Abstract
Background
Multimorbidity, the co-occurrence of two or more chronic medical conditions within a single individual, is increasingly becoming part of daily care of general medical practice. Literature-based discovery may help to investigate the patterns of multimorbidity and to integrate medical knowledge for improving healthcare delivery for individuals with co-occurring chronic conditions.
Objective
To explore the usefulness of literature-based discovery in primary care research through the key-case of finding associations between psychiatric and somatic diseases relevant to general practice in a large biomedical literature database (Medline).
Methods
By using literature based discovery for matching disease profiles as vectors in a high-dimensional associative concept space, co-occurrences of a broad spectrum of chronic medical conditions were matched for their potential in biomedicine. An experimental setting was chosen in parallel with expert evaluations and expert meetings to assess performance and to generate targets for integrating literature-based discovery in multidisciplinary medical research of psychiatric and somatic disease associations.
Results
Through stepwise reductions a reference set of 21 945 disease combinations was generated, from which a set of 166 combinations between psychiatric and somatic diseases was selected and assessed by text mining and expert evaluation.
Conclusions
Literature-based discovery tools generate specific patterns of associations between psychiatric and somatic diseases: one subset was appraised as promising for further research; the other subset surprised the experts, leading to intricate discussions and further eliciting of frameworks of biomedical knowledge. These frameworks enable us to specify targets for further developing and integrating literature-based discovery in multidisciplinary research of general practice, psychology and psychiatry, and epidemiology.
Keywords: literature based discovery, text mining, multimorbidity, primary care research, disease profiles, disease combinations
Introduction
Multimorbidity—the occurrence of two or more chronic diseases within a single individual—is common and may be a heavy burden on the patient, affecting quality of life, and leading to physical and social disability and increasing use of healthcare services.1–3 General practitioners are increasingly confronted with multimorbidity, resulting in complex healthcare conditions, where one condition might cause, maintain, or exacerbate other conditions.4 5 This requires developing new methods of investigating determinants of multimorbidity and the detection of populations at risk.1 5
Most multimorbidity studies focus on the identification of clinically relevant specific disease combinations in patient populations, based on one index disease and additional diseases, or the epidemiology of disease combinations that are found among patients, either in general or specific population-based studies or in administrative databases.4–7 The results of epidemiology directed studies, however important, have to be interpreted within a body of medical knowledge, concerning various aspects of clinical practice: etiology, diagnosis, the natural course of disease, and therapy.1 Further, susceptibility to multiple pathologies may transcend disease specific etiologies addressing a broader range of factors— for example, not only biological but also psychological and social mechanisms, and which may operate at multiple levels, that is, both at the level of the individual and at a population level.3 8 9
Investigating all possible combinations of diseases is an arduous and laborious task, given the large number of diseases, the dynamic character of biomedical knowledge, and the vast amounts of available biomedical information. In order to cope with this, data mining studies of comorbidity are receiving increasing attention in biomedicine.10–12 In contrast, text mining studies of comorbidity are scarce. Although multiple definitions coexist, text mining is directed at the retrieval, extraction, and synthesis of information from texts, with a special emphasis on gaining new knowledge.13 14 Swanson was the first to demonstrate by using Medline that text mining can lead to the discovery of new knowledge, that is, the treatment of Raynaud disease by fish oil.15 Medline was also used to make connections between seemingly dissociated entities— for example, the connection between migraine and magnesium deficiency—and to identify new uses for drugs and other substances in the treatment of diseases.16–19 In the past decade research on literature-based discovery (LBD) has intensified and generated new techniques for identifying concept relationships that have not yet been explicitly described in the (medical) literature.20–26 An important field of application has become the analysis of DNA microarray data.27 In microarray experiments, hundreds of genes can be identified and the interpretation of such gene lists depends on the examination of hundreds and even thousands of articles for each single gene.27 28 Text mining facilitates traversing the huge corpus of biomedical literature.29 In the course of time, text mining studies regarding disease-specific knowledge (disease–drug, disease–gene, etc.) appeared, but there have been few studies focused on disease–disease associations.30–34 These include an exploratory study of free-text clinical reports on comorbidity35 and a manual text-based classification of disease combinations in a cohort study on multimorbidity.8
We decided to investigate whether LBD is useful in assessing the association between diseases in an interdisciplinary study on the co-occurrence of psychiatric and somatic disorders, their physical and psychosocial determinants, as well as their medical and social consequences.3 8 9 36 37 Psychiatric and somatic diseases frequently co-occur,38 39 leading to adverse health effects, non-adherence to care, improper use of medication, and prolonged recovery time.39 40 New tools are required to analyze the enormous amount of possible disease combinations given the large spectrum of chronic psychiatric and somatic diseases.3 5 8 9 11 12
The present study is aimed at exploring the usefulness of LBD in establishing associations between psychiatric and somatic disorders relevant to general practice in a large biomedical literature database (Medline). We report a comparative analysis of the evaluation of associations between psychiatric and somatic diseases by LBD and by experts involved in multimorbidity studies.
Methods
The methods relate to five issues relevant for the design of the study: (1) the use of Medline and the LBD tool Anni; (2) the generation of a reference set of 21 945 disease combinations to determine the threshold for the association of a pair of diseases; (3) the study set of disease combinations (N=166) between psychiatric diseases and somatic diseases; (4) the experimental setting and expert meeting; and (5) the model used to compare the LBD assessment with the evaluation by the biomedical experts.
The use of Medline and the LBD tool Anni
The main repository of published biomedical literature is Medline, containing references to medical, nursing, dental, veterinary, healthcare, and preclinical sciences journal articles published since 1948.41
In this study Anni, an LBD tool developed by the Erasmus Medical Center, which provides an ontology-based interface to Medline, was used.42 Anni uses three sources: (1) an ontology composed of the 2006AC version of the Unified Medical Language System (UMLS) using Aronson's adaptation of the ULMS Metathesaurus for efficient natural language processing,43 and a gene thesaurus derived from various databases, for example, the NCBI's Homologene database44; (2) a database with indexed references to ontology concepts in Medline abstracts (from 1980 on); and (3) a database with concept profiles based on the Medline indexation and covering the full scope of the UMLS Metathesaurus.42 From all documents related to a concept (eg, a disease) the co-occurring concepts (eg, social demographic variables, neurotransmitters, genes, drugs, etc.) are retrieved and compiled into a weighted so-called concept profile. For example, the diseases diabetes mellitus (DM) and depression (D) might be represented by a concept profile, such as {DM: concept-1, concept-2, … concept-n}, respectively {D: concept-1, concept-2, … concept-m}. Concept profiles are vectors in a high-dimensional concept space.45 For all concepts a ‘best’ position in this space is calculated based on the matching score between the concept profiles, which reflects the strength of the relationship. The weights in the concept profile were derived by means of the symmetric uncertainty coefficient, which is a standard measure for stochastic (in)dependence.46 The matching score of two concept profiles can be computed by taking the inner product of the weights between the shared concepts, where the inner product increases with increasing overlap in concept profiles (a typical example is presented in the appendix). Further technical details of Anni and its application in mining microarray expression data by literature profiling and the discovery of gene–disease networks are described elsewhere.25 27 42 45–48
The reference set of disease combinations and the threshold for the association of diseases
This study starts with a limited scope regarding multimorbidity, namely the pairwise association of diseases, leaving the more complex task of assessing combinations of three or more diseases as a challenge for future research. A broad range of diseases was chosen representing the comorbidities, which had a prevalence of ≥5 in the database (N=87 838 patients) of the Registration Network Family Practices (RNH), in which 70 general practitioners (GPs)in the South of the Netherlands are participating.49 The RNH is a continuously updated database with a population comparable to the Dutch population.13 49 Health problems are registered in a standardized fashion, according to the International Classification of Primary Care (ICPC), which is related to ICD-9 and ICD-9 derived systems.50
ICPC classifies diseases into 16 organ systems, so-called ICPC chapters, for example cardiovascular diseases (ICPC chapter K) and gastrointestinal diseases (ICPC chapter D). The ICPC codes in each chapter represent disease entities such as ‘myocardial infarction’ (K75) and ‘diabetes mellitus’ (T90), but also more general codes, which were excluded from the analysis:
Codes referring to postoperative complications or other side effects of medical treatment (A85, A87).
Codes containing a too broad spectrum of diseases (eg, those ending at ‘99’, indicating ‘other diseases of an organ system’), which are too vague for corresponding MeSH terms.8
In total, 214 ICPC codes representing a broad spectrum of prevalent diseases were selected. The terms of these codes were imported as the seed concepts for the biomedical text mining and were mapped to all the UMLS concepts and all the concept profiles in Anni. The result was a set of 21 945 disease combinations with a matching score for each pair of diseases.
Setting the threshold for finding new disease combinations: the discovery zone
The matching scores of the disease combinations as found by Anni were heterogeneously distributed as expected since the disease concepts were mapped to the Medline abstracts covering all fields in biomedicine.
The set of 21 945 disease combinations can be split into a set of 17 642 disease combinations for which a matching score (MC) could be calculated (MC>00001) and a set of 4303 combinations lower than the Anni minimum score (MC<0.0001). The small set was studied separately, whereas the large set was used to determine the threshold for Anni's matching score. As expected, this set is very skewed and a box plot was created to determine the main features of the non-parametric distribution (see online supplementary appendix).51 We used the upper hinge (MC=0.007081) as the threshold for Anni's matching score because that area can be considered as the area of (severe) outliers and, hence, of relatively very high matching scores: disease combinations with a matching score exceeding this threshold were considered as having a relevant degree of concept overlap, thus forming what we call a zone of discovery (see box plot, online supplementary appendix).
The study set of combinations (N=166) between psychiatric and somatic diseases
From the reference set of 21 945, a limited set of disease combinations was chosen. For clinical and investigational reasons of interest, the research team selected six psychological conditions (with respective ICPC codes between brackets)—schizophrenia (P72), affective psychosis (P73), depressive disorder (P76), suicide attempt (P77), personality disorder (P80), and mental retardation (P85)—and 98 somatic disorders from six ICPC chapters: D (digestive system; 19 included ICPC codes), K (circulatory system; 20 ICPC codes), L (musculoskeletal system; 17 ICPC codes), N (neurological system; 14 ICPC codes), R (respiratory system; 16 ICPC codes), and T (endocrine, metabolic, and nutritional system; 12 ICPC codes). This resulted in a set of 166 disease combinations—relatively prevalent in the RNH database, each combination representing five or more patients; this number of 166 disease combinations was considered to be feasible for expert evaluation.
Experimental setting and expert meeting
Two authors (RV and SA), a doctor of philosophy and medicine, and epidemiology PhD student, performed the actual discovery process: SA, together with EvM, generated the reference set of 21 945 disease combinations, and the study set of 166 disease combinations; RV performed the basic analysis (supervised by MvdA) of the concept mapping and expert evaluations. The 166 disease combinations of psychiatric and somatic conditions were scored by a psychiatry–psychology expert (FV) in four categories (0: no causal association; 1: possible causal relation denoting that a ‘pathophysiological or pathopsychological’ explanation can be given; 2: probable causal relation denoting that an association can be expected; and 3: otherwise related denoting a coincidental relation, eg, that a disease combination is highly prevalent in specific age groups, or high risk patients are more prone to diagnostic tests). The results were explored with the experts (MvB, JM, FV) and discussed in an expert meeting (FV, JM).
In the expert meeting the separate disease combinations were assessed, as were the different frameworks for categorizing disease combinations. A framework is defined as the development of a conceptual model representing the decision problem and the relevant argumentation structure.52 53 In this study the focus was on the cognitive aspect of framing; that is, the kind of arguments activated for assessing relationships between diseases. Content analysis was performed to identify the arguments used by the experts.54 An example is when experts put forward arguments to elucidate specific causal mechanisms, for example, ‘… involving serotonergic pathways …’, possibly involved in some disease combination; hence the framework of causality is addressed. These types of arguments were analyzed in an exploratory manner without formal coding and iterative analysis. The expert discussions provided background and biomedical perspective to the generated disease combinations as presented in the results and discussion of this study.
Model of comparison of LBD matching and evaluation by the biomedical experts
LBD presupposes ‘discovery’, but this also assumes that LBD is able to distinguish between ‘unknown, but potentially relevant’ and well-known, established knowledge. In this respect two discovery situations can be distinguished: the situation where a strong concept profile match exists between two concepts that also appear in a single abstract in Medline; and the situation where a strong match is calculated without two concepts ever being co-mentioned in one abstract.
The following model was used, which presumes two dimensions are important in establishing an association between diseases: on the one hand, the availability of evidence, for example scientific information coming from randomized clinical trials, or experimental or observational studies; on the other hand, a scientific judgment based on some kind of decision making procedure to accept—or to reject—the association between two diseases.
As shown in the horizontal axis of figure 1, we operationalized scientific evidence as the number of quotations in Medline concerning the co-occurrence of the diseases. A number of zero quotations (N=0) implies there is no explicit connection between two diseases, in contrast to when there are publications present (N>0) which make such an explicit connection.
Figure 1.
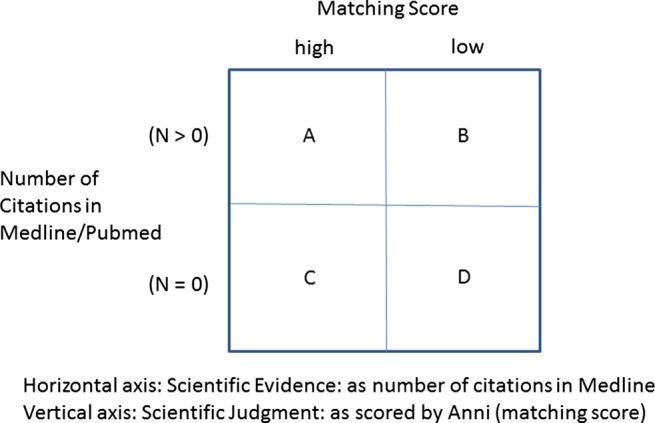
Innovation grid of association of disease combinations: matching score and available scientific information.
As shown in the vertical axis of figure 1, the scientific judgment about an association between the two diseases is represented by the result of Anni's profile matching (‘matching score’). A high matching score indicates a high overlap in the concept profiles of two diseases. Concepts with a high matching score but no Medline co-occurrences could indicate a new discovery: a relationship between concepts implicit in the literature but not yet explicitly described in biomedical research articles as has been shown by previous research.12 26 31 32
Thus, any method in LBD should identify (see figure 1) quadrants A and B as ‘established knowledge’ confirming or rejecting the association of two diseases, exclude D as non-relevant (medical) knowledge, and generate C as a source of potentially ‘novel’ biomedical information on relevant but unknown disease combinations.
In a more detailed analysis, the number of citations in Medline was mapped to four categories: N=0, N=1–9, N=10–49, and N ≥ 50 citations in Medline. The rationale is to see whether potential discoveries might be hidden not only in quadrant C (N=0), but perhaps also in the very small and modest subsets of medical literature (N<50) in quadrant A.
Results
Distribution of the cases in the innovation grid (N=166)
A little less than half of the disease combinations (N=75) have a high matching score, whereas 91 combinations score low (figure 2). About half (N=82) of the disease combinations have no (N=0) citations in Medline. While there are 61 cases with a high matching score and a number of citations (N>0) in Medline, only 23 combinations received much attention in the medical literature, but scored low on the matching score. The lower-left quadrant—the quadrant of potential new information—counts 14 cases.
Figure 2.

Innovation grid—distribution of disease combinations (N=166).
Distribution of cases and the kind of expert judgments
Of the 166 cases, 55 disease combinations were categorized by the expert as plausible—grouping together the expert judgments type 1: possible causal relation; type 2: probable causal relation; and type 3: otherwise related—and 111 disease combinations were categorized as not plausible (see figure 3, panels I and II; see also table 2 which summarizes all the results). In both panels the cases are fairly distributed over the quadrants A, B, and D. As expected, the smallest set is in quadrant C (the upper panel I: 2 cases; the lower panel II: 12 cases).
Figure 3.

Distribution of disease combinations (N=166) and the kind of expert judgments.
Table 2.
Distribution of disease combinations and expert judgments (N=166)
| Expert judgment | |||||
|---|---|---|---|---|---|
| Citations (N) | Type 0 | Type 1 | Type 2 | Type 3 | Total |
| Matching score=high | |||||
| ≥50 | 4 | 1 | 3 | 1 | 9 |
| 10–49 | 9 | 6 | 3 | 4 | 22 |
| 1–9 | 22 | 4 | 1 | 3 | 30 |
| 0 | 12 | 0 | 0 | 2 | 14 |
| Total | 47 | 11 | 7 | 10 | 75 |
| Matching score=low | |||||
| ≥50 | 0 | 0 | 0 | 2 | 2 |
| 10–49 | 1 | 0 | 0 | 0 | 1 |
| 1–9 | 9 | 3 | 3 | 5 | 20 |
| 0 | 54 | 5 | 2 | 7 | 68 |
| Total | 64 | 8 | 5 | 14 | 91 |
The subset of 14 potentially new disease combinations (high matching score, no citations)
Two cases (panel I, figure 3) have been evaluated by the expert as non-causally probable, that is, ‘otherwise’ related, and 12 cases (panel II, figure 3) were evaluated as having no causal possible relationship. However, from these 12 cases a distinctive pattern emerges (see figure 4).
Figure 4.

Histogram of 12 cases (no citations in Medicine/PubMed and judged not plausible). ICPC, International Classification of Primary Care.
Of the 12 combinations, 10 cases concern combinations between two psychiatric disorders, that is, suicide attempt (P77) and personality disorders (P80) on the one hand and somatic disorders of the gastrointestinal (D) and locomotor (L) system on the other hand. More specifically, the gastrointestinal disorders concern appendicitis (D88) and inguinal hernia (D89), the locomotor disorders relate to osteoarthritis (of hip or knee, L89 and L90), and femoral fractures (L75). The two other cases concern schizophrenia (P72) with osteoarthritis (of hip, L89), and suicide attempt (P77) with angina pectoris (K74).
This is a specific pattern, as is also shown by the analysis of the subset of 4303 disease combinations which scored below the minimum threshold of the matching score. In this subset 35 cases are combinations of psychiatric and somatic disorders, two of which concern the psychiatric disorders P77 (N=1) and P80 (N=1)— each with a digestive disorder (D) (N=2). In addition, four cases also concern P77 (N=2) and P80 (N=2)—although in combination with cardiovascular disorders (K) (N=4). Thus, concept profiling of P77 and P80 generates cases with a positive matching score, but also selectively cases of P77 and P80 with a negative matching score.
Quadrant A: hidden discoveries and type 1 expert judgment
If we look at the cases, which have been classified as causally possible (type 1 judgment) by the expert panel, an interesting pattern emerges (see figure 5). Whereas there are no cases in the category N=0 (figure 5), 10 of the 11 cases which the experts find causally possible associations are prominent in the categories of very low and modest number of citations in Medline. In terms of selective profiling, these 10 cases now concern the whole range of psychiatric disorders (table 1) and the four somatic disease classes other than the gastrointestinal (D) and locomotor (L) system.
Figure 5.
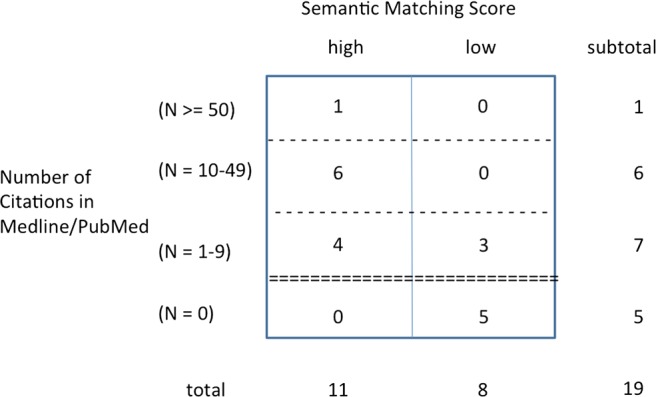
Distribution type 1 expert judgments (N=19).
Table 1.
Overview of the combinations of psychiatric and somatic disorders ranked high by the matching score and scored as ‘possible’ (type 1 judgment) by the medical experts
| Psychiatric disorder | ICPC-1 | Somatic disorder | ICPC-2 | Citations (N) |
|---|---|---|---|---|
| Suicide attempt | P77 | Brain concussion | N79 | 1–9 |
| Personality disorder | P80 | Pulmonary emphysema | R95 | 1–9 |
| Suicide attempt | P77 | Stroke | K90 | 1–9 |
| Depression | P76 | Pulmonary emphysema | R95 | 1–9 |
| Mental retardation | P85 | Heart murmur | K81 | 10–49 |
| Affective disorder | P73 | Epilepsy | N88 | 10–49 |
| Schizophrenia | P72 | Heart failure | K77 | 10–49 |
| Personality disorder | P80 | Migraine disorders | N89 | 10–49 |
| Mental retardation | P85 | Heart failure | K77 | 10–49 |
| Suicide attempt | P77 | Epilepsy | N88 | 10–49 |
| Depression | P76 | Diabetes mellitus | T90 | ≥50 |
ICPC, International Classification of Primary Care.
Table 2 summarizes all the results and shows ‘one wins and one loses’. Although two classes of possible new disease combinations are picked out, it is also clear that expert judgments (N=27) are ‘missed’, namely eight cases of type 1, five cases of type 2, and 14 cases of type 3 judgment were scored as less relevant by the matching score. On the other hand, more than half of these cases had no citations in Medline (type 1: five cases; type 2: two cases; type 3: seven cases). Moreover, 14 cases were of type 3, that is, links that were not considered causal by the expert.
Expert meeting: exploring the elicited hypotheses and hidden discoveries
The discussion focused on the major findings of this study. In particular subsets of diseases combinations from the various parts of the quadrants A, B, C, and D were discussed and explored, confirming the patterns found. Three topics, however, stand out.
First, the subset of quadrant A of cases in the categories of 1–9, 10–49, and ≥50 citations, scored high by Anni and evaluated as possibly causal by the expert, were accepted at face value; these cases were considered of clinical importance and interesting for further research. Among these, one combination, namely between depression and diabetes (table 1), is an emerging subject of research, and is also of interest for members of the research team.39 55 56 Additionally, personality disorders and comorbidity of somatic diseases recently received attention in psychiatry.57 58 In this case, the team also discussed the possibility that general practitioners might code a diagnosis of ‘personality disorder’ (ie, ICPC code P80) because of personality character changes after a cerebrovascular accident (ICPC code K90) (frame B of complex patient profiles, box 1). Thus, two disease entities might simultaneously be at stake—a specific psychiatric diagnostic category, and a broader disease category of ‘changes in personal character’, associated with cardiovascular disease, encountered in general practice, and to be distinguished in further research (frame A of disease categories, box 1).
Box 1. Discovery zone and frames of linking diseases.
-
Frames of disease categories
Aspects related to etiology, diagnosis, and natural course of disease, for example psychiatry, or general practice-directed views on diseases, for example personality disorder as a psychiatric category or personality change concurrent with dementia or cerebrovascular accident
-
Frames of causality
Biological causal mechanisms, for example linkage of diseases through serotonergic, dopaminergic, or other kind of biological pathways, but also psychological and social mechanisms, for example weakness of social support triggering a cascade of disease events
-
Frames of indirect pathways
Disease transcending mechanisms, for example pain, stress, and anxiety connecting different somatic and/or psychiatric diseases such as depression after a cardiovascular event or the other way around
-
Frames of complex patient profiles
Relation of disease conditions to demographic variables, medical history of patient, for example patients with different types of multimorbidity or with high risk comorbidity as with cancer in elderly patients
-
Frames of healthcare processes
Healthcare needs related to healthcare delivery, communication between health professionals, for example different views on medical problems by various specialists
Consequently, the cases of quadrant A inspired the explorations on mechanisms and processes linking the diseases involved in the respective disease combinations. The experts generated different frames for qualifying the found disease associations, hence placing diseases and their interrelationships in the body of biomedical knowledge (box 1). The box shows a broad scope on ‘mechanisms’, processes’, and ‘pathways’ as potential candidates for connective structures between diseases, which may be expected considering the intricate complexities of the interaction between psychiatric and somatic disorders.
From these frames hypotheses were generated, accepted as plausible, or rejected as uninteresting during the meeting. The aim of the discussion was not to find an explanation per se, but to see whether a variety of arguments could be given to support plausibility for the generated set of potential discoveries.
Second, the ‘mirror’ subset of the first type of discovery above, namely the cases scored low by Anni, but evaluated as causally possible by the expert—the eight cases in quadrants A and B, that is, the five cases in category N=1–9 and three cases in category N=0 (see table 2, summarizing all results)—were considered more uncertain and less interesting than the discovery set of table 1. The same pattern emerged comparing cases evaluated as causal probable—type 2 judgment—which were scored high by Anni versus those which were scored low(er) by Anni.
Third, the second set of potential discovery, that is, the 12 cases scored high by Anni having no citations in Medline, and evaluated as no association by the expert, left the experts perplexed and, in a sense, lost. No immediate hypotheses were generated, but the back and forth processing of the various frames elicited some speculative links, for example neurophysiological pathways (frame B of causality, box 1), food as a causative agent (frame B of causality and frame C of indirect pathways, box 1), or social processes such as despair and family disruption leading to multiple disease (frame C of indirect pathways, respectively, frame E of healthcare processes, box 1).
Discussion
The present study shows that LBD tools such as Anni are able to find specific patterns of combinations between psychiatric and somatic diseases out of a set of 166 combinations of psychiatric and somatic disorders. In combination with information on the occurrence of citations in Medline regarding disease combinations, it was possible to generate two different sets of disease combinations. One set correlated well with the expert judgments, whereas the other set differed quite substantially, literally perplexing the experts. Some pros and cons of the differences could be explained in further discussions. It is interesting to see that both sets formed a source of inspiration for discussing possible ‘chains of events’ between diseases: causal pathways between biological factors, but also the intricate complexities of psychological and social factors as represented by the biopsychosocial model of disease.8 59 Further, different frameworks underlying the appraisal of complex patient profiles could be specified as a result of the experimental setting with parallel expert evaluation and expert meeting.
Nevertheless, we want to propose a careful attitude here, not so much with the use of the concept of profile matching per se considering the well known and generally appreciated way to assess (dis)similarities between concept networks.13 14 24–26 28 42 However, the matching score is a relational assessment, not an absolute score. The threshold of a ‘higher’ and ‘lower’ degree of matching has to be considered from the perspective of the problem situation at hand. In the case of assessing associations between disease combinations, there was no a priori information available to consider some level as the right yardstick. Thus, the reference set of disease combinations was used to set the threshold.
In further research it might be promising to use more selective filters for finding potential candidates as connective structures between diseases. These filters can be constructed according to the different frames as used by the experts in evaluating the relevancy of disease associations and generating hypotheses for intermediate processes and pathways. In this respect it is worthwhile to differentiate within the frames themselves. Even within the frame of causality different models of causality can be used, from a biological but also from a psychosocial perspective.8 59 In addition, it might be possible to enhance the success of LBD by enriching the seed case: in this study merely the disease terms as denoted by the ICPC codes—and comparable MeSH headings—of each pair of diseases were used as an input for text mining, but this input can be augmented by adding concepts representing relevant clinical information, such as diagnostic or laboratory features. This augmentation approach needs to be evaluated in the most recent versions of the UMLS Thesaurus and the additional gene, chemicals, and toxicity thesauri as used in Anni.
Further research is needed to see whether, and if so, in which ways, LBD might be useful in assessing the associations between disease combinations to generate new patterns of multimorbidity, also for combinations of three or more diseases. In future work it is worthwhile to use different LBD tools as well as to expand the methodology of eliciting frames as structures of medical knowledge and as targets for text mining. A careful attitude is asked for because of the complexity of determining the grounds for a ‘true’ association between diseases. This requires a two-way, interactive process between the application of text mining and LBD tools and the cooperation of and evaluation by medical experts.8 13 14 18 19
Footnotes
Contributors: RV was responsible for the conception, design, and analysis, and was the primary author and revisor of this manuscript. SA was responsible for the design of the study and the preliminary analysis of the data. EvM was responsible for data analysis using ANNI and critical reading of the manuscript. JM was member of the expert panel and has critically reviewed the manuscript. MPvB was member of the expert panel and has critically reviewed the manuscript. FV was member of the expert panel and has critically reviewed the manuscript. MvdA was responsible for the conception, design, and supervision of the project and critical reading of the manuscript. All the authors were members of the research team that discussed progress of the research in the various phases of the study. All the authors have made a substantial contribution in drafting and revising up to the last version of the manuscript. All authors have approved the final version of the manuscript to be published.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Fortin M, Soubhi H, Hudon C, et al. Multimorbidity's many challenges. BMJ 2007;334:1016–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.France EF, Wyke S, Gunn JM,, et al. Multimorbidity in primary care: a systematic review of prospective cohort studies. Br J Gen Practice 2012;62:e297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van den Akker M, Buntinx F, Metsemakers JF, et al. Multimorbidity in general practice: prevalence, incidence, and determinants of co-occurring chronic and recurrent diseases. J Clin Epidemiol 1998;51:367–75 [DOI] [PubMed] [Google Scholar]
- 4.Marengoni A, Rizzuto D, Wang HX, et al. Patterns of chronic multimorbidity in the elderly population. J Am Geriatr Soc 2009;57:225–30 [DOI] [PubMed] [Google Scholar]
- 5.Barnett K, Mercer SW, Norbury M, et al. Epidemiology of multimorbidity and implications for health care, research, and medical education: a cross-sectional study. Lancet 2012;380:37–43 [DOI] [PubMed] [Google Scholar]
- 6.Wong A, Boshuizen HC, Schellevis FG, et al. Longitudinal administrative data can be used to examine multimorbidity, provided false discoveries are controlled for. J Clin Epidemiol 2011;64:1109–17 [DOI] [PubMed] [Google Scholar]
- 7.Hanauer DA, Rhodes DR, Chinnaiyan AM. Exploring clinical associations using -omics’ based enrichment analyses. PloS One 2009;4:e5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van den Akker M, Vos R, Knottnerus JA. In an exploratory prospective study on multimorbidity general and disease related susceptibility could be distinguished. J Clin Epidemiol 2006;59:934–9 [DOI] [PubMed] [Google Scholar]
- 9.Aarts S, Van den Akker M, Tan FE, et al. Influence of multimorbidity on cognition in a normal aging population: a 12-year follow-up in the Maastricht Aging Study. Int J Geriatr Psychiatry 2010;26:1046–53 [DOI] [PubMed] [Google Scholar]
- 10.Hh Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012;13:395–405 [DOI] [PubMed] [Google Scholar]
- 11.Ilgen MA, Downing K, Zivin K, et al. Identifying subgroups of patients with depression who are at risk for suicide. J Clin Psychiatry 2009;70:1495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roque FS, Jensen PB, Schmock H, et al. Using electronic patient records to discover disease correlations and stratify patient cohorts. Plos Comput Biol 2011;7:e1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rebholz-Schuhmann D, Oellrich A, Hoehndorf R. Text-mining solutions for biomedical research: enabling integrative biology. Nat Rev Genet 2012;13:829–39 [DOI] [PubMed] [Google Scholar]
- 14.Rzhetsky A, Seringhaus M, Gerstein M. Seeking a new biology through text mining. Cell 2008;134:9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson DR. Fish oil, Raynaud's syndrome, and undiscovered public knowledge. Perspect Biol Med 1986;30:7–18 [DOI] [PubMed] [Google Scholar]
- 16.Smalheiser NR, Swanson DR. Using Arrowsmith: a computer-assisted approach to formulating and assessing scientific hypotheses. Comp Meth Progr Biomed 1998;57:149–53 [DOI] [PubMed] [Google Scholar]
- 17.Srinivasan P, Libbus B. Mining Medline for implicit links between dietary substances and diseases. Bioinformatics 2004;20(Suppl. 1):i290–6 [DOI] [PubMed] [Google Scholar]
- 18.Weeber M, Vos R, Klein H, et al. Generating hypotheses by discovering implicit associations in the literature: a case report of a search for new potential therapeutic uses for thalidomide. J Am Med Inform Assoc 2003;10:252–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal P, Gearls DB. Can literature analysis identify innovative drivers in drug discovery. Nat Rev Drug Discovery 2009;8:865–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smalheiser NR. Literature based discovery: beyond the ABCs. JASIST 2012;63:218–24 [Google Scholar]
- 21.Weeber M, Klein H, Aronson AR, et al. Text-based discovery in biomedicine: the architecture of the DAD-system. Proceedings. AMIA Symposium 2000:903–7 [PMC free article] [PubMed] [Google Scholar]
- 22.Weeber M, Klein H, De Jong-van den Berg LTW, et al. Using concepts in literature based discovery. JASIST 2001;52:548–57 [Google Scholar]
- 23.Van Mulligan EM, van der Eijk C, Kors JA, et al. Research for research: tools for knowledge discovery and visualization. Proc AMIA Symp 2002:835–9 [PMC free article] [PubMed] [Google Scholar]
- 24.Rebholz-Schuhman D, Cameron G, Clark D, et al. SYMBIOmatics: synergies in medical informatics and bioinformatics—exploring current scientific literature for emerging topics. BMC Bioinformatics 2007;8(Suppl 1):S1–S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hettne KM, Weeber M, Laine ML, et al. Automatic mining of the literature to generate new hypotheses for the possible link between periodontitis and atherosclerosis: lipopolysaccharide as a case study. J Clin Periodontology 2007;34:1016–24 [DOI] [PubMed] [Google Scholar]
- 26.Cohen T, Schvaneveldt R, Widdows D. Reflective random indexing and indirect inference: a scalable method for discovery of implicit connections. J Biomed Inform 2010;43:240–56 [DOI] [PubMed] [Google Scholar]
- 27.Faro A, Giordano D, Spampinato C. Combining literature text mining with microarray data: advances for system biology modelling. Brief Bioinform 2011;13:61–82 [DOI] [PubMed] [Google Scholar]
- 28.Tiffin N, Kelso JF, Power AR, et al. Integration of text- and data-mining using ontologies successfully selects disease candidate genes. Nucleic Acids Res 2005;33:1544–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat Rev Genet 2006;7:119–29 [DOI] [PubMed] [Google Scholar]
- 30.Hristovski D, Stare J, Peterlin B, et al. Supporting discovery in medicine by association rule mining in Medline and UMLS. Stud Health Technol Inform 2001;84(Pt 2):1344–8 [PubMed] [Google Scholar]
- 31.Srinivasan P. MeSHmap: a text mining tool for MEDLINE. Proceedings. AMIA Symposium 2001:642–6 [PMC free article] [PubMed] [Google Scholar]
- 32.Srinivasan P, Rindflesch T. Exploring text mining from MEDLINE. Proceedings. AMIA Symposium 2002:722–6 [PMC free article] [PubMed] [Google Scholar]
- 33.Chen ES, Hripcsak G, Xu H, et al. Automated acquisition of disease drug knowledge from biomedical and clinical documents: an initial study. J Am Med Inform Assoc 2008;15:87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen AM, Hersch WR. A survey of current work in biomedical text mining. Brief Bioinformatics 2005;6:57–71 [DOI] [PubMed] [Google Scholar]
- 35.Ambert KH, Cohen AM. A system for classifying disease comorbidity status from medical discharge summaries using automated hotspot and negated concept detection. J Am Med Inform Assoc 2009;16:590–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jolles J, Houx PJ, Van Boxtel MPJ, et al. eds. Maastricht aging study: determinants of cognitive aging. Maastricht: Neuropsych Publishers, 1995 [Google Scholar]
- 37.Van Boxtel MPJ, Buntinx F, Houx PJ, et al. The relation between morbidity and cognitive performance in a normal aging population. J Gerontology 1998;53A:M146–54 [DOI] [PubMed] [Google Scholar]
- 38.Andrade LH, Viana MC Bensenor, et al. Clustering of psychiatric and somatic illnesses in the general population: multimorbidity and socioeconomic correlates. Braz J Med Biol Res 2010;43:483–91 [DOI] [PubMed] [Google Scholar]
- 39.Li C, Ford ES, Strine TW, et al. Prevalence of co-morbid depression among U.S. adults with diabetes: findings from the 2006 behavioral risk factor surveillance system. Diabetes Care 2008;31:105–7 [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez JS, Safren SA, Cagliero E, et al Depression, self-care and medication adherence in type 2 diabetes: relationships across the full range of symptom severity. Diabetes Care 2007;30:2222–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipscomb CE. Medical Subject Headings (MeSH). Bull Med Libr ASSoc 2000;88:265–6 [PMC free article] [PubMed] [Google Scholar]
- 42.Jelier R, Schuemie MJ, Veldhoven A, Kors JA, et al. Anni 2.0: a multipurpose text-mining tool for the life sciences. Genome Biol 2008;9:R86 See http://genomebiology.com/2008/9/6/R86 for additional data file with available overview of published text-mining tools, including Anni 2.0, and their functionality [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aronson AR. Filtering the UMLS metathesaurus for MetaMap. Technical Report. National Library of Medicine, 2006. http://skr.nlm.nih.gov/papers/references/filtering06.pdf
- 44.Wheeler DL, Barret Tç, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2007;35[Database issue]:D5–D12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van der Eijk C, Van Mulligen E, Kors JA, et al. Constructing an associative concept space for literature based discovery. JASIST 2004;55:436–44 [Google Scholar]
- 46.Jelier R, Schuemie MJ, Roes PJ, et al. Literature-based concept profiles for gene annotation: the issue of weighting. Int J Med Inform 2008;77:354–61 [DOI] [PubMed] [Google Scholar]
- 47.Schuemie M, Chicester C, Lisacek F, et al. Assignment of protein function and discovery of novel nucleolar proteins based on automatic analysis of Medline. Proteomics 2007;7:921–31 [DOI] [PubMed] [Google Scholar]
- 48.Van Haagen HH, 't Hoen PA, Bovo A Botelho, et al. Novel protein-protein interactions inferred from literature context. PLos ONE 2009;4:e7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metsemakers JF, Knottnerus JA, van Schendel GJ, et al. Unlocking patients’ records in general practice for research, medical education and quality assurance: the Registration Network Family Practices. Int J Biomed Comput 1996;42:43–50 [DOI] [PubMed] [Google Scholar]
- 50.Lamberts H, Wood M. eds. International classification of primary care. Oxford: Oxford University Press, 1987 [Google Scholar]
- 51.Sheskin DJ. Handbook of parametric and nonparametric statistical procedures. 4th edn. Boca Raton: Chapman &Hall/CRC, 2007 [Google Scholar]
- 52.Tversky A, Kahneman D. The framing of decisions and the psychology of choice. Science 1981;211:453–8 [DOI] [PubMed] [Google Scholar]
- 53.Higgins ET. Knowledge activation: accessibility, applicability, and salience. In: Higgins ET, Kruglanski AW. Social psychology—handbook of basic principles. New York: Guilford Press, 1996:133–68 [Google Scholar]
- 54.Wieringa NF, Pesschar JL, Denig P, et al. Connecting pre-marketing clinical research and medical practice: core issues and proposed changes in drug regulation. Int J Technol Assess Health Care 2003;19:202–19 [DOI] [PubMed] [Google Scholar]
- 55.Kunitz SJ. Holism and the idea of general susceptibility to disease. Int J Epidemiol 2002;31:722–9 [DOI] [PubMed] [Google Scholar]
- 56.Golden SH, Lazo M, Carnethon M, et al. Examining a bidirectional association between depressive symptoms and diabetes. JAMA 2008;229:2751–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aarts S, Van den Akker M, Van Boxtel MP, et al. Diabetes mellitus type II as a risk factor for depression: a lower than expected risk in general practice wetting. Eur J Epidemiol 2009; 24:641–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Douzenic A, Tsopelas C, Tzeferakos G. Medical comorbidity of cluster B personality disorders. Curr Opin Psychiatry 2012;25:398–404 [DOI] [PubMed] [Google Scholar]
- 59.Reich J, Schatzberg A. Personality traits and medical outcome of cardiac illness. J Psychiatr Res 2010;44:1017–20 [DOI] [PubMed] [Google Scholar]


