Abstract
The maternal nucleolus is required for proper activation of the embryonic genome (EGA) and early embryonic development. Nucleologenesis is characterized by the transformation of a nucleolar precursor body (NPB) to a mature nucleolus during preimplantation development. However, the function of NPBs and the involved molecular factors are unknown. We uncover a novel role for the pluripotency factor LIN28, the biological significance of which was previously demonstrated in the reprogramming of human somatic cells to induced pluripotent stem (iPS) cells. Here, we show that LIN28 accumulates at the NPB and the mature nucleolus in mouse preimplantation embryos and embryonic stem cells (ESCs), where it colocalizes with the nucleolar marker B23 (nucleophosmin 1). LIN28 has nucleolar localization in non-human primate (NHP) preimplantation embryos, but is cytoplasmic in NHP ESCs. Lin28 transcripts show a striking decline before mouse EGA, whereas LIN28 protein localizes to NPBs at the time of EGA. Following knockdown with a Lin28 morpholino, the majority of embryos arrest between the 2- and 4-cell stages and never develop to morula or blastocyst. Lin28 morpholino-injected embryos arrested at the 2-cell stage were not enriched with nucleophosmin at presumptive NPB sites, indicating that functional NPBs were not assembled. Based on these results, we propose that LIN28 is an essential factor of nucleologenesis during early embryonic development.
Keywords: LIN28, Mouse, Nucleolus, Preimplantation embryo, Non-human primate, Marmoset
INTRODUCTION
After fertilization, embryos drive their initial development using a stockpile of RNAs and proteins originating from the oocyte, until transcription of the embryonic genome becomes gradually activated. The time of embryonic genome activation (EGA) is species specific. In mice, a minor degree of transcription occurs in the zygote and a major burst at the late 2-cell stage (Schultz, 2002). Polymerase activity at the 2-cell stage is indispensable for subsequent divisions, which has been connected to the importance of embryonic genome activation (Golbus et al., 1973; reviewed by Minami et al., 2007). After five cleavage divisions, with ∼32 cells, mouse embryos form early blastocysts. Blastocysts continue to expand, while differentiating into trophectoderm (TE) and inner cell mass (ICM). As functional nucleoli are only assembled when the embryonic genome has been activated (Kopecny, 1989; Svarcova et al., 2009), it is speculated that early protein synthesis uses maternally stored ribosomal proteins and RNAs. The primary function of the nucleolus involves ribosome-subunit biogenesis in eukaryotic cells. Maintaining a sufficient supply of ribosome subunits is important to support protein synthesis during cell proliferation and divisions (Boisvert et al., 2007), such as during early embryonic development.
In somatic mammalian cells, the nucleolus contains three distinct functional and structural compartments. The fibrillar center harbors ribosomal (rRNA) genes and is the place where their transcription occurs. The dense fibrillar material contains nascent rRNA and the enzymes necessary for its processing. The granular component is the location of early ribosome assembly (Németh and Längst, 2011). The nucleolus in fully grown oocytes and early cleavage stage embryos consists only of the dense fibrillar material with mainly RNA and is thus termed the nucleolar precursor body (NPB) (Fléchon and Kopecný, 1998). NPBs are inactive and start to transform into fully differentiated, mature, functional nucleoli at different embryonic developmental stages, specific to a given species (e.g. 2-cell stage in the mouse). This transformation occurs at the time of the transition from maternal to embryonic genome control. Simultaneously, dramatic changes in the nuclear architecture occur during this period of transition. Chromocenters representing major heterochromatic domains form nucleoplasmic clusters after dissociation from the periphery of NPBs (Martin et al., 2006). The requirement of a functional nucleolus for proper embryonic development was demonstrated when enucleolated mouse oocytes were fertilized or parthenogenetically activated (Ogushi et al., 2008). The resulting embryos arrested after one or two cleavages, suggesting that the embryonic genome cannot be activated in the absence of the material of the maternal nucleolus. Whereas differentiated nucleoli have additional functions next to ribosome biogenesis and are involved in the regulation of other cellular processes, e.g. mitosis, cell cycle progression and stress response (Boisvert et al., 2007), the function of NPBs is largely unknown.
During the transition from the maternal to the embryonic genome, a dramatic reprogramming in the pattern of gene expression occurs. Novel embryonic transcripts, which are not expressed in the oocyte, are synthesized (Latham et al., 1991; Wang et al., 2010). This indicates the presence of factors and mechanisms to (re-)activate genes in oocytes and early embryos. Nuclear transfer experiments of differentiated somatic cell nuclei into enucleated oocytes ultimately demonstrated that the process of cell differentiation can be reversed and that the factors required for reprogramming gene expression are present in the oocyte (Yamanaka and Blau, 2010). Recent studies show that differentiated mouse and human somatic cells can be transduced to revert to pluripotent stem cells (also called induced pluripotent stem or iPS cells) with properties similar to embryonic stem cells (ESCs) by a defined number of key reprogramming factors (Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). The role of the reprogramming factors OCT4 (POU5F1) and SOX2 has been studied during early mouse development (Avilion et al., 2003; Foygel et al., 2008; Keramari et al., 2010). Whereas maternal OCT4 facilitates embryonic genome activation (Foygel et al., 2008), maternal SOX2 is essential for TE formation (Keramari et al., 2010).
Unlike other reprogramming factors, LIN28 is not a transcription factor, but belongs to the class of RNA-binding proteins with two unique RNA-binding domains, a cold-shock domain (CSD) and two retroviral-type CCHC zinc fingers (Moss et al., 1997). LIN28/LIN-28 was first described as a heterochronic gene and a regulator of developmental timing in Caenorhabditis elegans (Moss et al., 1997). Predominantly cytoplasmic with some nucleolar localization (Balzer and Moss, 2007; Moss et al., 1997), LIN28 is highly conserved in other mammalian genomes (Moss and Tang, 2003). In the mouse, LIN28 is abundantly expressed in undifferentiated ESCs, embryonal carcinoma cells and early embryonic tissue, but declines in expression and becomes tissue-restricted as the animal proceeds in development (Moss and Tang, 2003; Yang and Moss, 2003). A decrease in expression has also been reported in human ESCs during differentiation (Darr and Benvenisty, 2009).
The aim of this study was to characterize LIN28 expression temporally, spatially and functionally during mouse preimplantation development. We show that LIN28 protein is predominantly localized in the nucleolus and its precursor bodies of mammalian preimplantation embryos with expression commencing at a time corresponding with EGA. We found that knockdown of LIN28 at the zygote stage leads to a developmental arrest at the 2-cell stage to 4-cell stage transition and failure to acquire markers specific for maturing nucleolar precursor bodies. These results suggest that LIN28 is required for nucleologenesis during early mouse development, where its function can be related to the maternal-embryonic transition.
MATERIALS AND METHODS
Oocyte and embryo collection and culture
Oocytes and embryos were obtained from CD1 (bred in the European Neuroscience Institute Goettingen) or F1 (C57BL/6xCBA/Tar) (bred in the Faculty of Biology in Warsaw) females. Both strains of mice were maintained in a lighting regime of 12 hours light and 12 hours darkness.
Fully grown oocytes were obtained from the ovaries of 4- to 6-week-old females. To prevent spontaneous maturation of oocytes, the ovaries were placed in M2 medium containing dibutyrylic cAMP (M2+dbcAMP; 150 μg/ml; Sigma). Oocytes obtained by puncturing of the largest follicles were collected in the same medium. After a 1 hour incubation in M2+dbcAMP under paraffin oil, at standard culture conditions (37.5°C, 5% CO2), oocytes were transferred into Pronase (0.5% in Ringer’s Solution) to remove their zonae pellucidae. Subsequently, they were washed in M2+dbcAMP and fixed for immunocytochemistry.
Ovulated (metaphase II, MII) oocytes were obtained from F1 (C57BL/6/xCBA/Tar) females induced to superovulate with 10 IU of pregnant mare’s serum gonadotropin (PMSG; Folligon, Intervet, The Netherlands) and 10 IU human chorionic gonadotropin (hCG; Chorulon, Intervet, The Netherlands) 48-50 hours later. Females were sacrificed 16 hours after hCG injection and the cumuluses were released from the ampullae of oviducts into hyaluronidase (300 μg/ml, Sigma) to disperse the follicular cells. Oocytes were collected in M2 medium.
Zygotes (1-cell) and cleaving embryos were obtained from spontaneously ovulated females (CD1) or females induced to superovulate (F1). Prior to caging with males, spontaneously ovulated females were selected for estrous by visual inspection of the vagina. Detection of a copulation plug was performed to confirm successful mating. Zygotes at 0.5 days post coitum (dpc) were released from ampullae of oviducts and cumulus cells were removed by hyaluronidase treatment. To obtain cleaving embryos, the isolated oviducts were cut into pieces in M2 medium. Blastocysts were flushed from the uteri. Two-cell and 4-cell embryos were collected at 1.5 dpc, 8-cell and morula embryos at 2.5 dpc, blastocysts at 3.5 dpc. For in vitro culture, embryos were placed in 20 μl drops of KSOM medium (Millipore) under mineral oil (Irvine Scientific) in an atmosphere of 5% CO2 in air, at 37°C, in groups of 10-15 per drop.
F1 (C57BL/6xCBA/Tar) females were induced to superovulate as described for MII oocytes and were mated with F1 males immediately after hCG administration. The successful mating was confirmed by the presence of plugs. Time in hours after hCG injection (hours phCG) was a reference point for estimating the progression of the cell cycle (G1, S or G2) and embryonic development (1-, 2-, 4- or 8-cell embryos and blastocysts). The embryos were obtained as described above and collected in M2 medium.
Marmoset studies
Marmosets were obtained from a self-sustaining breeding colony of ∼500 animals at the German Primate Center (DPZ, Göttingen, Germany). The animals were kept at 26°C with 65% humidity and illumination for 12 hours per day, in full compliance with German animal protection laws. Experiments were approved and conducted under reference number AZ 33.42502-066/06. Minimally invasive transabdominal embryo collection method was applied as described (Hanazawa et al. 2012). Animals were anesthetized with diazepam (5 mg/ml) and the so-called Göttinger mixture consisting of 5 ml 100 mg/ml ketamine, 1 ml 10% xylazine, 0.1 ml 1% atropine and 3.9 ml sterile water; 0.5 ml per kg body weight of the monkey was injected intramuscularly. Anesthesia was monitored carefully by an experienced veterinarian.
ESC culture
For immunolocalization of LIN28 in mouse ESCs, H2B-GFP mouse ESC line (Hadjantonakis and Papaioannou, 2004) was used. ESCs were cultured on inactivated mouse embryonic fibroblasts (MEFs) in ESC culture medium consisting of 80% Knockout DMEM (Invitrogen) supplemented with 15% FBS (Invitrogen), 2 mM l-glutamine (Sigma-Aldrich), 0.1 mM MEM non-essential amino acids (Sigma-Aldrich), 0.1 mM β-mercaptoethanol (Sigma), 50 IU/ml penicillin and 50 μg/ml streptomycin sulfate (Invitrogen). ESCs were passaged every other day, culture medium was changed every 24 hours. For immunostaining, cells were passaged into 24-well dishes with sterile cover glasses at the bottom covered with inactivated MEF feeder layer and cultured for further 48 hours. Then, they were fixed in preparation for immunofluorescence staining.
Culture of the NHP ESC line cjes001 was carried out as previously described (Müller et al., 2009). In brief, ESCs were cultured on an MEF feeder layer. ESC culture medium contained 80% Knockout DMEM (Invitrogen) supplemented with 20% Knockout Serum Replacement (Invitrogen), 1 mM l-glutamine, 0.1 mM MEM non-essential amino acids, 0.1 mM β-mercaptoethanol (Sigma), 100 IU/ml penicillin and 100 μg/ml streptomycin sulfate. The cells were seeded on coverslips and grown on MEF cells for 2-5 days. Subsequently, they were fixed in preparation for immunofluorescence staining.
mRNA isolation and qRT-PCR
MII oocytes and embryos at desired stages of development were rinsed in PBS without Ca2+ and Mg2+ (Biomed, Lublin, Poland) and placed in a lysis buffer (five embryos or oocytes/20 μl buffer; Dynabeads mRNA DIRECT Micro Kit; Invitrogen) and frozen at –80°C. Additionally, 2.5 pg (0.25 pg/μl) of in vitro synthesized and polyadenylated cyc3GFP mRNA (see below) was added to each sample. mRNA was isolated with Dynabeads mRNA DIRECT Micro Kit (Invitrogen). After rinsing beads with Tris-HCl, RNA was eluted by 10 minute incubation in 10 μl water. Then, it was hybridized with 1 μl oligo(dT)12-18 (Invitrogen) and reverse transcribed with Super Script II Reverse Transcriptase (4 μl 5× first strand buffer, 2 μl 0.1 M DTT, 1 μl 10 mM dNTPs mix, 1 μl RNase out, 1 μl Reverse Transcriptase Superscript II; Invitrogen). Subsequently, cDNA was diluted 1:1 (v/v) with H2O and used for a qPCR amplification in triplicate with FastStart Universal SYBR Green Master with ROX (Roche Diagnostics) in a final volume of 10 μl per reaction: 5 μl SYBR Green Mix, 2.5 μl cDNA, 2.0 μl H2O, 0.25 μl 12 μM primer forward, 0.25 μl 12 μM primer reverse). The following primers were used: lin28a (forward: 5′- CCAAAAGGGAAGAACATGCAGAA-3′; reverse: 5′-GTTGATGCTTTGGCAAAAGTGG-3′; designed with Primer3 software); cyc3GFP (forward: TTCCATGGCCAACACTTGTCA; reverse: AGTGCGTTCC - TGTACATAACCTTCG; Primer3 software). The reaction was performed in a StepOne Real-Time PCR System (Applied Biosystems) by polymerase activation at 95°C for 10 minutes and then subsequent amplification was performed using 50 cycles consisting of denaturation at 95°C for 15 seconds and annealing, extension and measurement at 60°C for 45 seconds. Dissociation curve and gel electrophoresis were performed after qPCR to confirm specificity of primers.
In vitro synthesis of cyc3GFP mRNA
pcDNA3.1/NT-GFP plasmid (Invitrogen) was linearized with XbaI (Fermentas, Thermo Fisher Scientific) and, after cleaning with Axyprep PCR Clean-up Kit (Axygen Biosciences, Union City, CA, USA), it was used for in vitro transcription and subsequent polyadenylation with mMessage mMachine T7 Kit and Poly(A) Tailing Kit, respectively (both from Ambion). Polyadenylated mRNA was phenol/chloroform cleaned and precipitated with isopropanol. The pellet was dried and resuspended in water.
Microinjection of morpholino oligonucleotides
Antisense morpholino oligonucleotides (MOs; 25 nucleotides) were purchased from Gene Tools, LLC (Philomath, OR, USA). The specific morpholino blocks protein translation of the corresponding mRNA by targeting the 5′UTR through the initial 25 bases of the coding sequence. The sequence for these morpholino are as follows: Lin28A-MO 5′- ACTGCTGGTTGGACACCGAGCCCAT-3′; Control-MO 5′-ACTCCTCGTTCGACAGCCAGCCCAT-3′. Either Lin28A-MO or Control-MO (5-10 pl of 1 mM stock) was injected into the cytoplasm of zygotes on an inverted microscope (Zeiss Observer Z1) equipped with a semi-automatic micromanipulation system (TransferMan NK2, Eppendorf, Hamburg, Germany). Holding pipettes and microinjection capillaries (Type Pronucleus 1.6) were obtained from BioMedical Instruments (Zoellnitz, Germany). For each condition (Lin28A-MO, Control-MO, uninjected) 10-15 embryos were used in every experiment, which was conducted at least three times.
Microinjection of mRNA for Lin28
A full-length open reading frame (ORF) Lin28 DNA construct tagged to the coding red fluorescent protein tdTomato was cloned into pBluescript RN3P plasmid (Lemaire et al., 1995). As control, the ORF of tdTomato was cloned into pBluescript RN3P plasmid. Capped mRNAs were transcribed in vitro under the control of a T3 promoter with mMessage mMachine (Ambion). RNA was recovered using MEGAclear (Ambion), quantified by UV spectroscopy and analyzed by electrophoresis to confirm size. To overexpress LIN28 fusion protein, in vitro transcribed mRNAs were injected into zygotes.
Immunofluorescence staining and confocal microscopy
Oocytes and embryos were fixed in 4% paraformaldehyde for 20 minutes at 37°C. Following fixation, they were washed in PBS containing 3 mg/ml polyvinylpyrolidone (PBS/PVP), permeabilized for 30 minutes in PBS/PVP containing 0.25% Triton X-100 and blocked in blocking buffer (0.1% BSA, 0.01% Tween 20 and 2% donkey serum) prior to immunofluorescence staining (Nichols et al., 2009). For LIN28, a rabbit antibody (Cell Signaling) at 1:50 and AlexaFluor 488-conjugated anti-rabbit antibody (Invitrogen) at 1:50 were used. Specificity of the LIN28 antibody has been previously demonstrated (Aeckerle et al., 2012). For nucleophosmin (NPM), a mouse antibody (B23; Sigma) at 1:200 and AlexaFluor 594-conjugated (Invitrogen) or TRITC-conjugated (Millipore) anti-mouse antibodies at 1:200 were used. For HP1β, rat antibody (Abcam) at 1:100 and AlexaFluor 594-conjugated anti-rat antibody at 1:200 were used. Oocytes and embryos were incubated with primary antibodies (diluted in 2% BSA/PBS, without Mg2+ and without Ca2+) at 4°C overnight and with secondary antibodies for 1 hour at 37°C. Between incubations with antibodies, embryos were briefly rinsed and incubated in blocking buffer for an additional hour. Chromatin was counterstained with DAPI, DRAQ or propidium iodide before mounting embryos in CitiFluor (Electron Microscope Services, Munich, Germany) on poly-l-lysine-coated slides.
Colonies of mouse H2B-GFP ESC and NHP ESC cjes001 were fixed for 30 minutes in 4% paraformaldehyde followed by two washes with PBS. The cells were then permeabilized by incubating for 15 minutes with 0.1% Triton X-100 and then washed twice with PBS. Antibodies were diluted in PBS with 3% BSA. For LIN28, a rabbit antibody at 1:100 and AlexaFluor 488-conjugated (NHP ESC) or AlexaFluor 633-conjugated (H2B-GFP ESC) anti-rabbit antibodies at 1:200 were used. For nucleophosmin (NPM), a mouse antibody at 1:400 and AlexaFluor 594-conjugated or fluorescein-conjugated anti-mouse antibodies at 1:200 were used. Cells were incubated with primary and secondary antibodies for 1 hour at 37°C. Between antibody incubations, cells were washed twice with PBS. In case of NHP ESC cjes001, chromatin was additionally counterstained with DAPI in mounting media (Vectashield).
The following microscopes were used for imaging: PerkinElmer UltraView VoX spinning disc confocal microscope for DAPI staining; Zeiss LSM 5 Pascal confocal microscope for propidium iodide staining; Zeiss LSM 500 confocal microscope for imaging of H2B-GFP ESC and DRAQ staining. Images were further analyzed and processed using ImageJ software.
Statistical analysis
Statistical analysis was performed by the χ2 test with Yates’ correction.
RESULTS
LIN28 localizes to the nucleolus and its precursor body during mouse oogenesis and preimplantation development
To understand LIN28 function during early mouse development, we first determined its subcellular localization by immunofluorescence. In murine fully grown oocytes, germinal vesicles (GVs) exhibiting two different types of chromatin configurations can be distinguished: (1) GVs with non-condensed chromatin not surrounding the nucleolus [non-surrounded-nucleolus (NSN) oocyte, Fig. 1D]; and (2) GVs with condensed chromatin mostly surrounding the nucleolus [surrounded-nucleolus (SN) oocyte, Fig. 1H] (Debey et al., 1993; Zuccotti et al., 1995). In NSN oocytes, LIN28 localized at the periphery of nucleoli (Fig. 1A). Double staining with an antibody recognizing B23 (nucleophosmin, NPM), a protein used as a nucleolar marker and also involved in late rRNA processing, confirmed colocalization of LIN28 and NPM at the periphery of nucleoli (Fig. 1B,C). During transition from NSN to SN configuration, both LIN28 and NPM gradually disappeared from the surface of nucleoli and were only diffusely distributed in the nucleoplasm in SN oocytes (Fig. 1E-H).
Fig. 1.
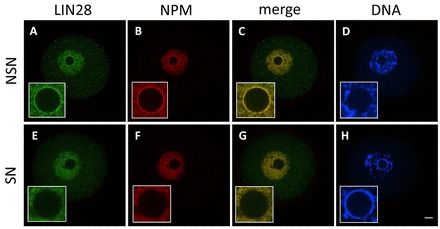
Cellular localization of LIN28 protein in fully grown mouse CD1 oocytes. Confocal immunofluorescence analysis of endogenous LIN28 in whole-mount oocytes. B23 (NPM) was used as a nucleolar marker. DNA was counterstained with DAPI. In NSN oocytes (A-D) LIN28 and NPM localize at the periphery of the nucleolus. During transition from NSN to SN configuration, both LIN28 and NPM gradually disappear from surface of the nucleolus and they are not detected there in SN oocytes (E-H). Insets show higher magnification of nucleus. Scale bar: 20 μm.
In zygotes, LIN28 remained in the nucleoplasm of both pronuclei (Fig. 2A). A weak LIN28 signal at the periphery of NPBs started to appear at the late 2-cell stage (Fig. 2E). Double staining with NPM confirmed its colocalization with LIN28 at NPBs (Fig. 2F,G). However, we observed LIN28 protein at NPBs of late 2-cell embryos, but never during earlier stages of this cell cycle (supplementary material Fig. S1Aa-d). Instead, LIN28 was diffusively distributed in the nucleoplasm with a weaker signal in the cytoplasm. Concomitantly, NPM, which colocalized with LIN28 at the periphery of NPBs in late 2-cell embryos, was enriched in the nucleoplasm in zygotes and in early 2-cell embryos (Fig. 2B; supplementary material Fig. S1Ab). The LIN28 signal increased in strength as it continued to be present at NPBs at the 4- and 8-cell stages (Fig. 2I-P) and in mature nucleoli at the morula and blastocyst stage (Fig. 2Q-X), where it was present in both TE and ICM cells (Fig. 2U-X; supplementary material Fig. S3). As ESCs are derived from the ICM of blastocyst embryos, we next determined the localization of LIN28 in a mouse ESC line expressing GFP fused with histone H2B. Double staining for LIN28 and NPM showed that they colocalize in nucleoli of these cells (supplementary material Fig. S1B).
Fig. 2.
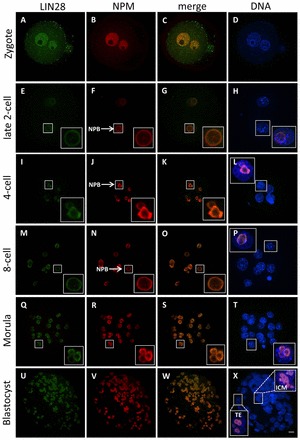
Cellular localization of LIN28 protein during mouse preimplantation development. Confocal immunofluorescence analysis of endogenous LIN28 in whole-mount embryos. B23 (NPM) was used as a nucleolar marker. DNA was counterstained with DAPI. An enlarged view of the boxed regions show that LIN28 (green) colocalizes with NPM (red). (A-D) In zygotes, homogenous nuclear signals of both LIN28 and NPM were observed. No enrichment at the periphery of nucleolar precursor bodies (NPBs) was observed for either marker. (E-P) At the early and mid 2-cell stage, LIN28 localizes at the periphery of nucleolar precursor bodies (NPBs) (arrow), where it colocalizes with NPM (E-H). LIN28 remains colocalized with NPM at NPBs (arrow) at the 4-cell (I-L) and 8-cell (M-P) stages. (Q-T) As NPBs have transformed to mature nucleoli in morula embryos, LIN28 remains nucleolar, as confirmed by the colocalization with NPM. (U-X) At the blastocyst stage, the localization of LIN28 in the nucleolus is concordant with NPM. LIN28 was present in both TE and ICM cells. Scale bar: 20 μm.
We also studied the subcellular localization of LIN28 by injecting mRNA coding for a LIN28-Tomato fusion protein into zygotes. Whereas control embryos injected with mRNA coding the red fluorescent protein tdTomato showed Tomato accumulation throughout the cytoplasm and in blastomeres’ nuclei at various cleavage stages (Fig. 3A-H), a weak LIN28-Tomato signal was observed in active NPBs at the late 2-cell stage for the first time, which is in accordance with the presence of native LIN28 demonstrated by immunofluorescence. Fig. 3M-P depicts the focal plane of one blastomere of a late 2-cell embryo. It clearly shows a peripheral NPM signal in one blastomere (Fig. 3N, arrow), indicative of an actively transcribing NPB, which is in strong contrast to the nucleoplasmic distribution in the second blastomere (Fig. 3M′). Only the NPM-positive nucleolar precursor body shows an accumulation of the LIN28-Tomato signal (Fig. 3O, arrow) suggesting a link between the recruitment of the rRNA processing factor NPM and LIN28 accumulation at active NPBs in late 2-cell embryos. This is further supported by the immunofluorescence analysis described above, showing that LIN28 only localizes to the periphery of those NPBs that exhibit presence of NPM protein. Before LIN28-Tomato appeared in the NPB of late 2-cell embryos, distinct cytoplasmic foci formed around the nucleus (Fig. 3I, arrows). As development proceeded, LIN28-Tomato became enriched in nucleoli in early morula embryos (Fig. 3U-X), as confirmed by the colocalization with NPM.
Fig. 3.
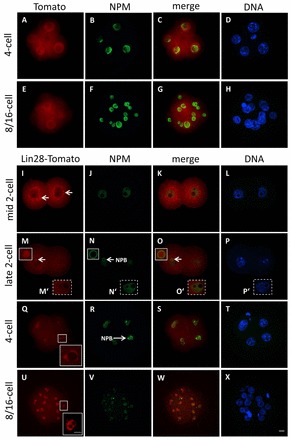
Cellular localization of a LIN28-Tomato fusion protein in early mouse embryos. Confocal immunofluorescence analysis of LIN28-Tomato fusion protein in whole-mount embryos following microinjection of mRNA, in vitro culture and fixation. B23 (NPM) was used as a nucleolar marker. DNA was counterstained with DAPI. As control, zygotes were microinjected with mRNA coding only for tdTomato protein. (A-H) Tomato expression is throughout the cytoplasm and nuclei in 4-cell embryos (A-D) and in 8/16 embryos (E-H). (I-L) At the 2-cell stage, the LIN28-Tomato fusion protein signal appears at distinct cytoplasmic foci that are formed around the nucleus (arrows). NPM staining is restricted to the nucleoplasm. (M-P) Focal plane of one blastomere of a late 2-cell embryo, where LIN28-Tomato localizes to one NPB, which is stained by NPM at the periphery (arrows). (M′-P′) Focal plane of second blastomere of the same late 2-cell embryo, where LIN28-Tomato signal is absent at presumptive NPBs, which lack NPM staining at periphery. (Q-T) At the 4-cell stage, LIN28-Tomato colocalizes with NPM at NPBs (an enlarged view of the LIN28-Tomato signal is shown in the boxed region). (U-X) In 8/16 embryos, LIN28-Tomato associates with mature, differentiated nucleoli. Scale bars: 20 μm in A-X; 10 μm in inset in U.
Lin28 mRNA is present in the oocyte and early mouse embryo
Using immunofluorescence we could detect the first clear localization of nucleolar LIN28 protein in blastomeres’ nuclei at the late 2-cell stage. However, in zygotes and in early 2-cell embryos we observed nuclear (but not nucleolar) signal that was significantly higher than the cytoplasmic signal. We reasoned that an early pool of Lin28 mRNA might exist before the late 2-cell stage. Therefore, we determined the presence of Lin28 mRNA in oocytes and early embryos using quantitative RT-PCR (qRT-PCR) (Fig. 4). For this purpose, embryos were obtained from superovulated F1(C57BL/6xCBA/Tar) females mated with F1 males and the timing of each cell cycle progression was estimated based on the time that passed from hCG injection (in hours post hCG). Such precise timing for this particular strain of mice has been established previously (Teperek-Tkacz et al., 2010) and confirmed (Meglicki et al., 2012). To exclude possible artifacts resulting from using a different mouse strain or a different method of obtaining embryos, we first determined whether localization of LIN28 in embryos derived from superovulated F1 and spontaneously ovulated CD1 mice was similar. The immunofluorescence analysis revealed that there is no difference in the localization of LIN28 in both strains of mice, irrespective of whether the embryos originated from spontaneous or induced ovulation (compare Fig. 2 and supplementary material Fig. S2). Additionally, we showed that dissociation of LIN28 and NPM during transition from NSN to SN configuration in fully grown oocytes occurs in F1 oocytes as well (compare Fig. 1 and supplementary material Fig. S2). Next, we performed qRT-PCR with normalization to an exogenous standard (cyc3GFP), which was added to each sample as in vitro synthesized and polyadenylated cycGfp mRNA, prior to mRNA isolation from the samples. Normalization using exogenous standard gives information about mRNA isolation and RT reaction efficiency and removes errors resulting from changes in mRNA level of endogenous standard. We found that Lin28 is abundantly present in MII arrested oocytes, 1-cell and early 2-cell embryos (Fig. 4), indicating that Lin28 mRNA must have a maternal origin at the beginning of mouse embryo development. Lin28 mRNA levels decrease and reach the minimum at late S phase of 2-cell stage. This suggests maternal mRNA degradation. Subsequently, it is followed by a gradual increase starting at G2 of the 4-cell stage. It should be noted that owing to normalization with standard these results show the level of mRNA per embryo and not per cell or nucleus. The data presented here provides evidence that (1) maternal Lin28 mRNA exists and (2) Lin28 mRNA levels change dynamically during the first embryonic cleavage stages.
Fig. 4.
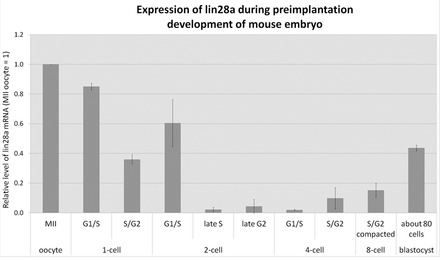
Expression of Lin28 mRNA during mouse preimplantation development. High levels of Lin28 are detectable in MII-arrested oocytes. As development proceeds, Lin28 mRNA levels start to decrease in zygotes reaching low levels at late G2 of the 2-cell stage. An increase of Lin28 expression is detected again at the late 4-cell stage and subsequent cleavage stages.
LIN28 localizes to the nucleolus in non-human primate (NHP) preimplantation embryos, but is predominantly cytoplasmic in NHP ESCs
The presence of LIN28 protein in nucleoli of mouse preimplantation embryos raised the question of whether the nucleolar localization of LIN28 could be extended to other mammalian species. Therefore, we obtained different cleavage-staged embryos from a non-human primate (NHP) model, the common marmoset monkey (Callithrix jacchus), as previously described (Müller et al., 2009), and performed immunofluorescence. LIN28 was enriched in the nucleoplasm in 11-cell embryos (Fig. 5A). By contrast, NPM was already present at the periphery of presumptive nucleolar precursor bodies (Fig. 5B). At the morula stage, LIN28 appeared for the first time in nucleoli (supplementary material Fig. S4A). However, a more detailed analysis of different z-sections revealed that LIN28 localized only to the nucleoli of outer blastomeres (supplementary material Fig. S4B, Z=13+61, arrows), whereas it was absent in inner blastomeres (supplementary material Fig. S4B, Z=49). It has been shown that inner blastomeres are precursors of the ICM and outer blastomeres are the precursors of the TE (reviewed by Johnson and McConnell, 2004; Yamanaka et al., 2006). The continued presence of nucleolar LIN28 in TE cells (Fig. 5E,G) and its lack in ICM cells (Fig. 5E,G), confirmed the commitment of outer and inner precursors to these first two lineages at the blastocyst stage. To distinguish the TE and ICM, different z-sections of the boxed region of the full projection (Fig. 5H) are displayed in supplementary material Fig. S5. Whereas the z-section showing ICM cells contains only NPM-positive nucleoli (Z=14), the z-section showing TE cells contain both NPM- and LIN28-positive nucleoli (Z=34).
Fig. 5.
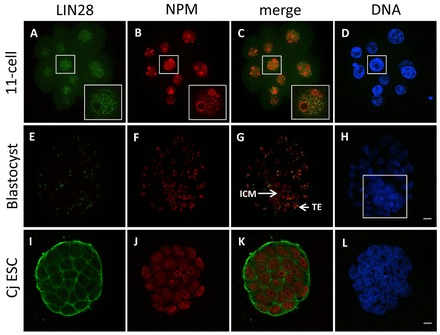
Cellular localization of LIN28 protein in early embryos and ESCs of the common marmoset monkey. Confocal immunofluorescence analysis of endogenous LIN28. B23 (NPM) was used as a nucleolar marker. DNA was counterstained with DAPI. (A-D) In 11-cell embryos, LIN28 is enriched in the nucleoplasm and not at the periphery of NPBs, whereas NPM is already present at the periphery of NPBs. An enlarged view of the boxed region shows that LIN28 (green) does not colocalize with NPM (red). (E-H) In blastocyst embryos, LIN28 colocalizes with NPM at the nucleolus, but only in trophoblast (TE) and not ICM cells. ICM cells only show a NPM signal (red). Boxed region in H shows full projection of ICM and TE cells, which are displayed as z-sections in supplementary material Fig. S5. (I-L) In ESCs, LIN28 is predominantly cytoplasmic and absent from the nucleolus. Scale bars: 20 μm.
Next, we examined the localization of LIN28 in an NHP ESC line (Müller et al., 2009). NPM clearly stained the nucleoli of ESCs (Fig. 5J). However, LIN28 is predominantly localized to the cytoplasmic region (Fig. 5I). We conclude that nucleolar LIN28 is also expressed in other mammalian species, e.g. the common marmoset monkey, during preimplantation development, but appears to be distributed differently than in mouse embryos.
Depletion of LIN28 from zygotes affects mouse development at the transition of the 2-cell stage to the 4-cell stage
The presence of an early pool of Lin28 mRNA in oocytes and zygotes raised the possibility of an early developmental function. Therefore, we investigated the role of LIN28 during early embryonic development using MO-mediated gene knockdown, which has been established as an efficient method for highly specific gene knockdown in mouse zygotes (Foygel et al., 2008; Jedrusik et al., 2010).
We found that following microinjection of 1 mM Lin28A-MO into zygotes, the rate of developmental arrest at the transition of the 2-cell to the 4-cell stage was dramatically increased in comparison with zygotes injected with control morpholino (Control-MO) (Fig. 6A,B). After 48 hours of culture, 59.5±3.0% of embryos injected with Lin28A-MO and only 16.7±1.9% of embryos injected with Control-MO (P<0.001, Fig. 6A,B) arrested at the 2-cell stage. None of the Lin28A-MO-injected embryos developed to morulae or blastocysts after 96 hours of culture, compared with the respective rates of 61.5±8.8% and 38.5±7.9% in Control-MO embryos (Fig. 6A,B). We confirmed the efficacy of the knockdown by immunofluorescence of morpholino-injected embryos. LIN28 protein accumulation was indeed reduced in Lin28A-MO-injected embryos at the 4-cell stage compared with Control-MO-injected and uninjected embryos, which show LIN28 protein localized to the nucleolus (Fig. 6C).
Fig. 6.
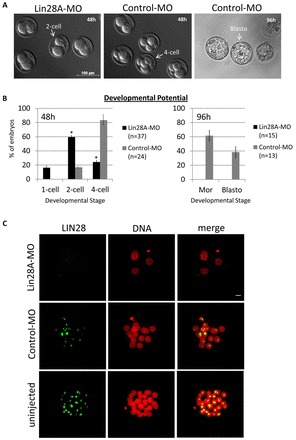
LIN28 depletion after fertilization results in a developmental arrest. (A,B) A significant number of Lin28A-MO-injected embryos arrest at the 2- or 4-cell stage after 48 hours of in vitro culture in comparison with controls (P<0.001). Lin28A-MO-injected embryos never develop to morula or blastocyst stage after 96 hours of in vitro culture, unlike the majority of control embryos. Error bars represent mean ± s.e.m. from at least three independent sets of experiments. (C) Immunostaining confirmed that nucleolar LIN28 expression is absent in Lin28A-MO-injected embryos, arrested at the 4-cell stage, but present in Control-MO-injected and uninjected embryos. DNA was counterstained with propidium iodide. Scale bar: 20 μm.
To gain more insight into the underlying reason for such a prominent developmental arrest at the 2-cell stage upon LIN28 depletion, we first analyzed whether active or functional NPBs in arrested embryos were assembled. Our previous observations that the timing of LIN28 accumulation at NPBs is correlated with the time (at the late 2-cell stage) at which functional NPB assembly is initiated (Fig. 2E-H; Fig. 3I-P) suggested a link between these two processes. We therefore reasoned that in the absence of functional LIN28, activation of NPBs in 2-cell embryos might be impaired. Our analysis revealed that, indeed, functional NPBs failed to assemble in all analyzed 2-cell-arrested embryos (n=15). In all of them, NPM staining remained restricted to the nucleoplasm (Fig. 7Ad-f). Furthermore, we assessed the nuclear organization in arrested embryos. Heterochromatin, brightly stained by the DNA-specific dye DAPI (Fig. 7A, arrow) and enriched in heterochromatin protein 1β (HP1β), detaches from the surface of NPBs and forms chromocenters in mouse late 2-cell embryos (Martin et al., 2006). We did not observe any abnormalities in chromocenter formation in LIN28-depleted embryos arrested at the 2-cell stage. The numbers of heterochromatic foci not associated and associated with presumptive NPB regions were similar in LIN28-depleted (Fig. 7Aa-c, arrow) and control embryos (Fig. 7Ag,h, arrow). The chromocenters were strongly enriched with HP1β protein (Fig. 7Ba-c, arrow). The results of LIN28 knockdown provide further evidence that its timely accumulation at NPBs is required for nucleologenesis and normal early mouse development.
Fig. 7.
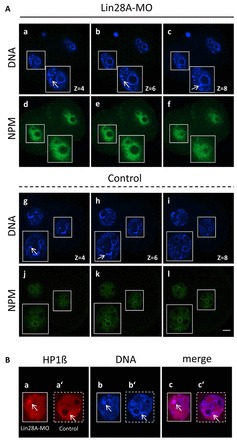
NPM expression and chromocenter formation following LIN28 depletion. Confocal immunofluorescence analysis of NPM (green) and HP1 (red) in whole-mount embryos. DNA was counterstained with DAPI. (Ad-f) Different optical slices illustrate that NPM is distributed throughout the nucleoplasm and is not enriched at the periphery of presumptive NPBs in 2-cell arrested embryos. (Aj-l) In control late 2-cell embryos, NPM is enriched at the periphery of NPBs. (Aa-c,g-i) Heterochromatin and chromocenter clustering is brightly stained by DAPI (arrow). Heterochromatic foci are outside (Aa,b,g) or in association (Ac,h) with NPBs (arrow). (Ba-c′) Chromocenters were strongly enriched in HP1. An enlarged view of a single optical slice shows that HP1 (red) colocalizes with condensed DNA (arrow) in 2-cell arrested (Ba-c) and control (Ba′-c′) embryos. Scale bar: 20 μm.
DISCUSSION
The conversion of mouse fibroblasts to pluripotent cells (iPS) by four key factors, namely OCT4, SOX2, KLF4 and MYC, opened the most recent chapter in the history of nuclear reprogramming (Takahashi and Yamanaka, 2006). Human iPS cells were obtained by inducing overexpression of the same (Takahashi et al., 2007) or a different combination of factors (Yu et al., 2007). One of these new reprogramming factors in the latter publication was LIN28. Whereas OCT4 and SOX2 are reported to play important roles during mouse preimplantation development (Avilion et al., 2003; Foygel et al., 2008; Keramari et al., 2010), the function of LIN28 is unknown in early mouse embryos.
In this study, we show that LIN28 localizes to the nucleolar precursor body (NPB) and to fully matured nucleoli in the early mouse embryo as revealed by immunostaining and expression of a LIN28-Tomato fusion protein. Nucleolar accumulation during mammalian preimplantation development appears to be a general feature of LIN28, as we also observed nucleolar LIN28 in embryos of other mammalian species, such as the common marmoset monkey and pig (porcine data not shown). Nucleologenesis in cleaving mammalian embryos involves the morphogenic transformation of the NPB to a fully functional nucleolus (Fléchon and Kopecný, 1998). Our results are in accordance with a previous report that rRNA processing factors such as B23/NPM are recruited to the periphery of NPBs, which are active for rDNA transcription, at the late 2-cell stage (Zatsepina et al., 2003). Remarkable, however, is our observation that LIN28 accumulation at the nucleolar precursor occurs concomitantly with the recruitment of NPM. Inactive NPBs not associated with peripheral NPM are also devoid of LIN28. We therefore hypothesized a link between LIN28 accumulation and the formation of active NPBs at the late 2-cell stage. To test this hypothesis, we knocked down LIN28 expression using specific morpholino oligonucleotides. The absence of NPM staining at the periphery of NPBs in arrested 2-cell embryos following Lin28 morpholino injection strongly supports the hypothesis that LIN28 contributes to the formation of active nucleolar precursors at the late 2-cell stage.
Previous studies demonstrated that the rapid knockdown of both maternal and embryonic transcripts using the morpholino approach revealed much earlier and more severe phenotypes in comparison with knockout studies of the same gene (Foygel et al., 2008; Jedrusik et al., 2010; Nichols et al., 1998; Strumpf et al., 2005). The developmental arrest following morpholino-mediated knockdown together with evidence about the presence of maternal Lin28 mRNA (Fig. 4) and the protein (Fig. 1) in fully grown oocytes as well as in the pronuclei of zygotes (Fig. 2) suggest that this early pool of Lin28 is required for normal development at a very early stage, i.e. is specific to the maternal-embryonic transition, which takes place at the late 2-cell stage in the mouse (Minami et al., 2007). In that context, it is worth noting that Lin28 transcript levels decline significantly before this major wave of EGA, whereas LIN28 protein localizes to NPBs for the first time during/after the major wave of EGA. Because LIN28 is deposited in the nucleoplasm of zygotes and early 2-cell embryos, we envision a translocation of the maternal protein from the nucleoplasm to the periphery of active NPBs at the late 2-cell stage. As functional nucleoli are assembled when the embryonic genome has been activated (Kopecny, 1989; Svarcova et al., 2009), the transformation of NPBs to mature nucleoli also reflects the transition from the maternal to embryonic control. In contrast to mice, EGA in non-human primate embryos occurs between the 8- and 16-cell stages (Niu et al., 2008). It is therefore noteworthy that LIN28 appeared at nucleoli of marmoset embryos following EGA, whereas it was still enriched in the nucleoplasm at the 11-cell stage. Thus, our results suggest a strong functional relationship between the spatial expression of LIN28 and EGA during early embryonic development.
RNA transcript profiling during EGA identified upregulated genes, which were categorized into functional networks (Zeng and Schultz, 2005). These over-represented networks included rRNA processing, ribosome biogenesis and protein synthesis, which are directly or indirectly related to nucleolar function. Previous studies in knockout mice or knockdown embryos of genes involved in rRNA processing and ribosome biogenesis demonstrated their importance during early embryonic development at either the post- or preimplantation stage. Knockout of the gene encoding the rRNA-processing factor NPM1 resulted in embryonic lethality between embryonic day (E) 11.5 and E12.5 (Grisendi et al., 2005). Phenotypes of preimplantation lethality have been reported for the two rRNA processing factors EMG1 (essential for mitotic growth) and RBM19 (RNA-binding motif protein 19) (Wu et al., 2010; Zhang et al., 2008). Similar preimplantation arrests were also observed for genes involved in ribosome biogenesis, e.g. Pescadillo, fibrillarin and SURF6 (Lerch-Gaggl et al., 2002; Newton et al., 2003; Romanova et al., 2006). These studies clearly show that the absence of functional nucleoli or nucleolus-associated factors grossly hampers development of mammalian embryos after EGA.
The requirement of a functional nucleolus for proper activation of the embryonic genome has been previously demonstrated when development of fertilized, enucleolated mouse oocytes ceased after one or two cleavage divisions (Ogushi et al., 2008). Re-injection of nucleoli into enucleolated oocytes could rescue the phenotype with embryos developing normally and giving rise to healthy offspring (Ogushi et al., 2008). It is not known which molecular factors are contributed by the nucleolar material to support EGA and proper embryonic development. Interestingly, nucleoli originating from a somatic cell or an ESC could not substitute for the original oocyte nucleolar material (Ogushi et al., 2008), suggesting significant differences in the origin of the nucleolar material. The absence of LIN28 in the nucleoli of two human somatic cell lines (our unpublished results) and NHP ESCs supports the view that the nucleolar material differs among species (mouse versus marmoset) and cell type (embryonic versus differentiated somatic cell) with respect to LIN28 content.
In conclusion, we propose a novel and essential role for the pluripotency factor LIN28 in the mammalian nucleolus. Formation of nucleoli via its precursor body appears to require LIN28 to successfully pass the maternal-embryonic transition.
Supplementary Material
Acknowledgments
We thank Dr M. Zernicka-Goetz (Gurdon Institute) for providing the pRN3P vector; Drs A. K. Hadjantonakis (Memorial Sloan-Kettering Cancer Center) and M. A. Ciemerych-Litwinienko (University of Warsaw) for the H2B-GFP mouse ESC line; Dr C. Drummer for embryo retrieval from common marmoset monkeys; and Dr K. Debowski for construct design of LIN28 fusion protein.
Footnotes
Funding
Funding was provided by the German Primate Center, which is a Leibniz Institute financed by the Bundesrepublik Deutschland and the Bundesländer. Deposited in PMC for immediate release.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.083279/-/DC1
References
- Aeckerle N., Eildermann K., Drummer C., Ehmcke J., Schweyer S., Lerchl A., Bergmann M., Kliesch S., Gromoll J., Schlatt S., et al. (2012). The pluripotency factor LIN28 in monkey and human testes: a marker for spermatogonial stem cells? Mol. Hum. Reprod. 18, 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003). Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer E., Moss E. G. (2007). Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 4, 16–25 [DOI] [PubMed] [Google Scholar]
- Boisvert F. M., van Koningsbruggen S., Navascués J., Lamond A. I. (2007). The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 [DOI] [PubMed] [Google Scholar]
- Darr H., Benvenisty N. (2009). Genetic analysis of the role of the reprogramming gene LIN-28 in human embryonic stem cells. Stem Cells 27, 352–362 [DOI] [PubMed] [Google Scholar]
- Debey P., Szöllösi M. S., Szöllösi D., Vautier D., Girousse A., Besombes D. (1993). Competent mouse oocytes isolated from antral follicles exhibit different chromatin organization and follow different maturation dynamics. Mol. Reprod. Dev. 36, 59–74 [DOI] [PubMed] [Google Scholar]
- Fléchon J. E., Kopecný V. (1998). The nature of the ‘nucleolus precursor body’ in early preimplantation embryos: a review of fine-structure cytochemical, immunocytochemical and autoradiographic data related to nucleolar function. Zygote 6, 183–191 [DOI] [PubMed] [Google Scholar]
- Foygel K., Choi B., Jun S., Leong D. E., Lee A., Wong C. C., Zuo E., Eckart M., Reijo Pera R. A., Wong W. H., et al. (2008). A novel and critical role for Oct4 as a regulator of the maternal-embryonic transition. PLoS ONE 3, e4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbus M. S., Calarco P. G., Epstein C. J. (1973). The effects of inhibitors of RNA synthesis (alpha-amanitin and actinomycin D) on preimplantation mouse embryogenesis. J. Exp. Zool. 186, 207–216 [DOI] [PubMed] [Google Scholar]
- Grisendi S., Bernardi R., Rossi M., Cheng K., Khandker L., Manova K., Pandolfi P. P. (2005). Role of nucleophosmin in embryonic development and tumorigenesis. Nature 437, 147–153 [DOI] [PubMed] [Google Scholar]
- Hadjantonakis A. K., Papaioannou V. E. (2004). Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 4, 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanazawa K., Mueller T., Becker T., Heistermann M., Behr R., Sasaki E. (2012). Minimally invasive transabdominal collection of preimplantation embryos from the common marmoset monkey (Callithrix jacchus). Theriogenology 78, 811–816 [DOI] [PubMed] [Google Scholar]
- Jedrusik A., Bruce A. W., Tan M. H., Leong D. E., Skamagki M., Yao M., Zernicka-Goetz M. (2010). Maternally and zygotically provided Cdx2 have novel and critical roles for early development of the mouse embryo. Dev. Biol. 344, 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. H., McConnell J. M. (2004). Lineage allocation and cell polarity during mouse embryogenesis. Semin. Cell Dev. Biol. 15, 583–597 [DOI] [PubMed] [Google Scholar]
- Keramari M., Razavi J., Ingman K. A., Patsch C., Edenhofer F., Ward C. M., Kimber S. J. (2010). Sox2 is essential for formation of trophectoderm in the preimplantation embryo. PLoS ONE 5, e13952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecny V. (1989). High-resolution autoradiographic studies of comparative nucleologenesis and genome reactivation during early embryogenesis in pig, man and cattle. Reprod. Nutr. Dev. 29, 589–600 [DOI] [PubMed] [Google Scholar]
- Latham K. E., Garrels J. I., Chang C., Solter D. (1991). Quantitative analysis of protein synthesis in mouse embryos. I. Extensive reprogramming at the one- and two-cell stages. Development 112, 921–932 [DOI] [PubMed] [Google Scholar]
- Lemaire P., Garrett N., Gurdon J. B. (1995). Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell 81, 85–94 [DOI] [PubMed] [Google Scholar]
- Lerch-Gaggl A., Haque J., Li J., Ning G., Traktman P., Duncan S. A. (2002). Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J. Biol. Chem. 277, 45347–45355 [DOI] [PubMed] [Google Scholar]
- Martin C., Beaujean N., Brochard V., Audouard C., Zink D., Debey P. (2006). Genome restructuring in mouse embryos during reprogramming and early development. Dev. Biol. 292, 317–332 [DOI] [PubMed] [Google Scholar]
- Meglicki M., Teperek-Tkacz M., Borsuk E. (2012). Appearance and heterochromatin localization of HP1α in early mouse embryos depends on cytoplasmic clock and H3S10 phosphorylation. Cell Cycle 11, 2189–2205 [DOI] [PubMed] [Google Scholar]
- Minami N., Suzuki T., Tsukamoto S. (2007). Zygotic gene activation and maternal factors in mammals. J. Reprod. Dev. 53, 707–715 [DOI] [PubMed] [Google Scholar]
- Moss E. G., Tang L. (2003). Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 258, 432–442 [DOI] [PubMed] [Google Scholar]
- Moss E. G., Lee R. C., Ambros V. (1997). The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88, 637–646 [DOI] [PubMed] [Google Scholar]
- Müller T., Fleischmann G., Eildermann K., Mätz-Rensing K., Horn P. A., Sasaki E., Behr R. (2009). A novel embryonic stem cell line derived from the common marmoset monkey (Callithrix jacchus) exhibiting germ cell-like characteristics. Hum. Reprod. 24, 1359–1372 [DOI] [PubMed] [Google Scholar]
- Németh A., Längst G. (2011). Genome organization in and around the nucleolus. Trends Genet. 27, 149–156 [DOI] [PubMed] [Google Scholar]
- Newton K., Petfalski E., Tollervey D., Cáceres J. F. (2003). Fibrillarin is essential for early development and required for accumulation of an intron-encoded small nucleolar RNA in the mouse. Mol. Cell. Biol. 23, 8519–8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998). Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. (2009). Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136, 3215–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Yang S., Yu Y., Ding C., Yang J., Wang S., Ji S., He X., Xie Y., Tang X., et al. (2008). Impairments in embryonic genome activation in rhesus monkey somatic cell nuclear transfer embryos. Cloning Stem Cells 10, 25–36 [DOI] [PubMed] [Google Scholar]
- Ogushi S., Palmieri C., Fulka H., Saitou M., Miyano T., Fulka J., Jr (2008). The maternal nucleolus is essential for early embryonic development in mammals. Science 319, 613–616 [DOI] [PubMed] [Google Scholar]
- Romanova L. G., Anger M., Zatsepina O. V., Schultz R. M. (2006). Implication of nucleolar protein SURF6 in ribosome biogenesis and preimplantation mouse development. Biol. Reprod. 75, 690–696 [DOI] [PubMed] [Google Scholar]
- Schultz R. M. (2002). The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum. Reprod. Update 8, 323–331 [DOI] [PubMed] [Google Scholar]
- Strumpf D., Mao C. A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. (2005). Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102 [DOI] [PubMed] [Google Scholar]
- Svarcova O., Dinnyes A., Polgar Z., Bodo S., Adorjan M., Meng Q., Maddox-Hyttel P. (2009). Nucleolar re-activation is delayed in mouse embryos cloned from two different cell lines. Mol. Reprod. Dev. 76, 132–141 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- Teperek-Tkacz M., Meglicki M., Pasternak M., Kubiak J. Z., Borsuk E. (2010). Phosphorylation of histone H3 serine 10 in early mouse embryos: active phosphorylation at late S phase and differential effects of ZM447439 on first two embryonic mitoses. Cell Cycle 9, 4674–4687 [DOI] [PubMed] [Google Scholar]
- Wang S., Kou Z., Jing Z., Zhang Y., Guo X., Dong M., Wilmut I., Gao S. (2010). Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. USA 107, 17639–17644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Sandhu S., Patel N., Triggs-Raine B., Ding H. (2010). EMG1 is essential for mouse pre-implantation embryo development. BMC Dev. Biol. 10, 99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S., Blau H. M. (2010). Nuclear reprogramming to a pluripotent state by three approaches. Nature 465, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y., Ralston A., Stephenson R. O., Rossant J. (2006). Cell and molecular regulation of the mouse blastocyst. Dev. Dyn. 235, 2301–2314 [DOI] [PubMed] [Google Scholar]
- Yang D. H., Moss E. G. (2003). Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr. Patterns 3, 719–726 [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zatsepina O., Baly C., Chebrout M., Debey P. (2003). The step-wise assembly of a functional nucleolus in preimplantation mouse embryos involves the cajal (coiled) body. Dev. Biol. 253, 66–83 [DOI] [PubMed] [Google Scholar]
- Zeng F., Schultz R. M. (2005). RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev. Biol. 283, 40–57 [DOI] [PubMed] [Google Scholar]
- Zhang J., Tomasini A. J., Mayer A. N. (2008). RBM19 is essential for preimplantation development in the mouse. BMC Dev. Biol. 8, 115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccotti M., Piccinelli A., Giorgi Rossi P., Garagna S., Redi C. A. (1995). Chromatin organization during mouse oocyte growth. Mol. Reprod. Dev. 41, 479–485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


