Cell type–specific expression of the splicing regulator SLM1 provides a mechanism for shaping the molecular repertoires of synaptic adhesion molecules in neuronal populations in vivo.
Abstract
The unique functional properties and molecular identity of neuronal cell populations rely on cell type–specific gene expression programs. Alternative splicing represents a powerful mechanism for expanding the capacity of genomes to generate molecular diversity. Neuronal cells exhibit particularly extensive alternative splicing regulation. We report a highly selective expression of the KH domain–containing splicing regulators SLM1 and SLM2 in the mouse brain. Conditional ablation of SLM1 resulted in a severe defect in the neuronal isoform content of the polymorphic synaptic receptors neurexin-1, -2, and -3. Thus, cell type–specific expression of SLM1 provides a mechanism for shaping the molecular repertoires of synaptic adhesion molecules in neuronal populations in vivo.
Introduction
Understanding the molecular mechanisms that direct the differentiation and connectivity of neuronal cells in the brain remains one of the major challenges in cell biology (Shen and Scheiffele, 2010; Zipursky and Sanes, 2010). Neuronal cell types are characterized by unique morphological and functional properties that shape signal processing in neuronal networks (Masland, 2004; Okaty et al., 2011). The remarkable diversity of neuronal properties is achieved by cell type–specific gene expression programs. Alternative splicing greatly amplifies the coding capacity of the genome and, thereby, provides a powerful mechanism controlling molecular and functional diversity. For example, alternative splicing programs control abundance, identity, transport, and turnover of certain neuronal mRNAs (Darnell, 2013; Zheng and Black, 2013). Ultimately, these RNA regulatory mechanisms contribute to the control of selective cell surface interactions, ion channel properties, and neuronal signaling (Siddiqui et al., 2010; Beck et al., 2012; Gehman et al., 2012; Lipscombe et al., 2013). It is an attractive hypothesis that cell type–specific alternative splicing factors are used to shape the molecular repertoires of functionally and morphologically defined sub-classes of neuronal cells. However, splicing factors that are linked to a genetically defined subsets of neurons and that are essential to sculpt cell type–specific neuronal gene expression are only beginning to emerge.
The KH domain–containing RNA-binding protein SAM68 (Src-associated in mitosis of 68 kD protein, Khdrbs1) is a critical regulator of RNA transport and neuronal activity–regulated alternative splicing (Iijima et al., 2011; Klein et al., 2013). SAM68 is broadly expressed in neuronal and nonneuronal cells and regulates alternative splicing of Nrxn1, which encodes a synaptic cell surface receptor (Missler and Südhof, 1998; Dean et al., 2003; Craig and Kang, 2007; Südhof, 2008; Iijima et al., 2011). Cerebellar Sam68KO neurons fail to increase exon skipping at the Nrxn1 alternatively spliced segment 4 (AS4) upon neuronal depolarization. In wild-type neurons, this SAM68-dependent exon skipping results in production of NRX protein variants with altered ligand interactions (Boucard et al., 2005; Chih et al., 2006; Graf et al., 2006; Uemura et al., 2010; Iijima et al., 2011; Matsuda and Yuzaki, 2011; Aoto et al., 2013). Consistent with an important function for SAM68 in vivo, there is a corresponding reduction in the skipped AS4(−) transcript in Sam68KO brains. Global ablation of the closely related RNA-binding protein SLM2 (SAM68-like mammalian protein 2; alternate names: T-STAR, Khdrbs3; Di Fruscio et al., 1999; Venables et al., 1999) also results in a reduction in exon skipping at Nrxn AS4, which correlates with the regional expression levels of SLM2 in the brain (Ehrmann et al., 2013). These studies established SAM68 and SLM2 in alternative splicing regulation in the mouse brain. However, it is not known whether the activity of these proteins is essential to generate cell type–specific gene expression programs in defined neuronal cell populations.
In this work, we uncover that SLM2 and the closely related SLM1 are expressed in highly selective and largely nonoverlapping populations of neurons in the central nervous system of mice. In the hippocampus, SLM1 is abundant in glutamatergic dentate granule cells but also in a specific set of cholecystokinin–calbindin double-positive (CCK+ calbindin+) inhibitory interneurons. By contrast, SLM2 is confined to glutamatergic pyramidal cells and GABAergic parvalbumin+, calretinin+, and somatostatin+ interneurons. We demonstrate that SLM1 differs from SAM68 in its ability to regulate alternative splicing of different Nrxn mRNAs at AS4 in vitro. Slm1KO mice and Sam68:Slm1DKO exhibit a severe reduction in Nrxn1 AS4(−) transcripts as well as defects in cerebellar morphogenesis. Finally, we demonstrate that cell type–specific conditional ablation of SLM1 disrupts cell type–specific generation of Nrxn splice variants. Thus, SLM1 is a critical RNA-binding protein that shapes cell type–specific alternative splicing programs in vivo.
Results
SLM1 and SLM2 are expressed in largely segregated neuronal populations
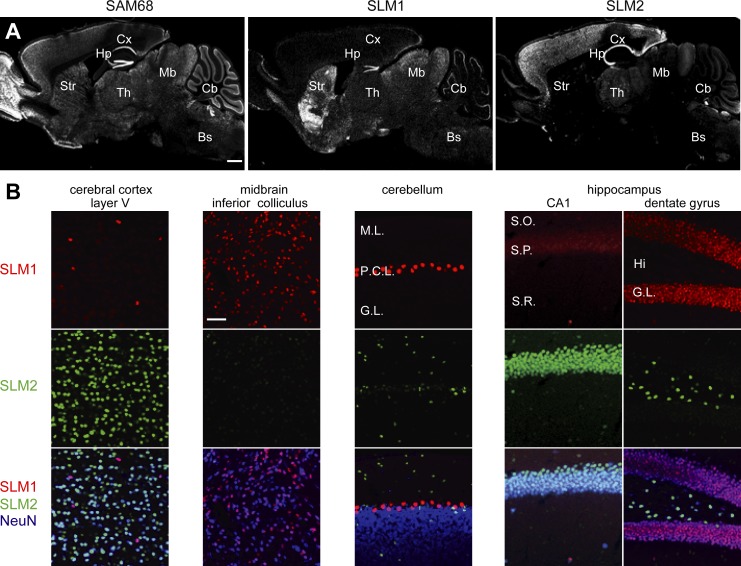
Western blotting analysis of different mouse brain areas with SAM68, SLM1, and SLM2 antibodies indicates that these proteins are detectable across all brain regions examined (Fig. S1 A). To explore whether SLM proteins are confined to specific anatomically and molecularly defined neuronal populations, we performed a detailed analysis using SLM1- and SLM2-specific antibodies. SLM1 and SLM2 were detected in largely nonoverlapping cell populations, whereas SAM68 is more widely expressed (Fig. 1 A; Fig. S1, B and C). In the cortex, SLM1 marks a sparse population of cells in layers 2–3 and layer 5, whereas SLM2 is expressed in the majority of NeuN-positive cortical cells (Fig. 1 B; unpublished data). By contrast, SLM2 is largely absent from midbrain neurons of the superior and inferior colliculus where the majority of cells express SLM1 (Fig. 1 B). In the cerebellum, SLM1 is highly concentrated in Purkinje cells, whereas SLM2 marks interneurons in the inner granular and molecular layer. Thus, SLM1 and SLM2 are restricted to sub-populations of neurons in the mouse brain.
Figure 1.
Differential distribution of SLM1 and SLM2 proteins in the mouse brain. (A) Gross expression patterns of STAR family proteins in adult brain. Cx, cortex; Hp, hippocampus; Str, striatum; Th, thalamus; Mb, midbrain; Cb, cerebellum; Bs, brain stem. Bar, 1 mm. (B) High magnification images of SLM expression in various brain areas. I–III, cortical layer I–III; M.L., molecular layer; P.C.L., Purkinje cell layer; G.L., granular layer. S.O., stratum oriens; S.P., stratum pyramidale; S.R., stratum radiatum. Hi, hilus. Bar, 50 µm.
A particularly interesting segregation of SLM1 and -2 is seen in the mature hippocampus. In principal cells, SLM2 is detectable exclusively in CA neurons, whereas SLM1 is highly expressed in dentate granule cells (Fig. 1 B; Fig. S2 A; Stoss et al., 2004). Notably, both proteins are also highly expressed in nonoverlapping populations of inhibitory interneurons (Fig. S2 B). SLM2 is concentrated in interneurons in the hilus (Fig. 1 B, dentate gyrus; Fig. S2 A). Within area CA1, the majority of SLM1-positive (SLM1+) cells are located in stratum radiatum (S.R.) and its border to the stratum lacunosum moleculare (S.L.M.; Fig. 2, A–C). We applied a combination of interneuron markers to directly define this population. 43% of all SLM1+ cells were marked with the interneuron marker calbindin and 54% were immuno-positive for CCK (Fig. 2 D). Conversely, ∼70% of all calbindin+ and 70% of all CCK+ interneurons in S.R. and S.L.M. expressed SLM1 (Fig. 2, E and F). SLM1 was largely absent from calretinin+, vasoactive intestinal peptide (VIP)+, parvalbumin+, and somatostatin+ interneurons. Calbindin and CCK immunoreactivity overlap in two specific interneuron populations in S.R./S.L.M. designated Schaffer collateral–associated (SCA) interneurons and perforant path–associated (PPA) interneurons (Lawrence, 2008; Klausberger, 2009). Interestingly, 93% of these CCK/calbindin double-positive interneurons showed SLM1 immunoreactivity. These results identify SCA interneurons and PPA interneurons as a major site of SLM1 expression (Fig. 2 F; Fig. S2 C). Calbindin and CCK-positive interneurons largely lack detectable SLM2 immunoreactivity, which instead was observed in the vast majority (>90%) of calretinin and VIP-positive interneurons in S.R. and S.L.M. Moreover, in stratum oriens (S.O.) SLM2 was highly expressed in parvalbumin+ and somatostatin+ interneurons (Fig. 2, E and F). These experiments uncover a striking nonoverlapping distribution of SLM1 and SLM2 in vivo. The quantitative analysis with cell type–specific markers demonstrates that the respective SLM1- and SLM2-positive cell populations are not due to stochastic expression or detection of the proteins in subsets of cells. By contrast, expression is tightly linked to molecularly defined cell identity. Thus, SLM1 and -2 are well suited to regulate neuronal alternative splicing programs in a cell type–specific manner.
Figure 2.
Hippocampal interneuron subclass-specific expression of SLM1 and SLM2. (A) Schematic illustration of expression of SLM proteins in principal cells of the hippocampal area. (B) SLM1-positive interneurons in hippocampal area CA1. Bar, 100 µm. S.R., stratum radiatum; S.L.M., stratum lacunosum moleculare; S.P., stratum pyramidale; S.O., stratum oriens. (C) Distribution of SLM1-positive interneurons in CA1. The percentage of SLM1+ cells that are located in each stratum was quantified. (D) Percentage of SLM1-positive interneurons that are immuno-positive for various inhibitory interneuron markers within stratum radiatum and stratum lacunosum moleculare of CA1 (right; n = 3 animals). (E) Mutually exclusive expression of SLM1 and SLM2 in hippocampal interneurons immuno-positive for calbindin (CB), calretinin (CR), parvalbumin (PV), and somatostatin (SOM). Coronal sections (from 2–3-mo-old mice) were co-immunostained with anti-SLM antibodies and inhibitory interneuron markers. Bar, 20 µm. (F) Fraction of SLM1- and SLM2-positive cells within immunohistochemically defined inhibitory interneuron populations in area CA1 (n = 3 animals).
SLM1 regulates Nrxn2 alternative splice reporters in heterologous cells
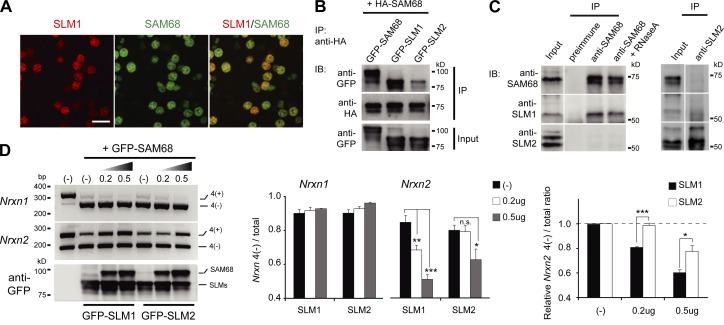
In previous work we demonstrated that SAM68, SLM1, and SLM2 can regulate Nrxn1 splice reporters (“mini-genes”) when transfected into human embryonic kidney cells (HEK293; Iijima et al., 2011). Given the highly selective expression of SLM1, we asked whether its ability to regulate mRNA targets might differ from SAM68. When SAM68, SLM1, and SLM2 activity toward Nrxn1, Nrxn2, and Nrxn3 splice reporters was analyzed in HEK293 cells we observed that all three proteins could efficiently drive exon skipping at Nrxn1 and Nrxn3 AS4. Importantly, SAM68 did not exhibit significant activity toward a Nrxn2 AS4 reporter, whereas SLM1 and SLM2 did induce exon skipping in Nrxn2 (Fig. 3, A and B). Under the same conditions, hnRNPH and A1 did not show activity toward any of the Nrxn AS4 reporters (Fig. 3 B). The specific activity of SLM proteins toward Nrxn2 reporters was further confirmed in assays with increasing amounts of the alternative splicing factors (Fig. 3, C and D).
Figure 3.
Function of SLM proteins in Nrxn1, Nrxn2, and Nrxn3 alternative splicing regulation in vitro. (A) Splice reporters for Nrxn1, Nrxn2, and Nrxn3 AS4. Nrxn splice reporter constructs contain AS4 with constitutive exons (dark gray), alternative exon 20 (light gray), and introns shown as lines. Intron sizes are indicated (drawings not to scale). (B) Splice reporter assays with various RNA-binding proteins. Reporter expression vectors were cotransfected into HEK293T cells with epitope-tagged expression constructs for GFP-SAM68, GFP-SLM1, GFP-SLM2, YFP-hnRNPA1, or YFP-hnRNPH1. Alternative splice isoform choice was measured by semi-quantitative RT-PCR with primers flanking the alternatively spliced segment. Fragment sizes for AS4(+) and AS4(−) isoforms are 318 bp and 228 bp (Nrxn1), 270 bp and 180 bp (Nrxn2), and 354 bp and 264 bp (Nrxn3). Expression of transfected RNA-binding proteins was confirmed by Western blotting with anti-GFP antibodies. (C) Dose-dependent activity of STAR family proteins toward Nrxn, Nrxn2, and Nrxn3 AS4. Splice reporter processing was assessed in experiments with increasing amounts of DNA encoding RNA-binding proteins transfected (DNA amounts used indicated in micrograms). (D) Alternative splicing was assayed by semi-quantitative RT-PCR. Expression levels of GFP-tagged proteins were detected by immunoblot. (E) Schematic drawing of domain organization of SAM68 and SLM1 and chimeric mutants proteins. An HA-tag was attached to the C-terminal end of each open reading frame. (F) Splice reporter assays with chimeric proteins using Nrxn1 and Nrxn2 AS4 reporters. Alternative splicing was assayed by semi-quantitative RT-PCR. Expression of the HA-tagged proteins was confirmed by immunoblotting.
To obtain insight into the molecular underpinnings of the target specificity of SAM68 versus SLM1 we generated chimeric forms of these RNA-binding proteins (Fig. 3 E). SAM68 differs from SLM1 and SLM2 in an extended N-terminal region of 96 amino acids that is absent in the SLM proteins. Fusion of this N-terminal domain to SLM1 did not alter its ability to regulate Nrxn2, indicating that this domain is not sufficient to abolish alternative splicing regulation toward Nrxn2 (Fig. 3 F). Replacement of the SLM1 RNA-binding domain with corresponding sequences from SAM68 rendered this protein inactive toward Nrxn2. By contrast, replacement of the C-terminal domain with SAM68 sequences did not modify activity toward Nrxn2. Importantly, all chimeric proteins retained activity toward processing of the Nrxn1 reporter, thereby confirming appropriate folding and subcellular targeting of the chimeric proteins (Fig. 3 F). These experiments demonstrate that SLM1 differs from SAM68 in that it exhibits activity toward Nrxn2 and that this activity arises from specific sequences in the SLM1 RNA-binding domain.
Co-expression and complex formation of SLM1 and SAM68
Whereas SLM1 and SLM2 expression are largely segregated at the cellular level, SLM1 and SAM68 are coexpressed in individual cells (note that given the broad distribution of SAM68 also most SLM2-positive cells co-express SAM68; Fig. 4 A; Fig. 1 A; unpublished data). Previous in vitro studies demonstrated that SLM1 and SAM68 form protein complexes that are not detected for SLM2 (Di Fruscio et al., 1999; Rajan et al., 2009). We confirmed SAM68–SLM1 association in coimmunoprecipitation experiments from transfected HEK293 cells (Fig. 4 B). To further explore complex formation in brain tissue we performed coimmunoprecipitation experiments for the endogenous proteins and observed robust, RNA-independent association between SAM68 and SLM1 but not SLM2 (Fig. 4 C). Given that STAR proteins are thought to function as oligomers that bind to bi-partite RNA motifs (Galarneau and Richard, 2009; Meyer et al., 2010), we examined whether SAM68–SLM1 complex formation would alter activity toward Nrxn2. Co-expression of increasing amounts of SAM68 progressively inhibited the activity of SLM1 toward Nrxn2 AS4 without modifying the activity toward Nrxn1 (Fig. 4 D). In the same assay, SAM68 was much less potent in inhibiting SLM2 activity toward Nrxn2 (Fig. 4 D), consistent with a preferential complex formation between SAM68 and SLM1. Thus, substrate specificity of STAR family proteins can be modulated by hetero-oligomer formation.
Figure 4.
Different regulation of STAR proteins in alternative splicing regulation in vitro. (A) Co-expression of SAM68 and SLM1 in vivo. SAM68 and SLM1 proteins are detected coexpressed in a subset of midbrain neurons in the adult mouse brain. Bar, 20 µm. (B) Co-immunoprecipitation of HA-tagged SAM68 and GFP-tagged SAM68, SLM1, or SLM2 from HEK293 cells cotransfected with expression constructs. Immunoprecipitation was performed with the anti-HA antibody and proteins in input or immunoprecipitates were detected with anti-HA or anti-GFP antibodies. (C) Complex formation of SAM68 and SLM1 in brain extracts. Complexes were precipitated with either anti-SAM68 (left) or anti-SLM2 (right) antibodies and precipitates probed with anti-SAM68, anti-SLM1, or anti-SLM2 antibodies. When indicated, lysates were treated with Ribonuclease A before antibody addition to eliminate interactions that depend on integrity of cellular RNAs. (D) Alternative splicing of Nrxn1 and Nrxn2 AS4 was assayed by semi-quantitative RT-PCR. To confirm protein expression levels, the lysates of HEK293 cells expressing GFP-SLM1 or GFP-SLM2 and increasing amounts of GFP-SAM68 were subjected to the immunoblotting. Quantitation of alternative splice isoform choice is displayed on the right.
Anatomical alterations in Slm1KO and Sam68:Slm1DKO mice
To explore functions of SLM1 in vivo we generated a conditional Slm1 allele in mice. Exon 2 of the Khdrbs2 gene (which encodes SLM1) was flanked by loxP sites (Fig. S3 A). Global Slm1KO mice were subsequently generated by Cre-mediated recombination in the germline. Homozygous Slm1KO mice completely lacked SLM1 protein expression but did not show detectable alterations in SAM68 or SLM2 expression levels (Fig. 5, A–C). Knockout animals were born at Mendelian frequencies, were viable and fertile, and did not exhibit any obvious behavioral alterations (Fig. 5 D). An anatomical survey of adult Slm1KO brains did not reveal any gross anatomical defects (Fig. 5 A). Given that SAM68 and SLM1 have overlapping substrates and are largely coexpressed in neuronal populations, we further generated Sam68:Slm1 double-knockout mice (Sam68:Slm1DKO). These animals were viable but not fertile (consistent with the previously reported function for SAM68 in reproduction). We did not detect major alterations in the distribution of synaptic markers in the hippocampus or cerebellum of the double-knockout mice (Fig. S3, B and C). However, Sam68:Slm1DKO mice had slightly smaller brains than wild-type animals and cerebella of Slm1KO and Sam68:Slm1DKO were significantly reduced in weight (Fig. 5 E). Interestingly, the cerebella of Sam68:Slm1DKO exhibited a defect in foliation with loss of the cerebellar fissure separating lobules VIb and VII (Fig. 5 F, phenotype observed in four out of four Sam68:Slm1DKO animals but none of their single-knockout littermates or wild-type controls). This defect was not observed in either single-knockout model, although the depth of the fissure was slightly reduced in Sam68KO mice (Fig. 5 G). These findings demonstrate a redundant function of Slm1 and Sam68 in cerebellar morphogenesis. Within the fused region of lobules VIb and VII the Purkinje cell layer was disorganized, with ectopic Purkinje cells scattered in the molecular layer (Fig. 5 H). SLM1 is highly expressed in Purkinje cells. Thus, the foliation phenotype most likely originates from a specific developmental deficit in this cell population.
Figure 5.
Generation of Slm1KO and Sam68:Slm1DKO mice. (A) Immunohistochemistry on para-sagittal sections reveals loss of SLM1 immunoreactivity in Slm1KO tissue. Bar, 1 mm. (B) High magnification images of immunohistochemistry with anti-SLM1 (red), anti-NeuN (green), and anti-calbindin (blue) antibodies shows loss of SLM1 immunoreactivity from cerebellar Purkinje cells. Bar, 50 µm. (C) Immunoblot analysis for SAM68, SLM1, and SLM2 in total brain lysates demonstrates loss of anti-SLM1 immunoreactivity in Slm1KO tissue. (D) Slm1KO mice are born at Mendelian frequencies (50 pups from 4 litters). (E) Weight of whole brains and cerebella from wild-type, Slm1KO, and Sam68:Slm1DKO. Brain tissues from adult animals were analyzed (n = 3–4 animals per genotype). (F) Cerebellar foliation defect of Sam68:Slm1DKO. Para-sagittal sections of cerebellar vermis from wild-type, Slm1KO, and Sam68:Slm1DKO were stained with anti-calbindin antibody to visualize cerebellar foliation. Bar, 1 mm. (G) High magnification images of fissure between lobule VIb and lobule VII in F. Bar, 100 µm. (H) Co-immunohistochemistry with anti-RORalpha (red) and anti-calbindin (green) antibodies in cerebellar lobule VI revealed abnormal alignment of cerebellar Purkinje cell bodies. Bar, 50 µm.
Cooperation of SAM68 and SLM1 in alternative splicing regulation in vivo
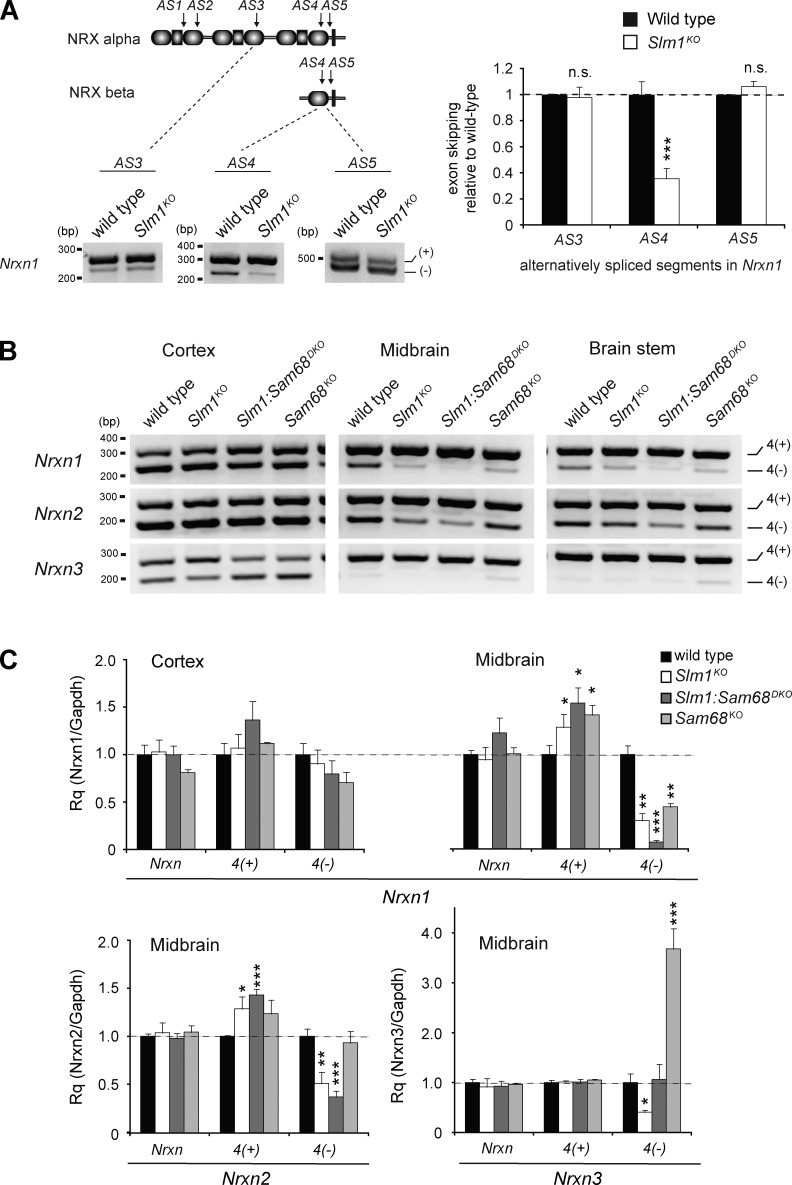
Considering the overlapping functions of SAM68 and SLM1 in the alternative splicing regulation of Nrxn1 and in cerebellar morphogenesis, we explored whether alternative splicing was modified in Slm1KO and Sam68:Slm1DKO mice. In Slm1KO brains, we detected a 70% reduction of the Nrxn1 AS4(−) isoforms in midbrain, an area with particularly broad SLM1 expression (Fig. 6, A and B). In the cortex, where only a small sub-population of neurons are SLM1-positive, no significant global change in Nrxn1 AS4(−) abundance was detected (Fig. 6, B and C). Notably, in the midbrain of Sam68:Slm1DKO mice exon skipping at AS4 was severely reduced, demonstrating that SAM68 and SLM1 have synergistic functions in Nrxn1 alternative splicing regulation in vivo. By contrast, Nrxn2 alternative splicing at AS4 was not altered in the midbrain of Sam68KO mice but selectively disrupted in Slm1KO mice (Fig. 6, B and C), consistent with the differential activity of SAM68 and SLM1 toward Nrxn2 observed in our cellular assays. Finally, we explored alternative splicing at Nrxn3 AS4 in the midbrain of single- and double-knockout mice. In Slm1KO brains skipping of the alternative exon at AS4 was significantly reduced, consistent with a critical function for SLM1 protein in suppressing incorporation of this alternative exon. By contrast, Sam68KO mice exhibit a substantial increase in Nrxn3 AS4 skipping, which was suppressed in the Sam68:Slm1DKO mice. This functional antagonism between SAM68 and SLM1 in vivo was surprising, as both proteins show similar activity toward a Nrxn3 AS4 reporter in heterologous cells (Fig. 3). Thus, our Sam68:Slm1DKO analysis reveals an unanticipated gene-specific interplay of SAM68 and SLM1 functions in the alternative splicing regulation of Nrxn3.
Figure 6.
Alternative splicing defects in SLM1KO mice. (A) Incorporation of alternative exons at AS3, -4, and -5 in midbrain cDNA samples was probed by semi-quantitative PCR with primers flanking the insertion site and analyzed by gel electrophoresis. (B) Representative images of semi-quantitative RT-PCR with Nrxn AS4 performed on cortex, midbrain, and brainstem from wild-type, Slm1KO, Sam68:Slm1DKO, and Sam68KO mice. (C) Quantitative RT-PCR performed on cortex and midbrain cDNA samples from wild-type, Slm1KO, Sam68:Slm1DKO, and Sam68KO mice (n = 3 animals per genotype).
Cell type–specific disruption of Nrxn alternative splicing
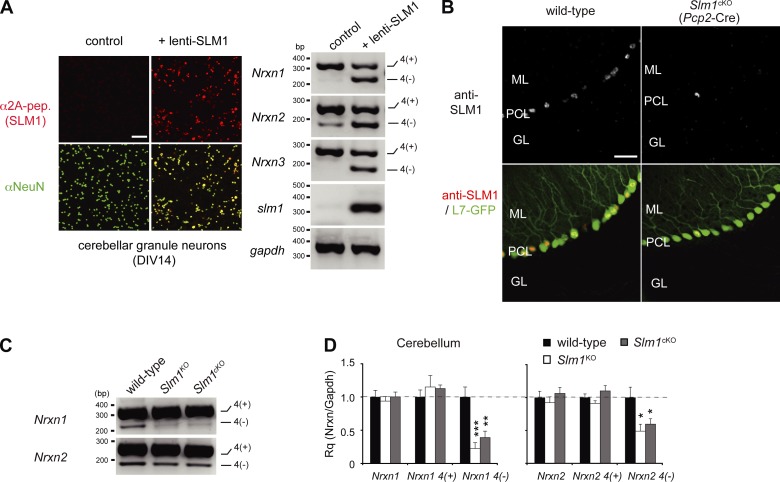
The SLM1 expression pattern suggests that SLM1 may regulate cell type–specific alternative splicing programs. We hypothesized that presence of SLM1 in a neuronal cell population might instruct Nrxn1 isoform choice. To test this, we mis-expressed SLM1 in cultured cerebellar cells, a largely homogeneous population of granule cells that lacks detectable SLM1 expression and shows predominant expression of Nrxn AS4(+) isoforms. Introduction of SLM1 into granule cells was sufficient to stimulate exon skipping and up-regulation of AS4(−) isoforms in Nrxn1,2,3 (Fig. 7 A). Thus, in this cellular context, SLM1 is sufficient to drive formation of AS4(−) isoforms.
Figure 7.
Cell type–specific alternative splicing of SLM1. (A) Shift in alternative splicing at Nrxn1, Nrxn2, and Nrxn3 AS4 in cerebellar granule cells expressing SLM1 ectopically with lentiviral infection. Expression of 2A-tagged SLM1 in lentivirus-infected cerebellar granule cells was confirmed by immunostaining with anti-2A antibody. Representative images of semi-quantitative RT-PCR with Nrxn AS4 in granule neurons with or without ectopic SLM1 expression. (B) High magnification views of SLM1 immunoreactivity in Purkinje cell–specific Slm1KO (Slm1cKO) mice. Purkinje cells are marked by transgenic expression of EGFP from the Purkinje cell–specific L7 promoter. Bar, 50 µm. (C) Representative images of semi-quantitative RT-PCR with Nrxn AS4 in cerebellum from wild-type, Slm1KO, and Slm1cKO mice. (D) Quantitative RT-PCR for Nrxn1 and Nrxn2 AS4 in wild-type, Slm1KO, and Slm1cKO mice (n = 4 animals per genotype).
Although the global Slm1KO analysis is consistent with a requirement for SLM1 in AS4(−) isoform choice in vivo, this analysis does not allow for conclusions about cell-autonomous function that dictates the molecular repertoire of a defined cell population. To address this issue, we generated conditional knockout mice (Slm1cKO) where SLM1 is ablated in Purkinje cells (Slm1flox/flox:: Pcp2cre; Fig. 7 B; Fig. S4). Note that SLM1 remains expressed in a subset of Purkinje cells, in particular in the caudal cerebellum, consistent with the heterogeneity in the activity of the cre driver (Saito et al., 2005). Importantly, analysis of cerebellar RNA from Slm1cKO mice reveals a severe reduction in the Nrxn1 AS4(−) mRNA, whereas total Nrxn1α mRNA levels are unchanged (Fig. 7, C and D). Thus, the vast majority of Nrxn1 AS4(−) and Nrxn2 AS4(−) isoforms in the cerebellum is derived from Purkinje cells where their expression depends on a cell-autonomous function of SLM1. Global ablation of SLM1 did not result in a significant further reduction of Nrxn1 AS4(−) levels, consistent with the conclusion that SLM1 functions specifically in Purkinje cells. Similarly, Nrxn2 AS4(−) was significantly reduced in Slm1cKO and Slm1KO mice (Fig. 7, C and D), further confirming a critical function of SLM1 in alternative splicing of Nrxn2 mRNA in vivo.
Discussion
Previous work has led to the identification of several families of neuronal RNA-regulatory proteins, including neuro-oncological ventral antigen (Nova-1, -2), neural poly-pyrimidine tract binding protein (nPTBP), and ELAV family proteins (CELFs), which regulate alternative exons in neuronal tissues (Darnell, 2013; Kuroyanagi et al., 2013). These factors direct generation of neuron-specific splice variants that are absent from neuronal precursor or nonneuronal cells. By contrast, RNA-binding proteins that drive the unique expression patterns of neuronal sub-populations, such as specific interneuron classes, are less well understood.
Role for SLM1 in the generation of neuron-specific molecular repertoires
In this study we demonstrate that ectopic expression of SLM1 in a neuronal population that normally is SLM1 deficient is sufficient to trigger production of specific neurexin splice variants. Moreover, conditional knock-out of SLM1 in an SLM-1–positive specific cell type results in a severe loss of specific neurexin splice variants in the cerebellum in vivo. This cell type–specific ablation of SLM1 also provides evidence that the lost splice variants were indeed selectively expressed in the cerebellar Purkinje cells. Therefore, our results begin to unravel cell type–specific neurexin isoform repertoires in neuronal populations in vivo. In the hippocampus, SLM1 is highly expressed in SCA and PPA interneurons. This class of GABAergic cells innervates CA1 pyramidal cell dendrites in the S.R. and is subject to specific forms of neuromodulatory regulation (Lawrence, 2008; Klausberger, 2009). In analogy to our findings in cerebellar neurons, we propose that selective expression of SLM1 drives a corresponding program of cell type–specific alternative splicing events in the hippocampus.
Synergistic and differential functions of SAM68 and SLM proteins
In a previous study, we reported a dynamic regulation of Nrxn1 AS4 by SAM68, a closely related alternative splicing factor (Iijima et al., 2011). However, expression and function of SAM68 and SLM1 differ in several fundamental aspects. SAM68 activity toward Nrxn1 AS4 depends on activation through neuronal signaling. Thus, the presence of SAM68 in a cell population is not predictive of Nrxn1 isoform choice (Iijima et al., 2011). Moreover, SAM68 is broadly expressed across neuronal and nonneuronal cells (Richard et al., 2005; Paronetto et al., 2009). By contrast, SLM1 exhibits a more restricted expression pattern. Our gain-of-function and loss-of-function studies demonstrate that SLM1 instructs cell type–specific alternative splicing choices. Moreover, our experiments uncover a complex interplay between SAM68 and SLM1 activities as the regulation of alternative splicing at AS4 differs significantly for Nrxn1, -2, and -3 transcripts. While SAM68 and SLM1 synergize in driving exon skipping at AS4 in Nrxn1, they antagonize each other at Nrxn3 AS4 in vivo. We demonstrated that specific sequences in the RNA-binding domain of SLM1 are required for regulation of Nrxn2 AS4, which is insensitive to SAM68. Finally, our in vitro experiments support a model where hetero-oligomerization of SAM68 with SLM1 provides one potential mechanism that gates substrate specificity of SLM1, and thereby a further mechanism for regulation. Interestingly, a recent study by Elliott and colleagues demonstrated a splicing regulatory function of SLM2 toward Nrxn2 that is similar to the function for SLM1 described here. Based on a comparison of regional SLM2 expression and estimates of Nrxn1 AS4(−) abundance they suggested that SLM2 is the primary regulator of Nrxn1 AS4(−) in vivo (Ehrmann et al., 2013). Considering the largely nonoverlapping expression patterns of SLM1 and SLM2 across cell populations as well as our global and cell type–specific knockout analysis of SLM1, we conclude that SLM2 is not the sole regulator of Nrxn1 AS4 in vivo. Instead, we demonstrate with gain-of-function and loss-of-function experiments that SLM1 as well as SAM68 have major, cell type–specific contributions to Nrxn alternative splice variant choices in vivo.
Functional relevance of SLM1 in the developing nervous system
Neuronal subtype–specific alternative splicing patterns might contribute to the unique functional properties of neuronal cell populations. Such a mechanism is particularly attractive for polymorphic receptor families such as the neurexins that may contribute to specific cellular interactions or specific synaptic functions (Missler and Südhof, 1998; Boucard et al., 2005; Chih et al., 2006; Graf et al., 2006; de Wit et al., 2009; Baudouin and Scheiffele, 2010; Futai et al., 2013). Further work will be needed to understand the respective contributions of SAM68 and SLM proteins in shaping cell type–specific neurexin isoform contents. Moreover, neurexins are likely to be only one of many targets for SLM-dependent alternative splicing regulation. Our analysis of Sam68:Slm1DKO animals uncovered the presence of ectopic Purkinje cells and defects in the foliation of the cerebellar cortex. These two aspects of the phenotype are most likely linked considering that Purkinje cell anchoring centers have been proposed to instruct the foliation process (Sudarov and Joyner, 2007). Specifically, we observed loss of the fissure between lobules VI and VII in the cerebellar vermis. Whereas the primary and secondary fissures flanking lobules VI–VIII form during late embryonic development, the separation of lobules VI and VII occurs during the first postnatal week (Altman and Bayer, 1997). The STAR protein pre-mRNA substrates relevant for this phenotype remain to be identified. Notably, similar foliation defects have been reported in knockout mice for several other neuronal signaling proteins (Sadakata et al., 2007; Lancaster et al., 2011). Definition of the complete alternative splicing programs for STAR proteins as well as the dissection of cellular phenotypes of SLM1-deficient neurons should provide important next steps in testing the links between cell type–specific alternative splicing programs and neuronal development.
Materials and methods
Antibodies and DNA constructs
Polyclonal antibodies to SAM68, SLM1, and SLM2 were raised in rabbits and guinea pigs using the following synthetic peptides (amino acids in brackets were added to improve solubility of the peptide and for coupling): RGVPPPPTVRGAPTPRAR-[C] (anti-SAM68), VNEDAYDSYAPEEWAT-[KKKC] (anti-SLM1), and PRARGVPPTGYRP-[C] (anti-SLM2; Iijima et al., 2011). Anti-GFP antibodies were raised in rabbits using recombinant GFP expressed in Escherichia coli as an antigen (Taniguchi et al., 2007). The following commercially available antibodies were used: mouse anti–β-tubulin (E7; DSHB), mouse anti-Fox3/NeuN (MAB377; EMD Millipore), rat anti-HA (clone 3F10; Roche), sheep anti-parvalbumin (R&D Systems), mouse anti-calbindin (Swant), goat anti-calretinin (Swant), rabbit anti-VIP (Immunostar), mouse anti-cholecystokinin (CCK8; Abcam), goat anti-RORalpha (C-16; Santa Cruz Biotechnology, Inc.), rabbit anti-vesicular glutamate transporter 1 (vGluT1, #1353303; Synaptic Systems), rabbit anti-vesicular glutamate transporter 2 (vGluT2, #135403; Synaptic Systems), guinea pig anti-vesicular GABA transporter (vGAT, #676780; EMD Millipore), and rabbit anti-2A peptide (EMD Millipore).
Somatostatin-positive interneurons were marked in Somatostatin-IRES-cre mice crossed to tdTomato reporter mice (Madisen et al., 2010; Taniguchi et al., 2011).
Expression vectors (pEGFP backbone, CMV promoter) for SAM68, SLM1, SLM2, hnRNPA1, hnRNPH, and Nrxn1 AS4 splice reporters were described previously (Di Fruscio et al., 1999; Fisette et al., 2010; Iijima et al., 2011). The Nrxn2 AS4 splice reporter contains a mouse Nrxn2 genomic cassette of AS4 from exon 19 to exon 21 containing 500 bp intronic sequence at each splice donor and acceptor site.
RNA isolation and alternative splicing assays
Human embryonic kidney (HEK293 and HEK293T) cells were cultured in DMEM (Invitrogen) supplemented with 10% FCS, l-glutamine (2 mM), penicillin, and streptomycin and grown in 5% CO2 at 37°C. For reporter assays, cells were cotransfected using Fugene 6 reagent (Roche) with expression vectors encoding the splice reporter and RNA-binding proteins. RNA was harvested 24–36 h after transfection using Trizol reagent (Invitrogen), followed by removal of contaminating DNA using Turbo DNA-free (RNase-free DNase; Ambion). 1 µg of total RNA was reverse transcribed using random hexamers and ImProm-II (Promega).
For semi-quantitative PCR, DNA fragment intensities were quantified by image analyzer (FAS-III; Toyobo) and ImageGauge software (Fujifilm). Oligonucleotide primers used for semi-quantitative PCR were described previously (Iijima et al., 2011).
Quantitative PCR was performed on a StepOnePlus qPCR system (Applied Biosystems). Custom and commercial gene expression assays (see Table 1) were used with TaqMan Master Mix (Applied Biosystems) and comparative CT method. The mRNA levels were normalized to that of Gapdh mRNA.
Table 1.
Oligonucleotide sequences of qPCR primer sets
| Primer set | Catalogue number/sequence |
| Nrxn1 alpha | Mm00660298m1 |
| Nrxn2 alpha | Mm01236851_m1 |
| Nrxn3 alpha | Mm00553213_m1 |
| Gapdh | Mm99999915g1 |
| Nrxn1 ex20-F | 5′-TAGTTGATGAATGGCTACTCGACAAA-3′ |
| Nrxn1 ex20-R | 5′-GACTCAGTTGTCATAGAGGAAGGCAC-3′ |
| Probe | 6FAM-CCGACCAGCCTCACATTCCCCACTAT-BBQ |
| Nrxn1 del-F | 5′-GCTACCCTGCAGGGCGT-3′ |
| Nrxn1 del-R | 5′-GAGGTGGACATCTCAGACTGCAT-3′ |
| Probe | 6FAM-CCGACCAGCCTCACATTCCCCACTAT-BBQ |
| Nrxn2 ex20-F | 5′-GACATTACAATTGATGAGCCCAAC-3′ |
| Nrxn2 ex20-R | 5′-CAGGCGCTCGTTATCAAAGTTT-3′ |
| Probe | 6FAM-CCATCGTGAGCGACGGCAAATATCAC-BBQ |
| Nrxn2 del-F | 5′-GACATTACAATTGATGAGCCCAAC-3′ |
| Nrxn2 del-R | 5′-GTCAGCTGGCGTCCTGC-3′ |
| Probe | 6FAM-CCATCGTGAGCGACGGCAAATATCAC-BBQ |
| Nrxn3 ex20-F | 5′-AAATACCACGTTGTGCGCTT-3′ |
| Nrxn3 ex20-R | 5′-TTGGAGGCGTTCATTATCAGTGTT-3′ |
| Probe | 6FAM-CCATCGTGAGCGACGGCAAATATCAC-BBQ |
| Nrxn3 del-F | 5′-GAACTCCTGTCAATGATGGCAAATAC-3′ |
| Nrxn3 del-R | 5′-TAGCTGCCGGCCTGTAGG-3′ |
| Probe | 6FAM-CCAGGAATGGGGGAAATGCTACACT-BBQ |
Biochemical procedures
Cells or brain tissues were lysed with RIPA buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 1% deoxycholate, and 0.1% SDS) containing protease inhibitor cocktail (Roche). For protein interaction studies the soluble fractions were subjected to immunoprecipitation for 24 h at 4°C and analyzed by immunoblotting. For visualization, HRP-conjugated secondary antibody and enhanced chemiluminescence (ECL) detection (Thermo Fisher Scientific) were used. Signals were acquired using an image analyzer (LAS-3000; Fujifilm).
Immunohistochemistry, image acquisition, and analysis
Animals were transcardially perfused with fixative (4% paraformaldehyde/15% picric acid in 100 mM phosphate buffer, pH 7.2). Tissues were sectioned at 50 µm in PBS on a vibratome (VT1000S; Leica) and floating sections were immunostained following standard procedures (fluorophores Alexa Fluor 488, Alexa Fluor 568, Cy2, Cy3, Cy5, and mounting medium Fluoromount G). Images were acquired at room temperature on a confocal microscope (LSM5; Carl Zeiss) using 4×, 40×, and 63× Plan Apochromat objectives (NA 0.2, 1.3, and 1.4, respectively) and controlled by Zen 2008 software (Carl Zeiss). Images were assembled using Photoshop and Illustrator software (Adobe). For quantitative assessment of interneuron association of SLM proteins, coronal sections from three animals were prepared as described above and confocal images were captured on a confocal system (LSM5; Carl Zeiss) from 5–10 separate fields per animal from the dorsal hippocampal CA1–CA2 region. All procedures related to animal experimentation were reviewed and approved by the Kantonales Veterinäramt Basel-Stadt.
Production of lentivirus
VSV-G pseudotyped lentiviral vectors provided by St. Jude Children’s Research Hospital (Memphis, TN; Hanawa et al., 2002) were used in this study. The pCL20c vectors were designed under the control of the MSCV promoter (Hawley et al., 1994). Viral supernatants were produced by cotransfection of HEK293T cells with a mixture of four plasmids using the calcium phosphate precipitation method. The four-plasmid mixture consisted of 6 µg of pCAG-kGP1R, 2 µg of pCAG-4RTR2, 2 µg of pCAG-VSV-G, and 10 µg of vector plasmid pCL20c MSCV-SLM1-2A-venus. Medium containing viral particles was harvested 40 h after transfection. The medium samples were filtered through 0.22-µm membranes and concentrated by centrifugation at 25,800 rpm for 90 min. The virus samples were suspended in cold PBS (pH 7.4), frozen in aliquots, and stored at −80°C.
Knockout mice
Sam68KO mice were provided by S. Richard (McGill University, Montreal, Quebec, Canada; Richard et al., 2005). An Slm1 conditional allele was generated by homologous recombination in mouse embryonic stem cells. In brief, a genomic DNA fragment containing exon 2 (ENSMUSE00000314986) was flanked by a LoxP site and a FRT-PGK-neo-LoxP cassette encoding neomycin phosphotransferase under control of the phosphoglycerate kinase 1 promoter. The targeting vector was electroporated into 129SvEvTac embryonic stem cells. Homologous recombination in G-418–resistant clones was confirmed and selected cells were blastocyst injected. Chimeric animals were crossed with ROSA-26 Flpe mice to remove PGK-neo sequences through Frt/Flp-mediated excision. The Slm1flox/+ mice were crossed with CMVcre deleter mice (Schwenk et al., 1995) to generate a germline deletion of Khdrbs2. Conditional ablation of Khdrbs2 in Purkinje was done using Pcp2cre knock-in mice (Saito et al., 2005). The Slm1flox allele was detected by PCR using primers 5′-CCCTGAGAGGCTGAGGTTAG-3′ (Lox gtF), 5′-AAGTGCAGTGCCACAAAATG-3′ (Lox gtR), 5′-CCACAAGCCATAAAATTGAGC-3′ (Frt gtF), and 5′-GCCAACAACATTTGGCTAGAG-3′ (Frt gtR).
Sam68:Slm1DKO mice were generated by intercrossing of the individual mutant mice. The resulting homozygous mutant mice were viable.
Statistical analysis
Pairwise comparisons were performed using Student’s t test. For multiple comparisons, analysis of variance (ANOVA) followed by Bonferroni or Dunnett test was used. Data are represented as the mean ± SEM. Significance is indicated as follows: ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Online supplemental material
Fig. S1 shows Western blot and immunohistochemical data on region-specific expression of SAM68 and SLM proteins. Fig. S2 shows high-resolution views of neuronal cell type–specific expression of SAM68 and SLM proteins in mouse hippocampus. Fig. S3 illustrates targeting strategy for generation of Slm1KO mice and morphological analysis. Fig. S4 shows Purkinje cell–specific Slm1KO mice. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201310136/DC1.
Supplementary Material
Acknowledgments
We thank S. Baudouin and D. Schreiner for advice and comments on the manuscript, and S. Richard and N. Suzuki for generously sharing Sam68KO and Pcp2cre mice, respectively. We are grateful to E. Sylwestrak for generously sharing reagents, and C. Bornmann, L. Burklé, A. Yamaguchi, and Y. Doi for experimental support.
This work was supported by funds to P. Scheiffele from the Swiss National Science Foundation and the Kanton Basel-Stadt. T. Iijima was supported by the Uehara Memorial Foundation, KANAE Foundation for the Promotion of Medical Science, and Astellas Foundation for Research on Metabolic Disorders.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AS4
- alternatively spliced segment 4
- CCK
- cholecystokinin
- nPTBP
- neural poly-pyrimidine tract binding protein
- NRX
- neurexin protein
- Nrxn
- Neurexin gene
- PPA
- perforant path–associated
- SAM68
- Src-associated in mitosis of 68-kD protein
- SCA
- Schaffer collateral associated
- SLM
- SAM68-like mammalian protein
- S.L.M.
- stratum lacunosum moleculare
- S.O.
- stratum oriens
- S.R.
- stratum radiatum
- VIP
- vasoactive intestinal peptide
References
- Altman J., Bayer S.A. 1997. Development of the cerebellar system: In Relation to Its Evolution, Structure, and Functions CRC Press, Boca Raton, FL: 230–251 [Google Scholar]
- Aoto J., Martinelli D.C., Malenka R.C., Tabuchi K., Südhof T.C. 2013. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 154:75–88 10.1016/j.cell.2013.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudouin S., Scheiffele P. 2010. SnapShot: Neuroligin-neurexin complexes. Cell. 141:908. [DOI] [PubMed] [Google Scholar]
- Beck E.S., Gasque G., Imlach W.L., Jiao W., Jiwon Choi B., Wu P.S., Kraushar M.L., McCabe B.D. 2012. Regulation of Fasciclin II and synaptic terminal development by the splicing factor beag. J. Neurosci. 32:7058–7073 10.1523/JNEUROSCI.3717-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard A.A., Chubykin A.A., Comoletti D., Taylor P., Südhof T.C. 2005. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 48:229–236 10.1016/j.neuron.2005.08.026 [DOI] [PubMed] [Google Scholar]
- Chih B., Gollan L., Scheiffele P. 2006. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 51:171–178 10.1016/j.neuron.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Craig A.M., Kang Y. 2007. Neurexin-neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 17:43–52 10.1016/j.conb.2007.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell R.B. 2013. RNA protein interaction in neurons. Annu. Rev. Neurosci. 36:243–270 10.1146/annurev-neuro-062912-114322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J., Sylwestrak E., O’Sullivan M.L., Otto S., Tiglio K., Savas J.N., Yates J.R., III, Comoletti D., Taylor P., Ghosh A. 2009. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 64:799–806 10.1016/j.neuron.2009.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C., Scholl F.G., Choih J., DeMaria S., Berger J., Isacoff E., Scheiffele P. 2003. Neurexin mediates the assembly of presynaptic terminals. Nat. Neurosci. 6:708–716 10.1038/nn1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fruscio M., Chen T., Richard S. 1999. Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc. Natl. Acad. Sci. USA. 96:2710–2715 10.1073/pnas.96.6.2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrmann I., Dalgliesh C., Liu Y., Danilenko M., Crosier M., Overman L., Arthur H.M., Lindsay S., Clowry G.J., Venables J.P., et al. 2013. The tissue-specific RNA binding protein T-STAR controls regional splicing patterns of neurexin pre-mRNAs in the brain. PLoS Genet. 9:e1003474 10.1371/journal.pgen.1003474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisette J.F., Toutant J., Dugré-Brisson S., Desgroseillers L., Chabot B. 2010. hnRNP A1 and hnRNP H can collaborate to modulate 5′ splice site selection. RNA. 16:228–238 10.1261/rna.1890310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai K., Doty C.D., Baek B., Ryu J., Sheng M. 2013. Specific trans-synaptic interaction with inhibitory interneuronal neurexin underlies differential ability of neuroligins to induce functional inhibitory synapses. J. Neurosci. 33:3612–3623 10.1523/JNEUROSCI.1811-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau A., Richard S. 2009. The STAR RNA binding proteins GLD-1, QKI, SAM68 and SLM-2 bind bipartite RNA motifs. BMC Mol. Biol. 10:47 10.1186/1471-2199-10-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman L.T., Meera P., Stoilov P., Shiue L., O’Brien J.E., Meisler M.H., Ares M., Jr, Otis T.S., Black D.L. 2012. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 26:445–460 10.1101/gad.182477.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf E.R., Kang Y., Hauner A.M., Craig A.M. 2006. Structure function and splice site analysis of the synaptogenic activity of the neurexin-1 beta LNS domain. J. Neurosci. 26:4256–4265 10.1523/JNEUROSCI.1253-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa H., Kelly P.F., Nathwani A.C., Persons D.A., Vandergriff J.A., Hargrove P., Vanin E.F., Nienhuis A.W. 2002. Comparison of various envelope proteins for their ability to pseudotype lentiviral vectors and transduce primitive hematopoietic cells from human blood. Mol. Ther. 5:242–251 [DOI] [PubMed] [Google Scholar]
- Hawley R.G., Lieu F.H., Fong A.Z., Hawley T.S. 1994. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1:136–138 [PubMed] [Google Scholar]
- Iijima T., Wu K., Witte H., Hanno-Iijima Y., Glatter T., Richard S., Scheiffele P. 2011. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 147:1601–1614 10.1016/j.cell.2011.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T. 2009. GABAergic interneurons targeting dendrites of pyramidal cells in the CA1 area of the hippocampus. Eur. J. Neurosci. 30:947–957 10.1111/j.1460-9568.2009.06913.x [DOI] [PubMed] [Google Scholar]
- Klein M.E., Younts T.J., Castillo P.E., Jordan B.A. 2013. RNA-binding protein Sam68 controls synapse number and local β-actin mRNA metabolism in dendrites. Proc. Natl. Acad. Sci. USA. 110:3125–3130 10.1073/pnas.1209811110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H., Watanabe Y., Hagiwara M. 2013. CELF family RNA-binding protein UNC-75 regulates two sets of mutually exclusive exons of the unc-32 gene in neuron-specific manners in Caenorhabditis elegans. PLoS Genet. 9:e1003337 10.1371/journal.pgen.1003337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Gopal D.J., Kim J., Saleem S.N., Silhavy J.L., Louie C.M., Thacker B.E., Williams Y., Zaki M.S., Gleeson J.G. 2011. Defective Wnt-dependent cerebellar midline fusion in a mouse model of Joubert syndrome. Nat. Med. 17:726–731 10.1038/nm.2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J.J. 2008. Cholinergic control of GABA release: emerging parallels between neocortex and hippocampus. Trends Neurosci. 31:317–327 10.1016/j.tins.2008.03.008 [DOI] [PubMed] [Google Scholar]
- Lipscombe D., Andrade A., Allen S.E. 2013. Alternative splicing: functional diversity among voltage-gated calcium channels and behavioral consequences. Biochim. Biophys. Acta. 1828:1522–1529 10.1016/j.bbamem.2012.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland R.H. 2004. Neuronal cell types. Curr. Biol. 14:R497–R500 10.1016/j.cub.2004.06.035 [DOI] [PubMed] [Google Scholar]
- Matsuda K., Yuzaki M. 2011. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur. J. Neurosci. 33:1447–1461 10.1111/j.1460-9568.2011.07638.x [DOI] [PubMed] [Google Scholar]
- Meyer N.H., Tripsianes K., Vincendeau M., Madl T., Kateb F., Brack-Werner R., Sattler M. 2010. Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain. J. Biol. Chem. 285:28893–28901 10.1074/jbc.M110.126185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M., Südhof T.C. 1998. Neurexins: three genes and 1001 products. Trends Genet. 14:20–26 10.1016/S0168-9525(97)01324-3 [DOI] [PubMed] [Google Scholar]
- Okaty B.W., Sugino K., Nelson S.B. 2011. Cell type-specific transcriptomics in the brain. J. Neurosci. 31:6939–6943 10.1523/JNEUROSCI.0626-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto M.P., Messina V., Bianchi E., Barchi M., Vogel G., Moretti C., Palombi F., Stefanini M., Geremia R., Richard S., Sette C. 2009. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J. Cell Biol. 185:235–249 10.1083/jcb.200811138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan P., Dalgliesh C., Bourgeois C.F., Heiner M., Emami K., Clark E.L., Bindereif A., Stevenin J., Robson C.N., Leung H.Y., Elliott D.J. 2009. Proteomic identification of heterogeneous nuclear ribonucleoprotein L as a novel component of SLM/Sam68 Nuclear Bodies. BMC Cell Biol. 10:82 10.1186/1471-2121-10-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S., Torabi N., Franco G.V., Tremblay G.A., Chen T., Vogel G., Morel M., Cléroux P., Forget-Richard A., Komarova S., et al. 2005. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet. 1:e74 10.1371/journal.pgen.0010074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadakata T., Kakegawa W., Mizoguchi A., Washida M., Katoh-Semba R., Shutoh F., Okamoto T., Nakashima H., Kimura K., Tanaka M., et al. 2007. Impaired cerebellar development and function in mice lacking CAPS2, a protein involved in neurotrophin release. J. Neurosci. 27:2472–2482 10.1523/JNEUROSCI.2279-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Tsumura H., Otake S., Nishida A., Furukawa T., Suzuki N. 2005. L7/Pcp-2-specific expression of Cre recombinase using knock-in approach. Biochem. Biophys. Res. Commun. 331:1216–1221 10.1016/j.bbrc.2005.04.043 [DOI] [PubMed] [Google Scholar]
- Schwenk F., Baron U., Rajewsky K. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080–5081 10.1093/nar/23.24.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K., Scheiffele P. 2010. Genetics and cell biology of building specific synaptic connectivity. Annu. Rev. Neurosci. 33:473–507 10.1146/annurev.neuro.051508.135302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui T.J., Pancaroglu R., Kang Y., Rooyakkers A., Craig A.M. 2010. LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J. Neurosci. 30:7495–7506 10.1523/JNEUROSCI.0470-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoss O., Novoyatleva T., Gencheva M., Olbrich M., Benderska N., Stamm S. 2004. p59(fyn)-mediated phosphorylation regulates the activity of the tissue-specific splicing factor rSLM-1. Mol. Cell. Neurosci. 27:8–21 10.1016/j.mcn.2004.04.011 [DOI] [PubMed] [Google Scholar]
- Sudarov A., Joyner A.L. 2007. Cerebellum morphogenesis: the foliation pattern is orchestrated by multi-cellular anchoring centers. Neural Dev. 2:26 10.1186/1749-8104-2-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof T.C. 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 455:903–911 10.1038/nature07456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H., Gollan L., Scholl F.G., Mahadomrongkul V., Dobler E., Limthong N., Peck M., Aoki C., Scheiffele P. 2007. Silencing of neuroligin function by postsynaptic neurexins. J. Neurosci. 27:2815–2824 10.1523/JNEUROSCI.0032-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H., He M., Wu P., Kim S., Paik R., Sugino K., Kvitsiani D., Fu Y., Lu J., Lin Y., et al. 2011. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 71:995–1013 10.1016/j.neuron.2011.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Lee S.J., Yasumura M., Takeuchi T., Yoshida T., Ra M., Taguchi R., Sakimura K., Mishina M. 2010. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 141:1068–1079 10.1016/j.cell.2010.04.035 [DOI] [PubMed] [Google Scholar]
- Venables J.P., Vernet C., Chew S.L., Elliott D.J., Cowmeadow R.B., Wu J., Cooke H.J., Artzt K., Eperon I.C. 1999. T-STAR/ETOILE: a novel relative of SAM68 that interacts with an RNA-binding protein implicated in spermatogenesis. Hum. Mol. Genet. 8:959–969 10.1093/hmg/8.6.959 [DOI] [PubMed] [Google Scholar]
- Zheng S., Black D.L. 2013. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends Genet. 29:442–448 10.1016/j.tig.2013.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipursky S.L., Sanes J.R. 2010. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell. 143:343–353 10.1016/j.cell.2010.10.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.