Abstract
Deletion of tumor-suppressor genes as well as other genomic rearrangements pervade cancer genomes across numerous types of solid tumor and hematologic malignancies. However, even for a specific rearrangement, the breakpoints may vary between individuals, such as the recurrent CDKN2A deletion. Characterizing the exact breakpoints for structural variants (SVs) is useful for designating patient-specific tumor biomarkers. We propose AmBre (Amplification of Breakpoints), a method to target SV breakpoints occurring in samples composed of heterogeneous tumor and germline DNA. Additionally, AmBre validates SVs called by whole-exome/genome sequencing and hybridization arrays. AmBre involves a PCR-based approach to amplify the DNA segment containing an SV's breakpoint and then confirms breakpoints using sequencing by Pacific Biosciences RS. To amplify breakpoints with PCR, primers tiling specified target regions are carefully selected with a simulated annealing algorithm to minimize off-target amplification and maximize efficiency at capturing all possible breakpoints within the target regions. To confirm correct amplification and obtain breakpoints, PCR amplicons are combined without barcoding and simultaneously long-read sequenced using a single SMRT cell. Our algorithm efficiently separates reads based on breakpoints. Each read group supporting the same breakpoint corresponds with an amplicon and a consensus amplicon sequence is called. AmBre was used to discover CDKN2A deletion breakpoints in cancer cell lines: A549, CEM, Detroit562, MOLT4, MCF7, and T98G. Also, we successfully assayed RUNX1–RUNX1T1 reciprocal translocations by finding both breakpoints in the Kasumi-1 cell line. AmBre successfully targets SVs where DNA harboring the breakpoints are present in 1:1000 mixtures.
Cancer develops through a series of genetic mutations, with tumor cells acquiring pernicious mutations that eventually lead to metastatic disease. The DNA mutations contributing to oncogenesis are not limited to point mutations, but include large chromosomal rearrangements, duplications, and deletions. It has been suggested that recurring mutations are the likely drivers for cancer and might be viable biomarkers for disease detection and prognosis. For instance, a translocation occurs between chromosomes 21 and 8 that fuses RUNX1 and RUNX1T1 genes in 12% of acute myeloid leukemia (AML) cases (Xiao et al. 2001). The fusion results in a chimeric oncoprotein. The chimeric protein contributes to initial leukemia cell growth mostly through transcriptional repression of wild-type RUNX1 targets (Downing 1999). Alternatively, the loss of DNA may also contribute to cancer progression. For example, many human cancers frequently delete the chromosome 9p21-22 locus containing MTAP, CDKN2A, and CDKN2B genes. The locus encodes INK4 proteins (p15INK4B, p16INK4A) that inhibit cyclin-dependent kinases, CDK4 and CDK6, and p14ARF, which inactivates MDM2, thereby regulating TP53. Thus, expression of these proteins is responsible for G1 cell cycle arrest and independently signaling apoptosis (Wessely 2010; Kim et al. 2012). Homozygous deletions frequent the 9p21-22 locus, in particular, CDKN2A, which encodes both p16INK4A and p14ARF, as the single event diminishes expression of multiple proteins—each with unique tumor-suppressor activity.
In a clinical setting, driver DNA lesions can be used to (1) detect tumor DNA in individuals and (2) monitor tumor burden during or after treatment. Michor et al. (2005) and Bartley et al. (2010) demonstrated how identification of the BCR–ABL1 gene fusion at the DNA level in leukemia patients leads to a more sensitive test for measuring tumor burden than current BCR–ABL1 mRNA tests. Measuring changes in tumor burden during therapeutic treatment is critical for checking therapy effectiveness and deciding to continue treatment. Their approach focuses on the frequent translocation of BCR–ABL1 in leukemia and has not been applied to solid tumors. In a more recent study, circulatory biomarkers were assessed in their ability to monitor metastatic breast cancer (Dawson et al. 2013). The researchers applied a variety of sequencing methods to identify point mutations in PIK3CA and TP53 and other somatic structural variations for use as circulatory tumor DNA markers. They found that circulatory tumor DNA had the highest correlation with tumor burden and greater dynamic range than current standard of care CA 15-3 biomarker and circulatory tumor cell counting.
These studies all focused on tumor burden monitoring after the specific lesion had been fully characterized. While monitoring is easy for point mutations and structural variants with known breakpoints, it is very difficult when the breakpoint of the structural variation is not known. At the same time, large variants are potentially much more specific for tumor detection and monitoring, and a test that could identify them reliably would have higher sensitivity for monitoring tumor burden. Reliable and sensitive identification of breakpoints in tumor DNA could also serve as a diagnostic for early detection.
Whole-genome sequencing experiments (analyzed with appropriate tools like BreakDancer [Chen et al. 2009], Pindel [Ye et al. 2009], and SVDetect [Zeitouni et al. 2010]) have the potential to identify point mutations and structural variations in individual samples. However, clinical tumor samples are a mixture of tumor cells and normal cells and require ultradeep sequencing to analyze tumor DNA.
Therefore, current approaches apply ultradeep sequencing after targeted amplification of select genes (Harismendy et al. 2011). Unfortunately, these methods are unable to reliably identify structural variation with uncertain breakpoints. Alternatively, DNA hybridization microarrays (SNP arrays), which are still widely used in clinics, are capable of calling copy number variation, from which deletions and gene amplifications can be inferred. However, the technology is only reliable with homogeneous samples and only reports low-resolution boundary estimates (Greenman et al. 2010), insufficient for performing tumor burden monitoring assays. Thus, a challenge remains: how to detect DNA markers, specifically, somatic structural variations, in a complex patient sample containing a mixture of tumor DNA and germline DNA. This is particularly challenging when the exact breakpoints are needed for quantitative DNA assays.
To identify unknown DNA breakpoints associated with known translocations and deletions, we describe a pipeline, AmBre (Amplification of Breakpoints), which builds on the PAMP approach (Liu and Carson 2007). PAMP is a PCR assay, developed to selectively amplify the tumor DNA sequence containing a structural variation. To illustrate how PAMP works, consider a deletion on chr9 (CDKN2A locus) with unknown breakpoints located around the CDKN2A gene. Illustrated in Figure 1, a tiling of evenly spaced forward (blue arrows) primers and reverse primers (red arrows) is selected around the CDKN2A gene. The spacing between primers is ∼1 kb apart. The innermost forward and reverse primers are distantly spaced such that they will not amplify sequence from germline DNA.
Figure 1.
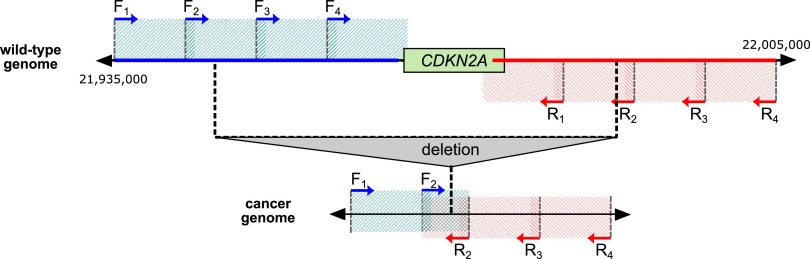
PAMP tiling design for capture of CDKN2A deletions. CDKN2A upstream and downstream breakpoint regions are defined on a germline genome, blue and red lines, respectively. Tiled forward primers (blue arrows) and reverse primers (red arrows) are spaced ≈1 kb apart (width of hashed boxes; not to scale with reference). Overlap of blue box and red box on tumor DNA indicates that a forward and reverse primer pair is <2 kb apart and will lead to amplification of tumor DNA harboring CDKN2A deletion breakpoints.
All tiling primers are used in a single multiplex PCR. Any CDKN2A deletion in the tumor DNA will lead to a forward and reverse primer being proximally located (<2 kb) on the tumor DNA, resulting in a targeted DNA amplification of the tumor DNA harboring the deletion, but not germline DNA. This strategy takes advantage of polymerases having a limited amplifying length and genomic rearrangements within tumor DNA resulting in novel adjacencies of germline DNA sequences for selective and sensitive amplification of tumor DNA over germline DNA.
Although it has potential, PAMP has challenges. In the multiplexed reaction, all primers must be evenly spaced so as to amplify any deletion in the region, and primer pairs cannot dimerize. In a large (say, 100 kb) region, this implies that we need to find a design of 100 applicable primers from a large candidate set of more than 5000 potential primers. An exhaustive search of all candidate primer combinations is infeasible (5000 candidate primers and 50–100 primers desired would result in searching  combinations). Bashir et al. (2007) formulated PAMP primer tiling as a computational problem and defined a cost associated with each subset of candidate primers. Furthermore, the investigators showed that simulated annealing (Kirkpatrick 1984) could efficiently find low-cost PAMP primer designs for contiguous breakpoint regions. Even with these improvements, PAMP is limited to recurrent structural variations where breakpoints appear in short breakpoint regions (<40 kb), as a large number of primers in a single reaction inevitably leads to loss of sensitivity with off-target DNA synthesis and increased spurious primer–primer interactions. Finally, PAMP detects the amplified product and identifies breakpoints via DNA hybridization arrays (Bashir et al. 2010), which had the additional challenge of designing probes that match the primer designs.
combinations). Bashir et al. (2007) formulated PAMP primer tiling as a computational problem and defined a cost associated with each subset of candidate primers. Furthermore, the investigators showed that simulated annealing (Kirkpatrick 1984) could efficiently find low-cost PAMP primer designs for contiguous breakpoint regions. Even with these improvements, PAMP is limited to recurrent structural variations where breakpoints appear in short breakpoint regions (<40 kb), as a large number of primers in a single reaction inevitably leads to loss of sensitivity with off-target DNA synthesis and increased spurious primer–primer interactions. Finally, PAMP detects the amplified product and identifies breakpoints via DNA hybridization arrays (Bashir et al. 2010), which had the additional challenge of designing probes that match the primer designs.
Results
Overview of AmBre
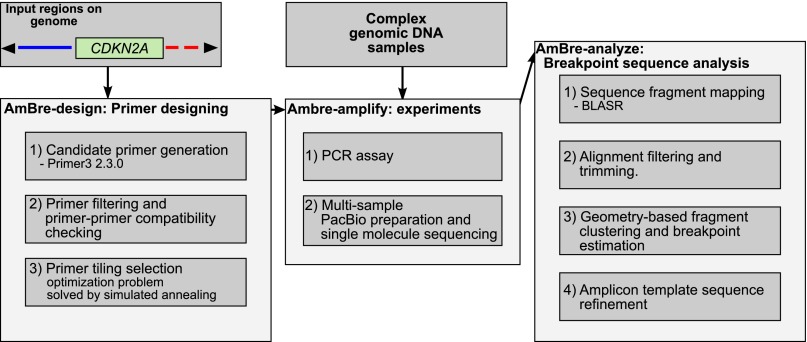
AmBre resolves these issues with a three-phase approach (Fig. 2). The first (AmBre-design) involves a revised computational approach to designing multiplex primers on discontiguous DNA regions, ignoring regions known to not contain breakpoints. This requires some changes to the optimization function and results in a more flexible design with better performance on sparse regions. The output of this phase is a collection of primers that can be mixed in a single multiple primer reaction.
Figure 2.
AmBre pipeline with primer designing and PacBio long fragment sequence analysis.
In the second, experimental phase (AmBre-amplify), long-range PCR amplifies target amplicons, which reduces the number of primers required in a single reaction. For example, PAMP, using their proposed traditional PCR, would require 600 primers to cover a 600-kb region, with more than 180,000 putative interactions. In contrast, to cover the same region, AmBre would need less than 100 primers with only 5000 possible interactions, which improves reliable amplification from proposed designs. In AmBre, the amplified products are sequenced using the Pacific Biosciences RS (PacBio) platform (English et al. 2012). Our analysis allows us to mix the amplicons prior to sequencing, with computational separation of breakpoints in the third phase.
The final, computational phase (AmBre-analyze) involves a customized analysis of sequenced reads to identify DNA breakpoints for each tumor genome. The analysis involves clustering of split mapped reads followed by error correction, and sequence reconstruction around the breakpoint regions. We demonstrated that AmBre can successfully detect targeted structural variations (potential tumor DNA biomarkers) by identifying CDKN2A deletion breakpoints in the cancer cell lines A549, CEM, Detroit562, MCF7, MOLT4, and T98G. AmBre resolved breakpoints for MCF7 and T98G, which had not been previously discovered by other studies. Furthermore, AmBre easily extends to identify translocations and inversions, which is demonstrated here with RUNX1–RUNX1T1 translocation in the cancer cell line Kasumi-1.
Designing primers
The input to AmBre-design is a collection of genomic intervals for the forward region, denoted by F, a collection of genomic intervals for the reverse region (R), and parameter d. The output is a collection of forward primers in F and reverse primers located in R spaced apart by approximately d. AmBre-design has the following steps:
Candidate primer generation from target breakpoint regions, where oligonucleotides are selected according to thermodynamic properties. Primers with significant self-dimerization are eliminated. Primer pairs that are likely to dimerize or cause off-target amplifications are marked as incompatible (Methods).
The list of candidate primers and incompatible primer pairs is used to design an optimal set of primers based on the considerations outlined below.
Denote a primer design P as a subset of candidate primers numbered according to the order of genomic start locations l1, l2, l3, …, ln. Set E to denote incompatible primer pairs. We associate a cost C(P) with each design and seek to find designs with minimum cost. Our formulation of cost differs from Bashir et al. (2007) to accommodate sparser primer designs and targeting discontiguous regions (see Supplemental Fig. S1). The parameter d is set to be half the maximum feasible PCR amplicon size. Thus, for the long-range polymerases used here, we use d = 6500, corresponding to a desirable amplicon size ≤13 kb. The cost of the design is a sum of incompatibility costs for each pair and coverage costs.
For the coverage, let Δi(P) = li+1 − li denote the gap between adjacent pairs. If Δi(P) > d, we run the risk of the product being too long to be amplified. On the other hand, if Δi(P) ≪ d, we have a design with extra primers that greatly decrease the efficiency of the reaction. Let parameter ρ, with 0 < ρ ≤ 1, describe a target density 1 + ρ of primers every d bp, corresponding to a primer every  bp. Ideally, the distance between adjacent primers is bounded by (1 − ρ)d ≤ Δi(P) ≤ d. A design is penalized if the distances violate these constraints. Formally,
bp. Ideally, the distance between adjacent primers is bounded by (1 − ρ)d ≤ Δi(P) ≤ d. A design is penalized if the distances violate these constraints. Formally,
Experiments revealed that even a single incompatible pair severely diminishes the multiple primer reaction (Bashir et al. 2007). Therefore, we set wp = ∞ for our designs. We empirically choose ρ = 0.2. Similar to Bashir et al. (2007), simulated annealing is used to find low-cost primer designs by applying our cost function (Fig. 3; Methods). The algorithm explores the large space of all primer designs by initiating a random primer subset and improving the primer subset with iterative addition or removals of primers. Since the algorithm involves randomization and has parameters governing convergence to low-cost designs, simulated annealing is repeated multiple times under different rates of convergence. The lowest-cost primer design from all simulated annealing runs is used as the final primer tiling design (Fig. 3).
Figure 3.
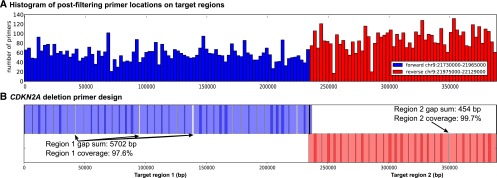
Designing AMBRE-68. (A) Candidate primers are uniformly distributed in CDKN2A locus, suggesting that good primer designs are possible. AmBre-design is tasked to capture CDKN2A deletion upstream and downstream breakpoints in regions chr9: 21,730,000–21,965,000 and chr9: 21,975,000–22,129,000 (GRCh37 coordinates), respectively. (B) Final low-cost 68-primer design to capture CDKN2A deletions in 380-kb breakpoint region. The solution has a 97.6% and a 99.7% coverage of breakpoint regions. The fraction of break pairs captured by the design (resulting in amplicon length <13 kb) is 99.99%.
Design results
To test AmBre-design, we analyzed cell-line copy number data to identify a large clustering of deletions in the CDKN2A region (Greenman et al. 2010). We identified a 380-kb region surrounding the CDKN2A gene, 230 kb upstream and 150 kb region downstream of CDKN2A that captures breakpoints in 55 of the 109 CDKN2A deletion cell lines considered. We chose d = 6500, as 13-kb products can be reliably amplified with LongAmp Taq DNA polymerase (New England Biolabs, NEB).
The candidate primer generation and primer filtering stages resulted in 5181 candidate primers. As shown in Figure 3A, the candidate primers are uniformly spread across breakpoint regions, suggesting that good tiling primer designs may exist. The simulated annealing algorithm is repeated for 12 different rates of convergence, with the fastest convergence rate having a 10-min average runtime and slowest convergence rate having an 864-min average runtime (Supplemental Fig. S2). When d = 6500, the lowest-cost solution (AMBRE-68) requires only 68 primers with 99.99% in silico capture of simple CDKN2A deletions that may occur in the 380-kb breakpoint region (Fig. 3B).
Sequencing amplified sequences harboring SVs
Sequencing the AmBre-amplify DNA confirms capture of CDKN2A deletions. We used PacBio RS technology due to its long reads, ideal for structural variation calling, and throughput, appropriate for medium sized experiments. Using computation, we correct for the high inherent error in PacBio sequencing.
Furthermore, if different samples do not share breakpoints (for example, all amplicons are of different sizes and amplify from different primer pairs within the design), the samples can be mixed and sequenced on a single run without additional barcoding. We employed this strategy with CDKN2A deleted samples on a single SMRT cell and relying on computation to deconvolute the breakpoints.
Define a breakpoint as a pair of disjoint coordinates a and b on a reference, and a nontemplate sequence s (of length ℓ) such that the sample sequence brings a and b together, separated only by the insertion of s. The objective of AmBre-analyze is to take as input a collection of PacBio sample sequences aligned to the reference genome and output a collection of breakpoints along with the sequence around each breakpoint. The code for this tool is stand-alone and can be used in the analysis of PacBio reads for SV detection. AmBre-analyze works by (1) alignment trimming (defined below), (2) breakpoint clustering of fragments, and (3) consensus sequence generation around each breakpoint (Fig. 2; see Methods).
Alignment trimming
Denote a local alignment (Chaisson and Tesler 2012) as a pair of intervals from the fragment and reference that can be aligned with a small number of edits. A split mapped fragment F supports a breakpoint (a, b, s) with two local alignments [denoted as (Fa, Ga), (Fb, Gb)]. In the ideal case, Ga ends at a and Gb begins at b, while the fragment segment between Fa and Fb is exactly the inserted sequence s (Methods). However, in real data, a fragment can span multiple breakpoints, sequence errors can result in spurious incorrect alignments, and the alignments output by standard tools like BLASR will have inaccurate boundaries. Specifically, inaccurate boundaries might result in overlapping consecutive segments Fa, Fb. AmBre-analyze resolves these errors by choosing the optimal alignment segments covering the fragment F. For a fragment F, the input is a chain of local alignments F = (Fa, Ga), (Fb, Gb), …. The output is a subset  of
of  , with alignment boundaries trimmed so (1) none of the fragment segments
, with alignment boundaries trimmed so (1) none of the fragment segments  overlap, (2) the number of distinct alignments is minimized, and (3) most of fragment F is covered. The second and third objectives reinforce the notion that a typical fragment covers a small number of breakpoints and is mostly well aligned except for nontemplate insertion sequence. The first objective helps to narrow down the breakpoint coordinates. To clarify, consider a trimmed reference interval
overlap, (2) the number of distinct alignments is minimized, and (3) most of fragment F is covered. The second and third objectives reinforce the notion that a typical fragment covers a small number of breakpoints and is mostly well aligned except for nontemplate insertion sequence. The first objective helps to narrow down the breakpoint coordinates. To clarify, consider a trimmed reference interval  that ends at x and a consecutive interval
that ends at x and a consecutive interval  beginning at y, while the gap between corresponding fragment segments is L. Then, we expect that a > x, b < y, and
beginning at y, while the gap between corresponding fragment segments is L. Then, we expect that a > x, b < y, and
Thus, the fragment constrains the location of the breakpoint (a, b) to lie in a small region between x, y. In the next section, we use information from multiple fragments to further narrow the breakpoint location. Given these three distinct objectives, the alignment trimming algorithm works by combining them into a single objective function and uses a dynamic programming approach to identify the optimal trimming (Methods).
Fragment clustering
Consider a two-dimensional (2D) representation of the genomic space with F and R being the vertical and horizontal axes, respectively. In this representation, a true breakpoint (a, b) is represented by a point, and each split-mapped read (x, y, L) is represented by a triangle of possible breakpoints (a, b) that satisfy (a − x) + (y − b) ≤ L (Methods). Multiple reads supporting the same breakpoint represent multiple triangles whose intersection reduces the uncertainty in breakpoint determination. Furthermore, if reads from multiple AmBre-amplify experiments are combined, the split-mapped reads will cluster according to overlap, revealing breakpoints for each experiment sample. We develop a fast, customized method to recover the aggregated read clusters for each breakpoint (Methods). The method took 2.5 min on a single desktop core to analyze all local alignments from 52,000 reads from a single PacBio SMRT cell experiment.
Consensus sequence determination
Predicted amplicon sequences are generated from the breakpoint estimates. In turn, these templates are supplied as reference sequences into PacBio's SMRT Analysis Resequencing protocol. The analysis protocol calls consensus amplicon sequences by correcting the predicted templates.
Identifying CDKN2A deletion given DNA break clustering
AmBre exploits the fact that variable breakpoints aggregate along fragile regions of the chromosome by designing primers around the fragile regions. We used this idea to produce a single design for five cancer cell lines: A549, CEM, Detroit562, MCF7, and T98G. Breakpoints were estimated by copy number changes for four cancer cell lines (A549, CEM, MCF7, and T98G) from SNP-array data (Supplemental Fig. S3; Table 1; Greenman et al. 2010), and the breakpoint was given for a fifth cell line (Detroit562) from prior studies. The error in breakpoint estimation for SNP-array data is roughly 10 kb. Thus, to generate cluster target regions, each breakpoint estimate was expanded to be a 10-kb interval, and overlapping intervals were merged. This created four regions (F) upstream of CDKN2A and three downstream regions (R), and the target regions were used as input for AmBre-design (d = 6500 bp). AmBre-design outputs a high-quality 16-primer design (AMBRE-16) with primers spaced apart by ∼6 kb to cover the 100-kb input region. The design was used by AmBre-amplify on DNA samples from each cell line. The experiment successfully amplified DNA from each cell line (Supplemental Fig. S4), where each line produced a unique-sized amplicon even though each reaction uses the same set of 16 primers.
Table 1.
Five cell lines with CDKN2A deletion breakpoints in GRCh37

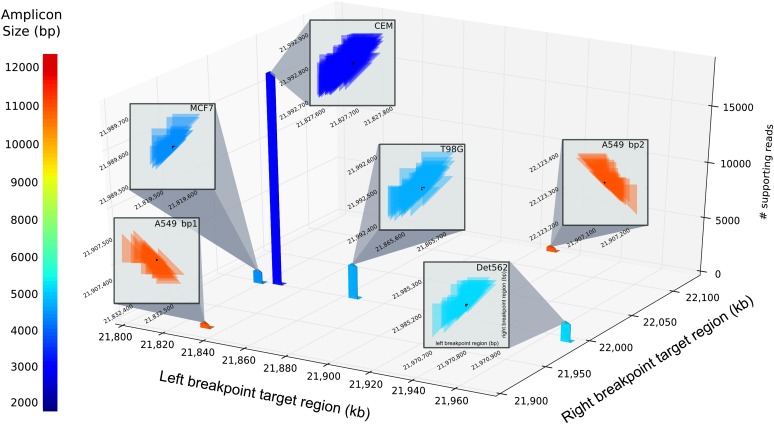
PCR products were mixed together for simultaneous preparation and sequencing on a single SMRT cell. The sequence data were the input to AmBre-analyze. The tool BLASR (Chaisson and Tesler 2012) identified 52k alignable fragments. After clustering in AmBre-analyze, we retrieved deep coverage of every breakpoint (although with six clusters instead of five; see below), with A549 having the lowest coverage of 400 fragments and CEM having the highest coverage of 18,000 fragments (Fig. 4). The difference in coverage is due to different amplicon sizes, where shorter amplicons are easier to load onto a PacBio SMRT cell than longer amplicons. Newer PacBio instrumentation is expected to normalize for this sequencing bias (Mason and Elemento 2012).
Figure 4.
Aggregates of breakpoints from each PacBio fragments after sweep line clustering. Target amplicons are strongly supported by fragments and breakpoints are well separated. Only breakpoints with L < 1 kb are displayed for inset boxes. The height of each cluster corresponds with number of fragments supporting the breakpoint (depth of breakpoint coverage).
AmBre-analyze generated consensus sequence for each cell line. A549, CEM, and Detroit562 breakpoints (Supplemental Figs. S5, S6) are concordant with previous studies (Kitagawa et al. 2002; Sasaki et al. 2003; Bashir et al. 2010). The A549 harbors a complex structural variation where in addition to a large DNA segmental loss including CDKN2A, there is a 325-bp internal inversion occurring at the deletion breakpoint junction. AmBre-analyze resolved the complex event as two separate breakpoints. The A549 amplicon template was created by ordering the reference segments corresponding to the two breakpoints. After template refinement, the A549 amplicon sequence matched the sequence found by Bashir et al. (2010).
To our knowledge, the nucleotide sequence for MCF7 and T98G had not been previously characterized in spite of previous efforts, including whole-genome sequencing of the MCF7 cell line. The ease of the discovery in our experiment attests to the value of a targeted approach to SV detection. Both MCF7 and T98G sequences were confirmed using Sanger sequencing. Interestingly, the SNP-array estimate for the MCF7 breakpoint is 15 kb away from the AmBre-detected breakpoint. The difference may be due to SINE and LINE repeats that mark the region of the upstream MCF7 breakpoint, a fact confirmed by the Sanger reads (Supplemental Fig. S5). Repetitive sequences are known to confound structural variation analysis and possibly explain why previous genome sequencing studies of MCF7 have not annotated the CDKN2A deletion breakpoints (Hampton et al. 2009, 2011).
We analyzed the physical properties of DNA around the breakpoints of CDKN2A deletions using the BreakSeq pipeline (Lam et al. 2009). All five deletion events were predicted to result from nonhomologous end joining (NHEJ). According to Lam et al. (2009), a characteristic of NHEJ is lower DNA duplex stability near the breakpoints of a structural variation. They assessed DNA duplex stability based on predictions of helix stability (average dissociation free energy of overlapping dinucleotides) and DNA flexibility (average twist angle of overlapping dinucleotides). We found no strong association to lower DNA duplex stability in CDKN2A deletion breakpoints, albeit we are analyzing much fewer structural variations (Supplemental Fig. S7). Alternatively, Kitagawa et al. (2002) suggested that the CDKN2A deletion in CEM is due to illegitimate V(D)J recombination, which is evidenced by V(D)J recombination motifs discovered near the deletion breakpoints.
Characterizing CDKN2A deletion assuming no DNA break clustering
Also, AmBre applies to contiguous break regions. We developed a 68-primer design to capture CDKN2A deletions with breaks in a 380-kb region (AMBRE-68) (Fig. 3).
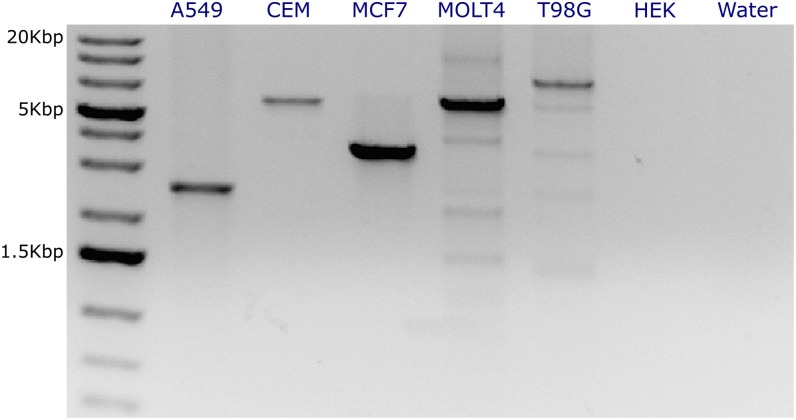
In AmBre-amplify experimentation, we observed that the high amount of multiplexing, and larger amplicon lengths (>4 kb) reduce amplification efficiency. Using all AMBRE-68 primers in a single reaction resulted in amplification of only the 2.2-kb A549 CDKN2A deletion loss (data not shown). To mitigate this effect, subsampling of primers from a design and performing multiple reactions per sample using different primer sets improved amplification results. To test whether the AMBRE-68 primers selected were viable at some level of subsampling, we sampled the nearest forward and reverse primer in AMBRE-68 to each CDKN2A break in cell lines: A549, CEM, Detroit562, MCF7, MOLT4, T98G. This resulted in a nine-primer subset, which again captures the CDKN2A deletion in each cell line. Of these cell lines, five lines resulted in amplicons ranging in lengths from 2.2 kb to 7.5 kb (Fig. 5). The Detroit562 breakpoints did not fall within the target breakpoint region given to AmBre-design, and the expected amplicon size using the closest AMBRE-68 primers is 16 kb. Thus, Detroit562 did not amplify with the nine-primer subset. For each remaining cell line, the observed amplicon length matched the spacing between CDKN2A breakpoints and nearest primers in AMBRE-68 design. Thus, a universal primer design divided into multiple primer subset experiments can be used to identify SVs.
Figure 5.
Subsampling of nine primers from the complete AMBRE-68 tiling design results in clean amplification of CDKN2A loss DNA fragments in six cell lines. (From left to right) Lanes contain 1 kb of Plus GeneRuler DNA ladder, PCR products from samples A549 (2.2 kb), CEM (5.8 kb), MCF7 (3.6 kb), MOLT4 (6.8 kb), T98G (7.5 kb), HEK, and water. The expected lengths of each amplicon according to AMBRE-68 design are listed in parentheses. HEK cells (no CDKN2A deletion) and H2O are negative controls.
Characterizing RUNX1–RUNX1T1 translocations
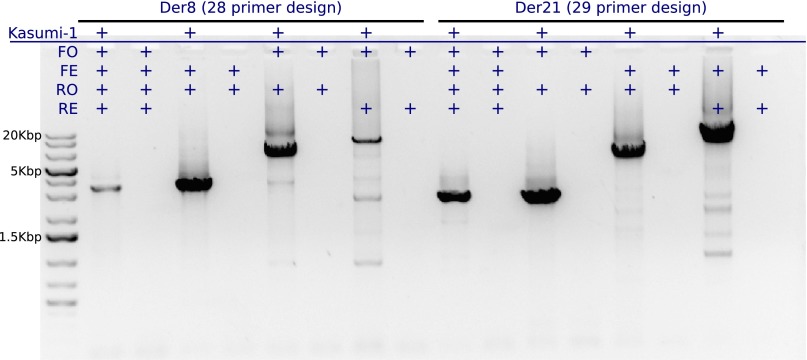
AmBre also captures more complex rearrangements like interchromosomal translocations. This was demonstrated with an experiment characterizing RUNX1–RUNX1T1 gene fusion, the result of a translocation between chr21 and chr8. In the tumor genome, breakpoint ends lie within a 30-kb region chr21: 36,205,000–36,235,000 in the RUNX1 intron, and a 55-kb region chr8: 93,030,000–93,085,000 in RUNX1T1, and the derivative chromosome 8 (Der8) encodes a fusion oncoprotein. In some cases, the translocation is balanced and also generates a fusion of RUNX1T1–RUNX1 on a derivative chromosome 21 (Der21). To capture the translocation producing Der8, we used AmBre to design 10 reverse primers in the RUNX1 region and 18 forward primers in the RUNX1T1 region with ∼3-kb primer spacing. Similarly, to capture Der21 breakpoints, 10 forward and 19 reverse primers were designed in the RUNX1 and RUNX1T1 regions, respectively. Recall, an ∼3-kb primer spacing supposes the maximum product size is ∼6 kb. The primer designs were tested on Kasumi-1, which carries the balanced translocation with both Der8 and Der21 breakpoints characterized (Xiao et al. 2001). AmBre spaced the primers in the two regions unaware of the true Kasumi-1 breakpoints, and we assayed the Der8 and Der21 chromosomes in two independent reactions using the respective 28 and 29 primers. The primers closest to the breakpoints produce a 3.5-kb and 2.7-kb amplicon from Der8 and Der21, respectively (Fig. 6). Both reactions resulted in a strong signal and virtually no background noise, despite there being close to 30 primers in each reaction.
Figure 6.
Characterizing RUNX1–RUNX1T1 balanced translocation in Kasumi-1. Lanes 1, 2, 4, 6, and 8 contain 1 kb of Plus GeneRuler DNA ladder, PCR products from Kasumi-1 Der8 with all 28 primers (3.5 kb), 14 primer FE ∪ RO (3.5 kb), 14 primer FO ∪ RO (6.8 kb), and 14 primer FO ∪ RE (10.1 kb). Lanes 3, 5, 7, and 9 contain matching water controls, which show no contamination. Lanes 10, 12, 14, and 16 contain PCR products from Kasumi-1 Der21 with all 29 primers (2.7 kb), 15 primer FO ∪ RO (2.7 kb), 15 primer FE ∪ RO (6.1 kb), and 14 FE ∪ RE (8.1 kb). The gel was loaded with 2 μL for lanes 2–5 and 10–13, and 4 μL for remaining volumes. Reactions with shorter amplicons amplified extremely well, and lesser volumes were used for visualization on the gel. The expected amplicon lengths according to the Der8 and Der21 design are listed in parentheses.
Furthermore, we investigated subsampling of primers and efficacy in generating longer amplicons. For each primer design, we divided the forward and reverse primers based on index parity when sorted by chromosome position. Thus, there are four primer sets: forward odd (FO), forward even (FE), reverse odd (RO), and reverse even (RE), with primers spaced by ∼6 kb. The forward and reverse primer sets make four combinations: FO ∪ RO, FO ∪ RE, FE ∪ RO, and FE ∪ RE, primers for capturing target breakpoints. These combinations can be treated as four new primer designs, each with a maximum product size of 12 kb, but half as many primers. This gives us the opportunity to assess amplification efficiency across different amplicon lengths and primer density per reaction using the same DNA template. In the original 28-primer design, the Kasumi-1 breakpoints for Der8 were generated by the sixth forward and ninth reverse primer. Thus, trying the 14 primer designs FE ∪ RO, FO ∪ RO, and FO ∪ RE produces 3.5-kb, 6.8-kb, and 10.1-kb amplicons (Fig. 6). Similarly, the 29-primer design for Der21 was subsampled into three reactions. Each reaction resulted in a strong signal band at the expected amplicon size, and all six amplicons were confirmed to span the Der8 and Der21 breakpoints via Sanger sequencing (Supplemental Fig. S8). From each reaction, a general trend of better amplification for shorter amplicon lengths is observed. However, there was no significant difference in amplification efficiency between using all primers and half the primers to generate the shortest amplicons. Longer amplicons had a strong signal, but weaker false products were visible. This effect is not seen with the shorter amplicons, and false products may be more prevalent in reactions with a greater number of primers and longer amplicons.
Dealing with tumor heterogeneity
The AmBre assay, unlike other methods, can target DNA with an SV in the context of high background of germline DNA. This feature is important for sensitive detection of tumor DNA and establishing a patient-specific tumor DNA marker for monitoring tumor burden. We successfully amplified a 2.2-kb CDKN2A deletion sequence from A549 and a 3.6-kb deletion sequence from MCF7 starting with A549 and MCF7 genomic DNA mixed with HEK genomic DNA (Supplemental Fig. S9). Each reaction starts with a heterogeneous mixture of ∼400 ng with tumor to wild-type gDNA mixture ratios of 1:1, 1:10, 1:100, and 1:1000. In a realistic application for AmBre, each reaction contains numerous primers where only two primers are responsible for amplification. In the experiment, each reaction contains 16 primers sampled from AMBRE-68 around CDKN2A deletion breakpoints for each cell line. In the heterogeneity experiment of A549, strong amplification is observed for each mixture ratio, whereas for MCF7 there is clearly a reduction of amplification efficiency as the fraction of starting cancer cell line gDNA decreases (Supplemental Fig. S9). Amplification of longer amplicons with AmBre in the complex gDNA sample is also possible, however, with reduced sensitivity (Supplemental Fig. S10). The sensitivity for the AmBre assay is largely dependent on expected amplicon length. CDKN2A deletion breakpoints corresponding to a smaller amplicon in a particular AmBre primer design are more easily amplified.
Discussion
AmBre addresses the challenge of highly sensitive SV targeting in complex DNA mixtures. This is accomplished with a careful design of tiling primers that enables amplification of DNA harboring the SV if present in the mixture and a specialized PacBio analysis pipeline to confirm SV breakpoints. AmBre was used to discover breakpoints associated with CDKN2A deletion in cancer cell lines MCF7 and T98G. In addition, we demonstrated that amplification occurs even in a complex DNA mixture where one in every 1000 DNA molecules contains the CDKN2A deletion. These features of AmBre are clinically important. An SV breakpoint specific to a cancer patient could serve as a personalized biomarker, where a quantitative PCR assay could accurately measure the patient's tumor burden (Michor et al. 2005; Bartley et al. 2010). With advancements in microfluidics and droplet PCR, quantifying one to three copies of tumor DNA in a complex sample is possible (Hatch et al. 2011).
If the problem is to simply observe an SV, there are numerous high-throughput methods: SNP hybridization arrays (SNP-array), whole-exome sequencing (WES), and whole-genome sequencing (WGS). However, these methods are not ideal for a clinical application in tumorburden monitoring. SNP arrays and WES give copy number readouts of DNA, which hint at the presence of SVs and a low-resolution estimate of corresponding breakpoints. Without a high-accuracy breakpoint estimate, a quantitative PCR assay specific to tumor DNA cannot be designed. WGS is capable of breakpoint calling but would require an exorbitant amount of deep sequencing to capture SVs occurring in a low fraction of DNA. Harismendy et al. (2011) reported the extent of this sequencing challenge, where more than 1500× coverage of cancer mutational hotspots (71.1-kb region) was necessary to capture single nucleotide variants (SNVs) occurring with prevalence >5% in the sample.
Therefore, a targeted approach for mutation detection is preferred to a high-throughput untargeted mutation discovery for clinical practice. A high-throughput method captures numerous SVs and SNVs where follow-up functional analysis is required for each mutation to determine its potential as a cancer driver or passenger mutation. Alternatively, there are numerous targetable SVs known to drive cancer progression, and they are being used in clinical laboratories to confirm cancer diagnosis and guide therapy. The most notable example, CML patients with the BCR–ABL1 translocation, are treated with tyrosine kinase inhibitors. The patient's response to therapy can be reliably tracked by measuring tumor DNA containing BCR–ABL1 gene fusion from blood samples (Michor et al. 2005; Bartley et al. 2010). Unfortunately, such success in tumor burden monitoring has not been observed for patients with solid tumors.
In this study, we present AmBre's application to capture RUNX1–RUNX1T1 translocations in AML cases and CDKN2A deletions, which are prevalent in many types of cancer. Using the accompanying software, this approach can be easily extended to target other SVs, like BCR–ABL1 in chronic myeloid leukemia, EML4–ALK in lung cancer, and TMPRSS2–ERG in prostate cancer. For EML4–ALK and TM-PRSS2–ERG, DNA breaks within introns and rearrangement of the chromosome fuse the genes together, similar to RUNX1–RUNX1T1 gene fusion. The remaining challenge for AmBre is a limited targetable breakpoint region. We presented a design capturing breakpoints falling within 100 kb and proposed a multiple primer subset strategy for encompassing a 380-kb breakpoint region. Further development is necessary to capture SVs with breakpoints appearing in a >1-Mb range. AmBre is a first step to a sensitive tumor DNA monitoring test for solid tumors. Extending the approach with improvements of applying multiple primer designs to target the same SV or the use of microfluidic devices may lead to an ultrasensitive assay capable of minimally invasive early cancer detection.
Methods
AmBre: Primer generation and filtering
Primer3 2.3.0 (Rozen and Skaletsky 2000) was used with long-range PCR-specific parameters to identify 31-bp candidate AmBre primers that were capable of amplification under the same thermocycling conditions. To minimize the chance of off-target amplification, candidate primers were aligned to the reference human assembly (GRCh37) using BLAT (Kent 2002). Define an end-aligning match as an exact match of length >18 between the 3′ end of a primer and an off-target location. Primers with >10 end-alignments were removed as having a high chance for off-target amplification. Second, pairs of primers that have compatible end-alignments within a 2d-long off-target region were marked as incompatible. Finally, each pair (including a self-pair) was tested for dimerization using MultiPlx (Kaplinski et al. 2005). Primers with self-dimerization (maximum binding energy ΔG < −8.0 kcal/mol for any region) were removed, and pairs with high binding affinity (maximum binding energy ΔG < −4.0 kcal/mol for primer–primer 3′-end binding or −8.0 kcal/mol for any region of primers) were marked as incompatible. The remaining candidate primers and incompatibilities formed the input to AmBre primer selection.
AmBre: Primer selection with simulated annealing
A final AmBre primer design was selected from a filtered list of candidate primers (PU) and primer–primer compatibilities. To compute an optimal primer design, a low-cost P according to C(P), we applied a simulated annealing (Kirkpatrick 1984) procedure. We computed an initial design P using a random subset of six primers. Define the neighboring design of P, N(P), as either the removal of a single primer from P or the addition of a single primer p ∉ P to P followed by removal of all primers p′ ∈ P s.t. (p, p′) ∈ E. The simulated annealing procedure described in Algorithm 1 was used to compute low-cost designs.
Algorithm 1.
Simulated annealing algorithm

The temperature schedule, T1, T2, T3, …, linearly decreases depending on intercept and slope parameters m and b. Parameters tested for T were combinations of m = 1, 0.1, 0.01, 0.001 and b = 104, 105, 106. The maximum number of iterations run was determined by the temperature schedule,  , and constrained to be at least 106 and at most 108 iterations. Each parameter set was repeated three times. The lowest-cost primer design of all runs was used as the final design. Supplemental Figure S2 demonstrates convergence to design minima under different parameters of T for a target CDKN2A breakpoint region of length 380 kb.
, and constrained to be at least 106 and at most 108 iterations. Each parameter set was repeated three times. The lowest-cost primer design of all runs was used as the final design. Supplemental Figure S2 demonstrates convergence to design minima under different parameters of T for a target CDKN2A breakpoint region of length 380 kb.
AmBre-analyze: PacBio sequence analysis
Alignment trimming
BLASR-computed local alignments between the PacBio reads and human reference assembly were provided as input to alignment trimming. An alignment pair (Fa, Ga), (Fb, Gb) with a ≪ b between a fragment F and reference G implies a breakpoint. The goal of alignment trimming is to trim the ends of each alignment for each fragment F, so that (1) each segment of F participates in a single alignment and (2) F is maximally covered.
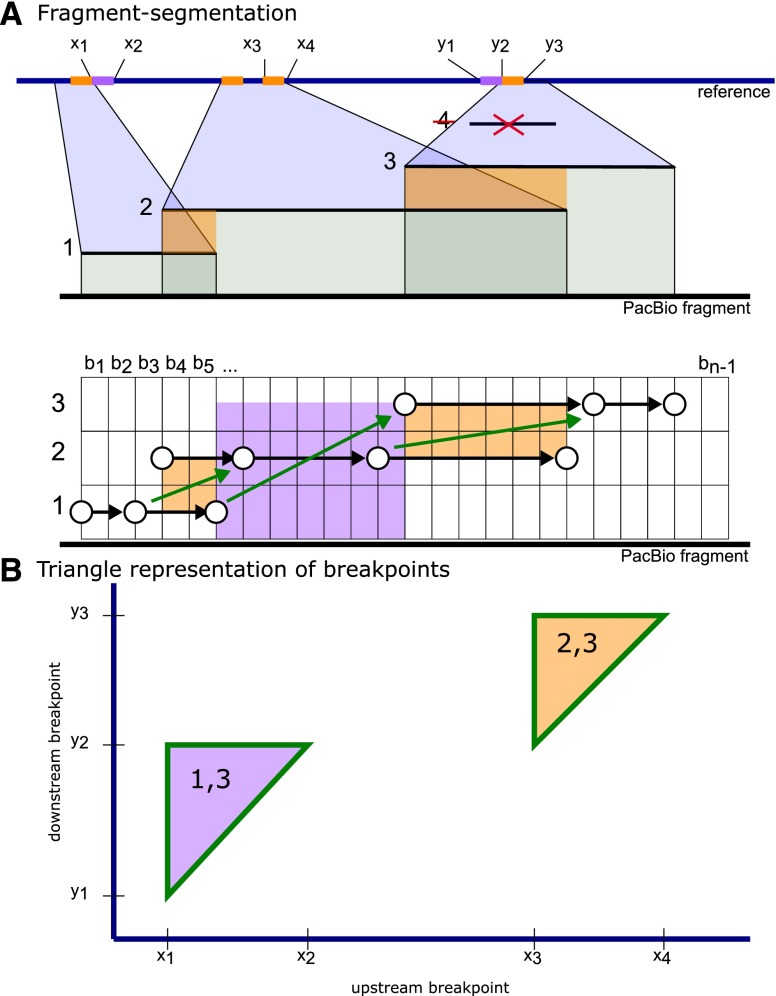
We first remove local alignments encompassed by other alignments (e.g., 4 in Fig. 7). We sort the remaining alignments by their location on the fragment, so that alignment i starts before alignment j if and only if i < j. Let bs(i) and be(i) denote the fragment breaks before the beginning and after the end of alignment i.
Figure 7.
(A) Fragment-segmentation example for local alignments 1, 2, 3, and 4 along a PacBio fragment. (B) Triangle representation of adjacent alignments 1, 2, and 3 on G × G plane.
We represent alignments on a grid with alignments as rows and fragment positions as columns (Fig. 7). An alignment is a series of breaks on the fragment [i.e., (1, b1) to (1, b5) in Fig. 7]. Alignments are chained together to cover a portion of F exactly once. To chain adjacent alignments, for each alignment j with an alignment i that terminates before j starts, add a jump from (i, be(i)) to (j, bs(j)) [for instance (1, be(1)) to (3, bs(3))]. Also, for each alignment j overlapping an earlier alignment i on the fragment, add a jump from (i, bs(j)) to (j, be(i)) [for instance (2, be(3)) to (3, bs(2))] if i spans bs(j) and j spans be(i). By this process, any alignment chain covers positions exactly once.
 |
An alignment chain is scored by summing local alignment scores (Aln[i, u, v] for alignment i for fragment coordinates u to v) and penalizing for jumps between alignments [J(u, v) for alignment u to v]. A high-scoring alignment chain corresponds to trimmed alignments that align well and cover most of the fragment. The score of a chain is computed using dynamic programming. Let S(j, v) denote the score of the best chain ending at (j, v). Then,
In the recursion, (i, u) is the start of alignment j, start of a jump to (j, v) [i.e., if (j, v) = (3, be(2)) then (i, u) could be (2, bs(3))], or previous position on alignment j where a jump ends [i.e., if (j, v) = (2, be(2)) then (i, u) = (2, be(1))]. By not computing the score for each alignment and fragment position on the grid, the optimal trimmed alignment chain is quickly found.
Along the maximum scoring chain, each jump,  , represents a breakpoint estimate
, represents a breakpoint estimate  . For example, the jump from 1 to 3 corresponds with breakpoint estimate (x1, y2, 6).
. For example, the jump from 1 to 3 corresponds with breakpoint estimate (x1, y2, 6).
In this formulation, two alignments that overlap may contribute to a high score since the overlap segment is scored as the average of both alignment scores. Above, for a breakpoint estimate from overlapping alignments, we use boundaries around the overlap and do not resolve a tighter breakpoint within the overlap segment. Finding a tighter breakpoint estimate would require computing S for all breaks within overlap intervals, which is inefficient for thousands of fragments. In any case, the conservative breakpoint estimates are improved with downstream clustering and refinement steps.
Breakpoint clustering
Breakpoint estimates from all fragments supporting the same breakpoint are aggregated into groups using a sweep line algorithm. Sindi et al. (2009) applied a similar geometric approach to efficiently identify structural variations using discordant paired-end reads.
For a breakpoint estimate (x, y, L), the true breakpoint junctions (a, b) in reference G lies between x … x + L and y − L … y, respectively, subject to a − x + y − b < L. Here, we assume that L, a spacing length on F, is a reasonable estimate for breakpoint uncertainty on G and the effect of sequencing deletion errors at the breakpoint junction is minimal. On a G × G plane, each breakpoint estimate x, y, and L with the above constraints defines a triangle that contains the true breakpoint (a, b) (Figs. 4, 7).
A line sweeps the plane and tracks when breakpoint triangles overlap along the sweep line. Here, a cluster is a collection of triangles where each triangle overlaps one or more triangles in the cluster. The consensus breakpoint (a, b) for the cluster is the mode of (x, y) estimates (see Fig. 4).
Accounting for reverse orientation alignments
With a slight modification, we can account for alignments in the reverse complement orientation to capture structural variations with inversions and bidirectional PacBio reads. PacBio reads DNA amplicons in both directions, in particular, a read in the forward direction produces an alignment chain (Fx, Gx), (Fy, Gy) and in the reverse direction [Hy, RC(Gy)], [Hx, RC(Gx)], where RC reverse-complements the sequence G. This is resolved by relabeling reverse complement alignments by a −, such that H supports the breakpoint (−y, −x).
The relabeling applies naturally to the sequence analysis pipeline. Alignment trimming relies only on projections on sequenced fragments and therefore does not change. Each DNA amplicon containing a breakpoint is associated with two breakpoint estimates (x, y) generated from forward reading and (−y, −x) from reverse reading.
In addition, the constraints of −y, −x, L in relation to −a, −b remain the same; therefore, both forward and reverse direction breakpoint estimates have the same triangle orientation on the G × G plane. All forward and reverse breakpoints are simultaneously recovered with the sweep line algorithm.
Using reverse complement alignments, breakpoints associated with inversions, like A549, are captured. In this case, a breakpoint corresponds with (−x, y) and (−y, x) or (x, −y) and (y, −x).
Breakpoint reconstruction
In the final step, predicted amplicon templates for each cluster are created by joining reference sequence G(6500 − a, a) and G(b, b + 6500). The PacBio SMRT Analysis 1.4 pipeline for Resequencing is performed to refine the amplicon template predictions using all fragments generated from the SMRT cell (Supplemental Fig. S6). The Resequencing protocol involves running BLASR for mapping followed by Quiver for consensus sequence calling. The protocol accurately recovered the sequence around breakpoints; the consensus amplicon sequence starting at aligned 25 − a and ending at b + 25 matched either sequencing from previous studies or independent Sanger sequencing chromatogram (Fig. 5). For clusters with L > 0, adding L “N” nucleotides at the breakpoint junction of the predicted amplicon template had no effect on the PacBio Resequencing protocol. In both cases, the correct amplicon breakpoint junction sequence was found.
A549, CEM, Detroit562, and T98G cells were thawed from Moore's Cancer Biorepository. MCF7, HeLa, and HEK (293T) cells were collected from the Rosenfeld laboratory. Standard DNAzol protocol was used for DNA extraction and DNA was quantified with NanoDrop 2000 spectrophotometer. DNA products are visualized on 1% agarose gels with EtBr. Gel images are either color value inverted or color curve adjusted uniformly across the image for visual enhancement. All PCRs were performed on a Bio-Rad iCycler instrument.
All PCR experiments used the following thermocycling conditions: initial denaturation for 3 min at 95°C, 10 cycles for 20 sec at 94°C, 30 sec at 64°C, 15 min at 66°C, 28 cycles for 5 sec at 94°C, 30 sec at 64°C, 15 min + 20 sec for each cycle at 66°C, final extension for 45 min at 64°C, and 4°C hold.
AMBRE-16 experiment
See the Supplemental Material for primer sequences. The standard protocol for NEB Crimson LongAmp Taq is used for 50-μL PCR reactions with the following changes. The same mix of 16 primers was used in each reaction where each primer is present with a final concentration of 0.2 μM. The starting genomic DNA for each cell line reaction is 10 ng. The QIAquick PCR purification kit (Qiagen) was used to clean up PCR samples. Samples were quantified, and 2 μg of the A549 reaction sample was mixed with 1 μg of each remaining cell line reaction sample and submitted for PacBio sequencing at the UCSD BioGem Core facility. Loading of DNA samples onto a PacBio SMRT cell is biased toward sequencing smaller amplicons, and increasing the amount of A549 reaction sample containing an 11-kb DNA fragment was necessary to sufficiently sequence the A549 DNA fragment.
AMBRE-68 experiment
See the Supplemental Material for primer sequences. The standard protocol for NEB Crimson LongAmp Taq is used for 50-μL PCR reactions with the following changes. The same mix of nine primers was used in each reaction, where each primer is present with a final concentration of 0.4 μM. The starting genomic DNA for each cell line reaction is 20 ng.
RUNX1–RUNX1T1 experiment
See the Supplemental Material for primer sequences. The standard protocol for NEB Crimson LongAmp Taq is used for 25-μL PCR reactions with the following changes. All primers at 0.4 μM PCR experiments were under the following conditions: initial denaturation for 1 min at 95°C, 10 cycles for 20 sec at 94°C, 30 sec at 63°C, 2 min at 68°C, 28 cycles for 5 sec at 94°C, 30 sec at 61°C, for 2 min + 5 sec for each cycle at 66°C, final extension for 30 min at 64°C, and 4°C hold. Subsampling experiments used the same primer concentration and thermocycling conditions except extension time for the first phase is 7 min and the second phase is 7 min with 10 sec increase per cycle.
Tumor:wild-type genomic DNA heterogeneity experiment
See the Supplemental Material for primer sequences. The standard protocol for NEB Crimson LongAmp Taq is used for 50-μL PCR reactions with the following changes. Each primer has a final concentration of 0.4 μM. Each reaction contains ≈400 ng of gDNA, with the following tumor-to-normal DNA ratios: 200 ng:200 ng, 40 ng:400 ng, 4 ng:400 ng, and 0.4 ng:400 ng. Normal DNA is derived from HEK cells.
MCF7 and T98G PCR validation
Primer pair sequences were generated using Primer3 2.3.0 given a short genomic sequence around the MCF7 and T98G breakpoints as determined by PacBio sequencing and analysis. See the Supplemental Material for primer sequences. The standard protocol for NEB Standard Taq is used for 50-μL PCR reactions starting with 250 ng of genomic DNA.
Data access
The sequencing data have been deposited at the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number SRX353044. The AmBre software is available at http://bix.ucsd.edu/AmBre.
Acknowledgments
This work was supported in part by grants 5RO1-HG004962 and U54 HL108460 from the National Institutes of Health and NSF-CCF-1115206 and IIS1318386 from the National Science Foundation. The core technology described in the present paper is covered by a pending PCT patent application (PCT/US2013/63539). Also, we thank the Rosenfeld laboratory for supplying MCF7 and HEK cell lines.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.161497.113.
References
- Bartley P, Ross D, Latham S, Martin-Harris M, Budgen B, Wilczek V, Branford S, Hughes T, Morley A 2010. Sensitive detection and quantification of minimal residual disease in chronic myeloid leukaemia using nested quantitative PCR for BCR–ABL DNA. Int J Lab Hematol 32: e222–e228 [DOI] [PubMed] [Google Scholar]
- Bashir A, Liu Y, Raphael B, Carson D, Bafna V 2007. Optimization of primer design for the detection of variable genomic lesions in cancer. Bioinformatics 23: 2807–2815 [DOI] [PubMed] [Google Scholar]
- Bashir A, Lu Q, Carson D, Raphael BJ, Liu YT, Bafna V 2010. Optimizing PCR assays for DNA based cancer diagnostics. J Comput Biol 17: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Tesler G 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinformatics 13: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. 2009. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat Methods 6: 677–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S-J, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin S-F, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. 2013. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368: 1199–1209 [DOI] [PubMed] [Google Scholar]
- Downing JR 1999. The AML1–ETO chimaeric transcription factor in acute myeloid leukaemia: Biology and clinical significance. Br J Haematol 106: 296–308 [DOI] [PubMed] [Google Scholar]
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. 2012. Mind the gap: Upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE 7: e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Bignell G, Butler A, Edkins S, Hinton J, Beare D, Swamy S, Santarius T, Chen L, Widaa S, et al. 2010. PICNIC: An algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics 11: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton O, Den Hollander P, Miller C, Delgado D, Li J, Coarfa C, Harris R, Richards S, Scherer S, Muzny D, et al. 2009. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res 19: 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton O, Miller C, Koriabine M, Li J, Den Hollander P, Carbone L, Nefedov M, Ten Hallers B, Lee A, De Jong P, et al. 2011. Long-range massively parallel mate pair sequencing detects distinct mutations and similar patterns of structural mutability in two breast cancer cell lines. Cancer Genet 204: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Schwab R, Bao L, Olson J, Rozenzhak S, Kotsopoulos S, Pond S, Crain B, Chee M, Messer K, et al. 2011. Detection of low prevalence somatic mutations in solid tumors with ultra-deep targeted sequencing. Genome Biol 12: R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch AC, Fisher JS, Tovar AR, Hsieh AT, Lin R, Pentoney SL, Yang DL, Lee AP 2011. 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab Chip 11: 3838–3845 [DOI] [PubMed] [Google Scholar]
- Hinrichs A, Karolchik D, Baertsch R, Barber G, Bejerano G, Clawson H, Diekhans M, Furey T, Harte R, Hsu F, et al. 2006. The UCSC genome browser database: Update 2006. Nucleic Acids Res 34: D590–D598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplinski L, Andreson R, Puurand T, Remm M 2005. MultiPLX: Automatic grouping and evaluation of PCR primers. Bioinformatics 21: 1701–1702 [DOI] [PubMed] [Google Scholar]
- Kent W 2002. BLAT: The BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Deluna A, Mungrue I, Vu C, Pouldar D, Civelek M, Orozco L, Wu J, Wang X, Charugundla S, et al. 2012. Effect of 9p21 coronary artery disease locus neighboring genes on atherosclerosis in mice. Circulation 126: 1896–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick S 1984. Optimization by simulated annealing: Quantitative studies. J Stat Phys 34: 975–986 [Google Scholar]
- Kitagawa Y, Inoue K, Sasaki S, Hayashi Y, Matsuo Y, Lieber MR, Mizoguchi H, Yokota J, Kohno T 2002. Prevalent involvement of illegitimate V(D)J recombination in chromosome 9p21 deletions in lymphoid leukemia. J Biol Chem 277: 46289–46297 [DOI] [PubMed] [Google Scholar]
- Lam H, Mu X, Stütz A, Tanzer A, Cayting P, Snyder M, Kim P, Korbel J, Gerstein M 2009. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol 28: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Carson D 2007. A novel approach for determining cancer genomic breakpoints in the presence of normal DNA. PLoS ONE 2: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason C, Elemento O 2012. Faster sequencers, larger datasets, new challenges. Genome Biol 13: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michor F, Hughes T, Iwasa Y, Branford S, Shah N, Sawyers C, Nowak M 2005. Dynamics of chronic myeloid leukaemia. Nature 435: 1267–1270 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Sasaki S, Kitagawa Y, Sekido Y, Minna JD, Kuwano H, Yokota J, Kohno T 2003. Molecular processes of chromosome 9p21 deletions in human cancers. Oncogene 22: 3792–3798 [DOI] [PubMed] [Google Scholar]
- Sindi S, Helman E, Bashir A, Raphael B 2009. A geometric approach for classification and comparison of structural variants. Bioinformatics 25: i222–i230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessely R 2010. Atherosclerosis and cell cycle: Put the brakes on!: Critical role for cyclin-dependent kinase inhibitors? J Am Coll Cardiol 55: 2269–2271 [DOI] [PubMed] [Google Scholar]
- Xiao Z, Greaves M, Buffler P, Smith M, Segal M, Dicks B, Wiencke J, Wiemels J 2001. Molecular characterization of genomic AMLI–ETO fusions in childhood leukemia. Leukemia 15: 1906–1913 [DOI] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z 2009. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitouni B, Boeva V, Janoueix-Lerosey I, Loeillet S, Legoix-Né P, Nicolas A, Delattre O, Barillot E 2010. SVDetect: A tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics 26: 1895–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]