Abstract
After export from the nucleus, ribosomes need to be distributed throughout the entire cell so that protein synthesis can occur even at distant sites. In the elongated hyphal cell of the fungus Ustilago maydis, Higuchi et al. (2014. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201307164) now demonstrate that polysomes associate with early endosomes that undergo kinesin 3– and dynein-dependent long-range motility. The bidirectional movement of early endosomes randomly distributes polysomes, which may ensure the even distribution of the translation machinery across the entire cell.
In large cells, such as oocytes and neurons, the nucleus, where ribosomes are assembled, is distant from regions of the cell where local protein synthesis takes place. So, how is the distribution of ribosomes within the entire cell achieved? In neurons, ribosomal subunits are found in RNA granules, large macromolecular structures that contain RNAs and serve as motile units that translocate these transcripts to remote regions of the cell (Krichevsky and Kosik, 2001; Kanai et al., 2004; Elvira et al., 2006). Also in neurons, ribosomes associate with a cell surface receptor (the netrin receptor DCC), which has led to a model for localization of protein synthesis based on the interaction of the translation machinery with membrane receptors (Tcherkezian et al., 2010). Lastly, evidence suggest that ribosomes can even translocate into myelinated axons from Schwann cells (Court et al., 2008). In this issue, Higuchi et al. identify another mechanism of ribosome transport; they find that in the fungus Ustilago maydis, polysomes are actively transported on early endosomes (Fig. 1).
Figure 1.
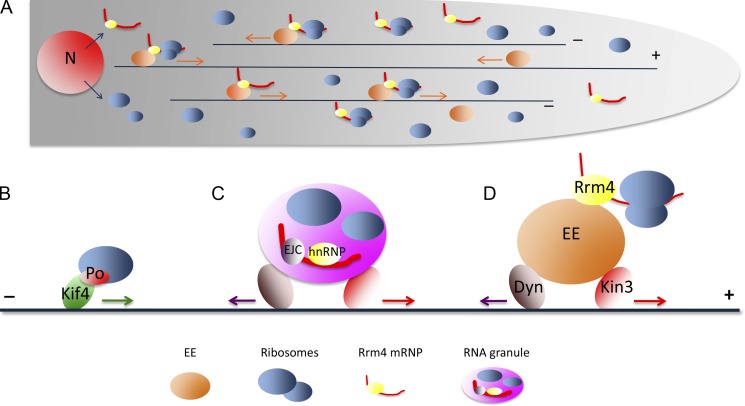
Modes of ribosome distribution. In U. maydis, ribosomes are distributed in the hyphal cell on microtubule tracks (black lines) by loading onto Rrm4 RNPs that are themselves hitchhiking on mobile early endosomes (A and D). This transport of ribosomes on early endosomes is the major mechanism for ribosome distribution in these cells, but it is unknown how conserved this mode of transport is in other systems. In addition to diffusion, and most likely facilitated diffusion by motor-mediated cytoplasmic streaming, ribosomes can also be distributed in the cell directly by motors (B; Bisbal et al., 2009; the ribosomal protein Po interacts with the motor Kif4) or as part of transported RNA granules (C; see main text). Ribosomes also interact with MTs through the motor protein Eg5 (Bartoli et al., 2011), although the possible link of this interaction with ribosome transport has not yet been explored. Dyn, Dynein; EJC, exon–exon junction complex; EE, early endosome; hnRNP, representing various hnRNPs bound to mRNAs; Kin3, kinesin 3; N, nucleus.
U. maydis is an interesting model to study the issue of localized translation. In addition to division by budding, U. maydis also undergoes dikaryotic hyphal growth, during which the hyphae become elongated with the nucleus positioned 50 µm behind the growing tip. Hyphae contain both unipolar and antiparallel microtubule (MT) arrays, and kinesin 3 (kin3) and dynein (dyn) are major motor proteins for the transport of many cargoes (for review see Egan et al., 2012). To understand how ribosomes are distributed within the hyphae, Higuchi et al. (2014) examined the movement of fluorescently tagged ribosomal proteins Rpl25 and Rps3. Through their measurements of ribosome movement in hyphae combined with mathematical modeling of ribosome allocation, they determined that diffusion alone could not support the observed distribution of ribosomal subunits. In fact, directed movement of particles containing both fluorescently tagged Rpl25 and Rps3 was occasionally detected at speeds of 2 µm/s, which suggests active transport on MTs. This is further supported by the fact that the inclusion of both diffusion and active transport in the model results in a distribution of the ribosomal subunits that match the experimental results. Moreover, in kin3 and dyn mutant cells, Rpl25 accumulates in the region close to the nucleus, and its distribution does not extend toward the tip, in a pattern that resembles the distribution of ribosomes from the mathematical model when only diffusion but not active transport is included.
Given that kin3 and dyn also transport RNAs associated with Rrm4, an RNA-binding protein required for mRNA transport (Baumann et al., 2012), the authors analyzed whether ribosomes might also travel with these Rrm4 RNPs. They observed that fluorescently tagged Rps3 and Rrm4 comigrated, and rrm4 mutant cells show reduced Rpl25 motility and partial depletion of ribosomes from the growing tip. Interestingly, these transported Rrm4 RNPs associate with organelles that carry the endosomal protein Yup1 (Baumann et al., 2012), raising the possibility that moving ribosomes also associate with a membrane-bound vesicle or compartment. In fact, Baumann et al. (2014) recently showed that septin mRNA, septin protein, and ribosomal proteins colocalize with shuttling endosomes, and that endosomal trafficking is important for delivery of septin protein to filaments at growth poles. Now, Higuchi et al. (2014) show that Rpl25 colocalizes with Rab5 on the moving organelles. Rab5 is an endosomal marker (Bucci et al., 1992), and in fungi, kin3- and dyn-dependent motile Rab-5 endosomes have been described previously (Wedlich-Söldner et al., 2002; Fuchs et al., 2006; Lenz et al., 2006; Abenza et al., 2009; Zhang et al., 2010; Schuster et al., 2011). This work by Higuchi et al. (2014) confirms the identity of these unusually mobile Rab5 vesicles as early endosomes, as shown by pulse-chase experiments with FM4-64, their colocalization with Rab4 (another early endosome marker), their enrichment of the lipid PtdIns(3)P, and their lack of colocalization with Rab7. More importantly, the reduction of early endosome numbers in yup1 mutants results in an impairment of Rpl25 motility, and defects in Rpl25 distribution. Collectively, these observations strongly suggest that ribosomes comigrate with early endosomes across the entire cell in a complex formed by the loading of ribosomes onto Rrm4 RNPs, and the interaction of Rrm4 with early endosomes (Fig. 1).
Is translation active on these early endosome-associated ribosome-containing RNPs? Baumann et al., (2014) showed that the presence of the septin mRNA is crucial for the entry of the encoded protein into the Rrm4-positive endosomal compartment, which suggests that local translation on endosomes takes place. Now, Higuchi et al. (2014) provide strong evidence supporting this idea. First, they estimate that there are several ribosomes per individual endosome (by comparing the intensity of each signal with a fluorescent nucleoporin). Second, exposing cells to mild stress conditions, which inhibit translation due to polysome disassembly, reduces Rpl25 association with early endosomes and ribosome motility, without affecting early endosome movement. Importantly, motility of Rpl25 is almost abolished when cells are treated with inhibitors of translation initiation. Third, nascent GFP-Rho3 (Rho3 mRNA is one of the transcripts in Rrm4 RNPs; König et al., 2009) comigrates with 2.76% of the Rab5 endosomes. All these results together support the notion that early endosome–associated polysomes are translationally active.
In kin3 and dyn mutants, early endosomes do not distribute along the cell, and ribosomes accumulate close to the nucleus, as they do in rrm4 mutant cells. Thus, an important function for this Rrm4-mediated association of polysomes to early endosomes might be to spread out the translation machinery across the cell. If this were the case, then it would be expected for ribosomes to hop on and off the moving early endosomes so that an even cytoplasmic distribution of the translation machinery can be achieved. Translation elongation inhibitors significantly increased the amount of time Rpl25 traveled on early endosomes, which suggests an impaired turnover of the ribosomal subunits. Furthermore, the mean distance traveled by the early endosomes is longer than the mean distance traveled by Rpl25 or Rrm4, which suggests that polysomes and the RNPs “fall off” the moving early endosomes. In fact, Rrm4- and Rpl25-GFP signals frequently separated from moving early endosomes, and remained relatively stationary after deposition in the cytoplasm.
This elegant work using an extremely resourceful model system has allowed the intriguing finding that ribosomes can be moved by “hitchhiking” on endosomes. Many new questions emerge from this study: Do other RNA-binding proteins interact with early endosomes or other endosomal compartments? Is the on/off rate regulated, and if so, how? And more importantly, is this mechanism for ribosome distribution used by any other organisms? Rrm4 homologues are restricted to the basidiomycete fungi, raising the question of whether that early endosome/Rrm4-mediated ribosome movement is widely conserved. However, other RNA-binding proteins might mediate the interaction of RNPs with membranes in higher eukaryotes. For example, in budding yeast, association of localized mRNAs with the ER requires She2p, an RNA-binding protein that can also specifically bind to ER-derived membranes in a membrane curvature–dependent manner (Genz et al., 2013). This new role of the early endosomes in spreading out the translation machinery by motor-mediated bidirectional movement will inspire researchers to further explore interactions between the translational apparatus and cellular organelles.
Acknowledgments
Support was provided by the Department of Zoology and School of Biology, University of Cambridge.
The author declares no competing financial interests.
References
- Abenza J.F., Pantazopoulou A., Rodríguez J.M., Galindo A., Peñalva M.A. 2009. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic. 10:57–75 10.1111/j.1600-0854.2008.00848.x [DOI] [PubMed] [Google Scholar]
- Bartoli K.M., Jakovljevic J., Woolford J.L., Jr, Saunders W.S. 2011. Kinesin molecular motor Eg5 functions during polypeptide synthesis. Mol. Biol. Cell. 22:3420–3430 10.1091/mbc.E11-03-0211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann S., Pohlmann T., Jungbluth M., Brachmann A., Feldbrügge M. 2012. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J. Cell Sci. 125:2740–2752 10.1242/jcs.101212 [DOI] [PubMed] [Google Scholar]
- Baumann S., Konig J., Koepke J., Feldbrugge M. 2014. Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep. 15:94–102 10.1002/embr.201338037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbal M., Wojnacki J., Peretti D., Ropolo A., Sesma J., Jausoro I., Cáceres A. 2009. KIF4 mediates anterograde translocation and positioning of ribosomal constituents to axons. J. Biol. Chem. 284:9489–9497 10.1074/jbc.M808586200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C., Parton R.G., Mather I.H., Stunnenberg H., Simons K., Hoflack B., Zerial M. 1992. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 70:715–728 10.1016/0092-8674(92)90306-W [DOI] [PubMed] [Google Scholar]
- Court F.A., Hendriks W.T., MacGillavry H.D., Alvarez J., van Minnen J. 2008. Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J. Neurosci. 28:11024–11029 10.1523/JNEUROSCI.2429-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M.J., McClintock M.A., Reck-Peterson S.L. 2012. Microtubule-based transport in filamentous fungi. Curr. Opin. Microbiol. 15:637–645 10.1016/j.mib.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira G., Wasiak S., Blandford V., Tong X.K., Serrano A., Fan X., del Rayo Sánchez-Carbente M., Servant F., Bell A.W., Boismenu D., et al. 2006. Characterization of an RNA granule from developing brain. Mol. Cell. Proteomics. 5:635–651 10.1074/mcp.M500255-MCP200 [DOI] [PubMed] [Google Scholar]
- Fuchs U., Hause G., Schuchardt I., Steinberg G. 2006. Endocytosis is essential for pathogenic development in the corn smut fungus Ustilago maydis. Plant Cell. 18:2066–2081 10.1105/tpc.105.039388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genz C., Fundakowski J., Hermesh O., Schmid M., Jansen R.P. 2013. Association of the yeast RNA-binding protein She2p with the tubular endoplasmic reticulum depends on membrane curvature. J. Biol. Chem. 288:32384–32393 10.1074/jbc.M113.486431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y., Ashwin P., Roger Y., Steinberg G. 2014. Early endosome motility spatially organizes polysome distribution. J. Cell Biol. 204:343–357 10.1083/jcb.201307164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., Hirokawa N. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 43:513–525 10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- König J., Baumann S., Koepke J., Pohlmann T., Zarnack K., Feldbrügge M. 2009. The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. EMBO J. 28:1855–1866 10.1038/emboj.2009.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A.M., Kosik K.S. 2001. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 32:683–696 10.1016/S0896-6273(01)00508-6 [DOI] [PubMed] [Google Scholar]
- Lenz J.H., Schuchardt I., Straube A., Steinberg G. 2006. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 25:2275–2286 10.1038/sj.emboj.7601119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Kilaru S., Fink G., Collemare J., Roger Y., Steinberg G. 2011. Kinesin-3 and dynein cooperate in long-range retrograde endosome motility along a nonuniform microtubule array. Mol. Biol. Cell. 22:3645–3657 10.1091/mbc.E11-03-0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J., Brittis P.A., Thomas F., Roux P.P., Flanagan J.G. 2010. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 141:632–644 10.1016/j.cell.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Straube A., Friedrich M.W., Steinberg G. 2002. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J. 21:2946–2957 10.1093/emboj/cdf296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhuang L., Lee Y., Abenza J.F., Peñalva M.A., Xiang X. 2010. The microtubule plus-end localization of Aspergillus dynein is important for dynein-early-endosome interaction but not for dynein ATPase activation. J. Cell Sci. 123:3596–3604 10.1242/jcs.075259 [DOI] [PMC free article] [PubMed] [Google Scholar]