Abstract
Background
Physician-coded verbal autopsy (PCVA) is the most widely used method to determine causes of death (CODs) in countries where medical certification of death is uncommon. Computer-coded verbal autopsy (CCVA) methods have been proposed as a faster and cheaper alternative to PCVA, though they have not been widely compared to PCVA or to each other.
Methods
We compared the performance of open-source random forest, open-source tariff method, InterVA-4, and the King-Lu method to PCVA on five datasets comprising over 24,000 verbal autopsies from low- and middle-income countries. Metrics to assess performance were positive predictive value and partial chance-corrected concordance at the individual level, and cause-specific mortality fraction accuracy and cause-specific mortality fraction error at the population level.
Results
The positive predictive value for the most probable COD predicted by the four CCVA methods averaged about 43% to 44% across the datasets. The average positive predictive value improved for the top three most probable CODs, with greater improvements for open-source random forest (69%) and open-source tariff method (68%) than for InterVA-4 (62%). The average partial chance-corrected concordance for the most probable COD predicted by the open-source random forest, open-source tariff method and InterVA-4 were 41%, 40% and 41%, respectively, with better results for the top three most probable CODs. Performance generally improved with larger datasets. At the population level, the King-Lu method had the highest average cause-specific mortality fraction accuracy across all five datasets (91%), followed by InterVA-4 (72% across three datasets), open-source random forest (71%) and open-source tariff method (54%).
Conclusions
On an individual level, no single method was able to replicate the physician assignment of COD more than about half the time. At the population level, the King-Lu method was the best method to estimate cause-specific mortality fractions, though it does not assign individual CODs. Future testing should focus on combining different computer-coded verbal autopsy tools, paired with PCVA strengths. This includes using open-source tools applied to larger and varied datasets (especially those including a random sample of deaths drawn from the population), so as to establish the performance for age- and sex-specific CODs.
Keywords: Causes of death, Computer-coded verbal autopsy (CCVA), InterVA-4, King-Lu, Physician-certified verbal autopsy (PCVA), Random forest, Tariff method, Validation, Verbal autopsy
Background
Verbal autopsy (VA) is used in areas with limited medical certification of death to obtain information on causes of death (CODs) [1-3]. VA tools typically consist of a structured survey administered to a close relative or associate of the deceased by a trained field worker, to record the signs and symptoms that occurred before death. This information is used to assign the most probable COD, most often via physician-certified verbal autopsy coding (PCVA).
PCVA has limitations in inter- and intra-observer differences in coding [3], but remains widely useful, particularly in establishing population-based estimates of the major CODs [1]. There has been interest in the use of computer-coded VA (CCVA) methods to automate COD assignment [3]. CCVA methods are, in theory, cheaper, faster and more consistent over time - but their performance against PCVA and against each other has not yet been assessed reliably.
Here, we compare the performance of four of the most promising CCVA methods - InterVA-4, King-Lu (KL), open source random forest (ORF) and open source tariff method (OTM) - across VA studies in several countries, covering more than 24,000 deaths, including community- and hospital-based deaths (Table 1). We define performance by their ability to replicate physician coding.
Table 1.
Dataset specifications
| Variable | China | Institute for Health Metrics and Evaluation | Million Death Study | Agincourt | Matlab |
|---|---|---|---|---|---|
| Region |
China |
N/Aa |
India |
South Africa |
Bangladesh |
| Sample size |
1,502 |
1,556 |
12,225 |
5,823 |
3,270 |
| Ages |
15+ years |
15 to 105 years |
1 to 59 months |
15 to 64 years |
20 to 64 years |
| Number of CODs |
31 |
32 |
15 |
17 |
17 |
| Population |
Hospital deaths |
Hospital deaths |
Community deaths |
Community deaths |
Community deaths |
| Proportion ill-defined deathsb |
0% |
0% |
3% |
12% |
2% |
| Physician coding | Coding by a panel of three physicians assisted with medical records and diagnostic tests | Coding by one physician assisted with medical records and diagnostic tests | Dual, independent coding of VA records, disagreements resolved by reconciliation, and for remaining cases by adjudication by a third physician | Dual, independent coding of VA records, disagreements resolved by third physician. | Single physician re-coding of VA records after initial coding by another physician. |
All VA data in the Million Death Study, Agincourt and Matlab studies were collected by non-medical field staff, and coded by medical staff. aThe full IHME hospital-based dataset includes 12,000 VA records from India, Philippines, Tanzania and Mexico and was released after this paper went to press; correspondence with the study team suggested these data were from Bangladesh but the full details of the 1,556 deaths are not published. bIll-defined deaths are International Classification of Diseases-10 codes R95-R99. VA, verbal autopsy.
Methods
Datasets
Table 1 summarizes important features of the five VA datasets. The datasets from the Matlab study in Bangladesh [4], from China [5], from Agincourt, South Africa [6], and a published study of the Institute for Health Metrics and Evaluation (IHME) [7,8] comprised adult deaths. The Indian Million Death Study (MDS) [9,10] included only child deaths from ages 1 to 59 months. Each study used different field procedures, although with similar collection of major symptoms for each death. Physician coding guidelines and procedures also varied but generally involved at least one doctor examining each record. The China and IHME datasets involved physician coding of hospital-based deaths with additional information on medical histories and diagnostic tests. The four CCVA methods were tested on all five datasets with each study’s PCVA assignment as the reference standard. We could not test InterVA-4 on the China and IHME data due to the unavailability of a data dictionary at the time of analysis.
Computer-coded verbal autopsy methods
InterVA-4
InterVA-4 assigns CODs using a Bayesian model with a priori probabilities based on expert consensus. InterVA-4 version 4.02 was used in this study, and the program with a full description of its logic can be freely obtained online [11].
Open-source random forest
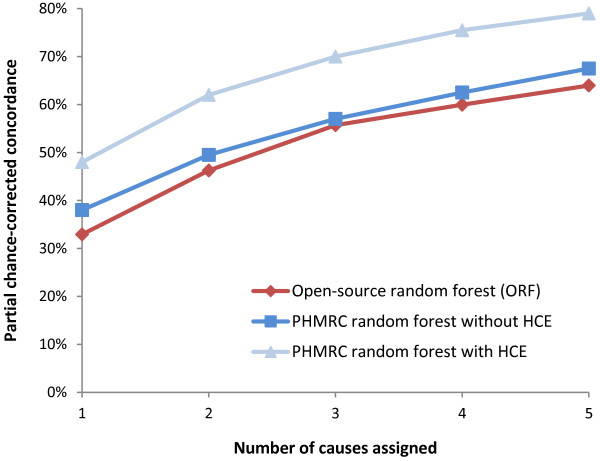
The ORF is a data-driven, probabilistic method that builds upon a similar tool published by IHME [12]. Random forest and tariff methods have been described as having unrivaled performance against all other VA coding methods [12]. However, at the time of writing, these two methods were not publicly available, and their results have not yet been independently replicated. We thus ‘reverse-engineered’ these two methods into open-source tools (details are in Additional file 1). An independent programming expert reviewed the algorithm to assess replication of the IHME method (to the extent of the published details), and we compared the ORF performance on the IHME data available to us to the published results of the IHME methods (Figure 1). The ORF showed very similar trends to those from the IHME random forest, though differences were to be expected due to the unavailability of the full IHME hospital-based dataset [7]. In addition, 96 symptom indicators were used by ORF whereas the IHME method used only the top 40 most-predictive symptoms, the details of which were unavailable [12].
Figure 1.
Comparison of open-source random forest to IHME random forest. The IHME random forest was tested on a set of IHME hospital-based data, both with and without health care experience (HCE) variables. HCE variables are binary questions on previous medically diagnosed conditions (including high blood pressure, tuberculosis, cancer), and details transcribed from the respondents’ medical records. Our IHME subset contained some, but not all, HCE variables. The ORF performance was similar to the IHME random forest method on the full hospital-based dataset without HCE variables, but performed less well when HCE variables were included [12]. HCE, health care experience; IHME, Institute for Health Metrics and Evaluation; ORF, open-source random forest.
Open-source tariff method
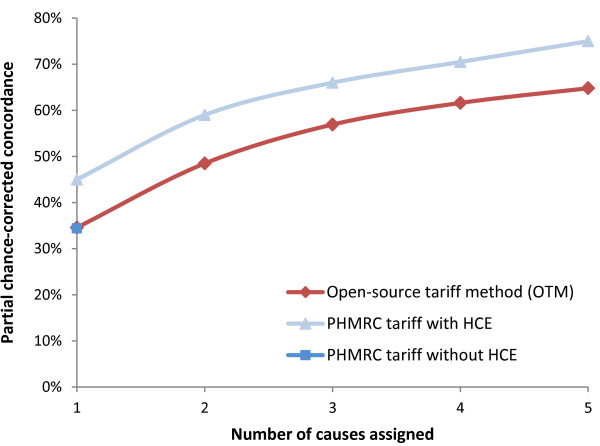
The OTM is a data-driven, probabilistic method that builds upon that published by IHME [13]. The OTM performance on the IHME data available to us was comparable to the results of the IHME method (Figure 2). The resulting differences may be due to similar factors as those mentioned in the ORF description above.
Figure 2.
Comparison of open-source tariff method to IHME tariff method. The IHME random forest was tested on a set of IHME hospital-based data, both with and without health care experience (HCE) variables. The ORF was tested on a subset of the full IHME data, containing some, but not all, HCE variables. The OTM performed almost exactly as the similar IHME method on the full hospital-based dataset without HCE variables (for the top cause), but less well than the same IHME analysis with HCE variables. Note that results for the full IHME dataset without HCE were only available for the top assigned cause [13]. HCE, health care experience; IHME, Institute for Health Metrics and Evaluation; OTM, open-source tariff method.
King-Lu method
The KL method directly estimates cause-specific mortality fractions (CSMFs) without assigning individual CODs. Version 0.9-2.12 was used, for which a full description of the method and discussions of its performance have been published [5], and which is available for download [14].
Testing
Dataset splits and resampling
InterVA-4 uses pre-assigned Bayesian probabilities to assign the COD and thus does not require a training component. The remaining three methods required a training component, consisting of a subset of the original data with assigned CODs, from which the method learned the cause-specific symptom profiles. The trained algorithm was then used to predict CODs in the test dataset.
Table 2 describes the training and testing samples used in the comparisons. Three sample sizes of the datasets were used to highlight changes in performance based on varying dataset sizes (training/testing splits of 1,100/400 and 1,100/1,100 cases, and a split of the full dataset into an equal number of training and testing cases, whose size varied by dataset.) The IHME and China datasets were relatively small (approximately 1,500 cases), which only allowed for the 1100/400 split. Each of the split sizes above were randomly repeated (random splits of the original dataset into the specified number of training and testing cases) 30 times. However, the performance of the methods converged (that is, did not alter by more than 0.5% compared to the average of any previous resamples [15]) well before the full 30 resamples were run.
Table 2.
Description of testing on multiple computer-coded verbal autopsy methods and datasets
|
Dataset |
Training/testing cases |
Number of diagnostic indicators |
|||
|---|---|---|---|---|---|
| King-Lu | Open-source random forest | Open-source tariff method s | InterVA-4 | ||
|
China |
1100 / 400 |
48 |
48 |
48 |
N/A |
|
Institute for Health Metrics and Evaluation |
1100 / 400 |
96 |
96 |
96 |
N/A |
|
Million Death Study |
1100 / 400 |
89 |
89 |
89 |
N/A |
| |
1100 / 1100 |
89 |
89 |
89 |
N/A |
| |
6100 / 6100a |
89 |
89 |
89 |
245 |
|
Agincourt |
1100 / 400 |
104 |
104 |
104 |
245b |
| |
1100 / 1100 |
104 |
104 |
104 |
245 |
| |
2900 / 2900 |
104 |
104 |
104 |
245 |
|
Matlab |
1100 / 400 |
224 |
224 |
224 |
245 |
| |
1100 / 1100 |
224 |
224 |
224 |
245 |
| 1600 / 1600 | 224 | 224 | 224 | 245 | |
Only the numbers of test cases are applicable for the InterVA-4 analyses, as this method does not require any training cases. Additionally, InterVA-4 requires the input of 245 diagnostic indicators, however as many of these were not available in the given datasets, the number of useable variables was lower than 245. aThe MDS dataset used for InterVA-4 contained 552 cases, in which we extracted additional InterVA-4 indicators from the narratives. bEach CCVA method ran 30 resamples for each training/testing split within each dataset, except InterVA-4, which used the following number of re-samples: 1 for MDS data; 8, 7, 6 for Agincourt data splits of 400, 1100, and 2900 test cases; and 10, 10, 10 for Matlab data splits of 400, 1100, and 1600 test cases, respectively.
The required format of input data varied by assignment method. Two slightly modified versions of each original dataset were created; one version to be used for the data-driven methods (ORF, OTM, KL), and another for InterVA-4, which only uses specific indicators [16]. InterVA-4 testing on the MDS dataset used 552 child deaths, which had additional details extracted from the MDS narratives to match the required InterVA-4 inputs. Resampling was not done on these 552 records due to the small sample size.
Performance metrics
Positive predictive value (PPV) and partial chance-corrected concordance (PCCC) were used to measure agreement for individual COD assignment. PPV is the proportion of assigned CODs that were the ‘true’ COD (hereafter called reference COD), and is a common metric in hospital-based VA comparison studies [17]. PCCC is a variation on the PPV, meant to account for correct COD assignments made purely by chance [15]. As the CCVA methods could assign each VA record several CODs with varying probabilities, we calculated PPV and PCCC for agreement between the reference COD and the CCVA’s most probable COD, and for the three most probable CODs. These two measures were simply interpreted as whether the reference COD matched the CCVA method’s most probable cause, or matched one of the three most probable causes.
At the population level, accuracy was measured using CSMF absolute accuracy and error. CSMF error is the absolute difference between the estimated and reference CSMFs, summed across all causes. CSMF accuracy, as proposed by Murray et al., is determined by dividing the CSMF error by 2(1-minimum(CSMFtrue)), and subtracting this term from one [15]. This is meant to account for the variation in number of CODs across comparisons, and the resulting values are relative to the worst possible performance in a dataset (that is, coding every death incorrectly) [15]. CSMF accuracy and CSMF error are interpreted in opposite directions: good performance yields high CSMF accuracy and low CSMF error. We retained PCCC and CSMF accuracy for comparability to previous IHME publications. Additional file 2 summarizes the equations for the four metrics.
Results
Individual-level agreement on cause of death
In comparison to physician-assigned causes, the agreement (as measured by PPV) for all CCVA methods for the most probable COD averaged 43% to 44% for all datasets, with the highest PPV being 58% for ORF, followed by 52% for OTM, both on the MDS data (Table 3). The average PPV improved for the top three most probable CODs, with greater improvements for ORF (69%) and OTM (68%) than for InterVA-4 (62%). Similar results were seen using PCCC (Table 4): the average PCCC for the most probable COD across the datasets, using ORF, OTM and InterVA-4, was 41%, 40% and 41%, respectively. The average PCCC improved for the top three most probable CODs to 67%, 62% and 58%, respectively.
Table 3.
Positive predictive values of computer-coded verbal autopsy methods versus physician-coded verbal autopsy reference standards
| |
Test cases |
Open-source random forest |
Open-source tariff method |
InterVA-4 |
Average for top cause (%) |
|||
|---|---|---|---|---|---|---|---|---|
| Dataset | Top (%) | Top 3 (%) | Top (%) | Top 3 (%) | Top (%) | Top 3 (%) | ||
|
China |
400 |
35 |
57 |
36 |
70 |
N/A |
N/A |
36 |
|
Institute for Health Metrics and Evaluation |
400 |
33 |
55 |
34 |
53 |
N/A |
N/A |
34 |
|
Million Death Study |
6100 |
58 |
82 |
52 |
76 |
42a |
63a |
51 |
|
Agincourt |
2900 |
45 |
77 |
42 |
69 |
42 |
58 |
43 |
|
Matlab |
1600 |
49 |
74 |
52 |
74 |
48 |
64 |
50 |
| Average | 44 | 69 | 43 | 68 | 44 | 62 | ||
Top cause represents accuracy of the CCVA method’s most probable cause matching the cause assigned by PCVA; Top 3 represents whether CCVA’s three most probable causes contain the cause assigned by PCVA. Averages calculated across CCVA methods only use results for the top cause. aThe Million Death Study dataset used for InterVA-4 contained a sample of 552 cases, in which we extracted additional InterVA-4 indicators from the narratives.
Table 4.
Partial chance-corrected concordance of computer-coded verbal autopsy methods versus physician-coded verbal autopsy reference standards
|
Dataset |
Test cases |
Open-source random forest |
Open-source tariff method |
InterVA-4 |
|
Average for top cause (%) |
||
|---|---|---|---|---|---|---|---|---|
| Top (%) | Top 3 (%) | Top (%) | Top 3 (%) | Top (%) | Top 3 (%) | |||
|
China |
400 |
33 |
55 |
32 |
64 |
N/A |
N/A |
33 |
|
Institute for Health Metrics and Evaluation |
400 |
31 |
54 |
32 |
48 |
N/A |
N/A |
32 |
|
Million Death Study |
6100 |
55 |
81 |
48 |
70 |
38a |
60a |
47 |
|
Agincourt |
2900 |
42 |
75 |
38 |
62 |
39 |
56 |
40 |
|
Matlab |
1600 |
45 |
72 |
48 |
68 |
45 |
59 |
46 |
| Average | 41 | 67 | 40 | 62 | 41 | 58 | ||
Top cause represents accuracy of the CCVA method’s most probable cause matching the cause assigned by PCVA; Top 3 represents whether CCVA’s three most probable causes contain the cause assigned by PCVA. Averages calculated across CCVA methods only use results for the top cause. aThe Million Death Study dataset used for InterVA-4 contained a sample of 552 cases, in which we attempted to extract additional InterVA-4 indicators from the narratives.
The values of PPV and PCCC rose with larger training and testing datasets, suggesting that their results were partly dependent on having a sufficient number of training cases for each COD. The confidence intervals for these metrics were narrow as they mostly represented random resampling, and did not express the true underlying uncertainty in the data arising from misclassification of causes. Additional file 3 provides detailed results for each of the four metrics, including the confidence intervals.
Population-level agreement on cause-specific mortality fraction
KL had the best average CSMF accuracy across all five datasets (91%), followed by InterVA-4 (72% across three datasets), ORF (71%) and OTM (54%). Except for KL, the remaining CCVA methods traded best performance by dataset, with no clear trend (Table 5). CSMF error yielded similar results, with KL having the lowest error scores (Additional file 3).
Table 5.
Cause-specific mortality fraction accuracy of computer-coded verbal autopsy methods versus physician-coded verbal autopsy reference standards
| Datasets | Test cases | King-Lu (%) | Open-source random forest (%) | Open-source tariff method (%) | InterVA-4 (%) | Average (%) |
|---|---|---|---|---|---|---|
|
China |
400 |
84 |
79 |
75 |
N/A |
79 |
|
Institute for Health Metrics and Evaluation |
400 |
88 |
73 |
63 |
N/A |
75 |
|
Million Death Study |
6100 |
96 |
64 |
33 |
70a |
66 |
|
Agincourt |
2900 |
94 |
72 |
38 |
75 |
70 |
|
Matlab |
1600 |
95 |
69 |
59 |
72 |
74 |
| Average | 91 | 71 | 54 | 72 | ||
aThe Million Death Study dataset used for InterVA-4 contained a sample of 552 cases, in which we attempted to extract additional InterVA-4 indicators from the narratives.
Using the MDS data, KL had the closest similarity to the ranking of population-level CODs as compared to PCVA, with the top three causes in children under 5 years being the same (acute respiratory infection, diarrheal diseases, and other and unspecified infections; Additional file 3). ORF ended up with the same top three, but ranked other and unspecified infections ahead of acute respiratory infections, and ahead of diarrheal diseases. In the Agincourt data, KL performed better than ORF, matching the top three causes but not in the same ranking as PCVA. By contrast, ORF, somewhat inexplicably, ranked maternal deaths as the second most common COD. In the Matlab data, both KL and ORF showed similar performance in ranking CODs, accurately matching the top three causes. Results for InterVA-4 varied across the comparisons.
Discussion
This is the largest comparison study yet done of CCVA and PCVA methods. We found that, at an individual level, ORF, OTM and InterVA-4 replicated the coding of physicians comparably, but that the average agreement level for the leading COD was about 50%. Agreement with the physician-assigned code rose substantially for all CCVA methods if the three most probable CODs were considered, and generally improved with larger subsets within a given study. On a population level, the KL method performed best in terms of CSMF accuracy and error, and replicated the CSMF distribution of PCVA in the original datasets fairly well. ORF did not outperform KL, even on the IHME dataset, and did not perform better than InterVA-4, despite claims to the contrary [18]. At the individual level, InterVA-4, which does require training on a dataset, produced broadly comparable results to the methods that do require training.
Comparison to physician coding as a reference standard poses several methodological challenges. Importantly, our study focused on CCVA replication of physician codes (and errors), and not whether the underlying COD assigned by the physician was correct. Validation of PCVA is limited by the lack of a true reference standard in countries where verbal autopsy is performed [1-3,9]. Nonetheless, PCVA with good quality control can yield useful results on COD distributions in countries where medical certification remains uncommon [1]. The studies we included in the comparisons had physician coding done with reasonably good levels of quality control [4-7,9] as shown by a low proportion of ill-defined deaths. Physician coding that contains large amounts of random errors would reduce agreement on a COD in the dual-physician coding system, and would tend to increase the number of unspecified CODs in the International Classification of Diseases (ICD-10) [19], such as ‘senility’ (ICD-10 code R54) or ‘cause not determined’ (ICD-10 code R99). This would in turn make it harder for CCVA methods to identify specific causes. Moreover, the size of the errors or biases in CCVA methods depends on the inherent errors and biases of PCVA results. This is particularly relevant for machine learning, as its accuracy requires learning on “true” class labels. High misclassification rates in the training set will also affect performance in the testing set across datasets, as noted recently on the full IHME dataset [20]
The performance of each CCVA method at individual assignment improved when trained and tested on a larger number of cases, most likely due to a greater number of cases from which to learn the distinct relationships between specific symptom profiles and CODs. The differences in the field and coding methods across studies would tend to reduce the observed differences in PCVA and CCVA between the various comparisons (and more likely so at the individual level than at the population level). This might have contributed to the observed comparability of the results for the four CCVA methods. With larger studies and more standardized field and physician coding methods, any real, underlying differences between various CCVA methods may become apparent. Finally, we note that InterVA-4 has a threshold of probability for designating the most probable cause (that is, the most probable cause must also have a probability above 50%, otherwise the death is classified as indeterminate), whereas ORF and OTM select the most probable causes without applying any thresholds.
On an individual level, no single method was able to replicate physician assignment of COD more than about half the time. At the population level, the King-Lu method was the best method to estimate CSMFs, though it does not assign individual CODs. However, good population-level agreement accuracy does not guarantee good individual agreement [21,22]. A key methodological feature is the need to avoid the false gold standard of hospital-based deaths [1,3]. Reliance on hospital or urban-based deaths for training of automated methods may lead to learning of symptom patterns and other features that are not representative of populations without medical attention. Indeed, the CSMFs between home and hospital deaths are dissimilar, as demonstrated in India [23].
Our study is the largest cross-country comparison of current CCVA methods versus PCVA, covering about twice as many deaths as an earlier multi-country study [7], and including a mix of various ages, and community and hospital deaths. Nonetheless, we faced certain limitations. First, we could not compare the original IHME random forest and tariff algorithms, though the original methods were re-created to the best of our abilities from the published descriptions [12,13], yielding broadly similar results (Figures 1 and 2). Second, access during the analyses phase to the full IHME hospital-based dataset of 12,000 records would have allowed more robust comparisons. Similarly, the China dataset was also somewhat limited by the small sample size.
Conclusions
Different CCVA methods have various strengths and weaknesses depending on the study scenario and study objective. An ideal solution could involve a combination of automated methods to obtain robust individual- and population-level estimates. In the medium term, it appears unwise and certainly premature to recommend that automated systems replace physicians in coding VAs. CCVA methods could be used in parallel with physician coding to increase speed, efficiency and quality of coding. Future work may focus on the performance of a combination of various automated methods, and must extend to larger datasets and explore specifics for important age groups (children, maternal, adult), by gender, and across various settings of home- and hospital-based deaths. Future studies need to also place specific emphasis on testing computer based methods on a random sample of deaths in countries, as this would be much more useful in determining the true underlying CSMF at the population level [24].
Abbreviations
CCVA: computer-coded verbal autopsy; COD: cause of death; CSMF: cause-specific mortality fraction; HCE: health care experience; ICD-10: International Classification of Diseases-10; IHME: Institute for Health Metrics and Evaluation; KL: King-Lu verbal autopsy method; MDS: Million Death Study; ORF: open-source random forest; OTM: open-source tariff method; PCCC: partial chance-corrected concordance; PCVA: physician-certified verbal autopsy; PPV: positive predictive value; VA: verbal autopsy.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ND, LA and PM conducted the analysis. All authors contributed to data interpretation and critical revisions of the paper. All authors read and approved the final manuscript.
Supplementary Material
Details of the open-source random forest and tariff methods. Explanation of the major logical steps of the open-source random forest and tariff methods.
Description of comparison metrics. Formulas and explanation of positive predictive value, partial chance-corrected concordance, CSMF error and CSMF accuracy.
Full results of CCVA comparisons on several dataset splits. Results presented by CCVA method, dataset, dataset splits, and top and top-three cause of death predictions.
Contributor Information
Nikita Desai, Email: nikita.desai@mail.utoronto.ca.
Lukasz Aleksandrowicz, Email: aleksandrowl@smh.ca.
Pierre Miasnikof, Email: MiasnikofP@smh.ca.
Ying Lu, Email: ying.lu@nyu.edu.
Jordana Leitao, Email: leitaojordana@googlemail.com.
Peter Byass, Email: peter.byass@epiph.umu.se.
Stephen Tollman, Email: Stephen.Tollman@wits.ac.za.
Paul Mee, Email: paul.mee@wits.ac.za.
Dewan Alam, Email: dsalam@icddrb.org.
Suresh Kumar Rathi, Email: RathiS@smh.ca.
Abhishek Singh, Email: abhishek@iips.net.
Rajesh Kumar, Email: dr.rajeshkumar@gmail.com.
Faujdar Ram, Email: fram@iips.net.
Prabhat Jha, Email: Prabhat.jha@utoronto.ca.
Acknowledgements
We thank Khaled Hasan for help with data formatting, and Michael Palmer for editorial assistance and Abraham Flaxman for guidance on IHME datasets.
References
- Jha P. Counting the dead is one of the world’s best investments to reduce premature mortality. Hypothesis. 2012;10:e3. [Google Scholar]
- Setel PW, Sankoh O, Rao C, Velkoff VA, Mathers C, Gonghuan Y, Hemed Y, Jha P, Lopez AD. Sample registration of vital events with verbal autopsy: a renewed commitment to measuring and monitoring vital statistics. Bull World Health Organ. 2005;83:611–617. [PMC free article] [PubMed] [Google Scholar]
- Fottrell E, Byass P. Verbal autopsy: methods in transition. Epidemiol Rev. 2010;32:38–55. doi: 10.1093/epirev/mxq003. [DOI] [PubMed] [Google Scholar]
- Health and Demographic Surveillance Unit. Health and Demographic Surveillance System: Matlab. Volume 36: Registration of Health and Demographic Events 2003. Dhaka: ICDDR, B; 2005. [Google Scholar]
- King G, Lu Y. Verbal autopsy methods with multiple causes of death. Statistical Science. 2008;23:78–91. doi: 10.1214/07-STS247. [DOI] [Google Scholar]
- Kahn K, Collinson MA, Gómez-Olivé FX, Mokoena O, Twine R, Mee P, Afolabi SA, Clark BD, Kabudula CW, Khosa A, Khoza S, Shabangu MG, Silaule B, Tibane JB, Wagner RG, Garenne ML, Clark SJ, Tollman SM. Profile: Agincourt health and socio-demographic surveillance system. Int J Epidemiol. 2012;41:988–1001. doi: 10.1093/ije/dys115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CJ, Lopez AD, Black R, Ahuja R, Ali SM, Baqui A, Dandona L, Dantzer E, Das V, Dhingra U, Dutta A, Fawzi W, Flaxman AD, Gómez S, Hernández B, Joshi R, Kalter H, Kumar A, Kumar V, Lozano R, Lucero M, Mehta S, Neal B, Ohno SL, Prasad R, Praveen D, Premji Z, Ramírez-Villalobos D, Remolador H, Riley I. et al. Population Health Metrics Research Consortium gold standard verbal autopsy validation study: design, implementation, and development of analysis datasets. Popul Health Metr. 2011;9:27. doi: 10.1186/1478-7954-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ST, Flaxman AD. AAAI Spring Symposium on Artificial Intelligence for Development (AI-D): March 22-24 2010; Stanford. Stanford: Stanford University; 2010. Machine learning methods for verbal autopsy in developing countries. [ https://github.com/aflaxman/verbal-autopsy-challenge] [Google Scholar]
- Jha P, Gajalakshmi V, Gupta PC, Kumar R, Mony P, Dhingra N, Peto R. Prospective study of one million deaths in India: rationale, design, and validation results. PLoS Med. 2006;3:e18. doi: 10.1371/journal.pmed.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassani DG, Kumar R, Awasthi S, Morris SK, Paul VK, Shet A, Ram U, Gaffey MF, Black RE, Jha P. Million Death Study Collaborators. Causes of neonatal and child mortality in India: a nationally representative mortality survey. Lancet. 2010;376:1853–1860. doi: 10.1016/S0140-6736(10)61461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byass P, Chandramohan D, Clark SJ, D'Ambruoso L, Fottrell E, Graham WJ, Herbst AJ, Hodgson A, Hounton S, Kahn K, Krishnan A, Leitao J, Odhiambo F, Sankoh OA, Tollman SM. Strengthening standardised interpretation of verbal autopsy data: the new InterVA-4 tool. Glob Health Action. 2012;5:1–8. doi: 10.3402/gha.v5i0.19281. Available for download at http://www.interva.net. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaxman A, Vahdatpour A, Green S, James S, Murray C. for the Population Health Metrics Research Consortium (PHMRC) Random forests for verbal autopsy analysis: multisite validation study using clinical diagnostic gold standards. Popul Health Metr. 2011;9:29. doi: 10.1186/1478-7954-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S, Flaxman A, Murray C. and the Public Health Metrics Research Consortium. Performance of the tariff method: Validation of a simple additive algorithm for analysis of verbal autopsies. Popul Health Metr. 2011;9:31. doi: 10.1186/1478-7954-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G, Lu Y, Shibuya K. Designing verbal autopsy studies. Popul Health Metr. 2010;8:19. doi: 10.1186/1478-7954-8-19. Available for download at http://gking.harvard.edu/va. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C, Lozano R, Flaxman AD, Vadhatpour A, Lopez AD. Robust metrics for assessing the performance of different verbal autopsy cause assignment methods in validation studies. Popul Health Metr. 2011;9:28. doi: 10.1186/1478-7954-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitao JC, Chandramohan D, Byass P, Jakob R, Bundhamcharoen K, Choprapowan C, de Savigny D, Fottrell E, França E, Frøen F, Gewaifel G, Hodgson A, Hounton S, Kahn K, Krishnan A, Kumar V, Masanja H, Nichols E, Notzon F, Rasooly MH, Sankoh O, Spiegel P, AbouZahr C, Amexo M, Kebede D, Alley WS, Marinho F, Ali M, Loyola E, Chikersal J. et al. Revising the WHO verbal autopsy instrument to facilitate routine cause-of-death monitoring. Glob Health Action. 2013;6:21518. doi: 10.3402/gha.v6i0.21518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitao J, Desai N, Aleksandrowicz L, Byass P, Miasnikof P, Tollman S, Alam D, Lu Y, Rathi SK, Singh A, Suraweera W, Ram F, Jha P. Comparison of physician-certified verbal autopsy with computer-coded verbal autopsy for cause of death assignment in hospitalized patients in low- and middle-income countries: systematic review. BMC Med. 2014;12:22. doi: 10.1186/1741-7015-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaxman A, Vahdatpour A, James S, Birnbaum J, Murray C. for the Population Health Metrics Research Consortium (PHMRC) Direct estimation of cause-specific mortality fractions from verbal autopsies: multisite validation study using clinical diagnostic gold standards. Popul Health Metr. 2011;9:35. doi: 10.1186/1478-7954-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) International Statistical Classifications of Diseases and Related Health Problems, Volume 1. 10. Geneva, Switzerland: World Health Organization; 2008. [Google Scholar]
- Byass P. Usefulness of the Population Health Metrics Research Consortium Gold Standard Verbal Autopsy Data for General Verbal Autopsy Methods. BMC Med. 2014;12:23. doi: 10.1186/1741-7015-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anker M. The effect of misclassification error on reported cause-specific mortality fractions from verbal autopsy. Int J Epidemiol. 1997;26:1090–1096. doi: 10.1093/ije/26.5.1090. [DOI] [PubMed] [Google Scholar]
- Maude GH, Ross DA. The effect of different sensitivity, specificity and cause-specific mortality fractions on the estimation of differences in cause-specific mortality rates in children from studies using verbal autopsies. Int J Epidemiol. 1997;26:1097–1106. doi: 10.1093/ije/26.5.1097. [DOI] [PubMed] [Google Scholar]
- Aleksandrowicz L, Malhotra V, Dikshit R, Gupta PC, Kumar R, Sheth J, Rathi SK, Suraweera W, Miasnikofi P, Jotkar R, Sinha D, Awasthi S, Bhatia P, Jha P. Performance criteria for verbal autopsy‒based systems to estimate national causes of death: development and application to the Indian Million Death Study. BMC Med. p. 21. [DOI] [PMC free article] [PubMed]
- Jha P. Reliable direct measurement of causes of death in low- and middle-income countries. BMC Med. 2014;12:19. doi: 10.1186/1741-7015-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of the open-source random forest and tariff methods. Explanation of the major logical steps of the open-source random forest and tariff methods.
Description of comparison metrics. Formulas and explanation of positive predictive value, partial chance-corrected concordance, CSMF error and CSMF accuracy.
Full results of CCVA comparisons on several dataset splits. Results presented by CCVA method, dataset, dataset splits, and top and top-three cause of death predictions.