Abstract
Obesity is associated with the development of asthma and considerable asthma-related healthcare utilization. To understand the immunological pathways that lead to obesity-associated asthma, we fed mice a high fat diet for 12 weeks, which resulted in obesity and the development of airway hyperreactivity (AHR), a cardinal feature of asthma. This AHR depended on innate immunity, since it occurred in obese Rag−/− mice, and on IL-17A and the NLRP3 inflammasome, since it did not develop in obese Il17−/− or Nlrp3−/− mice. The AHR was also associated with the presence in the lungs of CCR6+ innate lymphoid cells producing IL-17A (ILC3 cells), which could by themselves mediate AHR when adoptively transferred into Rag2−/− Il2rγ−/− mice. IL-1β played an important role by expanding the ILC3 cells, and treatment to block the function of IL-1β abolished obesity-induced AHR. Since we found ILC3-like cells in the bronchoalveolar lavage fluid of human patients with asthma, we suggest that obesity-associated asthma is facilitated by inflammation mediated by NLRP3, IL-1β and ILC3 cells.
Keywords: airway hyperreactivity, asthma, obesity, innate lymphoid cells, IL-17, NLRP3, ILC3
Introduction
The prevalence of obesity in industrialized countries has increased dramatically over the last two decades, such that obesity now affects 36% of all adults in the US (66% of all adults are classified as overweight or obese)1. The chronic complications from obesity, including type 2 diabetes mellitus, cardiovascular disease, liver disease and some forms of cancer, represent some of the greatest threats to human health2. Recent studies have also shown that obesity is a major risk factor for the development of asthma3–5. Although obesity itself can alter lung mechanics independent of asthma, the incidence of asthma increases with body mass index (BMI) in both men and women, in children and adults, and across multiple racial backgrounds. Substantial weight loss in obese asthmatic individuals can lead to significant improvements in asthma symptoms, reinforcing the relationship between obesity and asthma6. Unfortunately, asthma in obese individuals responds poorly to typical asthma medications, including corticosteroids, and leads to greater asthma-related healthcare utilization and to reduced quality of life3,7. The lack of effective therapies for asthma in obese individuals perhaps reflects the poor understanding of the specific immunological and metabolic mechanisms by which nutritional excess and adiposity lead to asthma8,9.
Although the mechanisms that cause asthma in the face of nutritional excess remain undefined, obesity is widely recognized as a cause of chronic inflammation that leads to type 2 diabetes, atherosclerosis and liver disease10. Immunologically, obesity is associated with a reduction in anti-inflammatory regulatory T (Treg) cells11, an increase in CD8 T cells12 and a transition from M2 to M1 macrophages in adipose tissue10,13. In the lungs however, the role of inflammation in obesity has been controversial, since little correlation of airway eosinophils with body mass index (BMI) has been observed, although increases in sputum neutrophils in obese asthmatics have been reported recently14. Increased circulating adipokines (leptin, adiponectin and resistin), as well as increased IL-6, TNF-α and oxidative stress3,8 are all associated with asthma in obese individuals but not with allergic asthma, the most common form of asthma. Moreover, the association between asthma and obesity is stronger in non-atopic than in atopic individuals15, and weight loss reduces airway hyperreactivity (AHR), a cardinal feature of asthma, in non-atopic and not in atopic obese asthmatics6. These observations suggest that asthma associated with obesity is an entity or phenotype distinct from that of allergic asthma16.
To clarify the mechanisms by which obesity might cause asthma, we examined a mouse model of diet-induced obesity. Our results using this model demonstrate a novel molecular and cellular pathway for the development of obesity-induced asthma, and suggest specific therapeutic modalities for this growing clinical problem.
Results
High fat diet results in obesity and airway hyperreactivity
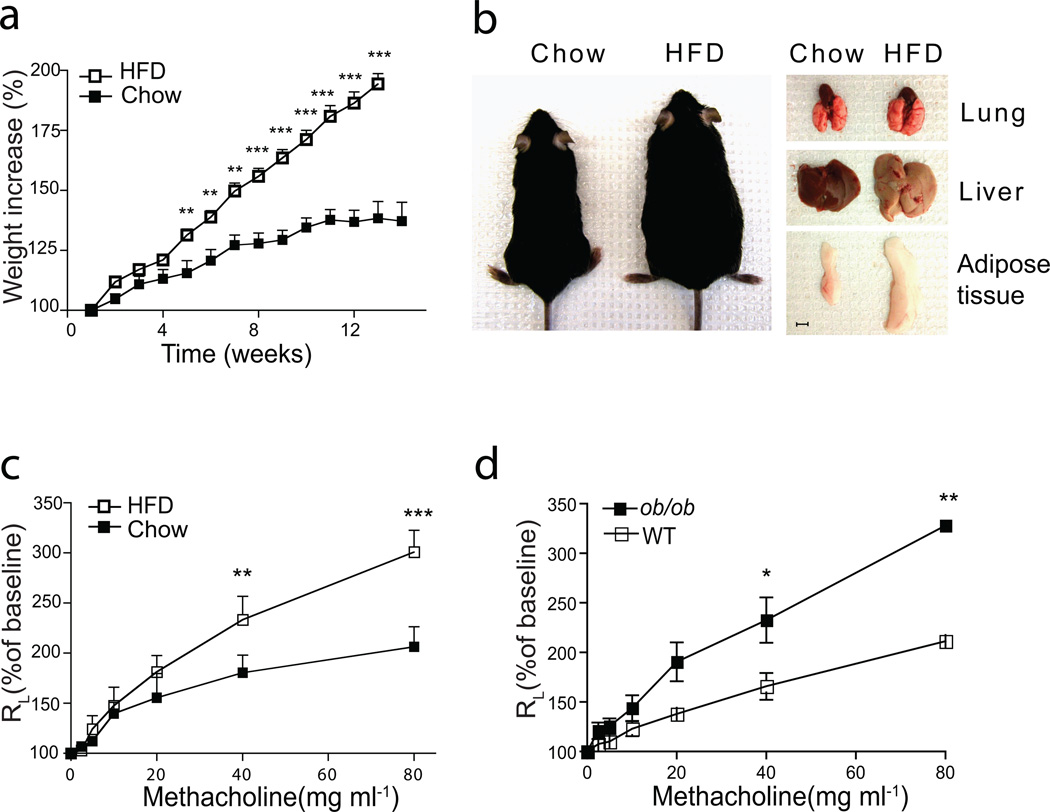
We studied airways disease in wildtype (WT) mice fed a high fat diet (HFD) for 14 wks starting at 3–4 wks of age. On a HFD, mice gained weight rapidly (Fig. 1a) and developed hepatic steatosis and increased epididymal fat volumes compared to control mice on a standard diet (chow) (Fig. 1b). Importantly, the obese mice on the HFD developed AHR (Fig. 1c), whereas mice on the standard diet had normal airway reactivity. Similarly, leptin deficient ob/ob mice, which rapidly developed obesity on normal chow, also expressed AHR (Fig. 1d), as previously reported17,18. These results demonstrate that diet-induced obesity and obesity associated with leptin-deficiency are both associated with the development of AHR.
Figure 1. High fat diet feeding induces AHR in wildtype mice.
(a–c). Wild type mice fed normal chow or 60% HFD, starting at weaning for 12–14 weeks.
a. Comparison of bodyweight gain in wild-type C57BL/6 mice on normal chow or the HFD. ***p<0.001, HFD group compared to chow fed group. (Student’s t-test)
b. Representative picture of mice and organs after chow or HFD feeding. Scale bar indicates 5mm
c. Wild-type mice develop AHR, as assessed by the response to increasing doses of methacholine. Results represent the changes in lung resistance (RL) as a measure of AHR. **p<0.01, and ***p<0.001, compared to chow fed group. (Two-way ANOVA).
Data are representative of at least three independent experiments. Error bars represent SEM.
d. 7 wks old ob/ob mice fed on normal chow diet develop AHR. Results represent the changes in lung resistance (RL) as a measure of AHR. *p<0.05, and **p<0.01, compared to wild type mice. (Two-way ANOVA).
IL-17A is increased in the lungs and is required for HFD-induced AHR
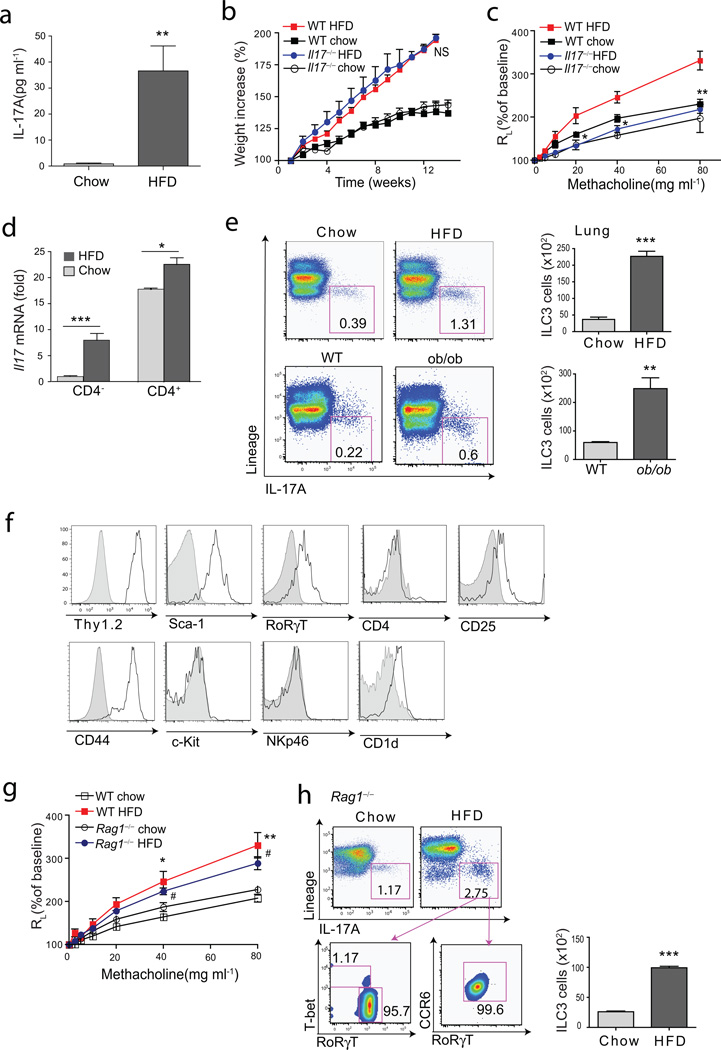
Since obesity-associated AHR is not linked strongly with atopy18, we asked if it was dependent on IL-17A. Indeed, IL-17A production was greatly increased in the lungs of the obese mice, as assessed in cultured lung cells (Fig. 2a). Moreover, Il17−/− mice on the HFD developed obesity (Fig. 2b), but failed to develop obesity-associated AHR (Fig. 2c), indicating a requirement for IL-17A. In the lungs of the obese WT mice, IL-17A mRNA levels were significantly increased in both CD4− and CD4+ lymphocyte fractions (which included both T and non-T cells) (Fig. 2d). Surprisingly, the major producers of IL-17A in the lungs of both obese WT and ob/ob mice were non-T, non-B, CD4−, lineage− cells, which are characteristics of innate lymphoid cells that produce IL-17A (ILC3 cells)19–22 (Fig. 2e), although some Lin+ cells (e.g., CD4+ Th17 cells and γδ cells) also produced IL-17A (Suppl Fig. 1). The ILC3-like cells expressed high levels of Thy1.2, Sca-1, RORγt, CD44, but not c-Kit (Fig. 2f), and did not produce IL-13 (Suppl Fig. 1), consistent with the features of IL-17+ ILC3 cells previously described in the intestines of mice and humans in the setting of IBD19,20, and distinct from LTi ILC3 cells23–25, IL-22+ ILC3 cells26,27 and lung ILC2 cells (also called nuocytes or natural helper cells) producing IL-13 and IL-5 and expressing variable amounts of c-Kit28–31.
Figure 2. HFD induced AHR requires the presence of IL-17A.
a. Cells were isolated from lungs of mice fed on normal chow or on the HFD, and supernatants were analyzed for IL-17A by ELISA.
b. Comparison of bodyweight gain between WT and Il17−/− mice on either chow or HFD. NS, WT mice on HFD vs Il17−/− mice on HFD (Student’s t-test).
c. AHR was measured using mice represented in Fig. 2b. Graph represents the changes in lung resistance (RL). *p<0.05, and **p<0.01, Il17−/− mice on HFD compared to WT mice on HFD (Two-way ANOVA).
d. Total lung cells from mice fed normal chow or HFD were sorted for CD4+ or CD4− cell fraction and analyzed for Il17A mRNA expression by qRT-PCR. ***p<0.001 (chow CD4− vs HFD CD4− fraction), and *p<0.05 (chow CD4+ vs HFD CD4+ fraction), (Student’s t-test).
e. ILC3 cells (CD45+Lineage−IL-17A+cells) from HFD fed WT mice (upper panel) and normal chow fed ob/ob mice (lower panel). Graph (right) represents total number of ILC3 cells in the lungs (upper panel; wild type chow vs HFD, ***p<0.001, lower panel; chow fed wild type vs ob/ob, **p<0.01).
f. ILC3 cells in Figure 2e were further characterized. Grey histograms represent isolype control staining.
g. The changes in lung resistance (RL) as a measure of AHR in Rag1−/− mice fed a high fat diet. *p<0.05, and **p<0.01, comparing normal chow fed and high diet fed groups (Two-way ANOVA).
h. ILC3 cells in the lungs of Rag1−/− mice. Graph (right) represents total number of ILC3 cells (chow vs HFD groups, ***p<0.001).
To better understand the role of Th17 and ILC3 cells in HFD-induced AHR, we placed Rag1−/− mice on the HFD for 12 wks before assessing AHR. Obese Rag1−/− mice, which develop type 2 diabetes presumably through mechanisms that are independent of adaptive immunity and Th17 cells32, also developed AHR associated with an increase in the lungs of ILC3 cells expressing IL-17, CCR6, RORγT (Fig. 2g, 2h), but little T-bet or IL-22 and no IFN-γ (Suppl.Fig. 1). This suggested that ILC3 cells were sufficient for the development of pulmonary inflammation and AHR in the obese mice, and that Th17 cells or γδ T cells were not required. Consistent with this idea, C57BL/6 mice from both Jackson Labs and Taconic Farms developed obesity-induced AHR and an increase in IL-17+ ILC3 cells (Suppl.Fig 2), but segmented filamentous bacteria (SFB), essential for the development of Th17 cells33,34, were detected only in the stools of mice obtained from Taconic Farms (data not shown)33. These results together demonstrate for the first time that ILC3 cells are present in the lungs, and suggest that ILC3 cells, which could develop in the absence of SFB, might play an important pathogenic role in the airways.
The inflammation in the lungs of obese mice requires the NLRP3 inflammasome
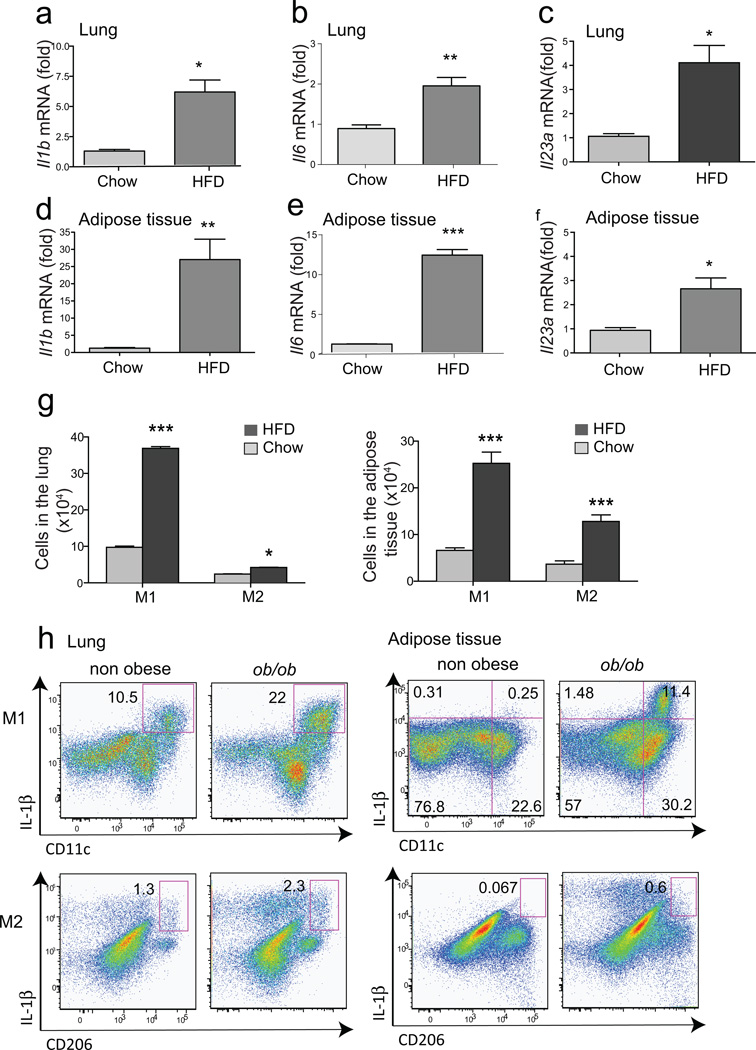
We examined other cytokines in the lungs and adipose tissue of obese mice, and found that IL-1β, IL-6 and IL-23 (known to induce IL-17A production in Th17 cells)35 were increased in the lungs and in the adipose tissue of the obese mice, as determined by RT-PCR (Fig. 3a–f). The inflammatory cells in the lungs and adipose tissue included macrophages, particularly M1 (F4/80+CD11c+CD206−) macrophages, although M2 macrophages (F4/80+CD206+) were also slightly increased after 12 wks on the HFD (Fig. 3g, Suppl.Fig.3). Moreover, M1, and to a much lesser extent M2 macrophages, from obese WT mice and obese ob/ob mice expressed IL-1β, as determined by intracellular cytokine staining (Suppl.Fig.3, Fig. 3h). In addition, in the lungs and adipose tissue of mice on the HFD, there was a reduction in Treg cells and NKT cells (data not shown), as previously reported11,36.
Figure 3. IL-1β production and M1 macrophages are increased in the lungs of obese mice.
(a–f). Total RNA was extracted from the lung and adipose tissue for qRT-PCR. Fold induction of Il1b (a,d), Il6 (b,e), and Il23 (c,f) were calculated based on GAPDH expression. *p<0.05, and **p<0.01, mRNA expression from HFD group was compared to chow group.
g. Expression of M1/M2 macrophage markers, CD206 (M2) and CD11c (M1), were analyzed after gating on of CD45+F4/80+ cells. Graph represents the total numbers of macrophages in the lung (left, *p<0.05 and ***p<0001) and adipose tissue (right, ***p<0.001). (Student’s t-test)
h. IL-1β production from M1/M2 macrophages. Cells were taken from lung or adipose tissue and IL-1β producing M1 (CD11c+, left panel) or M2 (CD206+, right panel) macrophages were assessed by flow cytometry.
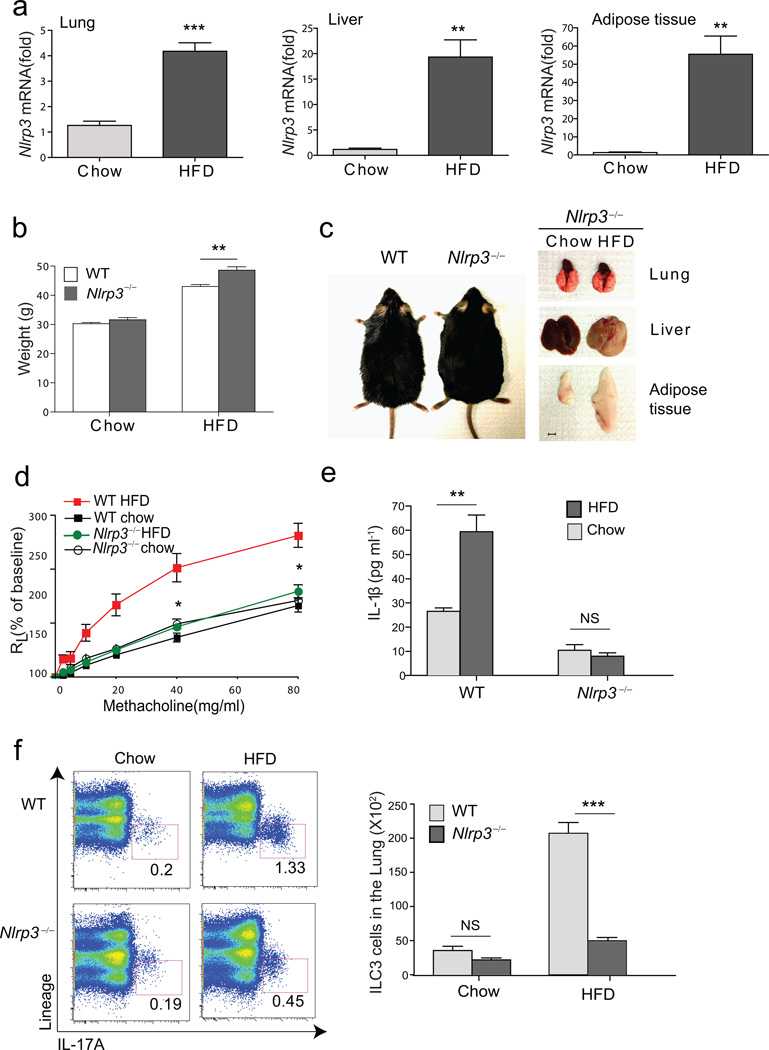
Since adipose tissue macrophages in obese mice have been shown to produce IL-1β in an inflammasome-dependent manner37,38, we examined Nlrp3 mRNA expression in the lungs, and found it was increased, as it was in the liver and adipose tissues of obese mice (Fig. 4a). Moreover, NLRP3 was required for obesity-induced AHR, as Nlrp3−/− mice fed the HFD rapidly gained weight, developed some degree of hepatic steatosis and increased adipose tissue volumes (Fig. 4b, 4c), but failed to develop AHR (Fig 4d). Further, Nlrp3−/− mice on a HFD failed to develop an increase in lung IL-1β production (Fig 4e), and had a significantly reduced number of pulmonary ILC3 cells compared to obese WT mice (Fig 4f). In contrast, the number of Lin+IL-17+ (Th17 and γδ cells) cells was only slightly increased in the lungs of obese WT and Nlrp3−/− mice, compared to the chow fed WT and Nlrp3−/− mice, respectively (Fig. 4f), supporting the idea that Th17 cells were not required. Therefore, the development of AHR in obese mice correlated with the activation of NLRP3, the production of IL-1β and with the expansion of IL-17+ ILC3 cells in the lungs.
Figure 4. HFD increases NLRP3, which is required for AHR.
a. Total RNA was extracted from the lung, liver and adipose tissue and assessed for Nlrp3. Fold induction of Nlrp3 mRNA was calculated based on GAPDH expression. **p<0.01 and ***p<0.001, mRNA level from mice fed on HFD compared to chow group. (Student’s t-test)
b. Graph represent grams of weight gain in WT or Nlrp3−/− mice. **p<0.01, WT mice on HFD compared to Nlrp3−/− mice on HFD group. (Student’s t-test)
c. WT and Nlrp3−/− mice 3 months after HFD (left). Representative pictures of lung, liver and epididymal adipose tissue from Nlrp3−/− mice fed on chow or HFD (right). Scale bar indicates 5mm
d. Development of obesity induced AHR was compared in WT and Nlrp3−/− mice. Graph represents the changes in lung resistance (RL). *p<0.05, Nlrp3−/− mice on HFD were compared to WT mice on HFD group (Two-way ANOVA).
e. IL-1β levels in the culture supernatant of lung cells from WT and Nlrp3−/− mice. The data shown are means ± SEM. **p<0.01, supernatants from HFD group were compared to chow group. (Student’s t-test)
f. Lungs were taken from chow or HFD fed mice and stimulated with PMA/ionomycin for 5 hr. −IL-17A+ ILC3 cells in WT mice (upper panel) or Nlrp3−/− mice (lower panel) were assessed by FACS. Graph represents the total number of ILC3 cells in the lung. ***p<0.001, ILC3 cells in WT mice were compared to Nlrp3−/− mice. (Student’s t-test).
Administration of IL-1β directly causes AHR by inducing IL-17A production
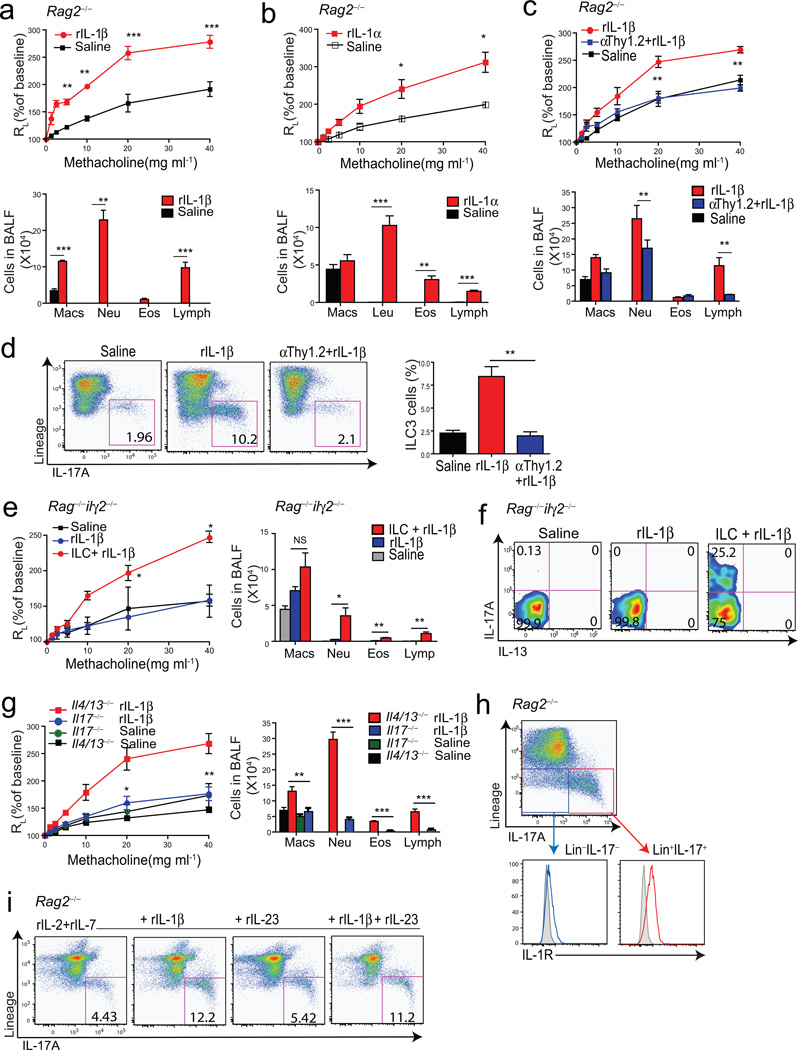
We hypothesized that in obese mice, IL-1β, produced by lung macrophages, induced the development of ILC3 cells in a process that was independent of adaptive immunity. Indeed, the administration of rIL-1β into the lungs of Rag2−/− mice rapidly induced a robust AHR response (Fig. 5a). Treatment of Rag2−/− mice with rIL-1α and rIL-23 but not rIL-6 also resulted in the development of AHR, though the effect with IL-23 was not as robust as with IL-1β (Fig. 5b and Suppl Fig.4). The IL-1β-induced AHR response required ILC3 cells, since it was abolished by depletion of ILC3 cells by in vivo treatment with anti-Thy1.2 mAb (Fig. 5c), which directly reduced the number of ILC3 cells (Fig. 5d), and resulted in a decrease in other inflammatory cells in the BAL fluid (Fig. 5c). The IL-1β treatment induced IL-17A producing ILCs that expressed CCR6, but only low levels of IL-17F, GM-CSF or T-bet as shown by intracellular staining (Suppl Fig. 4). Furthermore, treatment with IL-1β induced AHR in Nlrp3−/−, IL-6−/− and δ TCR−/− mice (lacking γδ T cells), which was associated with an increase in IL-17 production in ILCs in these mice, but not in Rorc−/− mice (Suppl Fig. 5), indicating that ILC3 cells required RORγt, but could develop and induce AHR in the absence of NLRP3, IL-6 or γδ T cells. Moreover, ILC3 cells by themselves were sufficient for the development of AHR, since adoptive transfer of the ILC3 cells (Lin−CCR6+) into Rag2−/−Il2rγ−/− mice restored IL-1β-induced AHR (Fig. 5e–f). The induction of AHR and airway inflammation by rIL-1β required IL-17A but not IL-4 or IL-13 producing cells (e.g., Th2 or ILC2 cells), since treatment of Il4/13−/− mice, but not Il17−/− mice, with rIL-1β resulted in robust AHR (Fig. 5g). Although treatment of WT mice with rIL-17A alone induced only minimal or no AHR (Suppl Fig. 6), treatment of the Il17−/− mice with both IL-1β and IL-17A resulted in robust AHR and airway inflammation (Suppl Fig. 6), confirming a critical role for IL-17A in IL-1β-induced AHR. Further, administration of rIL-1β to Rorc−/− mice (lacking RORγt) failed to induce AHR, and anti-IL-17A mAb administration blocked IL-1β-induced AHR in Rag1−/− mice (Suppl Fig. 6), confirming a requirement for ILC3 cells (which require Rorc for development31) and IL-17A and in the development of IL-1β-induced AHR. Finally, we showed that ILC3 cells could directly respond to IL-1β, by showing that ILC3 cells taken from IL-1β treated Rag2−/− mice expressed IL-1R (Fig. 5h), and produced more IL-17 when cultured in vitro with rIL-1β (along with IL-7 and IL-2) (Fig. 5i). Although IL-23 in the lungs of obese mice might contribute to ILC3 cell expansion, these studies together indicate that IL-1β can induce AHR independently of adaptive immunity, IL-4 or IL-13, by expanding lung ILC3 cells, which can directly cause AHR.
Figure 5. IL-17 producing innate lymphoid cells are required for the development of AHR.
Graphs represent the changes in lung resistance (RL) (upper panels) and BAL fluid cell counts (lower panels) after rIL-1β (a) or rIL-1α (b) treatment. For AHR, **p<0.05, and ***p<0.001, rIL-1β or rIL-1α treated mice were compared to saline treated mice (Two-way ANOVA). For BAL fluid, **p<0.01, and ***p<0.001, rIL-1β vs saline (a), **p<0.01, rIL-1α vs saline (b) (Student’s t-test).
c. Changes in lung resistance (RL) (upper panel) and BAL fluid cell counts (lower panel) after depletion of ILCs. For AHR, **p<0.01, rIL-1β treated mice were compared to anti-Thy1.2 mAb treated mice (Two way ANOVA). For BAL fluid, **p<0.01, and ***p<0.001, rIL-1β vs anti-Thy1.2 mAb+rIL-1β (Student’s t-test).
d. ILC3 cells in the lung (left panel), and total number of ILC3 cells in each group (right panel). **p<0.01, rIL-1β treated mice were compared to anti-Thy1.2 mAb treated mice (Student’s t-test).
e. Changes in lung resistance (RL) in Rag2−/−Il2rγ−/− mice after adoptive transfer of ILC3 cells (Lin−CCR6+). ***p<0.001, comparing Rag2−/−Il2rγ−/− mice treated with rIL-1β receiving or not receiving ILC3 cells (Two-way ANOVA). Cells in BAL fluid (right panel). *p<0.05, and ***p<0.001, ILC3 cells +rIL-1β vs rIL-1β group (Student’s t-test).
f. IL-17A and IL-13 production from adoptively transferred ILC3 cells.
g. Changes in lung resistance (RL) in Il4/Il13−/− and Il17−/− mice after rIL-1β treatment. *p<0.05, and **p<0.01, rIL-1β treated Il4/Il13−/− mice were compared to rIL-1β treated Il17−/− mice (Two way- ANOVA). Cells in BAL fluid (right panel). ** p<0.01, and ***p<0.001, rIL-1β treated Il4/Il13−/− mice were compared to rIL-1β treated Il17−/− mice (Student’s t-test).
h. IL-1R expression on ILC3 cells. Lin−IL-17+ cells (red) express higher level of IL-1R compared to Lin−IL-17− cells (blue). Shaded histogram: isotype control.
i. Cells were taken from rIL-1β treated Rag2−/− mice then cultured in vitro to induce ILC3 cells.
Blockade of IL-1β signaling prevents obesity induced AHR
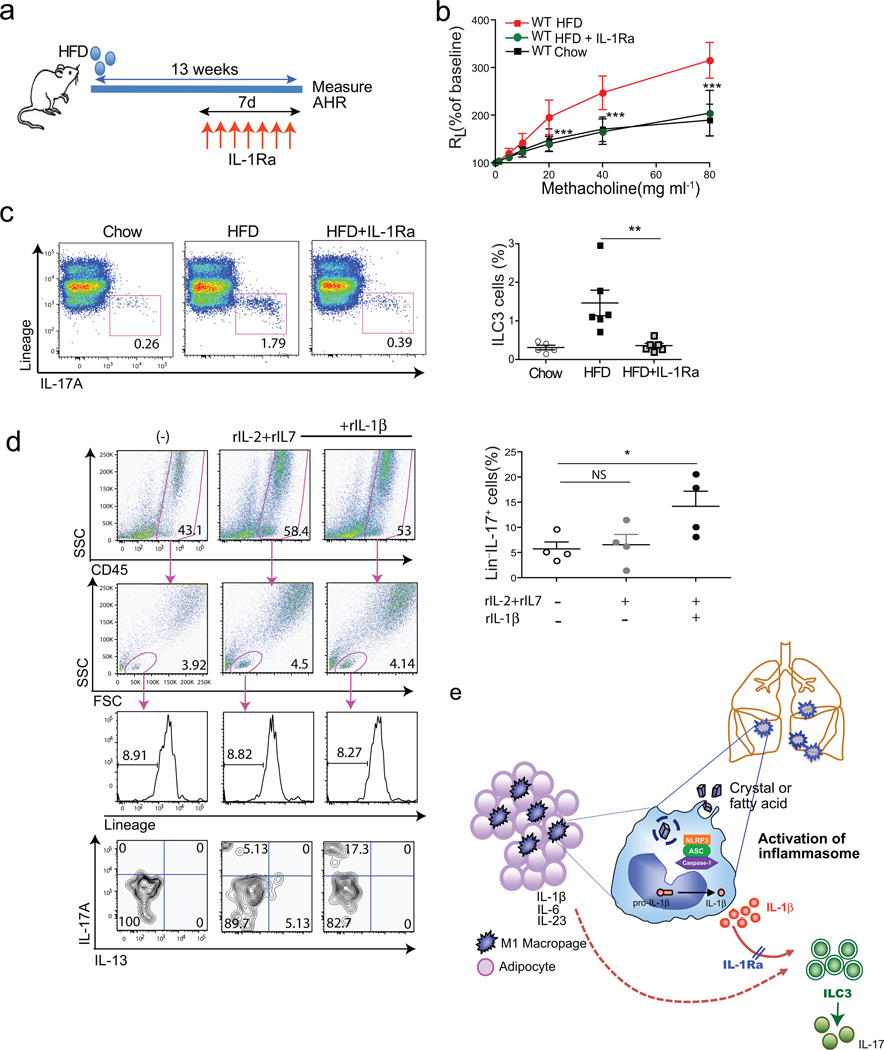
The important role of IL-1β in HFD-induced AHR was confirmed by blocking IL-1β signaling using IL-1Ra (anakinra), which has been shown previously to improve glycemia in obesity-induced type 2 diabetes in humans39. Treatment with anakinra for just 7 days abolished AHR in obese mice fed a HFD for 13 weeks (Fig. 6a, b), and greatly reduced the number of pulmonary ILC3 cells (Fig. 6c). These results indicate that chronic inflammation in obese mice triggers IL-1β production in the lung (presumably through NLRP3), which likely drives the expansion of ILC3 cells and the development of obesity-induced AHR.
Figure 6. Treatment of obese mice with anti-IL-1, prevents AHR.
a. Schedule of feeding and anti-IL1R antagonist treatment. To block IL-1β (and IL-1α), obese mice were treated with IL1R antagonist (50 mg/kg) subcutaneously for 7 days before measuring AHR.
b. AHR was measured 24hr after last IL1R antagonist treatment. Graph represents the changes in lung resistance (RL). ***p<0.001, IL1R antagonist treated obese mice were compared to HFD fed obese mice. (Two-way ANOVA).
c. ILC3 cells were analyzed after PMA/Ionomycin stimulation (left panel). Graph represents the percentage of IL-17A producing ILC3 cells of the total lung lymphocytes in each group (right panel).
d. Cells from BAL fluid were cultured in vitro in the presence of rIL-2, rIL-7 and rIL-1β for 72hr. To measure IL-17 production from Lin− population, cells were re-stimulated with PMA and ionomycin for 5hr. Graph represents the percentage of IL-17 producing CD45+Lin− lymphocytes in each group (bottom), *p<0.05 (Student’s T-test).
e. Possible mechanisms of obesity induced AHR. High fat diet results in the activation of the NLRP3 inflammasome, through fatty acids or cholesterol crystals in macrophages in adipose tissue and in the lungs. This results in IL-1β production, which drives the development of ILC3 cells in the lungs. Treatment with IL-1R antagonist blocks the effects of IL-1β (and IL-1α), and blocks the development of pulmonary ILC3 cells.
Human ILC3 cells in the lungs of patients with asthma
ILC3 cells have not been shown previously to be present in human lungs. However, in 10 consecutive bronchoalveolar lavage (BAL) fluid samples examined from patients with pulmonary disease (Suppl Table 1), we identified significant numbers of innate lymphoid cells (Lin−IL-7R+ cells), many of which expressed IL-17 (Suppl. Table 1, two right-most columns; Suppl Fig. 7). The Lin−IL-17+ cells appear to be comparable to murine ILC3 cells, and patients with severe asthma had more of these cells than patients with mild asthma or no asthma (2.1% versus 0.2%, p=0.013) (Suppl. Table 1). In vitro culture of BAL fluid cells with rIL-1β (along with IL-7 and IL-2) also increased the number of IL-17 producing Lin− cells (Fig. 6d). These studies demonstrate that IL-17 producing ILCs can be found in the lungs of humans, and potentially could play a pathological role in asthma.
Discussion
In these studies, we showed for the first time that diet-induced obesity leads to the development of AHR through a pathway that required NLRP3 and IL-17, and that was facilitated by pulmonary IL-17+ ILC3 cells, a cell type that has not been linked previously with airways disease (Fig. 6e). AHR did not develop in obese Nlrp3−/− mice or in obese Il17−/− mice, and was associated with IL-1β production by lung M1 macrophages in obese mice. Furthermore, IL-17+ ILC3 cells could by themselves induce AHR, since in vivo treatment of Rag2−/− mice with rIL-1β rapidly induced the expansion of airway ILC3 cells and the development of AHR, and since adoptive transfer of CCR6+ ILC3 cells into Rag2−/−Il2rγ−/− mice restored IL-1β-induced AHR. These results suggest that obesity-induced AHR develops through a unique pathway that involves an ILC3–IL-1β–NLRP3 inflammasome axis that is activated in the lungs in the face of nutritional excess37,38,40. Indeed, treatment of obese mice with a short course of the IL-1R antagonist, anakinra, abolished AHR associated with HFD-induced obesity, corroborating a novel mechanistic pathway by which obesity causes airway inflammation and AHR associated with an expansion of pulmonary IL-17+ ILC3 cells.
The association between obesity and asthma has been known for several years5, but the specific mechanisms by which obesity causes asthma have been undefined up to now. Although obesity in mice has been shown to induce AHR41,42, to enhance allergen-induced AHR43,44, and to enhance ozone-induced AHR17, and although obesity has been recognized as an inflammatory disease that causes type 2 diabetes, atherosclerosis and liver disease10, the precise mechanisms that link obesity with airway inflammation and AHR have not been understood. Previous studies have suggested that adipokines (leptin, adiponectin and resistin), as well as IL-6, TNF-α and oxidative stress3,8 could play important roles in mediating asthma in obese individuals. We now suggest that the inflammation in obesity, which has been recently shown to arise from NLRP3 activation and excess production of IL-1β37,38,40, also underlies the asthma that is associated with obesity.
In diet-induced obese WT and Rag−/− mice and in leptin-deficient obese ob/ob mice, we demonstrated the presence of a significant increase in pulmonary IL-17+ ILC3 cells. A pathogenic role for the ILC3 cells in the lungs was demonstrated by the rapid increase in the number of lung ILC3 cells in Rag−/− mice treated with IL-1β or IL-1α, which was associated with the development of AHR, and was suggested by the expansion of ILC3 cells in obese Rag−/− mice that developed AHR. Although Th17 cells, which can cause airway inflammation and AHR45–48, and γδ T cells producing IL-17, which can inhibit the development of AHR49, were also present in the lungs of WT obese mice, ILC3 cells greatly outnumbered the Th17 and γδ T cells in these mice, and were absent in obese Rag−/− mice and in Rag−/− mice treated with IL-1β, both of which developed significant AHR. In addition, mice obtained from either Taconic Farms (which contain SFB) or Jackson Labs (lacking SFB)33 developed obesity-induced and IL-1β-induced AHR, supporting the idea that IL-17+ ILC3 cells and HFD-induced AHR could develop independently of both Th17 cells and SFB (required for the development of Th17 cells). Moreover, AHR induced by IL-1β required RORγt since it failed to occur in Rorc−/− mice, and occurred independently of IL-4 and IL-13 in Il4/Il13−/− mice, indicating that ILC3 cells could function independently of ILC2 cells, which produce IL-13 and IL-5, and which cause AHR during influenza infection28 or airway inflammation in response to protease-containing allergens29. Additionally, the ILC3 cells associated with AHR produced little or no IL-17F, IL-22, GM-CSF or IFN-γ, consistent with previous studies showing that IL-17, but not IL-17F or IL-22, can induce AHR45, and strongly suggesting that IL-17F, IL-22 and IFN-γ have little or no role in inducing obesity-associated AHR. Furthermore, we confirmed that IL-17+ ILC3 cells, which developed in Il6−/− mice, express the IL-1 receptor and respond to IL-1β (and IL-1α), as was previously shown in studies of intestinal ILC3 cells21 that promote the pathology in inflammatory bowel disease19,20. Finally, adoptive transfer of CCR6+ ILC3 cells into Rag2−/−Il2rγ−/− mice restored IL-1β-induced AHR, indicating that ILC3 cells alone were sufficient for the development of AHR.
We also demonstrated an important role for IL-1β in obesity-induced AHR by showing that treatment of obese mice for 7 days with the IL-1R antagonist, anakinra, abolished obesity-induced AHR, which failed to develop in Nlrp3−/− mice. The fatty acid palmitate37, cholesterol crystals, LDL38 and ceramide (generated from fatty acids)40, which are all increased during nutritional excess, can each activate NLRP3, resulting in increased IL-1β production. In obese mice, an important source of IL-1β is the adipose tissue macrophage, which is thought to convert from M2 to M1 forms in association with the activation of NLRP3 and ASC40. In the lungs of obese mice, we showed that NLRP3 is indeed activated, and that M1 macrophages produced much of the IL-1β. Moreover, in both diet-induced and in ob/ob obese mice, activation of this obesity-NLRP3-IL-1β pathway resulted in the expansion and activation of pulmonary IL-17+ ILC3 cells, which facilitated AHR. However, blocking IL-1 signaling with anakinra abolished this pathway, suggesting that in the lung, IL-1β may be particularly important in regulating IL-17 production, as has been shown in the skin50, although other factors such IL-23, may contribute to this process. This NLRP3-IL-1β inflammatory pathway, in addition to mediating type 2 diabetes mellitus, non-alcoholic liver disease and cardiovascular disease10, plays a key role in autoinflammatory conditions, e.g., in cryopyrin-associated periodic syndromes (CAPS)51, suggesting that all of these metabolic diseases are in some respects autoinflammatory. Our studies suggest that obesity-induced AHR also has features of, and might be considered related to, autoinflammatory diseases.
In asthma, previous studies of the pathological role of IL-1β and NLRP3 have generated conflicting results. In these previous studies however, NLRP3 has been studied primarily in the context of allergic asthma. Several investigators found significant attenuation of allergen-induced airway inflammatory responses in Nlrp3−/− mice52,53, but others have found no defects in allergic inflammation, the development of Th2 cells, airway eosinophilia and AHR in Nlrp3−/− mice54–56. While part of this controversy has focused on the role of alum as an adjuvant that might activate NLRP3 during allergic sensitization, we propose instead that the NLRP3 pathway plays a critical role in asthma associated with obesity by mediating the development of ILC3 cells that cause AHR.
Although ILC3 cells have been found in the intestines of humans with IBD19–22 and at low frequencies in inflamed human tonsil57, ILC3 cells have not been found previously in the context of human lung disease or AHR. Nevertheless, our studies, which demonstrate for the first time that ILC3 cells responding to IL-1β can induce the development of AHR in mice, independent of ILC2 cells and adaptive immunity (Th17 cells or Th2 cells), are consistent with clinical observations in humans showing an association of obesity with elevated systemic levels of IL-17 and IL-1β58,59, particularly in non-atopic obese individuals18, 19. An important role for IL-17+ ILC3 cells in asthma, which likely are recruited to and expand in the lungs, and which may be somewhat resistant to corticosteroids, is also suggested by our studies showing that ILC3-like cells are present in the lungs of humans, and that these cells respond to IL-1β. However, as the number of ILC3 cells varied considerably in the BAL fluid of the 10 patients examined, the precise role of ILC3 cells in human lung disease, including asthma associated with obesity, will require clinical studies with much larger numbers of patients. In any case, our studies confirm that asthma is indeed heterogeneous, and suggest that an obesity-associated asthma phenotype may exist that involves an NLRP3-IL-1β-ILC3 cell axis and that is distinct from previously described pathways for asthma3,16,60.
In summary, we showed in a mouse model of asthma associated with obesity that AHR develops through a pathway involving NLRP3, IL-1β and pulmonary IL-17+ ILC3 cells. These studies are the first to show a role for ILC3 cells in the lungs, and may explain why asthma associated with obesity appears to be distinct from other forms of asthma.
On-line Materials and Methods
Mice
Wildtype C57BL/6 mice, Rag1−/− mice, δTCR−/−, IL-6−/− mice, and Rorc−/− mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and bred in our animal facility. Rag2−/− mice on the BALB/c background, and Rag2−/−Il2rγ−/− mice were purchased from Taconic. Il17−/− mice on the C57BL/6 background were generated as previously described 61. Nlrp3−/− mice on the 129 background were generated by Millennium Pharmaceuticals 62, but backcrossed to C57BL/6 background. Il4/Il13−/− mice were generated as previously described 63. The Animal Care and Use Committee of Boston Children’s Hospital approved all animal protocols.
Patients with asthma
Applicable human subject protocols were approved at the institutional review boards at Boston Children’s Hospital, the Brigham and Women’s Hospital and Connelly Hospital. All subjects provided informed consent. The characteristics of each subject are provided in Supplemental Table 1. All patients underwent bronchoscopy and bronchoalveolar lavage using standard protocols.
Diet Intervention
Starting at 3–4 wks of age, male mice were fed normal chow or a HFD for 12–16 wk, providing 60% of energy in the form of fat (D12492; Research Diets). Total body weight and food intake were measured twice a week.
Antibodies and reagents
Recombinant mouse IL-1α, IL-1β, IL-6 and IL-23 were purchased from BioLegend. 7wk-old Rag2−/− mice were treated with 0.1µg of recombinant proteins in saline intranasally for 3 consecutive days (day 1, 2, and 3). To deplete ILCs, mice were treated with 0.5mg of anti-Thy1.2 mAb (clone 30H12, BioXcell) i.p. at days −3, 0 and 2. To deplete IL-17A, mice were treated with anti-mouse IL-17A mAb (Clone TC11-18H10.1, BioLegend) before treatment with IL-1β. To block IL-1 signaling, obese mice were treated with 50 mg/kg of anakinra (soluble IL-1R antagonist, Amgen) daily for 7days before measuring AHR.
Measurement of AHR
Mice were anesthetized with 50 mg/kg pentobarbital and tracheostomized, intubated, and mechanically ventilated at a tidal volume of 0.2 ml and a frequency of 150 breath/min, as previously described 64. Lung resistance (RL) was measured using invasive BUXCO (BUXCO Electronics) in response to increasing doses (0.25 to 80 mg/ml) of aerosolized acetyl-β-methylcholine chloride methacholine (Sigma-Aldrich).
Tissue preparation
Lung tissues and epididymal white adipose tissue (WAT) was minced and digested with 1.6 mg/ml of collagenase type 4 (Worthington, Lakewood, NJ) and 0.1% of DNase I (fraction IX; Sigma) at 37 °C for 1h. Total cells were lysed for RBCs and stained for FACS analysis.
Flow Cytometry
Single cell suspensions were pre-incubated with anti-FcγR blocking mAb (2.4G2) and washed before staining. Cells were stained with fluorescein isothiocyanate-conjugated anti-CD3 (17A2 (555274); BD Biosciences), anti-CD19 (6D5 (115506); BioLegend), anti-CD11b (M1/70 (101206); BioLegend), anti-CD11c (HL3 (553801); BD Biosciences), anti-CD49b (DX5 (11-5971-82); eBioscience), anti-F4/80 (BM8 (123108); BioLegend) or anti-FcεR1 (MAR-1 (134306); BioLegend) (these FITC conjugated antibodies were used as Lineage markers); phycoerythrin-Texas red-conjugated antibody to mouse CD45 (MCD4517; Invitrogen); phycoerythrin-conjugated anti-CD1d (1B1 (123509); BioLegend); phycoerythrin-conjugated anti-CD44 (1M7 (103009); BioLegend); phycoerythrin-conjugated anti-CD121 (35F5 (557589); BD Biosciences); phycoerythrin-conjugated anti-IL-1β (NJTEN3 (12-7114-80); eBioscience); phycoerythrin-conjugated anti-IL-17F (ebio18F10 (12-7471-80); eBioscience); phycoerythrin-conjugated anti-IL-22 (poly5164 (516404); BioLegend); phycoerythrin- cyanine 7-conjugated anti-IFN-γ (XMG1.2 (505826); BioLegend); allophycocyanin-conjugated anti-c-Kit (2B8 (105812); BioLegend); allophycocyanin-conjugated anti-T-bet (4B10 (644813); BioLegend); allophycocyanin-conjugated anti-CD196 (CCR6) (29-2L17 (129812); BioLegend); allophycocyanin-cyanine 5.5-conjugated anti-CD25 (PC61 (551071); BD Biosciences); allophycocyanin-cyanine 5.5-conjugated anti-NKp46 (29A1.4 (137609); BioLegend); anti-CD90.2 (anti-Thy-1.2; 53-2.1 (17-0902-81); eBioscience); peridinin chlorophyll protein-conjugated anti-c-Kit (2B8 (105812); BioLegend); Alexa Fluor 647-conjugated anti-CD206 (MR5D3 (123010); BioLegend); Alexa Fluor 700–conjugated anti-Sca-1 (D7 (56-5981-82); eBioscience); Alexa Fluor 700-conjugated anti-CD4 (GK1.5 (100430); BioLegend); Alexa Fluor 780-conjugated anti-CD11c (N418 (47-0114-82); eBioscience). Human BAL fluid cells were stained with phycoerythrin-Texas red-conjugated to anti-human CD45 (MHCD4517; Invitrogen); fluorescein isothiocyanate-conjugated to anti-CD3 (anti-CD3; UCHT1 (555332); BD Biosciences), anti-CD19 (H1B19 (302206); BioLegend), anti-CD11b (ICRF44 (11-0118-42); eBioscience), anti-CD11c (3.9 (301604); BioLegend), anti-CD49b (AK-7 (314306); BioLegend), anti-CD14 (HCD14 (325604); eBioscience) or anti-FcεR1 (MAR-1 (134306); BioLegend), Alexa Fluor 647-conjugated anti-CD127 (eBioRDR5 (51-1278-73); eBioscience). For intracellular staining, cells were permeabilized (Cytofix/Cytoperm kit; BD Biosciences) and incubated with Alexa Fluor 700-conjugated anti-IL-17A (eBioTC11-18H10.1 (51-7172-80); eBioscience), Alexa Fluor 647-conjugated anti-IL-17A (eBio64CAP17 (51-7178-71); eBioscience), phycoerythrin-conjugated conjugated anti-RoRγT (B2D (12-6981-80); eBioscience) or the respective isotype-matched control antibody (rat IgG1 κ-chain; 51-4301; eBioscience). Cells were analyzed on a FACSCanto (BD Biosciences) with FlowJo 8.3.3 software (TreeStar).
Real-Time PCR
Total RNA was isolated from frozen tissues using TRI reagent (Sigma-Aldrich), and cDNA was synthesized using iScript™ cDNA synthesis kit (Invitrogen, CA) according to manufacturer’s instruction. 0.5–1 µg of cDNA were amplified in the presence of TaqMan universal master mix (Applied Biosystems), gene-specific TaqMan probe, and water. Gene-specific PCR products were measured using an Applied Biosystems 7500 Sequence Detection System. The levels of target gene expression were normalized to GAPDH expression using the 2−ΔΔCt method. The following primers and probes were purchased from Applied Biosystems: mouse GAPDH (4352339E), IL-1β (Mm00434228_m1), IL-6 (Mm00446190_m1), IL-23p19 (Mm01160011_g1), and NLRP3 (Mm00840904_m1).
ELISA
Total IL-17A and IL-1β production from lung cells were measured by ELISA assay kits according to manufacturer’s protocol (eBioscience).
Statistical Analysis
Data are given as mean ± SEM, and were analyzed by ANOVA or unpaired student’s t-tests (two-tailed) (Prism 4; GraphPad Software Inc.). Significance for all statistical tests was shown in figures for p<0.05(*), p <0.01 (**) and p<0.001 (***).
Supplementary Material
Acknowledgments
We thank the NIH tetramer facility, Emory University, Atlanta, GA, for providing CD1d tetramers. This work was supported by grants RO1AI068085, RO1HL62348 and ES013307 from the National Institutes of Health, and with funds from the Dave and Denise Bunning Food Allergy Project.
Abbreviations
- AHR
airway hyperreactivity
- AM
alveolar macrophages
- BAL
bronchoalveolar lavage
- BMI
body mass index
- DCs
dendritic cells
- ILCs
innate lymphoid cells
- HFD
high fat diet
- NKT
natural killer T
- NLRP3
NACHT LRR and PYD domains-containing protein 3
- SFB
segmented filamentous bacteria
- WT
wild type
Footnotes
Author contributions
H.Y.K. designed the study, did the experiments, analyzed the data and wrote the manuscript; H.J.L. did SFB experiments and analyzed the data; Y-J.C. helped with the experiments of human cells; M.P. did experiments; S.A.S analyzed the data and edited the manuscript; K.A.F. and Y.I. provided Nlrp3−/− and Il17−/− mice; E.I, K.B, and J.F provided human BAL fluid samples and information; R.H.D provided reagent support and edited the manuscript and D.T.U. designed the study and wrote the manuscript.
References
- 1.Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999–2010. JAMA. 2012;307:491–497. doi: 10.1001/jama.2012.39. [DOI] [PubMed] [Google Scholar]
- 2.Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18:363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- 3.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 4.Holguin F, et al. Obesity and asthma: an association modified by age of asthma onset. J Allergy Clin Immunol. 2011;127:1486–1493. e1482. doi: 10.1016/j.jaci.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camargo CA, Jr., Weiss ST, Zhang S, Willett WC, Speizer FE. Prospective study of body mass index, weight change, and risk of adult-onset asthma in women. Arch Intern Med. 1999;159:2582–2588. doi: 10.1001/archinte.159.21.2582. [DOI] [PubMed] [Google Scholar]
- 6.Dixon AE, et al. Effects of obesity and bariatric surgery on airway hyperresponsiveness, asthma control, and inflammation. J Allergy Clin Immunol. 2011;128:508–515. e501–e502. doi: 10.1016/j.jaci.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutherland ER, Lehman EB, Teodorescu M, Wechsler ME. Body mass index and phenotype in subjects with mild-to-moderate persistent asthma. J Allergy Clin Immunol. 2009;123:1328–1334. e1321. doi: 10.1016/j.jaci.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shore SA. Obesity, airway hyperresponsiveness, and inflammation. J Appl Physiol. 2010;108:735–743. doi: 10.1152/japplphysiol.00749.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farah CS, Salome CM. Asthma and obesity: a known association but unknown mechanism. Respirology. 2012;17:412–421. doi: 10.1111/j.1440-1843.2011.02080.x. [DOI] [PubMed] [Google Scholar]
- 10.Kanneganti TD, Dixit VD. Immunological complications of obesity. Nat Immunol. 2012;13:707–712. doi: 10.1038/ni.2343. [DOI] [PubMed] [Google Scholar]
- 11.Feuerer M, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura S, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15:914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 13.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 14.Scott HA, Gibson PG, Garg ML, Wood LG. Airway inflammation is augmented by obesity and fatty acids in asthma. Eur Respir J. 2011;38:594–602. doi: 10.1183/09031936.00139810. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Dales R, Jiang Y. The association between obesity and asthma is stronger in nonallergic than allergic adults. Chest. 2006;130:890–895. doi: 10.1378/chest.130.3.890. [DOI] [PubMed] [Google Scholar]
- 16.Moore WC, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181:315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu FL, et al. Increased pulmonary responses to acute ozone exposure in obese db/db mice. Am J Physiol Lung Cell Mol Physiol. 2006;290:L856–L865. doi: 10.1152/ajplung.00386.2005. [DOI] [PubMed] [Google Scholar]
- 18.Arteaga-Solis E, et al. Inhibition of leptin regulation of parasympathetic signaling as a cause of extreme body weight-associated asthma. Cell Metab. 2013;17:35–48. doi: 10.1016/j.cmet.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geremia A, et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med. 2011;208:1127–1133. doi: 10.1084/jem.20101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coccia M, et al. IL-1beta mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4(+) Th17 cells. J Exp Med. 2012;209:1595–1609. doi: 10.1084/jem.20111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klose CS, et al. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 23.Sun Z, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 24.Sawa S, et al. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 25.Cupedo T, et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10:66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 26.Sanos SL, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luci C, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 28.Chang Y-J, et al. Innate lymphoid cells mediate influenza-induced airway hyperreactivity independent of adaptive immunity. Nature Immunol. 2011;12:631–638. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halim TY, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity. 2012;36:451–463. doi: 10.1016/j.immuni.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 30.Klein Wolterink RG, et al. Pulmonary innate lymphoid cells are major producers of IL-5 and IL-13 in murine models of allergic asthma. Eur J Immunol. 2012;42:1106–1116. doi: 10.1002/eji.201142018. [DOI] [PubMed] [Google Scholar]
- 31.Spits H, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 32.Winer S, et al. Normalization of obesity-associated insulin resistance through immunotherapy. Nat Med. 2009;15:921–929. doi: 10.1038/nm.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 35.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Lynch L, et al. Adipose tissue invariant NKT cells protect against diet-induced obesity and metabolic disorder through regulatory cytokine production. Immunity. 2012;37:574–587. doi: 10.1016/j.immuni.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen H, et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol. 2011;12:408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duewell P, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsen CM, et al. Interleukin-1-receptor antagonist in type 2 diabetes mellitus. N Engl J Med. 2007;356:1517–1526. doi: 10.1056/NEJMoa065213. [DOI] [PubMed] [Google Scholar]
- 40.Vandanmagsar B, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. 2011;17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnston RA, et al. Diet-induced obesity causes innate airway hyperresponsiveness to methacholine and enhances ozone-induced pulmonary inflammation. J Appl Physiol. 2008;104:1727–1735. doi: 10.1152/japplphysiol.00075.2008. [DOI] [PubMed] [Google Scholar]
- 42.Williams AS, et al. Obesity and airway responsiveness: Role of TNFR2. Pulmonary pharmacology & therapeutics. 2012 doi: 10.1016/j.pupt.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnston RA, et al. Allergic airway responses in obese mice. Am J Respir Crit Care Med. 2007;176:650–658. doi: 10.1164/rccm.200702-323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calixto MC, et al. Obesity enhances eosinophilic inflammation in a murine model of allergic asthma. British journal of pharmacology. 2010;159:617–625. doi: 10.1111/j.1476-5381.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kudo M, et al. IL-17A produced by alphabeta T cells drives airway hyper-responsiveness in mice and enhances mouse and human airway smooth muscle contraction. Nat Med. 2012;18:547–554. doi: 10.1038/nm.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKinley L, et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J Immunol. 2008;181:4089–4097. doi: 10.4049/jimmunol.181.6.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wakashin H, et al. IL-23 and Th17 cells enhance Th2-cell-mediated eosinophilic airway inflammation in mice. Am J Respir Crit Care Med. 2008;178:1023–1032. doi: 10.1164/rccm.200801-086OC. [DOI] [PubMed] [Google Scholar]
- 48.Lajoie S, et al. Complement-mediated regulation of the IL-17A axis is a central genetic determinant of the severity of experimental allergic asthma. Nat Immunol. 2010;11:928–935. doi: 10.1038/ni.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murdoch JR, Lloyd CM. Resolution of allergic airway inflammation and airway hyperreactivity is mediated by IL-17-producing {gamma}{delta}T cells. Am J Respir Crit Care Med. 2010;182:464–476. doi: 10.1164/rccm.200911-1775OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naik S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gross O, Thomas CJ, Guarda G, Tschopp J. The inflammasome: an integrated view. Immunol Rev. 2011;243:136–151. doi: 10.1111/j.1600-065X.2011.01046.x. [DOI] [PubMed] [Google Scholar]
- 52.Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Besnard AG, et al. NLRP3 inflammasome is required in murine asthma in the absence of aluminum adjuvant. Allergy. 2011;66:1047–1057. doi: 10.1111/j.1398-9995.2011.02586.x. [DOI] [PubMed] [Google Scholar]
- 54.Allen IC, et al. Analysis of NLRP3 in the development of allergic airway disease in mice. J Immunol. 2012;188:2884–2893. doi: 10.4049/jimmunol.1102488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kool M, et al. An unexpected role for uric acid as an inducer of T helper 2 cell immunity to inhaled antigens and inflammatory mediator of allergic asthma. Immunity. 2011;34:527–540. doi: 10.1016/j.immuni.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 56.Marichal T, et al. DNA released from dying host cells mediates aluminum adjuvant activity. Nat Med. 2011;17:996–1002. doi: 10.1038/nm.2403. [DOI] [PubMed] [Google Scholar]
- 57.Crellin NK, et al. Regulation of cytokine secretion in human CD127(+) LTi-like innate lymphoid cells by Toll-like receptor 2. Immunity. 2010;33:752–764. doi: 10.1016/j.immuni.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 58.Winer S, et al. Obesity predisposes to Th17 bias. Eur J Immunol. 2009;39:2629–2635. doi: 10.1002/eji.200838893. [DOI] [PubMed] [Google Scholar]
- 59.Fantuzzi G. Adipose tissue, adipokines, and inflammation. J Allergy Clin Immunol. 2005;115:911–919. doi: 10.1016/j.jaci.2005.02.023. quiz 920. [DOI] [PubMed] [Google Scholar]
- 60.Kim HY, DeKruyff RH, Umetsu DT. The many paths to asthma: phenotype shaped by innate and adaptive immunity. Nat Immunol. 2010;11:577–584. doi: 10.1038/ni.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakae S, et al. Antigen-specific T cell sensitization is impaired in IL-17-deficient mice, causing suppression of allergic cellular and humoral responses. Immunity. 2002;17:375–387. doi: 10.1016/s1074-7613(02)00391-6. [DOI] [PubMed] [Google Scholar]
- 62.Sutterwala FS, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 63.McKenzie GJ, Fallon PG, Emson CL, Grencis RK, McKenzie AN. Simultaneous disruption of interleukin (IL)-4 and IL-13 defines individual roles in T helper cell type 2-mediated responses. J. Exp. Med. 1999;189:1565. doi: 10.1084/jem.189.10.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akbari O, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nature Medicine. 2003;9:582–588. doi: 10.1038/nm851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.