Abstract
The crystal structure of a pentameric α7 ligand-binding domain chimaera with bound α-btx (α-bungarotoxin) showed that of the five conserved aromatic residues in α7, only Tyr184 in loop C of the ligand-binding site was required for high-affinity binding. To determine whether the contribution of Tyr184 depends on local residues, we generated mutations in an α7/5HT3A (5-hydroxytryptamine type 3A) receptor chimaera, individually and in pairs, and measured 125I-labelled α-btx binding. The results show that mutations of individual residues near Tyr184 do not affect α-btx affinity, but pairwise mutations decrease affinity in an energetically coupled manner. Kinetic measurements show that the affinity decreases arise through increases in the α-btx dissociation rate with little change in the association rate. Replacing loop C in α7 with loop C from the α-btx-insensitive α2 or α3 subunits abolishes high-affinity α-btx binding, but preserves acetylcholine-elicited single channel currents. However, in both the α2 and α3 construct, mutating either residue that flanks Tyr184 to its α7 counterpart restores high-affinity α-btx binding. Analogously, in α7, mutating both residues that flank Tyr184 to the α2 or α3 counterparts abolishes high-affinity α-btx binding. Thus interaction between Tyr184 and local residues contributes to high-affinity subtype-selective α-btx binding.
Keywords: crystal structure, inter-residue coupling, molecular recognition, neurotoxin, nicotinic acetylcholine receptor
INTRODUCTION
Long chain α-neurotoxins, such as α-btx (α-bungarotoxin) and α-ctx (α-cobratoxin), are renowned for their high-affinity slowly reversible binding to nicotinic AChRs (acetylcholine receptors) [1,2]. The physiologically relevant AChR is located within the post-synaptic membrane of voluntary muscle, and contains two acetylcholine-binding sites coupled to an intrinsic ion channel. Binding of either α-btx or α-ctx to the AChR prevents binding of nerve-released acetylcholine, resulting in paralysis. Onset of paralysis is slow, owing to a toxin-association rate several orders of magnitude lower than that of a diffusion-limited reaction, but recovery is prolonged due to a very slow rate of dissociation [3,4]. The twin properties of high affinity and low dissociation have made α-neurotoxins valuable labels for tracking and quantifying AChRs [5]. Furthermore, because binding of α-neurotoxins and small-molecule agonists or antagonists is mutually exclusive, radiolabelled α-neurotoxins have been employed to determine fractional occupancy by unlabelled ligands [4,6,7].
Within the pentameric AChR, each ligand-binding site is formed at the interface between a principal and a complementary subunit [8–10]. Together, the principal and complementary subunits contribute seven binding-site loops, each containing several consecutive residues from a different section of the aminoacid sequences of the subunits. The principal subunit contributes loops A–C, whereas the complementary subunit contributes loops D–G. Naturally occurring sequence variants within loop C are present in animals resistant to α-neurotoxins [11,12], whereas short peptides derived from loop C bind to α-neurotoxins [13–15]. Furthermore, mutational analyses showed that loop C was the most important of the seven binding site loops in conferring high affinity for α-btx [16,17], and replacing loop C from the muscle AChR for the analogous structure in the pentameric bacterial channel GLIC (Gloeobacter violaceus pentameric ligand-gated ion channel) yielded high-affinity α-btx binding [18]. The key importance of loop C was confirmed by crystal structures of α-neurotoxins bound to receptor or receptor-like targets where loop C lodged between two of the three fingers of the α-neurotoxin [19–21]. The complex between α-btx and an α7/AChBP (acetylcholine-binding protein) chimaera revealed a large area of inter-protein contact, but only a single focal interaction was essential for high-affinity binding [21]. The stabilizing residues included a conserved tyrosine residue within loop C of α7 and conserved arginine and phenylalanine residues of α-btx. Remarkably, none of the four remaining canonical aromatic residues of the α7 ligand-binding site were essential for high-affinity binding.
In addition to binding with high affinity, long-chain α-neurotoxins are noted for their target selectivity [22–24]. The AChR family is diverse, comprising ten subtypes of α-subunits, four types of β-subunits, and γ-, ε- and δ-subunits. Some subunits form homopentamers, such as α7, or more commonly, heteropentamers, of which there are many subunit combinations. α-Neurotoxins bind tightly to homo- or hetero-pentameric AChRs that contain α1, α7, α8, α9 or α10 subunits, but they do not bind to hetero-pentamers containing α2, α3 or α4 subunits. This target selectivity occurs despite the fact that all AChR α-subunits contain a signature loop C in which five of 12 residues are conserved (Figure 1). Insensitivity of α2–α4 receptors to α-neurotoxins may potentially arise from differences in the variable residues of loop C, analogous to receptors in α-neurotoxin-resistant animals.
Figure 1. Sequences of loop C in subtypes of human nicotinic receptor α-subunits.

Tyr184 is highlighted in bold.
To identify structural elements that contribute to α-neurotoxin high affinity and target selectivity, we designed mutations in an α7 AChR based on the crystal structure of the complex between α-btx and a pentameric α7 ligand-binding domain chimaera [21]. In particular, we sought to determine whether the contribution of the essential tyrosine residue in loop C of α7 depends on nearby residues defined in the structure. The results show that, in contributing to high affinity and subtype-selective α-btx binding, the essential tyrosine residue depends on nearby conserved and non-conserved residues.
EXPERIMENTAL
Expression of AChRs in HEK (human embryonic kidney)-293 cells
A cDNA encoding the high-conductance form of the α7/5HT3A (5-hydroxytryptamine type 3A) chimaeric receptor, subcloned in the expression vector pRBG4 [25], was transfected into HEK-293 cells using the calcium phosphate precipitation method as described previously [26]. Mutations were generated using the QuikChange® site-directed mutagenesis kit (Stratagene) and confirmed by sequencing the entire coding region.
Concentration dependence of 125I-labelled α-btx binding
Transfected cells were harvested by gentle agitation in PBS, centrifuged at 1000 g for 1 min and resuspended in potassium Ringer’s solution (140 mM KCl, 5.4 mM NaCl, 1.8 mM CaCl2, 1.7 mM MgCl2, 25 mM Hepes, pH 7.4). Cell suspensions were incubated with specified concentrations of 125I-labelled α-btx (PerkinElmer) for 4 h at 21 °C and then toxin–receptor complexes were separated from free toxin using a Brandel M-48T cell harvester. Non-specific binding could not be determined by incubation in the presence of a competitive ligand, because many of the mutant AChRs impaired binding of small-molecule agonists and antagonists. Also, unlabelled α-btx is not suitable because it can displace the chemically similar 125I-labelled α-btx from non-specific sites, and its affinity is markedly reduced by several mutations. Thus non-specific binding was determined by applying identical procedures to cells transfected with an equal amount of the pRBG4 vector containing the muscle AChR δ-subunit cDNA, which induces protein expression but does not form AChRs. At a saturating concentration of 30 nM 125I-labelled α-btx, non-specific binding averaged 3.4 %of that obtained for the control α7/5HT3A receptor [21]. After subtracting non-specific binding, the amount of bound 125I-labelled α-btx was expressed as a percentage of the maximum binding obtained for the control non-mutant α7/5HT3A receptor, and the Hill equation was fitted to percentage occupancy as a function of 125I-labelled α-btx concentration using GraphPad Prism 5 software.
Kinetics of 125I-labelled α-btx association
To measure the time course of α-btx association, a cell suspension prepared as described above was mixed with a specified concentration of 125I-labelled α-btx at 21 °C, and aliquots of the suspension were rapidly filtered through type A/E glass fibre filters (Gelman Sciences) at specified times. To determine non-specific binding, identical procedures were applied to cells transfected with the pRBG4 vector containing the muscle AChR δ-subunit cDNA. For each type of AChR, separate binding time courses were determined for one to three concentrations of 125I-labelled α-btx and analysed with Dynafit software [27].
Kinetics of 125I-labelled α-btx dissociation
To measure the time course of α-btx dissociation, a cell suspension, prepared as described above, was mixed with a saturating concentration of 125I-labelled α-btx (150–300 nM) and incubated for 1 h at 21 °C. The suspension was then centrifuged at 1000 g for 1 min, the supernatant was removed, and the cell pellet was resuspended in 30 ml of potassium Ringer’s solution. Aliquots of the suspension were then rapidly filtered through type A/E glass fibre filters (Gelman Sciences) at specified times. To determine non-specific binding, identical procedures were applied to cells transfected with the pRBG4 vector containing the muscle AChR δ-subunit cDNA. After subtracting non-specific binding, a single exponential decay plus a variable plateau was fitted to the data using GraphPad Prism 5 software.
Patch-clamp recordings
Single-channel recordings were obtained in the cell-attached patch configuration [28] at a membrane potential of −70 mV at 21 °C, essentially as described before [29,30]. The bath solution contained 142 mM KCl, 5.4 mM NaCl, 0.2 mM CaCl2 and 10 mM Hepes, pH 7.4, whereas the pipette solution contained 80 mM potassium fluoride, 40 mM potassium aspartate, 2 mM MgCl2, 1 mM EGTA and 10 mM Hepes, pH 7.4. Patch pipettes were pulled from 7052 capillary tubes (King Precision Glass) and coated with Sylgard (Dow Corning). Single-channel currents were recorded using an Axopatch 200 B patch-clamp amplifier (Molecular Devices), digitized at 5 μs intervals with the PCI-6111E interface (National Instruments), and detected by the half amplitude threshold criterion using the program TAC 4.0.10 (Bruxton Corporation) at a final bandwidth of 10 kHz [29,30]. Open-time histograms were plotted using a logarithmic abscissa and a square root ordinate [31] and fitted to the sum of exponentials by maximum likelihood using the program TACFit (Bruxton).
RESULTS
The complex between α-btx and α7/AChBP
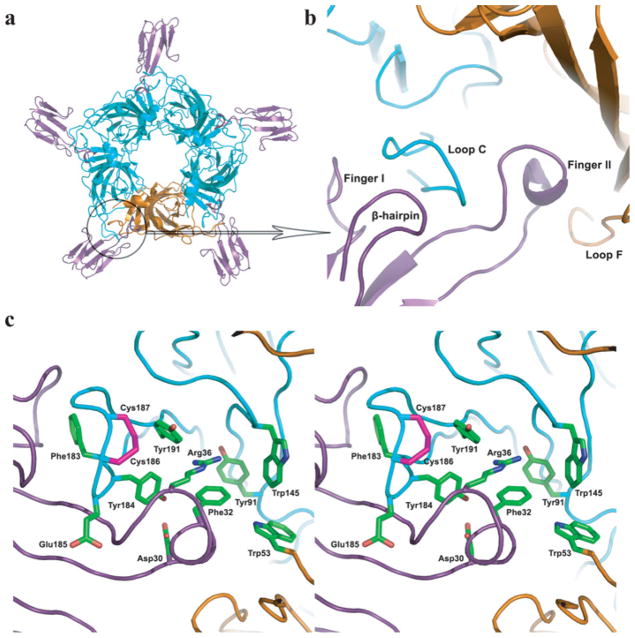
Mutational analyses were based on the crystal structure of α-btx bound to a pentameric α7 ligand-binding domain chimaera in which all residues within loop C and those that line the aromatic-rich binding cavity originate from the native human α7 receptor [21,32]. Despite a large area of contact in which the two binding partners exhibit shape and chemical complementarity, only a single aromatic residue, Tyr184 in loop C of α7, is required for high-affinity binding of α-btx [21]. Located deep within the binding cleft, Tyr184 interacts edge-to-face with Arg36 and Phe32 of α-btx that form a π-cation stack at the tip of toxin finger II (Figure 2). Furthermore, Phe183 of α7 inserts into a hydrophobic pocket between toxin fingers I and II, whereas Tyr191 from the C-terminus of loop C establishes an aromatic-cation interaction with Arg36. To determine whether the contribution of Tyr184 to high-affinity α-btx binding depends on local residues, we combined mutations of Tyr184 with mutations of either Phe183 or Tyr191.
Figure 2. Crystal structure of the complex between α-btx (purple) and the α7/AChBP chimaera (cyan and orange).
PDB code 4HQP [21]. (a) Top view of the complex with the complementary subunit of one binding site highlighted in orange. (b) Close-up view as in (a) showing the principal (cyan) and complementary (orange) subunits. (c) Stereo view of key residues between the tip of finger II of α-btx (purple) and the principal (cyan) and complementary (orange) subunits of the α7/AChBP chimaera.
Mutational analyses of single and pairwise mutations
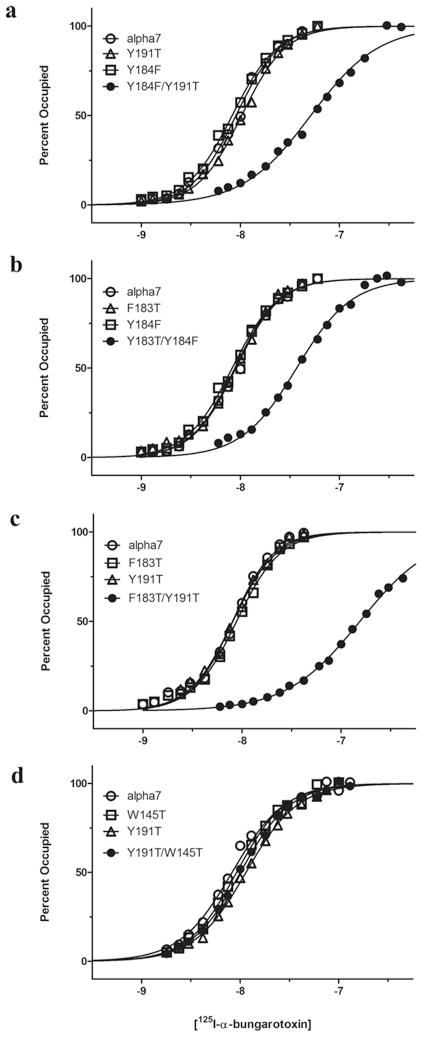
To assess the contributions of candidate residues to α-btx binding, we generated mutations in an α7/5HT3 receptor chimaera that contains human α7 sequence from the N-terminus to the start of the first transmembrane domain, followed by 5HT3 sequence to the C-terminus. This chimaeric AChR expresses robust quantities of cell-surface receptors in HEK-293 cells and exhibits single-channel currents in response to agonists [26,30]. For consistency with the α7/AChBP crystal structure, residue positions in α7/5HT3 are numbered according to those in the crystal structure rather than native α7. To determine how the mutations affect α-btx binding, intact cells expressing receptors were incubated with specified concentrations of 125I-labelled α-btx, and then bound complexes were separated from the free toxin using a cell harvester. Non-specific binding was determined by applying identical procedures to cells transfected with equal amounts of the pRBG4 vector containing the muscle AChR δ-subunit cDNA as described in the Experimental section. For the non-mutant α7/5HT3 receptor, specific binding of α-btx is saturable and exhibits an apparent Kd of approximately 8 nM and a Hill coefficient approaching two (Figure 3 and Table 1). Both parameters probably overestimate the true values because the incubation time of 4 h may not have been sufficient to reach equilibrium. Nevertheless, the apparent Kd is only 2-fold greater than that determined from the measurements of α-btx association kinetics described below.
Figure 3. Steady-state binding of 125I-labelled α-btx to α7/5HT3 receptors and the indicated mutants expressed in HEK-293 cells.
See the Experimental section for more details. Curves through the data are fits of the Hill equation with parameters given in Table 1. alpha7, α7/5HT3.
Table 1. Binding free energy and inter-residue coupling free energy determined from steady-state 125I-labelled α-btx binding.
For each fitted or calculated value, mean ± S.E.M. is given.
| Mutant | Kd (nM) | nH | ΔG (kcal/mol) | ΔΔG (kcal/mol) |
|---|---|---|---|---|
| α 7/5HT3 | 8.7 ± 0.56 | 1.96 ± 0.10 | 0.0 ± 0.017 | – |
| Y184F | 7.8 ± 0.36 | 1.63 ± 0.05 | 0.066 ± 0.022 | – |
| Y191T | 9.7 ± 0.44 | 1.81 ± 0.07 | −0.065 ± 0.021 | – |
| Y91T | 7.9 ± 0.46 | 1.87 ± 0.10 | 0.058 ± 0.025 | – |
| W145T | 9.3 ± 0.50 | 1.73 ± 0.06 | −0.04 ± 0.024 | – |
| W53T | 11.5 ± 0.60 | 1.66 ± 0.04 | −0.17 ± 0.025 | – |
| F183T | 7.7 ± 0.44 | 1.90 ± 0.09 | 0.073 ± 0.023 | – |
| Y191T/F183T | 154 ± 5.3 | 1.96 ± 0.02 | −1.72 ± 0.020 | −1.73 ± 0.028 |
| Y191T/Y184F | 51.2 ± 0.52 | 1.27 ± 0.07 | −1.06 ± 0.033 | −1.06 ± 0.038 |
| Y184F/F183T | 28.8 ± 2.6 | 1.51 ± 0.09 | −0.72 ± 0.029 | −0.86 ± 0.036 |
| F183T/W53T | 51 ± 5.4 | 1.57 ± 0.11 | −1.06 ± 0.034 | −0.85 ± 0.041 |
| Y184F/W53T | 32 ± 2.7 | 1.63 ± 0.09 | −0.78 ± 0.028 | −0.56 ± 0.036 |
| Y191T/W53T | 30 ± 3.4 | 1.37 ± 0.09 | −0.74 ± 0.036 | −0.38 ± 0.043 |
| Y91T/W53T | 12.8 ± 1.7 | 1.55 ± 0.06 | −0.23 ± 0.025 | 0.24 ± 0.036 |
| W145T/W53T | 15.7 ± 3.3 | 1.60 ± 0.10 | −0.35 ± 0.030 | 0.12 ± 0.039 |
| Y191T/W145T | 10.0 ± 0.35 | 1.72 ± 0.04 | −0.084 ± 0.020 | 0.022 ± 0.032 |
| Y191T/Y91T | 15.4 ± 1.4 | 1.72 ± 0.11 | −0.34 ± 0.029 | −0.34 ± 0.037 |
| Y91T/F183T | 11.4 ± 0.66 | 1.68 ± 0.07 | −0.16 ± 0.026 | −0.29 ± 0.036 |
| Y91T/W145T | 7.4 ± 0.49 | 1.44 ± 0.06 | 0.097 ± 0.025 | 0.079 ± 0.035 |
| Y91T/Y184F | 11.4 ± 0.31 | 1.68 ± 0.023 | 0.160 ± 0.024 | 0.37 ± 0.037 |
| W145T/F183T | 9.2 ± 0.35 | 1.80 ± 0.05 | −0.034 ± 0.020 | −0.067 ± 0.031 |
| W145T/Y184F | 8.1 ± 0.50 | 1.44 ± 0.06 | 0.043 ± 0.024 | 0.017 ± 0.033 |
To evaluate the contributions of aromatic residues at the ligand-binding site, we chose threonine as the replacement residue because it removes the aromatic ring, but retains a hydroxyl group. However, the Y184T mutation eliminates high-affinity α-btx binding [21], so instead Tyr184 was replaced with phenylalanine. The single residue mutations Y184F and Y191T do not affect the apparent Kd for α-btx (Figure 3a and Table 1). However, when both mutations are present in the same receptor, the apparent Kd increases approximately 6-fold. Analogously, the single residue mutations Y184F and F183T do not affect the apparent Kd (Figure 3b), but the pairwise mutations again increase the Kd. Mutant cycle analyses [33] applied to the pairs Y184F/Y191T and Y184F/F183T reveal coupling free energies of −1.0 and −0.8 kcal/mole (1 cal≈4.184 J) respectively (Table 1). Thus the contribution of Tyr184 to high-affinity α-btx binding depends on the two proximal residues Tyr191 and Phe183.
Because Tyr191 and Phe183 are energetically coupled with Tyr184, we asked whether the two residues are coupled with each other. Neither Y191T nor F183T affect α-btx affinity, as described above, but the pairwise mutation produces the largest decrease in affinity observed so far (Figure 3c). Mutant cycle analysis yields a large coupling free energy of −1.7 kcal/mole (Table 1). Thus, in view of the crystal structure of the complex (Figure 2), the observation of strong inter-residue energetic coupling suggests that Phe183 and Tyr191 are essential to the proper alignment of Tyr184 with the π -cation stack formed by Arg36 and Phe32 of α-btx.
Within the principal face of the ligand-binding site, two more conserved aromatic residues, Trp145 and Tyr91, interact with Arg36 of α-btx; the guanidinium group of Arg36 hydrogen bonds with the main chain carbonyl of Trp145 and the aromatic hydroxyl of Tyr91 (Figure 2). The individual mutations W145T and Y91T do not affect α-btx affinity, neither do any of the four pairwise combinations with Y191T and F183T (Figure 3d and Table 1). Similarly, combining either W145T or Y91T with Y184F does not affect α-btx affinity (Table 1). Thus, although Trp145 and Tyr91 contact the key Arg36 of α-btx, neither of these canonical residues contributes to affinity alone or in combination with Tyr184, Tyr191 or Phe183.
A fifth conserved aromatic residue, Trp53 from the complementary face of the ligand-binding site, contacts Phe32 of α-btx (Figure 2). The mutation W53T modestly reduces α-btx affinity, but the combination of W53T and F183T decreases affinity more than either mutation alone, yielding a coupling free energy of −0.85 kcal/mole. Also, the combination of W53T and Y184F yields a coupling free energy of −0.56 kcal/mole. On the other hand, Trp53 shows weak or no coupling to Tyr191, Tyr91 or Trp145 (Table 1). Thus Trp53 couples energetically with Tyr184 and Phe183, perhaps through the intervening Phe32 of α-btx. Notably, in the α7/AChBP crystal structure, Phe183, Tyr184, Trp53 and Phe32 span a distance of 24 Å (1 Å = 0.1 nm) between the principal and complementary subunits, and all four residues are co-linear.
Kinetics of 125I-labelled α-btx association
To determine how interactions among residues Tyr184, Tyr191 and Phe183 affect the rate constants underlying binding of α-btx, we measured time courses of radiolabelled α-btx binding to α7/5HT3 receptors without and with pairwise mutations. For the control and pairwise mutant receptors, time courses of α-btx binding were determined in the presence of two or three concentrations of α-btx, and the program Dynafit [27] was used to fit a reversible bimolecular binding mechanism simultaneously to the global set of time courses for each mutant. In this mechanism, the toxin (T) binds to the receptor (R) to form a reversible toxin–receptor complex (TR), with an association rate constant k+1 and a dissociation rate constant k−1.
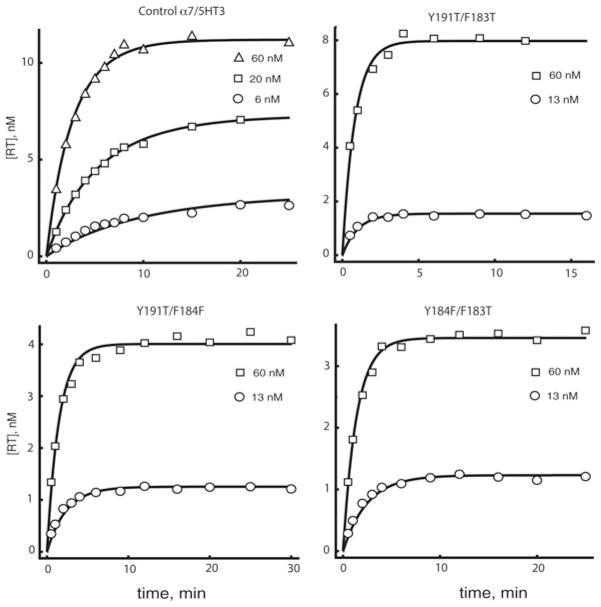
For the control α7/5HT3 receptor, simultaneous fitting describes the time courses for three concentrations of α-btx (Figure 4), and yields estimates of the association and dissociation rate constants (Table 2). The ratio of dissociation to association rate constants yields a binding dissociation constant of 4.8 nM, which is 2-fold smaller than that obtained from steady-state binding measurements (Figure 3 and Table 1). Furthermore, the association rate constant is approximately three orders of magnitude lower than that of a diffusion limited reaction, similar to previous determinations [3,4], whereas the dissociation rate constant yields a time constant of bound α-btx of approximately 33 min.
Figure 4. Binding time courses of 125I-labelled α-btx at the indicated concentrations to α7/5HT3 receptors and the indicated mutants expressed in HEK-293 cells.
See the Experimental section for more details. Curves through the data are global fits of the bimolecular binding mechanism (see the main text) to the data with fitted parameters given in Table 2. RT, receptor–toxin complex.
Table 2. Binding free energy and inter-residue coupling free energy determined from the kinetics of 125I-labelled α-btx binding.
In the first two columns, k+1 and k−1 are the values obtained by Dynafit analyses of the data in Figure 4. In the third and fourth columns, k−1 and Plateau (percentage of highly stable binding) were determined by analyses of the data in Figure 5 (see the main text). For each fitted rate constant a standard error is expressed as a percentage of the fitted value. nd, not determined.
| Mutant | k+1 (M −1·s−1) fitted | k−1 (s−1) fitted | k−1 (s−1) measured | Plateau | Kd (nM) | ΔG (kcal/mole) | ΔΔG (kcal/mole) |
|---|---|---|---|---|---|---|---|
| α 7/5HT3 | 9.2×104 (2.7 %) | 8.8×10 −4 (7.2 %) | 3.5×10 −4 (8.6 %) | 78 ± 1.1 | 3.8 ± 0.34 | 0.0 ± 0.05 | – |
| Y191T | 1.04×105 (2.8 %) | nd | 8.1×10 −4 (4.4 %) | 55 ± 1.7 | 7.8 ± 0.41 | 0.43 ± 0.032 | – |
| F183T | 9.6×104 (4.7 %) | nd | 7.5×10 −4 (12.5 %) | 71 ± 0.7 | 7.8 ± 1.04 | 0.43 ± 0.08 | – |
| Y184F | 1.5×105 (1.8 %) | nd | 5.7×10 −4 (2.2 %) | 51 ± 1.9 | 3.8 ± 0.11 | 0.0 ± 0.02 | – |
| Y191T/F183T | 6.2×104 (7.0 %) | 1.3×10 −2 (5.9 %) | 1.4×10 −2 (5.8 %) | 8.2 ± 0.6 | 210 ± 19 | 2.41 ± 0.05 | −1.54 + 0.12 |
| Y191T/Y184F | 9.5 ×104 (5.0 %) | 5.3×10 −3 (5.8 %) | 6.8×10 −3 (6.7 %) | 17 ± 1.5 | 56 ± 4.3 | 1.61 ± 0.05 | −1.18 ± 0.08 |
| Y184F/F183T | 1.2×105 (3.8 %) | 4.7×10 −3 (4.8 %) | 6.2×10 −3 (6.5 %) | 15 ± 1.5 | 39 ± 2.4 | 1.40 ± 0.04 | −0.97 ± 0.11 |
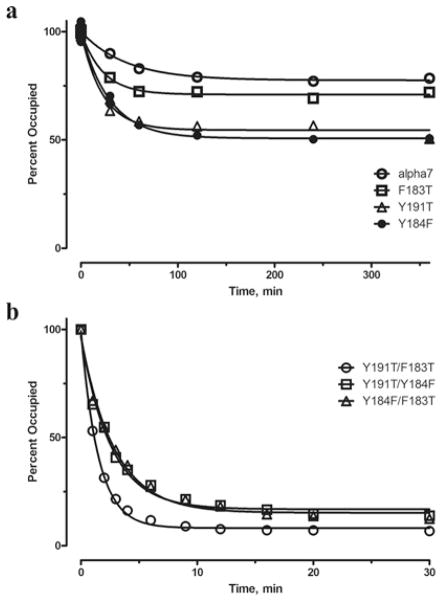
To measure the α-btx dissociation rate constant directly, cells expressing the α7/5HT3 receptor were incubated with a saturating concentration of radiolabelled α-btx, unbound toxin was removed, and bound toxin was measured as a function of time. The dissociation time course shows an exponential decay, with a time constant of 48 min, which reaches a stable plateau of 78 % of the initial toxin–receptor complex (Figure 5a and Table 2). The significance of the stable binding plateau is uncertain, but a possible interpretation is that the toxin–receptor complex is initially reversible, but it subsequently converts to a very stable complex. Despite the presence of a stable plateau, the directly measured decay rate constant is similar to the dissociation rate constant measured from fitting the reversible bimolecular association mechanism to the time course of toxin association (Table 2). A stable plateau was not evident from the kinetics of α-btx association, presumably because the association time course is measured over 25 min, whereas the stable complex is evident after approximately 60 min.
Figure 5. Time courses of dissociation of 125I-labelled α-btx from α7/5HT3 receptors and the indicated mutants expressed in HEK-293 cells.
See the Experimental section for more details. Curves through the data are fits of a single exponential plus a variable plateau to the data with fitted parameters in Table 2.
For receptors with pairwise mutations of coupled residues, the time courses of α-btx association are well described by the reversible bi-molecular association mechanism (Figure 4). The fitted association rate constant is unchanged from control for the pairs Y184F/Y191T and Y184F/F183T, but is decreased almost 2-fold for the pair Y191T/F183T (Table 2). By contrast with the association rate constants, the dissociation rate constants increase markedly compared with control for all three pairwise mutations (Table 2). Direct measurements of radiolabelled α-btx dissociation again show an exponential decay followed by a stable plateau (Figure 5b), but for the pairwise mutants, the plateau is much smaller than that observed for the control or any of the single residue mutations (Figure 5a). Because the pairwise mutations increase the rate of dissociation, the smaller plateau suggests that its magnitude is determined by the stability of the initial toxin–receptor complex. Moreover, the directly measured dissociation rate constants are similar to those determined from fitting the reversible bi-molecular association mechanism to the time courses of toxin association (Table 2). Thus, for these three residue pairs, energetic coupling arises primarily from increases in the rate of α-btx dissociation from the initial toxin–receptor complex.
For the three pairwise mutants, the ratio of dissociation to association rate constants yields kinetically determined dissociation constants that are similar to those determined from measurements of steady-state binding of radiolabelled α-btx (Table 2). Mutant cycle analyses, based on the kinetically determined dissociation constants, yield significant inter-residue coupling free energies (Table 2). These were similar to those determined from steady-state binding measurements (Table 1), and the kinetic and steady-state measurements exhibit the same rank order of coupling free energies for the three sets of pairwise mutants. Thus for the pairs Y184F/Y191T, Y184F/F183T and Y191T/F183T, the results from kinetic measurements agree with the results from steady-state measurements.
Contributions of inter-residue coupling with α-btx selectivity for nicotinic receptor subtypes
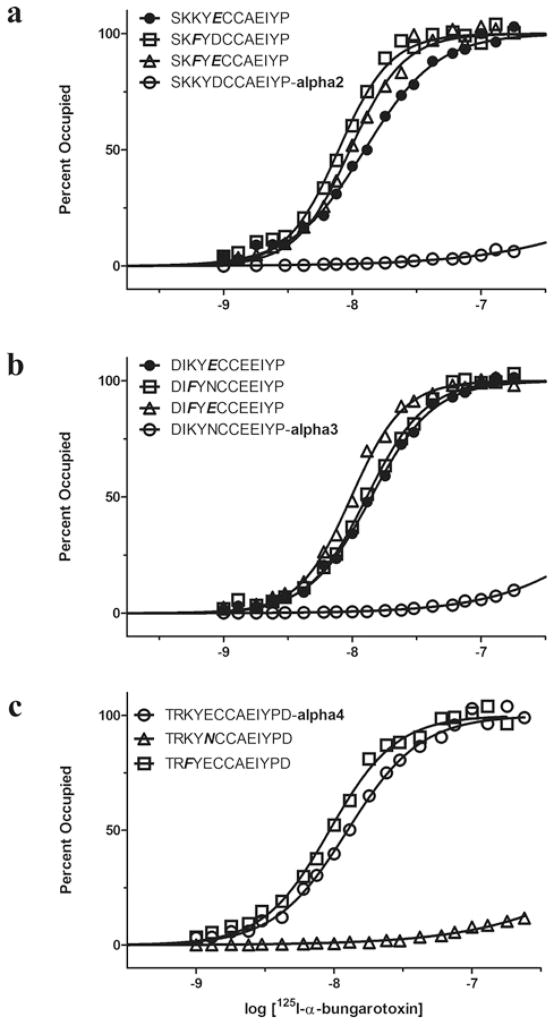
The results so far show that three local residues within loop C of α7 contribute interdependently to high-affinity α-btx binding. However, neuronal AChRs containing α2–α4 subunits are insensitive to α-btx [22,23], and residues in loop C of these subunits differ from those in α-subunits that confer sensitivity to α-btx (Figure 1). To determine whether differences in loop C contribute to subtype selectivity for α-btx, we replaced loop C in the α7/5HT3 receptor with loop C from either the α2 or α3 subunits, and measured steady-state binding of radiolabelled α-btx. Replacing loop C from either α2 or α3 abolishes high-affinity α-btx binding (Figures 6a and 6b); a small amount of specific binding is observed at α-btx concentrations greater than approximately 30 nM, suggesting that affinity is markedly reduced. Remarkably, in both the α2 and α3 constructs, replacing either of the residues that flank Tyr184 with the equivalent residue from α7 restores high-affinity α-btx binding (Figures 6a and 6b). Because the single mutants show high-affinity binding, and the double mutants show immeasurably low-affinity binding, the two flanking residues in α2 and α3 are interdependent in contributing to high-affinity α-btx binding. However, the inter-residue coupling free energy, although undoubtedly large, cannot be quantified.
Figure 6. Determinants of target selectivity of α-btx determined from steady-state 125I-labelled α-btx binding to α7/5HT3 receptors with the indicated loop C replacements expressed in HEK-293 cells.
See the Experimental section for more details. Curves through the data are fits of the Hill equation with parameters given in Table 3. Bold italic letters indicate substitutions.
Analogously, replacing loop C from the muscle α1 subunit into the α7/5HT3 receptor yields high-affinity α-btx binding, but mutating the two residues that flank Tyr184 to their α3 counterparts abolishes high-affinity binding (Table 3). In loop C of α1, the C-terminal half diverges from that in α3, but substituting the C-terminal half of loop C of α3 maintains high-affinity binding (Table 3). Thus sequence differences in the C-terminal half of loop C do not contribute to the subtype selectivity of α-btx.
Table 3. 125I-labelled α-btx binding and dissociation for mutant α7/5HT3A receptors with different substitutions of loop C.
Columns 1–3 give analyses of data from steady-state 125I-labelled α-btx binding as in Figure 5, whereas columns 4 and 5 give analyses of data from 125I-labelled α-btx dissociation as in Figure 6, where τdiss is the time constant for fast α-btx dissociation, and Plateau is the percentage of highly stable binding (see text). For each fitted value a standard error is given. Residues in bold and italic indicate mutations. nd, not defined.
| Loop C sequence | Kd (nM) | nH | Kmut/Kwt | τdiss (min) | Plateau |
|---|---|---|---|---|---|
| ERFYECCKEPYP (α7) | 10.1 ± 0.6 | 1.6 ± 0.08 | 1.0 | 43 ± 3.3 | 78 ± 0.4 |
| ERFYNCCKEPYP | 15.8 ± 1.0 | 1.4 ± 0.05 | 1.6 | – | – |
| ERKYECCKEPYP | 15.9 ± 1.0 | 1.6 ± 0.06 | 1.6 | 16 ± 1.1 | 29 ± 1.0 |
| ERKYNCCKEPYP | 1010 ± 630 | 1.0 ± 0.06 | 100 | – | – |
| SKKYDCCAEIYP (α2) | 7200 ± 5800 | 0.7 ± 0.05 | 712 | – | – |
| SKKYECCAEIYP | 12.5 ± 0.6 | 1.5 ± 0.05 | 1.2 | 14 ± 0.4 | 19 ± 0.5 |
| SKFYDCCAEIYP | 7.8 ± 0.5 | 2.1 ± 0.12 | 0.77 | – | – |
| SKFYECCAEIYP | 9.6 ± 0.6 | 2.1 ± 0.11 | 0.95 | 57 ± 17 | 83 ± 1.3 |
| DIKYNCCEEIYP (α3) | 2000 ± 350 | 0.9 ± 0.03 | 198 | – | – |
| DIKYECCEEIYP | 14.0 ± 0.5 | 1.8 ± 0.05 | 1.4 | 28 ± 1.7 | 39 ± 0.7 |
| DIFYNCCEEIYP | 12.9 ± 0.6 | 1.9 ± 0.07 | 1.3 | – | – |
| DIFYECCEEIYP | 9.8 ± 0.4 | 2.1 ± 0.08 | 0.97 | 189 ± 172 | 83 ± 7.3 |
| WVFYSCCPTTPYL (α1) | 4.8 ± 0.4 | 1.3 ± 0.06 | 0.48 | – | – |
| WVKYNCCPTTPYL | >5200 | nd | >515 | – | – |
| WVFYSCCEEI.YP | 3.9 ± 0.3 | 1.9 ± 0.11 | 0.39 | – | – |
| TRKYECCAEIYPD (α4) | 12.5 ± 0.6 | 1.6 ± 0.06 | 1.2 | 17 ± 0.6 | 19 ± 0.5 |
| TRFYECCAEIYPD | 9.3 ± 0.5 | 1.8 ± 0.8 | 0.92 | – | – |
| TRKYNCCAEIYPD | 2600 ± 800 | 0.8 ± 0.04 | 257 | – | – |
Finally, in α7, replacing the two residues that flank Tyr184 to their α3 counterparts abolishes high-affinity α-btx binding, but replacing just one of the flanking residues maintains high-affinity binding (Table 3). Thus, within loop C of the α2, α3, α1 and α7 subunits, Tyr184 and its two flanking residues are strongly interdependent in contributing to subtype-selective high-affinity α-btx binding.
A departure from the trend described above is observed when loop C from the α4 subunit is substituted into the α7/5HT3 receptor. Steady-state binding measurements reveal that α-btx binds with high affinity to the α4 construct (Table 3). Notably, loop C of α4 has a glutamate residue at position 185, like α7, whereas α2 and α3 have aspartate and asparagine respectively (Figure 1). However, replacing Glu185 with asparagine within loop C of the α4 construct abolishes high-affinity α-btx binding, analogous to the results for the α2 and α3 loop C replacements (Table 3). Thus α-btx insensitivity of α4-containing nicotinic receptors does not arise solely from residue differences within loop C, suggesting other residues in α4 or the complementary β-subunit contribute. Nevertheless, the findings from substituting loop C of α4 into α7 confirm the importance of the residues flanking Tyr184 in contributing to subtype-selective α-btx binding.
Steady-state binding of α-btx results from the combination of association and dissociation rate constants and a plateau of stable binding. Because steady-state measurements may not sufficiently sense changes in reaction steps underlying α-btx binding, we directly measured time courses of α-btx dissociation from α7/5HT3 receptors with various loop C replacements. The construct with loop C from α4 shows accelerated dissociation of α-btx and a much lower plateau of binding compared with the control α7/5HT3 receptor (Table 3). Similarly, constructs with the glutamate residue substituted at position 185 of either α2 or α3, while retaining the lysine residue at position 183, show accelerated dissociation of α-btx and a lower plateau (Table 3). On the other hand, in both the α2 and α3 constructs, substituting the phenylalanine residue at position 183 and the glutamate residue at position 185 decreases the rate of α-btx dissociation and increases the plateau, mimicking the control α7/5HT3 receptor (Table 3). These findings confirm that residues flanking the essential Tyr184 contribute both to the stability of the initial toxin–receptor complex and the magnitude of the stable plateau.
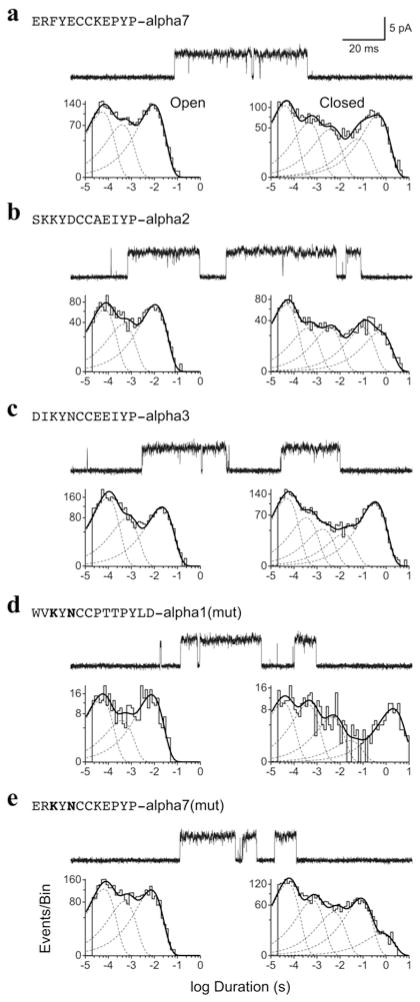
Four of the constructs in Table 3 abolish high-affinity α-btx binding, and the small amount of specific binding observed at high α-btx concentrations suggests markedly reduced affinity. Alternatively, the mutations may have impaired folding or subunit assembly, reducing or preventing expression of AChRs on the cell surface. To determine whether the four mutant receptors express on the cell surface, we used patch-clamp electrophysiology to assess cell-surface expression, and also to look for changes in functional properties. For all four mutants that lack high-affinity α-btx binding, acetylcholine elicits single-channel currents that are very similar to those observed for the control α7/5HT3 receptor (Figure 7). The open time histograms, fitted by the sum of three exponentials, are similar between the control α7/5HT3 receptor and each of the four mutants. The closed time histograms contain five exponential components, the three briefest of which are similar between the control α7/5HT3 receptor and the four mutants; time constants for the longest components differ among the four mutants, but one and possibly both long components depend on the number of receptors in the patch and thus are expected to vary among different patches. Thus the four α-btx-insensitive mutant receptors express on the cell surface and exhibit kinetic properties very similar to those of the control α7/5HT3 receptor.
Figure 7. Single-channel currents through α7/5HT3 receptors with the indicated loop C replacements expressed in HEK-293 cells.
See the Experimental section for more details. For each panel, a trace of single channel currents elicited by 1 mM acetylcholine is shown filtered at a bandwidth of 10 kHz with channel opening corresponding with upward deflections. Underneath each trace are open (left) and closed (right) time histograms plotted on a logarithmic time scale and fitted by the sum of exponentials; individual components are indicated by dashed curves and the sum by solid curves.
DISCUSSION
Atomic-resolution structural data, mutagenesis, radioligand binding and electrophysiological measurements reveal structural contributions to the high-affinity target-selective interaction between a long-chain α-neurotoxin and a nicotinic AChR. The results resolve an apparent paradox that, within a large contact surface area between α-btx and the α7 AChR, only one of five conserved aromatic residues that contact the toxin is essential for the high-affinity interaction [21]. In particular, the contribution of the essential Tyr184 in α7 depends on the proximal residues Tyr191 and Phe183, and these complementary residues also contribute to binding in a mutually interdependent manner. Furthermore, the concept of proximal interdependent residues explains α-btx insensitivity of neuronal AChRs containing α2 and α3 subunits. For these neuronal AChRs, the two residues that flank Tyr184 differ from those in α7 and are interdependent in conferring low affinity for α-btx. For constructs containing loop C from the α-btx-insensitive α4 subunit, high affinity is maintained, whereas the rate and extent of α-btx dissociation is increased, indicating that additional structural determinants contribute to α-btx insensitivity of AChR subtypes containing α4. The overall findings highlight the importance of interdependent residue networks in contributing to high-affinity protein–protein interactions.
High-affinity binding of long-chain α-neurotoxins to nicotinic AChRs arises from very low rate constants of both association and dissociation [3,4]. Association of α-btx with α7 occurs at a rate some three orders of magnitude lower than that of a diffusion-controlled reaction, suggesting the rate-limiting steps include conformational selection of each binding partner. In α7, loop C adopts a range of conformations [21,32], and a fully open conformation may be required to allow enough room to accommodate finger II of α-btx. In solution, the Arg36 and Phe32 side chains of α-btx adopt a range of conformations [14,15], and the guanidinium and aromatic moieties may need to come into close register to stably interact with Tyr184. The interdependent residue networks identified in the present study contribute little to the rate of α-btx association, suggesting that the networks do not affect the large-scale conformational dynamics of the receptor target.
Dissociation of α-btx from α7 occurs in two steps, one progressing over tens of minutes, and a second over more than several hours. We find that the interdependent residue networks affect the rate of the fast dissociation step, with the paired mutations increasing the dissociation rate as much as 40-fold, whereas they do not affect the rate of the slow dissociation step. Furthermore, the paired mutations increase the magnitude of the fast relative to the slow dissociation step, indicating that the stability of the initial complex determines the extent of formation of the stable complex. Thus, whereas the inter-residue interactions described in the present study determine the stability of the initial toxin–receptor complex, subsequent conformational rearrangements may be required to form the very stable complex. These slower rearrangements may occur subsequent to locking of loop C in its most outward conformation.
Previous studies established that sequence differences in loop C accounted for α-btx insensitivity in certain animals [11,12]. In particular, a glycosylation site at the position equivalent to Phe183 accounts for invulnerability of the snake to α-btx, and glycosylation at a position equivalent to Glu181 accounts for the invulnerability of the mongoose. On the other hand, loop C from the hedgehog does not contain a glycosylation site, yet the hedgehog is partially resistant to α-btx [34]. In loop C of the hedgehog, isoleucine and alanine flank the essential Tyr184, in contrast with phenylalanine and serine in animals most sensitive to α-btx. By altering the inter-residue network encompassing Tyr184, these naturally occurring replacements apparently produce a subtle structural change that is nearly as effective as a glycosylation site in reducing toxin sensitivity.
An analogous interdependent residue network underlies α-btx insensitivity of α2 and α3 containing neuronal AChRs, which exhibit sequence variability at the positions flanking the essential Tyr184 (Table 3). Because our studies were conducted with homomeric receptors with replacements in loop C, our conclusions require confirmation in heteromeric neuronal AChRs. However, previous studies of neuronal α3 AChRs suggest our results may extend to heteromeric AChRs. When the sequence YKHEIKYN, within loop C of the α3 subunit, was replaced by the sequence WKHWVYYT from Torpedo californica (residue equivalent to Tyr184 is bold and underlined), and the construct co-expressed with neuronal β-subunits, heteromeric AChRs formed that were sensitive to α-btx [16,35]. Thus previous studies in heteromeric AChRs containing α3 are consistent with our findings.
After loop C of α4 was substituted into α7, high-affinity α-btx binding remained, but the rate and extent of fast dissociation increased. Thus, in AChRs containing α4, structures in addition to loop C further reduce sensitivity to α-btx. Contributions of the complementary subunits to α-btx binding were observed in muscle AChRs [36], and interactions between residues on adjacent β-strands of the complementary subunits contributed interdependently to α-conotoxin M1 binding [37]. Thus additional contributions to α-btx insensitivity of heteromeric α4 AChRs may arise from within the α4 subunit or the neuronal β2–β4 subunits.
The structural principles elucidated in the present study clarify structural mechanisms of molecular recognition of AChRs by three-finger α-neurotoxins, including long-chain α-neurotoxins, κ-neurotoxins, short chain α-neurotoxins and non-conventional α-neurotoxins (Figure 8). Long-chain α-neurotoxins contain a disulfide bond within finger II, and share residues identical or chemically similar to Arg36, Phe32 and Asp30 of α-btx (Figure 8), suggesting that high-affinity binding of these α-neurotoxins depends on Tyr184 and flanking residues in loop C. κ-Neurotoxins bind selectively to neuronal AChRs containing the α3 subunit [38], contain a disulfide bond within finger II, and also share residues identical or chemically similar to Arg36, Phe32 and Asp30 of α-btx (Figure 8). Although κ-neurotoxins form non-covalent dimers [39], finger II from each monomer extends from opposing ends of the dimer [40], and could interact with an AChR subunit interface. Although dimerization of κ-neurotoxins likely contributes to their affinity for neuronal AChRs [41], specialized residues flanking Tyr184 (Figure 1) probably also contribute to κ-neurotoxin selectivity for AChRs containing α3. Short α-neurotoxins bind with high affinity to muscle, but not α7 AChRs [42], and lack both the disulfide bond in finger II and the four intervening residues. Finger II, though shorter, contains three residues that are either identical or chemically similar to the triad Arg36, Phe32 and Asp30 in α-btx (Figure 8). The structure of erabutoxin a (PDB code 1QKD) shows that the three equivalent residues occupy positions similar to the residue triad in α-btx; this explains why short α-neurotoxins bind to muscle AChRs, whereas their inability to bind to α7 AChRs may arise from structural specialization between fingers I and II. In non-conventional α-neurotoxins, the amino acid sequences of finger II vary widely and usually lack the residue triad found in α-btx, which explains the low potency of the weak α-neurotoxin from Naja kaouthia [43]. However another non-conventional α-neurotoxin, candoxin, binds to both muscle and α7 AChRs [44], and its structure (PDB code 1JGK) shows that the arginine and glutamate residues (Figure 8) project from finger II and occupy positions similar to Arg36 and Phe32 in α-btx. In summary, within the large surface area of contact between an α-neurotoxin and the AChR ligand-binding site, the interplay among a few residues, conserved and nonconserved, contributes to both high-affinity and subtype-selective molecular recognition.
Figure 8. Sequence alignment of Finger II in 3-finger α-neurotoxins.

Key residues are bold. α-Neurotoxin sequences are from [38,44,45].
Acknowledgments
We thank Chris Free for technical contributions and Dr Ariel Caride for assistance with the Dynafit software.
FUNDING
This work was supported by the National Institutes of Health [grant numbers NS031744 (to S.M.S.) and GM064642 (to L.C.)].
Abbreviations used
- AChR
acetylcholine receptor
- AChBP
acetylcholine-binding protein
- α-btx
α-bungarotoxin
- α-ctx
α-cobratoxin
- HEK
human embryonic kidney
- 5HT3A
5-hydroxytryptamine type 3A
Footnotes
AUTHOR CONTRIBUTION
Steven Sine, Lin Chen, Shu-Xing Li and Sun Huang designed the research. Steven Sine, Sun Huang and Corrie daCosta performed the research. Steven Sine and Lin Chen supervised the research and Steven Sine wrote the paper.
References
- 1.Chang CC, Lee CY. Isolation of neurotoxins from the venom of Bungaris multicinctus and their modes of neuromuscular blocking action. Arch Int Pharmacodyn Ther. 1963;144:241–257. [PubMed] [Google Scholar]
- 2.Lee CY. Chemistry and pharmacology of polypeptide toxins in snake venoms. Annu Rev Pharmacol. 1972;12:265–286. doi: 10.1146/annurev.pa.12.040172.001405. [DOI] [PubMed] [Google Scholar]
- 3.Weber M, Changeux JP. Binding of Naja nicricollis (3H) α-toxin to membrane fragments from Electropherus and Torpedo electric organs. I Binding of the tritiated α-neurotoxin in the absence of effector. Mol Pharmacol. 1974;10:1–14. [PubMed] [Google Scholar]
- 4.Weiland G, Georgia B, Wee VT, Chignell CF, Taylor P. Ligand interactions with cholinergic receptor-enriched membranes from Torpedo: influence of agonist exposure on receptor properties. Mol Pharmacol. 1976;12:1091–1105. [PubMed] [Google Scholar]
- 5.Anderson MJ, Cohen MW. Nerve-induced and spontaneous redistribution of acetylcholine receptors on cultured muscle cells. J Physiol. 1977;268:757–773. doi: 10.1113/jphysiol.1977.sp011880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber M, Changeux JP. Binding of Naja nigricollis (3H) α-toxin to membrane fragments from Electrophorus and Torpedo electric organs. II Effect of cholinergic agonists and antagonists on the binding of the tritiated α-neurotoxin. Mol Pharmacol. 1974;10:15–34. [PubMed] [Google Scholar]
- 7.Quast U, Schimerlik M, Lee T, Witzemann V, Blanchard S, Raftery MA. Ligand-induced conformation changes in Torpedo californica membrane-bound acetylcholine receptor. Biochemistry. 1978;17:2405–2414. doi: 10.1021/bi00605a024. [DOI] [PubMed] [Google Scholar]
- 8.Sine SM. The nicotinic receptor ligand binding domain. J Neurobiol. 2002;53:431–446. doi: 10.1002/neu.10139. [DOI] [PubMed] [Google Scholar]
- 9.Taylor P, Talley TT, Radic Z, Hansen SB, Hibbs RE, Shi J. Structure-guided drug design: conferring selectivity among neuronal nicotinic receptor and acetylcholine-binding protein subtypes. Biochem Pharmacol. 2007;74:1164–1171. doi: 10.1016/j.bcp.2007.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson AJ, Lester HA, Lummis SCR. The structural basis of function in Cys-loop receptors. Quart Rev Biophys. 2010;43:449–499. doi: 10.1017/S0033583510000168. [DOI] [PubMed] [Google Scholar]
- 11.Barchan D, Kachalsky S, Neumann D, Vogel Z, Ovadia M, Kochva E, Fuchs S. How the mongoose can fight the snake: the binding site of the mongoose acetylcholine receptor. Proc Natl Acad Sci USA. 1992;89:7717–7721. doi: 10.1073/pnas.89.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krienkamp H, Sine SM, Maeda R, Taylor P. Glycosylation sites selectively interfere with α-toxin binding to the nicotinic acetylcholine receptor. J Biol Chem. 1994;269:8108–8114. [PubMed] [Google Scholar]
- 13.Neuman D, Barchan D, Fridkin M, Fuchs S. Analysis of ligand binding to the synthetic dodecapeptide 185–196 of the acetylcholine receptor α subunit. Proc Natl Acad Sci USA. 1986;83:9250–9253. doi: 10.1073/pnas.83.23.9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scherf T, Balass M, Fuchs S, Katchalski-Katzir E, Anglister J. Three-dimensional structure of the complex of α-bungarotoxin with a library-derived peptide. Proc Natl Acad Sci USA. 1997;94:6059–6064. doi: 10.1073/pnas.94.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng H, Moise L, Grant MA, Hawrot E. The solution structure of the complex formed between α-bungarotoxin and an 18-mer cognate peptide derived from the α1 subunit of the nicotinic acetylcholine receptor from Torpedo californica. J Biol Chem. 2001;276:22930–22940. doi: 10.1074/jbc.M102300200. [DOI] [PubMed] [Google Scholar]
- 16.Levandoski MM, Lin Y, Moise L, McLaughlin JT, Cooper E, Hawrot E. Chimeric analysis of a neuronal nicotinic acetylcholine receptor reveals amino acids conferring sensitivity to α-bungarotoxin. J Biol Chem. 1999;274:26113–2619. doi: 10.1074/jbc.274.37.26113. [DOI] [PubMed] [Google Scholar]
- 17.Fruchardt-Gaillard C, Gilquin B, Antil-Delbeke S, Le Novere N, Tamiya T, Corringer PJ, Changeux JP, Menez A, Servent D. Experimentally based model of a complex between a snake toxin and the α7 nicotinic receptor. Proc Natl Acad Sci USA. 2002;99:3216–3221. doi: 10.1073/pnas.042699899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang HL, Cheng X, Sine SM. Intramembrane proton binding site linked to activation of a bacterial pentameric ion channel. J Biol Chem. 2012;287:6482–6489. doi: 10.1074/jbc.M111.305839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourne Y, Talley TT, Hansen SB, Taylor P, Marchot P. The crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake α-neurotoxins and nicotinic receptors. EMBO J. 2005;24:1512–1522. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Crystal structure of the extracellular domain of nAChR a1 bound to α-bungarotoxin at 1.94Å resolution. Nat Neurosci. 2007;10:953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- 21.Huang S, Li S, Bren N, Cheng K, Gomoto R, Chen L, Sine SM. Complex between α-bungarotoxin and an α7 nicotinic receptor ligand-binding domain chimaera. Biochem J. 2013;454:303–310. doi: 10.1042/BJ20130636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loring RH. The molecular basis of curarimimetic snake neurotoxin specificity for neuronal nicotinic receptor subtypes. J Toxicol, Toxin Rev. 1993;12:105–153. [Google Scholar]
- 23.Boulter J, Connolly J, Deneris E, Goldman D, Heinemann S, Patrick J. Functional expression of two neuronal nicotinic acetylcholine receptors from cDNA clones identifies a gene family. Proc Natl Acad Sci USA. 1987;84:7763–7767. doi: 10.1073/pnas.84.21.7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoepfer R, Conroy WG, Whiting P, Gore M, Lindstrom J. Brain α-bungarotoxin binding protein cDNAs and mAbs reveal subtypes of this branch of the ligand-gated ion channel superfamily. Neuron. 1990;5:35–48. doi: 10.1016/0896-6273(90)90031-a. [DOI] [PubMed] [Google Scholar]
- 25.Lee BS, Gunn RB, Kopito RR. Functional differences among non-erythroid anion exchangers expressed in a transfected human cell line. J Biol Chem. 1991;266:11448–11454. [PubMed] [Google Scholar]
- 26.Bouzat C, Bartos M, Corradi J, Sine SM. Binding-pore interface of homomeric Cys-loop receptors governs open channel lifetime and rate of desensitization. J Neurosci. 2008;28:7808–7819. doi: 10.1523/JNEUROSCI.0448-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 28.Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Arch. 1981;391:5–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 29.Bouzat C, Gumilar F, Spitzmaul G, Wang HL, Rayes D, Hansen SB, Taylor P, Sine SM. Coupling of agonist binding to channel gating in an ACh-binding protein linked to an ion channel. Nature. 2004;430:896–900. doi: 10.1038/nature02753. [DOI] [PubMed] [Google Scholar]
- 30.Rayes D, Spitzmaul G, Sine SM, Bouzat C. Single-channel kinetic analysis of chimeric α7-5HT3A receptors. Mol Pharmacol. 2005;68:1475–1483. doi: 10.1124/mol.105.015438. [DOI] [PubMed] [Google Scholar]
- 31.Sigworth F, Sine SM. Data transformations for improved display and fitting of single-channel dwell time histograms. Biophys J. 1987;52:1047–1054. doi: 10.1016/S0006-3495(87)83298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S, Huang S, Bren N, Noridomi K, Dellisanti CD, Sine SM, Chen L. Crystal structures of the ligand binding domain of a pentameric α7 nicotinic receptor chimera and its complex with agonist. Nat Neurosci. 2011;14:1253–1259. doi: 10.1038/nn.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horovitz A, Fersht AR. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J Mol Biol. 1990;214:613–617. doi: 10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]
- 34.Barchan D, Ovadia M, Kochva E, Fuchs S. The binding site of the nicotinic acetylcholine receptor in animal species resistant to α-bungarotoxin. Biochemistry. 1995;34:9172–9176. doi: 10.1021/bi00028a029. [DOI] [PubMed] [Google Scholar]
- 35.Caffery PM, Krishnaswamy A, Sanders T, Liu J, Hartlaub H, Klysik J, Cooper E, Hawrot E. Engineering neuronal nicotinic acetylcholine receptors with functional sensitivity to α-bungarotoxin: a novel α3-knock-in mouse. Eur J Neurosci. 2009;30:2064–2076. doi: 10.1111/j.1460-9568.2009.07016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sine SM. Identification of equivalent residues in the γ, δ, and ε subunits of the nicotinic receptor that contribute to α-bungarotoxin binding. J Biol Chem. 1997;272:23521–23527. doi: 10.1074/jbc.272.38.23521. [DOI] [PubMed] [Google Scholar]
- 37.Sine SM, Kreienkamp HJ, Bren N, Maeda R, Taylor P. Molecular dissection of subunit interfaces in the acetylcholine receptor: identification of determinants of α-conotoxin M1 selectivity. Neuron. 1995;15:205–211. doi: 10.1016/0896-6273(95)90077-2. [DOI] [PubMed] [Google Scholar]
- 38.Chiappinelli V, Weaver W, McLane K, Conti-Fine B, Fiordalisi J, Grant G. Binding of native κ-neurotoxins and site-directed mutants to nicotinic acetylcholine receptors. Toxicon. 1996;34:1243–1256. doi: 10.1016/s0041-0101(96)00110-9. [DOI] [PubMed] [Google Scholar]
- 39.Chiappinelli VA, Lee JC. κ-Bungarotoxin: self-association of a neuronal nicotinic receptor probe. J Biol Chem. 1985;260:6182–6186. [PubMed] [Google Scholar]
- 40.Dewan JC, Grant GA, Sacchettini JC. Crystal structure of κ-bungarotoxin at 2.3 Å resolution. Biochemistry. 1994;33:13147–13154. doi: 10.1021/bi00248a026. [DOI] [PubMed] [Google Scholar]
- 41.Osipov AV, Kasheverov IE, Makarova YV, Starkov VG, Vorontsova OV, Ziganshin RK, Andreeva TV, Serebryakova MV, Benoit A, Hogg RC, et al. Naturally occurring disulfide-bound dimers of three-fingered toxins: a paradigm for biological activity diversification. J Biol Chem. 2008;283:14571–14580. doi: 10.1074/jbc.M802085200. [DOI] [PubMed] [Google Scholar]
- 42.Servent D, Winckler-Dietrich V, Hu HY, Kessler P, Drevet P, Bertrand D, Ménez A. Only snake curaremimetic toxins with a fifth disulfide bond have high affinity for the neuronal α7 nicotinic receptor. J Biol Chem. 2008;272:24279–24286. doi: 10.1074/jbc.272.39.24279. [DOI] [PubMed] [Google Scholar]
- 43.Utkin YN, Kukhtina VV, Kryukova EV, Chiodini F, Bertrand D, Methfessel C, Tsetlin VI. Weak toxin from Naja kaouthia is a nontoxic antagonist of α7 and muscle-type nicotinic acetylcholine receptors. J Biol Chem. 2001;276:15810–1581. doi: 10.1074/jbc.M100788200. [DOI] [PubMed] [Google Scholar]
- 44.Nirthanan S, Charpantier E, Gopalakrishnakone P, Gwee MC, Khoo HE, Cheah LS, Bertrand D, Kini RM. Candoxin, a novel toxin from Bungarus candidus, is a reversible antagonist of muscle (αβγδ) but a poorly reversible antagonist of neuronal α7 nicotinic acetylcholine receptors. J Biol Chem. 2002;277:17811–17820. doi: 10.1074/jbc.M111152200. [DOI] [PubMed] [Google Scholar]
- 45.Nirthanan S, Gwee M. Three-finger α-neurotoxins and the nicotinic acetylcholine receptor, forty years on. J Pharmacol Sci. 2004;94:1–17. doi: 10.1254/jphs.94.1. [DOI] [PubMed] [Google Scholar]