Abstract
Objective
To determine whether the knowledge contained in a rich corpus of local terms mapped to LOINC (Logical Observation Identifiers Names and Codes) could be leveraged to help map local terms from other institutions.
Methods
We developed two models to test our hypothesis. The first based on supervised machine learning was created using Apache's OpenNLP Maxent and the second based on information retrieval was created using Apache's Lucene. The models were validated by a random subsampling method that was repeated 20 times and that used 80/20 splits for training and testing, respectively. We also evaluated the performance of these models on all laboratory terms from three test institutions.
Results
For the 20 iterations used for validation of our 80/20 splits Maxent and Lucene ranked the correct LOINC code first for between 70.5% and 71.4% and between 63.7% and 65.0% of local terms, respectively. For all laboratory terms from the three test institutions Maxent ranked the correct LOINC code first for between 73.5% and 84.6% (mean 78.9%) of local terms, whereas Lucene's performance was between 66.5% and 76.6% (mean 71.9%). Using a cut-off score of 0.46 Maxent always ranked the correct LOINC code first for over 57% of local terms.
Conclusions
This study showed that a rich corpus of local terms mapped to LOINC contains collective knowledge that can help map terms from other institutions. Using freely available software tools, we developed a data-driven automated approach that operates on term descriptions from existing mappings in the corpus. Accurate and efficient automated mapping methods can help to accelerate adoption of vocabulary standards and promote widespread health information exchange.
Keywords: automated mapping, LOINC, local laboratory tests, health information exchange, supervised machine learning, information retrieval
Background and significance
Health information technology has the potential to improve the quality and efficiency of care.1 However, the clinical data needed to make care decisions are often unavailable to providers at the right time and place.2 Whereas our patients seek care across many settings and institutions,3 the purview of our clinical information systems is usually curbed at organizational boundaries. Even within a single institution, the laboratory, radiology, pharmacy, and clinical note writing systems may function like data ‘islands’. Efficiently moving and aggregating patient data creates an important foundation for many tools and processes with the capability of improving healthcare delivery. The Health Information Technology for Economic and Clinical Health (HITECH) Act considerably increases the prospect of widespread electronic health record systems (EHRs) with health information exchange capabilities.4 HITECH requires that providers and hospitals demonstrate that EHR information exchange is eligible for the Medicare and Medicaid incentive payments.
A central barrier to efficient health information exchange is the unique local names and codes for the same clinical test or measurement performed at different institutions. When integrating many data sources, the only practical way to overcome this barrier is by mapping local terms to a vocabulary standard. Logical Observation Identifiers Names and Codes (LOINC) is a universal code system for identifying laboratory and clinical observations.5 When LOINC is used together with messaging standards such as HL7, independent systems can create interfaces with semantic interoperability for electronically reporting test results. LOINC has been adopted both in the USA and internationally by many organizations, including large reference laboratories, healthcare organizations, insurance companies, regional health information networks, and national standards.6–8 Within the USA, one recent and notable adoption of LOINC is as the standard for laboratory orders and results in the standards and certification criteria of the centers for Medicare and Medicaid services EHR ‘meaningful use’ incentive program.9
Before care organizations can realize the benefit of using vocabulary standards like LOINC, they must first map their local codes to terms in the standard. Unfortunately, this process is complex. It requires considerable domain expertise and is resource-intensive.8 10–12 Reducing the effort required to accurately map local terms to LOINC would accelerate interoperable health information exchange and would be especially helpful for resource-challenged institutions.
The Regenstrief LOINC mapping assistant (RELMA), a desktop program freely distributed with LOINC (http://loinc.org), is widely used by domain experts to map their local terms to LOINC one by one.12–15 It also contains a feature called the RELMA Auto Mapper that processes a set of local terms in batches and identifies a ranked list of candidate LOINC codes for each local term in the collection. Although RELMA's automated mapping feature has accurately mapped radiology report terms,16 17 laboratory terms present special challenges because of their characteristically short and ambiguous test names.8 10 18
Previous studies have described several methods and tools for mapping laboratory terms to LOINC. Lau et al19 used parsing and logic rules in conjunction with synonyms, attribute relationships, and mapping frequency data to map local laboratory test names to LOINC. This paper was a descriptive analysis and did not include an evaluation of its accuracy. Zollo and Huff used extensional definitions of laboratory concepts generated from actual test result data to map between two laboratories using a common dictionary that was also linked to LOINC.20 An extensional definition for a given laboratory term is a profile of fields created from the test result instance data. Zollo and Huff used fields such as the mean result value, centile for frequency within the dataset, units of measure, and an array of co-occurring concepts. The automated matching software that leveraged these extensional definitions correctly identified 75% of the possible matches. In addition to establishing new mappings, extensional definitions have also been used for auditing and characterizing the degree of interoperability of existing local laboratory terms to LOINC mappings.11 21 Sun and Sun evaluated the performance of an automated lexical mapping program on terms from three institutions to LOINC.22 The overall best lexical mapping algorithm identified the correct LOINC code for between 63% and 75% of local terms. Kim et al18 described an approach for augmenting local term names that modestly improved mapping results using RELMA for term-by-term mapping. Lastly, Khan et al15 developed an automated tool that used a master file of mapped local terms from several sites within the Indian health service. The local terms at these sites shared a common heritage, but had diverged over time in their naming conventions. Compared with a ‘gold standard’ mapping established by a term-by-term search with RELMA, the automated method correctly mapped 81% of the test terms.
Over the past 18 years, Regenstrief has mapped local terms from many institutions to a common dictionary as part of the process of creating and expanding the Indiana network for patient care (INPC), a comprehensive regional health information exchange.23 Thus, the INPC dictionary now represents a rich corpus of local terms mapped to LOINC. Like Lau et al and Khan et al, we hypothesized that the knowledge contained in this corpus of mappings could be leveraged to help map local terms from other institutions.
To test this corpus-based approach, we developed two models based on supervised machine learning and information retrieval using open-source tools. Our data-driven approach relies exclusively on a rich corpus of local term descriptions and does not directly reference the LOINC terminology. In this study we present the process of creating and validating these models and testing their performance on a set of local laboratory terms from three institutions. We also compare the performance of these models with that of the recently improved Lab Auto Mapper feature within RELMA.
Methods
Establishing the gold standard and normalizing the corpus
We compiled a corpus of all local terms from 104 different institutional code sets that were mapped to LOINC through the INPC common dictionary between 1997 and 2012. Each local term from these sets had been mapped by domain experts at Regenstrief through manual review, assisted by the use of RELMA and other locally developed tools. For all analyses, these existing LOINC mappings from the operational health information exchange served as our gold standard. A description of how Regenstrief performs and maintains the mappings in the INPC has been published previously.12 We did not perform additional auditing of the mappings as part of this analysis.
For each local term in the corpus, the set of words constituting its description (eg, the laboratory test name) was normalized using Apache Lucene's V.3.0.3 StandardAnalyzer.24 25 The Lucene StandardAnalyzer generates a set of tokens from the local term descriptions using lexical rules to recognize alphanumeric characters, convert strings to lowercase, and remove stop words. It splits strings at punctuation characters and removes the punctuation, except for a few cases. A dot that is not followed by whitespace is considered part of a token. Input strings are split into tokens at hyphens unless there is a number in the token, in which case the whole token is interpreted as a product number and is not split. For example, the local term descriptions ‘CSF CELL COUNT/DIFF’ and ‘GLU (TOL) UR-5 HR’ are normalized (tokenized) to each yield four tokens ‘csf’, ‘cell’, ‘count’, ‘diff’ and ‘glu’, ‘tol’, ‘ur-5’, ‘hr’, respectively.
Creating a model based on supervised machine learning—Maxent
We used Apache's OpenNLP Maxent V.3.0.126 to create a maximum entropy-based statistical algorithm for supervised machine learning. The principle of maximum entropy provides a probability distribution that is as uniform as possible by assuming nothing about what is unknown.27 The probability distribution derived from human specified constraints in training data is then used to predict the probability of a random set of constraints in test data.
To create a Maxent model each local term in the training set was considered as a separate event with its normalized description used as predicates and the mapped LOINC code used as outcome. When normalized local terms from the test set were applied against this model, Maxent calculated a probability score between zero and one for each LOINC code (outcome) contained in the corpus. The LOINC codes with the highest score (top 1) and those with the highest five scores (top 5) were noted for each local term.
Creating a model based on information retrieval—Lucene
We used Apache's Lucene V.3.0.324 to create an information retrieval-based model. Lucene is a popular information retrieval library that creates documents with indexed fields for fast searching. Its scoring formula matches the similarity between indexed fields and search terms for each document.25 Lucene's approach combines the Boolean model of information retrieval28 and the vector space model28–30 of information retrieval. Briefly, documents "approved" by the Boolean model are scored by the vector space model. In the vector space model, documents and queries are represented as weighted vectors in a multidimensional space, where each distinct index term is a dimension, and weights are the commonly used term frequency, inverse document frequency (TF-IDF) values.29 30
To create a Lucene model we created separate documents for every unique LOINC code in the training set. Each document then contained the normalized description from all local terms mapped to that LOINC code as its indexed field. When normalized local terms from the test set were queried against this model, Lucene calculated a score for each LOINC code (document) contained in the corpus. This score was based on the number of times queried words co-occurred with that document and the total number of documents associated with those words. The Lucene score ranged from zero with no fixed upper bound value.
An example of the models created by Maxent and Lucene
To illustrate use of the Maxent and Lucene models, consider a corpus that contains only five terms with manually mapped LOINC codes as shown in table 1. The data from this corpus are used to create a Maxent model with five events and three outcomes as shown in table 2. It is also used to create a Lucene model with three documents and corresponding indexed fields as shown in table 3. Note that the Lucene model concatenates all the term descriptions from different institutions mapped to the same LOINC code. Now suppose that a test institution contains five unmapped terms ‘indirect’, ‘direct’, ‘coombs’, ‘bilirubin’ and ‘direct, test’. The test terms are first normalized using the same StandardAnalyzer that was used to normalize the corpus. The first four test terms are each converted into one token ‘indirect’, ‘direct’, ‘coombs’ and ‘bilirubin’, respectively, whereas the last test term is converted to two tokens ‘direct’ and ‘test’. When these normalized (tokenized) term descriptions are applied against Maxent and Lucene, each model returns a set of three scores that represents the likelihood of that test term being mapped to each of the three LOINC codes contained in the corpus (table 4).
Table 1.
Hypothetical corpus containing five local terms
| Local code | Term description | Normalized term description | Mapped LOINC code |
|---|---|---|---|
| 12802 | Indirect AGT | indirect agt | 1003-3 |
| IAT | Indirect Coombs’ test | indirect coombs | 1003-3 |
| DCTG | Direct Coombs’ test | direct coombs test | 1006-6 |
| 18231 | Direct Coombs’ IgG Ab | direct coombs igg ab | 1006-6 |
| BILID | Bilirubin, Direct | bilirubin direct | 1968-7 |
Table 2.
Representation of the Maxent model based on the corpus shown in table 1
| Event # | Predicates (normalized term description) | Outcome |
|---|---|---|
| 1 | indirect agt | 1003-3 |
| 2 | indirect coombs | 1003-3 |
| 3 | direct coombs igg ab | 1006-6 |
| 4 | direct coombs test | 1006-6 |
| 5 | bilirubin direct | 1968-7 |
Table 3.
Representation of the Lucene model based on the corpus shown in table 1
| Document ID | Indexed field (normalized term description) |
|---|---|
| 1003–3 | indirect agt indirect coombs |
| 1006–6 | direct coombs igg ab direct coombs test |
| 1968–7 | bilirubin direct |
Table 4.
Maxent and Lucene scores for each LOINC from the corpus in table 1 when local term descriptions are queried against both models
| Maxent model scores | Lucene model scores | |||||
|---|---|---|---|---|---|---|
| Normalized term description/LOINC code | 1003–3 | 1006–6 | 1967–7 | 1003–3 | 1006–6 | 1967–7 |
| ‘indirect’ | 0.9134 | 0.0432 | 0.0432 | 1.9876 | 0.0000 | 0.0000 |
| ‘direct’ | 0.1352 | 0.4558 | 0.4090 | 0.0000 | 1.4142 | 1.0000 |
| ‘coombs’ | 0.5019 | 0.3704 | 0.1277 | 1.0000 | 1.4142 | 0.0000 |
| ‘bilirubin’ | 0.0241 | 0.0241 | 0.9517 | 0.0000 | 0.0000 | 1.4054 |
| ‘direct’, ‘test’ | 0.0136 | 0.9451 | 0.0412 | 0.0000 | 1.9651 | 0.2899 |
Evaluation approach
To characterize how well these models performed in mapping local terms to LOINC, we conducted three sets of analyses that are described in detail in the following sections. In each case, the top five scoring LOINC codes were compared with the LOINC code assigned by manual mapping (our gold standard). We chose to limit the list of LOINC codes returned by the analyses to the top five based on our practical experience with mapping and preliminary analyses that showed it was rare for the correct LOINC code to appear in the next few rankings. Domain experts can quickly review a short list of ranked candidate LOINC codes to determine which, if any, of the LOINC codes is the correct match. A longer list is more cumbersome to review, and our experience has been that mappers prefer an interactive search interface like RELMA for reviewing a long list of candidate codes.
In contrast to conventional information retrieval analyses, our mapping context used a very strict definition of ‘relevance’ in that one and only one ‘document’ (the gold standard LOINC code) is ever deemed relevant (or correct). When we report results for when the correct LOINC code is ranked first, this is equivalent to the traditional measure of precision (true positives/(true positives+false positives)). Because our models were very greedy at returning some LOINC codes from the training set as ‘positive’ (even if this was not correct), we opted not to report the recall (true positives/(true positives+false negatives)) for our models.
Validating the models using 80/20 splits
We validated the predictive performance of both models using a random subsample method (80% for training and 20% for testing) that was repeated 20 times. For each of the iterations, 80% of local terms from our normalized corpus were randomly selected as the training set to create Maxent and Lucene models as described above. Normalized local term descriptions from the remaining 20% that served as the test set were then queried against both models. We chose this approach to cross-validation to help prevent the models from being over fitted. Splitting the corpus at the term level (rather than at the level of a whole set of terms from an institution) demonstrates the prediction of the models for a heterogeneous set of terms with varying naming conventions. The top five scoring LOINC codes resulting from each model were compared with the LOINC code assigned by manual mapping (our gold standard).
Evaluating the models’ performance using local terms from three test institutions and comparison with Lab Auto Mapper
We determined the performance of our models in mapping an entire set of local laboratory terms from three test institutions. The INPC contains data from a variety of healthcare facilities, including large hospital systems, referral laboratories, diagnostic imaging centers, etc. Because we wanted to characterize the performance of our models for a ‘typical’ laboratory catalog, we selected as our test institutions three convenient community hospital laboratories. These three institutions were geographically dispersed across the state, not part of the same health system, and like most (but not all) institutions had separated their laboratory codes from other clinical results (radiology reports, dictated notes, etc). In this case, training sets comprising all terms in our corpus minus those belonging to the three test institutions were used to create Maxent and Lucene models as described above. Normalized local terms descriptions from the corresponding test sets containing all laboratory terms from the three institutions were then applied against both models. This approach simulates the typical mapping scenario of integrating all the terms from a new institution's laboratory system. Each institution code set covers the set of tests performed by typical community hospital laboratory, and reflects the idiosyncratic naming conventions established by that institution. The top five scoring LOINC codes resulting from each model were compared with the LOINC code assigned by manual mapping (our gold standard).
We also compared the performance of the models with RELMA's Lab Auto Mapper for local terms from these three test institutions. For this analysis we used the most recent publicly available version, RELMA v5.6.31 The Lab Auto Mapper uses a series of algorithms optimized for laboratory terms to generate a list of candidate LOINC codes. In addition to using words contained in a local term's description, it can also leverage information from battery terms, units of measures, common tests, and the synonymy contained in LOINC. Its score is based on the number and proportion of words that are matched between the local term and the fully specified LOINC name.32 We followed the recommended procedures for loading local terms into RELMA and running the Lab Auto Mapper as described in the RELMA Users’ Manual and LOINC and RELMA tutorial produced by Regenstrief Institute.32 33
Lastly, we investigated whether a threshold Maxent score could serve as a useful cut-off score for always identifying the correct LOINC code. We first plotted the rank of the correct LOINC code among the top five against its Maxent score for each term in the test set, and then evaluated the Maxent score above which the correct LOINC code was always ranked first.
Evaluating the models’ performance with incremental growth in corpus size
To determine our models’ performance against an incrementally growing corpus we again used the test set of all local laboratory terms from the three institutions as above. However, this time 12 training sets were used, each containing local terms from the corpus (minus those in the test set) in chronological order based on the time stamp when they were manually mapped. Each training set contained terms in 6400 increments. Thus, the first training set contained the first 6400 local terms created in the corpus; the second training set contained the first 12 800 local terms; and the twelfth and last training set contained all the local terms. Our choice of creating 12 training sets (each with 6400 additional terms) was arbitrary, but illustrates how the models perform as the corpus grows when new terms are added.
Normalized local term descriptions from the test set were applied against Maxent and Lucene models created from each of the 12 training sets. The top five scoring LOINC codes resulting from each model were compared with the LOINC code assigned by manual mapping (our gold standard).
Results
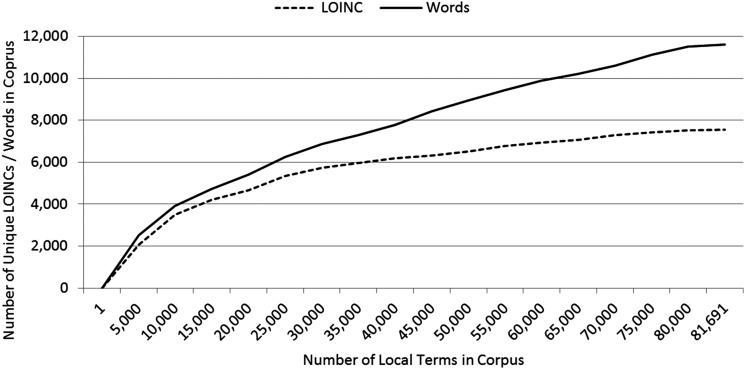
Our corpus from 104 institutional code sets contained 81 691 local terms, each associated with a description and mapped to a LOINC code. These local terms were mapped to 7565 unique LOINC codes and contained 244 405 total words and 11 620 unique words in their descriptions (test names). This corpus was built from 1997 to 2012 as a byproduct of the INPC expansion. New local terms were added to the INPC master dictionary and mapped to LOINC both because new institutions began to participate in the health information exchange and because participating institutions created new local terms. Figure 1 shows the growth in number of unique LOINC codes and number of unique words associated with all local terms as the corpus has expanded with new local terms.
Figure 1.
Growth of unique LOINC codes mapped to local terms and unique words in local term descriptions with an incrementally growing corpus.
Results of validating the models using 80/20 splits
In each of the 20 iterations of random subsampling from our corpus into 80% for training and 20% for testing, there were 65 361 local terms in the training set and 16 330 local terms in the test set. The number of unique LOINC codes to which these local terms were mapped varied between 7115 and 7190 for the training set and between 4391 and 4493 for the test set.
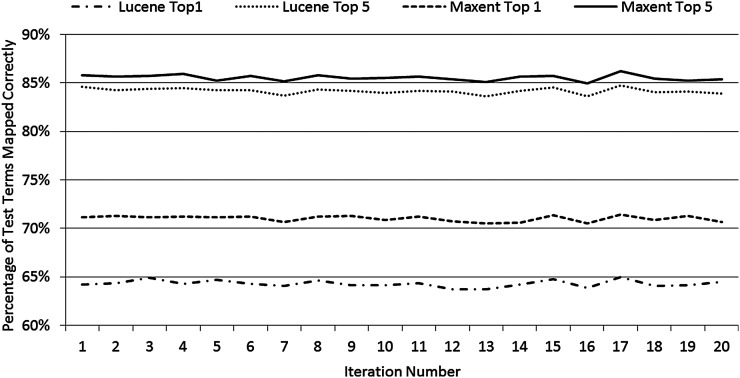
Maxent ranked the correct (manually mapped) LOINC code first for 11 513 to 11 661 (70.5–71.4%, mean 71.0%) local terms in the test sets and ranked the correct LOINC code among the top five for 13 871 to 14 073 (84.9–86.2%, mean 85.5%). Lucene ranked the correct LOINC code first for 10 407 to 10 610 (63.7–65.0%, mean 64.3%) of local terms in the test sets and ranked the correct LOINC code among the top five for 13 649 to 13 841 (83.6–84.8%, mean 84.2%). These results for each of the 20 iterations are shown in figure 2.
Figure 2.
Results of 20 iterations of repeated random subsampling validation showing the percentage of test terms with manually mapped LOINC codes ranked first (top 1) and among the top 5 by Maxent and Lucene.
Results of Maxent, Lucene and Lab Auto Mapper using laboratory terms from three test institutions
The first test institution contained 1099 local laboratory terms mapped to 573 unique LOINC codes and has 2539 total words and 667 unique words. The second test institution contained 1705 local laboratory terms mapped to 757 unique LOINC codes and has 3582 total words and 898 unique words. Finally, the third test institution contained 838 local laboratory terms mapped to 328 unique LOINC codes and has 1431 total words and 428 unique words. The results of applying these test sets against the Maxent model, the Lucene model, and the Lab Auto Mapper are shown in table 5. Averaging the performance across the three test institutions, the three mapping methods ranked the correct LOINC code first for 78.9%, 71.9%, and 50.3% and ranked among the top five for 91.4%, 90.0%, and 68.6% of local terms when applied against Maxent, Lucene, and Lab Auto Mapper, respectively.
Table 5.
Percentage of local laboratory terms from each test institution that when applied against Maxent, Lucene and Lab Auto Mapper had the correct LOINC code ranked highest (top 1) and among the highest five (top 5)
| Institution 1 | Institution 2 | Institution 3 | ||||
|---|---|---|---|---|---|---|
| (n=1099) | (n=1705) | (n=838) | ||||
| % | n | % | n | % | n | |
| Maxent top 1 | 78.6 | (864) | 73.5 | (1253) | 84.6 | (709) |
| Lucene top 1 | 72.6 | (798) | 66.5 | (1133) | 76.6 | (642) |
| Lab Auto Mapper top 1 | 49.6 | (545) | 46.8 | (798) | 54.5 | (457) |
| Maxent top 5 | 90.5 | (995) | 88.8 | (1514) | 94.7 | (794) |
| Lucene top 5 | 89.8 | (987) | 86.0 | (1466) | 94.3 | (790) |
| Lab Auto Mapper top 5 | 71.8 | (789) | 66.9 | (1140) | 67.1 | (562) |
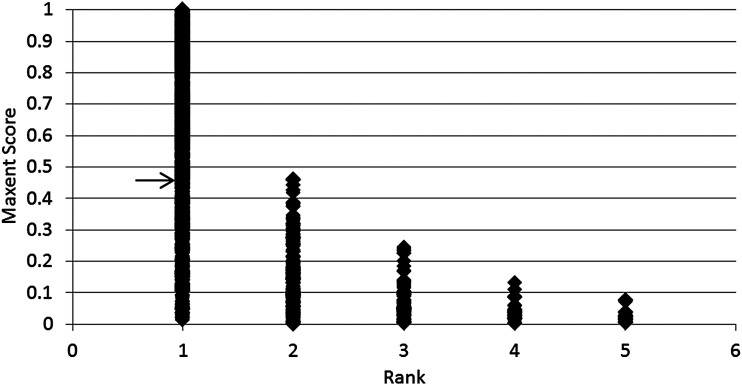
For the 3642 local terms in the three test sets, ranks of the correct LOINC codes among the top five were plotted against their Maxent scores. As illustrated in figure 3, this plot shows that when the score was above 0.46 the correct LOINC code was always ranked first by the model. Using this cut-off score to separate a high- certainty top rank, Maxent ranked the correct LOINC code first for 2099 (57.6%) of the local terms.
Figure 3.
Rank of correct LOINC codes and their Maxent score for local laboratory terms from three test institutions.
Results of the models’ performance with an incrementally growing corpus
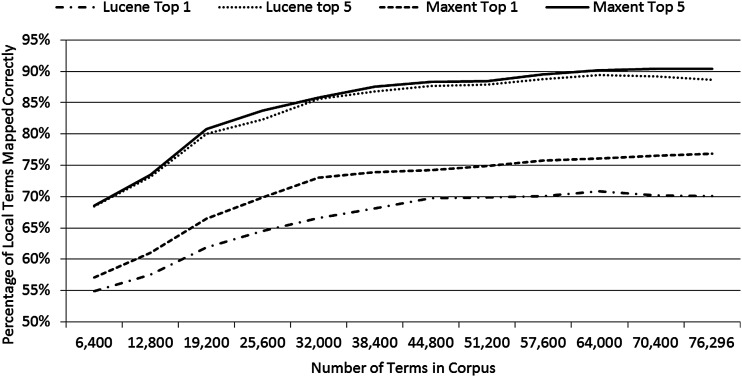
Figure 4 illustrates the performance of both models on the test set containing all local terms from three institutions using a series of training sets that represent an incrementally growing corpus. The training sets in this analysis organized the local terms in the corpus in chronologic order by increments of 6400 terms. The results show a gradual leveling off in Maxent's performance and a slight decrease in Lucene's performance as the number of terms in the corpus reached its maximum.
Figure 4.
Performance of Maxent and Lucene when applying the test set against an incrementally growing corpus.
Discussion
Our study shows that a rich corpus of local terms mapped to LOINC can help to map terms from other institutions. Overall, the supervised machine learning based Maxent model ranked the correct LOINC code first for 79% and the information retrieval based Lucene model for 72% of local laboratory terms from our three test institutions. These results are similar in accuracy to results of the best reported automated techniques from prior studies of laboratory test mapping. Our approach has the advantages of using freely available tools and only requiring local term descriptions as the data substrate.
Rationale for using Maxent and Lucene models
Given a rich corpus of existing mappings established by domain experts, we wanted to explore the validity and performance of a purely data-driven approach to automated LOINC mapping. We used Apache's Maxent to create a supervised machine learning model and Apache's Lucene to create an information retrieval model, as these tools are freely available, offer good performance on typical personal computer hardware, and are relatively easy to deploy.
The usual application of Maxent models involves a binary outcome, such as natural language processing tasks like sentence detection and part of speech tagging. In this study, we created a Maxent model with thousands of outcomes represented by all unique LOINC codes contained in the training corpus. We are not aware of prior studies that have used Maxent in this manner or in the context of automated mapping. Maxent has been used successfully in other informatics applications such as cTAKES,34 which is a natural language processing system for information extraction from electronic medical record clinical free-text. cTAKES uses Maxent for sentence detection, tokenizing, part-of-speech tagging, and chunking.
Lucene is widely used in a variety of applications for document indexing and search engine functions.35 Websites like Wikipedia, LinkedIn, and Twitter all use Lucene in their search features. Since V.5.0 (released December 2010), the search functionality in RELMA has implemented Lucene, including the Lab Auto Mapper. Previous studies have demonstrated that RELMA is a very capable tool for mapping local terms to LOINC.15–18 36 37 Our application of Lucene differs from RELMA in that we did not directly query the LOINC terminology at all. Whereas RELMA queries against the stylized LOINC names and synonyms included in LOINC, both the Lucene and Maxent models in our approach only queried against words from local term descriptions mapped to LOINC codes. We had hypothesized that the idiosyncratic variation present in a large corpus of local term descriptions might help overcome the challenge of relying on the synonymy in LOINC. Although the synonymy in LOINC is quite good for common abbreviations, the standards development process cannot possibly keep up with all the permutations of abbreviations seen in local term names. For example, just a few of the variants for ‘Neisseria gonorrhoeae’ present in our corpus include: ‘N.GONORRHOEA’, ‘N. GONORRHEAE’, ‘N.GONO’, ‘Gono’, ‘N. GONORR.’, ‘NEISS GONORR’, and ‘NEISSERIA GONORR’.
As is typical for information retrieval and machine learning applications, we normalized the strings of the local term descriptions to reduce the inherent variability. We used the freely available Lucene StandardAnalyzer to achieve this abstraction because it provides fast, basic normalization for European-language strings and is commonly used wherever Lucene is employed. We used the same normalization technique for use with both Maxent and Lucene so that the normalization process would not be a source of variability between the methods. The normalization performed by the StandardAnalyzer is quite simple. We recognize that it is possible that other normalization tools may perform better for this purpose. For example, the Norm program that is part of the UMLS Lexical tools38 creates an abstract representation of text strings in lower case, without punctuation, genitive markers, or stop words, diacritics, ligatures, with each word in its uninflected form, the words sorted in alphabetical order, and normalizes non-ASCII Unicode characters to ASCII. Moreover, other stemming algorithms39 or methods for term40 or concept41 identification may perform better at providing inputs into Maxent or Lucene. Alternatively, more aggressive normalization schemes may lose important information from the term descriptions. For example, an approach that stripped plural forms may normalize ‘amphetamines’ to ‘amphetamine’, which would lose the distinction of whether the drug screen was testing a class of compounds (eg, methamphetamine, amphetamine, MDMA (ecstasy), MDEA (Eve), etc) or the single chemical species ‘amphetamine’. Nevertheless, we believe that development and evaluation of advanced normalization schemes in combination with scoring techniques like Maxent and Lucene are important areas for future study.
Our approach with the Maxent and Lucene models is relatively simple compared with the processing algorithm of the RELMA Lab Auto Mapper or the drug-centric token matching approach employed by Peters et al42 in mapping drug name variants to RxNorm. The models in our approach did not attempt to interpret the semantics of tokens in the term descriptions. The Lab Auto Mapper has functions that try to identify the specimen (eg, cerebrospinal fluid or serum) and uses the units of measure associated with the test to limit candidate LOINC codes to those with a property attribute consistent with those units. For example, based on an internal mapping table, the Lab Auto Mapper would only return LOINC codes with a property of mass concentration if the local term had associated units of μg/dl. Similarly, the drug-centric token matching approach used by Peters et al42 attempts to identify and perform special processing on the drug name in a local string that is not performed on the tokens that may represent other components of the name like strength or dose form. An advantage of our data-driven approach is that it did not require any domain-specific tailoring.
Comparing the performance of Maxent and Lucene
Maxent performed better than Lucene in ranking the correct LOINC code first owing to Maxent's tendency to over fit the model. Maxent thus computes high scores for local terms with words that match very closely with those in training sets. However, both models ranked the correct LOINC code among the top five for more than 90% of local terms from the three test institutions.
Over the 20 iterations of random subsampling using 80/20 splits, Maxent on average identified the correct LOINC code for 2.9% (473) of local terms that Lucene failed to score among the top five. Conversely, Lucene on average identified 1.5% (251) of local terms that Maxent failed to score among the top five. For our analyses on test sets from three institutions, Maxent identified the correct LOINC code for 2.8% (101) of local terms that Lucene failed to score among the top five, whereas Lucene identified 2.4% (89) of local terms that Maxent failed to score among the top five. The relatively small number of terms ranked correctly by one model but not the other illustrates that they perform well on similar kinds of test descriptions.
One important advantage of Maxent over Lucene and Lab Auto Mapper is its normalized score. We used this normalized score to determine a helpful threshold above which only the correct LOINC code was ranked first. Using this cut-off score, we found that over 57% of local terms in our three test institutions could be ranked with a high degree of certainty. Such a cut-off score is valuable in separating local terms that can be mapped with little (or no) human review from those that need more extensive review.
Corpus growth and variability in mapping results across institutions
We probed the robustness of our corpus-based approach by analyzing several different test sets and evaluating performance with incremental growth of the corpus. These aspects are potentially relevant in deciding whether a corpus has reached critical mass to be used effectively for modeling. We observed slightly more variation in accuracy when considering entire term sets from each of our three test institutions than in our random 80/20 splits of the corpus. This suggests that institutions’ particular naming patterns can alter the mapping success even when the corpus is large. As local term mappings were added to our corpus, the growth rate in unique LOINC codes decreased more than the growth rate in unique words in term descriptions. This is a favorable pattern as it indicates a growth in diversity of words associated with LOINC codes already present in the corpus. Our results showed that Maxent's performance was not affected by the incremental growth in our corpus, but there was a slight decrease in Lucene's performance.
Limitations of a data-driven paradigm and potential future research
The primary drawback of our approach is that its success is limited by the relative completeness of the underlying training corpus. Of the 3642 local terms in our three test institutions, 46 were mapped to LOINC codes with no training data, 10 had words not associated with any LOINC code and 69 had words not associated with the correct LOINC code in the training set. While neither Maxent nor Lucene was capable of ranking the correct LOINC for these 125 (3.4%) local terms owing to limitations in the corpus or because their term descriptions were completely new, Lab Auto Mapper ranked the correct LOINC code first for 35 (28%) and among the top five for 45 (36%) of these local terms.
RELMA's Lab Auto Mapper succeeded where our models failed by directly querying the LOINC terminology. It also uses additional information such as the units of measure associated with a local term in its algorithm, and others20 have illustrated how extended profiles built from actual test results can be useful in mapping. Our corpus-based approach solely depends on matching words in term descriptions, and thus a global test name enhancement process such as that described by Kim et al18 may be beneficial. In contrast to the name enhancement process, a major benefit of our approach is that it requires little domain expertise at the front end. Evaluating the combined strengths of these different approaches; exploring the value in adding other axes such as units of measure to the data models; testing alternate algorithms for supervised machine learning; and using information retrieval models like ‘fuzzy search’ would be valuable future research.
Our study has some other important limitations. We used a single corpus of mapped local terms from institutions in a broad but geographically based area. Naming conventions used in other institutions may differ from those in our corpus in important ways that lower the accuracy of mapping with Maxent and Lucene. For instance, we have seen some institutions that use semantically meaningless descriptions such as ‘1001’ in lieu of something that resembles a test name. Clearly, an automated mapping approach like ours would fail to map such local terms. Moreover, significant differences in naming conventions may compromise the ability to normalize term descriptions from training and test data uniformly. We deliberately chose three community hospital laboratories as our test institutions to illustrate performance of the models on exemplar code sets from a typical laboratory, but the naming conventions of small laboratories may vary in important ways from other facilities such as referral laboratories or tertiary care centers. Additionally, since our corpus and test sets contained predominantly laboratory terms, we do not know how well data-driven models would generalize to other important clinical measurement variables.
Considerations for practical application
Expert review is a high cost resource in mapping. By identifying a short, accurate, ranked list of candidate LOINC codes for each local term we can optimize the process of human review. In settings where a large corpus of existing mappings is available, the Maxent model performed the best of those we evaluated and would be our recommendation for producing this ranked list. By choosing a high Maxent cut-off score (eg, >0.46), more than half of the local terms could probably be mapped with little or no human review. If human review of the ranked list reveals that a matching LOINC code is not present, the reviewer can default back to the typical term-specific search using interactive functions of RELMA.
Although the core software tools we used in this study (Maxent and Lucene) are available at no cost under open-source licenses, the corpus of local term descriptions mapped to LOINC from the INPC is not available publicly. Encouraged by the results of this study, the Regenstrief LOINC team recently announced a project to build a shared repository of local terms mapped to LOINC.43 Because it is open to contributions from the global LOINC community, this new repository has the potential to serve as an important data substrate for future analyses.
Conclusion
Our study shows that a rich corpus of local terms mapped to LOINC contains collective knowledge that can help to map terms from different institutions. We developed an automated mapping approach based on supervised machine learning and information retrieval using Apache's Maxent and Lucene, which are available at no cost. Our approach operates on term descriptions from existing mappings in the corpus. Overall, Maxent ranked the correct LOINC code first for 79% and Lucene for 72% of local terms from our three test institutions. Using a cut-off score of 0.46 would allow Maxent to identify over 57% of local terms that always had the correct LOINC code ranked first. Mapping local terms to a vocabulary standard is a necessary, but resource-intensive part of integrating data from disparate systems. Accurate and efficient automated mapping methods can help to accelerate adoption of vocabulary standards and promote widespread health information exchange.
Footnotes
Contributors: MF and DJV conceived and designed the study, collected the data, evaluated the results, and wrote and edited the manuscript. MF created the models and performed the analyses.
Funding: This work was supported, in part, by grant 5T 15 LM007117-14 and contract HHSN2762008000006C from the National Library of Medicine and performed at the Regenstrief Institute, Indianapolis, IN, USA.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Chaudhry B, Wang J, Wu S, et al. Systematic review: impact of health information technology on quality, efficiency, and costs of medical care. Ann Intern Med 2006;144 [DOI] [PubMed] [Google Scholar]
- 2.Smith PC, Araya-Guerra R, Bublitz C, et al. Missing clinical information during primary care visits. JAMA 2005;293:565–71 [DOI] [PubMed] [Google Scholar]
- 3.Finnell JT, Overhage JM, Grannis S. All health care is not local: an evaluation of the distribution of emergency department care delivered in Indiana. AMIA Annu Symposium Proceedings 2011;2011:409–16. [PMC free article] [PubMed] [Google Scholar]
- 4. 111th Congress of the United States of America. American Recovery and Reinvestment Act of 2009. [Google Scholar]
- 5.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem 2003;49:624–33 [DOI] [PubMed] [Google Scholar]
- 6.International LOINC downloads, linguistic variants in RELMA and translating LOINC. http://loinc.org/international/ (accessed 30 Apr 2012).
- 7.Vreeman DJ, Chiaravalloti MT, Hook J, et al. Enabling international adoption of LOINC through translation. J Biomed Inform 2012;45:667–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baorto DM, Cimino JJ, Parvin CA, et al. Combining laboratory data sets from multiple institutions using the logical observation identifier names and codes (LOINC). Int J Med Inform 1998;51:29–37 [DOI] [PubMed] [Google Scholar]
- 9.Department of Health and Human Services. 45 CFR Part 170. Health information technology: Initial set of standards, implementation specifications, and certification criteria for electronic health record technology; Final Rule; published July 28, 2010. [PubMed]
- 10.Lin MC, Vreeman DJ, McDonald CJ, et al. Correctness of voluntary LOINC mapping for laboratory tests in three large institutions. AMIA Annu Symp Proc 2010;2010:447–51 [PMC free article] [PubMed] [Google Scholar]
- 11.Lin MC, Vreeman DJ, Huff SM. Investigating the semantic interoperability of laboratory data exchanged using LOINC codes in three large institutions. AMIA Annu Symp Proc 2011;2011:805–14 [PMC free article] [PubMed] [Google Scholar]
- 12.Vreeman DJ, Stark M, Tomashefski GL, et al. Embracing change in a health information exchange. AMIA Annu Symp Proc 2008:768–72 [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Tokars JI, Lipskiy N, et al. An efficient approach to map LOINC concepts to notifiable conditions. Adv Dis Surveill 2007;4:172 [Google Scholar]
- 14.Dugas M, Thun S, Frankewitsch T, et al. LOINC codes for hospital information systems documents: a case study. J Am Med Inform Assoc 2009;16:400–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan AN, Griffith SP, Moore C, et al. Standardizing laboratory data by mapping to LOINC. J Am Med Inform Assoc 2006;13:353–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vreeman DJ, McDonald CJ. Automated mapping of local radiology terms to LOINC. AMIA Annu Symp Proc 2005:769–73 [PMC free article] [PubMed] [Google Scholar]
- 17.Vreeman DJ, McDonald CJ. A comparison of Intelligent Mapper and document similarity scores for mapping local radiology terms to LOINC. AMIA Annu Symp Proc 2006:809–1 [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H, El-Kareh R, Goel A, et al. An approach to improve LOINC mapping through augmentation of local test names. J Biomed Inform 2012;45:651–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau LM, Johnson K, Monson K, et al. A method for the automated mapping of laboratory results to LOINC. Proc AMIA Symp 2000:472–6 [PMC free article] [PubMed] [Google Scholar]
- 20.Zollo KA, Huff SM. Automated mapping of observation codes using extensional definitions. J Am Med Inform Assoc 2000;7:586–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin MC, Vreeman DJ, McDonald CJ, et al. Auditing consistency and usefulness of LOINC use among three large institutions—using version spaces for grouping LOINC codes. J Biomed Inform 2012;45:658–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun JY, Sun Y. A system for automated lexical mapping. J Am Med Inform Assoc 2006;13:334–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald CJ, Overhage JM, Barnes M, et al. The Indiana network for patient care: a working local health information infrastructure. Health Aff (Millwood) 2005;24:1214–20 [DOI] [PubMed] [Google Scholar]
- 24.Apache Lucene http://lucene.apache.org/ (accessed Apr 30 2012).
- 25.McCandless M, Hatcher E, Gospodnetic O. Lucene in action. Stamford: Manning Publications, 2010 [Google Scholar]
- 26.Apache OpenNLP http://opennlp.apache.org/ (accessed 30 Apr 2012).
- 27.Berger LA, Della Pietra VJ, Della Pietra SA. A maximum entropy approach to natural language processing. Comput Linguist 1996;22:39–71 [Google Scholar]
- 28.Belkin NJ, Croft WB. Information filtering and information retrieval: two sides of the same coin? Commun ACM 1992;35:29–38 [Google Scholar]
- 29.Salton G, McGill MJ. Introduction to modern information retrieval. McGraw-Hill, 1983 [Google Scholar]
- 30.Salton G, Buckley C. Global text matching for information retrieval. Science 1991;253:1012–5 [DOI] [PubMed] [Google Scholar]
- 31.Regenstrief LOINC Mapping Assistant, version 5.6. http://loinc.org/ (accessed Apr 30 2012).
- 32.RELMA version 5.6 Users’ Manual. http://loinc.org/downloads/relma (accessed Apr 30 2012).
- 33.Case JT. Using RELMA. Or…in search of the missing LOINC. http://loinc.org/slideshows/lab-loinc-tutorial (accessed 30 Apr 2012).
- 34.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.PoweredBy—Lucene-java Wiki. http://wiki.apache.org/lucene-java/PoweredBy (accessed 26 Sep 2012).
- 36.Abhyankar S, Demner-Fushman D, McDonald CJ. Standardizing clinical laboratory data for secondary use. J Biomed Inform 2012;45:642–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zunner C, Bürkle T, Prokosch HU, et al. Mapping local laboratory interface terms to LOINC at a German university hospital using RELMA V.5: a semi-automated approach. J Am Med Inform Assoc 2013;20:293–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.National Library of Medicine Lexical Tools. http://lexsrv3.nlm.nih.gov/LexSysGroup/Home/index.html (accessed 21 Dec 2012). [DOI] [PubMed] [Google Scholar]
- 39.Lovins JB. Development of a stemming algorithm. Mech Transl Comput Linguist 1968;11:22–31 [Google Scholar]
- 40.Krauthammer M, Nenadic G. Term identification in the biomedical literature. J Biomed Inform 2004;37:512–26 [DOI] [PubMed] [Google Scholar]
- 41.Huang Y, Lowe HJ, Klien D, et al. Improved identification of noun phrases in clinical radiology reports using a high performance statistical natural language parser augmented with the UMLS Specialist Lexicon. JAMIA 2005;12:275–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters L, Kapusnik-Uner JE, Nguyen T, et al. An approximate matching method for clinical drug names. AMIA Annu Symp Proc 2011;2011:1117–26 [PMC free article] [PubMed] [Google Scholar]
- 43.Regenstrief launches Community Mapping Repository and Asks for Contributions of Existing Mappings to LOINC. http://loinc.org/resolveuid/5fff22576bc94db371020ed12dbc5c34 (accessed 26 Sep 2012).