Abstract
Objective
Epilepsy encompasses an extensive array of clinical and research subdomains, many of which emphasize multi-modal physiological measurements such as electroencephalography and neuroimaging. The integration of structured, unstructured, and signal data into a coherent structure for patient care as well as clinical research requires an effective informatics infrastructure that is underpinned by a formal domain ontology.
Methods
We have developed an epilepsy and seizure ontology (EpSO) using a four-dimensional epilepsy classification system that integrates the latest International League Against Epilepsy terminology recommendations and National Institute of Neurological Disorders and Stroke (NINDS) common data elements. It imports concepts from existing ontologies, including the Neural ElectroMagnetic Ontologies, and uses formal concept analysis to create a taxonomy of epilepsy syndromes based on their seizure semiology and anatomical location.
Results
EpSO is used in a suite of informatics tools for (a) patient data entry, (b) epilepsy focused clinical free text processing, and (c) patient cohort identification as part of the multi-center NINDS-funded study on sudden unexpected death in epilepsy. EpSO is available for download at http://prism.case.edu/prism/index.php/EpilepsyOntology.
Discussion
An epilepsy ontology consortium is being created for community-driven extension, review, and adoption of EpSO. We are in the process of submitting EpSO to the BioPortal repository.
Conclusions
EpSO plays a critical role in informatics tools for epilepsy patient care and multi-center clinical research.
Keywords: Epilepsy and Seizure Ontology, Patient Data Capture, Clinical Free Text Processing, Clinical Data Integration
Objectives
The management of epilepsy faces many of the challenges described in the 2010 report on health information technology by the President's Council of Advisors on Science and Technology, including: (a) lack of consistent terminology used in data annotation; (b) use of disparate mechanisms to capture patient information at the point of care; (c) little support for data interoperability; and (d) significant challenges in the secondary use of clinical data.1 Epilepsy is the most common serious neurological disorder, affecting 50–60 million individuals worldwide.2 Data management challenges in epilepsy are increasing with improvements in neuroimaging techniques, the integration of data from high-throughput ’omics pipelines, and the increasing availability of multi-channel neurophysiologic datasets.3
Epilepsy data, such as patient's diagnosis or electrophysiological signals, are often generated in a wide range of settings using heterogeneous protocols, and use disparate terminology to describe the same information or identical terms to describe distinct information (also called ‘semantic heterogeneity’ in database research4). For example, a seizure with alteration of consciousness may be termed a complex partial seizure, a dialeptic seizure, or a focal dyscognitive seizure by different epilepsy experts. Reconciliation of semantic heterogeneity is essential for data interoperability and facilitating the integration of patient data for multi-center clinical research. Manual curation of heterogeneous data is a costly and non-scalable approach for studies involving thousands of patients. To address this issue, ontologies are increasingly being used as common terminological resources to automatically reconcile data heterogeneity and implement large-scale, distributed data management systems.5
An ontology is a formal representation of knowledge in a given domain that allows both human users and software applications to consistently and accurately interpret domain terminology.6–8 Ontologies are now commonly used across biomedical domains for addressing data management challenges, including integration of heterogeneous data,9 supporting intuitive user interfaces,10 and representation of provenance metadata for ensuring data quality.11 In this paper, we describe the development of an epilepsy ontology and its application to support multi-center clinical studies, such as the ongoing project to study sudden unexpected death in epilepsy (SUDEP).
The Prevention and Risk Identification of SUDEP Mortality Project
The Prevention and Risk Identification of SUDEP Mortality (PRISM) project is a multi-center study funded by the National Institute of Neurological Disorders and Stroke (NINDS) to examine the risk factors underlying SUDEP. SUDEP is defined as a sudden, unexpected, witnessed/un-witnessed, non-traumatic, and non-drowning death of persons with epilepsy.12 Its causes are not well understood and there is no effective prevention.13 PRISM is part of a ‘SUDEP Center Without Walls’ initiative funded in part because the low annual incidence (1% in high risk populations) of SUDEP that requires a multi-center study. The project involves epilepsy monitoring units (EMU) at the University Hospital Case Medical Center (UH-CMC, Case Western Reserve University, Cleveland), the Ronald Reagan Medical Center (University of California, Los Angeles), the Northwestern Memorial Hospital (NMH, Chicago), and the National Hospital for Neurology and Neurosurgery (NHNN, London). The project required the development of an informatics infrastructure to manage data from all participating EMUs, including:
A standardized tool to enter patient information at different points of care;
An integrated signal processing tool that allows clinicians to seamlessly interface between signal data and patient information;
An epilepsy-focused natural language processing (NLP) tool to extract structured information from clinical free text in patient records; and
A secure query environment to identify patient cohorts across multiple study centers.
These informatics tools are underpinned by a domain ontology that is also closely aligned with existing epilepsy classification systems.
Epilepsy and seizure classification systems
The International League Against Epilepsy (ILAE) Classification and Terminology Commission (CTC) has created classification systems to promote consistent use of epilepsy and seizure terms across different platforms.14 15 But the existing classification systems are based on a schema proposed in 1961 and do not reflect recent advances in knowledge.3 There is broad consensus in the epilepsy community that the current system is inadequate and needs to be updated.16–18 In 2010, the ILAE CTC proposed a radical new approach that aims to incorporate recent advances in molecular biology, genetics, and neuroimaging techniques in a new classification system.3 Specifically, the CTC report described two key requirements for a new classification system, namely:
Flexibility to evolve with advances in diagnostic techniques and domain knowledge; and
Ability to provide dynamic classification of epilepsy types along multiple dimensions for different types of applications and users.
An ontology created using formal knowledge representation languages is well suited to meet these requirements and also to play an important role in the development of effective epilepsy focused informatics tools.
Related work
The two prior efforts to develop an epilepsy ontology use the old epilepsy classification system that was proposed in 1981 and do not reflect changes suggested by the 2010 ILAE CTC report. The first ontology integrates user-defined rules with a class hierarchy of epilepsy terms, but it has multiple modeling issues, for example ‘Reason’ and ‘History’ have overlapping extent and intent but are modeled as disjoint sibling classes.19 Similarly, the class ‘EEGPatient’ represents patients diagnosed with epilepsy after review of EEG data, but is defined to be disjoint with the class ‘Patient,’ which could potentially lead to the incorrect inference that a person classified as a patient cannot also be classified as a patient with epilepsy (after EEG review).
The second project called EPILEPSIAE aims to create devices that would allow patients to predict seizures and manage the risk factors for epileptic seizures.20 This ontology also misclassifies the domain terms, for example both ‘Electrode’ and ‘Spike’ are defined as subclasses of the ‘Electroencephalography’ class (a spike is an EEG pattern). Hence, after initial review of both these projects and on the recommendations of the epileptologists involved in the PRISM project, we created the Epilepsy and Seizure Ontology (EpSO) to address their limited coverage of the epilepsy domain.
Background and significance: role of an epilepsy ontology
Biomedical ontologies have significantly enhanced the use of standardized terminology across many disciplines. Gene ontology (GO) is a notable example that is widely used to consistently annotate gene related information.21 The use of GO annotations has not only helped in the sharing and integration of genetic data, but has also been used to create analytical software applications that can mine GO annotated data for knowledge discovery.22 23 We envisage a similar set of applications to be supported by an epilepsy ontology that can be divided into two categories.
Common epilepsy terminology for electronic health record systems
The 2009 Health Information Technology for Economic and Clinical Health (HITECH) Act provided $20 billion in new funding to create electronic health records (EHR) for patients by 2014.24 In addition, the national health IT strategic vision emphasizes the implementation of a secure and robust health information exchange (HIE) network infrastructure to facilitate easier sharing of health information across care providers.24 A well-defined domain ontology is central to the development of effective EHRs that can be shared over the HIE infrastructure. The computer science research community has long realized the importance of a common schema that incorporates ‘domain semantics’ to facilitate data interoperability,4 and an epilepsy domain ontology can play a similar role in the evolving EHR systems.
An epilepsy ontology used by informatics tools to capture patient information, laboratory data, and event annotations on electrophysiological signals (collected during patient monitoring in EMUs) can be added to the hospital EHR systems using existing Health Level Seven (HL7) messaging interfaces.25 Ontology-aware query interfaces that are integrated with EHR systems can subsequently leverage the ontology annotations to support extensive query answering functionalities.
Integration of data dictionaries and biomedical ontologies
NINDS initiated the common data element (NINDS CDE) project in 2004 to create a common set of terms for consistent collection and analysis of data in neurological diseases, including epilepsy, Parkinson's disease, and stroke.26 The epilepsy specific CDEs in particular represent nine categories of terms, covering imaging, neurological exam, neuropsychology, seizures, and syndromes. The epilepsy CDEs are expected to be used in clinical studies to represent patient data, hence it is important to incorporate these data elements into an epilepsy ontology (either as classes or as class annotations). For example, the ‘Classification of Seizures’ case report form, which is part of the ‘Classification’ subdomain, describes seven CDEs that can be modeled in an epilepsy ontology, including ‘Seizure type,’ ‘Generalized seizure type,’ and ‘Focal seizure type.’ Together with ILAE classification terms, the epilepsy CDEs will enable the epilepsy ontology to be used as a single domain model with multiple levels of abstraction that can be easily integrated with a variety of informatics tools.
Many biomedical domains have created ontologies to model their terminological systems27 and many of these ontologies model epilepsy related terms. For example, the NeuroElectroMagnetic Ontologies (NEMO)28 and the Systematized Nomenclature of Medicine—Clinical Terms (SNOMED CT) can be used to describe a wide variety of signal and clinical concepts in epilepsy.29 In addition, there is increasing integration of ’omics data with clinical data in epilepsy research in an effort to understand the genetics of epilepsy and genomic basis of drug effectiveness in epilepsy.30 An epilepsy ontology can import concepts from existing ontologies to model anatomy, drug information, electrophysiological data, and genetic information (eg, the use of GO terms to link juvenile myoclonic epilepsy with the EFHC1 gene31). Further, using ontology mapping and alignment algorithms, datasets annotated with epilepsy ontology could be interoperable with datasets annotated with other ontologies.32
In the next section, we describe the ontology engineering approaches used to construct the epilepsy ontology. In the ‘Results’ section we describe the use of the epilepsy ontology in a set of informatics tools, in the ‘Discussion’ section we discuss our work in creating an epilepsy ontology consortium (EOC) and related work, and conclude in the final section.
Methods
EpSO is an ‘application ontology’ that aims to complement the efforts of the ILAE CTC. The epilepsy community recognizes the classification of individual patient cases along well-defined dimensions that directly impact decisions regarding prognosis and treatment.33 We use the ‘four-dimensional classification of epileptic seizures and epilepsies,’ which is a common approach used in many epilepsy centers,34 35 as the overarching framework to identify the scope and terms to be modeled in the first version of the ontology. EpSO uses the World Wide Web Consortium (W3C) recommended Web Ontology Language (OWL2).36 OWL2 is a formal knowledge representation language based on description logic that supports automated reasoning and can be used in web-based software applications.
The four-dimensional classification of epileptic seizures and epilepsies concepts modeled in EpSO
The components of the four-dimensional classification system are described below.
Seizures
A seizure is the ‘occurrence of signs and/or symptoms’ in patients due to abnormal electrophysiological brain activity.37 Seizure features are important for identifying symptoms, accurate diagnosis, and the prescription of appropriate anti-epileptic medications.34 EpSO models the details of epileptic seizures as two classes: (a) Seizure (all EpSO terms are italicized and are defined in the http://www.case.edu/EpSO.owl namespace); and (b) SeizureFeature. The Seizure class has three subtypes of seizures, namely EpilepticSeizure, NonEpilepticSeizure (seizures mimicking epileptic seizures), and ParoxysmalEvent (seizures that cannot be classified with certainty as epileptic or non-epileptic seizures). Aura is a seizure feature that usually occurs at the start of the seizure. EpSO models eight different aura categories that help in classifying seizures, for example AbdominalAura where the patient experiences sensations in the abdomen is usually associated with TemporalLobeEpilepsy.38
In addition to Aura, EpSO models five other categories of epileptic seizures, including AutonomicSeizure, DialepticSeizure, MotorSeizure, and SpecialSeizure, and their subtypes. For example, the two types of MotorSeizure, namely ComplexMotorSeizure and SimpleMotorSeizure, allow annotation of patient data for correlation with electroclinical activity in a particular part of the brain. These annotations can enable clinicians to correlate SimpleMotorSeizure movements with response elicited from patients who receive stimulation in the Brodmann areas 4 and 6 brain motor areas. Seizures that cannot be classified into one of the four epileptic seizure types are modeled as SpecialSeizure and include the subcategories of different negative motor seizures, such as AtonicSeizure, HypomotorSeizure, and AphasicSeizure.
One of the two types of SeizureFeature is LateralizingSign that can be directly correlated to the patient's EpileptogenicZone (EZ) and enables the clinician to classify the patient as having focal or generalized epilepsy. The various types of lateralizing signs include PostictalToddsParalysis, SignofFour, and IpsilateralBlinking, which can be combined with epileptic seizure type to associate a specific brain region with the seizure. For example, a DialepticSeizure preceded by AbdominalAura and followed by PostIctalAphasia (as a lateralizing sign) will allow a clinician to infer a left hemispheric seizure.38 EpSO explicitly associates different body parts with lateralizing signs using OWL2 class-level restriction (figure 1 illustrates the restriction for PostIctalNoseWiping as using the left or right hand).
Figure 1.
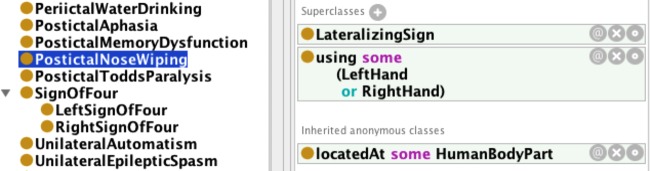
Lateralizing signs are associated with body parts using OWL2 existential restriction on the ontology class.
Location
Brain anatomy information associated with epilepsy is important for the treatment of patients. For example, anti-epileptic drugs have been found to be differentially effective in focal and generalized epilepsies, where the source of the seizure is localized or distributed across both hemispheres of the brain.33 The EZ is the primary concept associated with epilepsy location.39 EpSO models GeneralizedEpileptogenicZone and the different types of FocalEpileptogenicZone, such as BiLobarEpileptogenicZone, MultiFocalEpileptogenicZone, and HemisphericEpileptogenicZone. A ‘human-readable’ description of a concept and its syntactic variations, including synonyms and acronyms, are also modeled to support NLP tools.
EpSO comprehensively models the anatomical details of the human brain to enable accurate annotation of semiological seizure features, including EZ and epileptogenic networks. The Foundational Model of Anatomy (FMA) is a high quality anatomy reference ontology and its terms are extensively re-used in EpSO.40 For example, the subclasses of the fma:ImmaterialAnatomicalEntity and fma:MaterialAnatomicalEntity classes are used to model human brain anatomy and other body parts (‘fma’ refers to the http://sig.uw.edu/fma# namespace). Brain anatomy concepts are used to annotate EEG data, epileptogenic network and zone locations, seizure propagation patterns, and electrode placement. EpSO models both scalp electrodes and intracranial electrodes as well as the 10–20 and 10–10 placement schemes for scalp electrodes. Figure 2 illustrates the association of two ScalpElectrode subtypes with specific brain regions. The brain anatomy concepts include descriptions at a fine-level granularity allowing correlation of seizure patterns (recorded from the electrodes) to BrainGyrus and BrodmannArea.
Figure 2.
The location of scalp electrodes is defined in terms of specific brain regions to correlate EEG signals to anatomical location.
A semi-automated two-phase approach was used to add FMA concepts to EpSO. In the first phase, the relevant concepts from FMA were identified in consultation with the epileptologists and manually added to EpSO. In the second phase, all the OWL2 object properties defined for the FMA classes re-used in EpSO were automatically extracted and asserted in EpSO using an OWLAPI-based program.41
Etiology
The cause of epilepsy is important in the progression, treatment, and prognosis of the condition.16 The increased use of neuroimaging and genomic techniques have improved identification of Etiology, enabling clinicians to decide on therapy and evaluation prior to surgical intervention.33 EpSO models etiological subtypes according to ILAE CTC recommendations, which identified four etiology categories, namely GeneticDefect, MetabolicCause, StructuralCause, and UnknownCause.3 The GeneticDefect class includes epilepsy caused by GeneticMutation, such as mitochondrial encephalomyopathy, lactic acid, and stroke-like episodes (MELAS). The different structural causes for epilepsy, such as BirthInjury, CerebralPalsy, CerebroVascularDisease, and TraumaticBrainInjury, are modeled as subclasses of StructuralCause. EpSO also models the metabolic causes of epilepsy, for example, SubstanceAbuse and its subtype AlcoholAbuse.
Related medical conditions
This dimension factors in the presence of certain conditions that are relevant for treatment and follow-up perspective and allows a rapid, overall understanding of the patient's epilepsy (eg, neuropsychiatric problems and certain congenital neurological deficits). The different forms of NeuropsychiatricProblem (eg, Depression) and LearningDisability (eg, AutismSpectrumDisorder) are modeled in EpSO as types of Disease.
Additional ontology concepts—EEG patterns
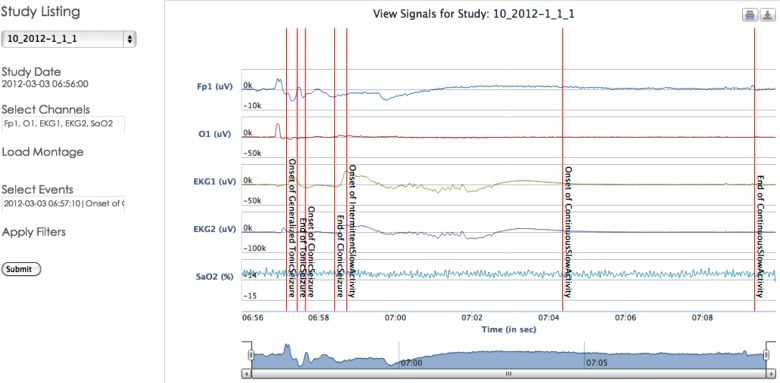
EEG signal data represent brain electrical activity and are used by clinicians to determine the location and extent of the EZ. EpSO models four categories of EEG patterns, namely AbnormalEEGPattern (eg, interictal spikes), BenignEEGPattern (eg, pyschomotor variant), NormalEEGPattern (eg, posterior head background), and SharpTransient (eg, wicket). Normal EEG data can be annotated with the subtypes of NormalEEGPattern, for example, BetaActivity, MuRhythm, TemporalSlowActivity, and LambdaWave. EEG patterns that cannot be classified as pathological but are not normal patterns are modeled as benign patterns with its subtypes, for example RhythmicBenignVariant. Figure 3 illustrates the use of EpSO terms to annotate multi-channel EEG signal data.
Figure 3.
Multi-channel signal data are annotated using EpSO classes to describe seizure and associated localized events.
Additional ontology concepts—drug information
Two categories of drug information are modeled in EpSO at present, namely anti-depressant and anti-epileptic drugs, using the RxNorm terminological system for clinical drugs.42 RxNorm includes mappings to other terminologies used in electronic medical record systems, such as the Veterans Health Administration National Drug File-Reference Terminology (NDF-RT), which facilitates the interoperability of drug information across different systems. The drug names were used together with the RxNav tool to navigate the RxNorm database and retrieve brand names as well as precise drug ingredients.43 At present, 70 ClinicalDrugComponents are modeled in EpSO, such as brand name (eg, Diastat) and its precise drug ingredient (eg, Diazepam).
Formal concept analysis for classification of epilepsy syndromes
Formal concept analysis (FCA) is a mathematical theory to identify concepts from a list of objects and arrange them in a concept lattice using their properties.44 45 OWL2 ontology classes can be interpreted as concepts in FCA using their extent and intent. The primary notion in FCA is a formal context (O, P, B) consisting of a set of objects (O), a set of properties (P), and a binary relation B⊂O × P. If o∈O and p ∈ P are in B, then (o, p) ∈ B, that is, object o has property p. FCA takes this binary relation and automatically generates a lattice hierarchy capturing the subclass relationship among the ‘formal concepts.’44 These formal concepts represent basic units of information by harmonizing subsets of intent and extent, which are compatible with classification approaches used by domain experts.
For EpSO, we enumerated all the semiological and anatomical properties of epilepsy syndromes in a two-dimensional matrix, which is a representation of the formal context and can be transformed into a concept lattice using an FCA algorithm.44 Figure 4 illustrates part of the concept lattice diagram generated from this matrix. The lattice diagram shows the classification of five epilepsy syndromes, namely TemporalLobeEpilepsy, BasalTemporalEpilepsy, HippocampalEpilepsy, AmygdalarEpilepsy, and LateralTemporalEpilepsy, based on their distinguishing attributes.
Figure 4.
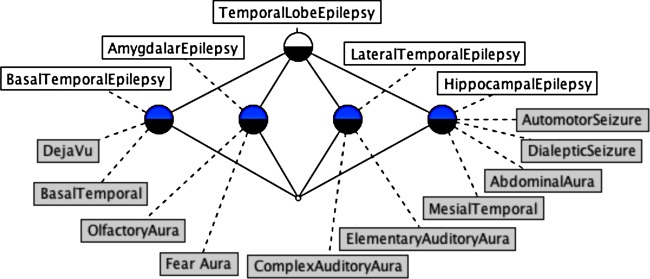
A fragment of the concept lattice for epilepsy syndromes (objects) and seizure semiology and anatomy (attributes).
The concept lattice classifies TemporalLobeEpilepsy as the parent concept of the other epilepsy syndromes because of the additional attributes associated with the subtypes. For example, BasalTemporalEpilepsy and LateralTemporalEpilepsy have the additional attributes of DejaVu and AuditoryAura, respectively, as semiological features that are not associated with the TemporalLobeEpilepsy concept. The result of the FCA algorithm is reviewed by epileptologists who can use the labeled terms in the concept lattice to identify incorrectly assigned or missing attributes.46 After verification by the epilepsy experts, the classification structure produced by FCA is added to EpSO.
Upper-level ontology: adopting the basic formal ontology classes in EpSO
Upper-level ontologies are reference terminologies with a common set of abstract classes and properties that facilitate interoperability between domain ontologies and support consistent modeling decisions. There are many upper-level ontologies, such as the Basic Formal Ontology (BFO)47 and the Descriptive Ontology for Linguistics and Cognitive Engineering (DOLCE).48 BFO in particular has been widely adopted in the development of biomedical domain ontologies.
BFO consists of two top-level classes called bfo:Continuant and bfo:Occurrent (‘bfo’ refers to the http://www.ifomis.org/bfo/1.1# namespace). The bfo:Continuant term represents entities that maintain their identity over time, for example, an electrode, while bfo:Occurrent represents entities that span a period of time, for example, a seizure. The top-level classes in EpSO were defined to be subclasses of the appropriate BFO classes based on the EpSO class characteristics. For example, BodilyProcess is modeled as a subclass of bfo:ProcessualEntity; Gender and EEGPattern are modeled as subclasses of bfo:DependentContinuant; and Electrode and BodilyFeature are modeled as subclasses of bfo:IndependentContinuant. The extension of BFO to model EpSO classes facilitates the interoperability of EpSO with other BFO compliant biomedical ontologies that use the same set of BFO classes as the parent of domain-specific classes.
Results
EpSO currently has more than 1000 classes (some with a seven-level-deep subclass structure) and is being used in three informatics tools being developed for the PRISM project.
The Ontology-driven Patient Information Capture system
The use of consistent terminology during the entry of patient data is important for ensuring the quality of data and their subsequent use in clinical research, which is especially relevant in the PRISM project with four participating EMUs. For example, the UH-CMC EMU used a Microsoft Word document that had multiple drawbacks, including frequent incorrect entries, missing information, and the need for manual processing of clinical notes to identify patient cohorts. We developed the Ontology-driven Patient Information Capture (OPIC) system that uses EpSO to implement flexible web-based forms to address the drawbacks of the earlier document-based approach.49
The OPIC forms use multi-level drop-down menus populated with EpSO classes that are structured to reflect the ontology class hierarchy, for example, users can navigate from a top-level class (SeizureSemiology) to its most specific descendant class (TonicClonicSeizure). OPIC also includes real time data validation and missing value checking to ensure that the user enters correct values (eg, numeric values for drug dosage) and essential data fields are not left blank (eg, patient age). OPIC is currently in use at the UH-CMC and is in the process of being deployed at NMH, Chicago. OPIC has streamlined the workflow for the entry of patient information, reduced the workload of EMU staff (eg, fellows and medical residents), and improved data quality.49
Epilepsy clinical free text processing using EpiDEA
The Epilepsy Data Extraction and Annotation (EPiDEA) tool is an ontology-driven clinical free text processing system that extends the clinical Text Analysis and Knowledge Extraction System (cTAKES)50 for analyzing epilepsy-specific clinical reports.51 Four cTAKES modules are used in EpiDEA for sentence detection, sentence tokenization, part-of-speech tagging, and shallow parsing. An EpSO-driven epilepsy named entity recognition module and a negation detection module process the output of these modules. EpSO is used in EpiDEA to support: (a) term disambiguation, (b) term normalization, and (c) query expansion using subsumption reasoning.51
EpiDEA has processed and created a patient database of 500 patient discharge summaries from a total of 1800 patient reports available in the UH-CMC EMU. Users can query these aggregated de-identified patient data using a web-based federated query environment called the Multi-Modality Epilepsy Data Capture and Integration System (MEDCIS).
MEDCIS: adapting VISAGE for epilepsy clinical research
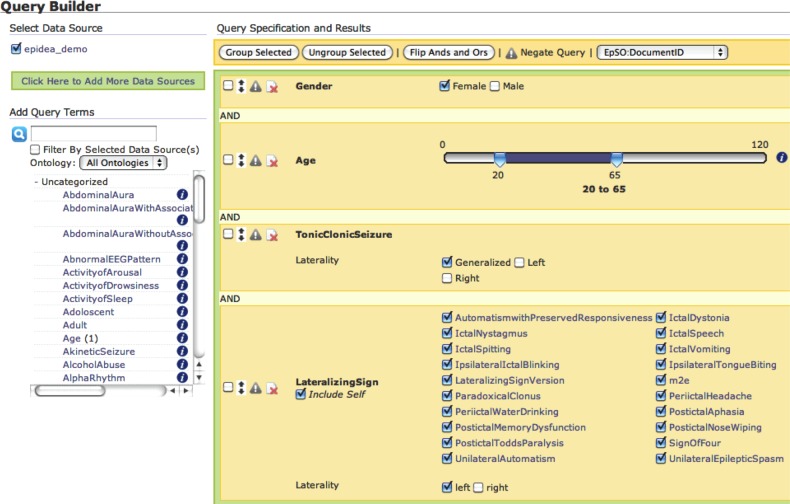
The outputs of the OPIC and EpiDEA systems are integrated into a single patient database that can be accessed through the MEDCIS query interface. MEDCIS extends the VISual AGgregator and Explorer (VISAGE)10 with the EpSO to implement a powerful and intuitive query interface for the PRISM project. MEDCIS consists of three modules that allow users to compose queries for clinical studies using visual query widgets, explore the query results in a tabular format, and store the queries in a query manager for future execution or sharing with other collaborators. Figure 5 illustrates the MEDCIS query builder.
Figure 5.
The Multi-Modality Epilepsy Data Capture and Integration System (MEDCIS) query builder with ontology terms in the left frame and visual query widgets in the right frame.
Users can select EpSO classes to incrementally build a query to identify patient cohorts. MEDCIS uses each ontology term to create a suitable query widget, for example a ‘slider bar’ is displayed for the Age term and multiple ‘check boxes’ are displayed for TonicClonicSeizure (figure 5). Users can traverse the EpSO class hierarchy (figure 5 illustrates the subclasses of the LateralizingSign class) and select multiple subclasses of a term. The user can explore the results of a query within the MEDCIS interface. Figure 6 illustrates the results of a patient cohort query that found five patients who satisfied the query constraints. The result explorer also includes links to the original patient discharge summary.
Figure 6.
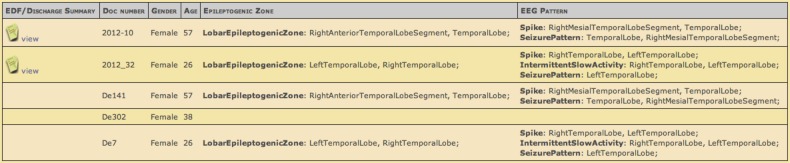
The results of a patient cohort identification query listing the de-identified patient records and links to original records in the left-most column.
Discussion
The current version of EpSO includes the relevant terms for supporting clinical studies on focal epilepsy patients with an emphasis on SUDEP patients for the PRISM project. We are in the process of establishing a consortium for EpSO (EOC) that brings together epileptologists, opinion leaders in epilepsy, and other domain experts to transform EpSO into a community resource. The EOC will enable community members to contribute terminological content, provide feedback on existing EpSO classes and properties, and collaborate on the addition of new terms to EpSO. EpSO is listed at the BioPortal ontology repository and also at http://prism.case.edu/prism/index.php/EpilepsyOntology). We are also implementing a new EpSO-based tool for epilepsy signal data analysis.
Electrophysiological signal analysis and visualization tool using EpSO
Electrophysiological signal data, such as EEGs, electrocardiograms, and blood oxygen levels, are often used for patient diagnosis, medication, and pre-surgical evaluation in epilepsy.39 Current stand-alone desktop-based signal visualization and analysis tools make it extremely difficult to support multi-center collaborative research involving interactive access to common sets of signal data. In addition, existing tools do not use a common terminology for event annotation on signal data, such as start or end of seizures. We are currently implementing a web-based signal visualization and analysis tool called Cloudwave that maps event annotations to EpSO classes and uses it to query for specific segments of signal data.52 In addition, we have integrated the Cloudwave signal processing module with the open source Hadoop platform for distributed data processing that will use the EpSO-based indexing scheme for faster processing of signal segments.
In addition to data integration and query answering, we believe that similar to other disease ontologies,53 EpSO will be increasingly used in data mining and knowledge discovery tasks over large scale semantically annotated datasets.
Conclusions
In this paper, we presented EpSO to underpin epilepsy-focused informatics tools for patient care and clinical research. We described the role of EpSO in bridging the gap between the epilepsy classification systems and NINDS CDE. EpSO was developed using the four-dimensional epilepsy classification system and used FCA to automatically create a taxonomy of epilepsy syndromes. In the PRISM project, EpSO is currently being used in the OPIC system to streamline the entry of patient information and significantly improve data quality. EpSO is also being used in EpiDEA to process clinical free text in patient records. The integrated patient data from OPIC and EpiDEA are accessed through the MEDCIS federated query platform allowing clinicians to create patient cohorts across multiple study centers. The EOC is expected to facilitate greater participation of the epilepsy community in the development, adoption, and maintenance of EpSO in a sustainable manner.
Footnotes
Contributors: SSS together with GQZ led the development of the ontology and the writing of the paper. SDL together with DKG and AB provided domain expertise, validated the structure and content of the ontology, and contributed to writing the paper. LC, MZ, and CJ integrated the ontology into EpiDEA, OPIC, and Cloudwave, respectively, and contributed to writing the paper and preparing the figures.
Funding: This work was supported by NIH/NINDS grant number 1-P20-NS076965-01 and NIH/NCATS grant number UL1TR000439.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: This work is available for download and use under a Creative Commons license at: http://prism.case.edu/prism/index.php/EpilepsyOntology.
References
- 1.Holdren JP, Lander E. Realizing the full potential of health information technology to improve healthcare for Americans: the path forward. In: Executive Office of the President, Washington, DC, 2010 [Google Scholar]
- 2.Epilepsy Foundation http://www.epilepsyfoundation.org/aboutepilepsy/whatisepilepsy/statistics.cfm (accessed 29 Mar 2013).
- 3.Berg AT, Berkovic SF, Brodie MJ, et al. Revised terminology and concepts for organization of seizures and epilepsies: Report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia 2010;51:676–85 [DOI] [PubMed] [Google Scholar]
- 4.Sheth AP. Changing focus on interoperability in information systems: from system, syntax, structure to semantics. In: Goodchild M, Egenhofer MJ, Fegeas R, Kottman C. Interoperating geographic information systems. Kluwer Academic Publishers, 1999:5–30 [Google Scholar]
- 5.Bodenreider O, Stevens R. Bio-ontologies: current trends and future directions. Brief Bioinform 2006;7:256–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodenreider O. Quality assurance in biomedical terminologies and ontologies. Technical report. Bethesda, MD: Lister Hill National Center for Biomedical Communications, National Library of Medicine, 2010 [Google Scholar]
- 7.Smith B, Ceusters W, Klagges B, et al. Relations in biomedical ontologies. Genome Biol 2005;6:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rector AL. Thesauri and formal classifications: terminologies for people and machines. Methods Inf Med 1998;37:501–9 [PubMed] [Google Scholar]
- 9.Sahoo SS, Bodenreider O, Rutter JL, et al. An ontology-driven semantic mashup of gene and biological pathway information: application to the domain of nicotine dependence. J Biomed Inform 2008;41:752–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang GQ, Siegler T, Saxman P, et al. VISAGE: a query interface for clinical research. In: AMIA Clinical Research Informatics Summit. 2010:76–80 [PMC free article] [PubMed] [Google Scholar]
- 11.Lebo T, Sahoo SS, McGuinness D. PROV-O: the PROV ontology. W3C Recommendation, http://www.w3.org/TR/prov-o/ (accessed 30 Apr 2013) [Google Scholar]
- 12.Nashef L, Hindocha N, Makoff A. Risk factors in sudden death in epilepsy (SUDEP): the quest for mechanisms. Epilepsia 2007;48:859–71 [DOI] [PubMed] [Google Scholar]
- 13.So EL. What is known about the mechanisms underlying SUDEP? Epilepsia 2008;49(Suppl 9):93–8 [DOI] [PubMed] [Google Scholar]
- 14.ILAE CTC Proposal for revised clinical and electrographic classification of epileptic seizures. Epilepsia 1981;22:489–501 [DOI] [PubMed] [Google Scholar]
- 15.ILAE CTC Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia 1989;30:389–99 [DOI] [PubMed] [Google Scholar]
- 16.Shorvon SD. The etiologic classification of epilepsy. Epilepsia 2011;52:1052–7 [DOI] [PubMed] [Google Scholar]
- 17.Panayiotopoulos CP. The new ILAE report on terminology and concepts for organization of epileptic seizures: a clinician's critical view and contribution. Epilepsia 2011;52:2155–60 [DOI] [PubMed] [Google Scholar]
- 18.Wong M. Epilepsy is both a symptom and a disease: a proposal for a two-tiered classification system. Epilepsia 2011;52:1201–3 [DOI] [PubMed] [Google Scholar]
- 19.Ghosh B, Ghosh PS, Sikder IU. Modeling a Classification Scheme of Epileptic Seizures Using Ontology Web Language. Int J Comput Models Algorithms Med 2010;1:45–60 [Google Scholar]
- 20.Almeida P, Gomes P, Sales F, et al. Ontology and knowledge management system on epilepsy and epileptic seizures. In: Paschke A, Burger A, Splendiani A, et al. Proceedings of the 3rd International Workshop on Semantic Web Applications and Tools for the Life Sciences 2010 [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hvidsten TR, Lægreid A, Komorowski J. Learning rule-based models of biological process from gene expression time profiles using Gene Ontology. Bioinformatics 2003;19:1116–23 [DOI] [PubMed] [Google Scholar]
- 23.Chiang J, Yu H. MeKE: discovering the functions of gene products from biomedical literature via sentence alignment. Bioinformatics 2003;19:1417–22 [DOI] [PubMed] [Google Scholar]
- 24.Office of the National Coordinator for Health Information Technology. http://healthit.hhs.gov/portal/server.pt/community/healthit_hhs_gov__home/1204 (accessed 29 Mar 2013).
- 25.Health Level Seven (HL7). http://www.hl7.org/ (accessed 29 Mar 2013).
- 26.Loring DW, Lowenstein DH, Barbaro NM, et al. Common data elements in epilepsy research: development and implementation of the NINDS epilepsy CDE project. Epilepsia 2011;52:1186–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The National Center for Biomedical Ontology. http://bioontology.org (accessed 29 Mar 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dou D, Frishkoff G, Rong J, et al. Development of NeuroElectroMagnetic Ontologies (NEMO): a framework for mining brain wave ontologies. Thirteenth International Conference on Knowledge Discovery and Data Mining (KDD2007). ACM New York. 2007:270–9 [Google Scholar]
- 29.SNOMED Clinical Terms. http://www.nlm.nih.gov/research/umls/Snomed/snomed_main.html (accessed 29 Mar 2013).
- 30.Mirza N, Vasieva O, Marson AG, et al. Exploring the genomic basis of pharmacoresistance in epilepsy: an integrative analysis of large-scale gene expression profiling studies on brain tissue from epilepsy surgery. Hum Mol Genet 2011;20:4381–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T, Delgado-Escueta AV, et al. Mutations in EFHC1 cause juvenile myoclonic epilepsy. Nat Genet 2004;36:842–9 [DOI] [PubMed] [Google Scholar]
- 32.Shvaiko P, Euzenat J. A survey of schema-based matching approaches. J Data Semantics 2005;4:146–71 [Google Scholar]
- 33.Lüders HO, Amina S, Baumgartner C, et al. Modern technology calls for a modern approach to classification of epileptic seizures and the epilepsies. Epilepsia 2012;53:405–11 [DOI] [PubMed] [Google Scholar]
- 34.Loddenkemper T, Kellinghaus C, Wyllie E, et al. A proposal for a five-dimensional patient oriented epilepsy classification. Epileptic Disord 2005;7:308–16 [PubMed] [Google Scholar]
- 35.Kellinghaus C, Loddenkemper T, Wyllie E, et al. Suggestion for a new, patient-oriented epilepsy classification. Nervenarzt 2006;77:961–9 [DOI] [PubMed] [Google Scholar]
- 36.Hitzler P, Krötzsch M, Parsia B, et al. OWL 2 Web Ontology Language Primer. In: W3C Recommendation. World Wide Web Consortium (W3C), 2009 [Google Scholar]
- 37.Fisher RS, Boas WE, Blume W, et al. Epileptic seizures and epilepsy: definitions proposed by the International League Against Epilepsy (ILAE) and the International Bureau for Epilepsy (IBE). Epilepsia 2005;46:470–2 [DOI] [PubMed] [Google Scholar]
- 38.Lüders H, Acharya J, Baumgartner C, et al. Semiological seizure classification. Epilepsia 1998;39:1006–13 [DOI] [PubMed] [Google Scholar]
- 39.Rosenow F, Lüders H. Presurgical evaluation of epilepsy. Brain 2001;124:1683–700 [DOI] [PubMed] [Google Scholar]
- 40.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform 2003;36:478–500 [DOI] [PubMed] [Google Scholar]
- 41.Horridge M, Bechhofer S. The OWL API: a Java API for OWL ontologies. Semantic Web J 2011;2:11–21 [Google Scholar]
- 42.Nelson SJ, Zeng K, Kilbourne J, et al. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc 2011;18:441–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bodenreider O, Peters L, Nguyen T. RxNav: browser and application programming interfaces for drug information sources. AMIA Annual Symposium, 2011:2129 [Google Scholar]
- 44.Ganter B, Stumme G, Wille R. Formal concept analysis: foundations and applications. Springer-Verlag, 2005 [Google Scholar]
- 45.Wille R. Restructuring lattice theory: an approach based on hierarchies of concepts. Dordrecht/Boston: Reidel, 1982:445–70 [Google Scholar]
- 46.Zhang GQ, Bodenreider O. Large-scale, exhaustive lattice-based structural auditing of SNOMED CT. AMIA Annual Symposium Proceedings;2010:922–6 [PMC free article] [PubMed] [Google Scholar]
- 47.Spear A. Ontology for the twenty-first century: an introduction with recommendations. Technical Report, 2006
- 48.Gangemi A, Guarino N, Masolo C, et al. Sweetening ontologies with DOLCE. 13th International Conference on Knowledge Engineering and Knowledge Management Ontologies and the Semantic Web: 2002. Springer Verlag, 2002:166–81 [Google Scholar]
- 49.Sahoo SS, Zhao M, Luo L, et al. OPIC: ontology-driven patient information capturing system for epilepsy. The American Medical Informatics Association (AMIA) Annual Symposium: 2012. AMIA, 2012 [PMC free article] [PubMed] [Google Scholar]
- 50.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui L, Bozorgi A, Lhatoo SD, et al. EpiDEA: extracting structured epilepsy and seizure information from patient discharge summaries for cohort identification The American Medical Informatics Association (AMIA) Annual Symposium: 2012. AMIA, 2012 [PMC free article] [PubMed] [Google Scholar]
- 52.Jayapandian CP, Chen CH, Bozorgi A, et al. Electrophysiological signal analysis and visualization using cloudwave for epilepsy clinical research. Copenhagen, Denmark: MedInfo, 2013 [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Coulet A, LePendu P, et al. FOCUS on clinical research informatics: Using ontology-based annotation to profile disease research. J Am Med Inform Assoc 2012;19(e1):e177–86 [DOI] [PMC free article] [PubMed] [Google Scholar]