% crystal violet for 1 min at 23°C, washed with PBS, and photographed for colony density. To quantify surviving cells, the crystal violet in stained colonies were extracted by incubation with 10% acetic acid for 20 min, absorbance at 590 nm was measured.
3H-Thymidine incorporation assay
Cells cultured in 24 well plates in triplicate were incubated with 5 μCi 3H-thymidine/well for 1 h and washed with PBS. One ml of ice cold 5% trichloroacetic acid (TCA) was added and incubated for 30 min at 4°C. The cells were washed with PBS and lysed in 1 ml of 0.5 N NaOH, 0.5% SDS for 30 min at 37 °C. The lysate (1 ml) was mixed with 4 ml of liquid scintillation cocktail (Beckman), and radioactivity was measured by liquid scintillation counting.
Supplementary Material
Acknowledgments
We would like to thank the Moffitt Molecular Genomics Core for DNA sequence analyses, and Analytic Microscopy Core for image quantification. H. Zheng is supported by a postdoctoral fellowship from the Florida Department of Health James and Esther King Biomedical Research Program. This work is supported by grants from the National Institutes of Health to J. Chen (CA141244, CA109636).
Footnotes
Author contribution
Hong Zheng and Lihong Chen designed and performed the experiments. W. Jack Pledger and Jia Fang provided important intellectual input and reagents. Hong Zheng and Jiandong Chen designed the experiments and wrote the manuscript.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Kastan MB, Lim DS. The many substrates and functions of ATM. Nat Rev Mol Cell Biol. 2000 Dec;1(3):179–86. doi: 10.1038/35043058. [DOI] [PubMed] [Google Scholar]
- 2.Kemp CJ, Wheldon T, Balmain A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat Genet. 1994 Sep;8(1):66–9. doi: 10.1038/ng0994-66. [DOI] [PubMed] [Google Scholar]
- 3.Donehower LA, Godley LA, Aldaz CM, Pyle R, Shi YP, Pinkel D, et al. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes & development. 1995 Apr 1;9(7):882–95. doi: 10.1101/gad.9.7.882. [DOI] [PubMed] [Google Scholar]
- 4.Harvey M, Sands AT, Weiss RS, Hegi ME, Wiseman RW, Pantazis P, et al. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993 Sep;8(9):2457–67. [PubMed] [Google Scholar]
- 5.Bischoff FZ, Yim SO, Pathak S, Grant G, Siciliano MJ, Giovanella BC, et al. Spontaneous abnormalities in normal fibroblasts from patients with Li-Fraumeni cancer syndrome: aneuploidy and immortalization. Cancer research. 1990 Dec 15;50(24):7979–84. [PubMed] [Google Scholar]
- 6.Bouffler SD, Kemp CJ, Balmain A, Cox R. Spontaneous and ionizing radiation-induced chromosomal abnormalities in p53-deficient mice. Cancer Res. 1995 Sep 1;55(17):3883–9. [PubMed] [Google Scholar]
- 7.Bishop AJ, Hollander MC, Kosaras B, Sidman RL, Fornace AJ, Jr, Schiestl RH. Atm-, p53-, and Gadd45a-deficient mice show an increased frequency of homologous recombination at different stages during development. Cancer research. 2003 Sep 1;63(17):5335–43. [PubMed] [Google Scholar]
- 8.Mekeel KL, Tang W, Kachnic LA, Luo CM, DeFrank JS, Powell SN. Inactivation of p53 results in high rates of homologous recombination. Oncogene. 1997 Apr 17;14(15):1847–57. doi: 10.1038/sj.onc.1201143. [DOI] [PubMed] [Google Scholar]
- 9.Helton ES, Chen X. p53 modulation of the DNA damage response. Journal of cellular biochemistry. 2007 Mar 1;100(4):883–96. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- 10.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nature reviews Molecular cell biology. 2005 Jan;6(1):44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 11.Yan W, Chen X. Characterization of functional domains necessary for mutant p53 gain of function. The Journal of biological chemistry. 2010 May 7;285(19):14229–38. doi: 10.1074/jbc.M109.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuddihy AR, Bristow RG. The p53 protein family and radiation sensitivity: Yes or no? Cancer metastasis reviews. 2004 Aug-Dec;23(3–4):237–57. doi: 10.1023/B:CANC.0000031764.81141.e4. [DOI] [PubMed] [Google Scholar]
- 13.Gudkov AV, Komarova EA. The role of p53 in determining sensitivity to radiotherapy. Nat Rev Cancer. 2003 Feb;3(2):117–29. doi: 10.1038/nrc992. [DOI] [PubMed] [Google Scholar]
- 14.Kuhne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Lobrich M. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer research. 2004 Jan 15;64(2):500–8. doi: 10.1158/0008-5472.can-03-2384. [DOI] [PubMed] [Google Scholar]
- 15.Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA repair. 2010 Dec 10;9(12):1273–82. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Puerto S, Ramirez MJ, Marcos R, Creus A, Surralles J. Radiation-induced chromosome aberrations in human euchromatic (17cen-p53) and heterochromatic (1cen-1q12) regions. Mutagenesis. 2001 Jul;16(4):291–6. doi: 10.1093/mutage/16.4.291. [DOI] [PubMed] [Google Scholar]
- 17.Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PloS one. 2007;2(10):e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, et al. Global chromatin compaction limits the strength of the DNA damage response. The Journal of cell biology. 2007 Sep 24;178(7):1101–8. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Molecular cell. 2008 Jul 25;31(2):167–77. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 20.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nature cell biology. 2006 Aug;8(8):870–6. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 21.Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011 Mar 4;144(5):732–44. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001 Nov 2;107(3):323–37. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 23.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. The EMBO journal. 2005 Feb 23;24(4):800–12. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nature genetics. 2004 Jan;36(1):94–9. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 25.Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS genetics. 2009 Mar;5(3):e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fodor BD, Kubicek S, Yonezawa M, O’Sullivan RJ, Sengupta R, Perez-Burgos L, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes & development. 2006 Jun 15;20(12):1557–62. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slee RB, Steiner CM, Herbert BS, Vance GH, Hickey RJ, Schwarz T, et al. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene. 2011 Nov 14; doi: 10.1038/onc.2011.502. [DOI] [PubMed] [Google Scholar]
- 28.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999 Aug;104(3):263–9. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. The EMBO journal. 2009 Dec 16;28(24):3857–67. doi: 10.1038/emboj.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nature reviews Cancer. 2008 Dec;8(12):957–67. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finelli P, Antonacci R, Marzella R, Lonoce A, Archidiacono N, Rocchi M. Structural organization of multiple alphoid subsets coexisting on human chromosomes 1, 4, 5, 7, 9, 15, 18, and 19. Genomics. 1996 Dec 15;38(3):325–30. doi: 10.1006/geno.1996.0635. [DOI] [PubMed] [Google Scholar]
- 32.Chen L, Li Z, Zwolinska AK, Smith MA, Cross B, Koomen J, et al. MDM2 recruitment of lysine methyltransferases regulates p53 transcriptional output. The EMBO journal. 2010 Aug 4;29(15):2538–52. doi: 10.1038/emboj.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosch-Presegue L, Raurell-Vila H, Marazuela-Duque A, Kane-Goldsmith N, Valle A, Oliver J, et al. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Molecular cell. 2011 Apr 22;42(2):210–23. doi: 10.1016/j.molcel.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 34.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. The EMBO journal. 2009 Jul 8;28(13):1878–89. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008 Oct 3;135(1):23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 36.Kondo Y, Issa JP. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. The Journal of biological chemistry. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] 2003 Jul 25;278(30):27658–62. doi: 10.1074/jbc.M304072200. [DOI] [PubMed] [Google Scholar]
- 37.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nature genetics. 1992 Apr;1(1):45–9. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 38.Lohr K, Moritz C, Contente A, Dobbelstein M. p21/CDKN1A mediates negative regulation of transcription by p53. The Journal of biological chemistry. 2003 Aug 29;278(35):32507–16. doi: 10.1074/jbc.M212517200. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi K, Kaneko I. Changes in nuclease sensitivity of mammalian cells after irradiation with 60Co gamma-rays. International journal of radiation biology and related studies in physics, chemistry, and medicine. 1985 Sep;48(3):389–95. doi: 10.1080/09553008514551391. [DOI] [PubMed] [Google Scholar]
- 40.Carrier F, Georgel PT, Pourquier P, Blake M, Kontny HU, Antinore MJ, et al. Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Molecular and cellular biology. 1999 Mar;19(3):1673–85. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubbi CP, Milner J. p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. The EMBO journal. 2003 Feb 17;22(4):975–86. doi: 10.1093/emboj/cdg082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mungamuri SK, Benson EK, Wang S, Gu W, Lee SW, Aaronson SA. p53-mediated heterochromatin reorganization regulates its cell fate decisions. Nature structural & molecular biology. 2012 Apr 1; doi: 10.1038/nsmb.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007 Aug 2;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohki S, Onda M, Fukushima T, Takita K, Suzuki S, Hoshino M, et al. Telomeric repeat-length alterations in colorectal carcinoma are associated with loss of heterozygosity and point mutation in p53 gene. International journal of molecular medicine. 2003 Apr;11(4):485–90. [PubMed] [Google Scholar]
- 45.Hiyama K, Ishioka S, Shirotani Y, Inai K, Hiyama E, Murakami I, et al. Alterations in telomeric repeat length in lung cancer are associated with loss of heterozygosity in p53 and Rb. Oncogene. 1995 Mar 2;10(5):937–44. [PubMed] [Google Scholar]
- 46.Dikomey E, Dahm-Daphi J, Brammer I, Martensen R, Kaina B. Correlation between cellular radiosensitivity and non-repaired double-strand breaks studied in nine mammalian cell lines. International journal of radiation biology. 1998 Mar;73(3):269–78. doi: 10.1080/095530098142365. [DOI] [PubMed] [Google Scholar]
- 47.Taneja N, Davis M, Choy JS, Beckett MA, Singh R, Kron SJ, et al. Histone H2AX phosphorylation as a predictor of radiosensitivity and target for radiotherapy. The Journal of biological chemistry. 2004 Jan 16;279(3):2273–80. doi: 10.1074/jbc.M310030200. [DOI] [PubMed] [Google Scholar]
- 48.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001 Feb 15;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 49.Scott SL, Earle JD, Gumerlock PH. Functional p53 increases prostate cancer cell survival after exposure to fractionated doses of ionizing radiation. Cancer research. 2003 Nov 1;63(21):7190–6. [PubMed] [Google Scholar]
- 50.Banath JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer research. 2004 Oct 1;64(19):7144–9. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- 51.Bertheau P, Turpin E, Rickman DS, Espie M, de Reynies A, Feugeas JP, et al. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS medicine. 2007 Mar;4(3):e90. doi: 10.1371/journal.pmed.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertheau P, Plassa F, Espie M, Turpin E, de Roquancourt A, Marty M, et al. Effect of mutated TP53 on response of advanced breast cancers to high-dose chemotherapy. Lancet. 2002 Sep 14;360(9336):852–4. doi: 10.1016/S0140-6736(02)09969-5. [DOI] [PubMed] [Google Scholar]
- 53.Kroger N, Milde-Langosch K, Riethdorf S, Schmoor C, Schumacher M, Zander AR, et al. Prognostic and predictive effects of immunohistochemical factors in high-risk primary breast cancer patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006 Jan 1;12(1):159–68. doi: 10.1158/1078-0432.CCR-05-1340. [DOI] [PubMed] [Google Scholar]
- 54.Jackson JG, Pant V, Li Q, Chang LL, Quintas-Cardama A, Garza D, et al. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer cell. 2012 Jun 12;21(6):793–806. doi: 10.1016/j.ccr.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palomera-Sanchez Z, Bucio-Mendez A, Valadez-Graham V, Reynaud E, Zurita M. Drosophila p53 is required to increase the levels of the dKDM4B demethylase after UV-induced DNA damage to demethylate histone H3 lysine 9. The Journal of biological chemistry. 2010 Oct 8;285(41):31370–9. doi: 10.1074/jbc.M110.128462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. The EMBO journal. 2003 Aug 15;22(16):4212–22. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang B, Deo D, Xia M, Vassilev LT. Pharmacologic p53 activation blocks cell cycle progression but fails to induce senescence in epithelial cancer cells. Molecular cancer research: MCR. 2009 Sep;7(9):1497–509. doi: 10.1158/1541-7786.MCR-09-0144. [DOI] [PubMed] [Google Scholar]
- 58.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003 Jun 13;113(6):703–16. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 59.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005 Aug 4;436(7051):660–5. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 60.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008 May 29;453(7195):682–6. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 61.Ayoub N, Jeyasekharan AD, Venkitaraman AR. Mobilization and recruitment of HP1: a bimodal response to DNA breakage. Cell cycle. 2009 Sep 15;8(18):2945–50. [PubMed] [Google Scholar]
- 62.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. The Journal of cell biology. 2009 May 18;185(4):577–86. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma SS, Ma L, Bagui TK, Forinash KD, Pledger WJ. A p27(Kip1) mutant that does not inhibit CDK activity promotes centrosome amplification and micronucleation. Oncogene. 2011 Dec 12;31(35):3989–98. doi: 10.1038/onc.2011.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ. c-Myc target gene specificity is determined by a post-DNAbinding mechanism. Proc Natl Acad Sci U S A. 1998 Nov 10;95(23):13887–92. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Batzer MA, Alegria-Hartman M, Deininger PL. A consensus Alu repeat probe for physical mapping. Genetic analysis, techniques and applications. 1994;11(2):34–8. doi: 10.1016/1050-3862(94)90058-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.