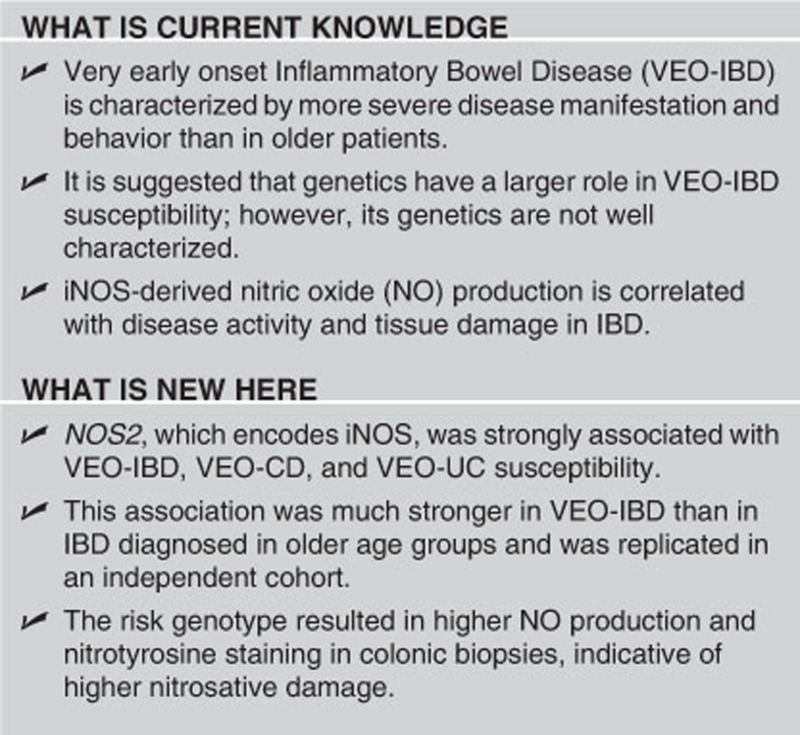
Abstract
OBJECTIVES:
The NOS2 gene encodes for the inducible nitric oxide synthase (iNOS), responsible for nitric oxide (NO) production, which contributes to antimicrobial and antipathogenic activities. Higher levels of both iNOS and NO-induced damage have been observed in inflammatory bowel disease (IBD) patients. NOS2 may have a role in a specific subset of IBD patients with severe and/or extensive colitis. Therefore, the aim of this study is to examine the role of NOS2 in such a subset, very early onset IBD (VEO-IBD).
METHODS:
Seventeen tag single nucleotide polymorphisms (SNPs) in the NOS2 gene were successfully genotyped in VEO-IBD patients. Genetic associations were replicated in an independent VEO-IBD cohort. Functional analysis for iNOS activity was performed on the most significantly associated functional variant.
RESULTS:
The NOS2 rs2297518 SNP was found to be associated in VEO-IBD in two independent cohorts. Upon combined analysis, a coding variant (S608L) showed the strongest association with VEO-IBD (Pcombined=1.13 × 10−6, OR (odds ratio)=3.398 (95% CI (confidence interval) 2.02–5.717)) as well as associations with VEO-Crohn's disease and VEO-ulcerative colitis (UC). This variant also showed an association with UC diagnosed between 11 and 17 years of age but not with adult-onset IBD (>17 years). B-cell lymphoblastoid cell lines genotyped for the risk variant as well as Henle-407 cells transfected with a plasmid construct with the risk variant showed higher NO production. Colonic biopsies of VEO-IBD patients showed higher immunohistochemical staining of nitrotyrosine, indicating more nitrosative stress and tissue damage.
CONCLUSIONS:
These studies suggest the importance of iNOS in genetic susceptibility to younger IBD presentation due to higher NO production.
INTRODUCTION
Very early onset inflammatory bowel disease (VEO-IBD), according to the Paris Modification to the Montreal classification, is described as IBD diagnosed in children under the age of 10 years.1 It is characterized by a higher tendency for disease manifestation in the colon, more disease extension, and a change in disease location over time.2, 3, 4, 5 Pediatric genome-wide association studies (GWAS) have not focused on VEO-IBD;6, 7 however, candidate gene studies have identified novel genetic variants that are associated exclusively with VEO-IBD indicating that very young children have unique genetic susceptibility (reviewed in ref. 8).
Although the pathogenesis of IBD is multi-factorial, genetic and functional studies have confirmed the critical role of the immune system in the pathogenesis of disease.9 Encoded by the NOS2 gene, inducible nitric oxide synthase (iNOS) produces nitric oxide (NO) in response to pro-inflammatory cytokines.10 Although NO is a weak free radical,11 it mediates cell-damaging and toxic effects by forming peroxynitrite, which contributes to DNA damage as well as protein damage by combining with tyrosine to form nitrotyrosine (NT).11, 12 Both iNOS expression and NT staining have previously been shown to be upregulated in the intestinal epithelium and inflamed colonic mucosa of IBD patients.10, 11
Here, we report a genetic association with the NOS2 single nucleotide polymorphisms (SNPs) and VEO-IBD, VEO-Crohn's disease (CD), and VEO-ulcerative colitis (UC). In addition, we found a strong age-biased association between the iNOS variant, rs2297518 (S608L), and VEO-IBD. Last, we explored the function of this SNP and found that it conferred higher NO production based on the risk allele.
METHODS
SNP analysis and genotyping
Eighteen tag SNPs providing complete genetic coverage of the NOS2 gene (chromosome 17, 26,083,792-26,127,555) were selected from the International HapMap Project (www.hapmap.org) Caucasian (CEU) phase II data Release 23a (minor allelic frequency >1%). The Illumina Goldengate Custom Chip (discovery cohort) as previously described13, 14 and Taqman (for replication of the two most significant SNPs from the discovery cohort) were used at the Centre for Applied Genomics, Hospital for Sick Children.
Subjects
All VEO-IBD subjects had a confirmed diagnosis of IBD before the age of 10 based on the Paris classification.1 Phenotypic information and DNA samples were obtained from study subjects with approval of the institutional review ethics board for IBD genetic studies at the Hospital for Sick Children and Mount Sinai Hospital in Toronto. Replication cohorts had ethics board approval for genetic and phenotypic studies at the individual institutions. Written informed consent was obtained from all participants.
The discovery cohort consisted of total of 1,072 subjects including 159 VEO-IBD patients (91 VEO-CD and 68 VEO-UC) and 913 healthy controls recruited from the Hospital for Sick Children and Mt. Sinai Hospital in Toronto. The replication cohort consisted of 736 subjects including 153 VEO-IBD patients (50 VEO-CD and 53 VEO-UC), and 480 healthy controls. The affected subjects were recruited from NEOPICS sites (www.NEOPICS.org) and healthy controls were obtained from the Centre for Applied Genomics (Ontario Population Genomics Platform (plates used: 1–5; a complete description of this control population can be found at http://www.tcag.ca/cyto_population_control_DNA.html)). For the analysis of older IBD groups, 498 IBD subjects (351 CD and 147 UC) diagnosed between 11 and 17 years of age and 918 IBD subjects (419 CD and 499 UC) diagnosed after 17 years of age were included. To conduct systematic quality control on the raw genotyping data, we examined 770 SNPs genotyped for the initial cohort only (18 SNPs genotyped in the initial cohort). Detailed quality control has been previously reported in detail elsewhere.13, 14 One NOS2 SNP, rs2297515, was excluded as it deviated significantly from Hardy–Weinberg equilibrium in the controls (P<0.001). No other NOS2 SNP was excluded due to a genotype call rate of <95%, or due to sex discrepancies based on the heterozygosity rate from SNPs on chromosome X.
Association analysis
Association analyses of the discovery and replication cohorts were used to test associations of the 17 NOS2 SNPs with VEO-IBD, VEO-CD, and VEO-UC vs. healthy controls. Logistic regression analysis was applied for an additive model and Pearson X2 tests were applied for dominant and recessive models. Models used to report P values are described in this report. This analysis was done using the Goldenhelix (SVS 7.6.4) program. The combined cohort analysis pooled population data from both cohorts and analyzed using the same protocol as for the individual cohorts.
Cell culture
Genotyped B-lymphoblastoid cell lines were obtained from Coriell Cell Repositories and cultured in RPMI-1640X with 15% fetal bovine serum at 2 × 105 cells/ml in an upright position at 5% CO2 and 37 °C. Supernatant was collected for Griess assay. In addition, Henle-407 cells transfected with the wild-type and S608L variant (designed with Agilent Technologies QuickChange II Site-Directed Mutagenesis Kit, according to the manufacturer's instructions) of the iNOS-pcDNA3 were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. The Griess assay was performed using a Promega Griess Assay kit (Madison, WI, USA), according to the manufacturer's instructions. Results were replicated. For the B-lymphoblastoid cell lines, absorption readings were taken at 570 nm as described by Uto et al.15 iNOS expression in B-lymphoblastoid cells was measured by western blot analysis using polyclonal Anti-Nitric Oxide Synthase 2 antibody (Boster Biological Technology, Pleasanton, CA, USA) and iNOS-myc expression in transfected Henle-407 cells was measured using monoclonal Anti-Myc Tag antibody (Millipore, Billerica, MA, USA; clone 4A6). Briefly cells were grown in a 10-cm dish, washed twice with 1 × phosphate-buffered saline and then lysed with standard protein lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM ethylene glycol tetraacetic acid, 10% glycerol, 1% Triton X-100), supplemented with 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 1 mM vanadate. A total of 50 ug of protein were used for western blot analysis.
Immunohistochemistry
Formalin-fixed, paraffin-embedded colonic biopsy samples from six VEO-IBD patients (three with major genotype and three with risk genotype) were obtained from the Department of Pathology at the Hospital for Sick Children with approval of the institutional review ethics board for IBD genetic studies at the Hospital for Sick Children. Location of biopsy samples and disease activity were matched between the two groups by a pathologist. Immunohistochemistry was performed with the SuperPicture 3rd Gen IHC Detection Kit from Invitrogen (Carlsbad, CA, USA). Briefly, paraffin sections were deparaffinized using Xylene and afterwards rehydrated with different percentages of ethanol. Antigen retrieval step was performed with high pressure-cooking, using 1 mM EDTA, pH 9 with 0.05% Tween 20 as an antigen retrieval buffer. Primary antibody incubation was performed at 4 °C overnight. Subsequently, the sections were washed three times with phosphate-buffered saline and incubated with peroxidase quenching solution for 5 min. The sections were quickly rinsed in distilled water and washed two times with phosphate-buffered saline. HRP Polymer conjugate incubation was 30 min followed by three times washing with phosphate-buffered saline, each time for 5 min. 3,3′-Diaminobenzidine (DAB) chromogen was added to the sections for 1–5 min. The sections were counterstained with Haematoxylin and mounted with Entelan. NT staining was performed using the polyclonal Anti-Nitrotyrosine antibody from Millipore (anti-rabbit) at a dilution of 1:100.
Slides were scanned using a Zeiss Mirax Digital Slide scanner. Acquired images were deconvolved into grayscale DAB and haematoxylin channels using the Ruifrok and Johnston method as described.16 The staining intensity in the DAB channel for each patient sample and control was evaluated using ImageJ software. Briefly, a technician blinded to the patient diagnoses defined regions of interest representing intestinal epithelia. Within each region of interest, the NT-positive areas were defined as regions containing pixels with values >50. These regions were determined algorithmically using the ImageJ ‘Threshold' function. Regions of interest were converted to binary (where NT-positive regions were defined as 1.0; unstained regions were 0.0) and the fraction of NT-positive area to total area (within each region of interest) was determined. This method was repeated on multiple sections for each patient.
RESULTS
NOS2 association with VEO-IBD
Seventeen tag SNPs in the NOS2 gene were successfully genotyped in the VEO-IBD discovery cohort of 159 VEO-IBD subjects and 913 healthy control individuals (Table 1, Supplementary Table 1). We found that rs2297518 (S608L) was associated with VEO-IBD, VEO-CD, and VEO-UC (PIBD=6.2 × 10−6, OR (odds ratio) (95% confidence interval)=4.4 (2.3–8.4)) (Supplementary Tables 2 and 3). This variant was also significantly associated with IBD diagnosed under the age of 6 years (Supplementary Table 4), a more homogeneous phenotype of IBD patients with a predominance of pancolitis.17 In addition, rs1137933 was associated with VEO-IBD and VEO-UC (P=7.43 × 10−4, OR=2.6 (1.5–4.6)). Both NOS2 variants showed a stronger association with VEO-IBD than adult IBD as observed here and in a previous study by Martin et al.18
Table 1. Discovery cohort association analyses of 17 NOS2 SNPs with VEO-IBD.
|
Recessive association |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr. | SNP | Position | MAF | FU | FA | P-value | OR | L95 | U95 |
| 17 | rs2297518 | 23120724 | 0.18 | 0.23 | 0.17 | 6.2 × 10−6 | 4.43 | 2.32 | 8.46 |
| 17 | rs1137933 | 23130059 | 0.21 | 0.27 | 0.21 | 7.4 × 10−4 | 2.64 | 1.50 | 4.65 |
| 17 | rs9906835 | 23113501 | 0.42 | 0.38 | 0.43 | 0.39 | 0.81 | 0.51 | 1.29 |
| 17 | rs944725 | 23133698 | 0.41 | 0.50 | 0.40 | 0.03 | 1.56 | 1.03 | 2.34 |
| 17 | rs3730013 | 23150045 | 0.34 | 0.29 | 0.34 | 0.19 | 0.67 | 0.36 | 1.22 |
| 17 | rs4795067 | 23130802 | 0.33 | 0.35 | 0.33 | 0.08 | 1.48 | 0.94 | 2.33 |
| 17 | rs2297516 | 23119857 | 0.42 | 0.38 | 0.42 | 0.91 | 1.02 | 0.66 | 1.58 |
| 17 | rs3794764 | 23135555 | 0.23 | 0.21 | 0.23 | 0.71 | 0.86 | 0.40 | 1.86 |
| 17 | rs8072199 | 23140975 | 0.44 | 0.51 | 0.44 | 0.482 | 1.16 | 0.76 | 1.75 |
| 17 | rs10459953 | 23151645 | 0.35 | 0.33 | 0.35 | 0.93 | 1.02 | 0.61 | 1.65 |
| 17 | rs2314809 | 23119505 | 0.40 | 0.36 | 0.40 | 0.92 | 0.97 | 0.62 | 1.53 |
| 17 | rs11080344 | 23128638 | 0.46 | 0.44 | 0.46 | 0.09 | 1.39 | 0.94 | 2.05 |
| 17 | rs3794756 | 23110756 | 0.01 | 0.02 | 0.01 | NA | NA | NA | NA |
| 17 | rs3729508 | 23133157 | 0.40 | 0.38 | 0.40 | 0.96 | 1.01 | 0.64 | 1.58 |
| 17 | rs11653716 | 23108659 | 0.032 | 0.05 | 0.03 | NA | NA | NA | NA |
| 17 | rs2314810 | 23128237 | 0.04 | 0.04 | 0.04 | 0.56 | 1.93 | 0.19 | 18.71 |
| 17 | rs3730017 | 23133229 | 0.020 | 0.02 | 0.02 | NA | NA | NA | NA |
FA, frequency affected; FU, frequency unaffected; L95 and U95, lower and upper 95th confidence interval; MAF, minor allelic frequency.
P values are presented as uncorrected and recessive modeling.
In an independent replication cohort, consisting of 153 VEO-IBD patients and 480 healthy controls, rs2297518 was also associated with VEO-IBD with a combined analysis of Pcombined=1.13 × 10−6; OR=3.4 (2.0–5.7). The association of rs1137933 with VEO-IBD did not replicate in this cohort; however, remained significant in the combined analysis (Table 2).
Table 2. Replication and combined cohort analyses of NOS2 SNP associated with VEO-IBD.
|
Replication analysis: VEO-IBD |
Combined analysis: VEO-IBD |
||||
|---|---|---|---|---|---|
| SNP | Diagnosis | P-value | OR (95% CI) | P-value | OR (95% CI) |
| rs2297518 | VEO-IBD | 0.04 | 2.4 (1.0–5.8) | 1.13 × 10−6 | 3.3 (2.0–5.7) |
| VEO-CD | 0.51 | 1.6 (0.3–7.6) | 1.19 × 10−4 | 3.5 (1.7–6.8) | |
| VEO-UC | 0.18 | 2.3 (0.6–8.5) | 4.81 × 10−4 | 3.3 (1.6–7.0) | |
| rs1137933 | VEO-IBD | 0.63 | 1.2 (0.4–3.3) | 6.13 × 10−3 | 1.9 (1.1–3.1) |
| VEO-CD | 0.71 | 1.31(0.2–5.9) | 0.02 | 2.0 (1.0–3.9) | |
| VEO-UC | 0.61 | 0.5 (0.7–4.5) | 0.04 | 2.0 (0.9–4.0) | |
CD, Crohn's disease; CI, confidence interval; IBD, inflammatory bowel disease; OR, odds ratio; SNP, single nucleotide polymorphism; UC, ulcerative colitis.
P values are presented as uncorrected and recessive modeling.
In the analysis of older children and adults with IBD (Supplementary Tables 5–10), NOS2 was associated with UC diagnosed between 11 and 17 years of age (A1b, Paris classification; rs2297518; P=9.65 × 10−4, OR=3.3 (1.6–6.8); Supplementary Table 10) and not associated with adult-onset IBD (>17 years age group). This analysis suggests that NOS2 variants are associated with VEO-IBD.
rs2297518 risk genotype is associated with higher production of NO
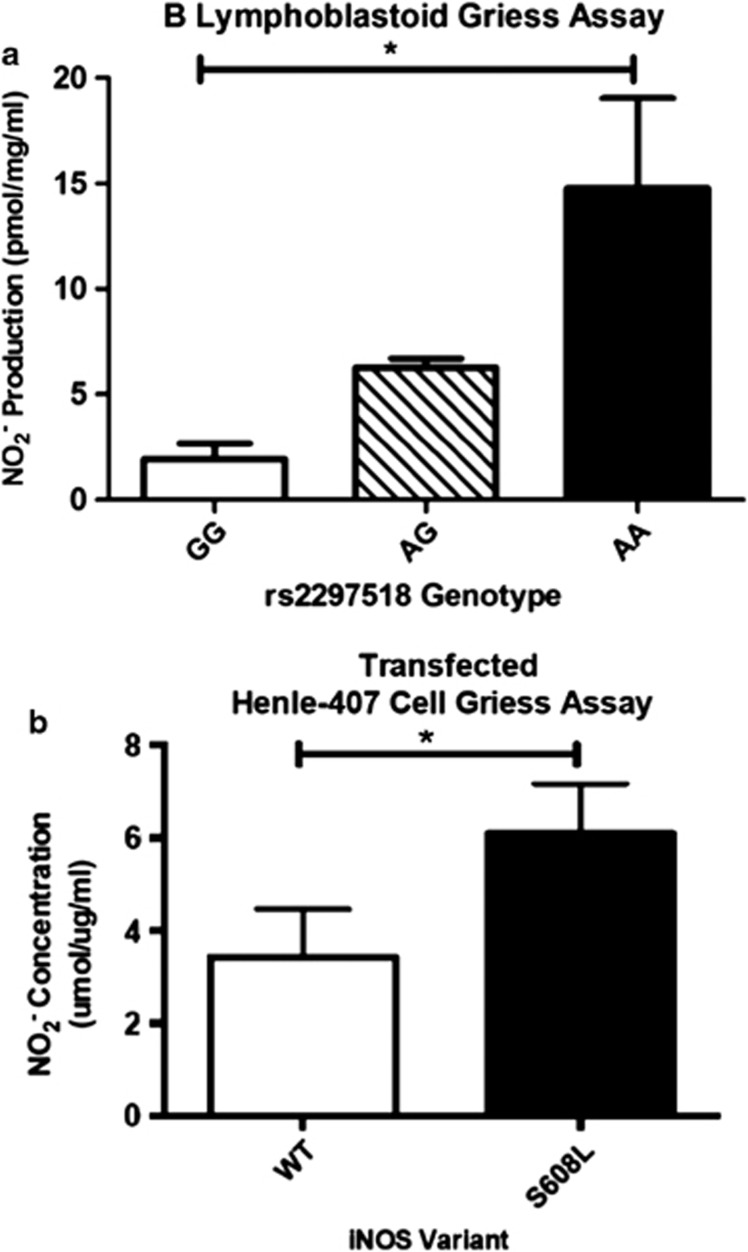
As B-lymphoblastoid cell lines have constitutive expression of iNOS,19 supernatants from B-lymphoblastoid cell lines genotyped for rs2297518, were collected. To measure the NO production, 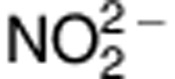 levels, indicative of NO production, were measured using the Griess assay. Levels were compared among the three genotypes of rs2297518, homozygous for the non-risk allele (G/G), homozygous for the risk allele (A/A), and heterozygous for both alleles (A/G). As shown in Figure 1a, cell lines homozygous for the risk allele had significantly higher production of NO (one-way ANOVA, F(2,8)=24.39; P=0.0026; Tukey's HSD (honestly significant difference) test, P<0.05) indicating that leukocytes in patients homozygous for the risk allele (iNOS S608L) have higher levels of NO production, when normalized with protein concentration. As shown by the imputation analysis (Supplementary Table 13), B-lymphoblastoid cell lines genotyped for the rs2297518 SNP do not have a significant change in iNOS expression.20 Western blot for iNOS (Supplementary Figure 1a), although expression varied between cell lines, demonstrated no trend of expression changes among the three genotypes.
levels, indicative of NO production, were measured using the Griess assay. Levels were compared among the three genotypes of rs2297518, homozygous for the non-risk allele (G/G), homozygous for the risk allele (A/A), and heterozygous for both alleles (A/G). As shown in Figure 1a, cell lines homozygous for the risk allele had significantly higher production of NO (one-way ANOVA, F(2,8)=24.39; P=0.0026; Tukey's HSD (honestly significant difference) test, P<0.05) indicating that leukocytes in patients homozygous for the risk allele (iNOS S608L) have higher levels of NO production, when normalized with protein concentration. As shown by the imputation analysis (Supplementary Table 13), B-lymphoblastoid cell lines genotyped for the rs2297518 SNP do not have a significant change in iNOS expression.20 Western blot for iNOS (Supplementary Figure 1a), although expression varied between cell lines, demonstrated no trend of expression changes among the three genotypes.
Figure 1.
(a) Griess assay results from the B-lymphoblastoid cell culture. G is the non-risk allele and A is the risk-allele (N=9). ANOVA testing (F(2,8)=24.39; P=0.0026) revealed statistically significant variance between the genotypes. Mean NO2 production was not significantly different between the AG and GG genotypes; however, both genotypes were significantly different from the AA genotype (Tukey's HSD test; P<0.05). (b) Griess assay results from the transfected Henle-407 cells, comparing constitutive expression of wild-type and S608L variants of iNOS (Student's t-test, n=6, P<0.05).
To explore the difference in the production of NO in human intestinal epithelial-derived cells, Henle-407 cells were used to study the difference in NO production between the major and risk variants of rs2297518. As shown in Figure 1b, cells transfected with the S608L variant of the iNOS-pcDNA3.1 construct showed higher constitutive production of 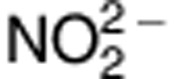 than the wild-type variant (P<0.05). This result further suggests that this coding variant alters iNOS protein function. Western blot for iNOS in transfected cells (Supplementary Figure 1b) confirmed that this difference was not due to an iNOS expression difference.
than the wild-type variant (P<0.05). This result further suggests that this coding variant alters iNOS protein function. Western blot for iNOS in transfected cells (Supplementary Figure 1b) confirmed that this difference was not due to an iNOS expression difference.
Patients with rs2297518 risk genotype show higher peroxynitrite levels in colonic biopsies
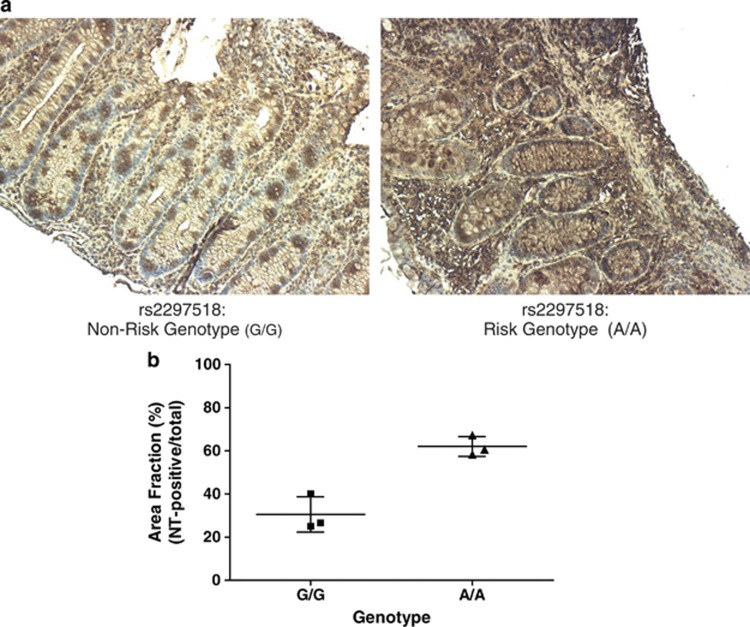
To assess any change in peroxynitrite levels in these patients, colonic biopsies from genotyped patients were stained with antibodies to NT, which is formed by nitrosylation of tyrosine residues due to peroxynitrite.12 Consistent with Griess assay results from the genotyped B-lymphoblastoid cell lines and transfected Henle-407 cells, VEO-IBD patients with the risk genotype of rs2297518 were shown to have higher levels of NT staining (Figure 2, Supplementary Figure 2). It is suggested that this is indicative of higher levels of nitrosative stress and damage in patients with the risk genotype. Although overall levels observed varied due to differences among individual patients and cases, NT staining was consistently more pronounced in both the epithelial (crypt cells and brush border) and leukocyte infiltrate.
Figure 2.
(a). Immunohistochemistry staining for NT of paraffin-embedded slides of colonic biopsies of VEO-IBD patients from the Hospital for Sick Children genotyped for the rs2297518 SNP. Shown is a representative image of VEO-IBD patients with non-risk (G/G) and risk genotypes (A/A). Images shown are taken at 10 × magnification. (b) Plot of area fraction (as %) describing NT-positive regions as a ratio of the total tissue section area in patients with non-risk (G/G) and risk (A/A) genotype (Student's t-test, n=3 per group, P=0.02).
DISCUSSION
iNOS is expressed in inflamed tissues and there is a known correlation between higher iNOS expression and colitis although the mechanism is unknown.10 Here we report that both rs2297518 and rs1137933 were found to be associated with VEO-IBD. Although previously reported in adults,18 this association was not replicated in recent adult-onset IBD GWAS and meta-analysis, where rs2297518 was not associated with either CD or UC, and rs1137933 was not included in the meta-analysis (http://medicine.yale.edu/intmed/ibdgc/index.aspx; Supplementary Tables 11 and 12).21, 22, 23 Furthermore, neither SNPs in LD (r2>0.8) with rs2297518 (Supplementary Table 13) nor other NOS2 SNP were not found to be associated with either CD or UC in the meta-analyses. However, an NOS2 SNP, rs2945412, was found to be associated with CD in the recent adult-onset IBD meta-analyses indicating that common variants in NOS2 may also be associated with adult-onset IBD.21 Current estimates suggest that adult-onset IBD GWAS have explained ∼23–33% of the inherited contribution to IBD risk.21, 22, 23 Therefore, there remains substantial ‘missing heritability' that will not be fully explained by simply expanding the GWAS approach with larger numbers of patients.22 Our candidate gene approach using a specific extreme IBD phenotype, VEO-IBD in this case, may allow for the identification of novel susceptibility loci not found in GWAS and allow for further understanding of the genetic basis of IBD. Here we observed associations between rs2297518 and VEO-IBD, VEO-CD, and VEO-UC as well as an association with UC between 11 and 17 years of age. This age-biased association indicates the functional significance of this variant as well as the importance of iNOS activity in the susceptibility to younger disease presentation. However, there are limitations to the study including the small sample size compared with adult IBD genetic studies and variability in inflammation and iNOS staining in our patient population.
A number of inflammatory diseases have overlapping genetic susceptibility with IBD24 and rs2297518 (S608L) has also been implicated in the susceptibility of type 1 diabetes, severe asthma, and atrophic gastritis.25 Interestingly, the above diseases all have inflammatory or autoimmune phenotypes, indicating that an increase in iNOS activity may contribute to the pathogenesis of these diseases. The rs2297518 risk allele results in a S608L amino-acid change that is located in the catalytic domain of iNOS and Johannesen et al.26 suggest that this S608L substitution may affect the catalytic activity of iNOS, as it is close in proximity to the Flavin mononucleotide-binding region. This risk genotype of rs2297518 also resulted in higher iNOS activity in both genotyped B-lymphoblastoid cell lines as well as plasmid vector-transfected Henle-407 cells, as measured by the Griess assay. The higher iNOS activity observed was further supported by higher levels of NT staining in colonic biopsies of VEO-IBD patients with the risk genotype, suggesting that the higher NO production from this risk variant contributes to higher nitrosative stress and peroxynitrite damage in the intestines of IBD patients. As younger pediatric IBD patients (both CD and UC) often present with severe pancolitis3, 4, 5 and iNOS expression is correlated to the severity of colitis in UC patients,10 the higher tissue damage in these young children may explain the association of rs2297518 with VEO-IBD as compared with adolescent- and adult-onset IBD.
In an expression analysis from imputed genotypes (http://www.sph.umich.edu/csg/liang/imputation/), it was shown that rs2297518 was associated with decreased ARG2 gene expression (P=6.0 × 10−5). Arg2 protein is an inhibitor of iNOS activity as it competes for L-arginine.27 With respect to gastrointestinal inflammation, it has been shown that Arg2 activity is induced in Helicobacter pylori-induced macrophages, which have reduced bacterial killing due to lower iNOS translation.28, 29 As such, the higher iNOS activity observed in VEO-IBD patients with the rs2297518 risk allele may be due to the lower expression of the Arg2 that has been shown to be associated with the NOS2 risk allele.
Internationally there is a documented rise in incidence of pediatric IBD, with 77.8% of studies investigating this trend reporting significant increases.30 In Ontario, this rise in pediatric IBD has been shown to be due to the specific increase in incidence of VEO-IBD, in contrast to the age group diagnosed between the ages of 11 and 17 years.31 Pediatric GWAS have not shown unique susceptibility loci compared with adult-onset IBD;6, 7 however, these studies have focused on older children with the disease diagnosed between 11 and 17 years of age and mostly excluded VEO-IBD. VEO-IBD is considered a unique group of patients with a distinct phenotype and novel genetic susceptibility.8 This has been demonstrated in recent studies showing that rare and private mutation in a number of genes result in susceptibility to VEO-IBD including NCF2,14 IL10R,32, 33 and ADAM17.34 As these examples have yielded the potential for personalized therapies for severe and complicated VEO-IBD cases, it is possible that VEO-IBD patients with higher activity of iNOS may benefit from advancements in therapeutic inhibition of NO production including specific iNOS inhibitors, such as N-[3-(aminomethyl) benzyl] acetamidine (1400W) and L-N6-(1-iminoethyl) lysine (L-NIL), which have been shown to have a protective effect in rat 2,4,6-trinitrobenzenesulfonic colitis models.11 Thus, we propose that NOS2 variants influencing higher activity of iNOS contribute to the genetic susceptibility to earlier onset of IBD due to NO-induced tissue damage and intestinal inflammation.
Study Highlights
Acknowledgments
We thank all IBD patients and their families who participated in this research. We also acknowledge the work of Karoline Fiedler and Maggie Zhang at the Hospital for Sick Children and Joanne Stempak at Mount Sinai. We also appreciate the input and guidance from Dr Ramzi Fattouh, Ziad Al Adham, and Zhen Zhao. Last, we acknowledge the assistance from the Department of Pathology at the Hospital for Sick Children for providing samples for immunohistochemistry.
Guarantor of the article: Aleixo M. Muise, MD, PhD.
Specific author contributions: S.S.D., L.A.M., and A.M.M. conceived and designed all experiments. A.M.M., T.W., A.M., D.M., M.S.S., H.H., S.B., and S.B.S. provided patient samples. S.S.D., A.E., W.X., and T.W. analyzed the data. S.S.D., L.A.M., C.T., C.G., and C.G. performed functional experiments under the supervision of A.M.M. with help and guidance from J.H.B. S.S.D. wrote the manuscript with L.A.M. and A.M.M. and contributions from all authors.
Financial support: A.M.M. is supported by an Early Researcher Award from the Ontario Ministry of Research and Innovation and a CDHNF/NASPGHAN George Ferry Young Investigator Development Award and funded by a Canadian Institute of Health Research—Operating Grant (MOP119457). Partial funding for this study was provided by the FLIBD (Future Leaders in IBD) Program through an unrestricted grant from Janssen Canada to A.M.M. S.B.S. is supported in part by the Wolpow Family Chair in IBD Treatment and Research.
Potential competing interests: None.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Supplementary Material
References
- Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis. 2011;17:1314–1321. doi: 10.1002/ibd.21493. [DOI] [PubMed] [Google Scholar]
- Van Limbergen J, Russell RK, Drummond HE, et al. Definition of phenotypic characteristics of childhood-onset inflammatory bowel disease. Gastroenterology. 2008;135:1114–1122. doi: 10.1053/j.gastro.2008.06.081. [DOI] [PubMed] [Google Scholar]
- Ruemmele FM, El Khoury MG, Talbotec C, et al. Characteristics of inflammatory bowel disease with onset during the first year of life. J Pediatric Gastroenterol Nutr. 2006;43:603–609. doi: 10.1097/01.mpg.0000237938.12674.e3. [DOI] [PubMed] [Google Scholar]
- Heyman MB, Kirschner BS, Gold BD, et al. Children with early-onset inflammatory bowel disease (IBD): analysis of a pediatric IBD consortium registry. J Pediatr. 2005;146:35–40. doi: 10.1016/j.jpeds.2004.08.043. [DOI] [PubMed] [Google Scholar]
- Heyman MB, Kirschner BS, Gold BD, et al. Children with early-onset inflammatory bowel disease (IBD): analysis of a pediatric IBD consortium registry. J Pediatr. 2005;146:35–40. doi: 10.1016/j.jpeds.2004.08.043. [DOI] [PubMed] [Google Scholar]
- Kugathasan S, Baldassano RN, Bradfield JP, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Baldassano RN, Griffiths A, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muise AM, Snapper SB, Kugathasan S. The age of gene discovery in very early onset inflammatory bowel disease. Gastroenterology. 2012;143:285–288. doi: 10.1053/j.gastro.2012.06.025. [DOI] [PubMed] [Google Scholar]
- Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guihot G, Guimbaud R, Bertrand V, et al. Inducible nitric oxide synthase activity in colon biopsies from inflammatory areas: correlation with inflammation intensity in patients with ulcerative colitis but not with Crohn's disease. Amino Acids. 2000;18:229–237. doi: 10.1007/s007260050020. [DOI] [PubMed] [Google Scholar]
- Cross RK, Wilson KT. Nitric oxide in inflammatory bowel disease. Inflamm Bowel Dis. 2003;9:179–189. doi: 10.1097/00054725-200305000-00006. [DOI] [PubMed] [Google Scholar]
- McCafferty DM. Peroxynitrite and inflammatory bowel disease. Gut. 2000;46:436–439. doi: 10.1136/gut.46.3.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muise AM, Walters T, Xu W, et al. Single nucleotide polymorphisms that increase expression of the guanosine triphosphatase RAC1 are associated with ulcerative colitis. Gastroenterology. 2011;141:633–641. doi: 10.1053/j.gastro.2011.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muise AM, Xu W, Guo CH, et al. NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut. 2012;61:1028–1035. doi: 10.1136/gutjnl-2011-300078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uto T, Contreras MA, Gilg AG, et al. Oxidative imbalance in nonstimulated X-adrenoleukodystrophy-derived lymphoblasts. Dev Neurosci. 2008;30:410–418. doi: 10.1159/000191212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23:291–299. [PubMed] [Google Scholar]
- Sherlock M, van den Heuvel M, Walters T, et al. Phenotypic evolution of pediatric inflammatory bowel disease using the new paris classification Gastroenterology 2011140S–90.S-91. [Google Scholar]
- Martin MC, Martinez A, Mendoza JL, et al. Influence of the inducible nitric oxide synthase gene (NOS2A) on inflammatory bowel disease susceptibility. Immunogenetics. 2007;59:833–837. doi: 10.1007/s00251-007-0255-1. [DOI] [PubMed] [Google Scholar]
- Mannick JB, Asano K, Izumi K, et al. Nitric oxide produced by human B lymphocytes inhibits apoptosis and Epstein-Barr virus reactivation. Cell. 1994;79:1137–1146. doi: 10.1016/0092-8674(94)90005-1. [DOI] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees CW, Barrett JC, Parkes M, et al. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- Karasneh JA, Darwazeh AM, Hassan AF, et al. Association between recurrent aphthous stomatitis and inheritance of a single-nucleotide polymorphism of the NOS2 gene encoding inducible nitric oxide synthase. J Oral Pathol Med. 2011;40:715–720. doi: 10.1111/j.1600-0714.2011.01039.x. [DOI] [PubMed] [Google Scholar]
- Johannesen J, Pie A, Pociot F, et al. Linkage of the human inducible nitric oxide synthase gene to type 1 diabetes. J Clin Endocrinol Metab. 2001;86:2792–2796. doi: 10.1210/jcem.86.6.7559. [DOI] [PubMed] [Google Scholar]
- Mori M. Regulation of nitric oxide synthesis and apoptosis by arginase and arginine recycling. J Nutr. 2007;137:1616S–1620S. doi: 10.1093/jn/137.6.1616S. [DOI] [PubMed] [Google Scholar]
- Lewis ND, Asim M, Barry DP, et al. Arginase II restricts host defense to Helicobacter pylori by attenuating inducible nitric oxide synthase translation in macrophages. J Immunol. 2010;184:2572–2582. doi: 10.4049/jimmunol.0902436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ND, Asim M, Barry DP, et al. Immune evasion by Helicobacter pylori is mediated by induction of macrophage arginase II. J Immunol. 2011;186:3632–3641. doi: 10.4049/jimmunol.1003431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchimol EI, Fortinsky KJ, Gozdyra P, et al. Epidemiology of pediatric inflammatory bowel disease: a systematic review of international trends. Inflamm Bowel Dis. 2011;17:423–439. doi: 10.1002/ibd.21349. [DOI] [PubMed] [Google Scholar]
- Benchimol EI, Guttmann A, Griffiths AM, et al. Increasing incidence of paediatric inflammatory bowel disease in Ontario, Canada: evidence from health administrative data. Gut. 2009;58:1490–1497. doi: 10.1136/gut.2009.188383. [DOI] [PubMed] [Google Scholar]
- Glocker EO, Kotlarz D, Boztug K, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran CJ, Walters TD, Guo CH, et al. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm Bowel Dis. 2012;19:115–123. doi: 10.1002/ibd.22974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaydon DC, Biancheri P, Di WL, et al. Inflammatory skin and bowel disease linked to ADAM17 deletion. N Engl J Med. 2011;365:1502–1508. doi: 10.1056/NEJMoa1100721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.