Abstract
This study aimed to reduce reliance on large training datasets in support vector machine (SVM)-based clinical text analysis by categorizing keyword features. An enhanced Mayo smoking status detection pipeline was deployed. We used a corpus of 709 annotated patient narratives. The pipeline was optimized for local data entry practice and lexicon. SVM classifier retraining used a grouped keyword approach for better efficiency. Accuracy, precision, and F-measure of the unaltered and optimized pipelines were evaluated using k-fold cross-validation. Initial accuracy of the clinical Text Analysis and Knowledge Extraction System (cTAKES) package was 0.69. Localization and keyword grouping improved system accuracy to 0.9 and 0.92, respectively. F-measures for current and past smoker classes improved from 0.43 to 0.81 and 0.71 to 0.91, respectively. Non-smoker and unknown-class F-measures were 0.96 and 0.98, respectively. Keyword grouping had no negative effect on performance, and decreased training time. Grouping keywords is a practical method to reduce training corpus size.
Keywords: Classification/methods; Medical Records Systems, Computerized; Natural Language Processing; Smoking
Objective
To explore practical methods to improve support vector machine (SVM) accuracy in automated patient smoking status extraction by reducing training dataset requirements.
Background and significance
Tobacco exposure is an important oncologic health status to determine. It is a known carcinogen,1 and may also be linked to the development of second cancers.2–4 Furthermore, it is associated with poorer outcomes after cancer therapy,5–7 and also increased long-term side effects.8
Automated smoking detection based upon SVM techniques has been shown to be a reliable and accurate method of clinical text analysis.9–12 However, its application to the ‘real world’ is hindered by the magnitude of training data requirements. For a given number of training samples, the number of features considered by the SVM correlates with training time and accuracy.
The clinical Text Analysis and Knowledge Extraction System (cTAKES) 2.513 smoking status detection package uses an analysis pipeline first described by Savova et al9 in response to the 2006 Informatics for Integrating Biology and the Bedside (i2b2) initiative (see https://www.i2b2.org/) challenge.14 It has subsequently been released under the Apache License 2.0, an open-source license.15 The pipeline was implemented at a large Australian cancer center, to facilitate structured documentation of smoking status within the electronic medical record. The aim of this study was to maximize pipeline performance using a limited number of training samples by grouping keywords into lexically similar categories.
Materials and methods
The Mayo smoking status detection pipeline and cTAKES components have been described extensively elsewhere.9 10 16 Sentence-based smoking status detection first employs a rule-based analytics layer to detect the presence of smoking-related keywords. A second layer is applied to any sentence with smoking information, and detects any negation terms relating to smoking information based upon the published NegEX-algorithm.17 A SVM document classifier then determines whether the sentence relates to past or current smoking (see figure 1). Final document-level smoking status is determined using a rule-based layer to resolve the classifications of multiple sentence-level classifications into a single document-level classification.
Figure 1.
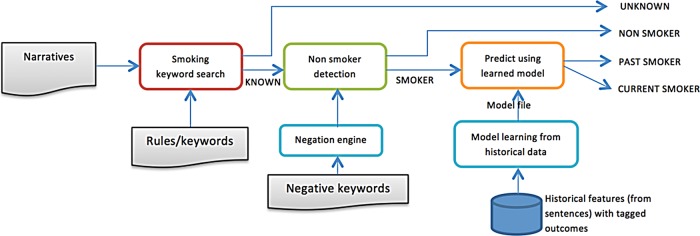
Mayo/Savova Smoking Classification schema.
Dataset
We manually annotated 709 patient narratives to create a corpus for training and validation. The patient narrative history generated from first contact with a patient was used, containing both unstructured and semistructured data. Narratives were selected randomly from clinics at Peter MacCallum Cancer Centre, a specialist cancer hospital. In this instance, the patient narrative relates to a free-text document dictated by a clinician, with variable organizational structure which could include headings. Documents were coded into past smoker (P), current smoker (C), non-smoker (N) or unknown (U). The extracted narratives were converted from HTML to UTF-8 format.
Processing optimization
The apostrophe character was not recognized by the default cTAKES installation, which led to non-detection of negation contractions (ie, she doesn't smoke). We altered the tokenizer to enable detection of common apostrophe characters used (Unicode characters \u2019, \u0027, and \u0092).
Sentence-level checking for the subject assertion was added, to discriminate for smoking-related information not related to the subject (eg, pertaining to family members). Header detection was also incorporated to process only relevant sentences. For example, the header, ‘Smoking History,’ should not be classified as past smoker.
Feature optimization
Grouping of keyword features by equivalent temporality and context was implemented, similar to categories employed by the Penn Treebank tag set18 but with smoking-related application. This reduced the number of features from 84 to 34, with a corresponding decrease in training time, and improved representation of less frequent keywords. We also added localized keywords that were not identified in the original North American feature sets. Table 1 shows a sample of the keyword list employed, which is included for illustrative purposes. We did not discard low-frequency keywords. The word ‘nil’ was added to the list of the negation finite state machine, which is not included in the initial NegEX algorithm.17
Table 1.
Sample of equivalent keyword list
| [Noun (smoker)] | smoker | ||||
| [Noun (past smoker)] | exsmoker | ex_smoker | ex-smoker | past_smoker | |
| [Verb base (smoking)] | smokes | smoke | |||
| [Verb base (now smoking)] | continues_to | still | continue_to | ||
| [Verb past (past smoker)] | smoked | ||||
| [Verb past (past smoker)] | discontinued | ceased | refrained | stopped | quit |
| [Verb past (past smoker)] | has_discontinued | has_ceased | has_quit | has_stopped | gave_up |
| [Verb past (past smoker)] | stopped | gave_up | ceased | quit | |
| [Verb present singular (future)] | will | ||||
| [Time noun (year)] | year | years | yrs | ||
| [Time noun (month)] | month | months | mths | mth | |
| [Time noun (week)] | week | weeks | wks | wk | |
| [Time noun (day)] | day | days | |||
| [Time adjective (current)] | currently | now | |||
| [Time adjective (past)] | previously | used_to | use_to | prior | |
| [Time adjective (used with time noun)] | ago | ||||
| [Time adjective ] | remote | distant | teenager | youth | |
| [Adjective (past smoker)] | former | reformed | past | previous | ex-heavy |
| [Verb base (past smoker)] | refrain | reform | refrains | avoids |
Optimization for local lexicon and data entry
Heading detection was implemented to deal with semistructured data. Information in the family history that was unlikely to be related to the patient was discarded. This worked synergistically with the subject assertion detection we implemented. Several patterns of heading dictation were seen, such as a heading followed by a short phrase (‘Smoking: Nil’) or a heading followed by several sentences of clinical detail. We corrected the latter case, in which the heading was detected as a non-negated smoking-related feature, and generated false-positive results.
SVM retraining and pipeline validation
SVM retraining for classifying past and current smokers was performed using LIBSVM V.3.17.19 Different kernels for SVM hyperplane segmentation were tested, including polynomial (the default kernel), radial basis function (RBF) and sigmoid. We also tested adjusted current smoker to past smoker weights of each model (from 1.0 to 2.5) with the intention of fine tuning the model to predict for more current smokers. To increase training dataset efficiency, 10-fold validation was performed. The corpus was first divided into 10 subsamples. Nine subsamples were used as training and one as validation. This process was repeated 10 times, with each subsample used once as validation data. Accuracy, precision, and F-measure were reported with their correspondent Clopper–Pearson 95% CI using the results from all subsamples.
Results
Our corpus of 709 narratives comprised 16.6% current smokers (C), 30.4% past smokers (P), 32.8% non-smokers (N), and 20.2% of unknown status (U).
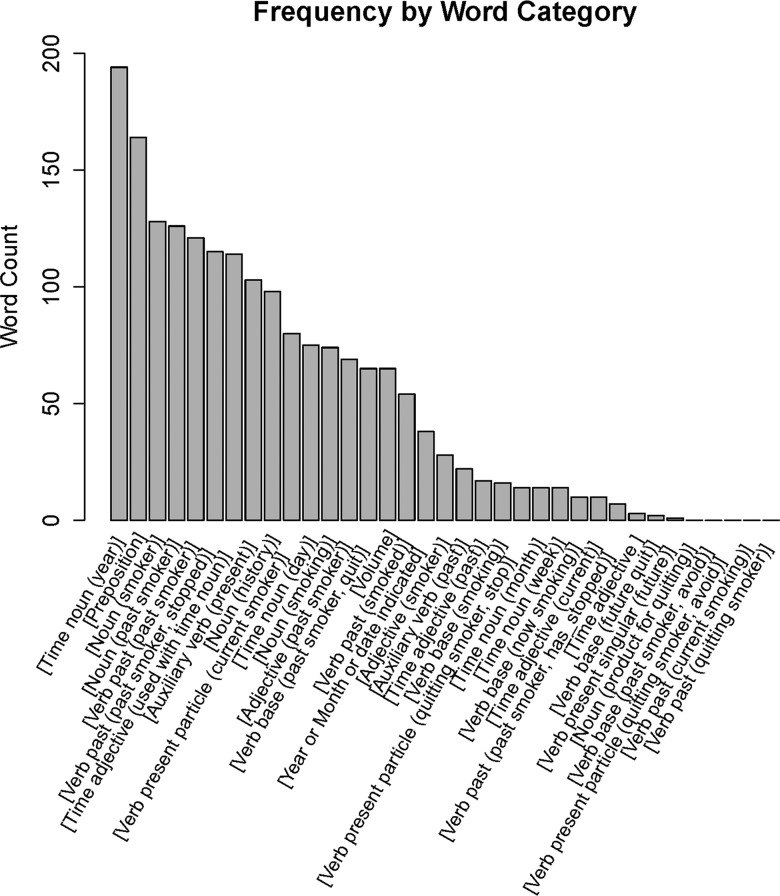
Keyword grouping resulted in 34 feature groups, comprising 84 individual features of interest. The distribution of feature events is shown in figure 2.
Figure 2.
Feature frequency by word category.
Initial accuracy of the cTAKES 2.5 smoking status package with 10-fold cross-validation was 0.69. Incorporating localization and keyword grouping improved system accuracy to 0.9 and 0.92, respectively. F-measures for C and P classes improved from 0.43 to 0.81 and 0.71 to 0.91, respectively. We also tested sigmoid and RBF kernels for SVM hyperplane determination, but these did not improve system performance in any circumstance over the polynomial kernel which is included in the default package. Adjusting the weights of current to past smoker classes of each model resulted in poorer performance for each of the three tested models. Subsequently, we have omitted these results for the sake of brevity. Overall system accuracy and precision, recall, and F-measure for SVM retrained classes are shown in table 2, with 95% CIs given in brackets. The precision, recall, and F-measure for the N were 0.96, 0.96, and 0.96, respectively. For the U class they were 0.95, 1.0, and 0.98.
Table 2.
System performance, and results of support vector machine (SVM) retraining using 10-fold validation
| SVM retraining results | |||||
|---|---|---|---|---|---|
| Model | Overall system accuracy (95% CI) | Class | Precision (95% CI) | Recall (95% CI) | F-measure (95% CI) |
| Original cTAKES, no changes, polynomial SVM | 0.69 (0.65 to 0.72) | C | 0.3 (0.24 to 0.35) | 0.81 (0.72 to 0.89) | 0.43 (0.36 to 0.51) |
| P | 0.87 (0.81 to 0.91) | 0.6 (0.54 to 0.66) | 0.71 (0.64 to 0.77) | ||
| Modified cTAKES rules, original keywords, polynomial SVM | 0.9 (0.87 to 0.92) | C | 0.66 (0.57 to 0.75) | 0.77 (0.68 to 0.85) | 0.71 (0.62 to 0.8) |
| P | 0.92 (0.88 to 0.95) | 0.83 (0.78 to 0.88) | 0.87 (0.83 to 0.91) | ||
| Modified rules, grouped keywords, polynomial SVM default weights | 0.92 (0.9 to 0.94) | C | 0.75 (0.66 to 0.82) | 0.89 (0.81 to 0.94) | 0.81 (0.72 to 0.88) |
| P | 0.97 (0.93 to 0.99) | 0.87 (0.82 to 0.91) | 0.91 (0.87 to 0.95) | ||
| Modified rules, grouped keywords, RBF SVM, default weights | 0.9 (0.88 to 0.92) | C | 0.81 (0.69 to 0.89) | 0.56 (0.45 to 0.66) | 0.66 (0.55 to 0.76) |
| P | 0.85 (0.81 to 0.89) | 0.93 (0.89 to 0.96) | 0.89 (0.85 to 0.92) | ||
| Modified rules, grouped keywords, sigmoid SVM, default weights | 0.84 (0.81 to 0.87) | C | 0.49 (0.39 to 0.59) | 0.53 (0.42 to 0.63) | 0.51 (0.4 to 0.61) |
| P | 0.82 (0.77 to 0.87) | 0.77 (0.72 to 0.82) | 0.8 (0.74 to 0.84) | ||
C, current smoker; cTAKES, clinical Text Analysis and Knowledge Extraction System; P, past smoker; RBF, radial basis function.
Discussion
We were able to achieve 0.92 system accuracy with a corpus of modest size and localization optimization. Initially, a number of errors occurred owing to erroneous detection of smoking-related information included in a thorough family or social history. We subsequently prioritized subject assertion filtering at a sentence and heading level to deal with this problem. Apart from this, however, few changes were made to the overall structure of the pipeline apart from bug fixes related to code additions.
Feature keyword grouping by Penn Treebank tag set categories resulted in modest overall improvements in accuracy. This implies that to construct an equally accurate model using grouped keywords would require fewer training data. The reduction in model complexity from grouping keywords did not impair system performance. Keyword grouping also has several other practical advantages, the most important of which is limiting the circumstances in which retraining of the SVM model is required. Specifically, expanding keyword lists with localized keywords can now be done without retraining the SVM module. This is pertinent considering that many clinical institutions may not possess the technical capabilities to perform retraining of the SVM classifier. The keyword list now expands in a horizontal fashion rather than vertically. Thus, retraining can be avoided except when a new keyword group is added. Words are also grouped by function and temporality, so that adding keywords is intuitive. Assuming that keyword grouping only groups interchangeable words with equivalent meaning, it should result in a lower reliance on corpus size to achieve the same effect. Furthermore, reducing the number of effective keywords also avoids discarding low-frequency, but potentially important, features, which is a common practice in SVM feature selection.
The performance of the SVM module improved out of proportion to overall system accuracy. For instance, the SVM retraining resulted in F-measure improvements for the C class from 0.71 to 0.81, while accuracy improved from 0.9 to 0.92. This is because SVM retraining only affects the classification of smokers into C or P classes (see figure 1 for pipeline schema). As system accuracy depends on the true positive detection of all four classes, the absolute gains from SVM retraining are consequently diluted.
Our document-level results are similar to those reported in other publications using this algorithm. Sohn and Savova10 extended the initial i2b2 entry, and reported final F-measures for detection of P, C, N, and U classes as 0.857, 0.706, 0.961, and 1.0, respectively. Liu et al12 deployed the Mayo algorithm on patient data from Vanderbilt University Hospital, and achieved F-measures for the P, C, and N classes of 0.92, 0.94, and 0.97. Notably, the datasets used in our study are all free text generated by human dictation, and are likely to be very different from the datasets used in previous studies.
The final accuracy is acceptable for deployment in a clinical setting, especially if real-time performance feedback methods are used to increase the training dataset over time. Interestingly, when examining incorrect predictions made by the final model, errors were often related to improper grammar that rendered the meaning of a sentence ambiguous. This suggests that providing real-time feedback might have synergistic benefits with text mining methods from increasing quality of text input as clinical personnel continually improve their grammar.
Our plan is to implement this pipeline at our clinical organization, and use live feedback from clinicians to further train the algorithm. If an acceptably accurate transferrable model can be developed, live training data could supplement the initial corpus and facilitate quicker deployment of the SVM-based algorithms in other centers.
Conclusion
Keyword grouping in our dataset can decrease training dataset requirements without sacrificing accuracy. It has practical advantages of both model expandability and localization.
Footnotes
Contributors: RK, FF, and GD devised the study, wrote the protocol. RK and WR created the validation dataset including manual tagging. W-KY, MB, and WR wrote the statistical plan and performed the analysis, programming, and validation. All authors were involved in drafting the manuscript for production, and approved the final version.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Gandini S, Botteri E, Iodice S, et al. Tobacco smoking and cancer: a meta-analysis. Int J Cancer 2008;122:155–64 [DOI] [PubMed] [Google Scholar]
- 2.Chaturvedi AK, Engels EA, Gilbert ES, et al. Second cancers among 104760 survivors of cervical cancer: evaluation of long-term risk. J Natl Cancer Inst 2007;99:1634–43 [DOI] [PubMed] [Google Scholar]
- 3.Salminen E, Pukkala E, Teppo L. Bladder cancer and the risk of smoking-related cancers during followup. J Urol 1994;152(5 Pt 1):1420–3 [DOI] [PubMed] [Google Scholar]
- 4.Travis LB, Gospodarowicz M, Curtis RE, et al. Lung cancer following chemotherapy and radiotherapy for Hodgkin's disease. J Natl Cancer Inst 2002;94: 182–92 [DOI] [PubMed] [Google Scholar]
- 5.Browman GP, Mohide E, Willan A, et al. Association between smoking during radiotherapy and prognosis in head and neck cancer: a follow-up study. Head Neck 2002;24:1031–7 [DOI] [PubMed] [Google Scholar]
- 6.Browman GP, Wong G, Hodson I, et al. Influence of cigarette smoking on the efficacy of radiation therapy in head and neck cancer. N Engl J Med 1993;328:159–63 [DOI] [PubMed] [Google Scholar]
- 7.Lassen P, Eriksen JG, Hamilton-Dutoit S, et al. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol 2009;27:1992–8 [DOI] [PubMed] [Google Scholar]
- 8.Lilla C, Ambrosone CB, Kropp S, et al. Predictive factors for late normal tissue complications following radiotherapy for breast cancer. Breast Cancer Res Treat 2007;106:143–50 [DOI] [PubMed] [Google Scholar]
- 9.Savova GK, Ogren PV, Duffy PH, et al. Mayo clinic NLP system for patient smoking status identification. J Am Med Inform Assoc 2008;15:25–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sohn S, Savova GK. Mayo clinic smoking status classification system: extensions and improvements. AMIA Annual Symposium Proceedings/AMIA Symposium AMIA Symposium, 2009, 2009:619–23 [PMC free article] [PubMed] [Google Scholar]
- 11.Heinze DT, Morsch ML, Potter BC, et al. Medical i2b2 NLP smoking challenge: the A-Life system architecture and methodology. J Am Med Inform Assoc 2008;15:40–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu M, Shah A, Jiang M, et al. A study of transportability of an existing smoking status detection module across institutions. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium, 2012, 2012:577–86 [PMC free article] [PubMed] [Google Scholar]
- 13. cTAKES 2.5. 2013 [cited 2013 01/03/2013]. https://wiki.nci.nih.gov/display/VKC/cTAKES+2.5.
- 14.Uzuner Ö, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008;15:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Open Health Natural Language Processing (OHNLP) Consortium 2013. [cited 6/1/2013]. http://www.ohnlp.org
- 16.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 18.Marcus MP, Marcinkiewicz MA, Santorini B. Building a large annotated corpus of English: the Penn Treebank. Comput Linguistics 1993;19:313–30 [Google Scholar]
- 19.Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2011;2:27 [Google Scholar]