Abstract
The incidence of human papillomavirus (HPV)-positive head and neck squamous cell carcinoma (HNSCC) has rapidly increased over the past 30 years prompting the suggestion that an epidemic may be on the horizon. Therefore, there is a clinical need to develop alternate therapeutic strategies to manage the growing number of HPV-positive HNSCC patients. High-risk HPV E6 inactivates p53 through two distinct mechanisms; association with E6AP to degrade p53 and association with p300 to block p300-mediated p53 acetylation and activation. In this study, we determined if targeting the E6-p300 interaction is an effective approach to reactivate p53 in HPV-positive HNSCC. Ectopic expression of the CH1 domain of p300 in HPV-positive HNSCC blocks the association between E6 and p300, increases total and acetylated p53 levels, and enhances p53 transcriptional activity. Moreover, expression of p21, miR-34a, and miR-200c are increased demonstrating functional p53 reactivation. CH1 overexpression in HPV-positive HNSCC has a global anti-cancer effect resulting in a decrease in cell proliferation and clonogenic survival and an increase in apoptosis. The in vivo tumor initiating ability of HPV-positive HNSCC is severely compromised with CH1 overexpression, in part through a reduction in the cancer initiating cell population. A novel small molecule CH1 inhibitor, CH1iB, reactivates p53 and potentiates the anti-cancer activity of cis-platinum in HPV-positive HNSCC cells. Our work shows that CH1 domain inhibitors represent a novel class of p53 reactivation therapeutics for managing HPV-positive HNSCC patients.
Keywords: human papillomavirus, anti-cancer therapeutics, p53, p300, head and neck cancer
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer with approximately 600,000 new cases worldwide (1). Human papillomavirus (HPV) infection is recognized as a major risk factor for the development of a subset of HNSCC, oropharyngeal SCC. HPV16 is the most prevalent subtype and accounts for ~90% of HPV-positive HNSCC (2, 3). Epidemiological data indicate that the prevalence of HPV-positive HNSCC has increased by ~3-fold in the past three decades in the United States and Europe (4–6). Data obtained from the Swedish Cancer Registry showed a 2.8-fold increase in the incidence of oropharyngeal SCC in the Stockholm area between 1970 and 2002. Interestingly, over the same time period, the incidence of HPV-positive oropharyngeal SCC increased by ~3-fold from 23% in the 1970s to 68% in the 2000s (7). Based on these alarming numbers, it has been suggested that an epidemic of HPV-positive HNSCC will emerge in the near future (6, 7).
There is concrete clinical data that the HPV vaccine, Gardasil, protects against HPV-positive cervical, vaginal, and vulvar carcinomas (8). It is assumed that the HPV vaccine will protect against HPV-positive HNSCC; however, there is no clinical evidence to support this expectation. The HPV vaccine uptake in females has been modest even though the Centers for Disease Control and Prevention issued a recommendation to vaccinate females, between the ages of 9 to 26, for high-risk HPV in 2006. A study using the 2010 National Health Interview Survey showed that only about 30% and 15% of eligible females received one dose and the full three-dose series of the HPV vaccine, respectively (9). Gardasil was approved for males, 9 to 26 years old, in 2009; however, vaccine uptake was reported to be extremely poor at 2% (10). It is clear that a significant number of age eligible females and males are not vaccinated and may remain unprotected against HPV-positive carcinomas, including HNSCC, over their lifetime.
Gardasil was shown to be highly effective to protect against cervical carcinoma for HPV-infection naïve individuals but provided much more limited benefit to individuals already exposed to high-risk HPV, including HPV16 (11, 12). HPV vaccination is not recommended for adults >26 years old since these individuals are likely to be exposed to high-risk HPV already. Therefore, several generations of individuals already exposed to high-risk HPV or are >26 years old will not be vaccinated routinely or even if vaccinated will have minimal protection against HPV-positive carcinomas, including HNSCC. In light of these points, there is a clinical need to develop alternative therapeutic strategies to manage an anticipated growing number of HPV-positive HNSCC patients.
In contrast to HPV-negative HNSCC, p53 is predominantly wildtype in HPV-positive HNSCC (13–15). However, high-risk HPV E6 inactivates p53 through two distinct mechanisms. E6 associates with E6AP to degrade p53 through the proteasome pathway and associates with p300 to block p300-mediated p53 acetylation (16–21). Acetylation of p53 enhances p53 stability, and transcriptional activity (19–23). Inactivation of p53 by E6 is indispensible for HPV-mediated tumorigenesis suggesting that reactivation of p53 may be a strategy to ablate HPV-positive carcinoma cells. Several genetic and chemical strategies to reactivate p53 have been demonstrated in HPV-positive cervical carcinomas. Most of these approaches focused on targeting E6 levels, E6AP levels, or E6-E6AP association to increase p53 stability and accumulation (24–29). In this study, we took a novel approach and functionally reactivated p53 in HPV-positive HNSCC by blocking the interaction between E6 and p300. Ectopic expression of the CH1 domain of p300 squelched E6 to disrupt E6-p300 association resulting in an increase in p53 acetylation, accumulation, and activity. Exogenous CH1 promoted a pleiotropic, anti-cancer effect in HPV-positive HNSCC partly due to a reduction in the cancer initiating cell (CIC) population. A small molecule CH1 domain inhibitor, CH1iB, reactivated p53 and dramatically potentiated the efficacy of cis-platinum in HPV-positive HNSCC. Taken together, our work revealed a novel druggable approach to reactivate p53 in HPV-positive HNSCC that may translate to other HPV-positive carcinomas.
Results
Exogenous CH1 reactivates p53 by blocking the association between HPV16 E6 and p300
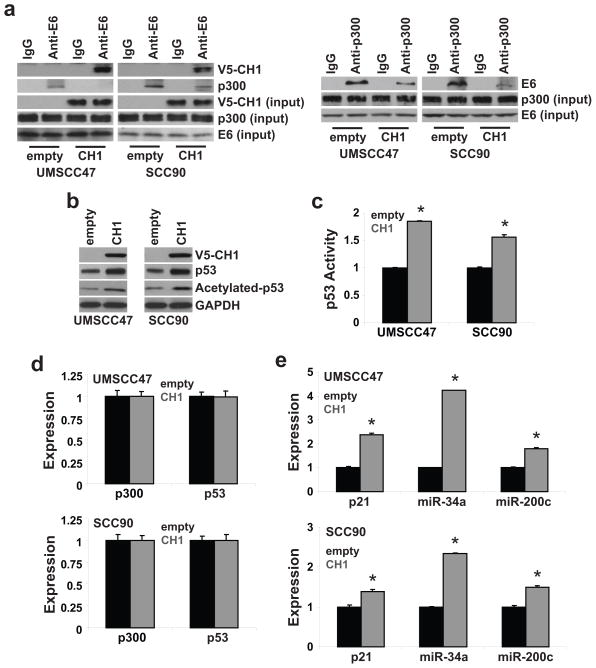
High-risk HPV E6 was reported to associate with p300 to inhibit p300-mediated p53 acetylation (19–21). Acetylation is a critical regulatory mechanism to control p53 stability and transcriptional activity (19–23). Therefore, targeting the E6-p300 interaction may be a novel approach to reactivate p53 in HPV-positive HNSCC. Published work showed that high-risk HPV E6 binds to the CH1, CH3, and C-terminal domain of p300 (20). Thus, we determined if targeting one of the contact sites, the CH1 domain, is a tractable approach to block the association between E6 and p300 and reactivate p53. As shown in Figure 1a, exogenous CH1 squelched E6 to reduce the association between E6 and p300 in UMSCC47 and UPCI:SCC090, two HPV16-positive HNSCC cell lines. An accumulation of total p53 and an increase in acetylated p53 was revealed in CH1-overexpressing UMSCC47 (UMSCC47/CH1) and UPCI:SCC090 (SCC090/CH1) cells (Figure 1b). p53 transcription activity was elevated by 85% (P<0.01) and 50% (P<0.01) in UMSCC47/CH1 and UPCI:SCC090/CH1 cells, respectively (Figure 1c). Overexpression of CH1 had no effect on p53 and p300 expression but enhanced the expression of three well-recognized p53 targets. p21, miR-34a, and miR-200c expression were increased by 114%, 323%, and 80% in UMSCC47/CH1 cells (P<0.01) and 39%, 134%, and 49% in UPCI:SCC090/CH1 cells (P<0.01), respectively (Figure 1e). These results demonstrate that blocking the E6-p300 interaction is an efficient approach to reactivate p53, through p53 accumulation and acetylation, in HPV-positive HNSCC.
Figure 1. Exogenous CH1 reactivates p53 by blocking the association between HPV16 E6 and p300.
Stable polyclonal UMSCC47/empty, UMSCC47/CH1, SCC90/empty, and SCC90/CH1 cells were generated by transfection and antibiotic selection. a. HPV16 E6-p300 association. Cell lysates were extracted, immunoprecipitated with anti-E6 or anti-p300 antibody, and immunoblotted with anti-V5, anti-E6, or anti-p300 antibody. Cell lysates were immunoblotted with anti-V5, anti-E6, or anti-p300 antibody for input control. b. Total and acetylated p53 levels. Cell lysates were immunoblotted with anti-p53 or anti-acetylated[K382]-p53 antibody. c. p53 transcriptional activity. Cells were co-transfected with a p53 Firefly luciferase reporter plasmid and a control Renilla luciferase plasmid. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency. Data were normalized to empty vector cells and presented as mean ± SEM. *P<0.01, n=4. d. p300 and p53 expression. e. p21, miR-34a, and miR-200c expression. mRNA expression was determined using qPCR with validated TaqMan assays. Data were normalized to empty vector control cells and presented as mean ± SEM. *P<0.01, n=3.
Exogenous CH1 has a pleiotropic anti-tumor effect in HPV16-positive HNSCC
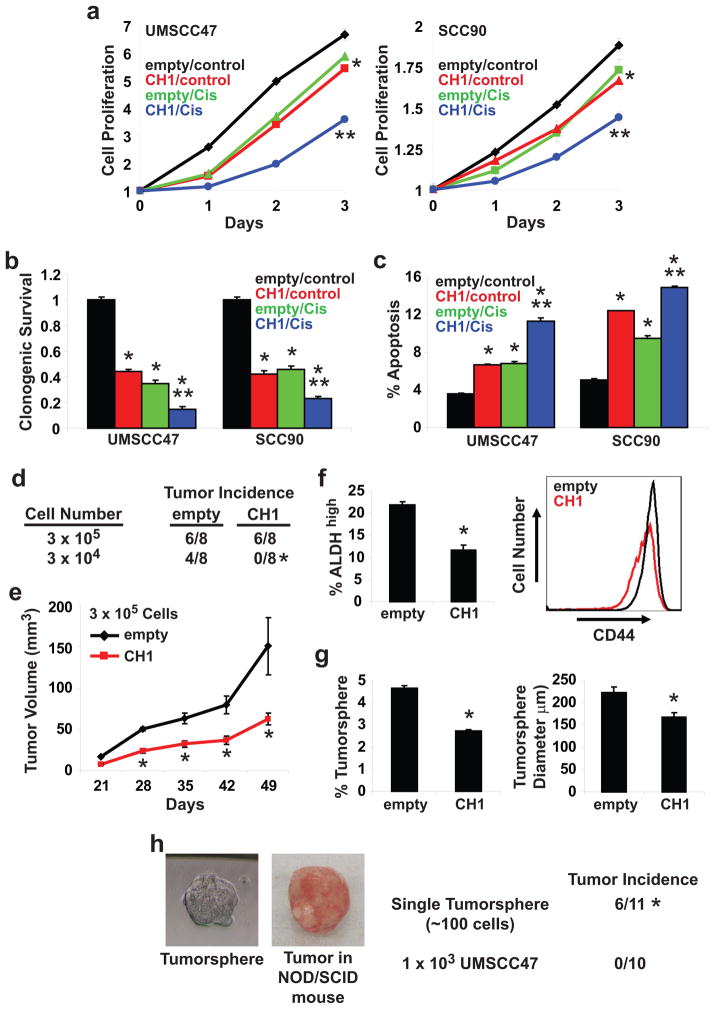
We determined if reactivation of p53 is sufficient to promote an anti-tumor response in HPV-positive HNSCC cells. Cell proliferation was reduced by 20% and 11% in UMSCC47/CH1 and UPCI:SCC090/CH1 cells at 72 hours, respectively (Figure 2a). Overexpression of CH1 in UMSCC47 and UPCI:SCC090 cells decreased clonogenic survival and enhanced apoptosis (Figures 2b and 2c). A similar response in cell proliferation, clonogenic survival, and apoptosis was observed for empty vector-transfected UMSCC47 and UPCI:SCC090 cells treated with single agent cis-platinum (10 μM). UMSCC47/CH1 and UPCI:SCC090/CH1 cells were dramatically more responsive to cis-platinum (10 μM) than UMSCC47/empty and UPCI:SCC090/empty cells. Overexpression of CH1 enhanced the effect of cis-platinum on cell proliferation (72 hours, P<0.01), clonogenic survival (P<0.01) and apoptosis (P<0.01). Our results indicate that reactivation of p53 was sufficient to promote a broad anti-tumor response and furthermore, enhanced the efficacy of cis-platinum in HPV-positive HNSCC cells.
Figure 2. Exogenous CH1 has a pleiotropic anti-tumor effect in HPV16-positive HNSCC cells.
a. Cell proliferation. Cells were treated with control (vehicle) or cis-platinum (10 μM) for 24, 48, and 72 hours. Data were normalized to Day 0 and presented as mean ± SEM. *P<0.01, control vs. CH1, cis-platinum, or CH1 + cis-platinum, **P<0.01, CH1 or cis-platinum vs. CH1 + cis-platinum, n=6. b. Clonogenic survival. Cells were treated with control (vehicle) or cis-platinum (10 μM). Colonies were stained with crystal violet. Data were normalized to empty/control cells and presented as mean ± SEM. *P<0.01, control vs. CH1, cis-platinum, or CH1 + cis-platinum, **P<0.01, CH1 or cis-platinum vs. CH1 + cis-platinum, n=3. c. Apoptosis. Cells were treated with control (vehicle) or cis-platinum (10 μM) for 24 hours. FACS was used to quantitate Annexin V-positive apoptotic cells. Data is presented as mean ± SEM. *P<0.01, control vs. CH1, cis-platinum, or CH1 + cis-platinum, **P<0.01, CH1 or cis-platinum vs. CH1 + cis-platinum, n=3. d. In vivo tumor incidence. Two different dilutions, 3×105 or 3×104, of UMSCC47/empty and UMSCC47/CH1 cells were implanted in the flanks of NOD/SCID mice. Tumor incidence was monitored for 49 days following tumor cell implantation. *P<0.02, n=8. e. In vivo tumor growth. Tumors were measured weekly using a digital caliper and tumor volumes were calculated. Data is presented as mean ± SEM. *P<0.01, n=6. f. ALDH and CD44. ALDHhigh cells were quantitated using the ALDEFLUOR assay. Data is presented as mean ± SEM. *P<0.01, n=3. CD44 intensity was determined using FACS with an anti-PE-CD44 antibody and presented as a histogram. g. Tumorsphere formation efficiency and diameter. Tumorsphere formation efficiency was calculated as the number of tumorspheres (≥ 50 μm in diameter) formed divided by the original number of cells seeded. Tumorsphere diameter was measured using NIS-Elements software. Data is presented as mean ± SEM. *P<0.01, n=6. h. In vivo tumor incidence of a single tumorsphere. NOD/SCID mice were implanted with a single UMSCC47 tumorsphere (mean diameter of 60–80 μm with ~100 cells) or 1×103 UMSCC47 cells. A representative UMSCC47 tumorsphere and the resulting tumor grown in NOD/SCID mice are shown. Tumor incidence was monitored over a 6 month period. *P<0.005, n=11 for single UMSCC47 tumorsphere and n=10 for 1×103 UMSCC47 cells.
Next, we determined if CH1 overexpression modulates the in vivo tumorigenicity of HPV-positive HNSCC cells. Two different dilutions, 3×105 or 3×104, of UMSCC47/empty and UMSCC47/CH1 cells were implanted in the flanks of athymic nude mice (Figure 2d). At a dilution of 3×105 cells, tumor incidence was the same between UMSCC47/empty and UMSCC47/CH1 cells however a difference (P<0.01, n=6) in tumor volume was observed. Mean tumor volume was 142 mm3 for UMSCC47/empty and 67 mm3 for UMSCC47/CH1 (Figure 2e). Interestingly, at a dilution of 3×104 cells, tumor incidence was 50% (4/8) for UMSCC47/empty but 0% (0/8) for UMSCC47/CH1 (P<0.02). This observation suggests that the CIC population may be compromised in HPV16-positive HNSCC following p53 reactivation. CICs are a sub-set of cancer cells within the tumor with the exclusive capacity to divide and expand the CIC pool or to differentiate into heterogeneous non-tumorigenic cells that constitute the bulk of the tumor. CICs are postulated to be the unique cells responsible for disease recurrence and/or metastasis. Therefore, elimination of CICs may be essential to optimally manage cancer patients. ALDH and CD44 are two markers used to identify the CIC population in HNSCC (30–32). As shown in Figure 2f, CH1 overexpression reduced the ALDHhigh population by 46% (P<0.01) and CD44high population by 31% in UMSCC47 cells (P<0.01). Moreover, FACS analysis showed that CD44 levels were reduced by 33% in UMSCC47/CH1 cells compared to UMSCC47/empty cells. Tumorsphere formation is an in vitro assay to assess the CIC population. Overexpression of CH1 in UMSCC47 cells inhibited tumorsphere formation efficiency by 42% (P<0.01) and reduced tumorsphere diameter by 25% (P<0.01) (Figure 2g). To confirm the tumor initiating potential of tumorspheres, NOD/SCID mice were implanted with a single tumorsphere (mean diameter of 60–80 μm with ~100 cells) and monitored for tumor incidence over a 6 month period (Figure 2h). Mice implanted with a single tumorsphere had a tumor incidence rate of 55% (6/11). In contrast, all the mice implanted with 1×103 UMSCC47 cells failed to develop tumors over a 6 month period. Our work demonstrate that reactivation of p53 suppress the in vivo tumorigenicity of HPV-positive HNSCC, in part through a reduction in the CIC population.
Exogenous CH1 has a pleiotropic anti-tumor effect in HPV-negative HNSCC
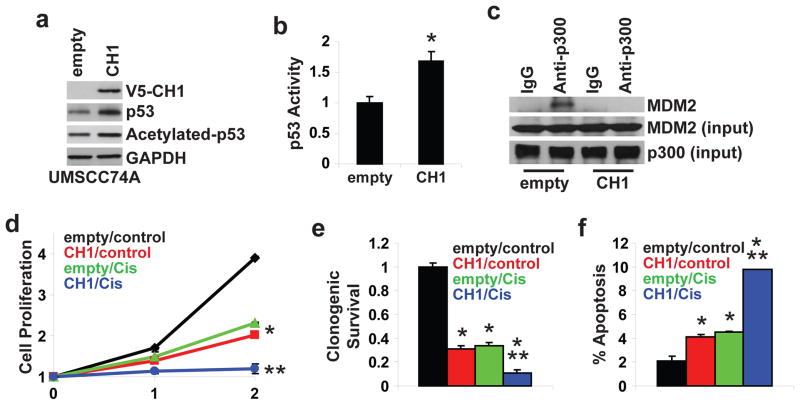
There is evidence that p300 is indispensable for MDM2-mediated p53 degradation (33, 34). MDM2 was shown to bind to the CH1 domain of p300 and overexpression of CH1 was sufficient to enhance p53 stability in p53 wildtype U2OS osteosarcoma cells (33, 34). In line with these observations, ectopic expression of CH1 increased total and acetylated p53 in p53 wildtype, HPV-negative UMSCC74A HNSCC cells (Figure 3a). p53 transcription activity was elevated by 68% (P<0.05) in UMSCC74A/CH1 compared to UMSCC74A/empty cells (Figure 3b). As shown in Figure 3c, the interaction between p300 and MDM2 in UMSCC74A cells was disrupted with the introduction of CH1. Overexpression of CH1 inhibited cell proliferation (72 hours, P<0.01), reduced clonogenic survival (P<0.01), and increased apoptosis (P<0.05) in UMSCC74A cells. In addition, UMSCC74A/CH1 cells were more responsive to the anti-tumor effects of cis-platinum (10 μM) than UMSCC74A/empty cells. These results show that exogenous CH1 blocked p300-MDM2 interaction, enhanced p53 activity, and promoted a broad anti-tumor response in HPV-negative HNSCC cells.
Figure 3. Exogenous CH1 has a pleiotropic anti-tumor effect in HPV-negative HNSCC.
Stable polyclonal UMSCC74A/empty and UMSCC74A/CH1 cells were generated by transfection and antibiotic selection. a. Total and acetylated p53 levels. Cell lysates were immunoblotted with anti-V5, anti-p53, or anti-acetylated[K382]-p53 antibody. b. p53 transcriptional activity. Cells were co-transfected with a p53 Firefly luciferase reporter plasmid and a control Renilla luciferase plasmid. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency. Data were normalized to empty vector cells and presented as mean ± SEM. *P<0.01, n=4. c. MDM2-p300 association. Cell lysates were extracted, immunoprecipitated with anti-p300 antibody, and immunoblotted with anti-MDM2. Cell lysates were immunoblotted with anti-MDM2 or anti-p300 antibody for input control. d. Cell proliferation. Cells were treated with control (vehicle) or cis-platinum (10 μM) for 24 and 48 hours. Data were normalized to Day 0 and presented as mean ± SEM. *P<0.01, control vs. CH1 or cis-platinum, **P<0.01, CH1 or cis-platinum vs. CH1 + cis-platinum, n=4. e. Clonogenic survival. Cells were treated with control (vehicle) or cis-platinum (10 μM). Colonies were stained with crystal violet. Data were normalized to empty/control cells and presented as mean ± SEM. *P<0.01, control vs. CH1, cis-platinum, or CH1 + cis-platinum, **P<0.01, CH1 or cis-platinum vs. CH1 + cis-platinum. f. Apoptosis. Cells were treated with control (vehicle) or cis-platinum (10 μM) for 24 hours. FACS was used to quantitate Annexin V-positive apoptotic cells. Data is presented as mean ± SEM. *P<0.01, control vs. CH1, cis-platinum, or CH1 + cis-platinum, **P<0.01, CH1 or cis-platinum vs. CH1 + cis-platinum.
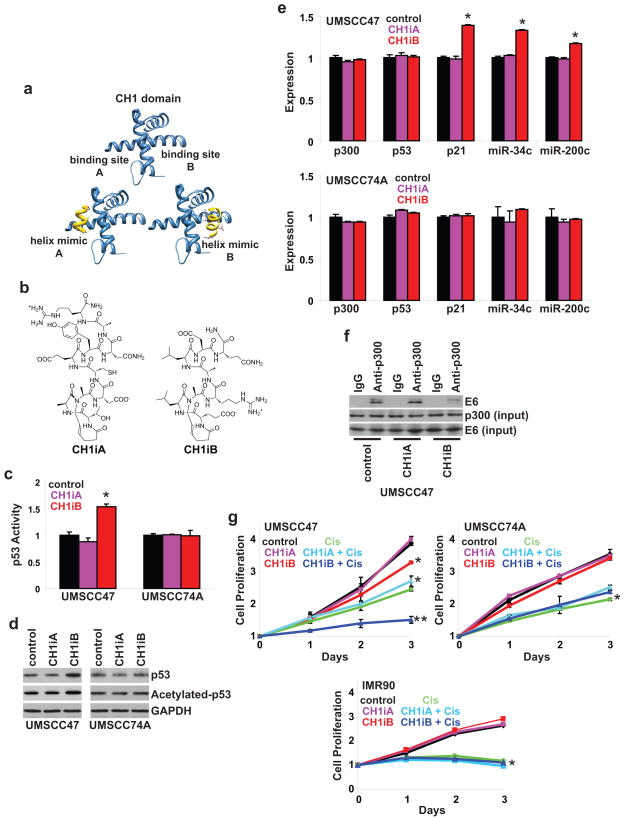
CH1iB, a small molecule CH1 inhibitor, preferentially reactivates p53 in HPV16-positive HNSCC
Our data indicates that exogenous CH1 reactivated p53 in HPV-positive and HPV-negative HNSCC. We determined if small molecule CH1 ligands can function as competitive inhibitors to mask the E6 and MDM2 binding sites on p300 and enhance p53 activity. HIF-1α recruits and binds to the CH1 domain of p300 to facilitate HIF-1α-mediated transcription of target genes (35, 36). A stabilized α-helical mimic, constrained by the hydrogen bond surrogate methodology (37), of HIF-1α (CH1iA) was reported to function as a CH1 inhibitor and compete with endogenous HIF-1α for p300 resulting in a reduction in HIF-1α-mediated transcription of vascular endothelial growth factor (38). The CH1 domain of p300 features multiple binding sites for individual α-helices (35, 36) suggesting that targeting a distinct CH1-binding partner may be a possibility. We tested the ability of synthetic helices that target binding site A and site B in CH1 in HPV-negative and HPV-positive HNSCC (Figure 4b). CH1iA and CH1iB did not modulate p53 activity and levels in UMSCC74A, a HPV-negative, p53 wildtype HNSCC cell line (Figures 4c and 4d). Expression of p300, p53, and p53-regulated genes were unchanged following CH1iA or CH1iB treatment in UMSCC74A cells (Figure 4e). In HPV-positive UMSCC47 cells, CH1iA had no effect whereas CH1iB enhanced p53 activity (71% increase, P<0.01), accumulation, and acetylation. A modest but significant increase in p21, miR-34a, and miR-200c expression was shown following CH1iB treatment (Figure 4e). Furthermore, the association between E6 and p300 was reduced with CH1iB but not with CH1iA treatment in HPV-positive HNSCC cells (Figure 4f). These results reveal that the critical binding contacts between E6 and CH1 are located within or in proximity to binding site B of the CH1 domain. In Figure 4g, CH1iA was inactive but CH1iB inhibited the proliferation of HPV-positive UMSCC47 cells as single-agent and potentiated the anti-proliferative efficacy of cis-platinum. CH1iA and CH1iB had no effect and did not augment the efficacy of cis-platinum on the proliferation of HPV-negative UMSCC74A cells and human normal IMR90 fibroblasts. Taken together, our work demonstrates that targeting binding site B in CH1 with CH1iB preferentially reactivates p53 in HPV-positive HNSCC cells by disrupting the association between E6 and p300.
Figure 4. CH1iB preferentially reactivates p53 in HPV16-positive HNSCC.
a. Targeting two distinct sites in CH1. HIF1-α/p300 structures were used as guides to design helix mimetics that target site A and site B on the CH1 domain of p300. b. Synthetic helices. CH1iA and CH1iB were designed to mimic two helices from the C-terminal domain of HIF-1α. The peptides were locked into the helical conformation by the hydrogen bond surrogate method. c. p53 activity. UMSCC47 and UMSCC74A cells were co-transfected with a p53 Firefly luciferase reporter plasmid and a control Renilla luciferase plasmid. After 24 hours, cells were treated with control (vehicle), CH1iA (10 μM), or CH1iB (10 μM) for 24 hours. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency. Data were normalized to control and presented as mean ± SEM. *P<0.05, n=3. d. Total and acetylated p53 levels. UMSCC47 and UMSCC74A cells were treated with control (vehicle), CH1iA (10 μM), or CH1iB (10 μM) for 24 hours. Cell lysates were immunoblotted with anti-p53 or anti-acetylated[K382]-p53 antibody. e. p300, p53, p21, miR-34a, and miR-200c expression. UMSCC47 and UMSCC74A cells were treated with control (vehicle), CH1iA (10 μM), or CH1iB (10 μM) for 24 hours. mRNA expression was determined using qPCR with validated TaqMan assays. Data were normalized to control cells and presented as mean ± SEM. *P<0.01, control vs. CH1iB, n=3. f. HPV16 E6-p300 association. UMSCC47 cells were treated with control (vehicle), CH1iA (10 μM), or CH1iB (10 μM) for 24 hours. Cell lysates were extracted, immunoprecipitated with anti-p300 antibody, and immunoblotted with anti-E6 antibody. Cell lysates were immunoblotted with anti-E6 or anti-p300 antibody for input control. g. Cell proliferation. UMSCC47, UMSCC74A, and IMR90 (human normal fibroblasts) cells were treated with control (vehicle), CH1iA (10 μM), CH1iB (10 μM), cis-platinum (10 μM), CH1iA (10 μM) and cis-platinum (10 μM), or CH1iB (10 μM) and cis-platinum (10 μM) for 24, 48, and 72 hours. Data were normalized to Day 0 and presented as mean ± SEM. *P<0.01, control vs. CH1iB or cis-platinum, **P<0.01, CH1iB or cis-platinum vs. CH1iB and cis-platinum, n=6.
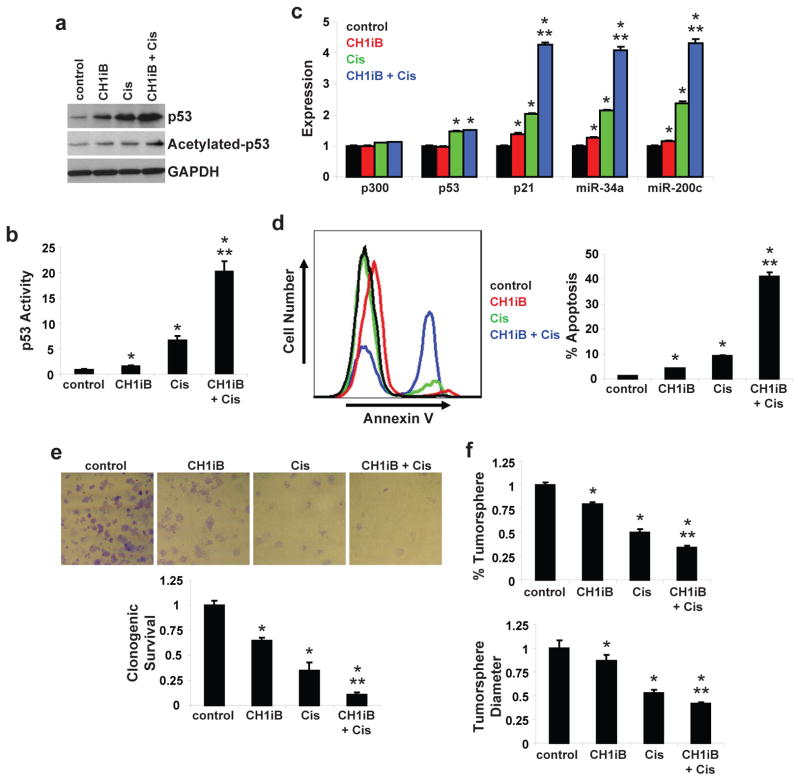
CH1iB potentiates the efficacy of cis-platinum in HPV16-positive HNSCC
Our results showed that introduction of CH1 potentiated the effects of cis-platinum on cell proliferation, clonogenic survival, and apoptosis in HPV-positive HNSCC. In addition, CH1iB enhanced the anti-proliferative action of cis-platinum in UMSCC47 cells. As shown in Figures 5a–b, CH1iB potentiated the effect of cis-platinum on p53 accumulation, acetylation, and activity. Expression of p21, miR-34a, and miR-200c was dramatically higher (P<0.01) with the combination treatment than with either single-agent treatment (Figure 5c). Compared to vehicle-treated UMSCC47 cells, CH1iB reduced clonogenic survival by 35% and tumorsphere formation by 20%, and enhanced apoptosis by 353% (P<0.01). Furthermore, the combination regimen was highly active and almost completely ablated the clonogenic survival (91% inhibition, P<0.01) of HPV16-positive HNSCC cells. Apoptosis induced by the combination treatment was increased by 984% (P<0.01) and 443% (P<0.01) compared to CH1iB and cis-platinum, respectively (Figure 5d). In addition, tumorsphere formation was suppressed by a greater extent with the combination regimen than single-agent CH1iB or cis-platinum (Figure 5f). CH1iB-mut, a designed specificity control for CH1iB in which one energetically important leucine residue is mutated to alanine, showed a minimal but significant increase (14% increase, P <0.05) in p53 activity but, importantly, had no effect as single-agent or in combination with cis-platinum to inhibit cell proliferation in UMSCC47 cells (Figure S1). These results indicate that CH1iB, a CH1 inhibitor, potentiates the anti-tumor activity of cis-platinum in HPV-positive HNSCC.
Figure 5. CH1iB potentiates the efficacy of cis-platinum in HPV16-positive HNSCC.
a. Total and acetylated p53 levels. UMSCC47 cells were treated with control (vehicle), CH1iB (10 μM), cis-platinum (10 μM), or CH1iB (10 μM) and cis-platinum (10 μM) for 24 hours. Cell lysates were immunoblotted with anti-p53 or anti-acetylated[K382]-p53 antibody. b. p53 transcriptional activity. UMSCC47 cells were co-transfected with a p53 Firefly luciferase reporter plasmid and a control Renilla luciferase plasmid. After 24 hours, cells were treated with control (vehicle), CH1iB (10 μM), cis-platinum (10 μM), or CH1iB (10 μM) and cis-platinum (10 μM) for 24 hours. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency. Data were normalized to control and presented as mean ± SEM. *P<0.01, control vs. CH1iB, cis-platinum or CH1iB + cis-platinum, **P<0.01, CH1iB or cis-platinum vs. CH1iB + cis-platinum, n=5. c. p300, p53, p21, miR-34a, and miR-200c expression. UMSCC47 cells were treated with control (vehicle), CH1iB (10 μM), cis-platinum (10 μM), or CH1iB (10 μM) and cis-platinum (10 μM) for 24 hours. mRNA expression was determined using qPCR with validated TaqMan assays. Data were normalized to control cells and presented as mean ± SEM. *P<0.01, control vs. CH1iB, cis-platinum or CH1iB + cis-platinum, **P<0.01, CH1iB or cis-platinum vs. CH1iB + cis-platinum, n=3. d. Apoptosis. UMSCC47 cells were treated with control (vehicle), CH1iB (10 μM), cis-platinum (10 μM), or CH1iB (10 μM) and cis-platinum (10 μM) for 24 hours. FACS was used to quantitate Annexin V-positive apoptotic cells. Data is presented as mean ± SEM. *P<0.01, control vs. CH1iB, cis-platinum or CH1iB + cis-platinum, **P<0.01, CH1iB or cis-platinum vs. CH1iB + cis-platinum, n=3. e. Clonogenic survival. UMSCC47 cells were treated with control (vehicle), CH1iB (10 μM), cis-platinum (10 μM), or CH1iB (10 μM) and cis-platinum (10 μM) at day 0 and colonies were stained with crystal violet at 14 days. Data were normalized to control and presented as mean ± SEM. *P<0.01, control vs. CH1iB, cis-platinum or CH1iB + cis-platinum, **P<0.01, CH1iB or cis-platinum vs. CH1iB + cis-platinum, n=3. f. Tumorsphere formation efficiency and diameter. UMSCC47 cells were seeded on low-attachment plates and treated with control (vehicle), CH1iB (10 μM), cis-platinum (3 μM), or CH1iB (10 μM) and cis-platinum (3 μM). Tumorsphere formation efficiency was calculated as the number of tumorspheres (≥ 50 μm in diameter) formed in 7 days divided by the original number of cells seeded. Tumorsphere diameter was measured using NIS-Elements software. Data were normalized to control and presented as mean ± SEM. *P<0.01, control vs. CH1iB, cis-platinum or CH1iB + cis-platinum, **P<0.01, CH1iB or cis-platinum vs. CH1iB + cis-platinum, n=8.
Discussion
High-risk HPV is recognized as an etiological agent for the pathogenesis of anogenital and head and neck squamous cell carcinomas. HPV E6 inactivates p53 through two distinct and independent pathways. It is well recognized that E6 complexes with E6AP to form an active E3-ubiquitin ligase to target p53 for proteasome-dependent proteolysis (18). A second but much more under-appreciated mechanism is that E6 associates with the p300-p53 complex to block p300-mediated acetylation and activation of p53 (21). p300 acetylates p53 at multiple lysine residues, including K370, 372, 381, and 382 (39). Acetylation was shown to control p53 function through multiple mechanisms, including an increase in protein stability, tetramerization, DNA binding, and co-activator recruitment (21, 23, 39).
Several groups reported that reactivation of p53 is achievable in HPV-positive cervical carcinomas using different strategies to reduce E6 or E6AP levels (24–29). Treatment with E6AP anti-sense oligonucleotides accumulated p53 but did not promote apoptosis (27). These authors suggest that a threshold level of p53 levels may be required for p53-mediated apoptosis. An alternate explanation is the ablation of E6AP may be inefficient to reactivate p53 since E6 is still available to suppress p300-mediated acetylation and activation of p53. Co-delivery of a HPV16 E6 antibody and E6 siRNA enhanced p53 levels and decreased clonogenic survival; however, an apoptotic response was not detected (26). An interesting study showed that siRNA-mediated ablation of E6 results in a transient increase in p53 protein and activity despite a sustained E6 knockdown suggesting that a compensatory p53 degradation and/or inactivation mechanism is quickly triggered in HPV-positive cervical carcinomas cells under these experimental conditions (28).
Disruption of E6-p300 association is an approach that has not be utilized to reactivate p53 in HPV-positive carcinomas. Restoration of p300-mediated acetylation of p53 may be an ideal strategy since acetylation controls p53 function through multiple mechanisms, including stability and transcriptional activation. Our results with stable CH1 overexpressing HNSCC cells indicate that targeting the E6-p300 interaction is sufficient to maintain elevated p53 accumulation, acetylation, and activity ad infinitum. Exogenous CH1 inhibits cell proliferation and clonogenic survival and enhances apoptosis in HPV-positive HNSCC. Importantly, the in vivo tumorigenicity of UMSCC47/CH1 cells is severely compromised in part through a reduction in the CIC population. Thus, our data showed that restoration of p300-mediated p53 acetylation induces a sustained p53 reactivation and anti-tumor response in HPV-positive HNSCC.
It was reported that p300 functions as a scaffold for MDM2 and p53 to facilitate MDM2-mediated degradation of p53 (33). Overexpression of the CH1 domain of p300 enhanced p53 accumulation in human U2OS osteosarcoma cells presumably by blocking the physical interaction between p300 and MDM2 (33). Also, binding of MDM2 to the p300-p53 complex blocked p300-mediated acetylation and activation of p53 (34). These results indicate that the MDM2-p300-p53 complex is intimately involved in p53 turnover, acetylation, and activation. In direct support, our results showed that exogenous CH1 disrupts MDM2-p300 association and increases p53 levels and activity in p53 wildtype, HPV-negative UMSCC74A HNSCC cells. In addition, CH1 overexpression sensitized UMSCC74A cells to the anti-tumor efficacy of cis-platinum. These results demonstrate that targeting the CH1 domain of p300 is a tractable approach to enhance p53 activity in HNSCC cells with wildtype p53, regardless of HPV status, albeit through distinct mechanisms. CH1iB, but not CH1iA, blocked E6-p300 association and reactivated p53 in HPV-positive HNSCC indicating that binding site B in the CH1 domain contains the critical contacts for E6 and p300 interaction. Interestingly, selective targeting of the CH1 domain with CH1iA and CH1iB did not enhance p53 accumulation and activity in UMSCC74A cells. The preferential activity of CH1iB for HPV-positive HNSCC over HPV-negative HNSCC suggests that the CH1-binding interface for E6 may be distinct from the CH1-binding interface for MDM2. Another possibility is that MDM2 may have a tighter binding association for p300 than E6 and thus, CH1iB and CH1iA were unable to successfully compete against MDM2 for p300 binding. Ongoing efforts in our laboratory are to further probe these possibilities by exploring structure-activity relationships of our synthetic CH1 domain ligands. In any event, our work reveals that CH1iB preferentially reactivates p53 activity in HPV-positive HNSCC cells providing initial evidence that discrete chemical targeting of the CH1 domain of p300 can be realized.
The role of p53 in normal stem cell regulation is established and is beginning to emerge for CICs. Inhibition of p53 dramatically enhanced the transformation efficiency of differentiated cells into induced pluripotent stem cells (40–42). Loss of p53 favored self-renewal, symmetric cell division, of mammary stem cells resulting in an expansion of the stem cell population (43). Two p53 targets, miR-34a and p21, were shown to contribute to p53 repression of induced pluripotent stem cells (40, 41, 44). Additionally, miR-34a blocked prostate CIC expansion (45). Loss of p53 in mammary epithelial cells led to reduced miR-200c expression resulting in an increase in EMT-associated CIC population (46). Our results are in line with these studies and further support the link between p53 and CICs. We show that reactivation of p53 in HPV-positive HNSCC increase the expression of p21, miR-34a, and miR-200c and reduce the CIC population. These observations suggest that the p53-p21/miR-34a/miR-200c circuitry to limit normal stem cell expansion, either through reprogramming or self-renewal, can be triggered in HPV-positive HNSCC to block CIC expansion through p53 reactivation. It is unclear at this time whether the reduction in the CIC population is due to a shift in favor of asymmetric CIC division and/or differentiation of CICs. Additional work will be necessary to address this question and to dissect the contributions of p21, miR-34a, and miR-200c in controlling the CIC population.
High-dose cis-platinum-based therapy is the standard of care for definitive treatment of HPV-positive but is associated with high toxicities and difficult for patients to tolerate (47). Treatment-associated toxicities from high-dose cis-platinum-based therapy are a major concern and have prompted considerable discussion whether alternate treatment or de-intensification of treatment should be offered for the HPV-positive HNSCC population. Considering there are limited clinical options for HPV-positive HNSCC at this time, alternative treatment strategies are critically needed. We showed that CH1iB, a CH1 inhibitor, reactivates p53 in HPV-positive HNSCC. Single-agent CH1iB exhibits broad anti-cancer activity to suppress cell proliferation and clonogenic survival and enhance apoptosis in UMSCC47 and UPCI:SCC090 cells. Interestingly, CH1iB potentiates cis-platinum-mediated p53 activity and anti-tumor efficacy. HPV-positive HNSCC cells are almost completely eliminated following treatment with the combination of CH1iB and cis-platinum. Based on these results, we speculate that fewer cycles or a tapered dose of cis-platinum may be sufficient in the presence of a CH1 inhibitor to effectively manage HPV-positive HNSCC patients with a better toxicity profile. Our data strongly supports further development of CH1 inhibitors as p53 reactivation therapeutics for HPV-positive HNSCC.
In conclusion, our results reveal that targeting the E6-p300 association is a novel approach to reactivate p53 in HPV-positive HNSCC. CH1iB, a small molecule CH1 inhibitor, reactivates p53 and potentiates the anti-tumor activity of cis-platinum in HPV-positive HNSCC cells. Our work indicates that the CH1 domain inhibitor platform represents a novel class of p53 reactivation therapeutics for HPV-positive HNSCC.
Materials and Methods
Cell lines
UMSCC47 and UMSCC74A were obtained from Dr. Thomas Carey at the University of Michigan. UPCI:SCC090 was provided by Dr. Susanne Gollin at the University of Pittsburgh (48). UMSCC47, UMSCC74A, and UPCI:SCC090 cells were grown in DMEM containing 10% FBS, 2 mM glutamine, 100 mg/mL streptomycin and 100 U/mL penicillin and maintained in a humidified atmosphere of 5% CO2 at 37°C.
Plasmid construction and transfection
The CH1 domain of p300 (nucleotides 332–417) was amplified by PCR (forward primer: 5′-GGATCCATGCCAGAGAAGCGCAAGCTCATCCAGC-3′; reverse primer: 5′-CTCGAGATCACCAGCATTTTTGAGGGGGAGACAC-3′) and inserted into pcDNA3.1 between the BamHI and Xhol restriction enzyme sites. UMSCC47, UMUPCI:SCC090, and UMSCC74A cells were transfected with pcDNA3.1/empty or pcDNA3.1/CH1 using Lipofectamine2000 (Invitrogen, Carlsbad, CA). Stable polyclonal populations were selected and maintained in the presence of G418 (Invitrogen).
Western blot
Whole cell lysates were mixed with Laemmli loading buffer, boiled, separated by SDS-PAGE, and transferred to a nitrocellulose membrane. Subsequently, immunoblot analyses were performed using antibodies specific to V5 (Invitrogen), p53 (sc-126, Santa Cruz Biotechnology, Santa Cruz, CA), or acetylated[K382]-p53 (2525, Cell Signaling Technology). The signal was developed using the SuperSignal Western Blotting Kit (Pierce, Rockford, IL).
p53 transcriptional activity
Cells were transfected with 100 ng of Cignal p53 reporter (SABiosciences, Valencia, CA) using Lipofectamine 2000. Cignal p53 reporter contains tandem repeats of the p53 consensus transcriptional response element. After 48 hours, cells were washed with cold PBS, lysed in passive lysis buffer (Promega), and measured for Firefly/Renilla dual luciferase activities in a luminometer using the Dual-Light System (Applied Biosystems, Foster City, CA). Renilla luciferase activity was normalized to Firefly luciferase activity to control for transfection efficiency. A modification to the protocol was used for compound treatment. UMSCC47 cells were transfected with 100 ng of Cignal p53 reporter. After 24 hours, cells were treated with vehicle, CH1iB (10 μM), cis-platinum (10 μM), or combination of CH1iB (10 μM) and cis-platinum (10 μM) and measured for Firefly/Renilla dual luciferase activities 24 hours post-treatment.
Quantitative real-time PCR
Cells were extracted for total RNA using the TRIzol® reagent (Invitrogen) or TaqMan PreAmp Cells-to-CT kit (Applied Biosystems). Expression of p300, p53, p21, miR-34a and miR-200c were determined using the Applied Biosystems 7900HT Fast Real-Time PCR System with validated TaqMan gene expression assays (Applied Biosystems). p53, p300, and p21 expression were normalized to GADPH and miR-34a and miR-200c expression were normalized to RNU44 using the ΔΔCt method.
Immunoprecipitation
Cells were lysed with NP buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, and 1.0% NP-40] containing 1× Protease Inhibitor Cocktail (Roche, Switzerland) at 4°C with gentle rocking for 15 min. The supernatant was pre-cleared to block nonspecific binding with 50 μL protein A/G Agarose beads (Pierce Biotechnology) that had been pre-washed with NP buffer before use. Equal amounts of anti-E6 antibody (Abcam), anti-p300 antibody (Millipore), or IgG antibody (Cell Signaling) were added to the respective samples. After 4 hours incubation at 4°C, 50 μL pre-washed protein A/G-agarose beads were added to each tube and immunoprecipitation was performed by rocking overnight at 4°C. The immunoprecipitated complexes were washed with NP buffer and then eluted using 2XSDS sample buffer. Eluted sample and 10% of input were resolved by SDS-PAGE for Western blot analysis with anti-E6, anti-p300, or anti-V5 antibodies.
Cell proliferation, clonogenic survival, and apoptosis
Cell proliferation was assessed using the MTT reagent (Roche Molecular Biochemicals, Nutley, NJ) to detect metabolic active cells. Absorbance was measured at 570nm in the Spectra Max 190 ELISA reader (Molecular Devices, Sunnyvale, CA) after overnight incubation. For clonogenic survival, 200 cells per well were plated in complete growth media and allowed to grow until visible colonies were formed (14 days). Cell colonies were fixed with cold methanol, stained with 0.25% crystal violet in 25% methanol, washed, and air dried. For apoptosis, cells were harvested, washed with cold PBS, and co-stained with Annexin V and propidium iodide according to the manufacturer’s protocol (ApoAlert Annexin V-FITC Apoptosis Kit; Clontech). Apoptotic cells were analyzed using BD FACS Calibur (BD Biosciences Corporation, Franklin Lakes, NJ) at The Ohio State University Comprehensive Cancer Center Analytical Cytometry Core.
Tumor incidence and growth in athymic nude mice
UMSCC47/empty and UMSCC47/CH1 cells were suspended in 50:50 DMEM:Matrigel and implanted subcutaneously into the left and right flanks of 6-week old athymic nude mice (8 mice/group), respectively. After 3 weeks, tumors were measured once a week using a digital caliper and tumor volumes were calculated using the formula d1 × d2 × d3 × 0.5236, where “d” represents the three orthogonal diameters. Tumor growth and incidence were monitored for 49 days following tumor cell implantation.
ALDH and CD44
Cells were assessed for ALDH activity using the ALDEFLUOR kit according to the manufacturer’s protocol (Stem Cell Technologies, British Columbia, Canada). Cells were suspended in ALDEFLUOR assay buffer containing ALDH substrate (bidipy-aminoacetaldehyde, 1 μM per 1 × 106 cells) and incubated for 40 minutes at 37°C. For each experiment, a sample of cells was incubated with 50 mM of diethylaminobenzaldehyde (DEAB), a specific ALDH inhibitor, to serve as the negative control. For CD44 expression, cells were harvested and resuspened in incubation buffer with PE-CD44 antibody (Abcam) or mouse PE-IgG (Abcam) for 50 minutes on ice. Suspensions were centrifuged at 300× g for 5 minutes at 4°C and resuspend in 0.5 mL of 1% paraformaldehyde solution for analysis. Fluorescence activated cell sorting (FACS) analyses were performed using BD FACS Calibur at The Ohio State University Comprehensive Cancer Center Analytical Cytometry Core.
Tumorsphere formation
Cells were harvested and seeded in a serum-free defined medium consisting of KSF medium supplemented with epidermal growth factor, basic fibroblast growth factor, insulin, and hydrocortisone in low-attachment plates (Corning Incorporated, Corning, NY) for tumorspheres. Tumorsphere formation efficiency was calculated as the number of tumorspheres (≥ 50 μm in diameter) formed in 7 days divided by the original number of cells seeded. Tumorsphere diameter was measured using NIS-Elements software.
Tumor incidence with a single tumorsphere
Tumorsphere derived from UMSCC47 cells were generated and measured using NIS-Elements software. A single tumorsphere (60–80 μm in diameter) was suspended in 50:50 KSF:Matrigel and implanted subcutaneously into the flank of 6-week old NOD/SCID mice (n=11). In a separate set of animals, parental UMSCC47 cells (1×103) were suspended in 50:50 DMEM:Matrigel and implanted subcutaneously into the flank of 6-week old NOD/SCID mice (n=10). Tumor incidence was monitored for 180 days following tumorsphere or tumor cell implantation.
Synthesis of inhibitors
Synthetic helices were synthesized as previously described (38, 49). Compounds were purified by reverse-phase HPLC and characterized by ESI-MS.
Statistical analysis
Data were analyzed by two-tailed Student’s t-test. P-values < 0.05 were considered significant.
Supplementary Material
Table S1. Mass spectroscopic characterization of CH1 inhibitors.
Figure S1. Effect of CH1iB-mut, an inactive analog of CH1iB, on p53 transcriptional activity and cell proliferation. a. p53 transcriptional activity. UMSCC47 cells were co-transfected with a p53 Firefly luciferase reporter plasmid and a control Renilla luciferase plasmid. After 24 hours, cells were treated with control (vehicle) or CH1iB-mut (10 μM) for 24 hours. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency. Data is presented as mean ± SEM. *P<0.05, n=5. b. Cell proliferation. UMSCC47 cells were treated with control (vehicle), CH1iB-mut (10 μM), cis-platinum (10 μM), or CH1iB-mut (10 μM) and cis-platinum (10 μM) for 24 and 48 hours. Data were normalized to Day 0 and presented as mean ± SEM. *P<0.01, control or CH1iB-mut vs. cis-platinum or cis-platinum + CH1iB-mut, n=6.
Figure S2. Analytical HPLC traces of CH1 inhibitors.
Acknowledgments
Work was supported in part by National Institutes of Health grants R01CA135096 (to QP) and R01GM073943 (to PSA); Mary E. and John W. Alford Cancer Research Endowment Fund; The Michelle Theado Memorial Grant from the Joan Bisesi Fund for Head and Neck Oncology Research; and Arthur G. James Cancer Hospital and Richard J. Solove Research Institute, The Ohio State University Comprehensive Cancer Center.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006 May 10;24(14):2137–50. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000 May 3;92(9):709–20. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 3.Klussmann JP, Gultekin E, Weissenborn SJ, Wieland U, Dries V, Dienes HP, et al. Expression of p16 protein identifies a distinct entity of tonsillar carcinomas associated with human papillomavirus. Am J Pathol. 2003 Mar;162(3):747–53. doi: 10.1016/S0002-9440(10)63871-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Licitra L, Rossini C, Bossi P, Locati LD. Advances in the changing patterns of aetiology of head and neck cancers. Curr Opin Otolaryngol Head Neck Surg. 2006 Apr;14(2):95–9. doi: 10.1097/01.moo.0000193170.23956.5f. [DOI] [PubMed] [Google Scholar]
- 5.Shiboski CH, Schmidt BL, Jordan RC. Tongue and tonsil carcinoma: increasing trends in the U.S. population ages 20–44 years. Cancer. 2005 May 1;103(9):1843–9. doi: 10.1002/cncr.20998. [DOI] [PubMed] [Google Scholar]
- 6.Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers? Cancer. 2007 Oct 1;110(7):1429–35. doi: 10.1002/cncr.22963. [DOI] [PubMed] [Google Scholar]
- 7.Hammarstedt L, Lindquist D, Dahlstrand H, Romanitan M, Dahlgren LO, Joneberg J, et al. Human papillomavirus as a risk factor for the increase in incidence of tonsillar cancer. Int J Cancer. 2006 Dec 1;119(11):2620–3. doi: 10.1002/ijc.22177. [DOI] [PubMed] [Google Scholar]
- 8.Group FIS. Quadrivalent vaccine against human papillomavirus to prevent high-grade cervical lesions. N Engl J Med. 2007 May 10;356(19):1915–27. doi: 10.1056/NEJMoa061741. [DOI] [PubMed] [Google Scholar]
- 9.Laz TH, Rahman M, Berenson AB. An update on human papillomavirus vaccine uptake among 11–17 year old girls in the United States: National Health Interview Survey, 2010. Vaccine. 2012 May 21;30(24):3534–40. doi: 10.1016/j.vaccine.2012.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reiter PL, McRee AL, Kadis JA, Brewer NT. HPV vaccine and adolescent males. Vaccine. 2012 Aug 5;29(34):5595–602. doi: 10.1016/j.vaccine.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munoz N, Kjaer SK, Sigurdsson K, Iversen OE, Hernandez-Avila M, Wheeler CM, et al. Impact of human papillomavirus (HPV)-6/11/16/18 vaccine on all HPV-associated genital diseases in young women. J Natl Cancer Inst. 2010 Mar 3;102(5):325–39. doi: 10.1093/jnci/djp534. [DOI] [PubMed] [Google Scholar]
- 12.Sigurdsson K, Sigvaldason H, Gudmundsdottir T, Sigurdsson R, Briem H. The efficacy of HPV 16/18 vaccines on sexually active 18–23 year old women and the impact of HPV vaccination on organized cervical cancer screening. Acta Obstet Gynecol Scand. 2009;88(1):27–35. doi: 10.1080/00016340802566770. [DOI] [PubMed] [Google Scholar]
- 13.Balz V, Scheckenbach K, Gotte K, Bockmuhl U, Petersen I, Bier H. Is the p53 inactivation frequency in squamous cell carcinomas of the head and neck underestimated? Analysis of p53 exons 2–11 and human papillomavirus 16/18 E6 transcripts in 123 unselected tumor specimens. Cancer Res. 2003 Mar 15;63(6):1188–91. [PubMed] [Google Scholar]
- 14.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011 Aug 26;333(6046):1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011 Aug 26;333(6046):1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991 Dec;10(13):4129–35. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993 Nov 5;75(3):495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 18.Talis AL, Huibregtse JM, Howley PM. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem. 1998 Mar 13;273(11):6439–45. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 19.Zimmermann H, Degenkolbe R, Bernard HU, O’Connor MJ. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol. 1999 Aug;73(8):6209–19. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999 Sep 15;18(18):5061–72. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas MC, Chiang CM. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol Cell. 2005 Jan 21;17(2):251–64. doi: 10.1016/j.molcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002 Nov 15;21(22):6236–45. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002 Dec 27;277(52):50607–11. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 24.Beerheide W, Bernard HU, Tan YJ, Ganesan A, Rice WG, Ting AE. Potential drugs against cervical cancer: zinc-ejecting inhibitors of the human papillomavirus type 16 E6 oncoprotein. J Natl Cancer Inst. 1999 Jul 21;91(14):1211–20. doi: 10.1093/jnci/91.14.1211. [DOI] [PubMed] [Google Scholar]
- 25.Beerheide W, Sim MM, Tan YJ, Bernard HU, Ting AE. Inactivation of the human papillomavirus-16 E6 oncoprotein by organic disulfides. Bioorg Med Chem. 2000 Nov;8(11):2549–60. doi: 10.1016/s0968-0896(00)00193-0. [DOI] [PubMed] [Google Scholar]
- 26.Courtete J, Sibler AP, Zeder-Lutz G, Dalkara D, Oulad-Abdelghani M, Zuber G, et al. Suppression of cervical carcinoma cell growth by intracytoplasmic codelivery of anti-oncoprotein E6 antibody and small interfering RNA. Mol Cancer Ther. 2007 Jun;6(6):1728–35. doi: 10.1158/1535-7163.MCT-06-0808. [DOI] [PubMed] [Google Scholar]
- 27.Beer-Romero P, Glass S, Rolfe M. Antisense targeting of E6AP elevates p53 in HPV-infected cells but not in normal cells. Oncogene. 1997 Feb 6;14(5):595–602. doi: 10.1038/sj.onc.1200872. [DOI] [PubMed] [Google Scholar]
- 28.Koivusalo R, Mialon A, Pitkanen H, Westermarck J, Hietanen S. Activation of p53 in cervical cancer cells by human papillomavirus E6 RNA interference is transient, but can be sustained by inhibiting endogenous nuclear export-dependent p53 antagonists. Cancer Res. 2006 Dec 15;66(24):11817–24. doi: 10.1158/0008-5472.CAN-06-2185. [DOI] [PubMed] [Google Scholar]
- 29.Zhao CY, Szekely L, Bao W, Selivanova G. Rescue of p53 function by small-molecule RITA in cervical carcinoma by blocking E6-mediated degradation. Cancer Res. 2010 Apr 15;70(8):3372–81. doi: 10.1158/0008-5472.CAN-09-2787. [DOI] [PubMed] [Google Scholar]
- 30.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007 Jan 16;104(3):973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clay MR, Tabor M, Owen JH, Carey TE, Bradford CR, Wolf GT, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010 Jan 13; doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen YC, Chen YW, Hsu HS, Tseng LM, Huang PI, Lu KH, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun. 2009 Jul 31;385(3):307–13. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 33.Grossman SR, Perez M, Kung AL, Joseph M, Mansur C, Xiao ZX, et al. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998 Oct;2(4):405–15. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 34.Kobet E, Zeng X, Zhu Y, Keller D, Lu H. MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins. Proc Natl Acad Sci U S A. 2000 Nov 7;97(23):12547–52. doi: 10.1073/pnas.97.23.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha/CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci U S A. 2002 Apr 16;99(8):5271–6. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, et al. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci U S A. 2002 Apr 16;99(8):5367–72. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patgiri A, Jochim AL, Arora PS. A hydrogen bond surrogate approach for stabilization of short peptide sequences in alpha-helical conformation. Acc Chem Res. 2008 Oct;41(10):1289–300. doi: 10.1021/ar700264k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henchey LK, Kushal S, Dubey R, Chapman RN, Olenyuk BZ, Arora PS. Inhibition of hypoxia inducible factor 1-transcription coactivator interaction by a hydrogen bond surrogate alpha-helix. J Am Chem Soc. 2010 Jan 27;132(3):941–3. doi: 10.1021/ja9082864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997 Aug 22;90(4):595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 40.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009 Aug 27;460(7259):1132–5. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009 Aug 27;460(7259):1140–4. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009 Aug 27;460(7259):1145–8. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009 Sep 18;138(6):1083–95. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 44.Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. Nov;13(11):1353–60. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. Feb;17(2):211–5. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. Mar;13(3):317–23. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan Q, Gorin MA, Teknos TN. Pharmacotherapy of head and neck squamous cell carcinoma. Expert Opin Pharmacother. 2009 Oct;10(14):2291–302. doi: 10.1517/14656560903136754. [DOI] [PubMed] [Google Scholar]
- 48.White JS, Weissfeld JL, Ragin CC, Rossie KM, Martin CL, Shuster M, et al. The influence of clinical and demographic risk factors on the establishment of head and neck squamous cell carcinoma cell lines. Oral Oncol. 2007 Aug;43(7):701–12. doi: 10.1016/j.oraloncology.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patgiri A, Menzenski MZ, Mahon AB, Arora PS. Solid-phase synthesis of short alpha-helices stabilized by the hydrogen bond surrogate approach. Nat Protoc. 2010 Nov;5(11):1857–65. doi: 10.1038/nprot.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Mass spectroscopic characterization of CH1 inhibitors.
Figure S1. Effect of CH1iB-mut, an inactive analog of CH1iB, on p53 transcriptional activity and cell proliferation. a. p53 transcriptional activity. UMSCC47 cells were co-transfected with a p53 Firefly luciferase reporter plasmid and a control Renilla luciferase plasmid. After 24 hours, cells were treated with control (vehicle) or CH1iB-mut (10 μM) for 24 hours. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency. Data is presented as mean ± SEM. *P<0.05, n=5. b. Cell proliferation. UMSCC47 cells were treated with control (vehicle), CH1iB-mut (10 μM), cis-platinum (10 μM), or CH1iB-mut (10 μM) and cis-platinum (10 μM) for 24 and 48 hours. Data were normalized to Day 0 and presented as mean ± SEM. *P<0.01, control or CH1iB-mut vs. cis-platinum or cis-platinum + CH1iB-mut, n=6.
Figure S2. Analytical HPLC traces of CH1 inhibitors.