Abstract
Background
Smaller studies have evaluated SLC6A4 5-HTTLPR and GNβ3 825C>T polymorphisms in IBS, and interactions between 5-HTT LPR with life events have been reported in the psychiatric literature, but gene-environment studies in IBS are lacking.
Aims
To assess the association of two polymorphisms with IBS and age of onset; and to assess whether there are gene-environment interactions with IBS.
Methods
Outpatients with IBS and controls completed a validated questionnaire and provided blood for DNA. Comparisons of genotype/allele frequencies between cases and controls were performed with logistic regression. Linear regression was used to evaluate the association between the variants and age of onset. Environmental variables tested included abuse, parental alcohol abuse, parental psychiatric disorders, and gastrointestinal infections.
Results
Genotyping was performed in 385 cases and 262 controls with median age of 50 yrs (range: 18.0–70.0) and 498 (77%) females. The IBS subtype distribution among cases was: 102 (26%) D-IBS, 40 (10%) C-IBS, 125 (32%) M-IBS, 118 (31%) other. No association was observed between IBS or age of onset and both variants. Significant interactions were observed between GI infection and the GNβ3 825T allele. For those reporting gastrointestinal infection, the OR for IBS was 3.9 (95%CI: 1.2–12.7) whereas the OR was 0.86 (95% CI: 0.65–1.13) for those without prior infection.
Conclusions
There was a significant interaction between the GNβ3 polymorphism and infection in the development of IBS, suggesting that its etiology is the result of a combination of specific genetic and environmental risk factors.
Keywords: Irritable bowel syndrome, genes, genetics, infection
INTRODUCTION
Irritable bowel syndrome (IBS) has been shown to aggregate in families [1–3], and twin studies support a modest genetic basis for IBS [4–7]. Various candidate genes have been studied (e.g. SLC6A4, IL10, etc.) [8], but the susceptibility locus for IBS has yet to be clearly identified. There has been great interest in gene discovery for IBS, as diagnostic tests for IBS are lacking and the molecular basis for IBS pathophysiology remains unclear. If a genetic risk factor for IBS were identified, a more confident diagnosis of IBS could be more easily made and specific treatments focusing on the underlying molecular abnormalities could be developed. In the gene hunt for functional gastrointestinal disorders, however, there has been a great deal of interest in the potential role of functional polymorphisms in the gene encoding the serotonin transporter protein (SLC6A4) in IBS and in the gene encoding the beta-3 subunit of the G-protein (GNβ3) in functional dyspepsia [9,10].
5-HTT LPR is a variable number tandem repeat polymorphism in the promoter region of SLC6A4. The presence or absence of 44 base pairs results in a long (L) transcript or a short (S) transcript that is either 528 base pairs or 472 base pairs long. The S allele has been linked to depression and anxiety [11,12], has been associated with differential brain activation in response to visceral pain responses in patients with IBS [13], and predicts response to alosetron and clonidine [14]. A meta-analysis of eight studies concluded that there is not an association between this variant and IBS and its subtypes [15]. Several psychiatric studies have also shown a gene-environment interaction between the 5-HTT LPR S allele and abuse where the allele may only increase risk of depression in the context of early maltreatment [11],[16], but these gene-environment interactions have not been tested in IBS.
825C>T is a single nucleotide polymorphism (SNP) on the GNβ3 gene that results in a shortened transcript. This SNP has been linked to depression, increased chemotaxis [17–19], and functional dyspepsia [10]. More recently, this polymorphism has been shown to predict response (rectal sensation, pain) to clonidine in patients with IBS [20], raising the question as to whether or not this genetic variant could potentially play a role in IBS pathophysiology. A recent study of 214 IBS patients demonstrated no association between 825 C>T with IBS [21], but no analyses to evaluate its association with IBS subtypes or evaluating gene-environment interactions was performed. Because both post-infectious IBS [22] and this genetic variant are both associated with psychological comorbidity and immune responses, we postulate that there may be an underlying gene-environment interaction between this SNP and gastrointestinal infections.
The aim of this study was to assess the association of these polymorphisms with IBS. Because the 5HTT-LPR S allele results in less transcription of the serotonin transporter and results in increased availability of synaptic serotonin, we hypothesized that it would be associated with IBS-D, and conversely, that the L allele would be associated with IBS-C. Because of the association of the 825T allele with dyspepsia, we hypothesized that it would also be associated with IBS. Our second aim was to assess whether there are significant gene-environment interactions in the development of IBS, specifically if there is an interaction between the 5HTT LPR S allele and abuse as well as interactions between the 825T allele with prior gastrointestinal infections. To this end, a case-control candidate gene association study of these two polymorphisms was performed in a sample of 385 cases and 262 controls.
METHODS
Study design
The study employed a case-control study design embedded in an ongoing family case-control study conducted at a major medical center in the Upper Midwest of the United States. This project was approved by the Mayo Clinic Institutional Review Board (IRB).
Subject recruitment
A total of 647 subjects—385 cases and 262 controls—were selected for this study. The cases of this study, prospectively recruited between July 2004 and June 2007, were adult outpatients with IBS, ages 18 to 70, seen at a single medical institution. Cases were identified by reviewing their indication for an appointment within the GI Division, by direct referral by a treating physician, through recruitment at an IBS patient education class, or by using a research database of medical records at Mayo Clinic that is updated daily and was programmed to search for patients with IBS listed among their final diagnoses during the recruitment period. Unrelated controls were frequency matched to cases on age-decile, gender, and race. Controls were prospectively recruited between February 2005 and July 2007 from patients seen in the Division of General Internal Medicine and via identification of the research database, with recruitment purposefully lagging behind cases to allow for frequency matching. Recruitment of case and control probands was performed in person if being seen in clinic or by mail if identified through the computerized database. Potential controls were also asked to complete an initial one-page screening questionnaire to identify exclusion criteria prior to signing consent. Case and control probands were excluded if they were under 18 years of age, had difficulty reading or speaking English, were non-U.S. residents, were prisoners, had dementia or mental retardation, had a condition requiring a legal guardian, or were adopted. Case and control probands were also excluded if they had a current diagnosis of cancer or had another gastrointestinal diagnosis (e.g. inflammatory bowel disease, celiac sprue, history of major abdominal surgery) that could produce IBS-like symptoms. Formal chart review of probands was performed to confirm the clinical diagnosis of IBS and rule out alternate diagnoses in cases, to exclude clinical diagnosis of IBS in controls, to screen for exclusion criteria, document relevant medical diagnoses, document social history, and review results of all gastrointestinal testing. Controls were excluded if they reported a diagnosis of IBS or if they met Rome I or II diagnostic criteria [23,24] for IBS on either the screening or study questionnaire. If consented, all probands were asked to complete a self-reported bowel symptom questionnaire and donate 20 ml of blood, in addition to providing contact information for first-degree relatives for the family portion of the study. Only cases and control subjects who met inclusion criteria and completed all aspects of the study were included in the final sample. Data from relatives were not included in this study.
Study questionnaires
The short screening questionnaire completed by controls prior to recruitment asked about a past diagnosis of IBS and validated questions pertaining to the Rome III diagnostic criteria [25]. The proband (case and control) study questionnaire contained items regarding demographics, gastrointestinal symptoms, social history, medical history, dietary history, somatization, anxiety and depression, and family history, and contained the following previously developed and validated survey instruments: Talley Bowel Disease Questionnaire [26], Psychosomatic Symptom Checklist [27], Beck Anxiety Index [28], Center for Epidemiological Studies Depression Scale [29], and IBS-Quality of Life (IBS-QoL) [30,31].
Medical chart review data
All patients seen at our institution completed standardized forms collecting information about their past medical history, social history, and family medical history for clinical use. Data collected by chart review for this study included: gastrointestinal diseases, abuse history, and psychiatric disorders (anxiety, depression, or mental illness).
Study definitions
IBS subtypes were based on the supportive symptom combination as suggested by the Rome II criteria [24]. Constipation-predominant IBS (C-IBS) was defined by meeting either Rome I or II criteria, but endorsing one or more constipation symptoms (less than 3 bowel movements per week, hard stools, straining) and no diarrhea symptoms (greater than 3 bowel movements per day, loose stools, urgent stools). Conversely, diarrhea-predominant IBS (D-IBS) was defined by meeting either Rome criteria, and endorsing one or more diarrhea symptoms but no constipation symptoms. Mixed-IBS (M-IBS) was defined as meeting either Rome but not meeting criteria for C-IBS or D-IBS. Functional constipation (FC), functional diarrhea (FD), and functional bloating (FB) were also based on Rome II definitions [24]. FC was defined as not meeting either Rome criteria for IBS, endorsing two or more constipation symptoms, and not endorsing loose stools. FD was defined as not meeting either Rome criteria for IBS, endorsing frequent loose stools, and no/rare abdominal pain. FB was defined as not meeting either Rome criteria for IBS, but endorsing frequent bloating or distension. Unspecified functional bowel disorder (UFBD) was reserved for any case who did not meet Rome criteria for IBS, endorsing some Rome symptoms, but did not meet criteria for FC, FD, or FB. Abuse was defined by reporting verbal or physical abuse on the study questionnaire. Gastrointestinal infection was defined by an endorsement of any of the following infections: Campylobacter, Salmonella, Shigella, Yersinia, E. coli, Clostridium difficile, Giardia, Microsporidia, or other. Depression and anxiety were defined by questionnaire reporting of these diagnoses in their medical history. Parental alcohol abuse and parental psychiatric disease were defined as a positive endorsement of the above on a standardized personal/family history form completed by all patients been seen as an outpatient or inpatient at our institution, as collected by medical chart review.
Genetic analysis
Lymphocyte DNA from cases and controls was extracted using a Gentra Autopure ®™ (Gentra Systems, Inc. Minneapolis, MN USA) automated DNA extractor, and DNA quantification was performed with a spectrophotometer (SPECTRAmax PLUS 384, Nanodrop Technologies, Wilmington, DE, USA). Genotyping of specimens for the 5-HTT LPR and GNβ3 825C>T polymorphisms was performed in the institutional Genotyping Shared Resource (GSR) facility. Participant identifiers were removed except for study identification numbers. Laboratory personnel were blinded to subject characteristics, DNA specimens were randomly plated, cases and control samples were assessed in the same assays, and positive and negative control DNA samples were included, with automated data entry.
Genotyping of the 5-HTT LPR and GNβ3 825C>T polymorphisms was performed using the Thermo Electron Hybrid MBS thermal cycler (Thermal Electron Corp., Waltham, MA, USA). The forward primer for the 5-HTT LPR polymorphism was 5′-GTT TCT TGA GGG ACT GAG CTG GAC AAC CAC-3′. The reverse primer used for this polymorphism was 5′-GGC GTT GCC GCT CTG AAT GC-3′ (VIC labeled). The Roche Expand Long Template PCR kit (Roche Applied Science, Indianapolis, IN) was used with 7.36μl H2O; 0.4μl DNTPs mix; 1μl Roche Expand Buffer no. 1; 0.1 μl each primer (25μmol l−1); 0.04 μl Roche Expand Taq 94°C, 3 min; 40 cycles of: 94°C, 30 s; 58°C, 30 s; and 72°C, 1 min; followed by 72°C 10 min. The samples were then processed on the Applied Biosystems 3100 16 capillary system (Applied Biosystems, Foster City, CA, USA). DNA fragments were used for base calling and precision sizing of samples. Allelic calls were done using Applied Biosystems GeneMapper v3.5 software (Applied Biosystems). For GNβ3 825C>T polymorphism, the forward primer used was 5′-GAC GCT TCC TGC CGC TGG T-3′; the reverse primer used was 5′-GCT GGC CCT TAC CCA CAC GCT-3′ (biotinylated); the sequencing primer used was 5′-GCG GTCA TCA CGT C-3′. Polymerase chain reaction substrates and conditions included 11.4 μl H2O, 1.6 μl MgCl2, 4 μl 5x ABI buffer, 0.4 μl each primer (25 μmol L−1), 0.2 μl Taq Gold (Applied Biosystems) 95°C, 12 min; 40 cycles of: 95°C, 30 s; 63°C, 30 s; and 72°C, 30 s; followed by 72°C, 10 min. The samples were processed on the PSQ™ 96 machine (Biotage, Uppsala, Sweden). SNP calls were done using PSQ 96 Evaluation Software.
Statistical analysis
Demographic comparisons used t-tests and chi-square tests for continuous and categorical data, respectively. Allele and genotype frequencies (%) were calculated for each polymorphism among cases and controls. Comparison of these frequencies between cases and controls was performed using chi-square analysis (or Fisher’s exact tests) and Armitage trend tests depending on the genetic model. Allele and genotype frequency comparisons were also performed with logistic regression for IBS (cases v. controls) and linear regression for age of onset. Ordinal (e.g 0=LL, 1=SL, 2=SS), dominant (e.g. SS or SL v. LL), and recessive (e.g. SS v. SL or LL) genetic effects as well as environmental effects and gene-environment interactions were evaluated in genetic models. SAS statistical software (SAS Institute Inc., Cary, NC, USA) was utilized for analyses.
RESULTS
Subject characteristics
As shown in Table 1, the median age of the 647 study participants was 50 years old (range 18–70), 498 (77%) were female, and 631 (98%) were Caucasian. Controls were similar to cases with respect to age, but there were more females in cases than controls (83% v. 67%, p<0.001). Overall, 267 (69%) of cases met Rome I or II criteria for IBS. When these individuals were broken down into IBS subtypes, 10% were IBS-C, 26% were IBS-D, and 32% were IBS-M. Among the 118 (31%) individuals who did not meet Rome criteria, 23 (6%) met criteria for functional constipation, and 1 (0%) met criteria for functional diarrhea. Cases were more likely than controls to have a higher somatization score (case median 1.1 v. control median 0.4, p<0.001), and to report an abuse history, a personal psychiatric disorder, a parent with a psychiatric disorder, and prior gastrointestinal infection (all p<0.001, Table 1). There was no association between a parental alcohol abuse history and IBS,
Table 1.
Subject Characteristics
| Cases n=385 |
Controls n=262 |
|
|---|---|---|
|
| ||
| Median age (range) | 49 (18–70) | 51 (19.6–69.5) |
|
| ||
| Female (%) | 320 (83%) | 178 (67%)* |
|
| ||
| Race | ||
| Caucasian, n (%) | 378 (98%) | 253 (97%) |
|
| ||
| Rome I or II criteria | ||
| Yes, n (%) | 267 (69%) | 0 (0%) |
| IBS-C, n (%) | 40 (10%) | |
| IBS-D, n (%) | 102 (26%) | |
| IBS-M, n (%) | 125 (32%) | |
| No, n (%) | 74 (19%) | |
| Functional constipation, n (%) | 23 (6%) | |
| Functional diarrhea, n (%) | 1 (0%) | |
| Unspecified bowel disorder, n (%) | 50 (25%) | |
| Insufficient responses, n (%) | 44 (11%) | |
|
| ||
| Median somatization level (range)* | 1.1 (0.0, 3.5) | 0.4 (0.4, 2.5) |
|
| ||
| Psychiatric Disorder, n (%) | 213 (55%) | 48 (18%)* |
|
| ||
| Abuse history, n (%) | 146 (38%) | 32 (13%)* |
|
| ||
| Parental alcohol abuse, n (%) | 68 (18%) | 36 (14%) |
|
| ||
| Parental psychiatric disorder, n (%) | 106 (28%) | 26 (10%)* |
|
| ||
| Prior gastrointestinal infection, n (%) | 79 (21%) | 15 (6%)* |
p<0.001
5-HTT LPR and GNβ3 825C>T results
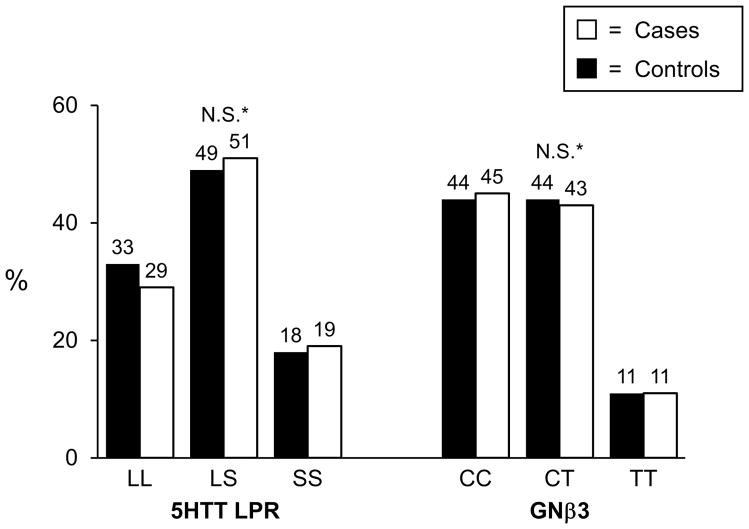
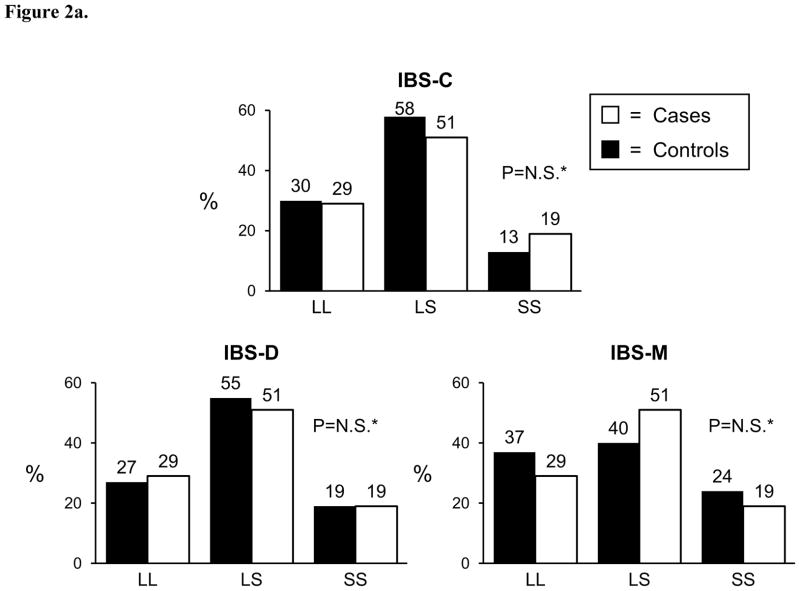
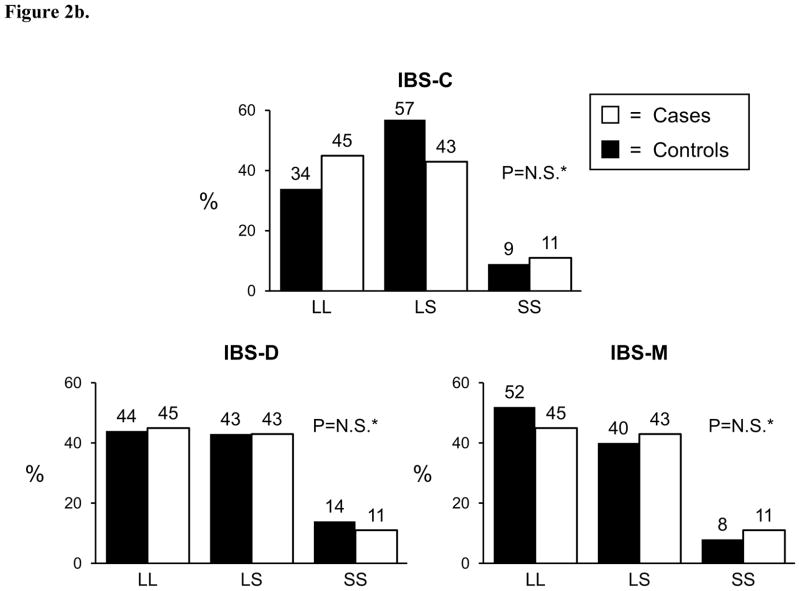
Genotyping was successfully performed for the 5-HTT LPR polymorphism in 374 (97%) cases and 262 controls; genotyping was successful for the GNβ3 825C>T polymorphism in 342 cases and 253 controls. Both polymorphisms were found to be in Hardy-Weinberg equilibrium among controls (p>0.05). The genotyping frequencies for both variants are shown in Figure 1 where no differences were observed between cases and controls. Similarly, allele frequencies for the 5-HTT LPR S allele was 43% in cases and 45% in controls (p=0.37), and the allele frequencies for the GNβ3 825T allele was 34% in cases and 33% in controls (p=0.85). When cases were divided into symptom predominant subtypes, again no genotype or allele frequency differences were observed between cases and controls (Figure 2a and 2b). Conclusions did not change when those with functional constipation were combined with C-IBS patients and compared with controls, or when those with functional diarrhea were combined with D-IBS patients and compared with controls (data not shown). Neither variant predicted age of onset of IBS, irrespective of genetic model tested. To ensure that racial differences were not affecting results, the analyses were repeated evaluating Caucasians only. Conclusions did not change with these restricted analyses.
Figure 1. IBS Cases vs. Controls.
*ordinal, dominant, recessive p –values > 0.05; N.S.=not significant -genotype frequencies in Hardy-Weinberg equilibrium among controls
Figure 2.
Figure 2a. 5-HTT LPR
*p-value for ordinal, dominant, recessive > 0.05; N.S.=not significant
Figure 2b. GNβ3
*p-value for ordinal, dominant, recessive > 0.05; N.S.=not significant
Gene-environment interactions
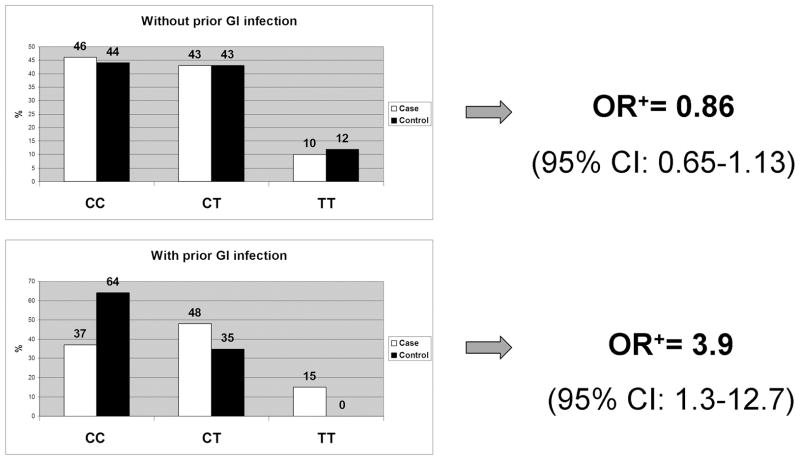
Four environmental risk factors were evaluated for gene-environment interactions with the two genetic variants of interest: history of abuse, parental history of alcohol abuse, parental history of psychiatric disorder, and history of intestinal infections. A significant interaction was observed between the GNβ3 825C>T polymorphism and prior gastrointestinal infections (p=0.02) (Figure 3). For example, among those without a prior infection, the odds ratio was 0.86 (95% CI: 0.65–1.13) for predicting IBS in a multivariate model adjusting for gender, abuse, parental psychiatric disorder, and infection. In contrast, among those with a prior infection, the polymorphism conferred a four-fold increased odds of IBS with an odds ratio of 3.9 (95% CI: 1.2–12.7). Prediction accuracy of the model with these variables was 72.7% (AUC=0.727). No gene-environment interactions were observed for either polymorphism with abuse, parental alcohol abuse, or parental psychiatric disorder. For example, for the multivariate model adjusting for age and gender evaluating gene-environment interactions between 5-HTTLPR and abuse, the OR was 0.74 (95%CI: 0.43–1.28) among those with abuse with a similar OR of 0.96 (95%CI: 0.73–1.26) among those without abuse (p>0.05).
Figure 3.
GxE interaction: GNβ3 825 C>T x GI infection. A significant interaction was observed between the GNβ3 825C>T polymorphism and prior gastrointestinal infections (p=0.02)
+adjusted for gender, abuse, parental psychiatric disorder, genotype, infection, and interaction term
DISCUSSION
Despite great interest in the role of 5-HTT LPR and GNβ3 825C>T polymorphisms in psychiatric disease and functional dyspepsia, this large case-control study of 385 patients with IBS and 262 controls found no association between the 5-HTT LPR and GNβ3 825C>T polymorphisms and IBS, nor the IBS subtypes. Furthermore, neither variant predicted age of onset of IBS—a genetic trait commonly observed in other disease susceptibility loci.
We anticipated that, like others have observed [15], there would be no association between the serotonin transporter promoter polymorphism with IBS overall, but speculated that the S allele may be associated with IBS-D and that the L allele would be associated with IBS-C. Mechanistically, one would anticipate that the less efficient transcription of the S allele would result in more serotonin in neuronal synapses, resulting in more post-synaptic stimulation of serotonin receptors. Conversely, one would have anticipated that the L allele would result in less synaptic serotonin, and thus, less serotonin receptor activation. Our findings did not support this hypothesis. Our sample size for each IBS subgroup was moderately large (n≥100), with the only exception being the IBS-C group. The allele and genotype frequencies observed in our control group for both polymorphisms are comparable to that reported by others [32,33], lending strength to the validity of our study results. This suggests that this genetic variant either plays no role in IBS pathophysiology, its effect on serotonin transporter function in the context of IBS are modest, or that there are other genetic, environmental, or physiological factors that attenuate its effect.
Furthermore, because IBS and functional dyspepsia appear to overlap significantly [34] and because the 825C>T polymorphism has been linked with dyspepsia, we hypothesized that the polymorphism would be associated with IBS as well. The lack of an association between this genetic variant and IBS and its subtypes suggest that there may be a different biological mechanisms between the two disorders despite the overlap. However, although no direct association was observed between this functional polymorphism and IBS, we did observe a statistically significant interaction between this variant and prior infections, whereby this polymorphism conferred a four-fold risk increase in the development of IBS. Because the T allele has been linked to increase lymphocytic chemotaxis [19,35], this interaction with intestinal infections raises questions as to whether this polymorphism, in the presence of infection and due to an altered or varied immune response, may somehow result in altered GI function and/or sensation in genetically susceptible individuals. One significant methodological concern is that we relied on self-report of infection in this study, which may have led to under- or over-reporting of events. Under-reporting may have resulted from either lack of health-care seeking to identify the specific organism during illness or due to poor recall of a remote illness. Over-reporting of infections, particularly by cases, due to recall bias (i.e. those with IBS are more likely to remember a previous gastrointestinal infection) is a potential confounder. Furthermore, as this was not a prospective study, we can not definitively know that the infection preceded IBS symptom onset. Ideally, this positive finding of gene-environment interaction should be reproduced in another separate cohort to determine whether this was simply a spurious finding or a biologically-based, real observation.
In contrast to the psychiatric literature that has repeatedly reported that there appears to be an interaction between the 5-HTT LPR polymorphism and life events such as stress, childhood adversity, and abuse [11,16], we did not observe a gene-environment interaction between the 5-HTT LPR polymorphism and abuse with IBS status. We were interested in investigating the same gene-environment interaction in IBS because psychiatric comorbidity is common in IBS [36,37], abuse is common in IBS [38], IBS has been observed in post-traumatic stress disorder in veterans [39], and some mental health experts include IBS as part of an affective spectrum disorder [40]. Our results suggest a different biological—or at least genetic-- mechanism for depression and anxiety compared with IBS. Nonetheless, we acknowledge that the method by which we collected information on abuse, parental alcoholism and psychiatric disease through self-report and medical chart review is likely imperfect with high risk for under-reporting, and thus, may in part explain our negative findings with respect to these specific gene-environment interactions. We are currently collecting data on early life trauma using a validated questionnaire, and will have the opportunity in the future to re-evaluate this specific methodological issue.
In summary, we found that two well-known, functional polymorphisms—5-HTT LPR and GNβ3 825C>T—did not appear to be directly associated with IBS, IBS subtypes or IBS age of onset in this large case-control study of patients with a clinical diagnosis of IBS. However, we did observe a statistically significant gene-environment interaction between the 825C>T polymorphism and prior bacterial infections that warrants further investigation to determine if our findings are confirmed. If the latter finding is a real finding, it suggests that IBS truly is a complex genetic disorder with both a genetic and environmental basis for symptom development and manifestations.
Acknowledgments
Grant Support: This study was supported in part by research grants from the National Institutes of Health (DK066271), the American Gastroenterological Association, and Solvay Pharmaceuticals.
The authors would like to thank Lori R. Anderson for her assistance in preparing this manuscript.
Footnotes
Conflict of Interest: None
Author Contributions
YAS: Designed the research study, provided oversight in execution and analysis of data, and wrote the paper.
JJL: Analyzed the data
EJA: Analyzed the data
ER: Analyzed the data.
AEA: Performed the research (e.g. recruitment, data entry, etc.)
GMP: Assisted with research design and analysis
NJT: Assisted with research design and assisted with writing and editing the manuscript.
References
- 1.Saito YA, Zimmerman JM, Harmsen WS, et al. Irritable bowel syndrome aggregates strongly in families: A family-based case-control study. Neurogastroenterol Motil. 2008;20:790–797. doi: 10.1111/j.1365-2982.2007.1077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Locke GR, 3rd, Zinsmeister AR, Talley NJ, Fett SL, Melton LJ., 3rd Familial association in adults with functional gastrointestinal disorders. Mayo Clin Proc. 2000;75:907–912. doi: 10.4065/75.9.907. [DOI] [PubMed] [Google Scholar]
- 3.Whorwell PJ, McCallum M, Creed FH, Roberts CT. Non-colonic features of irritable bowel syndrome. Gut. 1986;27:37–40. doi: 10.1136/gut.27.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohammed I, Cherkas LF, Riley SA, Spector TD, Trudgill NJ. Genetic influences in irritable bowel syndrome: A twin study. Am J Gastroenterol. 2005;100:1340–1344. doi: 10.1111/j.1572-0241.2005.41700.x. [DOI] [PubMed] [Google Scholar]
- 5.Morris-Yates A, Talley NJ, Boyce PM, Nandurkar S, Andrews G. Evidence of a genetic contribution to functional bowel disorder. Am J Gastroenterol. 1998;93:1311–1317. doi: 10.1111/j.1572-0241.1998.440_j.x. [DOI] [PubMed] [Google Scholar]
- 6.Bengtson MB, Ronning T, Vatn MH, Harris JR. Irritable bowel syndrome in twins: Genes and environment. Gut. 2006;55:1754–1759. doi: 10.1136/gut.2006.097287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy RL, Jones KR, Whitehead WE, Feld SI, Talley NJ, Corey LA. Irritable bowel syndrome in twins: Heredity and social learning both contribute to etiology. Gastroenterology. 2001;121:799–804. doi: 10.1053/gast.2001.27995. [DOI] [PubMed] [Google Scholar]
- 8.Saito YA, Petersen GM, Locke GR, 3rd, Talley NJ. The genetics of irritable bowel syndrome. Clin Gastroenterol Hepatol. 2005;3:1057–1065. doi: 10.1016/s1542-3565(05)00184-9. [DOI] [PubMed] [Google Scholar]
- 9.Camilleri CE, Carlson PJ, Camilleri M, et al. A study of candidate genotypes associated with dyspepsia in a U.S. Community. Am J Gastroenterol. 2006;101:593–595. doi: 10.1111/j.1572-0241.2006.00481.x. [DOI] [PubMed] [Google Scholar]
- 10.Holtmann G, Siffert W, Haag S, et al. G-protein beta 3 subunit 825 cc genotype is associated with unexplained (functional) dyspepsia. Gastroenterology. 2004;126:971–979. doi: 10.1053/j.gastro.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Vergne DE, Nemeroff CB. The interaction of serotonin transporter gene polymorphisms and early adverse life events on vulnerability for major depression. Curr Psychiatry Rep. 2006;8:452–457. doi: 10.1007/s11920-006-0050-y. [DOI] [PubMed] [Google Scholar]
- 12.Serretti A, Chiesa A, Calati R, Perna G, Bellodi L, De Ronchi D. Common genetic, clinical, demographic and psychosocial predictors of response to pharmacotherapy in mood and anxiety disorders. Int Clin Psychopharmacol. 2009;24:1–18. doi: 10.1097/YIC.0b013e32831db2d7. [DOI] [PubMed] [Google Scholar]
- 13.Fukudo S, Kanazawa M, Mizuno T, et al. Impact of serotonin transporter gene polymorphism on brain activation by colorectal distention. NeuroImage. 2009;47:946–951. doi: 10.1016/j.neuroimage.2009.04.083. [DOI] [PubMed] [Google Scholar]
- 14.Camilleri M, Atanasova E, Carlson PJ, et al. Serotonin-transporter polymorphism pharmacogenetics in diarrhea-predominant irritable bowel syndrome. Gastroenterology. 2002;123:425–432. doi: 10.1053/gast.2002.34780. [DOI] [PubMed] [Google Scholar]
- 15.Van Kerkhoven LA, Laheij RJ, Jansen JB. Meta-analysis: A functional polymorphism in the gene encoding for activity of the serotonin transporter protein is not associated with the irritable bowel syndrome. Aliment Pharmacol Ther. 2007;26:979–986. doi: 10.1111/j.1365-2036.2007.03453.x. [DOI] [PubMed] [Google Scholar]
- 16.Brown GW, Harris TO. Depression and the serotonin transporter 5-httlpr polymorphism: A review and a hypothesis concerning gene-environment interaction. J Affect Disord. 2008;111:1–12. doi: 10.1016/j.jad.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Rosskopf D, Busch S, Manthey I, Siffert W. G protein beta 3 gene: Structure, promoter, and additional polymorphisms. Hypertension. 2000;36:33–41. doi: 10.1161/01.hyp.36.1.33. [DOI] [PubMed] [Google Scholar]
- 18.Lee HJ, Cha JH, Ham BJ, et al. Association between a g-protein beta3 subunit gene polymorphism and the symptomatology and treatment responses of major depressive disorders. Pharmacogenomics J. 2004;4:29–33. doi: 10.1038/sj.tpj.6500217. [DOI] [PubMed] [Google Scholar]
- 19.Lindemann M, Virchow S, Ramann F, et al. The G protein beta3 subunit 825T allele is a genetic marker for enhanced T cell response. FEBS Lett. 2001;495:82–86. doi: 10.1016/s0014-5793(01)02339-0. [DOI] [PubMed] [Google Scholar]
- 20.Camilleri M, Busciglio I, Carlson P, et al. Candidate adrenergic, GMβ3, and serotonergic polymorphisms, endophenotype, pharmacogenetics of clonidine in irritable bowel syndrome and health. Gastroenterology. 2008;134:A-682. [Google Scholar]
- 21.Andresen V, Camilleri M, Kim HJ, et al. Is there an association between GMβ3-C825T genotype and lower functional gastrointestinal disorders? Gastroenterology. 2006;130:1985–1994. doi: 10.1053/j.gastro.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 22.Thabane M, Kottachchi D, Marshall J. Systematic review and meta-analysis: The incidence and prognosis of post-infectious irritable bowel syndrome. Aliment Pharmacol Ther. 2007;26:535–544. doi: 10.1111/j.1365-2036.2007.03399.x. [DOI] [PubMed] [Google Scholar]
- 23.Thompson WG, Creed FH, Drossman DA, Heaton KW, Mazzacca G. Functional bowel disease and functional abdominal pain. Gastrenterol Int. 1992;5:75–91. [Google Scholar]
- 24.Thompson WG, Longstreth GF, Drossman DA, Heaton KW, Irvine EJ, Muller-Lissner SA. Functional bowel disorders and functional abdominal pain. Gut. 1999;45:II43–47. doi: 10.1136/gut.45.2008.ii43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 26.Talley NJ, Phillips SF, Wiltgen CM, Zinsmeister AR, Melton LJ., 3rd Assessment of functional gastrointestinal disease: The bowel disease questionnaire. Mayo Clin Proc. 1990;65:1456–1479. doi: 10.1016/s0025-6196(12)62169-7. [DOI] [PubMed] [Google Scholar]
- 27.Attanasio V, Andrasik F, Blanchard EB, Arena JG. Psychometric properties of the sunya revision of the psychosomatic symptom checklist. J Behav Med. 1984;7:247–257. doi: 10.1007/BF00845390. [DOI] [PubMed] [Google Scholar]
- 28.Beck AT, Steer RA. Manual for the Beck Anxiety Inventory. San Antonio: Psychological Corporation; 1990. [Google Scholar]
- 29.Haringsma R, Engels GI, Beekman AT, Spinhoven P. The criterion validity of the center for epidemiological studies depression scale (ces-d) in a sample of self-referred elders with depressive symptomatology. Int J Geriatr Psychiatry. 2004;19:558–563. doi: 10.1002/gps.1130. [DOI] [PubMed] [Google Scholar]
- 30.Patrick DL, Drossman DA, Frederick IO, DiCesare J, Puder KL. Quality of life in persons with irritable bowel syndrome: Development and validation of a new measure. Dig Dis Sci. 1998;43:400–411. doi: 10.1023/a:1018831127942. [DOI] [PubMed] [Google Scholar]
- 31.Drossman DA, Patrick DL, Whitehead WE, et al. Further validation of the ibs-qol: A disease-specific quality-of-life questionnaire. Am J Gastroenterol. 2000;95:999–1007. doi: 10.1111/j.1572-0241.2000.01941.x. [DOI] [PubMed] [Google Scholar]
- 32.Lesch KP, Bengel D, Heils A, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 33.Siffert W, Forster P, Jockel KH, et al. Worldwide ethnic distribution of the g protein beta3 subunit 825t allele and its association with obesity in caucasian, chinese, and black african individuals. J Am Soc Nephrol. 1999;10:1921–1930. doi: 10.1681/ASN.V1091921. [DOI] [PubMed] [Google Scholar]
- 34.Locke GR, 3rd, Zinsmeister AR, Fett SL, Melton LJ, 3rd, Talley NJ. Overlap of gastrointestinal symptom complexes in a U.S. community. Neurogastroenterol Motil. 2005;17:29–34. doi: 10.1111/j.1365-2982.2004.00581.x. [DOI] [PubMed] [Google Scholar]
- 35.Virchow S, Ansorge N, Rubben H, Siffert G, Siffert W. Enhanced fMLP-stimulated chemotaxis in human neutrophils from individuals carrying the G protein beta3 subunit 825 T-allele. FEBS Lett. 1998;436:155–158. doi: 10.1016/s0014-5793(98)01110-7. [DOI] [PubMed] [Google Scholar]
- 36.Whitehead WE, Palsson O, Jones KR. Systematic review of the comorbidity of irritable bowel syndrome with other disorders: What are the causes and implications? Gastroenterology. 2002;122:1140–1156. doi: 10.1053/gast.2002.32392. [DOI] [PubMed] [Google Scholar]
- 37.Talley NJ, Howell S, Poulton R. The irritable bowel syndrome and psychiatric disorders in the community: Is there a link? Am J Gastroenterol. 2001;96:1072–1079. doi: 10.1111/j.1572-0241.2001.03741.x. [DOI] [PubMed] [Google Scholar]
- 38.Drossman DA. Irritable bowel syndrome and sexual/physical abuse history. Eur J Gastroenterol Hepatol. 1997;9:327–330. doi: 10.1097/00042737-199704000-00002. [DOI] [PubMed] [Google Scholar]
- 39.Gray GC, Reed RJ, Kaiser KS, Smith TC, Gastanaga VM. Self-reported symptoms and medical conditions among 11,868 Gulf War-era veterans: The Seabee Health Study. Am J Epidemiol. 2002;155:1033–1044. doi: 10.1093/aje/155.11.1033. [DOI] [PubMed] [Google Scholar]
- 40.Hudson JI, Mangweth B, Pope HG, Jr, et al. Family study of affective spectrum disorder. Arch Gen Psychiatry. 2003;60:170–177. doi: 10.1001/archpsyc.60.2.170. [DOI] [PubMed] [Google Scholar]