Abstract
In recent years there has been a large expansion in our understanding of SIRT6 biology, including its structure, regulation, biochemical activity and biological roles. SIRT6 functions as an ADP-ribosylase and NAD+-dependent deacylase of both acetyl groups and long-chain fatty acyl groups. Through these functions SIRT6 impacts cellular homeostasis by regulating DNA repair, telomere maintenance and glucose and lipid metabolism, thus affecting diseases such diabetes, obesity, heart disease, and cancer. Such roles may contribute to overall longevity and health of the organism. Until recently, much of the known functions of SIRT6 were restricted to the chromatin. In this article, we seek to describe and expand this knowledge with recent advances in understanding the mechanisms of SIRT6 action and their implications for human biology and disease.
Keywords: Sirtuin 6, Histonese deacetylase, Cancer, DNA repair, Metabolism, Aging and Inflammation
Sirtuins
The sirtuins are a family of highly conserved, nicotinamide adenine dinucleotide (NAD)+-dependent enzymes that are central regulators of lifespan in lower organisms. The founding member of the sirtuin family, Sir2, was originally discovered in Saccharomyces cervisiae in a screen for silencing factors (silencing information regulator) [1, 2]. Sir2 promotes longevity by suppressing the formation of toxic extrachromosomal ribosomal DNA circles in yeast [3–6]. Additionally, Sir2 functions in a complex with other Sir proteins to repress transcription at the silent mating type loci [7, 8] and subtelomeric sequences [8, 9]. Such repressive functions are dependent on Sir2 histone deacetylase activity [10–12].
Mammals have seven sirtuins (SIRT1-7) [13, 14], with broad cellular functions including energy metabolism, cellular stress resistance, genomic stability, aging and tumorigenesis (reviewed in [15]). Each family member has distinct functions and subcellular localizations. SIRT6 and SIRT7 are found in the nucleus, SIRT3, SIRT4 and SIRT5 are found in the mitochondria and SIRT1 and SIRT2 have been found in both the nucleus and the cytosol [16]. Since their discovery, the field of sirtuin biology has exploded, demonstrating the importance and diversity of roles of this important class of proteins in human biology and disease.
In this review, we will focus on SIRT6 and its diverse enzymatic activities including NAD+-dependent deacetylation and mono ADP-ribosylation. We will discuss how these enzymatic activities impart SIRT6 with unique biological functions in genomic stability/DNA repair, inflammation and glucose/lipid metabolism, and finally we will relate these findings to how SIRT6 impacts organismal function and disease with respect to heart disease, diabetes, obesity, cancer and aging.
Mouse knockout models for all of the sirtuins have been used as tools for exploring sirtuin function. In this regard, SIRT6 deficient mice develop normally for the first two weeks, but then undergo several acute degenerative processes, dying at around one month of age. Defects observed in these mice are severe hypoglycemia, low levels of serum insulin growth factor receptor-1 (IGF-1), loss of subcutaneous fat, a curved spine and lymphopenia, resembling a progeroid-like syndrome [17]. At the cellular level, SIRT6 deficiency leads to a switch in glucose metabolism, as discussed in detail below, and marked genomic instability, with hypersensitivity to ionizing radiation, methylmethanesulfonate, and hydrogen peroxide, which are all cellular phenotypes consistent with potential defects in base excision repair (BER). SIRT6 thus promotes resistance to DNA damage and oxidative stress and suppresses genomic instability, while playing a role in metabolic homeostasis [17]. These studies provided the first insight into the diverse functions of SIRT6 and highlight the importance of SIRT6 in aging, metabolism and cancer.
SIRT6 is tightly bound to chromatin [16, 17] and is best characterized as a NAD+-dependent deacetylase of histone H3 lysine 9 (H3K9) [18] and H3 lysine 56 (H3K56) [19, 20] (Box 1). Histone deacetylation is associated with a closed chromatin conformation and decreased chromatin accessibility. Thus, the discovery of this enzymatic activity instigated a series of studies that demonstrated roles for SIRT6 in regulating telomeric chromatin, the dynamic binding of DNA repair factors to chromatin and gene expression.
Genomic Stability and DNA Repair
Telomere maintenance
Loss of SIRT6 leads to the formation of dysfunctional telomeres with stochastic replication-associated telomere sequence loss, accumulation of telomeric DNA damage foci, and genomic instability with chromosomal end-to-end fusions that help drive the cell into premature senescence. SIRT6-mediated deacetylation of telomeric H3K9 [18] and H3K56 residues [19] during S-phase is required for efficient association of the Werner syndrome (WRN) protein with telomeric chromatin [18] (Figure 1). The WRN protein is a RECQ-like helicase that plays a major role in genome stability, particularly during DNA replication and telomere metabolism [21]. WRN may be required for proper capping of telomeres by the telosome/shelterin complex as well as for replication of lagging telomeric DNA [22]; therefore, the genomic instability observed when SIRT6 is lost could partly be explained by the loss of association between WRN and chromatin [18].
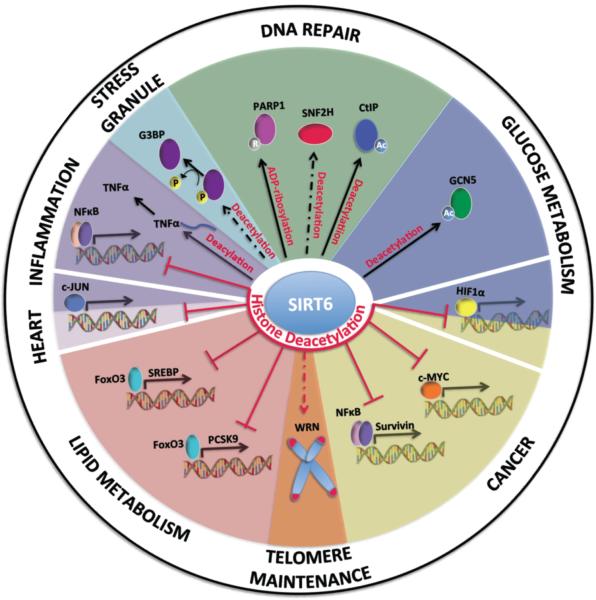
Figure 1. SIRT6 cellular functions and their impact on organismal biology and disease.
SIRT6 primarily functions as a H3K9 and H3K56 histone deacetylase that decreases chromatin accessibility for transcription factors such as nuclear factor kappa (NF-κB), c-JUN, sterol-regulatory element binding protein (SREBP), proprotein convertase subtilisin/kexin type 9 (PCSK9), Survivin, MYC and Hypoxia inducible factor 1α (HIF1α) to their respective promoters and thus inhibits expression of their target genes. SIRT6 can also regulate protein activity through direct deacetylation of GCN5 and CtIP and ADP-ribosylation of PARP1. SIRT6 has also been found to interact with SNF2H to enhance its recruitment to sites of damage thereby enhancing DSB repair. Additionally, SIRT6 associates with telomeric chromatin and deacetylates H3K9 and H3K56 leading to stabilization of WRN, which contributes to telomere maintenance. SIRT6 function has now been expanded to include cytosolic roles in stress granule formation through the promotion of G3BP dephosphorylation and TNF-α secretion by lysine deacylation. These cellular functions impact many aspects of health and disease such as DNA repair, telomere maintenance, stress granule formation, glucose/lipid metabolism, inflammation cardiac hypertrophy and cancer. Solid arrow, SIRT6 directly modifies the protein or directly affects histone deacetylation at the promoters of target genes. Dashed arrow, SIRT6 deacetylation activity is necessary, but is not direct. Red arrows, histone deacetylation. P (phosphorylation), Ac (acetylation) and R (ADP-ribosylation).
Base excision repair
From SIRT6 knockout mouse studies, SIRT6 was initially hypothesized to play a role in facilitating BER [17]. One mechanism may be simply that SIRT6 regulates chromatin to increase the accessibility of DNA to BER factors; another possibility is that lack of SIRT6 may increase levels of oxidative stress, given it's role in activating PARP1 following oxidative damage (see below); however such models awaits experimental confirmation.
Double-strand break repair
Evidence also points to a role for SIRT6 in double strand break (DSB) repair. Overexpression of SIRT6 improved the efficiency of non-homologous end joining (NHEJ) and homologous recombination (HR). Both deacetylation and mono-ADP-ribosylation activities of SIRT6 are required to stimulate DSB repair. More specifically, SIRT6 was found to interact with and ADP-ribosylate PARP1 and stimulate its poly-ADP-ribosylation activity [23] (Figure 1). Notably, the requirement for this SIRT6/PARP1 function in DSB repair was only seen upon oxidative stress, suggesting that the effect of SIRT6 on PARP1 may only play a role during DNA damage caused by oxidative stress [23]. As PARP1 is involved in both BER and DSB repair, the role of SIRT6 as an activator of PARP1 may also explain the BER defects observed in SIRT6 deficient cells.
Further evidence that SIRT6 is involved in DSB repair and more specifically HR is the finding that SIRT6 interacts with and deacetylates CtIP [C-terminal binding protein (CtBP) interacting protein] [24] (Figure 1). CtIP [25, 26] and BRCA1 [27] are responsible for DSB end resection, which results in single-stranded DNA (ssDNA). This ssDNA is bound by replication protein A (RPA), leading to the formation of a ssDNA-RAD51 nucleoprotein filament that mediates HR [28]. SIRT6 loss impaired the accumulation of RPA and ssDNA at DNA damage sites, reduced the rates of HR, and sensitized cells to DSB-inducing agents. SIRT6 thus promotes end resection and HR thereby improving genome stability [24]. Interestingly, CtIP is the first non-histone substrate discovered for SIRT6 deacetylase activity. The finding that SIRT6 can deacetylate cellular proteins has the potential to expand the substrates for SIRT6 and thus its biological roles.
SIRT6 is also interacts with the DNA–dependent protein kinase (DNA-PKc) holoenzyme macromolecular complex, which is comprised of repair factors such as DNA-PKcs and Ku70/80. This complex is known to promote DNA DSB repair by NHEJ in mammalian cells. In response to DSBs, the interaction between SIRT6 and chromatin is stabilized and global deacetylation of H3K9 is observed. Additionally, SIRT6 is required for the mobilization and stabilization of the DNA-PKs with chromatin in response to DSBs, however it remains unclear whether the latter requires SIRT6 enzymatic activity [29].
Finally, recent studies indicate that SIRT6 deacetylation of H3K56 and interaction with the ATP-dependent chromatin remodeler SNF2H accelerates the localization of SNF2H to sites of DSB damage and both of these events are required for efficient DSB repair [30] (Figure 1). Indeed, SNF2H increases local chromatin accessibility at DNA breaks and is required for proper DNA repair. Lack of SIRT6 profoundly impacts downstream recruitment of DNA repair factors both in vitro and in vivo, suggesting that both H3K56 deacetylation and SNF2H chromatin remodeling play critical roles in the DSB DNA repair pathway [30].
Overall, it is now clear that SIRT6 impacts genome integrity and DNA repair in multiple ways. Future studies will dissect how these different roles are coordinated and which of them are crucial for each type of DNA damage repair mechanism.
Metabolism
Glucose metabolism
SIRT6 is a central regulator of glucose homeostasis where SIRT6 function impacts both glycolysis and gluconeogenesis. As mentioned earlier, SIRT6 deficient mice exhibit severe hypoglycemia that ultimately leads to death by one month of age [17]. This was not due to defects in glucose absorption in the intestine or increased glucose secretion by the kidney. Strikingly, SIRT6 deficient mice were found to exhibit a pronounced increase in glucose uptake in both muscle and brown adipose tissue (BAT), potentially explaining the hypoglycemic phenotype of SIRT6-deficient mice. Furthermore, SIRT6-deficiency causes a specific and cell-autonomous increase in glucose uptake in multiple cell types in vivo and in vitro [31]. The increased uptake of glucose in the absence of SIRT6 correlated with an increase in membrane glucose transporter -1 (GLUT1) expression and enhanced glycolysis, whereas mitochondrial respiration was inhibited. SIRT6 was found to directly suppress expression of multiple glucose-metabolic genes [(pyruvate dehydrogenase kinase-1 (PDK1), lactate dehydrogenase (LDH), phosphofructokinase-1 (PFK1) and GLUT1] by interacting with hypoxia inducible factor-1α (HIF1α) and deacetylating H3K9 at the promoter of HIF1α target genes (Figure 1). HIF1α is known to modulate multiple genes in order to activate glycolysis and simultaneously repress mitochondrial respiration in a coordinated fashion. The importance of SIRT6-medited inhibition of HIF1α was validated in vivo where treatment of SIRT6-deficient mice with a HIF1α inhibitor rescued the hypoglycemic phenotype [31].
In addition to impacting glycolysis, Zhong et al. also found that expression of gluconeogenic genes were higher in SIRT6-deficient livers, suggesting that the liver was trying to compensate for the hypoglycemia. This observation was further investigated by Puigserver and colleagues, who found that SIRT6 plays a direct role in controlling gluconeogenesis in the liver [32]. SIRT6 controls gluconeogenesis through regulation of peroxisome proliferator-activated receptor-γ coactivator 1-α (PGC-1α), the main regulator of gluconeogenesis that stimulates hepatic gluconeogenesis in part by increasing the expression of gluconeogenic enzymes [33]. SIRT6 does this through its interaction with General Control Non-repressed Protein 5 (GCN5) (Figure 1), which enhances its activity. GCN5, in turn, is an acetyltransferase that catalyzes acetylation of PGC-1α. Acetylation of PGC-1α coincides with PGC-1α's relocalization away from the promoters of its gluconeogenic enzyme target genes, thus SIRT6 suppresses hepatic glucose production. In addition, hepatic SIRT6 was reduced in obese/diabetic mice, and expression of SIRT6 normalized blood glucose levels. Overall, these findings point to another non-histone substrate for SIRT6, namely GCN5, and have uncovered a SIRT6-mediated pathway for the control of PGC-1α activity and hepatic glucose production [32]. Interestingly, another study demonstrated that SIRT6 overexpression reduces gluconeogenic gene expression in the liver of wild type but not liver-specific Forkhead box O1 (FoxO1)/3/4 knockout mice [34]. Thus, it is possible that SIRT6 modulates hepatic gluconeogenesis through both PGC-1α and FoxO1, however it remains to be determined whether those functions are inter-dependent.
Lipid metabolism
An important finding is that the absence of SIRT6 results in accumulation of triglycerides (TG), which is associated with fatty liver disease or hepatic steatosis. SIRT6 deficiency resulted in increased expression of genes responsible for hepatic long-chain fatty acid uptake and reduced expression of genes for β-oxidation. SIRT6 deficiency also increased expression of several genes involved in multiple steps of TG synthesis. These data suggest that SIRT6 serves as a negative regulator of TG synthesis [35]. Additionally, SIRT6-overexpressing mice fed a high-fat diet were found to exhibit a decrease in visceral fat accumulation, improved blood lipid profile, glucose tolerance, and insulin secretion, and reduced expression of selected PPARγ-regulated genes which are involved in lipid metabolism, lipid transport and adipogenesis, thus dramatically affecting lipid homeostasis [36]. The role of SIRT6 histone deacetylase activity in this mouse model, however, remains to be clarified.
Additionally, SIRT6 was found to play an important role in the protective effects of rosiglitazone (RGZ), a thiazolidinedione that acts as an agonist of PPARγ and is a treatment for hepatic steatosis [37]. RGZ treatment ameliorated hepatic lipid accumulation and increased expression of SIRT6, PGC-1α and FoxO1 and phosphorylation levels and thus activation of liver kinase B1 (LKB1) and 5' adenosine monophosphate- activated protein kinase (AMPK) in rat livers. LKB1 is an upstream activating kinase of AMPK and they play a key role in the regulation of lipid metabolism by controlling fatty acid oxidation and synthesis. The knockdown of SIRT6 aggravated hepatocyte fat accumulation as shown by increased TG levels, and suppressed the favorable effects of RGZ on hepatocyte steatosis. At the molecular level, SIRT6 knockdown suppressed mRNA and protein expression of PGC-1α and FoxO1, and phosphorylation levels of LBK1 and AMPK. These results suggest that RGZ may act as a SIRT6 activator and a potential therapeutic strategy for fatty liver disease [37]. It is unclear why SIRT6 would on one hand increase the mRNA and protein expression of PGC-1α [37] and on the other hand inactivate PGC-1α through the enhanced activity of GCN5 [32]. Further work will have to be done to determine the predominant SIRT6 function in the different model systems used.
SIRT6 was also recently described as a critical histone deacetylase in the regulation of expression of the proprotein convertase subtilisin/kexin type 9 (PCSK9) gene and low density lipoprotein (LDL)-cholesterol homeostasis [38]. PCSK9 targets LDL receptor for degradation and the PCSK9 gene knockout in mice dramatically reduces LDL-cholesterol [39]. Hepatic knockout of SIRT6 led to elevated LDL-cholesterol levels and increased PCSK9 gene expression. FoxO3 also regulates the PCSK9 gene and SIRT6 and FoxO3 liver-specific knockout mice show similarly elevated PCSK9 gene expression and LDL-cholesterol levels, suggesting they might operate as a pathway to regulate cholesterol homeostasis. Indeed, FoxO3 works to recruit SIRT6 to the PCSK9 gene promoter in order to suppress its gene expression through H3K9 and H3K56 deacetylation (Figure 1). Finally, the converse is true, where SIRT6 overexpression in high fat diet fed mice reduces PCSK9 gene expression and serum LDL-cholesterol, thus demonstrating the importance of SIRT6 in regulating LDL-cholesterol homeostasis [38].
Hepatic knockout of SIRT6 also caused elevated serum cholesterol levels compared to control mice. Interestingly, the sterol-regulatory element binding protein (SREBP), which is a key regulator of cholesterol biosynthesis, was increased in SIRT6-deficient livers [40]. SIRT6 was found to repress SREBP by at least three mechanisms. First, SIRT6 is recruited by FoxO3 to the SREBP gene promoter where it represses the transcription levels of SREBP and that of its target genes by deacetylating H3K9 [40, 41] and H3K56 [40] at SREBP promoter regions. Second, SIRT6 inhibits the cleavage of SREBP1/SREBP2 into their active forms by decreasing the transcription of the SREBP1/SREBP2 protease complex (SCAP, S1P, and S2P)[41]. Third, SIRT6 activates AMPK by increasing the AMP/ATP ratio; AMPK phosphorylates SREBP1 on Ser372 thus inactivating SREBP1 by suppressing its cleavage and nuclear translocation[41]. Reciprocally, the microRNAs miR-33a and miR-33b are generated from the introns of SREBP2 and SREBP1, respectively, and they repress expression of both SREBP itself and SIRT6 [41, 42] (Figure 2). Hepatic levels of SIRT6 mRNA and protein were decreased in diet-induced or genetically obese mice and both precursor and nuclear SREBP2 proteins were significantly decreased. Remarkably, SIRT6 or FoxO3 overexpression decreased serum and hepatic cholesterol levels thus improving hypercholesterolemia [40]. Taken together, these findings could explain why mice overexpressing SIRT6 have improved cholesterol homeostasis.
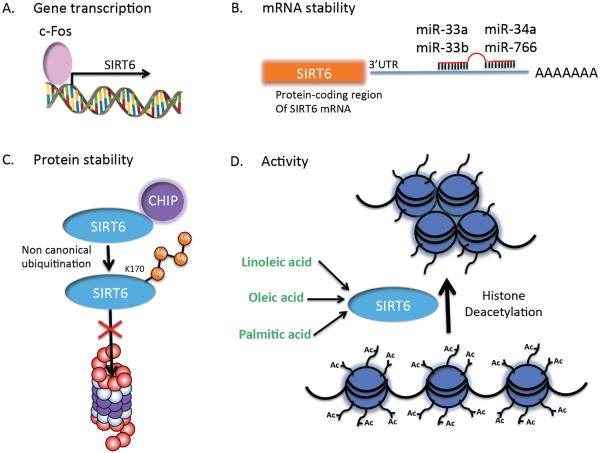
Figure 2. Mechanisms of SIRT6 regulation.
(A) c-FOS binds to an AP-1 binding site (TAAGTCA) at position −208 base pairs in the SIRT6 promoter and leads to c-FOS-dependent transactivation of SIRT6 gene expression. (B) Posttranscriptional regulation of SIRT6 by miR-33a, −33b, −34a and −766, which bind to the 3' untranslated region of the SIRT6 transcript leading to decreased SIRT6 expression. (C) Noncanonical ubiquitylation of SIRT6 at lysine 170 by the ubiquitin ligase CHIP leads to increased protein stability and reduced proteasome-mediated degradation of SIRT6. (D) SIRT6 histone deacetylase activity is enhanced upon the binding of FFA (oleic, linoleic and palmitic acid) leading to a closed chromatin conformation and decreased chromatin accessibility and thus gene transcription.
As described above in detail, many of the diverse functions carried out by SIRT6 are carried out by its deacetylation activity of H3K9 and H3K56 histone marks and different cellular proteins. This is interesting given that, originally, very low in vitro deacetylase activity was reported for SIRT6 [43–45]. Such weak deacetylase activity is suggested to be due to a splayed configuration between the Rossmann fold domain and the zinc-binding domain [44, 46]. However, recent studies indicate that FFAs can increase the in vitro deacetylase activity of SIRT6 basically to levels of other robust sirtuins [44] (Figure 2), indicating that the metabolism of lipids may dictate SIRT6 histone deacetylase activity in vivo, a model that remains to be tested (Box 2). In addition, SIRT6 was also found to hydrolyze long-chain fatty acyl groups, including myristoyl and palmitoyl groups, a process known as lysine deacylation [45]. In fact, long-chain deacylase activity was found to be an intrinsic activity of most sirtuins [44]. The hydrolysis of fatty acyl lysine modifications occurs more efficiently in vitro than deacetylation, at least when no FFAs are added to the reaction [45]. Acyl lysine modifications such as formylation, propionylation, butyrylation, crotonylation, malonylation, succinylation and myristoylation are posttranslational modifications that may occur to histones or other cellular proteins for which their removal could modify enzyme activity or chromatin regulation. In fact, the nutrient status of the cell may favor the addition of one acyl group over another and be dependent on the levels of acyl-CoA, which can be formed from intermediates of various metabolic pathways such as FA synthesis, β-oxidation, glycolysis and the tricarboxylic acid (TCA) cycle [43, 44]. Thus, the discovery of this novel substrate for SIRT6 suggests we are only grasping the tip of the iceberg on putative targets and biological roles for SIRT6, as well as the potential influence of metabolic states on SIRT6 activity, a point we will discuss later.
Inflammation
SIRT6 has both pro-inflammatory and anti-inflammatory roles depending on the context and cell type involved. The ability of SIRT6 to act as a lysine deacylase (as mentioned previously) has important pro-inflammatory consequences. More specifically, SIRT6 can catalyze the hydrolysis of myristoylated lysine 19 and 20 of tumor necrosis factor-α (TNF-α), which allows for TNF-α secretion from the cell [45] (Figure 1). TNF-α is a key pro-inflammatory cytokine that is known to play a major part in numerous inflammatory diseases. In vivo, TNF-α had lower lysine fatty acylation and was more efficiently secreted in SIRT6 wild type macrophages compared to SIRT6 knockout macrophages [45]. Notably, intracellular NAD+ concentration was also found to promote TNF mRNA translation efficiency by activated immune cells in a SIRT6-dependent manner, suggesting SIRT6 may regulate TNF ligand through multiple mechanisms [47].
Conversely, work by a separate group demonstrated that SIRT6 could also play an anti-inflammatory role by inhibiting TNF-α function downstream. TNF-α is known to activate nuclear factor kappaB (NF-κB), a potent proinflammatory cytokine. Work by Kawahara et al. demonstrated that SIRT6 is recruited to promoters of a subset of NF-κB target genes through a physical interaction with the NF-κB subunit RELA and functions as a corepressor of NF-κB, silencing NF-κB target genes through deacetylation of H3K9 at target gene promoters and decreasing NF-κB-dependent apoptosis and senescence [48]. Interestingly, RELA heterozygosity attenuates shortened life span and aging-related phenotypes of SIRT6 deficient mice. Together, these data suggest that excessive NF-κB-dependent gene expression contributes to the shortened life span and degenerative symptoms observed in SIRT6 deficient mice [48]. The group did not explore how these findings affect inflammation, but one could postulate that negative regulation of NF-κB is another immunosuppressive function of SIRT6. Other groups have found that SIRT6 and RELA indeed interact, however over expression of SIRT6 had no detectable effect on the nuclear/cytoplasmic ratio of RELA nor was the nuclear localization of RELA in response to TNF-α stimulation affected by overexpression of either wild type or catalytically inactive mutant SIRT6 [49]. These differences in findings could be explained by the fact that Grimley el al. used an overexpression system. The physiologic levels of SIRT6 expression could have already achieved the maximal effects of NF-κB subcellular localization and thus no further changes could be detected with SIRT6 overexpression. Additionally, the groups used different experimental readouts such as the functional effects of SIRT6 inactivation on chromatin and gene expression [48] versus an indirect readout of nuclear-cytoplasmic ratio of RELA [49]. Interestingly, SIRT6 did not affect NF-κB targets in knockout ES cells [31]. Further work will have to be done to resolve these seemingly disparate findings, which may result from differences in specific systems/cell types tested.
Further evidence of the anti-inflammatory role of SIRT6 was found in SIRT6 null 129/BlackSwiss/FVB mice that suffered from progressive and chronic inflammation of the liver leading to fibrosis. This phenotype was caused by SIRT6 deficiency in the lymphocytes and myeloid-derived cells and their subsequent aberrant activation. Mechanistically, SIRT6 interacts with c-JUN and deacetylates H3K9 at the promoter of proinflammatory genes such as monocyte chemotactic protein-1 (MCP-1), interleukin 6 (IL-6), and TNF-α. Thus, SIRT6 loss leads to hyperacetylation of H3K9 and increased occupancy of c-JUN at the promoter of these genes, leading to their elevated expression and enhanced production [50].
Heart disease
SIRT6 was found to play a role in the development of cardiac hypertrophy and failure. Decreased SIRT6 levels and activity were found in both human and mouse failing hearts [51, 52]. SIRT6 knockout mice spontaneously developed concentric cardiac hypertrophy at around 8–12 weeks of age, whereas SIRT6 overexpression blocks the cardiac hypertrophic response indicating that SIRT6 acts as a negative regulator of cardiac hypertrophy. At the cellular level, SIRT6 blocks IGF signaling by interacting with c-JUN and deacetylating H3K9 at IGF downstream targets. Inhibition of c-Jun or IGF signaling blocks hypertrophy of SIRT6-deficient hearts [51]. This suggests that SIRT6 activators could provide therapeutic benefits against cardiac hypertrophy.
Cancer
Proliferating tumor cells have unique metabolic requirements, characterized by enhanced cell-autonomous nutrient uptake and reorganization of metabolic pathways, supporting the biosynthesis of macromolecules needed for cell growth and division [53]. Enhanced glycolysis under aerobic conditions is the best-known example of metabolic reprogramming observed in cancer cells, as described decades ago by Otto Warburg (the Warburg Effect) [54]. As described in a previous section, loss of SIRT6 de-repressed HIF1α-dependent transactivation of glycolytic genes and led to enhanced uptake of glucose and increased glycolysis with concomitant inhibition of mitochondrial respiration [31]. This phenotype was reminiscent of aerobic glycolysis in tumor cells. Indeed, loss of SIRT6 in mouse embryonic fibroblasts (MEFs) led to tumor formation independent of oncogene activation and tumors exhibited enhanced aerobic glycolysis [55]. Strikingly, inhibition of glycolysis through PDK1 knockdown suppressed tumorigenesis in SIRT6 knockout cells. In addition to suppressing glycolysis through corepressing HIF1α, SIRT6 was also found to co-repress MYC transcriptional activity of ribosomal genes (Figure 1). Loss of SIRT6 in an in vivo model of colon cancer lead to a 3-fold increase in the number of adenomas when compared to control animals and these tumors exhibited enhanced glycolysis as measured by fluorodeoxyglucose - positron emission tomography (FDG-PET) and expression analysis of glycolytic genes. Importantly, pharmacological inhibition of PDK1 with dichloroacetate (DCA) specifically inhibited tumor formation in mice lacking SIRT6. Further, SIRT6 expression is downregulated in human pancreatic ductal adenocarcinoma and colorectal carcinomas compared to normal samples, and concomitantly, the expression of the HIF1α-target genes GLUT1, LDH, and PFK1 is significantly upregulated in these samples. Finally, levels of SIRT6 correlated with cancer progression and/or survival in colorectal cancer. Patients with low levels of nuclear SIRT6 had a shorter time to relapse and were more likely to relapse than those patients with high levels of nuclear SIRT6. Taken together, these results demonstrate that SIRT6 functions as a tumor suppressor, inhibiting the initiation and progression of colorectal cancer in vivo by repressing HIF1α and MYC thereby inhibiting aerobic glycolysis and ribosomal biogenesis gene expression respectively [55].
Recent studies have also shown that, in a mouse model specific for liver cancer initiation, survival of initiated cancer cells are controlled by c-JUN through suppression of c-FOS-induced SIRT6 transcription [56]. Mechanistically, c-FOS induces SIRT6 transcription (Figure 2), which represses survivin expression by reducing histone H3K9 acetylation and NF-κB activation (Figure 1). Importantly, increasing the level of SIRT6 or targeting the anti-apoptotic activity of survivin at the initiation stage markedly impairs cancer development. However, this pathway does not operate in advanced hepatocellular carcinomas [56]. These studies suggest that inhibiting this pathway may provide preventive strategies to treat premalignant liver lesions.
In addition to the above-mentioned malignancies, the expression levels of SIRT6 were also significantly down regulated at the RNA and protein level in head and neck cancer compared with noncancerous tissues. Of all the sirtuins that showed down regulation, only SIRT6 was not reduced in advanced stages compared to early stages, suggesting that reduced SIRT6 expression is an early event [57].
In contrast to the cancers already mentioned, the expression of SIRT6 seems to be up regulated in squamous cell carcinoma [57]. Recent studies demonstrate that microRNA-34a (miR-34a) expression is downregulated by promoter methylation in keratinocyte-derived SCC cell lines and tumors, whereas it is induced with differentiation. SIRT6 expression is downregulated by miR-34a and downregulation of miR-34a increases SIRT6 expression (Figure 2). Functionally, knockdown of SIRT6 was sufficient to reproduce the effects of increased miR-34a on differentiation in human keratinocytes as well as in squamous cell carcinoma cells [57].
Similarly, patients with chronic lymphocytic leukemia (CLL) show a 4-fold increase in SIRT6 expression compared to healthy volunteers. The study also found that higher SIRT6 expression values might be associated with poor prognosis [58]. Indeed, data from the cancer cell line encyclopedia (CCLE) shows that hematopoietic cancer lines are the only ones exhibiting gain of copies of SIRT6, instead of deletion of SIRT6, as observed in basically all other tumor types [55].
Taken together, these results demonstrate that in particular tumors, SIRT6 could act as an oncogene. One possibility could be that in these tumors, the effect of SIRT6 on metabolism may not be dominant, but rather overexpression of SIRT6 may offer protection against genomic instability. Conversely, it could be a matter of timing; perhaps SIRT6 expression is down regulated early in tumor formation leading to the increased genomic instability and metabolic reprogramming that increases the mutational rate and enhances biosynthesis of macromolecules needed for tumor cell proliferation. The increased rate of cell growth and genomic instability could allow subsequent mutations in oncogenes (e.g. KRAS), however at later stages, enhanced SIRT6 activity may protect against further mutagenesis, at stages where those could impact tumor growth in a negative way.
Lifespan
Sir2, the yeast homologue and founding member of the sirtuins, has been implicated in lifespan extension [59, 60]. Remarkably, in other model organisms (including C. elegans and Drosophila) Sir2 homologues are also implicated as a determinant of lifespan [61]. In both yeast and flies, Sir2 levels increased following caloric-restriction treatment [61]. Conversely, sirtuin-deficient yeast and mice abolished the extension of lifespan by caloric restriction [62, 63]. Notably, these studies were challenged by others indicating no effect or minor effects for sirtuins in lifespan extension in these organisms [64, 65]. Such discrepancies may indicate specific differences in protocols, levels of expression of the manipulated genes or strain differences. Undoubtedly, further work will be required to resolve this controversy. Although correlative data implies a beneficial effect on healthspan for several mammalian sirtuins, it was not until recently that, using transgenic mice, a direct role for SIRT6 and SIRT1 has been described in extension of lifespan [66, 67].
SIRT6 overexpression was found to extend the lifespan of males by 15% but not females, potentially by reducing insulin-like growth factor 1 (IGF1) signaling specifically in white adipose tissue [66]. Although no significant differences in tumor types or incidence were defined between wild type and SIRT6 overexpressing mice, analysis of the data suggest that, at least in part, a protection against tumors could be playing a role in lifespan extension in these animals [66].
The assembly and disassembly of stress granules in response to stress and during stress recovery are both important prosurvival mechanisms which are impaired during aging. Recent work demonstrates that SIR-2.4 (C. elegans) and its mammalian orthologue SIRT6 localize to cytoplasmic stress granules (SGs) in response to stress [9, 68]. SGs are cytoplasmic RNA-protein complexes that form in response to cellular stress such as heat shock, nutrient deprivation or oxidative damage [69]. SIRT6 translocates into the cytoplasm under stress and interacts with [9, 68] and promotes dephosphorylation of G3BP at serine 149 (Ser149) [68] (Figure1). G3BP is an evolutionarily conserved RNA-binding protein that was initially characterized through its interaction with a p120Ras-GTPase-activating protein [70, 71]. Importantly, G3BP is a positive regulator of SG assembly, and SIRT6 and RAS signaling contributes to this process by regulating G3BP dephosphorylation. Loss of SIRT6 or expression of a catalytically inactive SIRT6 in MEF cells disrupts SG formation and delays disassembly during recovery from stress [9, 68], while deficiency of SIR-2.4 diminishes maintenance of P granules (found in C. elegans and encompassing the properties of mammalian stress granules) and decreases survival of C. elegans under stress conditions. Based on these findings, it is tempting to speculate that SIRT6 affects age-related processes not only as a histone deacetylase but also in the cytoplasm by regulating the structure and dynamics of SGs [68].
More recently, the expression of SIRT6 in human dermal fibroblasts was shown to decrease with age [72]. Mechanistically, miR-766 negatively regulates SIRT6 expression posttranscriptionally (Figure 2) and expression of miR-766 is higher in older dermal fibroblasts. They also found that inclusion of SIRT6 or blocking miR-766 along with classical Yamanaka factors resulted in significant improvements in the reprogramming efficiency of dermal fibroblasts from older subjects [72].
Taken together, SIRT6 appears to function as a critical enzyme responsible for modulating life span and aging related diseases such as heart disease, diabetes, obesity, inflammation and cancer through control of genomic stability and metabolism.
CONCLUDING REMARKS
SIRT6 has emerged as a master nutrient sensor and regulator of cellular homeostasis. In particular, SIRT6 performs its diverse cellular functions through two distinct enzymatic reactions that include deacetylation of both acetyl groups and long-chain fatty acyl groups and ADP-ribosylation all of which use NAD+ as a cofactor (Box 1). These enzymatic reactions occur mostly in the nucleus at the level of chromatin, with the purpose of regulating gene expression (e.g. HIF1α, MYC, RelA/ NF-κB, c-JUN, Survivin, SREBP and PCSK9) or protein activity (GCN5, PARP1, CtIP) (Figure 1). Additionally, SIRT6 function has now been expanded to include cytosolic roles in stress granule formation and TNF-α secretion. These pleiotropic enzymatic activities give SIRT6 its far-reaching functions in genomic stability through regulating aspects of DNA repair and telomere maintenance, and in metabolism through the control of glycolysis, gluconeogenesis, triglyceride synthesis and LDL cholesterol homeostasis. The important biological roles of SIRT6 influence heart disease, diabetes, obesity, inflammation and cancer, thereby impacting healthy organismal aging. Thus, modulation of SIRT6 activity has the potential to affect multiple human diseases and possibly enhance longevity. As SIRT6 is poised at the intersection between chromatin modulation, metabolism and genome maintenance, understanding how SIRT6 regulates the crosstalk between these pathways will be highly intriguing (Box 3). The recent findings that SIRT6 has a novel substrate, namely long-chain fatty acyl groups, and that free-fatty acids activate its activity, open up a new realm of possible regulation and cellular functions. It will be interesting to determine the extent to which free fatty acids are involved in SIRT6 biology. It is clear that much more needs to be understood about the molecular mechanisms governing SIRT6 activity and function during tumorigenesis. With a better understanding of the biology, novel therapeutics can be designed that either activate SIRT6 or are synthetically lethal with SIRT6 downregulation. The pleiotropic functions of SIRT6 have expanded well past chromatin modulation and involve important roles in cellular homeostasis. It is likely that additional biological targets will be discovered in the future, setting the stage to fully define the importance of SIRT6 in human biology and disease.
Box 1: SIRT6 structure and enzymatic activity.
All sirtuins contain an approximately 275 amino acid conserved catalytic core region. However, they have variable N- and C-terminal extensions (NTE and CTE) that differ in length and sequence. The catalytic core region contains a large and structurally homologous Rossmann-fold domain for NAD+ binding and a more structurally diverse, smaller, zinc-binding domain [73].
Full-length human SIRT6 has 355 amino acids and is best characterized as a NAD+-dependent histone deacetylase. Deacetylation of lysine by SIRT6 is coupled to NAD+ hydrolysis yielding O-acetyl-ADP adenosine 5'-diphosphoribose, nicotinamide and a deacetylated substrate [74]. SIRT6, unlike all other studied sirtuins, can bind NAD+ in the absence of an acetylated substrate perhaps allowing it to act as an NAD+ sensor [46]. The nicotinamide product of this reaction is thought to allosterically inhibit SIRT6 activity. Interestingly, the deacetylase activity of purified protein in vitro is 1000-fold lower for SIRT6 compared to SIRT1. This can be explained structurally: SIRT6 exists in a more open conformation, containing a zinc-binding motif that is splayed from the larger Rossmann fold domain. Additionally, a hydrogen bond connects the zinc-binding motif to the Rossmann fold, stabilizing SIRT6 in this conformation [46]. Strikingly, it was recently discovered that free fatty acids (FFAs) act as endogenous activators of SIRT6 deacetylase activity in vitro [44] (Figure 2, Box 2). Although it remains to be seen whether FFAs could impact SIRT6 deacetylase activity in vivo, this novel observation provides an explanation for the previously “weak” in vitro deacetylase activity reported, and further implicates a new mechanism through which SIRT6 may sense changes in lipid metabolism and act upon it.
Additionally, SIRT6 has been recently described to not only remove single acetyl groups but also long-chain fatty acyl groups (myristoyl and palmitoyl) from lysine residues. The demyristoylation activity of SIRT6, for example, is approximately 300-fold higher than its deacetylation activity in vitro. The increased catalytic efficiency comes mainly from the decrease in Km. Similar to the deacetylation reaction, SIRT6 uses NAD+ to produce O-myristoyl- adenosine 5'-diphosphoribose and the deacylated substrate [45].
SIRT6 also has very weak ADP-ribosylation activity where it can use NAD+ as a substrate, however only poly ADP ribose polymerase (PARP1) and itself [23] described in more detail later, have been described as substrates to date.
Further characterization of the NTE and the CTE of SIRT6 revealed important functional roles. The CTE of SIRT6 contains the nuclear localization signal 345PKRVKAK351 and is critical for proper sub-cellular targeting, but is dispensable for enzymatic activity. The NTE of SIRT6, by contrast, is critical for chromatin association and intrinsic H3K9 and H3K56 deacetylase activity in cells. Deletion of the NTE reduces deacetylase activity through a direct defect in enzymatic activity, not because of a failure to properly localize to chromatin substrates or interact with critical binding partners. Finally, both the NTE and CTE of SIRT6 are important for nucleosome binding [75]. Given the diverse enzymatic activities of SIRT6 it is not difficult to understand the diverse biological roles attributed to this protein.
Box 2: Modulators of SIRT6 activity.
In recent years there has been a push to determine how SIRT6 function is regulated endogenously and to try to find chemical activators of SIRT6 expression and/or function.
One mechanism to increase the levels of SIRT6 is to reduce its protein degradation. Recently, SIRT6 was found to be regulated at the posttranslational level by CHIP (carboxyl terminus of Hsp70-interacting protein), which is a protein that exhibits both chaperone and ubiquitin ligase activities and is an integral component of protein quality control (PQC) [76]. CHIP ubiquitylates SIRT6 at K170 and prevents canonical ubiquitylation of SIRT6 by other ubiquitin ligases, which leads to its increased protein stability and reduced proteasome-mediated degradation (Figure 2). The reduced levels of SIRT6 expression in the absence of CHIP is associated with increased histone acetylation and increased gene transcription of SIRT6 regulated genes. Similar to SIRT6 knockout cells, cells lacking CHIP are hypersensitive to DNA-damaging agents and the resulting genomic defects are rescued by overexpressing SIRT6, thus further confirming that SIRT6 is an important target of CHIP [76].
SIRT6 exhibits poor deacetylase activity in vitro, despite a clear increase of histone acetylation in SIRT6 deficient cells in vivo. To try to explain this difference, Cohen and colleagues looked at the difference in SIRT6 activity when it was in the presence of histones versus nucleosomes. They found that SIRT6 did not associate with any of the core histones, however once packed as part of the nucleosome complex, histone H3 and SIRT6 were found to associate. Interestingly, the association between SIRT6 and the nucleosome significantly improved its enzymatic activity [77].
More recently, endogenous activators of SIRT6 deacetylase activity were found. Upon close examination of the crystal structure of SIRT6 bound to acylated-peptides, Denu and colleagues hypothesized that free fatty-acids (FFA) may bind to the same acyl-group binding pocket and induce closure of the Rossmann fold domain and the zinc-binding domain, thus allowing the canonical active-site conformation and increasing deacetylase activity [44]. This was indeed the case; they found that FFA increased the deacetylase activity in vitro up to 10-fold depending on the FFA length (Figure 2). The elevated deactylase activity was due to an increased affinity of SIRT6 for acetylated substrate. Interestingly, this appeared to be a characteristic specific to SIRT6 as SIRT1 deacetylase activity could not be activated by FFA [44]. These novel findings present a model of SIRT6 activity where in the absence of FFA it acts as a lysine deacylase and as the nutritional status of the cell changes to increase FFA, SIRT6 can switch to an activated deacetylase, in turn modulating glycolysis and lipogenesis, as described in the text.
Therapies that specifically activate or increase the expression of hepatic SIRT6 may be useful in suppressing the chronically active hepatic gluconeogenesis commonly found in insulin-resistant diabetes. Additionally, these therapeutics would also be able to potentially ameliorate hepatic lipid accumulation and reduce hepatic steatosis. In those tumors where SIRT6 expression is downregulated, activation or increased expression of SIRT6 may be beneficial, acting as an inhibitor of the Warburg effect.
Box 3: Outstanding questions
How is SIRT6 gene expression regulated? And under which physiological conditions is SIRT6 regulated?
How is the function of SIRT6 coordinated with the activities of other sirtuins? In particular, are the different sirtuins competing for the intracellular NAD pools, and if so, how?
Are there posttranslational modifications of SIRT6 that affect activity?
Does metabolism have an impact on DNA repair efficiency that is dependent on SIRT6 function?
How and when during tumorigenesis is SIRT6 downregulated? in different tumor types?
How much of SIRT6 biology is controlled by levels of FFAs?
What other biological roles does SIRT6 have that involve lysine deacylation?
HIGHLIGHTS
The multiple roles for the mammalian deacylase SIRT6
SIRT6 structure, enzymatic activity and regulation
SIRT6 function and impact on human disease
ACKNOWLEDGEMENTS
Work in the Mostoslavsky lab is supported in part by NIH grants GM093072-01, DK088190-01A1 and the National Pancreatic Foundation. SK is supported by a Canadian Institutes of Health postdoctoral fellowship. RM is the Kristine and Bob Higgins MGH Research Scholar and a Howard Goodman Awardee.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD- dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 2.Klar AJ, Strathern JN, Broach JR, Hicks JB. Regulation of transcription in expressed and unexpressed mating type cassettes of yeast. Nature. 1981;289(5795):239–244. doi: 10.1038/289239a0. [DOI] [PubMed] [Google Scholar]
- 3.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91(7):1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 4.Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11(2):255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 5.Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11(2):241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 6.Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56(5):771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 7.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116(1):9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66(6):1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 9.Simeoni F, Tasselli L, Tanaka S, Villanova L, Hayashi M, Kubota K, Isono F, Garcia BA, Michishita-Kioi E, Chua KF. Proteomic analysis of the SIRT6 interactome: novel links to genome maintenance and cellular stress signaling. Scientific reports. 2013;3:3085. doi: 10.1038/srep03085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 11.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97(11):5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci U S A. 2001;98(2):415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260(1):273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 14.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273(2):793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 15.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460(7255):587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily Conserved and Nonconserved Cellular Localizations and Functions of Human SIRT Proteins. Mol Biol Cell. 2005 doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 18.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452(7186):492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8(16):2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8(16):2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Huang S, Lee L, Davalos A, Schiestl RH, Campisi J, Oshima J. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging cell. 2003;2(4):191–199. doi: 10.1046/j.1474-9728.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- 22.Multani AS, Chang S. WRN at telomeres: implications for aging and cancer. Journal of cell science. 2007;120(Pt 5):713–721. doi: 10.1242/jcs.03397. [DOI] [PubMed] [Google Scholar]
- 23.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332(6036):1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329(5997):1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450(7169):509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You Z, Shi LZ, Zhu Q, Wu P, Zhang YW, Basilio A, Tonnu N, Verma IM, Berns MW, Hunter T. CtIP links DNA double-strand break sensing to resection. Molecular cell. 2009;36(6):954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. The Journal of biological chemistry. 2008;283(12):7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 28.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, Bohr VA, Chua KF. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 2009;1(1):109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, D'Urso A, Naar AM, Kingston R, et al. SIRT6 Recruits SNF2H to DNA Break Sites, Preventing Genomic Instability through Chromatin Remodeling. Molecular cell. 2013;51(4):454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140(2):280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominy JE, Jr., Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP, Ruan HB, Feldman J, Pierce K, Mostoslavsky R, Denu JM, Clish CB, Yang X, et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Molecular cell. 2012;48(6):900–913. doi: 10.1016/j.molcel.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423(6939):550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 34.Xiong X, Tao R, Depinho RA, Dong XC. Deletion of Hepatic FoxO1/3/4 Genes in Mice Significantly Impacts on Glucose Metabolism through Downregulation of Gluconeogenesis and Upregulation of Glycolysis. PloS one. 2013;8(8):e74340. doi: 10.1371/journal.pone.0074340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong WI, Park O, Ki SH, Gao B, Deng CX. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell metabolism. 2010;12(3):224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanfi Y, Peshti V, Gil R, Naiman S, Nahum L, Levin E, Kronfeld-Schor N, Cohen HY. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging cell. 2010;9(2):162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 37.Yang SJ, Choi JM, Chae SW, Kim WJ, Park SE, Rhee EJ, Lee WY, Oh KW, Park SW, Kim SW, Park CY. Activation of peroxisome proliferator-activated receptor gamma by rosiglitazone increases sirt6 expression and ameliorates hepatic steatosis in rats. PloS one. 2011;6(2):e17057. doi: 10.1371/journal.pone.0017057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao R, Xiong X, Depinho RA, Deng CX, Dong XC. FoxO3 Transcription Factor and Sirt6 Deacetylase Regulate Low Density Lipoprotein (LDL)-cholesterol Homeostasis via Control of the Proprotein Convertase Subtilisin/Kexin Type 9 (Pcsk9) Gene Expression. The Journal of biological chemistry. 2013;288(41):29252–29259. doi: 10.1074/jbc.M113.481473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lambert G, Sjouke B, Choque B, Kastelein JJ, Hovingh GK. The PCSK9 decade. Journal of lipid research. 2012;53(12):2515–2524. doi: 10.1194/jlr.R026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao R, Xiong X, Depinho RA, Deng CX, Dong XC. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. Journal of lipid research. 2013;54(10):2745–2753. doi: 10.1194/jlr.M039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elhanati S, Kanfi Y, Varvak A, Roichman A, Carmel-Gross I, Barth S, Gibor G, Cohen HY. Multiple Regulatory Layers of SREBP1/2 by SIRT6. Cell reports. 2013;4(5):905–912. doi: 10.1016/j.celrep.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328(5985):1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin H, Su X, He B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS chemical biology. 2012;7(6):947–960. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feldman JL, Baeza J, Denu JM. Activation of the Protein Deacetylase SIRT6 by Long-chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, Lin H. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496(7443):110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. The Journal of biological chemistry. 2011;286(16):14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Gool F, Galli M, Gueydan C, Kruys V, Prevot PP, Bedalov A, Mostoslavsky R, Alt FW, De Smedt T, Leo O. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nature medicine. 2009;15(2):206–210. doi: 10.1038/nm.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136(1):62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimley R, Polyakova O, Vamathevan J, McKenary J, Hayes B, Patel C, Smith J, Bridges A, Fosberry A, Bhardwaja A, Mouzon B, Chung CW, Barrett N, et al. Over expression of wild type or a catalytically dead mutant of Sirtuin 6 does not influence NFkappaB responses. PloS one. 2012;7(7):e39847. doi: 10.1371/journal.pone.0039847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao C, Wang RH, Lahusen TJ, Park O, Bertola A, Maruyama T, Reynolds D, Chen Q, Xu X, Young HA, Chen WJ, Gao B, Deng CX. Progression of chronic liver inflammation and fibrosis driven by activation of c-JUN signaling in Sirt6 mutant mice. The Journal of biological chemistry. 2012;287(50):41903–41913. doi: 10.1074/jbc.M112.415182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sundaresan NR, Vasudevan P, Zhong L, Kim G, Samant S, Parekh V, Pillai VB, Ravindra PV, Gupta M, Jeevanandam V, Cunningham JM, Deng CX, Lombard DB, et al. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nature medicine. 2012;18(11):1643–1650. doi: 10.1038/nm.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai Y, Yu SS, Chen SR, Pi RB, Gao S, Li H, Ye JT, Liu PQ. Nmnat2 protects cardiomyocytes from hypertrophy via activation of SIRT6. FEBS letters. 2012;586(6):866–874. doi: 10.1016/j.febslet.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 53.Metallo CM, Vander Heiden MG. Understanding metabolic regulation and its influence on cell physiology. Molecular cell. 2013;49(3):388–398. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. The Journal of general physiology. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, Cosentino C, Greenson JK, MacDonald AI, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151(6):1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen X, Chen L, Scheuch H, Zheng H, Qin L, Zatloukal K, Hui L, Wagner EF. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nature cell biology. 2012;14(11):1203–1211. doi: 10.1038/ncb2590. [DOI] [PubMed] [Google Scholar]
- 57.Lai CC, Lin PM, Lin SF, Hsu CH, Lin HC, Hu ML, Hsu CM, Yang MY. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34(3):1847–1854. doi: 10.1007/s13277-013-0726-y. [DOI] [PubMed] [Google Scholar]
- 58.Wang JC, Kafeel MI, Avezbakiyev B, Chen C, Sun Y, Rathnasabapathy C, Kalavar M, He Z, Burton J, Lichter S. Histone deacetylase in chronic lymphocytic leukemia. Oncology. 2011;81(5–6):325–329. doi: 10.1159/000334577. [DOI] [PubMed] [Google Scholar]
- 59.Kennedy BK, Austriaco NR, Jr., Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80(3):485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- 60.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13(19):2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guarente L, Picard F. Calorie restriction--the SIR2 connection. Cell. 2005;120(4):473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 62.Mercken EM, Hu J, Krzysik-Walker S, Wei M, Li Y, McBurney MW, de Cabo R, Longo VD. SIRT1 but not its increased expression is essential for lifespan extension in caloric restricted mice. Aging cell. 2013 doi: 10.1111/acel.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289(5487):2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 64.Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477(7365):E1–2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- 65.Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, Vazquez-Manrique RP, Orfila AM, Ackerman D, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477(7365):482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483(7388):218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 67.Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai S. Sirt1 Extends Life Span and Delays Aging in Mice through the Regulation of Nk2 Homeobox 1 in the DMH and LH. Cell metabolism. 2013;18(3):416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jedrusik-Bode M, Studencka M, Smolka C, Baumann T, Schmidt H, Kampf J, Paap F, Martin S, Tazi J, Muller KM, Kruger M, Braun T, Bober E. The sirtuin SIRT6 regulates stress granules formation in C. elegans and in mammals. Journal of cell science. 2013 doi: 10.1242/jcs.130708. [DOI] [PubMed] [Google Scholar]
- 69.Anderson P, Kedersha N. Stress granules. Current biology : CB. 2009;19(10):R397–398. doi: 10.1016/j.cub.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 70.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. The Journal of cell biology. 2003;160(6):823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Irvine K, Stirling R, Hume D, Kennedy D. Rasputin, more promiscuous than ever: a review of G3BP. The International journal of developmental biology. 2004;48(10):1065–1077. doi: 10.1387/ijdb.041893ki. [DOI] [PubMed] [Google Scholar]
- 72.Sharma A, Diecke S, Zhang WY, Lan F, He C, Mordwinkin NM, Chua KF, Wu JC. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. The Journal of biological chemistry. 2013;288(25):18439–18447. doi: 10.1074/jbc.M112.405928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feldman JL, Dittenhafer-Reed KE, Denu JM. Sirtuin catalysis and regulation. The Journal of biological chemistry. 2012;287(51):42419–42427. doi: 10.1074/jbc.R112.378877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97(26):14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tennen RI, Berber E, Chua KF. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech Ageing Dev. 2010;131(3):185–192. doi: 10.1016/j.mad.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ronnebaum SM, Wu Y, McDonough H, Patterson C. The ubiquitin ligase CHIP prevents SirT6 degradation through noncanonical ubiquitination. Molecular and cellular biology. 2013 doi: 10.1128/MCB.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gil R, Barth S, Kanfi Y, Cohen HY. SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic acids research. 2013;41(18):8537–8545. doi: 10.1093/nar/gkt642. [DOI] [PMC free article] [PubMed] [Google Scholar]