Abstract
Context
People meeting diagnostic criteria for anxiety or depressive disorders tend to score high on the personality scale of neuroticism. Studying this personality dimension can give insights into the aetiology of these important psychiatric disorders.
Objective
To undertake a comprehensive genome-wide linkage study of neuroticism, using large study samples that have been measured multiple times. To compare the results between countries for replication and across time within countries for consistency.
Design
Genome wide linkage scan.
Setting
Twin individuals and their family members from Australia (AU) and the Netherlands (NL).
Participants
19,635 sibling pairs completed self-report questionnaires for neuroticism up to five times over a period of up to 22 years. 5,069 sibling pairs were genotyped with microsatellite markers.
Methods
Non-parametric linkage analyses were conducted in Merlin-Regress for the mean neuroticism scores averaged across time. Additional analyses were conducted for the time specific measures of neuroticism from each country to investigate consistency of linkage results.
Results
Three chromosomal regions exceeded empirically-derived thresholds for suggestive linkage using mean neuroticism scores: 10p 5 cM (NL), 14q 103 cM (NL) and 18q 117 cM (AU & NL combined), but only 14q retains significance after correction for multiple testing. These regions all showed evidence for linkage in individual time-specific measures of neuroticism and one (18q) showed some evidence for replication between countries. Linkage intervals for these regions all overlap with regions identified in other studies of neuroticism or related traits and/or in studies of anxiety in mice.
Conclusions
Our results demonstrate the value of the availability of multiple measures over time and add to the optimism reported in recent reviews for replication of linkage regions for neuroticism. These regions are likely to harbour causal variants for neuroticism and its related psychiatric disorders and can inform prioritisation of results from genome-wide association studies.
Introduction
The personality trait of “neuroticism” is defined as a tendency to experience psychological distress. Individuals with high neuroticism scores are characterized by emotional instability, low self-esteem, and feelings of anxiety, depression, and guilt1. Neuroticism scores are found to be high in those suffering from psychiatric disorders such as major depression and anxiety disorders2 and this association appears to be reciprocal. Prospective studies demonstrate that neuroticism or neuroticism-like traits predict future major depression3–7 and not merely because of overlap with prodromal symptoms of major depression. Self-report questionnaires can be used to score neuroticism as a quantitative trait measurable on large population cohorts8, 9. Therefore, study of neuroticism in large populations is relatively easy and can give insights into the aetiology of important psychiatric disorders.
Neuroticism scores have been found to be robust measures with test-retest correlations of 0.7910 to > 0.909, 11 for scores measured up to two years apart, and approximately 0.60 for scores measured up to six years12 or 19 years11 apart. It is well established that neuroticism is partially under genetic control13, 14, with heritability estimates of 30%–54%8, 12, 15, 16. Twin studies have consistently shown no evidence for a shared common environmental component12, 15, 17. Genetic correlations between neuroticism scores taken over a six year period were above 0.88 for all age groups12. On average, women score higher for neuroticism than men, but heritability estimates are mostly consistent across sexes14–16. However, opposite sex sibling correlations16, 17 and mother-son correlations15 have been reported as lower, suggesting that different genes may be of importance in men and women. Estimates of the genetic correlation between neuroticism and depression or anxiety range from 0.4 to 0.817–20.
Four previous linkage studies of neuroticism have been published10, 16, 21, 22; three of these studies used a single measure of neuroticism and one10 used an average of two measures taken six months apart. For two of the studies, the linkage analyses for neuroticism were secondary to the analyses of the ascertainment criteria of their study cohorts, namely alcohol22 or nicotine21 dependence. Recent reviews14, 23 summarised the linkage analysis results from the three earliest published of these studies and from an additional 14 studies of psychiatric disorders considered to be genetically related to neuroticism and concluded that some consistency is starting to emerge across studies.
Examples of genetic linkage analysis of longitudinal data on any trait in adults are rare24 despite recognition that use of multiple measures can increase power by reducing between sib residual non-shared variance25. Consistency in linkage regions across repeated measures cannot be considered as a replication, as this requires identification of the same linkage region in independent data sets. Nonetheless, inconsistency in linkage regions identified from repeated measures might indicate type I error and biological implausibility of the putative region.
In this study, we report a linkage analysis of neuroticism from two large study samples of twin families from Australia and the Netherlands. Individuals in the Australian study have been measured up to four times over a 22 year period and on different scales. Individuals in the Dutch study sample have been measured up to five times over an 11 year period using the same scale. These data sets are independent between countries and therefore provide an opportunity to investigate replication of linkage results. Within countries, there are partly overlapping samples of participants at each measurement occasion, providing an opportunity to investigate consistency of linkage results.
Methods
Australian study sample: Participants and measures of neuroticism
All participants were adult twins and their families recruited through the Australian Twin Registry and were of North European ancestry. All provided written informed consent under study protocols approved by the Queensland Institute of Medical Research Human Research Ethics Committee. Participants completed one or more personality questionnaires: the Eysenck Personality Questionnaire revised 23-item (EPQ-R)26, the shortened 12-item subset (EPQ-R-S) or the NEO five factor inventory personality questionnaire27 which includes 12 items in the neuroticism domain and compared to the EPQ-R probes angry hostility, self-consciousness, impulsiveness and vulnerability as well as anxiety and depression. Each individual could have up to four measures of neuroticism measured at four different times, these (or their transformations, discussed below) are referred to as AU80 (EPQ-R), AU89 (EPQ-R-S), AU99 (EPQ-R) and AU02 (NEO) with these trait codes reflecting the approximate year in which the scores were collected. The participants contributing AU80, AU89 and AU99 measures are described in detail elsewhere8. Briefly, participants contributing AU80 or AU89 scores were ascertained solely on the status of being a twin registered through the Australian Twin Registry or, in the case of AU89, being a family member of a registered twin. The participants contributing AU99 measures were ascertained as siblings pairs selected for discordance or concordance with respect to extreme neuroticism or anxiety or depression scores: one sibling in the top or bottom decile, the other sibling in the top or bottom quintile, excluding monozygotic (MZ) twin pairs and allowing for selection of multiple siblings per family, in an Extreme Discordant and Concordant (EDAC) design28 (for full details see Kirk et al8). The EDAC design identifies the sibling pairs that are most informative for genetic studies29. The participants in the 1999 study had the opportunity to complete the EPQ-R by telephone interview and/or by mail; ~80% completed both within six months with a test-retest correlation of 0.98, 11. The two scores were averaged for analysis in this study. The long-term stability of the AU80, AU89 and AU99 measures are reported in Birley et al11 (in which the 1980, 1989 and 1999 studies are named Canberra, Alcohol cohorts (where “Alcohol” does not refer to any ascertainment criteria) and Anxiety studies). The participants contributing AU02 measures were ascertained as being extended twin families with a high incidence of smokers as part of an ongoing Nicotine Addition Genetics study30. Where possible, blood (or buccal) samples were obtained from the study participants and their parents.
Dutch study sample: Participants and measures of neuroticism
Families with adolescent and adult twins have been assessed roughly every two years since 1991 as part of an ongoing longitudinal survey study of the Netherlands Twin Register (NTR). Participants are of Dutch ancestry31 and were recruited under informed consent. Each survey, with the exception of the 1995 wave, collected information on personality and psychopathology31, 32 and was conducted under protocols approved by ethics committee of the Free University Hospital, Amsterdam. Consequently, each individual could have up to five measures of neuroticism measured at five different times, these (or their transformations, discussed below) are referred to as NL91, NL93, NL97, NL99 and NL02 with subscript codes reflecting the approximate year in which the scores were collected (corresponding to Waves 1,2 and 4–6 of data collection32). Neuroticism was measured using the Amsterdamse Biografische Vragenlijst (ABV)33, a self-report questionnaire similar in content to the EPQ-R34. The neuroticism scale comprises 30 questions with a 3-item response scale (no, don’t know, yes). The neuroticism score is a weighted sum of the item responses.
Neuroticism scores
Neuroticism scores are sum scores and such data typically deviate from normality by having heavy tails. The averaged angular transformation35 was used to normalise the distribution, as in other studies8, 11, 16, 36. The neuroticism scores used in the analysis were residuals from regression of the transformed neuroticism scores on age, sex and age*sex (and age2 and age2*sex for AU89) which were standardised separately for each sex. The mean AU89 score of those selected for measurement in the AU99 study sample was not significantly different from that of the full study group, but the variance was higher. Therefore, the AU99 measures were standardised using the variance of the AU89 cohort so that the higher variance of AU99 measures was maintained. Finally, an average neuroticism score was calculated for each person within each country, denoted by AU and NL. The number of measures contributing to each average was recorded and used as a weight in the repeated measures linkage analysis. Descriptions of the phenotype (all those measured) and genome scan (only those used in the linkage analysis) data sets are given in Tables 1 and 2.
Table 1.
Description of data sets. For inclusion in a) the phenotype data set, families needed to have at least two individuals with neuroticism scores and known age, only one of an MZ pair included and b) the linkage analysis data set, families size limited to 2–5 siblings with neuroticism scores, known age and genotype data on > 280 markers.
| Australia | Netherlands | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AU80 | AU89 | AU99 | AU02 | AU | NL91 | NL93 | NL97 | NL99 | NL02 | NL | AU&NL | |
| a) Phenotype | ||||||||||||
| # families | 2017 | 4253 | 938 | 576 | 4999 | 1661 | 2014 | 1283 | 1332 | 2069 | 3808 | 8807 |
| # sib-pairs | 2033 | 10513 | 1709 | 1589 | 12772 | 997 | 1148 | 3220 | 2691 | 2553 | 6863 | 19635 |
| Mean age | 33.4 | 35.1 | 43.1 | 40.2 | 17.7 | 19.2 | 26.6 | 30.9 | 32.8 | |||
| Sib-correlations | 0.18 | 0.14 | 0.274 | 0.10 | 0.14 | 0.25 | 0.28 | 0.16 | 0.20 | 0.17 | 0.20 | 0.16 |
| Female-female | 0.25 | 0.16 | 0.314 | 0.13 | 0.14 | 0.25 | 0.34 | 0.20 | 0.19 | 0.25 | 0.23 | 0.17 |
| Male-male | 0.18 | 0.13 | 0.304 | 0.15 | 0.18 | 0.27 | 0.34 | 0.07 | 0.18 | 0.13 | 0.17 | 0.18 |
| Female-Male | 0.12 | 0.12 | 0.244 | 0.07 | 0.11 | 0.23 | 0.21 | 0.18 | 0.21 | 0.12 | 0.19 | 0.14 |
| b) Linkage analysis | ||||||||||||
| # families | 1035 | 1634 | 802 | 306 | 1945 | 133 | 224 | 410 | 367 | 359 | 564 | 2509 |
| # individuals with G1 | 3209 | 5523 | 2862 | 1378 | 6522 | 558 | 834 | 1533 | 1405 | 1376 | 2030 | 8552 |
| # full sib-pairs2 | 1046 | 2988 | 1350 | 912 | 3870 | 133 | 226 | 825 | 678 | 613 | 1199 | 5069 |
| #female-female | 427 | 11191 | 512 | 325 | 1501 | 47 | 86 | 319 | 268 | 261 | 461 | 3542 |
| #male-male | 168 | 431 | 202 | 154 | 586 | 30 | 48 | 161 | 124 | 106 | 219 | 805 |
| Proportion of families | ||||||||||||
| 2 sibs | 1.00 | 0.75 | 0.75 | 0.52 | 0.73 | 1.00 | 1.00 | 0.65 | 0.69 | 0.72 | 0.62 | |
| 3 sibs | 0.00 | 0.17 | 0.19 | 0.24 | 0.17 | 0.00 | 0.00 | 0.23 | 0.22 | 0.21 | 0.23 | |
| 4+ sibs | 0.00 | 0.08 | 0.06 | 0.24 | 0.10 | 0.00 | 0.00 | 0.12 | 0.09 | 0.08 | 0.12 | |
| LOD suggestive3 | 1.6 | 1.7 | 1.7 | 1.7 | 1.7 | 1.5 | 1.5 | 1.7 | 1.7 | 1.7 | 1.6 | 1.7 |
| LOD significant3 | 2.9 | 3.0 | 3.0 | 3.5 | 3.1 | 2.7 | 2.8 | 3.1 | 3.4 | 3.3 | 3.1 | 3.1 |
| ELOD204 | 1.4 | 8.0 | 2.3 | 1.3 | 11.9 | 0.4 | 0.9 | 2.0 | 1.7 | 1.6 | 10.0 | 20.96 |
G =genome scan data;
MZ pairs excluded; a small number of families only contributed half-sib pairs;
empirical suggestive and significant LOD score thresholds from 1000 gene drop simulation genome scans.
Expected LOD of a QTL that accounts for 20% of the phenotypic variance.
the high full-sib correlation reflects the ascertainment for inclusion in the Australian 1999 study.
ELOD20 for AU&NL ≠ ELOD20 for AU + ELOD20 for NL, because of different heritabilities used.
Table 2.
Overlap in data sets used in linkage analysis: the proportion of full-sib pairs who both have the row measure of neuroticism that also both have the column measure of neuroticism.
| AU80 | AU89 | AU99 | AU02 | NL91 | NL93 | NL97 | NL99 | NL02 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| AU80 | 0.82 | 0.17 | 0.06 | NL91 | 0.81 | 0.62 | 0.53 | 0.50 | ||
| AU89 | 0.28 | 0.43 | 0.12 | NL93 | 0.48 | 0.65 | 0.56 | 0.52 | ||
| AU99 | 0.13 | 0.97 | 0.04 | NL97 | 0.09 | 0.15 | 0.59 | 0.51 | ||
| AU02 | 0.06 | 0.33 | 0.06 | NL99 | 0.09 | 0.16 | 0.74 | 0.64 | ||
| AU | 0.26 | 0.76 | 0.34 | 0.26 | NL02 | 0.10 | 0.17 | 0.71 | 0.71 | |
| NL | 0.10 | 0.17 | 0.71 | 0.57 | 0.51 |
Genotyping
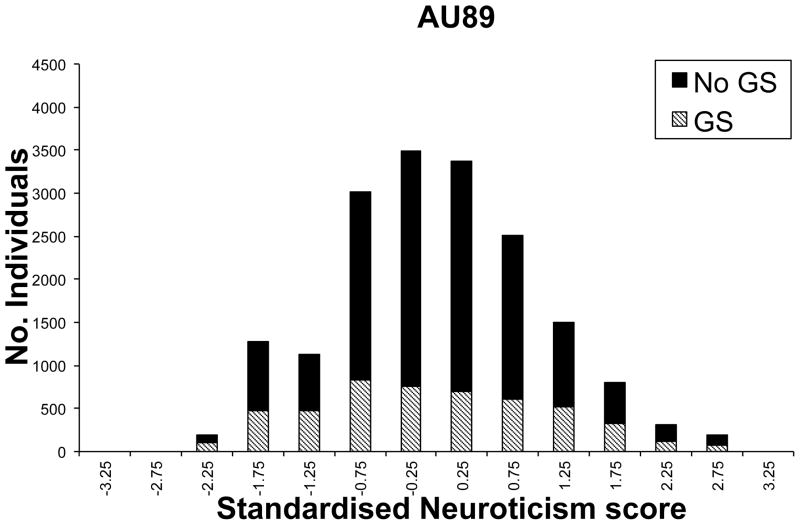
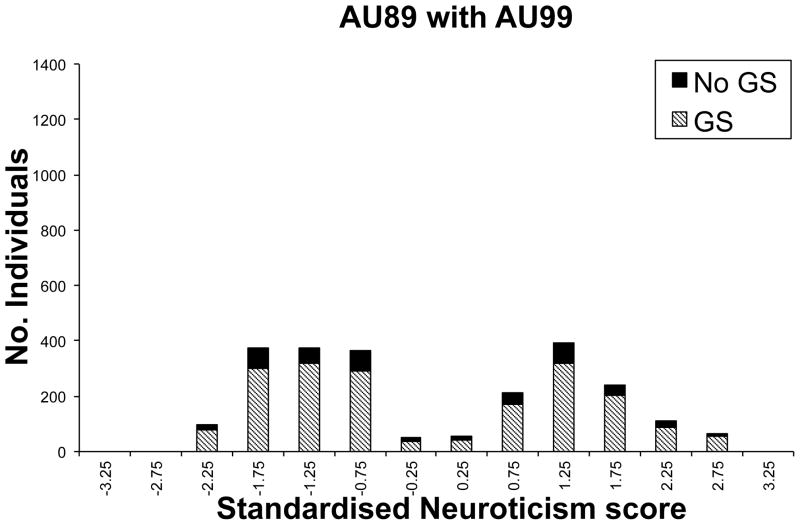
The genotypic data available for the Australian study resulted from submission of DNA samples to one or more of six genotyping centres, namely Gemini P/L (G), Sequana Therapeutics Inc (S), Leiden University Medical Centre(L), the Mammalian Genotyping Service, the Center for Mammalian Genetics at the Marshfield Clinic Research Foundation (M), the Australian Genotyping Research Facility, AGRF (A) and the Finnish Genome Center, University of Helsinki (H). A description of the G,L,M,S genotyping and the subsequent merging and cleaning of the marker data sets is described in detail elsewhere37, 38. Since then, additional M, A and H genotypes have been merged using the same protocol. Family members were submitted to the same genotyping facility. Participants with measure AU99 were submitted preferentially for genotyping ((Figure 1b), but this was not the sole criterion used to select families for genotyping and so the impact of the EDAC design was less marked for the AU89 measure which was available on the largest subset of samples (Figure 1a). Data cleaning based on Mendelian errors, unlikely genotypes and consistency of pedigree and marker relationships was undertaken as described by Cornes et al37.
Figure 1.
Distribution of a)standardised AU89 Neuroticism scores for those in the genome scan (GS) and those not in the genome scan (No GS) and b) the same but only for those selected to have AU99 scores.
Dutch samples were genotyped by the M or L laboratories. The genotype data from these screens were combined. Allele calling and binning were equalized between markers that were present in multiple scans, using ~ 30 control samples. Data cleaning based on Mendelian errors, unlikely genotypes and consistency of pedigree and marker relationships was undertaken as described by Middeldorp et al39. The distributions of the neuroticism measures for those with and without genome scan data were similar.
Map positions of all genotyped markers were estimated in Kosambi cM by locally weighted linear regression (http://www.qimr.edu.au/davidD) from the NCBI Build 35.1 physical map positions and published Decode and Marshfield genetic map positions40. Identical markers genotyped at different genotyping facilities were all included, separated by 0.001cM on the genetic map. Using markers genotyped in common, the Fst between the Australian and Dutch samples was estimated to be 0.30%, implying that these samples can be combined for joint genetic analysis41. Individuals were required to have genotypes on more than 280 markers resulting in an average distance of 8.2 cM (AU) and 11.0 cM (NL) between genotyped markers of sib pairs. 38% (AU) and 51% (NL) of parents were genotyped.
Preliminary Analyses
Phenotypic (test-retest) correlations between the EPQ measures AU80, AU89 and AU99 correlations range between 0.59–0.6211, 36. Test-retest correlations of these measures with AU02 are lower 0.46–0.5436 reflecting the different emphasis of some of the items included in the NEO personality inventory neuroticism domain. The average phenotypic correlation between the Dutch measures is 0.65, range 0.56–0.77, with higher correlations between consecutive measures. The highest sib-pair correlations (estimated in Sib-Pair42, Table 1) were for the youngest cohorts NL91 and NL93. The high sib-correlation for the AU99 is a reflection of the EDAC selection. The lowest sib-correlation was for AU02 scored on the NEO scale. Analyses of subsets of the Australian11, 15 and Dutch32 data have consistently shown no evidence for influence of common environmental effects. Genetic correlations were estimated in ASReml43 and ranged between 0.91–0.95 between the EPQ measures (AU80, AU89 and AU99) and ranged between 0.80 and 0.95 between these measures and the AU02 NEO measure. Formal testing showed that the measures can be considered as repeated measures of the same trait36. The genetic correlations between the five Dutch traits range from 0.84 to 0.95. Across all neuroticism measures, averaged estimates of heritability, phenotypic and genetic correlations were 0.32, 0.61 and 0.90 respectively. Preliminary linkage analyses conducted using a full multivariate model (not presented) suggested that there was little to be gained compared to the repeated measures model with genetic correlations of this magnitude.
Linkage analyses
Genetic linkage analysis of the autosomes was conducted in Merlin-Regress44 which regresses estimated identity-by-descent between relative pairs on the squared sums and squared differences of trait values of the pairs. Investigation of the properties of the method by simulation44 showed it to be powerful and efficient even for selected samples (EDAC designs). It requires phenotypic measures to be standardised in the unselected population sample and uses the population parameters (mean, variance, heritability) derived from the full population sample rather than the selected or genotyped sample. The method is also appropriate for general pedigrees including multiple sibs per family. However, simulation studies44 showed that, although large sibships can increase power, the distribution of the test statistic can become distorted if the contributions from families become highly skewed. For this reason sibships were limited to a maximum of 5, selecting sibs that maximised either the discordance or concordance of each family. Mean neuroticism scores were analysed in Merlin-Regress options –mean 0 –var 1, with heritabilities entered as twice the sib correlations (Table 1) and using the –testretest option with correlation of 0.61. Analyses were repeated using mean measures from only males and only females because other studies have reported sex-specific linkage regions (summarised in16). Analyses using scores of males or females only are denoted with subscripts m or f respectively. Linkage analysis for the X-chromosome was conducted in Merlin MINX. In all analyses, multipoint LOD scores for presence of a quantitative QTL were estimated every 5cM (a 1 cM grid was used to determine linkage region confidence intervals, as the region bounded by 1 LOD less than the maximum observed). Using the 5cM grid allows the linkage statistic to be collected over all families even when families were genotyped for different markers. Option –singlepoint was used to identify the individual marker contributing most to regions showing evidence of linkage. Linkage analyses were repeated using individual measures of neuroticism to allow examination of consistency in linkage signal between time-specific measures for each country.
Autosomal genome-wide empirical significance thresholds were derived from 1000 gene drop simulations as implemented in Merlin –simulate which utilises the allele frequencies, marker positions and missing genotype patterns of the real data set and simulates under a model which assumes random linkage between genotype and phenotypes. All phenotypes were analysed using the same simulated data sets, which maintains the correlation structure between phenotypic measures. The maximum LOD scores from each chromosome of each simulation replicate were retained and were used to derive the empirical LOD thresholds for Lander-Kruglyak45 suggestive linkage (1 LOD exceeding the threshold per genome scan) and significant linkage (1 LOD exceeding the threshold per 20 genome scans) for each neuroticism measure analysed and for the nine mean measures of neuroticism simultaneously to derive thresholds that account for multiple testing.
Within Merlin-Regress option –rankFamilies gives an ELOD20 score for each phenotypic measure. ELOD20 is the LOD expected given the data of a quantitative trait locus (QTL) that accounts for 20% of the phenotypic variance, assuming fully informative markers. Observed marker informativeness (I) was estimated as the average information content of the 5cM estimates across the autosomes. ELOD20 scores corrected for observed marker informativeness were calculated as ELOD20(I) = ELOD20*I. Both ELOD20(I) and ELOD10(I) scores were used to calculate the power of our study samples given the phenotypic and genotypic information to detect QTL that account for 20% and 10% of the total variance at the empirical significant 45 or suggestive45 thresholds for linkage using the ‘Probability Function Calculator’ of the ‘Genetic Power Calculator’46, where ELOD10(I) = ELOD20(I)/4.
Results
Empirically derived suggestive and significant LOD thresholds for samples with each level of neuroticism are listed in Table 1. The lowest thresholds are for the samples comprised predominantly of a single sib pair per family: AU80, NL91 and NL93. The empirical threshold for suggestive and significant linkage accounting for the multiple testing of the 9 mean measures of neuroticism are 2.5 and 4.1, respectively.
The mean ± standard deviation of the information content across the autosomes as calculated every 5cM in Merlin-Regress was 0.73 ± 0.08 (AU) and 0.51 ± 0.10 (NL), the difference reflecting the average distance of genotyped markers between sib-pairs. The ELOD20 scores are listed in Table 1. By accounting for the observed informativeness of the genotyped markers, we estimate that the study samples AU&NL, AU and NL have 100%, 99% and 86% power to detect a QTL that accounts for 20% of the total variance at the significant threshold of linkage. These samples have power of 60%, 27% and 9% to detect a QTL that accounts for 10% of the total variance at the significant thresholds, and of 89%, 65%, 37% at the suggestive thresholds. The power of sex specific analyses is much lower, as expected from the number of same sex sib-pairs contributing to the analysis. The sex specific AU&NL, AU and NL measures have, for females, 99%, 86% and 64% and, for males, 69%, 40% and 24% power to detect a QTL that accounts for 20% of the total variance at the suggestive threshold of linkage.
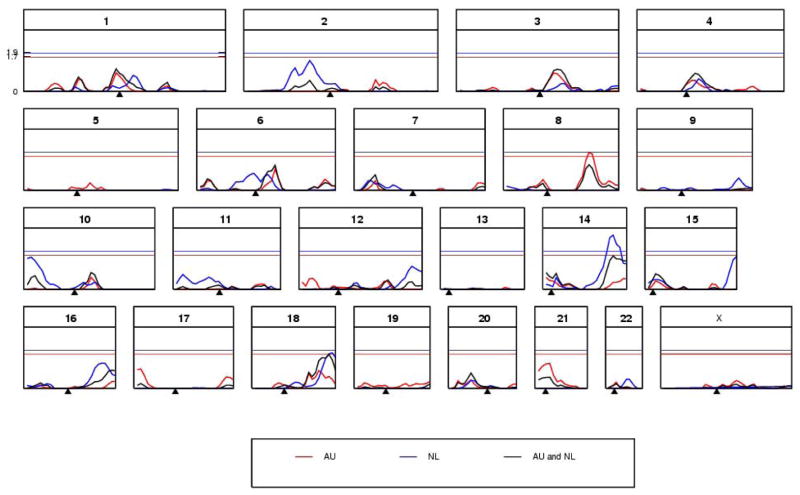
The genome-wide linkage plot for AU, NL and the joint analysis of AU&NL (Figure 2) show three regions that exceed the empirical threshold for suggestive linkage for their respective measures: 18q 116 cM for AU&NL, 14q 104cM for NL and 10p 5 cM for NL. An additional two regions just fail to reach the this threshold: 8q 132 cM for AU and 6cen 75 cM for NL. The chromosomal position with the maximum LOD score based on a 1 cM grid scan, linkage intervals, corresponding cytogenetic band and the marker with the maximum LOD score within the region based on a singlepoint analysis are listed in Table 3. To investigate these results, we looked for consistency in linkage signal in time-specific measures of neuroticism within the linkage intervals and found that for all five regions at least two individual measures achieved LOD >1 (Table 3). In contrast, an additional 10 chromosomal regions (NL97: Chr 14 22 cM; AU80: Chr16 56 cM, AU02: Chr 3 14 cM, AU80: Chr 6 178 cM, NL02: Chr 9 149 cM, NL97: Chr 16 124 cM, AU99: Chr 17 14 cM, NL93: Chr 18 60 cM, AU80: Chr19 104 cM, AU89 chr 21 21 cM) achieved LOD scores that exceeded the empirical suggestive threshold for significance for an individual measure of neuroticism and only the first two listed achieved a LOD >1 for any other individual measure within the 1 LOD drop confidence interval. Within country, some datasets were longitudinal (e.g., > 90% of those included in the AU80 or AU99 analysis were also included in the AU89 analysis, Table 2), whilst some data sets were largely independent (e.g., < 10% of participants with AU80 or AU99 scores were also scored for the AU02, Table 2). The most extreme example of inconsistency was for AU89 which achieved a LOD of 2.7 for 21p 21 cM, yet no evidence for linkage was found with the AU99 measure (maximum LOD within the region of 0.1), a difference which persisted when the analysis was limited to include only phenotypes of individuals who were measured in both studies. Examination of the sib-pair phenotypic scores and IBD sharing from families that contribute most to these linkage signals, showed nothing that could not reasonably be attributed to stochastic variation.
Figure 2.
Merlin-Regress linkage results of LOD score (y-axis) for each chromosome (1–22, X) based on a 5 cM grid (x-axis) for mean neuroticism score of the Australian (AU), Dutch (NL) and combined (AU&NL) data sets. Empirical thresholds for suggestive linkage were 1.7 (red horizontal) for AU and AU&NL and 1.9 (blue horizontal) for NL.
Table 3.
Chromosomal regions where the LOD >1.5 for either AU&NL, AU or NL. Evidence for consistency of signal for individual measures within country and evidence for support from other studies.
| Measurea | Chr | Position cMb | LODb | Linkage intervalb cM |
Linkage interval cytogenetic band | Singlepoint Markerc, LOD, position |
LOD > 1.5d | 1.0 < LOD < 1.5e | Fullerton14 identified regionf | Human region homologous mouse linkage regiong | Primary sourcs for human linkage studiesh |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NL | 6 | 75 | 1.5 | 59–111 | 6p21.2-6q21 | D6S2410, 2.4, 75 cM | NL99* NL02 |
AU&NL | 99-190 cM | ||
| AU | 8 | 134 | 1.6 | 125–145 | 8q24.12-8q24.21 | D8S592, 1.7, 124 cM | AU89 AU99, |
NL91 | 104-154 cM | ||
| NL | 10 | 5 | 2.0* | 0–29 | 10p15.3-10p14 | D10S1412, 1.8, 25 cM | NL99* NL97* NLf* NLm |
*Neuroticism 16 | |||
| NL | 14 | 103 | 2.6* | 94–118 | 14q32.12-14q32.31 | D14S1434, 1.7, 104 cM | NL99* AU&NL* |
NL02 | 76-134 cM | * Anxiety 105 cM39 R Anxiety 105 cM48 |
|
| AU&NL | 18 | 117 | 1.9* | 95–125 | 18q21.33-18qter | D18S61, 1.4, 99 cM | NL* | NL99 NL02 AU89 |
R 80 cM | 85-109 cM | ** Neuroticism Female 91 cM22 * RE-MDD or anxiety 73 cM 47 Neuroticism Female 115 cM22 R Harm avoidance 109 cM 54 Panic Disorder 104 cM55 |
| Sex specific | |||||||||||
| AUm &NLm | 2 | 112 | 1.6* | 94–118 | 2p13.2-2q11.2 | D2S1790, 1.6, 111 cM | NL02* | AUm NLm AU89 |
102-151 cM | ** Suicide & RE-MDD 100 cM | |
| NLm | 5 | 191 | 2.2* | 185–199 | 5q35.1-5q35.2 | D5S211, 1.4, 191 cM | NL99m* NL97m |
NL02m | |||
| AUm | 8 | 45 | 1.6* | 34–53 | 8p22-8p21.1 | D8S1771, 1.6, 49 cM | AU89m | M 50cM |
** Harm avoidance 60 cM 56 Suicide & RE-MDD 37 cM 50 * RE-MDD 60 cM Anxiety 21 cM 48 RE-MDD 25 cM Male pairs57 Neuroticism both sexes and male pairs14 |
||
| NLm | 10 | 175 | 1.7* | 164–175 | 10q26.3 | D10S212, 1.1, 173 cM | NL99m | 70-171 cM | |||
| NLf | 15 | 17 | 1.8 * | 0–35 | 15p11.2-15q14 | GTTTT001, 1.3, 24 cM | NL99f* | NL02f | |||
Mean neuroticism measure with highest LOD score in region based on 5cM grid search.
Maximum LOD score from 5cM grid search. Position and linkage interval (1 LOD less than the maximum LOD) based on 1 cM grid search of identified regions; those marked with * are significant at the suggestive threshold for linkage
Marker within interval which shows largest singlepoint LOD score.
other measures within linkage interval boundaries with LOD > 1.5, those marked with * are significant at the suggestive threshold for linkage.
other individual measures with 1< LOD <1.5 scores within linkage interval boundaries.
Regions identified by Fullerton14 as having support from multiple (M) studies or having reasonable (R) support for linkage from analyses of neuroticism, major depression, anxiety and panic disorder as listed in the primary sources for results where linkage intervals overlap (or are likely to overlap if not presented).
Regions identified by Smoller et al49 (S) as being homologous to linkage regions identified in studies of anxiety in mice.
Linkage studies used in the review by Fullerton45, plus additional studies22, 39, 48, 50, 55, 58. RE-MDD recurrent early onset major depression. ** significant, * suggestive (these include estimated suggestive regions from the linkage study of Fullerton et al16 who discussed in detail only significant linkage results; from their Figure 2 we have estimated which additional peaks may have exceeded a suggestive threshold for linkage, −logP >2.5), R reported (approx LOD > 1.5)
Five regions which exceed empirical suggestive thresholds of linkage are reported for analyses of single sex average neuroticism scores (Table 3). For the linkage interval of these regions, analyses were conducted for the relevant sex for the nine time-specific data sets. Of the five regions, three (2p, 5q and 15cen ) were supported by more than one sex specific individual measure with LOD > 1.0 (Table 3). Of the 8 regions listed in Table 3 that exceed the empirical suggestive threshold for significance only the 14q region exceeds the threshold that accounts for multiple testing of the 9 mean measures.
Discussion
We have performed a linkage analysis for neuroticism using two large independent study samples of North European descent. In total, 5,069 sibling pairs contributed to the linkage analysis which used mean neuroticism scores from both Australia and the Netherlands to maximise sample size and power. Linkage analyses of mean neuroticism score for each country separately allowed us to look for replication between independent data sets. The mean neuroticism measure of each participant could comprise between 1 and 5 individual measures and we used individual neuroticism scores to look for consistency of linkage results. Although subjects with more than one measure of neuroticism age over time, the high genetic correlations between measures would not lead us to expect different genetic variants to be identified in the linkage analysis of different measures. Using mean neuroticism score, we identified five regions where LOD > 1.5, for three of these the LOD score exceeded the empirical threshold for significance. All five regions showed some consistency in linkage scores for individual time-specific measures within country and two regions (8q 134 cM and 18q 117 cM) showed some evidence for replication between countries. Other studies that have reported linkage to these regions are listed in Table 3; we include studies reviewed by Fullerton14 plus a small number of additional, mostly subsequent, publications. Region 18q 117 cM overlaps the linkage intervals reported by three other studies: recurrent early onset and major depression47 73 cM, neuroticism in females22, 91 cM and 115 cM; harm avoidance22 109 cM. Region 14q 103 cM has previously been identified in a linkage analysis of the Dutch study samples for a broad anxiety phenotype39 but also in an independent study of extended families with a high occurrence of anxiety disorders48. Region 10p 5cM was estimated from the linkage graph for EPQ neuroticism presented by Fullerton et al16 to have exceeded the level of suggestive linkage. Only the confidence interval of the 18q region overlaps with a region considered to have “reasonable support for linkage” by Fullerton14 (10 regions were identified comprising ~9% of the genome). Also listed in Table 3 are human chromosomal regions homologous to linkage regions from studies of anxiety in mouse as summarised by Smoller et al49; 11 homologous human chromosomal regions were identified which totalled ~17% of the human genome. Linkage studies in mice are relevant because similar brain processes are likely to exist for anxiety in mice and neuroticism in humans13 and the powerful design of studies that are possible in mice can lead to highly significant linkage regions bounded by tight confidence intervals. Of the five regions we have identified (Table 3) 4 overlap with regions identified by Smoller, an overlap that exceeds chance expectations (Binomial p= 0.003)
Five sex specific linkage regions exceeded thresholds of suggestive linkage (Table 3) of which 2cen 112 cM (males) shows evidence for replication between countries and 5q 191 cM (males) shows evidence for consistency between the Dutch time-specific measures. Of these, region 8p has previously been identified in other linkage studies including two male specific reports (Table 3) and linkage with suicide and recurrent early major depression and been reported for 2p50. Two of the five sex specific regions overlap with homologous regions identified by Smoller49 from mouse linkage studies. Analysis of male and female mean scores separately had much reduced power compared to the joint sex analyses, particularly the male specific analyses and so we place less emphasis on the sex specific results.
For a study of its kind our sample size is large (Table 4), yet the number of linkage regions that we identified for AU&NL, AU and NL were 2, 0 and 3 respectively, not very different from the one per linkage scan expected by chance. Of the other linkage studies for neuroticism (Table 4), only the study of Fullerton et al16 has more power to detect a QTL. Based on observed phenotypic and marker information we had 100% (or 89%) power in the combined Australian and Dutch sample to detect a QTL that accounts for 20% (or 10%) of the total variance at the suggestive45 threshold for linkage. For a trait with a heritability of 30% these are perhaps optimistic power calculations, none-the-less the next largest neuroticism linkage study10 to date, assuming fully informative marker information, reports only 72% power for a QTL that accounts for 20% of the variance. We note that studies likely to have much less power to detect QTL have identified more suggestive and significant linkage regions (Table 5). Theoretically, sample sizes of more than 50 sib pairs should not result in a biased number of linkage statistics exceeding suggestive or significant linkage thresholds under the null hypothesis45. Although under the alternate hypothesis (when a QTL does exist) an inverse correlation between sample size and LOD score is expected51. One conclusion is that there simply are no variants that explain 10% or more of the genetic variance. When do our suggestive linkage peaks represent false positives and when does their low significance reflect variants of smaller effect size? It is not possible to answer this question but by considering multiple measures of neuroticism, we reduce the impact of the environmental noise surrounding chance extreme concordance or discordance of measures and therefore reduce one cause of the occurrence of false positive linkage signals. The examination of linkage analyses from the individual measures of neuroticism provides some evidence for the robustness of our results using mean score.
Table 4.
Summary of linkage studies of neuroticism ordered by number of sibpairs measured for neuroticism
| a. Country b. Study Name (if any) c. Reference d. Measure of neuroticism. e. No. of measures if >1 |
Ascertainment of base population | # sibpairs measured | Selection criterion for inclusion in linkage analysis | # sibpairs in linkage analysis. Inter-marker distance. |
Analysis | No. linkage peaks identified1 | |||
|---|---|---|---|---|---|---|---|---|---|
| SIG | SIGsex | SUG | SUGsex | ||||||
|
Community-based sample | 34,580 | Used neuroticism score to select 2.5% most discordant and 2.5% most concordant, 78%2 response rate | 629 10cM¶ |
Visscher-Hopper59 regression | 2 | 3 | 83 | 33 |
|
| |||||||||
|
See next 2 rows | 19,635 | See next 2 rows | 5,424 | Merlin-Regress44 | 0 | 0 | 2 | 1 |
|
| |||||||||
|
|
|
|
Merlin-Regress | 0 | 0 | 0 | 1 | |
|
| |||||||||
|
Twin individuals and their families | 6,863 | None. No difference in distribution of neuroticism scores for those with and without genotypes. | 1,358 11cM§ |
Merlin-Regress | 0 | 0 | 3 | 3 |
|
| |||||||||
|
Community-based sample | 4,824 | Most informative 10%, Response rate 65%2 | 297 9 cM |
Merlin-Regress | 0 | 1 | 04 | 2 |
|
| |||||||||
|
Sibships concordant for alcohol dependence | 714 | None | 714 4cM |
Merlin-Regress | 1 | 4 | 25 | 45 |
|
| |||||||||
|
Sibships concordant for nicotine dependence | 201 | None | 201 10cM¶ |
Merlin-Regress | 0 | N/A | 5 | N/A |
Exceed Lander and Kruglyak12 empirical thresholds for significant (SIG) or suggestive (SUG) linkage: One linkage test statistic of observed magnitude or greater expected by chance every twenty genome scans (SIG) or each genome scan (SUG). Sex specific linkage peaks, SIGsex, SUGsex.
Response rates are for individuals and reflect the success of the selection criterion for entry into the linkage study; response rate for sibpairs are expected to be the square of this number.
Estimated from their Figure 2 assuming threshold of −logP = 2.5.
One suggestive region identified but listed as significant sex specific region.
Estimated as those with LOD >1 (as empirical threshold for SIG was 1.29).
Calculated as the average distance between markers genotyped in both members of sib pairs, which is likely to be higher than the average distance between markers reported for the other studies.
estimated from number of markers
Limitations of our study include different measures of neuroticism, both between countries and within the Australian sample. The Dutch participants came from a younger cohort than the Australian participants. A recent study has suggested that subtle differences in the EPQ-R and NEO neuroticism instruments may be important for genetic studies52. However, the high genetic correlations between different measures suggest the different measurement instruments are probing the same underlying trait, at least within country. As in other studies, we have undertaken some multiple testing (sex dependent analyses, both between and within countries) which has not been accounted for in the empirical thresholds derived for each mean measure. The empirical threshold (LOD 2.5) derived to account for the multiple testing of the 9 mean neuroticism measures (including sex specific means) is exceeded only for 14q 103 cM. None-the-less, the robustness of our results as measured by consistency in linkage score between time-specific measures combined with the high rate of overlap with regions reported in other studies add to the optimism reported in recent reviews14, 47 for replication of linkage regions, even though the true effect sizes of underlying variants are unlikely to be large. A recent genome-wide association study of neuroticism using DNA pooling53 failed to identify any loci that explained more than 1% of the variance. It is unlikely that the consensus in linkage signals across studies is driven by single variants of such a small magnitude, but more likely implies allelic heterogeneity of causal variants within functionally important genes. Consistently identified regions from linkage analyses will be important in prioritisation of results from genome-wide association studies (GWAS). Time will tell if GWAS result in the identification of causal variants which account for the majority of the observed genetic variance. International collaborations compiling large family-based study samples for linkage analysis may well be necessary for identification of genes that contain multiple but rare causal variants.
Acknowledgments
We thank our interviewers and clerical and administrative support staff supervised by Dixie Statham. We thank David Smyth, Harry Beeby and Olivia Zheng for data management and computer support and our laboratory staff, especially Megan Campbell, Anjali Henders and Leanne McNeil, for sample processing and preparation. We acknowledge the role of Dr Andrew Heath and NIH grants (AA07535 and AA07728) in earlier projects in which selection variables were collected. Phenotype collection was funded by grants to NGM and JGA from the Australian National Health and Medical Research Council (971232, 339450) and by Gemini Genomics plc (now defunct). Additional funding was from MH059160 (Sullivan). For the genome scans we acknowledge and thank: the Mammalian Genotyping Service, Marshfield WI (Director: Dr James Weber) for genotyping conducted under grants awarded to Drs. Daniel T. O’Connor, David Duffy, Dale Nyholt and Patrick Sullivan; Dr Eline Slagboom (Leiden scan); Dr Peter Reed (Gemini scan); Dr Jeff Hall (Sequana); Dr Aarno Palotie (Finnish Genome Center, Helsinki); and Dr Sue Forrest (Australian Genome Research Facility). We thank Arthur Gilmour for his help with ASReml. Most of all, we thank the twins and their relatives for their willing participation in the study.
The Dutch study was supported by the Netherlands Organization for Scientific Research NWO/ZonMW (400-05-717, 911-03-016, 904-61-193, 985-10-002, 575-25-006), NIH R01 HL55976 and NHBLI Mammalian Genotyping Service (Marshfield). CM was supported by the Hersenstichting Nederland (13F05(2).47).
DIB and NGM take responsibility for the integrity of the data and NRW takes responsibility for the accuracy of the data analysis.
References
- 1.Eaves LJ, Eysenck HG, Martin NG. Genes, Culture, and Personality: An Empirical Approach. London: Oxford University Press; 1989. [Google Scholar]
- 2.Hirschfeld RM, Klerman GL. Personality attributes and affective disorders. Am J Psychiatry. 1979;136(1):67–70. doi: 10.1176/ajp.136.1.67. [DOI] [PubMed] [Google Scholar]
- 3.Angst J, Clayton PJ. Premorbid personality of depressive, bipolar, and schizophrenic patients with special reference to suicidal issues. Comprehensive Psychiatry. 1986;27:511–32. doi: 10.1016/0010-440x(86)90055-6. [DOI] [PubMed] [Google Scholar]
- 4.Nystrom S, Lindegard B. Predisposition for mental syndromes. Acta Psychiatrica Scandinavica. 1975;51:69–76. doi: 10.1111/j.1600-0447.1975.tb00214.x. [DOI] [PubMed] [Google Scholar]
- 5.Hirschfeld RMA, Klerman GL, Lavori PW, Keller MB, Griffith P, Coryell W. Premorbid personality assessments of first onset of major depression. Archives of General Psychiatry. 1989;46:345–50. doi: 10.1001/archpsyc.1989.01810040051008. [DOI] [PubMed] [Google Scholar]
- 6.Boyce P, Parker G, Barnett B, Cooney M, Smith F. Personality as a vulnerability factor to depression. British Journal of Psychiatry. 1991;159:106–14. doi: 10.1192/bjp.159.1.106. [DOI] [PubMed] [Google Scholar]
- 7.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. A longitudinal twin study of personality and major depression in women. Archives of General Psychiatry. 1993;50:853–62. doi: 10.1001/archpsyc.1993.01820230023002. [DOI] [PubMed] [Google Scholar]
- 8.Kirk KM, Birley AJ, Statham DJ, et al. Anxiety and depression in twin and sib pairs extremely discordant and concordant for neuroticism: prodromus to a linkage study. Twin Research. 2000;3(4):299–309. doi: 10.1375/136905200320565274. [DOI] [PubMed] [Google Scholar]
- 9.Martin N, Goodwin G, Fairburn C, et al. A population-based study of personality in 34,000 sib-pairs. Twin Res. 2000;3(4):310–5. [PubMed] [Google Scholar]
- 10.Nash MW, Huezo-Diaz P, Williamson RJ, et al. Genome-wide linkage analysis of a composite index of neuroticism and mood-related scales in extreme selected sibships. Hum Mol Genet. 2004;13(19):2173–82. doi: 10.1093/hmg/ddh239. [DOI] [PubMed] [Google Scholar]
- 11.Birley AJ, Gillespie NA, Heath AC, Sullivan PF, Boomsma DI, martin NG. Heritability and nineteen-year stability of long and short EPQ-R neuroticism scales. Personality and individual differences. 2006;40:737–47. [Google Scholar]
- 12.Viken RJ, Rose RJ, Kaprio J, Koskenvuo M. A developmental genetic analysis of adult personality: extraversion and neuroticism from 18 to 59 years of age. J Pers Soc Psychol. 1994;66(4):722–30. doi: 10.1037//0022-3514.66.4.722. [DOI] [PubMed] [Google Scholar]
- 13.Flint J. The genetic basis of neuroticism. Neurosci Biobehav Rev. 2004;28(3):307–16. doi: 10.1016/j.neubiorev.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Fullerton J. New approaches to the genetic analysis of neuroticism and anxiety. Behav Genet. 2006;36(1):147–61. doi: 10.1007/s10519-005-9000-4. [DOI] [PubMed] [Google Scholar]
- 15.Lake RI, Eaves LJ, Maes HH, Heath AC, Martin NG. Further evidence against the environmental transmission of individual differences in neuroticism from a collaborative study of 45,850 twins and relatives on two continents. Behav Genet. 2000;30(3):223–33. doi: 10.1023/a:1001918408984. [DOI] [PubMed] [Google Scholar]
- 16.Fullerton J, Cubin M, Tiwari H, et al. Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative-trait loci that influence variation in the human personality trait neuroticism. Am J Hum Genet. 2003;72(4):879–90. doi: 10.1086/374178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jardine R, Martin NG, Henderson AS. Genetic covariation between neuroticism and the symptoms of anxiety and depression. Genet Epidemiol. 1984;1(2):89–107. doi: 10.1002/gepi.1370010202. [DOI] [PubMed] [Google Scholar]
- 18.Fanous A, Gardner CO, Prescott CA, Cancro R, Kendler KS. Neuroticism, major depression and gender: a population-based twin study. Psychol Med. 2002;32(4):719–28. doi: 10.1017/s003329170200541x. [DOI] [PubMed] [Google Scholar]
- 19.Kendler KS, Gatz M, Gardner CO, Pedersen NL. Personality and major depression: a Swedish longitudinal, population-based twin study. Arch Gen Psychiatry. 2006;63(10):1113–20. doi: 10.1001/archpsyc.63.10.1113. [DOI] [PubMed] [Google Scholar]
- 20.Middeldorp CM, Cath DC, Van Dyck R, Boomsma DI. The co-morbidity of anxiety and depression in the perspective of genetic epidemiology. A review of twin and family studies. Psychol Med. 2005;35(5):611–24. doi: 10.1017/s003329170400412x. [DOI] [PubMed] [Google Scholar]
- 21.Neale BM, Sullivan PF, Kendler KS. A genome scan of neuroticism in nicotine dependent smokers. Am J Med Genet B Neuropsychiatr Genet. 2005;132(1):65–9. doi: 10.1002/ajmg.b.30095. [DOI] [PubMed] [Google Scholar]
- 22.Kuo PH, Neale MC, Riley BP, et al. A genome-wide linkage analysis for the personality trait neuroticism in the Irish affected sib-pair study of alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2007 doi: 10.1002/ajmg.b.30478. [DOI] [PubMed] [Google Scholar]
- 23.Camp NJ, Cannon-Albright LA. Dissecting the genetic etiology of major depressive disorder using linkage analysis. Trends Mol Med. 2005;11(3):138–44. doi: 10.1016/j.molmed.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Diego VP, Atwood L, Mathias RA, Almasy L. Consistency of genetic analyses in longitudinal data: observations from the GAW13 Framingham Heart Study data. Genet Epidemiol. 2003;25 (Suppl 1):S29–35. doi: 10.1002/gepi.10281. [DOI] [PubMed] [Google Scholar]
- 25.Sham PC, Cherny SS, Purcell S, Hewitt JK. Power of linkage versus association analysis of quantitative traits, by use of variance-components models, for sibship data. Am J Hum Genet. 2000;66(5):1616–30. doi: 10.1086/302891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eysenck SBG, Eysenck HJ, Barrett P. A revised version of the Pyschoticism scale. Personality and individual differences. 1985;6:21–30. [Google Scholar]
- 27.Costa PT, MacCrae RR. NEO PI-R Professional Manual. Florida: Psychological Assessment Resources, Inc; 1991. [Google Scholar]
- 28.Risch N, Zhang H. Extreme discordant sib pairs for mapping quantitative trait loci in humans. Science. 1995;268(5217):1584–9. doi: 10.1126/science.7777857. [DOI] [PubMed] [Google Scholar]
- 29.Purcell S, Cherny SS, Hewitt JK, Sham PC. Optimal sibship selection for genotyping in quantitative trait locus linkage analysis. Hum Hered. 2001;52(1):1–13. doi: 10.1159/000053350. [DOI] [PubMed] [Google Scholar]
- 30.Saccone SF, Pergadia ML, Loukola A, et al. Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. Am J Hum Genet. 2007;80(5):856–66. doi: 10.1086/513703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boomsma DI, de Geus EJ, Vink JM, et al. Netherlands Twin Register: from twins to twin families. Twin Res Hum Genet. 2006;9(6):849–57. doi: 10.1375/183242706779462426. [DOI] [PubMed] [Google Scholar]
- 32.Boomsma DI, Beem AL, van den Berg M, et al. Netherlands twin family study of anxious depression (NETSAD) Twin Res. 2000;3(4):323–34. doi: 10.1375/136905200320565300. [DOI] [PubMed] [Google Scholar]
- 33.Wilde GJS. Neurotische labiliteit gemeten volgens de vragenlijstmethode (The questionnaire method as a means of measuring neurotic instability) Amsterdam: Van Rossen; 1970. [Google Scholar]
- 34.Eysenck H, Eysenck S. Manual of the Eysenck Personality Questionnaire. London: Hodder and Stoughton; 1975. [Google Scholar]
- 35.Freeman MF, Tukey JW. Transformations Related to the Angular and the Square Root. Annals of Mathematical Statistics. 1950;21(4):607–11. [Google Scholar]
- 36.Wray NR, Birley AJ, Sullivan PF, Visscher PM, Martin NG. Genetic and phenotypic stability of measures of neuroticism over 22 years. Twin Research and Human Genetics. 2007 doi: 10.1375/twin.10.5.695. In press. [DOI] [PubMed] [Google Scholar]
- 37.Cornes BK, Medland SE, Ferreira MA, et al. Sex-limited genome-wide linkage scan for body mass index in an unselected sample of 933 Australian twin families. Twin Res Hum Genet. 2005;8(6):616–32. [PubMed] [Google Scholar]
- 38.Nyholt DR, Morley KI, Ferreira MA, et al. Genomewide significant linkage to migrainous headache on chromosome 5q21. Am J Hum Genet. 2005;77(3):500–12. doi: 10.1086/444510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Middeldorp C, Hottenga J, Slagboom P, et al. Linkage on chromosome 14 in a genomewide linkage study of a broad anxiety phenotype. Molecular psychiatry. 2007 doi: 10.1038/sj.mp.4002061. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duffy DL. An integrated genetic map for linkage analysis. Behav Genet. 2006;36(1):4–6. doi: 10.1007/s10519-005-9015-x. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan PF, Montgomery GW, Hottenga JJ, Wray NR, Boomsma DI, Martin NG. Empirical evaluation of the genetic similarity of samples from twin registries in Australia and the Netherlands using 359 STRP markers. Twin Res Hum Genet. 2006;9(4):600–2. doi: 10.1375/183242706778025026. [DOI] [PubMed] [Google Scholar]
- 42.Duffy D. Sib-Pair: A program for simple genetic analyses. 2006 http://www2qimreduau/davidD/sib-pairhtml.
- 43.Gilmour AR, Gogel BJ, Cullis BR, Welham SJ, Thompson R. ASReml User Guide Release 1.0. 2002. [Google Scholar]
- 44.Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet. 2002;71(2):238–53. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11(3):241–7. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 46.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–50. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 47.Camp NJ, Lowry MR, Richards RL, et al. Genome-wide linkage analyses of extended Utah pedigrees identifies loci that influence recurrent, early-onset major depression and anxiety disorders. Am J Med Genet B Neuropsychiatr Genet. 2005;135(1):85–93. doi: 10.1002/ajmg.b.30177. [DOI] [PubMed] [Google Scholar]
- 48.Kaabi B, Gelernter J, Woods SW, Goddard A, Page GP, Elston RC. Genome scan for loci predisposing to anxiety disorders using a novel multivariate approach: strong evidence for a chromosome 4 risk locus. Am J Hum Genet. 2006;78(4):543–53. doi: 10.1086/501072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smoller JW, Acierno JS, Jr, Rosenbaum JF, et al. Targeted genome screen of panic disorder and anxiety disorder proneness using homology to murine QTL regions. Am J Med Genet. 2001;105(2):195–206. doi: 10.1002/ajmg.1209. [DOI] [PubMed] [Google Scholar]
- 50.Zubenko GS, Maher BS, Hughes HB, 3rd, Zubenko WN, Scott Stiffler J, Marazita ML. Genome-wide linkage survey for genetic loci that affect the risk of suicide attempts in families with recurrent, early-onset, major depression. Am J Med Genet B Neuropsychiatr Genet. 2004;129(1):47–54. doi: 10.1002/ajmg.b.30092. [DOI] [PubMed] [Google Scholar]
- 51.Goring HH, Terwilliger JD, Blangero J. Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet. 2001;69(6):1357–69. doi: 10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmitz A, Hennig J, Kuepper Y, Reuter M. The association between neuroticism and the serotonin transporter polymorphism depends on structural differences between personality measures. Personality and Individual Differences. 2007;42(4):789–99. [Google Scholar]
- 53.Shifman S, Bhomra A, Smiley S, et al. Awhole genome association study of neuroticism using DNA pooling. Molecular psychiatry. 2007 doi: 10.1038/sj.mp.4002048. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cloninger CR, Van Eerdewegh P, Goate A, et al. Anxiety proneness linked to epistatic loci in genome scan of human personality traits. Am J Med Genet. 1998;81(4):313–7. doi: 10.1002/(sici)1096-8628(19980710)81:4<313::aid-ajmg7>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 55.Thorgeirsson TE, Oskarsson H, Desnica N, et al. Anxiety with panic disorder linked to chromosome 9q in Iceland. Am J Hum Genet. 2003;72(5):1221–30. doi: 10.1086/375141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dina C, Nemanov L, Gritsenko I, et al. Fine mapping of a region on chromosome 8p gives evidence for a QTL contributing to individual differences in an anxiety-related personality trait: TPQ harm avoidance. Am J Med Genet B Neuropsychiatr Genet. 2005;132(1):104–8. doi: 10.1002/ajmg.b.30099. [DOI] [PubMed] [Google Scholar]
- 57.Holmans P, Weissman MM, Zubenko GS, et al. Genetics of recurrent early-onset major depression (GenRED): final genome scan report. Am J Psychiatry. 2007;164(2):248–58. doi: 10.1176/ajp.2007.164.2.248. [DOI] [PubMed] [Google Scholar]
- 58.McGuffin P, Knight J, Breen G, et al. Whole genome linkage scan of recurrent depressive disorder from the depression network study. Hum Mol Genet. 2005;14(22):3337–45. doi: 10.1093/hmg/ddi363. [DOI] [PubMed] [Google Scholar]
- 59.Visscher PM, Hopper JL. Power of regression and maximum likelihood methods to map QTL from sib-pair and DZ twin data. Ann Hum Genet. 2001;65(Pt 6):583–601. doi: 10.1017/S0003480001008909. [DOI] [PubMed] [Google Scholar]