Abstract
Nucleosome occupancy plays a key role in regulating access to eukaryotic genomes. Although various chromatin regulatory complexes are known to regulate nucleosome occupancy, the role of DNA sequence in this regulation remains unclear, particularly in mammals. To address this problem, we measured nucleosome distribution at high temporal resolution in human cells at hundreds of genes during the reactivation of Kaposi's sarcoma–associated herpesvirus (KSHV). We show that nucleosome redistribution peaks at 24 h post-KSHV reactivation and that the nucleosomal redistributions are widespread and transient. To clarify the role of DNA sequence in these nucleosomal redistributions, we compared the genes with altered nucleosome distribution to a sequence-based computer model and in vitro–assembled nucleosomes. We demonstrate that both the predicted model and the assembled nucleosome distributions are concordant with the majority of nucleosome redistributions at 24 h post-KSHV reactivation. We suggest a model in which loci are held in an unfavorable chromatin architecture and “spring” to a transient intermediate state directed by DNA sequence information. We propose that DNA sequence plays a more considerable role in the regulation of nucleosome positions than was previously appreciated. The surprising findings that nucleosome redistributions are widespread, transient, and DNA-directed shift the current perspective regarding regulation of nucleosome distribution in humans.
The position and density of nucleosomes can regulate access to DNA, thereby influencing all nuclear processes, such as transcription, replication, recombination, and repair (Kornberg and Lorch 1999). Protein complexes that act in trans, by covalently modifying histones or by altering the relationship of the DNA to the histone octamer, are critical in the formation of these chromatin architectures (Kingston and Narlikar 1999). In addition, the DNA sequence acting in cis can affect the DNA conformation required to form a nucleosome and thereby regulate nucleosome occupancy (Drew and Travers 1985). This raises the question of what role cis- and trans-acting factors play in the redistribution of nucleosomes.
Recent studies have demonstrated that there is an interplay between chromatin remodeling and DNA-directed nucleosome occupancy in yeast (Fazzio and Tsukiyama 2003; Whitehouse and Tsukiyama 2006; Gkikopoulos et al. 2011). However, the extent to which DNA-directed nucleosome positioning signals play a role in the redistribution of nucleosomes, as families of human genes respond to a common stimulus, remains largely unexplored. DNA sequences containing naturally occurring nucleosome-positioning signals, such as the mouse mammary tumor virus long terminal repeat and the Xenopus 5S rDNA, have been investigated for years and have served as key models for our current understanding of the role of nucleosome position in transcription (Gottesfeld and Bloomer 1980; Richard-Foy and Hager 1987). Computational models have been derived that identify strong, DNA-directed (cis-acting) nucleosome-positioning signals (Segal et al. 2006; Gupta et al. 2008; Yuan and Liu 2008). DNA-encoded nucleosome position signals are static values, while nucleosome distributions are dynamic.
The role that DNA sequence plays in regulating in vivo nucleosome redistributions has been debated for several years (Segal et al. 2006; Albert et al. 2007; Kaplan et al. 2009, 2010; Zhang et al. 2009, 2010). In this study, we investigated the role that DNA sequence plays in nucleosome redistribution. Three formal possibilities relate DNA sequence to nucleosome redistributions. (1) The DNA-encoded nucleosome distribution will disagree with both basal and redistributed nucleosome architecture, suggesting that DNA sequence does not play a significant role in nucleosome distribution. (2) The DNA-encoded nucleosome distribution will agree with the basal architecture, suggesting that DNA sequence plays a role in the architecture of the basal state. (3) The DNA-encoded nucleosome distribution will agree with the redistributed architecture, suggesting that DNA sequence plays a role in the architecture of the redistributed state.
Here we use the well-studied reactivation of Kaposi's sarcoma–associated herpesvirus (KSHV) as a model for a set of genes responding to a common stimulus. The immune response is a well-characterized acute response in which certain genes are induced to undergo chromatin structural changes by pathogen infection or other environmental stresses (Agalioti et al. 2000; Ramirez-Carrozzi et al. 2006, 2009; Foster et al. 2007). KSHV is a human DNA tumor virus and the etiological agent of several human cancers, including Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease (Chang et al. 1994; Mesri et al. 2010). Upon infection, KSHV characteristically establishes latency, during which the viral genome is tethered to the host chromosome and replicated episomally by the host machinery. To spread within an individual or to new hosts, the virus must periodically reactivate to enter a lytic cycle, during which most viral genes are expressed, resulting in the production of infectious progeny and ultimately cell death. Despite its cell-destructive nature, the lytic cycle is believed to play key roles in the development of KSHV malignancies (Ganem 2010). The study of host responses to KSHV lytic replication is therefore crucial for understanding KSHV pathogenesis.
We measured the nucleosome distribution at high temporal resolution during KSHV lytic reactivation. Upon KSHV reactivation, there is a concerted, widespread, and transient nucleosomal redistribution that is facilitated by the DNA sequence, as nucleosome redistributions change in a manner predicted by DNA sequence. We therefore propose that a genetically directed “spring-loaded” redistribution of nucleosomes on these promoters facilitates the concerted action that is critical for the genomic response.
Results
Reactivation of KSHV resulted in widespread, transient nucleosome redistribution
To assess the relationship between nucleosome distribution and the reactivation of KSHV, we investigated the transcription start sites (TSSs) of 472 immunity-related genes (Supplemental Fig. 1; Supplemental Table 1). We chose to study the well-characterized iSLK system for the reactivation of KSHV (Vieira and O'Hearn 2004; Myoung and Ganem 2011). We used control cells (iSLK/mock), and cells with latent KSHV infection (iSLK.219), induced both cell lines with doxycycline, and measured nucleosome distribution at the TSS of these 472 genes at 6, 12, 24, and 48 h post-KSHV reactivation. We confirmed that maximal reactivation of the virus occurs 48 h post-induction by measuring protein levels of RTA, ORF45, and PF8 (Supplemental Figs. 2, 3).
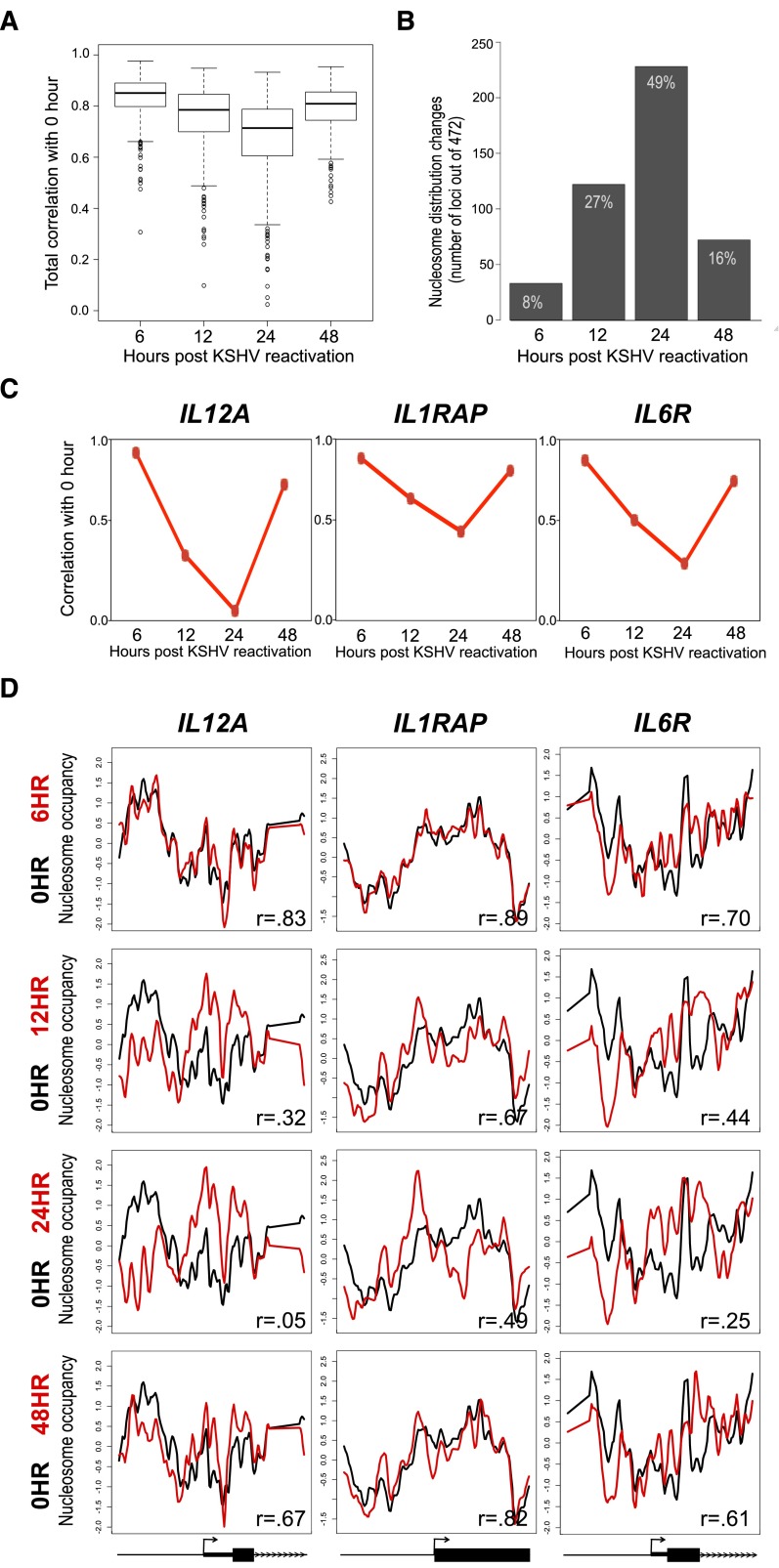
We identified widespread nucleosome distribution changes by calculating the correlation between the untreated cells (0 h) in their basal state and KSHV-reactivated time points (Fig. 1A). We used a correlation threshold (r = 0.7) to identify loci with nucleosome distribution changes at each time point (Supplemental Figs. 4–6; Girimurugan and Chicken 2013). Nucleosome distribution changes peaked at 24 h post-reactivation, where 226 genes (49%) had altered nucleosome distribution, showing that these changes are widespread (Fig. 1B). Remarkably, a majority of these changes were transient, with 158 of the 226 loci (70%) returning to the basal architectures at 48 h post-reactivation (Fig. 1B; Supplemental Fig. 7). Doxycycline induced no significant nucleosome distribution changes in the control cells (mock), verifying that the changes resulted solely from the reactivation of KSHV (Supplemental Figs. 8, 9).
Figure 1.
Reactivation of KSHV resulted in widespread, transient nucleosome redistribution. (A) Boxplot of the correlation values for the 472 loci nucleosome distributions between the 0-h time point and the time points following KSHV reactivation. (B) Number of loci with nucleosome distribution changes (r < 0.7) at 6, 12, 24, and 48 h post-reactivation of KSHV. The greatest number of nucleosome distribution changes occurred at 24 h, and the nucleosome redistributions transiently return to the basal state nucleosome distribution (0 h) at 48 h. (C) The correlation values between the nucleosome distributions of the 0-h and post-KSHV reactivation time points for the IL12A, IL1RAP, and IL6R TSSs. (D) The nucleosome distribution from latent (black lines) and reactivated (red lines) post-KSHV time points for three genes: IL12A, IL1RAP, and IL6R loci. The x-axis represents genomic position showing 2 kb centered on a TSS. The y-axis is the log2 ratio of nucleosomally protected DNA to genomic DNA signal at each probe on the microarray. Gene models from the RefSeq annotations are shown below each gene column.
Identification of these widespread and transient changes provided the unique opportunity to determine whether the nucleosome distribution at the TSS is altered in a concerted manner. To determine whether the changes in nucleosome distribution reflected a concerted response, we measured the range of nucleosome distribution values across each locus and analyzed the changes in nucleosome occupancy strength. An increase in the intensity of the nucleosome-free signals and concomitant increase in the intensity of the nucleosome occupied signals would suggest stronger nucleosome positioning. A significant increase in nucleosome occupancy occurs post-reactivation, with maximal occupancy occurring at 24 h (Supplemental Fig. 10). These data suggest that the nucleosomes are becoming more positioned at the 24-h time point.
To illustrate the widespread and transient changes, we show three of the loci showing the greatest degree of nucleosome redistribution (Fig. 1C,D; for exemplar unchanged loci please see IL7, REL, and TNFSF9 in Supplemental Fig. 1). We observed progressive nucleosome redistribution from 0 h (basal state), with the greatest difference occurring at 24 h (transient state). The nucleosome redistributions were transient and returned to their basal state nucleosome distributions by 48 h. We next focused our analyses on the underlying forces that influenced the nucleosome redistributions.
Nucleosome redistributions are temporally associated with induction of BRG1
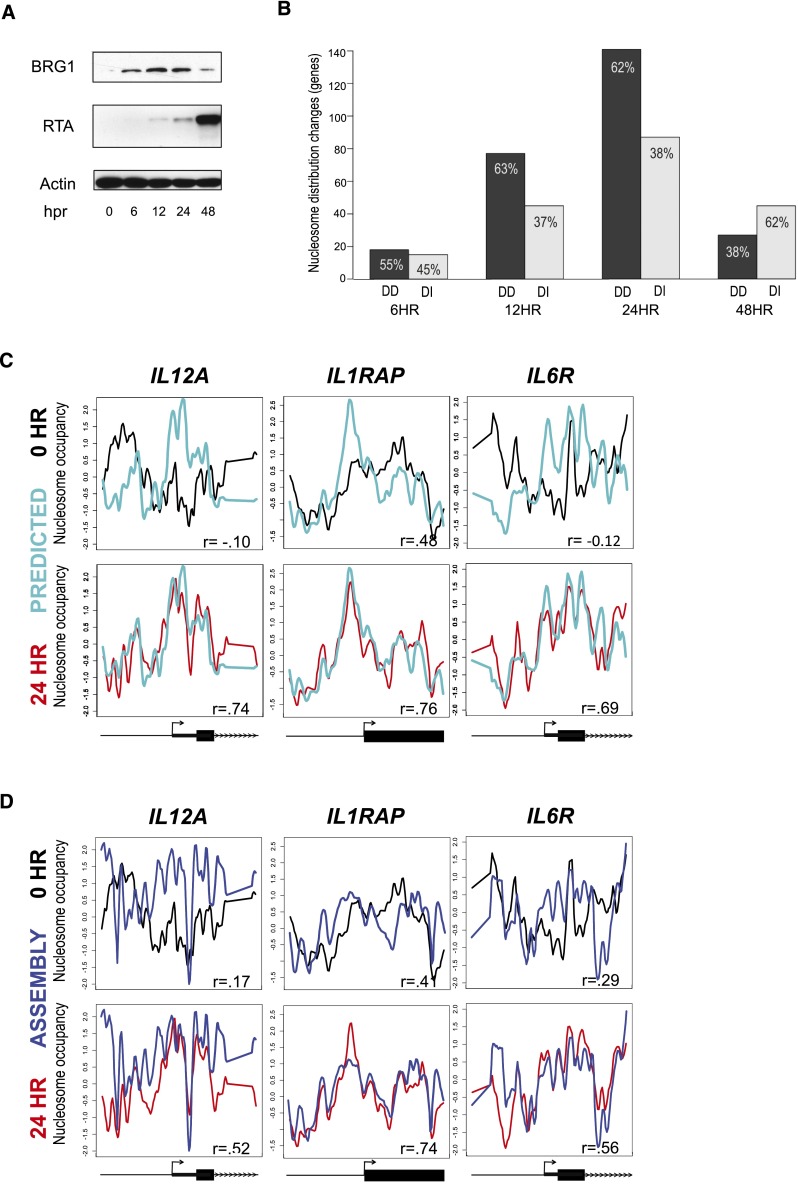
Nucleosome distribution is the result of the interplay between chromatin regulatory complexes, such as ATP-dependent remodelers, and features of the underlying DNA sequence (Gupta et al. 2008; Fincher and Dennis 2011). Because BRG1, the motor protein of the SWI/SNF ATP-dependent remodeling complex, is recruited to the promoter region of several genes investigated in this study (Agalioti et al. 2000; Ramirez-Carrozzi et al. 2006, 2009), we expected that chromatin remodelers would play a role in the nucleosome redistributions post-KSHV reactivation. Therefore, we measured levels of BRG1 to determine whether global levels of remodelers were altered in the reactivation of KSHV. BRG1 levels increased at 6 and 12 h post-reactivation, peaked at 24 h, and then fell to background levels by 48 h (Fig. 2A). Protein levels of BRG1 were concordant with the changes in nucleosome distributions.
Figure 2.
Nucleosome redistributions are determined by the underlying DNA sequence. (A) Western blots with the specified antibodies, at various times (in hours) after KSHV reactivation (hpr), of iSLK.219 cells treated with 0.2 µg/mL doxycycline. BRG1 protein levels peaked at 24 h. The RTA immunoblot is included to confirm KSHV reactivation. Beta-actin is included as a loading control. (B) Relative proportions of DNA-directed and DNA-independent nucleosome redistributions at 6, 12, 24, and 48 h post-KSHV reactivation. Approximately two-thirds of the genes with altered nucleosome occupancy at 12 h and 24 h were DNA-directed. (C) Observed and predicted (Gupta et al. 2008) nucleosome distributions. The nucleosome distributions from basal (black lines) KSHV, 24 h post-KSHV reactivation (red lines), and DNA-sequence-based prediction (cyan lines) for the same three genes shown in Figure 1D. The x-axis represents genomic position showing 2 kb centered on a TSS. The y-axis is the log2 ratio of nucleosomally protected DNA to genomic DNA signal at each probe on the microarray. These plots show strong agreement between the sequence-based model and the reactivated state. Gene models from the RefSeq annotations are shown below each gene column. (D) Measured and assembled nucleosome distributions. The nucleosome distributions from latent (black lines) KSHV, 24 h post-KSHV reactivation (red lines), and assembly (blue lines) for the same three genes shown in Figure 1D. The x-axis represents genomic position showing 2 kb centered on a TSS. The y-axis is the log2 ratio of nucleosomally protected DNA to genomic DNA signal at each probe on the microarray. These plots show strong agreement between the assembled and the reactivated state. Gene models from the RefSeq annotations are shown below each gene column.
Nucleosome redistributions are determined by the underlying DNA sequence
We next assessed the role of DNA sequence in determining the nucleosome distributions in the basal or in the transient state. Specifically, we compared our observed nucleosome distributions (in vivo) to the computationally predicted nucleosome occupancy scores based on DNA sequence (in silico), using a well-characterized and tested model (Gupta et al. 2008; Tanaka and Nakai 2009; Fincher et al. 2013). We calculated the difference of the correlation value between 24 h with the predicted model, and the correlation between 0 h with the predicted model. By definition, a positive value resulting from this calculation indicates greater agreement between 24 h and the predicted model (Supplemental Figs. 6, 11). Of the 226 loci showing maximal nucleosome redistributions at 24 h (Fig. 1B), 140 loci (62%) showed greater agreement with the predicted model, indicating that the changes at these loci are directed by the underlying DNA sequence. These 140 loci were classified as DNA-directed, while the remaining 86 loci were classified as DNA-independent (Fig. 2B; for exemplar DNA-independent loci, please see TNFRSF6B, IL13RA2, and CD1C in Supplemental Fig. 12). The predicted model and observed nucleosome distribution at 24 h are strikingly concordant for the 140 DNA-directed loci (Supplemental Fig. 12; Supplemental Table 2). To illustrate the DNA-directed nature of the nucleosome distribution changes, we plotted the nucleosome distribution at 0 and 24 h with the predicted distribution for the three loci previously shown in Figure 1D (Fig. 2C). To ensure that the similarities between the predicted model and the observed 24-h data were not exclusive to any particular in silico model, we compared our observed nucleosome distributions to three other models of DNA encoded nucleosome distribution and found the same agreement at 24 h (Supplemental Figs. 13, 14).
If DNA sequence is a major determinate of nucleosome redistribution, then the measured nucleosome distributions of a salt dialysis assembly (in vitro), in the absence of nuclear machinery, should reconstitute the nucleosome distributions observed at 24 h (in vivo). Salt gradient dialysis is known to produce nucleosomes near their sequence-determined locations on fragments of DNA (Lee and Narlikar 2001; Thastrom et al. 2004; Fincher and Dennis 2011; Gkikopoulos et al. 2011). Indeed, both the assembled and predicted nucleosome distribution profiles are concordant with the observed 24-h nucleosome distributions (Fig. 2D; Supplemental Fig. 15). These results support a substantial role for DNA sequence driving the genomic response, and led us to investigate the generality of this response.
The role of DNA sequence in the concerted, widespread, and transient redistribution of nucleosomes
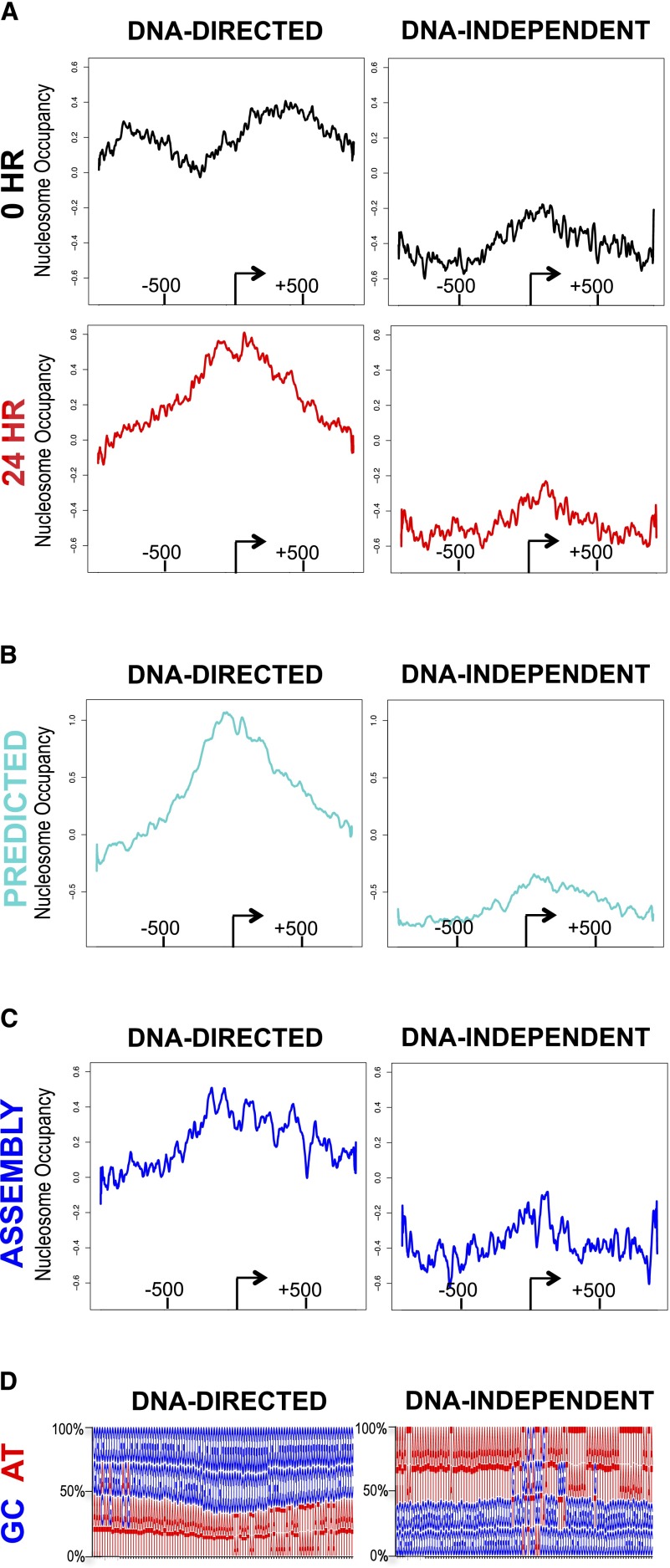
To reveal inherent overarching TSS architecture features, we averaged the observed nucleosome distribution of the 140 loci classified as DNA-directed for the 0-h and 24-h time points. At 0 h, DNA-directed loci were characterized by high overall nucleosome occupancy but showed a dip in nucleosome occupancy upstream of the TSS. At 24 h, nucleosome occupancy remained high and is centered on the TSS (Fig. 3A, left panels). We then averaged the observed nucleosome distribution of the 86 loci classified as DNA-independent for the 0-h and 24-h time points. In contrast to the DNA-directed loci, the 86 loci classified as DNA-independent were characterized by particularly low nucleosome occupancy (Fig. 3A, right panels). In addition, the average nucleosome occupancy for both the predicted and assembly agreed with the 24-h nucleosome occupancy profiles (Fig. 3B,C). Altogether, DNA-directed loci had relatively high nucleosome occupancy, DNA-independent loci had relatively low nucleosome occupancy, and loci that showed no changes had intermediate nucleosome occupancy (Supplemental Figs. 16, 17). This analysis confirms a concerted response in the loci classified as DNA-directed in which nucleosome occupancy increases over the TSS in the genomic response. These results reveal an important role for DNA sequence directing nucleosome distribution during a response to a stimulus.
Figure 3.
DNA sequence determined the concerted, widespread, transient redistribution of nucleosomes. (A) Average values for all genes identified as DNA-directed and DNA-independent, calculated by alignment of loci to the TSS for 0 h (black) and 24 h (red). (B) Average values for all genes identified as DNA-directed and DNA-independent, calculated by alignment of loci to the TSS for the predicted (cyan). (C) Average values for all genes identified as DNA-directed and DNA-independent, calculated by alignment of loci to the TSS for the assembly (blue). (D) Average GC (blue) and AT (red) content at each position is represented as a sequence logo below each column.
Features intrinsic to the DNA sequence that confer nucleosome forming or inhibitory characteristics are complex (Fincher and Dennis 2011), and the predictive model encompasses this complexity. Given this complexity, we next wanted to refine the potential sequence characteristics of DNA-directed and DNA-independent loci. We calculated the percentage of each nucleotide at each position for the 2000 bp surrounding the TSS. The DNA-directed loci showed enrichment in GC content; conversely, the DNA-independent loci showed enrichment in AT content (Fig. 3D). The degree to which GC content drives nucleosome distribution has generated substantial discussion, and our results support a role for this simplified regulatory code (Hughes and Rando 2009; Tillo and Hughes 2009). We show herein that loci with high GC content have higher nucleosome occupancy, which are directed by features intrinsic to the DNA sequence. In contrast, loci with low GC content show lower nucleosome occupancy and are not directed by features intrinsic to the DNA sequence (Fig. 3D).
Discussion
Upon KSHV reactivation, widespread nucleosome redistributions peaked at 24 h. These changes in nucleosome distribution were transient and returned to their basal state nucleosome distribution at 48 h. The widespread and transient nucleosome redistributions post-KSHV reactivation were directed by the DNA sequence. The results presented herein are consistent with the notion that some loci are held in a chromatin state that is unfavorable and different from the transient intermediate state directed by DNA sequence information. We suggest that a spring-loaded redistribution of nucleosomes in a DNA-directed manner is a fundamental step in response to a stimulus.
Nucleosome redistributions are widespread and transient
These results confirm that nucleosome redistributions are widespread and that the nucleosomes at the TSS deviate from their basal distributions only fleetingly. We suggest the nucleosome redistributions are not rare events, as prior literature would indicate, but widespread. What distinguishes the present study from the literature is the high-temporal resolution, which allows for the detection of the widespread and transient nucleosome redistribution events.
Despite decades of study, relatively little is known about the role that nucleosome distribution plays in gene regulation. The expectation that nucleosome distributions would accompany genome response has been informed by several single-locus studies including: PHO5, MMTV, IFNB1, and HOX (Richard-Foy and Hager 1987; Agalioti et al. 2000; Boeger et al. 2003; Reinke and Horz 2003; Woo et al. 2010). This expectation has been unfulfilled as whole-genome studies reveal that nucleosome distributions are remarkably similar between different cell types (Supplemental Fig. 18; Gaffney et al. 2012; Kundaje et al. 2012; Teif et al. 2012). Whole-genome studies have demonstrated that transcribing RNA polymerase is associated with lower nucleosomal occupancy and that conserved noncoding sequences may be sites of regulatory nucleosomes (Schones et al. 2008). It has also been shown that mammalian genomes have relatively few strongly positioned nucleosomes (Valouev et al. 2011). Our fundamentally distinct observation that nucleosome redistributions are widespread and transient is an important advance. The transient nature of the redistribution explains why advances in understanding and characterizing nucleosome distribution have lagged behind other measures of more enduring chromatin regulatory features, such as histone post-translational modifications and DNase I sensitivity (The ENCODE Project Consortium 2012). Our observation paves the way to transform nucleosome-mapping studies.
The DNA dependency of loci suggests a functional complementarity between cis- and trans-acting factors
This study makes steps toward reconciling models that invoke prevailing roles for either genetically encoded (cis) or ATP-dependent remodeler (trans)–driven nucleosome distribution. We propose a functional complementarity between these cis- and trans-acting factors. An interesting observation arising from our study is that loci classified as DNA-directed have higher GC content; while loci classified as DNA-independent have lower GC content. It has been previously demonstrated that GC content confers different requirements for ATP-dependent chromatin remodelers (Ramirez-Carrozzi et al. 2006, 2009; Moshkin et al. 2012).
SWI/SNF-independent genes have greater GC content around the TSS, whereas SWI/SNF-dependent genes have lower GC content. A majority of the genes previously characterized as SWI/SNF-independent were classified as DNA-directed in our assay, and the SWI/SNF-dependent genes are largely DNA-independent in our assay (Ramirez-Carrozzi et al. 2009). We speculate that the nucleosome positioning information content within the DNA sequence at the DNA-directed loci contains information that reduces the need for SWI/SNF activity; likewise, the lack of nucleosome positioning information content of the DNA sequence at the DNA-independent loci increases the need for SWI/SNF activity. This speculation is borne out in the well-characterized IFNB1 promoter enhancer, which has a transient association with BRG1 correlating with the remodeling of a promoter nucleosome (Agalioti et al. 2000). Consistent with this model, the IFNB1 locus was classified as DNA-independent in our assay.
Our results indicate that the loci that adopt DNA-directed positions have greater GC content and are SWI/SNF-independent. This observation is consistent with multiple studies that have investigated a role for the ISWI family of chromatin remodelers. Genomic targets of ISWI family members have increased GC content (Moshkin et al. 2012). Additionally, studies in yeast and flies have shown that in the absence of ISWI complexes, nucleosomes adopt DNA-encoded positions (Gkikopoulos et al. 2011; Moshkin et al. 2012). It is interesting to speculate a role for ISWI at the DNA-directed loci in which this ATP-dependent remodeler “pushes” nucleosomes away from genetically encoded positions in the basal state (Moshkin et al. 2012). Upon a regulatory stimulus, ISWI is lost, providing the opportunity for another class of remodelers to move the nucleosomes to DNA-directed positions. In aggregate, these results suggest a functional complementary between DNA sequence (cis) and ATP-dependent remodelers (trans) in regulating a genomic response.
A spring-loaded genome for a concerted, widespread, and transient genomic response
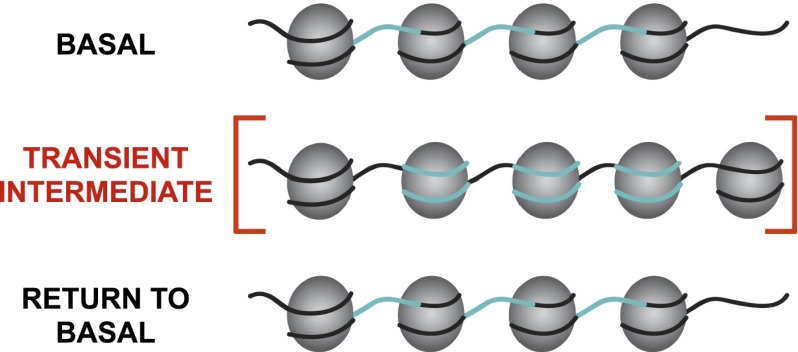
Our results show that some loci are held in a chromatin state different from the state directed by DNA sequence information. We hypothesize that chromatin regulatory complexes, such as ATP-dependent remodelers, acting in trans, maintain chromatin architecture. Upon KSHV reactivation, there is a concerted, widespread, and transient chromatin reorganization that appears to be facilitated by features intrinsic to the DNA sequence, as nucleosome occupancy changes in a manner predicted by DNA sequence. In fact, these results are in complete agreement with recent conclusions from a lung adenocarcinoma study in which widespread nucleosome distribution changes are limited to low-grade tumors and are not observed in high-grade tumors (Druliner et al. 2013). We therefore propose that a genetically directed “spring-loaded” repositioning of nucleosomes, from the basal state, to the transient intermediate state, and returning to the basal state, facilitates the concerted response (Fig. 4).
Figure 4.
Model of chromatin regulation in which nucleosome distributions move from a basal state architecture, to a transient intermediate state, then return to the basal architecture, in response to a common stimulus. The transient intermediate state's architecture is directed by features intrinsic to the underlying DNA sequence (cyan).
The widespread, concerted changes observed in the transient intermediate state suggest a possible hierarchical mechanism for genomic regulation. In this hierarchical organization, we speculate that a superset of genomic loci is made available for regulatory licensing in the transient intermediate state. Gene loci in the superset in a physiology with the appropriate regulatory machinery will be licensed, and those without the regulatory machinery will not be affected. This hierarchical organization maximizes the potential for a concerted response of multiple loci with a limited number of genomic architectures, particularly when one considers the DNA-directed nature of the response.
The conventional wisdom surrounding gene regulation involves chromatin regulatory machinery that responds to a physiological cue to move nucleosomes. Our results indicate that chromatin regulatory machinery acting in trans might hold nucleosome architecture in a state unfavorable to regulation. These results have allowed the development of a model in which DNA sequence features play a considerable role in directing nucleosome architecture in genome response (Fig. 4). Upon the appropriate physiological cue, mechanisms acting in cis—encoded by the DNA sequence—drive a concerted, widespread, and transient chromatin reorganization leading to architecture favorable for the necessary response. Thus, interplay between enzymatic complexes regulating nucleosome distribution and DNA sequences dictating nucleosome distribution drives the regulation of these loci. The surprise is that the transient intermediate state, not the basal state, is favored by DNA sequence. We propose that DNA sequence is more broadly used to direct nucleosome redistribution than previously described and that it can function to facilitate regulation in ways not previously envisioned.
Methods
Cell growth and KSHV reactivation
iSLK.219 (clone 10), derived from iSLK, was latently infected with a recombinant rKSHV.219 virus (Vieira and O'Hearn 2004) and cultured and maintained as described previously (Myoung and Ganem 2011). One day before induction of KSHV reactivation, 5 × 106 cells were seeded into a 150-mm dish. The next day, the medium was replaced with DMEM containing 1% FBS, and doxycycline was added to a final concentration of 0.2 µg/mL. Cells were harvested at the designated times after doxycycline addition. The 0 h was treated identically to the 6 h except that doxycycline was not added.
Cell harvest and nucleus purification
iSLK and iSLK.219 cell lines were harvested at 2.5 × 107 cells. The cells were cross-linked in 1% formaldehyde in PBS and incubated for 10 min at room temperature. The cross-linking reaction was stopped with 125 mM glycine. The nuclei were then isolated in nucleus isolation buffer (0.3 M sucrose, 2 mM MgOAc2, 1 mM CaCl2, 1% Nonidet P-40, 10 mM HEPES at pH 7.8) by centrifugation at 1000g for 5 min at 4°C.
In vitro assembly of nucleosomes
Nucleosome arrays were assembled using HeLa histones as previously described, except that total genomic DNA sonicated to an average length of 500 bp was used (Lee and Narlikar 2001).
MNase cleavage and mononucleosomal-protected DNA purification
All MNase nucleosome-mapping experiments were performed with two biological replicates from completely independent growths and treatments of cells (Supplemental Fig. 19). iSLK.219 nuclei or in vitro–assembled nucleosomes were digested with a titration of MNase (Worthington Biochemical Corp.) for nucleosome-distribution experiments. The concentrations for the MNase titration were 4 units/mL, 2 units/mL, 1 unit/mL, and 0.5 unit/mL of MNase in MNase cleavage buffer: 5 mM KCl, 4 mM MgCl2, 1 mM CaCl2, 12.5% glycerol, and 50 mM Tris-Cl (pH 7.4). The MNase reaction occurred at a titration of MNase concentration to ensure equivalent representation of the early- and late-digesting fractions of the genome. These reactions were performed for 5 min at 37°C, and the reactions were stopped with 50 mM EDTA. We then treated the MNase-digested nuclei with 0.2 μg/μL proteinase K, 1% sodium dodecyl sulfate, and incubated overnight at 60°C to reverse the protein–DNA cross-links.
Following the separation of nucleosomal ladder on a 2% agarose gel, mononucleosomally protected DNA (∼150 bp) was isolated by excision of the specific sample's band, and the DNA was purified by electroelution. For each sample, mononucleosomally protected DNA of all MNase concentrations were combined. Mononucleosomal DNA was extracted with phenol-chloroform, precipitated with alcohol, and dissolved in TE (10 mM Tris-Cl at pH 8.0, 0.1 mM EDTA). Bare genomic DNA (reference) was isolated from iSLK and iSLK.219 by resuspension of the cells in a 0.2 μg/μL proteinase K and 1% SDS solution and incubated overnight at 55°C.
Antibodies and Western blotting
The BRG1 antibody (#4081) was purchased from Abcam. Monoclonal antibodies against KSHV, ORF45, and PF8 were described previously (Zhu et al. 2005). A monoclonal antibody against beta-actin was purchased from Sigma-Aldrich. Whole-cell extract equivalent to 0.1 × 106 cells was resolved on SDS-PAGE 10% Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membranes. The membranes were blocked in 5% dried milk in PBS containing 0.2% Tween 20 (PBST) followed by an overnight incubation with diluted primary antibodies at 4°C. Anti-rabbit or anti-mouse immunoglobulin G antibody conjugated to horseradish peroxidase was used as the secondary antibody. The SuperSignal West Dura Chemiluminescent Substrate (Pierce) was used for detection of antibody–antigen complexes.
Microarray design and processing
We used a custom-designed high-resolution DNA microarray that covers 2 kb flanking the TSS 472 immunity- and inflammation-related genes. Only unique probes were printed on the microarray, and repeats were masked around the TSS. Both forward and reverse DNA strands were printed on the microarray. The 60-mer oligonucleotide probes were tiled with an average 47-bp overlap. We used NimbleGen's 12-plex and HD2 design for our microarrays. Each locus on the microarray contains ∼180 probes of a median spacing of 12.5 bp. Samples were labeled (nucleosomally protected DNA Cy3 and genomic DNA Cy5) and hybridized to the custom tiling microarray (Roche) according to manufacturer's instructions.
Data processing and analysis
The resulting microarray data were normalized, the log2 ratio of nucleosomally protected DNA to genomic DNA signals was computed for each probe, and replicate probes at each chromosomal position were averaged. Analysis of nucleosome distribution data was performed in the R environment for statistical computing and graphical output using our lab-developed software, drawGff.R. To calculate correlation on a per-locus basis, all probes were compared for each condition and a correlation value was calculated. To calculate nucleosome signal strength on a per locus basis, all probes were compared for each condition and an interquartile range (IQR) value was calculated.
Computer model scores
Computational model scores were derived from algorithms previously described (Gupta et al. 2008). We applied models derived from nucleosome distribution data from A375 human melanoma, THP1 human monocytic leukemia, and Zea mays tissue.
Data access
Microarray data have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSM1267162–GSM1267166.
Acknowledgments
We are grateful to M. Simon, S. Bowman, M. Charrel-Dennis, D. Gilbert, D. Houle, and A. Thistle for critical reading of the manuscript. We thank Drs. Jinjong Myoung and Don Ganem for providing iSLK.rKSHV219 cells. J.H.D. and R.E.K. were supported by National Institutes of Health Award R37 GM048405. J.H.D., J.Z., S.G., and B.S.S. were supported by National Institutes of Health Award R01 DA033775. S.G. and W.S.N. were supported by National Institutes of Health Award R01 GM071923. F.Z. and D.A. were supported by National Institutes of Health Award R01 DE016680.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.160150.113.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103: 667–678 [DOI] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11: 1587–1598 [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266: 1865–1869 [DOI] [PubMed] [Google Scholar]
- Drew HR, Travers AA 1985. DNA bending and its relation to nucleosome positioning. J Mol Biol 186: 773–790 [DOI] [PubMed] [Google Scholar]
- Druliner BR, Fincher JA, Sexton BS, Vera DL, Roche M, Lyle S, Dennis JH 2013. Chromatin patterns associated with lung adenocarcinoma progression. Cell Cycle 12: 1536–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Tsukiyama T 2003. Chromatin remodeling in vivo: Evidence for a nucleosome sliding mechanism. Mol Cell 12: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Fincher J, Dennis JH 2011. DNA sequence determinates of nucleosome position. In Epigenetics: A reference manual (ed. Craig JM, Wong NC), pp. 133–142. Caister Academic Press, Norfolk, UK [Google Scholar]
- Fincher JA, Vera DL, Hughes DD, McGinnis KM, Dennis JH, Bass HW 2013. Genome-wide prediction of nucleosome occupancy in maize reveals plant chromatin structural features at genes and other elements at multiple scales. Plant Physiol 162: 1127–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R 2007. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447: 972–978 [DOI] [PubMed] [Google Scholar]
- Gaffney DJ, McVicker G, Pai AA, Fondufe-Mittendorf YN, Lewellen N, Michelini K, Widom J, Gilad Y, Pritchard JK 2012. Controls of nucleosome positioning in the human genome. PLoS Genet 8: e1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D 2010. KSHV and the pathogenesis of Kaposi sarcoma: Listening to human biology and medicine. J Clin Invest 120: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girimurugan SB, Chicken E 2013. Wavelet Analysis of Variance (WANOVA). Technical Reports of the FSU. Department of Statistics M1012: 1–7. http://stat.fsu.edu/techreports/M1012.pdf.
- Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T 2011. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333: 1758–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld JM, Bloomer LS 1980. Nonrandom alignment of nucleosomes on 5S RNA genes of X. laevis. Cell 21: 751–760 [DOI] [PubMed] [Google Scholar]
- Gupta S, Dennis J, Thurman RE, Kingston R, Stamatoyannopoulos JA, Noble WS 2008. Predicting human nucleosome occupancy from primary sequence. PLoS Comput Biol 4: e1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A, Rando OJ 2009. Chromatin ‘programming’ by sequence—is there more to the nucleosome code than %GC? J Biol 8: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Hughes TR, Lieb JD, Widom J, Segal E 2010. Nucleosome sequence preferences influence in vivo nucleosome organization. Nat Struct Mol Biol 17: 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev 13: 2339–2352 [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98: 285–294 [DOI] [PubMed] [Google Scholar]
- Kundaje A, Kyriazopoulou-Panagiotopoulou S, Libbrecht M, Smith CL, Raha D, Winters EE, Johnson SM, Snyder M, Batzoglou S, Sidow A 2012. Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Res 22: 1735–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Narlikar G 2001. Assembly of nucleosomal templates by salt dialysis. Curr Protoc Mol Biol 54: 21.6.1–21.6.16 [DOI] [PubMed] [Google Scholar]
- Mesri EA, Cesarman E, Boshoff C 2010. Kaposi's sarcoma and its associated herpesvirus. Nat Rev Cancer 10: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkin YM, Chalkley GE, Kan TW, Reddy BA, Ozgur Z, van Ijcken WF, Dekkers DH, Demmers JA, Travers AA, Verrijzer CP 2012. Remodelers organize cellular chromatin by counteracting intrinsic histone–DNA sequence preferences in a class-specific manner. Mol Cell Biol 32: 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myoung J, Ganem D 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: Maintenance of tight latency with efficient reactivation upon induction. J Virol Methods 174: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST 2006. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev 20: 282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Horz W 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell 11: 1599–1607 [DOI] [PubMed] [Google Scholar]
- Richard-Foy H, Hager GL 1987. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J 6: 2321–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J 2006. A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Nakai K 2009. An assessment of prediction algorithms for nucleosome positioning. Genome Inform 23: 169–178 [PubMed] [Google Scholar]
- Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Hofer T, Rippe K 2012. Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol 19: 1185–1192 [DOI] [PubMed] [Google Scholar]
- Thastrom A, Bingham LM, Widom J 2004. Nucleosomal locations of dominant DNA sequence motifs for histone–DNA interactions and nucleosome positioning. J Mol Biol 338: 695–709 [DOI] [PubMed] [Google Scholar]
- Tillo D, Hughes TR 2009. G + C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A 2011. Determinants of nucleosome organization in primary human cells. Nature 474: 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J, O'Hearn PM 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325: 225–240 [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Tsukiyama T 2006. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol 13: 633–640 [DOI] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE 2010. A region of the human HOXD cluster that confers Polycomb-group responsiveness. Cell 140: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu JS 2008. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput Biol 4: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K 2009. Intrinsic histone–DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K 2010. Evidence against a genomic code for nucleosome positioning. Reply to ‘Nucleosome sequence preferences influence in vivo nucleosome organization’. Nat Struct Mol Biol 17: 920–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FX, Chong JM, Wu L, Yuan Y 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J Virol 79: 800–811 [DOI] [PMC free article] [PubMed] [Google Scholar]