Abstract
Air breathing is an essential motor function for vertebrates living on land. The rhythm that drives breathing is generated within the central nervous system and relayed via specialised subsets of spinal motor neurons to muscles that regulate lung volume. In mammals, a key respiratory muscle is the diaphragm, which is innervated by motor neurons in the phrenic nucleus. Remarkably, relatively little is known about how this crucial subtype of motor neuron is generated during embryogenesis. Here, we used direct differentiation of motor neurons from mouse embryonic stem cells as a tool to identify genes that direct phrenic neuron identity. We find that three determinants, Pou3f1, Hoxa5 and Notch, act in combination to promote a phrenic neuron molecular identity. We show that Notch signalling induces Pou3f1 in developing motor neurons in vitro and in vivo. This suggests that the phrenic neuron lineage is established through a local source of Notch ligand at mid-cervical levels. Furthermore, we find that the cadherins Pcdh10, which is regulated by Pou3f1 and Hoxa5, and Cdh10, which is controlled by Pou3f1, are both mediators of like-like clustering of motor neuron cell bodies. This specific Pcdh10/Cdh10 activity might provide the means by which phrenic neurons are assembled into a distinct nucleus. Our study provides a framework for understanding how phrenic neuron identity is conferred and will help to generate this rare and inaccessible yet vital neuronal subtype directly from pluripotent stem cells, thus facilitating subsequent functional investigations.
Keywords: Embryonic stem cell, Phrenic neuron, Transcriptional identity, Motor neuron differentiation
INTRODUCTION
Land vertebrates, including humans, use lungs to breathe air. The inspiratory and expiratory movements of the lungs are driven by a complex neural circuitry that consists of a central network in the brainstem that generates breathing rhythms and an output layer of motor neurons (MNs) that connect to respiratory muscles. These respiratory circuits develop prenatally and must become functional immediately after birth. Although significant progress has been made in understanding the central pattern generator itself (Champagnat et al., 2009; Fortin and Thoby-Brisson, 2009), relatively little is known about the formation of MNs that relay breathing rhythms from the CNS to the periphery. In mammals, respiration is driven by muscles that connect to the rib cage and thereby indirectly inflate and deflate the lungs. Arguably, the most important of these muscles is the diaphragm, which forms the boundary between the thoracic and abdominal cavities and contracts during inspiration. The diaphragm is innervated by the phrenic nucleus (PN), a population of MNs located in the mid-cervical spinal cord. During embryonic development, phrenic neurons emerge alongside other MNs from ventral progenitors (Arber et al., 1999; Briscoe et al., 2000; Thaler et al., 1999), send their axons through cervical ventral roots and then project caudally through the thoracic cavity to innervate the diaphragm muscle (Allan and Greer, 1997). We have only a partial understanding of the molecular cascade that establishes phrenic MN identity. It is, however, important that this pathway is defined as this would provide insights into how mammal-specific anatomical adaptations are patterned and allow us to model aspects of respiratory motor circuitry in neuronal cultures to study neuromuscular function and disease.
Spinal MNs segregate into distinct columns during embryogenesis. Each column connects to a specific set of muscles: the medial motor column (MMC) projects to epaxial muscles, the lateral motor column (LMC) innervates limb muscles (Lance-Jones and Landmesser, 1980; Tsuchida et al., 1994), and the hypaxial motor column (HMC) innervates body wall muscles (Dasen et al., 2003; Peljto and Wichterle, 2011). MNs acquire subtype transcriptional identities due to exposure to locally restricted morphogens (Marshall et al., 1992). For example, the expression of the MMC determinants Lhx3 and Lhx4 is sustained by floor plate-derived Wnt4 and Wnt5 (Agalliu et al., 2009). Likewise, brachial LMC fate depends on overlapping, segmentally restricted gradients of retinoic acid and Fgfs (Liu et al., 2001), which induce Hox6 paralogues and the accessory factor Foxp1 in presumptive LMC neurons in register with forelimbs (Dasen et al., 2008; Dasen et al., 2003). The HMC, by contrast, appears to lack specific determinants and might represent a ground state of MNs. Phrenic neurons are thought to be derivatives of the HMC (Rousso et al., 2008).
Some candidate determinants for early phrenic development have been identified, although their contribution to phrenic neuron specification is poorly understood: the transcription factor (TF) Pou3f1 is enriched in phrenic neurons, and Pou3f1 deficiency in mice leads to disorganisation of the PN (Bermingham et al., 1996). Absence of mid-cervical Hox5 paralogues affects the maintenance of phrenic neurons, but not their initial specification (Philippidou et al., 2012). Lastly, Foxp1 appears to negatively regulate the phrenic MN lineage, since Foxp1 mutants have increased numbers of phrenic MNs (Rousso et al., 2008). What is largely lacking at this point is an understanding of which potential effector genes are downstream of these factors, how these and other determinants interact and, in the case of Pou3f1, how the expression of the factor itself is initiated.
MN development can be recapitulated in vitro from mouse or human embryonic stem cells (ESCs), which will form functional spinal MNs under the appropriate culture conditions (Li et al., 2008; Miles et al., 2004; Wichterle et al., 2002). ESC-derivation of MNs depends on the same extrinsic and intrinsic cues that act during normal embryogenesis and has been repeatedly used to investigate subtype-specific developmental pathways in these cells (Jung et al., 2010; Peljto et al., 2010; Soundararajan et al., 2006). We set out to apply this approach to the acquisition of phrenic neuron identity.
To address how phrenic neuron fate is established in the developing spinal cord, we first identified candidate determinants in primary MNs sorted from mouse embryos, and then used a systematic in vitro gain-of-function (GOF) screening approach to test whether any given candidate approximates phrenic neuron transcriptional patterns when ectopically expressed in ESC-derived MNs (ESC-MNs). The aim was to define modules of effector genes downstream of the key determinants, as well as to understand how the determinants interact with each other. We found that the TFs Pou3f1, Hoxa5 and Notch intracellular domain (NICD) combine to regulate distinct sets of effector genes, which together comprise a large fraction of all phrenic neuron-specific genes. Moreover, expression of the receptors Cdh10, which is downstream of Pou3f1, or Pcdh10, a gene coordinately regulated by Hoxa5 and Pou3f1, is sufficient to mediate like-like clustering of MNs into aggregates in vitro, mimicking nucleus formation in vivo. Our findings suggest that local Notch-Delta interaction in the ventral spinal cord might be an early event in phrenic neuron specification and that a defined combination of intrinsic and/or extrinsic factors may be used to emulate phrenic neuron transcriptional identity and morphological features in MNs derived from pluripotent stem cells in vitro.
RESULTS
Isolation of phrenic neurons from E11.5 mouse embryos by flow cytometry
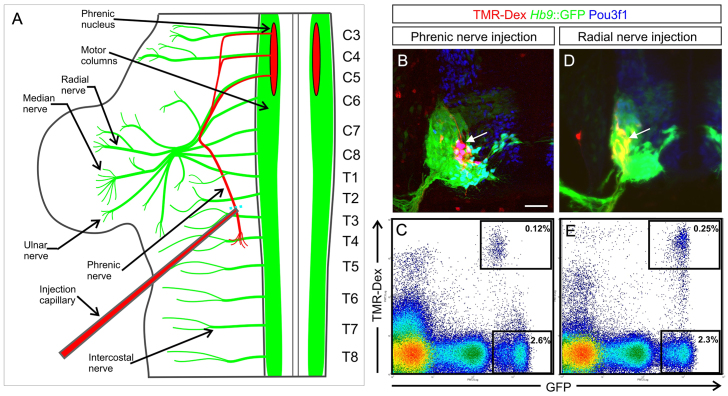
In the ventral embryonic spinal cord, phrenic neurons are generated alongside other MNs from ventral progenitors. To determine the transcriptional profile of embryonic phrenic neurons, and how they differ from other MNs, we isolated phrenic neurons from E11.5 Hb9::GFP mouse embryos. The genetic reporter labels axons and cell bodies of all spinal MNs (Wichterle et al., 2002). In cervical and thoracic trunk explants, the cell bodies of phrenic neurons were retrogradely labelled with the tracer TMR-dextran (Fig. 1A,B). Then, spinal cord segments C3-C5 were excised from the trunks, dissociated, and phrenic neurons were sorted as GFP+ TMR+ cells by flow cytometry (Fig. 1C). Immunohistochemistry confirmed that only Pou3f1+ phrenic MNs in the medial-dorsal part of the ventral horn were TMR positive (Fig. 1B). As a control population, non-phrenic MNs were isolated from the same cell suspension. Given their segmental origin, these MNs are likely to represent a mix of MMC, HMC and LMC neurons (Peljto et al., 2010). A second control population of pure LMC neurons was isolated by retrograde labelling through the radial nerve (Fig. 1D,E). Following isolation by flow cytometry, genome-wide transcriptional patterns of phrenic neurons and the two control populations of MNs were determined using Affymetrix arrays.
Fig. 1.
Retrograde labelling and purification of mouse E11.5 embryonic phrenic MNs by flow cytometry. (A) The phrenic nerve was injected with TMR-dextran in Hb9::GFP trunk explants. Spinal segments are numbered according to the ventral roots that emerge from them (labels on right). C, cervical; T, thoracic. (B) Mid-cervical spinal cord transverse section: phrenic neurons (arrow) are labelled with TMR-dextran (red). The Hb9::GFP transgene labels all MNs; phrenic neurons co-express Pou3f1. Scale bar: 50 μm. (C) TMR+ GFP+ phrenic neurons and GFP+ non-phrenic MNs were isolated from spinal cords (C3-C5 levels) by flow cytometry. (D) TMR+ GFP+ radial LMC neurons (arrow) in E11.5 cervical spinal cord (C6-C8 levels), following retrograde tracing through the radial nerve. (E) Purification of TMR+ GFP+ radial LMC neurons by flow cytometry.
Identification of phrenic neuron-specific genes
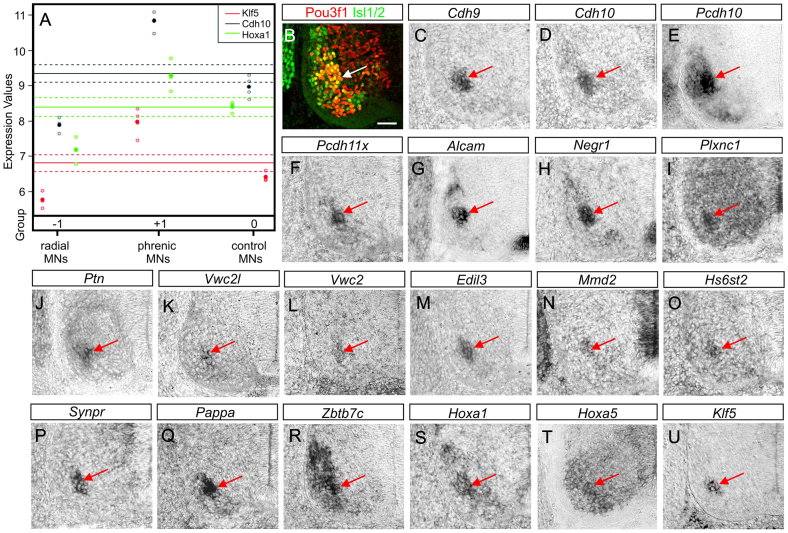
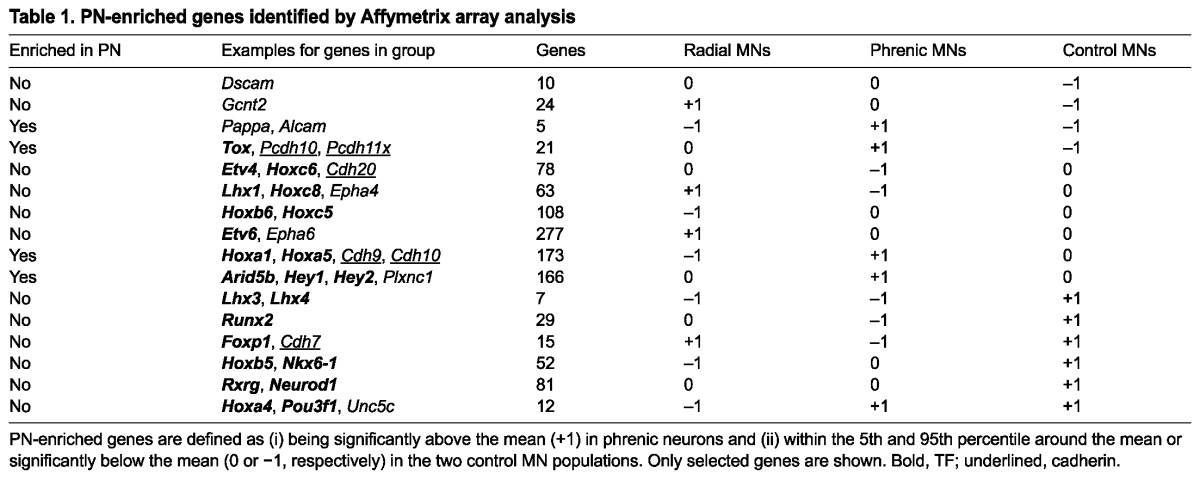
We next devised a scheme to systematically catalogue genes into groups based on their expression pattern in radial LMC MNs, phrenic MNs and non-phrenic control MNs derived from segments C3-C5, respectively. A logical value was attributed to each gene in each cell population: genes expressed significantly above the mean across the three MN populations received the value +1, genes expressed significantly below the mean -1, and genes with expression levels above the 5th and below the 95th percentile around the mean received the value 0. Hoxa1, Cdh10 and Klf5 are examples of phrenic neuron-enriched genes with an expression value of -1 in radial MNs, +1 in phrenic MNs, and 0 in control MNs (Fig. 2A). This pattern can be described with the row vector [-1 +1 0].
Fig. 2.
Identification of genes enriched in phrenic neurons by Affymetrix array analysis. (A) Logical values were attributed to each gene in each cell type based on whether it is enriched (+1), within the mean (0) or depleted (-1). Three examples for genes enriched in phrenic neurons (Klf5, Hoxa1, Cdh10) are shown. Expression values are shown in log2 scale. (B) The PN is identified by expression of Pou3f1 and Isl1/2 in E11.5 mid-cervical spinal cords (arrow). Scale bar: 50 μm. (C-U) Candidate mRNAs are enriched in phrenic MNs. Red arrows indicate PN.
A number of known MN subtype markers found in different groups of neurons suggest that our approach yields genuine expression patterns (Table 1). The MMC determinants Lhx3 and Lhx4 follow the pattern [-1 -1 +1] and are enriched in the C3-C5 control (Table 1), the only cell population with an MMC component. Pou3f1, one of the few established phrenic neuron markers (Bermingham et al., 1996; Rousso et al., 2008), is found both in phrenic neurons and C3-C5 control MNs. Subsequent expression analysis revealed that, at E11.5, Pou3f1 is also expressed in a subset of MMC neurons (Fig. 2B; supplementary material Figs S3, S4), which explains the pattern that we observed. A second factor required for PN development, Hoxa5 (Philippidou et al., 2012), is enriched in phrenic neurons as well. We were able to confirm phrenic neuron-specific expression for several candidate genes by in situ hybridisation (Fig. 2C-U). These genes fall into different functional categories and include cadherins (Fig. 2C-F), Ig superfamily receptors (Fig. 2G,H), BMP inhibitors (Fig. 2K,L), synaptic proteins (Fig. 2P) and TFs (Fig. 2R-U). The identification of phrenic MN-specific cadherins is significant, as adhesion molecules of this protein family are expressed in specific combinations on many, if not all, MN subsets. Cadherins mediate like-like clustering of MN cell bodies and drive the formation of topographic maps in the ventral spinal cord (Bello et al., 2012; Demireva et al., 2011; Price et al., 2002). In summary, we were able to identify a large number of genes potentially enriched in primary phrenic neurons and confirmed most of those analysed by histology.
Table 1.
PN-enriched genes identified by Affymetrix array analysis
An ESC-MN-based screening method for subtype determinants
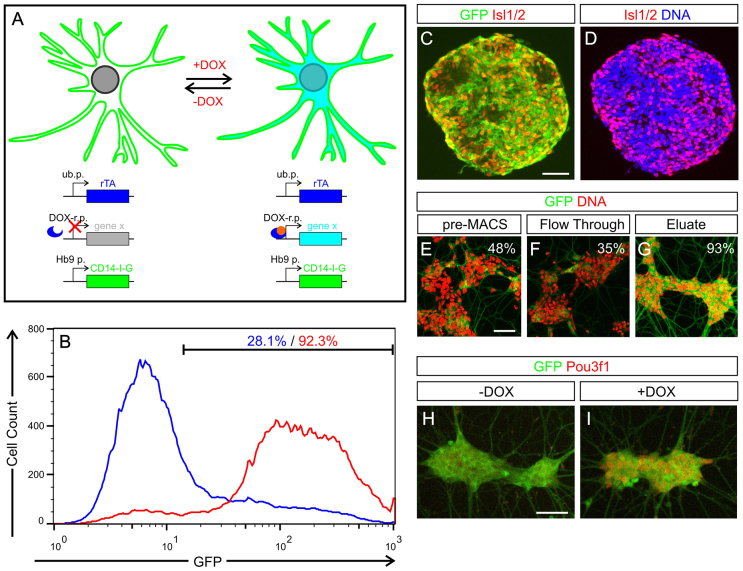
To assess the efficacy of these genes to assign PN identity, we adapted the ESC-MN culture system such that we could perform GOF screens for multiple factors in purified MNs. We chose a mouse ESC clone that allows the efficient insertion of DOX-inducible transgenes by Cre/loxP-mediated cassette exchange (Iacovino et al., 2011) and equipped it with an additional MN-specific reporter transgene that allows magnetic enrichment of this cell type (Fig. 3A). During the initial phase of the project, we used ESCs carrying an Hb9::CD2-GFP MN-specific reporter (supplementary material Fig. S5, Table S2). To optimise the sorting procedure, we generated an ESC subclone that carries the Hb9::CD14-IRES-GFP transgene (supplementary material Fig. S1), which resulted in an improved signal-to-background ratio (Fig. 3B) and sorting efficiency. The GFP MN reporter tightly correlates with the endogenous embryonic MN marker Isl1/2 (Fig. 3C,D), and, following MACS, yielded large numbers (105-106) of viable, enriched ESC-MNs (Fig. 3B,E-G). Furthermore, we were able to induce the expression of candidate transgenes with DOX in culture, as shown here for the candidate factor Pou3f1 (Fig. 3H,I).
Fig. 3.
Inducible expression of candidate phrenic determinants in sorted ESC-derived MNs. (A) GOF screen experimental setup for the identification of PN-specific transcriptional patterns. ESC-MNs were differentiated in vitro, isolated from mixed cultures with an MN-specific Hb9::CD14-IRES-GFP reporter gene and induced to express a given candidate gene with DOX. rTA, reverse Tet transactivator; ub.p., ubiquitous promoter; DOX-r.p., DOX-responsive promoter; Hb9 p., Hb9 promoter. (B) Flow cytometry analysis of ESC-MNs enrichment by anti-CD14 MACS. Blue, pre-MACS cell suspension (dissociated EBs); red, anti-CD14-enriched MACS eluate. (C,D) GFP reporter and Isl1/2 expression in day 6 EBs differentiated from Hb9::CD14-IRES-GFP ESCs. (E-G) GFP-labelled MNs were enriched by anti-CD14 MACS and cultured (G), compared with the input (E) or the flowthrough (F). Percentage: GFP+ cells. (H,I) DOX induction of iPou3f1 ESC-MNs cultured for 24 hours. Scale bars: 50 μm.
We next examined the subtype composition of parental ESC-MNs to determine the baseline identity in our assay. Primary mid-cervical MNs belong to three distinct subsets: HMC, MMC and PN (supplementary material Fig. S3A-G). When we compared the phenotypes of ESC-MNs with in vivo MNs, we found that they mostly belonged to the Pou3f1- Lhx3- HMC and the Pou3f1- Lhx3+ or Pou3f1+ Lhx3+ MMC subsets, with only a small percentage of Pou3f1+ Lhx3- phrenic-like neurons (supplementary material Fig. S3H-L), consistent with an earlier study (Peljto et al., 2010). The percentage of ESC-MNs with an MMC phenotype declines between day 5 and day 6 (supplementary material Fig. S3L). This might be explained by the fact that all MN progenitors express Lhx3 initially, but only MNs committed to the MMC fate sustain its expression (Sharma et al., 1998). The MN subtype composition does not depend on the ESC clone used in this study, as we have detected similar ratios of MN subtypes in cultures differentiated from a second, independently derived ESC clone (supplementary material Fig. S3M).
Our analysis of genes enriched in primary phrenic neurons pinpointed a cluster of known and putative TFs that would be good candidates as determinants of PN identity (Table 1; supplementary material Table S1). In cases in which two closely related genes were isolated, only one gene was investigated further. Our transcriptional profile also identified Notch signalling as a potential key player in this process, as we found that Notch targets, such as Hey1 and Hey2 (Table 1), are enriched in phrenic neurons. Furthermore, we included a dominant-negative mutant form of Lhx3 (DNLhx3) (West et al., 2004) because DNLhx3 suppresses Lhx3 target genes and might inhibit this key MMC determinant (Agalliu et al., 2009). Vwc2 was the only non-TF chosen for analysis, because it is a chordin-like BMP inhibitor and may modulate neural patterning. Finally, we selected Hoxc6 and wild-type Lhx3 as control TFs, as they represent known LMC and MMC determinants, respectively. We established 19 sets of ESC subclones carrying single candidate transgenes (supplementary material Table S2) and confirmed inducible candidate gene expression in embryoid bodies (EBs) (supplementary material Fig. S6).
Reconstruction of PN-specific transcriptional patterns in ESC-MNs
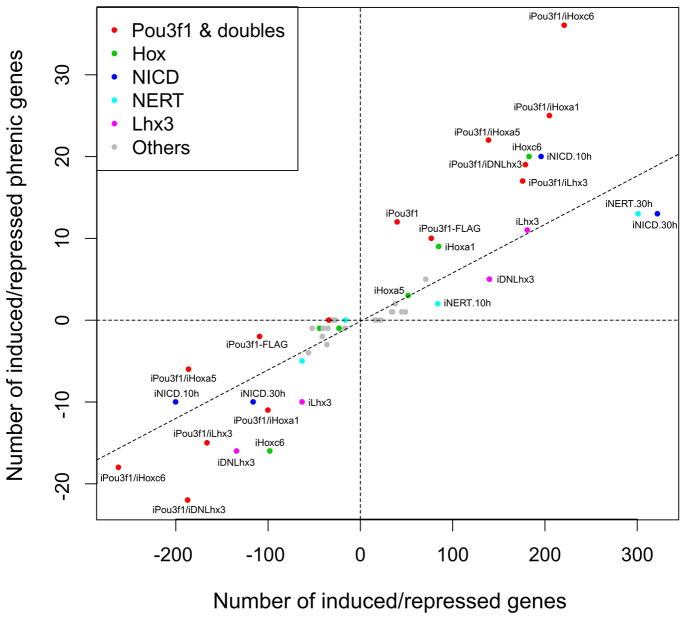
To address whether candidate determinants promote transcriptional profiles associated with phrenic neurons when ectopically expressed in ESC-MNs, we differentiated transgenic ESCs into MNs in EB cultures. MNs were magnetically sorted from dissociated EBs on day 5 and induced to express the transgene with DOX during the subsequent 30 hour culture period. In some experiments with Notch constructs, the ESC-MNs were only induced for the last 10 hours to limit the effect of the transgenes. RNA of ESC-MNs derived from two independent subclones for each candidate gene was isolated, and gene expression profiles determined using Affymetrix arrays. The expression data are represented as a scatter plot in Fig. 4, in which the number of PN-enriched genes (supplementary material Table S4) repressed/induced (y-axis) is plotted against all genes repressed/induced by a given candidate factor (x-axis). The mean value for each transgene is represented by two data points, one for repression (lower left quadrant) and one for induction (upper right quadrant). We would like to point out that the terms repression and induction do not necessarily imply a direct genetic interaction, as we measure mRNA levels and not promoter binding.
Fig. 4.
Identification of TFs that control parts of the PN-specific transcriptional pattern. MACS-sorted ESC-MNs were induced to express candidate determinants, and their transcriptional profiles were examined by Affymetrix array analysis. In the scatter plot, the number of PN-specific genes induced/repressed (y-axis) is plotted against the number of all genes induced/repressed (x-axis) by a given candidate factor or combination thereof. The diagonal line represents a robust fit through all the points shown, both induced and repressed.
Thus, the lower the y/x ratio for repression (R-ratio) and the higher the y/x ratio for induction (I-ratio) the more the transcriptional pattern evoked by a transgene approximates that of primary phrenic neurons. The P-values obtained using a hypergeometric distribution for over-representation of induced or repressed genes (supplementary material Table S5) provide a very similar picture to that offered by the R- and I-ratios, except that these results are more difficult to visualise.
Of all candidate determinants tested, ESC-MNs overexpressing Pou3f1 showed the most specific induction of PN-enriched genes (I-ratio=0.3; Fig. 4, upper right quadrant), whereas not a single specific gene is repressed (R-ratio=0). The genes upregulated by Pou3f1 include the confirmed PN-specific genes Cdh9, Cdh10 and Pcdh11x (Fig. 2C,D,F). NICD, when DOX-induced for the last 10 hours of ESC-MN culture, was identified as a second PN-candidate factor with a high I-ratio (0.102) and low R-ratio (0.05) (Fig. 4; supplementary material Fig. S7A). The set of PN-specific genes upregulated by iNICD includes Vwc2l (Fig. 2K) and is largely non-overlapping with that observed in inducible (i) Pou3f1 ESC-MNs (supplementary material Fig. S8A, Table S6), although a few target genes, such as Hs6st2, are shared. The timing and signal intensity of Notch activity appear to be critical, as iNICD expressed for 30 hours, or the attenuated mutant isoform iNERT (Schroeder et al., 2003) induced for 10 hours or 30 hours, do not evoke a PN-like pattern. iHoxc6, despite the absence of Hoxc6 from primary phrenic neurons (Table 1), has a high I-ratio (0.109), yet also a high R-ratio (0.163) (supplementary material Fig. S7B). However, although iHoxc6 does induce 20 PN-enriched genes, including Hoxa5 (supplementary material Table S6), it also upregulates two key LMC determinants, Foxp1 (Dasen et al., 2008) and Aldh1a2 (Sockanathan and Jessell, 1998) (supplementary material Table S7). Ectopic iHoxa5 expression in ESC-MNs, by contrast, does not induce these LMC markers, and positively regulates only three phrenic neuron-enriched genes, including Ptn (Fig. 2J; supplementary material Table S6). Thus, when activated in isolation, iHoxa5 does not emulate a PN transcriptional programme. Finally, iDNLhx3 does not evoke PN-like patterns (Fig. 4), but surprisingly shows the best I-ratio for LMC-like patterns (supplementary material Fig. S9, Tables S5, S7). This suggests that repression of Lhx3 targets unlocks parts of the LMC but not of the PN transcriptional programme in ESC-MNs.
Pou3f1 interacts with Hoxa5 to establish a phrenic-like transcriptional profile in vitro
To explore whether any of the candidate determinants interact to elicit PN-like transcriptional patterns, we combined the most promising candidate, Pou3f1, with Hoxa5, Hoxc6, Hoxa1 and DNLhx3 in an expression system that allows us to simultaneously induce two candidate genes in the same ESC-MNs (Bondue et al., 2011). In addition, we combined Pou3f1 with Lhx3 to investigate negative interactions, as we observed that Pou3f1 target genes, such as Cdh9 and Cdh10 (Fig. 2C,D), are restricted to the PN, whereas Pou3f1 itself is also expressed by a subpopulation of the MMC (supplementary material Figs S3, S4) (Rousso et al., 2008). Array analysis of double-transgenic ESC-MNs revealed that iPou3f1/iHoxa5 and iPou3f1/iHoxc6 DOX induction resulted in nearly identical I-ratios (0.158 versus 0.163; Fig. 4; supplementary material Fig. S7C). By contrast, the R-ratio for iPou3f1/iHoxa5 (0.032) indicates higher specificity than that for iPou3f1/iHoxc6 (0.069). As in the iHoxc6 single-transgenic ESC-MNs, iPou3f1/iHoxc6 expression upregulates the LMC determinants Foxp1 and Aldh1a2, whereas iPou3f1/iHoxa5 expression does not (supplementary material Table S7). Interestingly, the number of PN-specific genes activated above the twofold threshold in iPou3f1/iHoxa5 ESC-MNs (22 genes) is higher than the sum of the two target gene sets in single transgenic MNs (12+3 genes), which suggests that several PN target genes, such as Pcdh10, require the combined activity of both determinants (supplementary material Table S6). Comparison of transcriptional patterns of iPou3f1 and iPou3f1/iLhx3 ESC-MNs shows that four of 12 iPou3f1-induced PN-specific targets, including Cdh9, are downregulated more than twofold by co-expression of Lhx3 (supplementary material Fig. S7D, Table S6). These findings provide evidence that Pou3f1 can interact with other determinants to regulate the activation of PN-specific target genes, and that both positive (Hoxa5) and negative (Lhx3) combinatorial interactions can be observed in ESC-MNs. Furthermore, Hoxa5 and Hoxc6, despite considerable overlap in their target genes (supplementary material Fig. S8B), differ in that Hoxc6 initiates LMC-like transcriptional patterns in ESC-MNs, whereas Hoxa5 does not.
In order to validate the transcriptional patterns observed by array analysis, we tested the expression of selected candidate genes in iYFP, iHoxa5, iNICD (10 hour pulse), iPou3f1 and iPou3f1/iHoxa5 ESC-MNs by qRT-PCR (supplementary material Table S8). The qRT-PCR expression data are largely consistent with the array data, although some additional target genes were detected. Crucially, the qRT-PCR analysis showed that NICD upregulates the PN determinant Pou3f1.
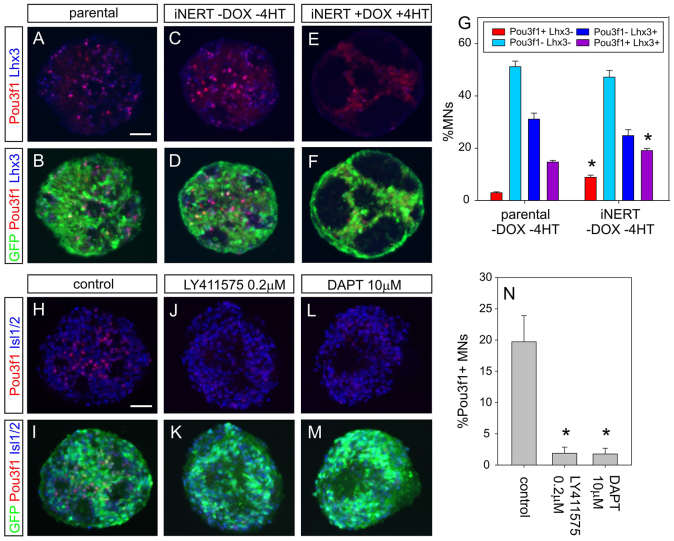
Notch signalling induces the PN determinant Pou3f1 in MNs in vitro and in vivo
Do the three positive phrenic determinants we identified simply act in parallel or is there a transcriptional hierarchy, i.e. does one of the candidate factors initiate phrenic neuron specification? As the qRT-PCR data suggested that NICD activity upregulates Pou3f1, we further examined a possible hierarchical relationship between the two factors. We compared EBs derived from the parental ESC line with those derived from iNERT ESCs, which express an attenuated, tamoxifen (4HT)-inducible version of NICD (Fig. 5A-F). Even in the absence of induction with DOX and 4HT, iNERT EBs contained a significantly higher proportion of both phrenic-like Pou3f1+ Lhx3- MNs and MMC-like Pou3f1+ Lhx3+ MNs (Fig. 5G), without a change in total MN numbers. This increase is likely to be caused by low-level transgene expression/activity in the absence of added inducers, which is a common phenomenon seen in inducible genetic systems (Howe et al., 1995; Schroeder et al., 2003). Full induction of the iNERT transgene led to a decrease in the number of MNs (Fig. 5E,F), consistent with previous findings when NICD was overexpressed in spinal progenitors in vivo (Dias et al., 2012). We next examined whether Notch signalling is required for Pou3f1 expression in a subset of normal ESC-MNs. We treated developing EBs with γ-secretase inhibitors, which block ligand-dependent Notch activation (De Strooper et al., 1999). Both of the inhibitors that we tested decreased the percentage of Pou3f1+ ESC-MNs by more than 90% (Fig. 5H-N), whereas they did not affect MN development in general. To test if Notch GOF upregulates additional PN markers, we examined the effect of a 10 hour pulse of DOX on the transcriptional patterns of iNICD EBs and cultured ESC-MNs, compared with normal controls. We found that NICD induces the PN-enriched genes Synpr (supplementary material Fig. S10), Cdh9, Cdh10, Pcdh11x and Ptn (supplementary material Table S8).
Fig. 5.
Notch activation regulates Pou3f1 expression in ESC-derived MNs. (A-F) GFP, Lhx3 and Pou3f1 staining in day 6 EBs derived from parental H14IG#E3 ESCs and iNERT ESCs (without and with DOX/4HT induction). (G) Percentages of Pou3f1+ Lhx3- (PN-like, red), Pou3f1- Lhx3-- (HMC-like, light blue), Pou3f1- Lhx3+ (dark blue) and Pou3f1+ Lhx3+ (purple) cells among ESC-derived GFP+ MNs in day 6 EBs. The EBs were derived from ESC clones H14IG#E3 and iNERT without DOX/4HT induction. (H-M) GFP, Pou3f1 and Isl1/2 labelling in day 6 EBs. EBs were either differentiated according to the standard protocol (H,I) or treated the γ-secretase inhibitors LY411575 (J,K) or DAPT (L,M) from day 2 onwards. (N) Percentages of Pou3f1+ MNs among all GFP+ MNs in the absence or presence of γ-secretase inhibitor. Error bars indicate mean values from three independent experiments ± s.e.m. (G,N). *P<0.05 (paired Student’s t-test). Scale bars: 50 μm.
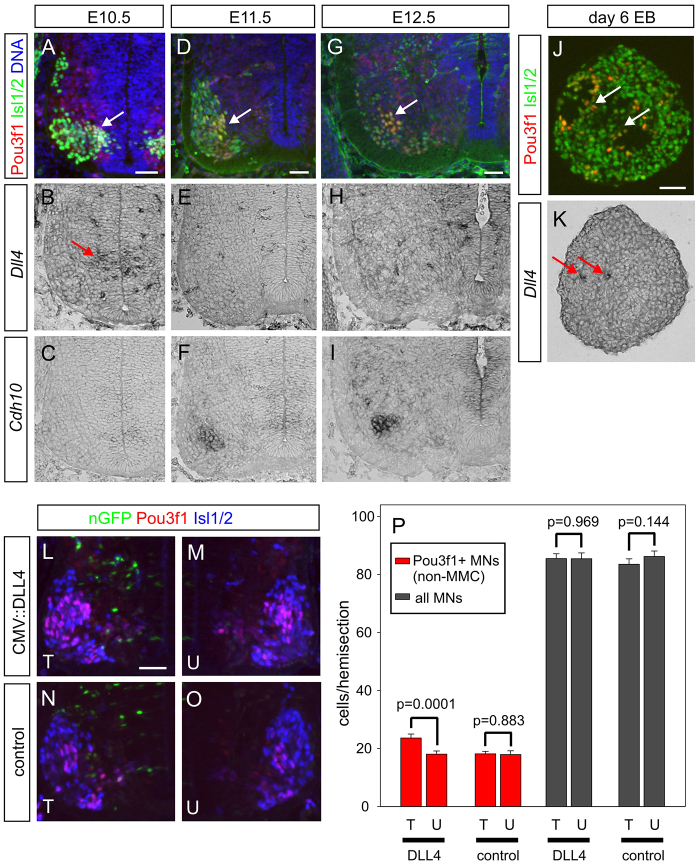
Given that Notch activity controls Pou3f1 in MNs in vitro, does the presence of a Notch ligand correlate with the appearance of Pou3f1+ MNs in vivo? Analysis of mid-cervical ventral spinal cord revealed that at E10.5, which is when MNs emerge from the pMN domain, a cluster of Dll4-positive cells is located just dorsal of nascent Pou3f1+ Isl1/2+ MNs (Fig. 6A,B). Previous studies have mapped Dll4 expression to the p2 progenitor domain (Del Barrio et al., 2007; Peng et al., 2007). Furthermore, Notch1 protein is detectable on the most immature, medial MNs, and Pou3f1 expression in MNs overlaps with that of the Notch target gene Hey1 (supplementary material Fig. S11). At E11.5, Pou3f1+ MNs have migrated laterally to settle in the PN and the MMC (Fig. 6D), a pattern that remains unchanged at E12.5 (Fig. 6G). Dll4 expression, by contrast, is transient and not seen at later time points (Fig. 6E,H), except in blood vessels. The expression of the Pou3f1 target gene Cdh10 is delayed compared with Pou3f1 itself and is first detected in the PN at E11.5 (Fig. 6C,F,I). The juxtaposition of Pou3f1 and Dll4 in vivo is mirrored by the pattern that we see in EBs, where patches of Pou3f1+ Isl1/2+ MNs are localised close to Dll4+ non-MNs (Fig. 6J,K).
Fig. 6.
The Notch ligand Dll4 is expressed adjacent to nascent Pou3f1+ MNs and induces Pou3f1 in primary embryonic MNs. (A,D,G) Expression of Pou3f1 and the pan-MN marker Isl1/2 in E10.5, E11.5 and E12.5 mid-cervical spinal cord. Phrenic neurons are labelled by both markers (arrow). (B,E,H) Red arrow indicates Dll4 expression at E10.5. At E11.5 and E12.5, few Dll4+ cells are visible, apart from blood vessels. (C,F,I) Cdh10, a Pou3f1 target gene, labels phrenic neurons at E11.5 and E12.5. (J) Day 6 EBs stained for Isl1/2 and Pou3f1; arrows indicate double-positive cells. (K) Expression of Dll4 in small patches of day 6 EBs (arrows). The sections in J and K are adjacent. (L-O) Mid-cervical spinal cords of mouse embryos transfected with expression vectors for human DLL4 and nGFP (L,M) or mock and nGFP (N,O) at ∼E9.75 and then allowed to develop in WEC for 40 hours. Sections were stained for nGFP, Isl1/2 and Pou3f1. (P) Analysis of mean numbers of Pou3f1+ Isl1/2+ MNs outside the MMC (red bars) and all Isl1/2+ MNs (black bars) in mid-cervical spinal cord hemi-sections of transfected embryos. Error bars indicate mean values from four (DLL4) or three (control) embryos ± s.e.m. (25 pairs of images each). P-values are by paired Student’s t-test. Similar significances were obtained using the Mann-Whitney test and logistic regression. T, transfected side; U, untransfected side. Scale bars: 50 μm.
To examine whether Dll4 protein is capable of inducing Pou3f1 expression in primary MNs, we used whole embryo culture (WEC) (Osumi and Inoue, 2001) to ectopically express this Notch ligand in cervical spinal cords of intact mouse embryos. Owing to the timing of the WEC cultures, many non-MNs in close proximity to Isl1/2+ MNs and ventral progenitors express the co-transfected reporter nGFP (Fig. 6L,N), whereas MNs themselves are mostly negative. Hence, any effect seen in MNs is likely to be non-cell-autonomous. Embryos developed normally in culture and formed a Pou3f1+ PN (Fig. 6L-O). In DLL4-transfected embryos (Fig. 6L,M), but not in controls (Fig. 6N,O), the transfected side of the spinal cord contains significantly more MNs of phrenic phenotype (Fig. 6P). At the same time, the total number of MNs is unchanged (Fig. 6P), suggesting that ectopic expression of DLL4 does not expand MN progenitors. Taken together, these findings show that Pou3f1 induction in embryonic MNs is controlled by Notch signalling both in ESC-MNs and in vivo, and suggest that Delta-Notch interaction is involved in the acquisition of phrenic neuron identity.
The PN-specific receptors Cdh10 and Pcdh10 mediate cell clustering of ESC-MNs
A key event in early MN patterning is the aggregation of MNs of the same subtype into clusters/nuclei within the ventral horn. This like-like recognition of cell bodies is mediated by receptors of the cadherin family. Expression profiling of primary MNs suggested that the cadherins Cdh10 and Pcdh10 are highly enriched in the PN (Fig. 2D,E). To examine whether these potential effector genes are sufficient to drive specific MN clustering, we ectopically expressed Cdh10 or Pcdh10 in ESC-MNs and mixed them with RFP-labelled control ESC-MNs (Fig. 7A-C). Near-neighbour analysis revealed that iCdh10 MNs and iPcdh10 MNs, but not RFP- control MNs, segregated from RFP+ control MNs in culture and formed nucleus-like aggregates (Fig. 7D-J).
Fig. 7.
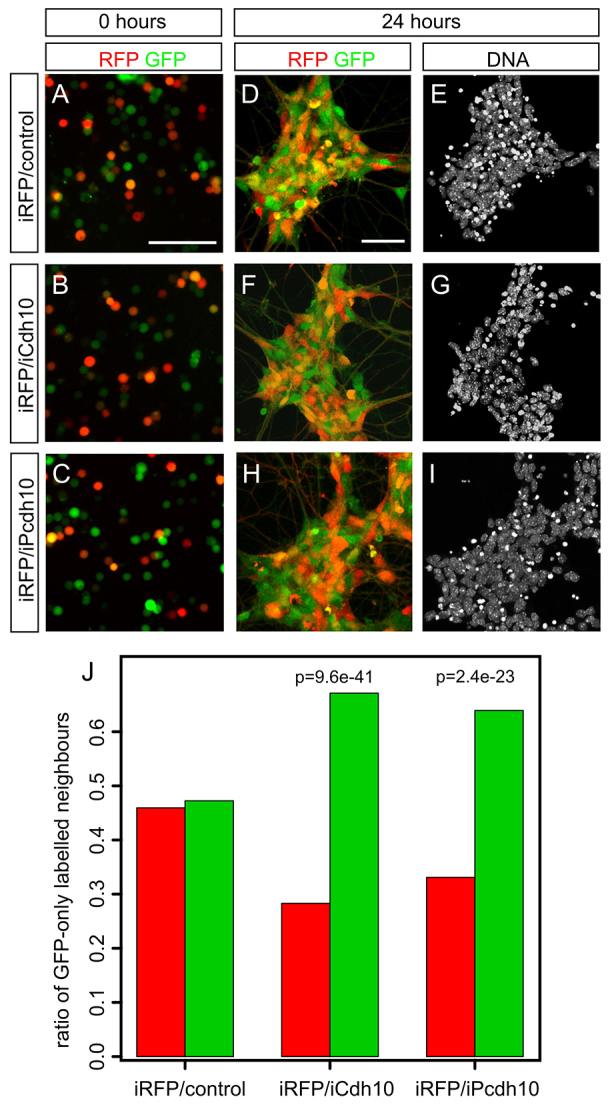
The PN-specific cadherins Cdh10 and Pcdh10 mediate specific clustering of ESC-MNs. (A-C) GFP-only and RFP+ GFP+ ESC-MNs are randomly mixed at 0 hours. (D-I) Mixed cultures of GFP-only and RFP+ GFP+ ESC-MNs aggregate within 24 hours. (J) Analysis of like-like clustering of ESC-MNs in mixed cultures (24 hours) by near-neighbour analysis. Bar colours indicate the label of the central cell: red, RFP+ GFP+ control MNs; green, GFP-only control, iCdh10 or iPcdh10 MNs. P-values were calculated based on the generalised linear model with random effects. The chart shows pooled data from three independent experiments. Eight to ten cell clusters per condition (2267 MNs total) were scored. Scale bars: 50 μm.
DISCUSSION
The development of phrenic neurons, which drive respiration in mammals, requires the integration of positional signals to restrict their specification to mid-cervical segments of the spinal cord, where they emerge in register with myoblasts destined to form their target muscle, the diaphragm (Babiuk et al., 2003). We performed expression profiling in primary embryonic neurons followed by the reconstruction of phrenic-like transcriptional patterns in transgenic ESC-MNs to identify factors that combine to establish PN identity along the dorsoventral and anteroposterior axes of the spinal cord. We found that three factors, Pou3f1, Hoxa5 and Notch, interact to control sets of putative PN-specific effector genes: Hoxa5 only evokes PN-like transcriptional patterns when combined with Pou3f1, and Notch signalling is upstream of Pou3f1. The latter interaction was observed both in ESC-MNs and in spinal cords of intact mouse embryos. Thus, our study suggests that Delta-Notch signalling at the dorsal margin of the nascent motor columns has to intersect with Hox5-dependent mid-cervical segmental identity for phrenic neurons to develop (Fig. 8). Given that NICD controls Pou3f1, why is there only a limited overlap between their target genes in ESC-MNs (supplementary material Fig. S8A)? The most likely explanation for this is the short time frame of iNICD induction (10 hours), which is sufficient to upregulate Pou3f1 itself (supplementary material Table S8) but not second-order Pou3f1 targets. Based on co-expression assays in ESC-MNs, we also predict that Lhx3 is a negative regulator of phrenic identity in Pou3f1+ MMC neurons. In addition to transcriptional determinants, we identified and validated several sets of putative PN effector genes downstream of these regulators of PN identity. Many of these genes are regulated in a modular fashion by single factors, whereas others require the combined activity of two determinants.
Fig. 8.
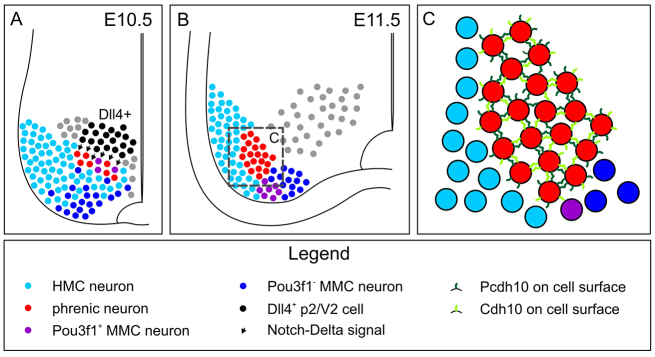
Model of Delta/Notch induction of phrenic neuron identity. (A) At E10.5, Dll4-positive p2 progenitors and V2 interneurons are located immediately dorsal of nascent MNs. MNs committed to MMC or HMC fate adjacent to Dll4+ territory fate are exposed to Notch signalling and upregulate the phrenic neuron determinant Pou3f1. Other Notch ligands expressed in the embryonic spinal cord might also contribute to this process. (B) Following their specification by Delta/Notch, Pou3f1+ phrenic neurons migrate laterally and settle into a medial position next to the HMC. (C) The Pou3f1 targets Cdh10 and Pcdh10 mediate like-like clustering of phrenic MNs and contribute to the formation of the PN.
The aggregation of MNs that connect to the same target muscle into nuclei and motor pools is a key event in early MN development and depends on MN subtype-specific expression of cadherins (Bello et al., 2012; Demireva et al., 2011; Price et al., 2002). This establishes a topographic map of MN cell bodies within the ventral horn and ensures that functionally related MNs receive the same sensory synaptic input (Sürmeli et al., 2011), as well as facilitating the formation of electrical synapses between MNs to coordinate activity (Personius et al., 2007). We found that the phrenic neuron-enriched receptors Cdh10, which is regulated by Pou3f1, and Pcdh10 (Hirano et al., 1999), which is regulated by Hoxa5 and Pou3f1, mediate like-like clustering of ESC-MN cell bodies in vitro (Fig. 7, Fig. 8C) and may contribute to the aggregation of phrenic neurons into the PN, probably in concert with other adhesion molecules.
Our findings on PN specification in ESC-MNs suggest that the coordinated expression of cadherins and other effector genes in phrenic neurons depends on the combinatorial activity of at least three positive and one negative determinant. Nevertheless, our understanding of PN gene regulation remains incomplete. For example, we do not know how the PN-enriched receptor genes Alcam and Plxnc1 are regulated. There are a number of possible explanations for the limitations to our approach: (1) additional, unknown transcriptional determinants might positively regulate additional sets of PN-selective genes; (2) ESC-MNs might contain more negative regulators, which mask aspects of induced PN identity; (3) peripheral cues encountered by outgrowing phrenic axons might control some PN target genes, and these cues would not be present in vitro; (4) some of the PN-enriched genes that are not yet confirmed by histology could be false positives, in particular those in the least stringent [0 +1 0] group (supplementary material Table S4).
The two cervical Hox genes Hoxa5 and Hoxc6 evoked strikingly similar transcriptional patterns when overexpressed in ESC-MNs as single transgenes (supplementary material Fig. S8B) or combined with Pou3f1 (supplementary material Table S7), consistent with the fact that different cervical Hox proteins can bind to the same consensus DNA site (Pellerin et al., 1994). Co-expression of iPou3f1 plus iHoxa5 revealed combinatorial effects, and the overall PN specificity of the pattern is higher than that of iPou3f1 plus iHoxc6. Crucially, iHoxc6 and iPou3f1 plus iHoxc6, but not iHoxa5 and iPou3f1 plus iHoxa5, induce the TF Foxp1. This key LMC determinant, which largely depends on Hox6 expression in vivo (Dasen et al., 2008; Lacombe et al., 2013), suppresses PN identity when ectopically expressed in cervical MNs (Rousso et al., 2008). Thus, the combination of iPou3f1 plus iHoxc6 induces an LMC-type programme that is similar to that of the PN, yet at the same time is incompatible with PN identity. An in vivo correlate to the Foxp1-positive iPou3f1/iHoxc6 ESC-MNs indeed exists: ulnar motor pools, which are caudal LMC subpopulations that innervate distal limb muscles in mice (Lacombe et al., 2013), and anatomically related flexor carpi ulnaris MNs in chick (Dasen et al., 2005) co-express Pou3f1 and Hoxc6 during its initial specification. Despite their different identities, aspects of the subtype-specific genetic programmes are shared between the PN and ulnar MNs, as both populations selectively express the Pou3f1 target genes Cdh9 and Cdh10 (Fig. 2B-D; supplementary material Fig. S12).
MN subtype identity has been linked to Notch signalling: in Gde2 (Gdpd5) null mutant mice, which have increased levels of activated Notch, MNs show a marked shift in columnar subtype composition (Sabharwal et al., 2011). Intriguingly, in their study, Notch affects MN fate at the level of progenitor cells. We first observe Pou3f1 upregulation in Isl1/2+ postmitotic MNs (Fig. 6A). Nevertheless, we do not know whether Notch induction of this key PN determinant occurs before or after cell cycle exit. Delta-Notch interaction as a mechanism to diversify V2a/V2b spinal interneurons appears to be conserved between fish, birds and mammals (Batista et al., 2008; Del Barrio et al., 2007; Peng et al., 2007). There are two scenarios of how the ancestral Dll4+ territory in the p2 progenitor domain, which drives this process, might relate to the emergence of the PN during mammalian evolution. In the first scenario, Dll4 expression might have acquired a novel role in establishing the mammal-specific PN. This would imply that phrenic neurons arose from cervical HMC neurons, which were respecified by Delta-Notch signalling, as the diaphragm evolved from cervical hypaxial muscle (Perry et al., 2010). Alternatively, the PN might be an evolutionary modification of an ancestral LMC subset that predates mammals. Hirasawa and Kuratani recently proposed that the diaphragm originated from limb muscle associated with the pectoral girdle (Hirasawa and Kuratani, 2013). If this scenario is correct, then phrenic neurons might have evolved from a Pou3f1+ LMC subset that lost its brachial Hox code due to a duplication of two cervical segments coupled with a caudal shift in brachial identity, an event that might have occurred during early mammalian phylogeny (Hirasawa and Kuratani, 2013). This view is supported by the fact that, in mammals, there are two Pou3f1+ MN subsets spaced by about two segments at cervical levels, one rostral of the LMC (the PN) and one caudal within the LMC (ulnar MNs).
In this study, we have demonstrated that MN derivation from ESCs is a viable screening tool to systematically dissect developmental pathways. Our experimental strategy was based on initially identifying candidate determinants in sorted primary embryonic neurons by transcriptional profiling and then validating the factors in the ESC-derived equivalent of these ex vivo cells. Both a highly reductionist approach (genome-wide expression analysis in ESC-MNs) and the more complex, heterogeneous EB culture system provided insights into how a network of TFs interact to establish PN identity. Similar ESC-based screens could be undertaken for other cell types that are difficult to access in large numbers. We think that the derivation of MNs from pluripotent stem cells will mature into a powerful tool to investigate not only prenatal developmental programmes, but also subtype-dependent features of adult MN function and connectivity, complementing existing animal models. Our long-term aim is to assemble adult-like phrenic ESC-MNs, as well as other defined cell types, into artificial neuromuscular circuits to simulate aspects of respiratory motor function in health and disease in vitro. The progress towards understanding embryonic phrenic neuron identity reported here will allow us to extend the investigation to later steps of neuronal maturation and circuit formation.
MATERIALS AND METHODS
Summary
Embryonic phrenic neurons were labelled by injecting a retrograde tracer (TMR-dextran) into the phrenic nerve in E11.5 trunk explants derived from mouse embryos carrying the Hb9::GFP transgene (Wichterle et al., 2002). Explants were cultured in an oxygenated bath for 2 hours and then dissociated with papain, and TMR+ GFP+ phrenic neurons, as well as GFP+ control MNs, isolated by flow cytometry (Fig. 1A-C). In separate experiments, radial LMC neurons were retrogradely labelled and isolated using the same methodology. Gene expression profiles of sorted primary MNs were determined by Affymetrix array analysis.
MNs were directly differentiated in vitro from mouse ESCs as described (Peljto et al., 2010). Briefly, A2.lox ESCs (Iacovino et al., 2011) carrying the MN-specific, magnetically sortable reporter genes Hb9::CD2GFP or Hb9::CD14-IRES-GFP (supplementary material Fig. S1) were grown for 2 days as EBs, induced with 1 μM retinoic acid and 0.5 μM smoothened agonist (SAG), and dissociated on day 5. MNs were then isolated by magnetically activated cell sorting (MACS). MACS-purified ESC-MNs were cultured for 30 hours on Matrigel, and the expression of PN-specific candidate genes (supplementary material Tables S1, S2) was induced with doxycycline (DOX). Then, transcriptional profiles of ESC-MNs were determined by Affymetrix array analysis and qRT-PCR (Spandidos et al., 2010) (supplementary material Table S3). ESC-MNs expressing the neutral transgene YFP were used as the baseline, as the DOX-inducible Cre recombinase present in the parental ESC line triggers a DNA damage response in MNs (supplementary material Fig. S2).
Detailed protocols are available in supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank Tom Jessell and Andrew Lumsden for support and encouragement; Noriko Osumi and Yoko Arai for advice on WECs; Ira Schieren, Kaity Miao and Fazal Oozeer for expert technical assistance; Jeremy Dasen, Fiona Watt, Robert Knight, Linda Greensmith, Christopher E. Henderson, QueeLim Ch’ng, Stephen Price and Anthony Graham for comments on the manuscript; Chris G. Tan for sharing unpublished observations; and Austin Smith, Esteban O. Mazzoni, Hynek Wichterle, Koichi Kawakami, Cedric Blainpain, Dafe A. Uwanogho, Shinji Hirano, Ursula Just, Malcolm Logan, Linda Greensmith, Matthew Grist, Uwe Drescher and Simon J. Rhodes for providing reagents.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
C.B.M., I.L., M.N., K.C.K. and P.K. generated DNA constructs. C.B.M., I.L., P.K., D.S., M.C. and M.N. established and characterised ESC subclones. C.B.M. and I.L. derived MNs from ESCs, MACS sorted them and analysed them by flow cytometry and microscopy. M.I. and M.K. contributed the A2lox.Cre ESC clone prior to its publication. I.L. isolated primary MN populations from embryos and D.C. determined their transcriptional patterns. K.C.K. contributed to the original concept of the study. E.B. developed the strategy for large-scale comparison of transcriptional patterns and performed statistical analysis. C.B.M., E.B. and I.L. wrote the manuscript and assembled the figures. I.L. developed the original concept, designed and oversaw the study.
Funding
Personnel and work were supported by the Medical Research Council [grant G0900585, I.L.]. Additional funding was provided by the Thierry Latran Foundation (I.L.); the Biotechnology and Biological Sciences Research Council [grant G1001234, I.L. and Britta Eickholt, KCL/Charité, Berlin, Germany]; Project ALS (K.C.K.) and the Tow Foundation (K.C.K.). K.C.K. was the recipient of a Kirschstein-NRSA Fellowship from NINDS, National Institutes of Health. Deposited in PMC for immediate release.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.097188/-/DC1
References
- Agalliu D., Takada S., Agalliu I., McMahon A. P., Jessell T. M. (2009). Motor neurons with axial muscle projections specified by Wnt4/5 signaling. Neuron 61, 708–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan D. W., Greer J. J. (1997). Embryogenesis of the phrenic nerve and diaphragm in the fetal rat. J. Comp. Neurol. 382, 459–468 [DOI] [PubMed] [Google Scholar]
- Arber S., Han B., Mendelsohn M., Smith M., Jessell T. M., Sockanathan S. (1999). Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron 23, 659–674 [DOI] [PubMed] [Google Scholar]
- Babiuk R. P., Zhang W., Clugston R., Allan D. W., Greer J. J. (2003). Embryological origins and development of the rat diaphragm. J. Comp. Neurol. 455, 477–487 [DOI] [PubMed] [Google Scholar]
- Batista M. F., Jacobstein J., Lewis K. E. (2008). Zebrafish V2 cells develop into excitatory CiD and Notch signalling dependent inhibitory VeLD interneurons. Dev. Biol. 322, 263–275 [DOI] [PubMed] [Google Scholar]
- Bello S. M., Millo H., Rajebhosale M., Price S. R. (2012). Catenin-dependent cadherin function drives divisional segregation of spinal motor neurons. J. Neurosci. 32, 490–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham J. R., Jr, Scherer S. S., O’Connell S., Arroyo E., Kalla K. A., Powell F. L., Rosenfeld M. G. (1996). Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 10, 1751–1762 [DOI] [PubMed] [Google Scholar]
- Bondue A., Tännler S., Chiapparo G., Chabab S., Ramialison M., Paulissen C., Beck B., Harvey R., Blanpain C. (2011). Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J. Cell Biol. 192, 751–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., Pierani A., Jessell T. M., Ericson J. (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–445 [DOI] [PubMed] [Google Scholar]
- Champagnat J., Morin-Surun M. P., Fortin G., Thoby-Brisson M. (2009). Developmental basis of the rostro-caudal organization of the brainstem respiratory rhythm generator. Philos. Trans. R. Soc. B 364, 2469–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen J. S., Liu J. P., Jessell T. M. (2003). Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature 425, 926–933 [DOI] [PubMed] [Google Scholar]
- Dasen J. S., Tice B. C., Brenner-Morton S., Jessell T. M. (2005). A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 123, 477–491 [DOI] [PubMed] [Google Scholar]
- Dasen J. S., De Camilli A., Wang B., Tucker P. W., Jessell T. M. (2008). Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell 134, 304–316 [DOI] [PubMed] [Google Scholar]
- De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., Mumm J. S., Schroeter E. H., Schrijvers V., Wolfe M. S., Ray W. J., et al. (1999). A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 [DOI] [PubMed] [Google Scholar]
- Del Barrio M. G., Taveira-Marques R., Muroyama Y., Yuk D. I., Li S., Wines-Samuelson M., Shen J., Smith H. K., Xiang M., Rowitch D., et al. (2007). A regulatory network involving Foxn4, Mash1 and delta-like 4/Notch1 generates V2a and V2b spinal interneurons from a common progenitor pool. Development 134, 3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demireva E. Y., Shapiro L. S., Jessell T. M., Zampieri N. (2011). Motor neuron position and topographic order imposed by β- and γ-catenin activities. Cell 147, 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias T. B., Yang Y. J., Ogai K., Becker T., Becker C. G. (2012). Notch signaling controls generation of motor neurons in the lesioned spinal cord of adult zebrafish. J. Neurosci. 32, 3245–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin G., Thoby-Brisson M. (2009). Embryonic emergence of the respiratory rhythm generator. Respir. Physiol. Neurobiol. 168, 86–91 [DOI] [PubMed] [Google Scholar]
- Hirano S., Yan Q., Suzuki S. T. (1999). Expression of a novel protocadherin, OL-protocadherin, in a subset of functional systems of the developing mouse brain. J. Neurosci. 19, 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa T., Kuratani S. (2013). A new scenario of the evolutionary derivation of the mammalian diaphragm from shoulder muscles. J. Anat. 222, 504–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J. R., Skryabin B. V., Belcher S. M., Zerillo C. A., Schmauss C. (1995). The responsiveness of a tetracycline-sensitive expression system differs in different cell lines. J. Biol. Chem. 270, 14168–14174 [DOI] [PubMed] [Google Scholar]
- Iacovino M., Bosnakovski D., Fey H., Rux D., Bajwa G., Mahen E., Mitanoska A., Xu Z., Kyba M. (2011). Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells 29, 1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Lacombe J., Mazzoni E. O., Liem K. F., Jr, Grinstein J., Mahony S., Mukhopadhyay D., Gifford D. K., Young R. A., Anderson K. V., et al. (2010). Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron 67, 781–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe J., Hanley O., Jung H., Philippidou P., Surmeli G., Grinstein J., Dasen J. S. (2013). Genetic and functional modularity of Hox activities in the specification of limb-innervating motor neurons. PLoS Genet. 9, e1003184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lance-Jones C., Landmesser L. (1980). Motoneurone projection patterns in the chick hind limb following early partial reversals of the spinal cord. J. Physiol. 302, 581–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. J., Hu B. Y., Jones S. A., Zhang Y. S., Lavaute T., Du Z. W., Zhang S. C. (2008). Directed differentiation of ventral spinal progenitors and motor neurons from human embryonic stem cells by small molecules. Stem Cells 26, 886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. P., Laufer E., Jessell T. M. (2001). Assigning the positional identity of spinal motor neurons: rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and retinoids. Neuron 32, 997–1012 [DOI] [PubMed] [Google Scholar]
- Marshall H., Nonchev S., Sham M. H., Muchamore I., Lumsden A., Krumlauf R. (1992). Retinoic acid alters hindbrain Hox code and induces transformation of rhombomeres 2/3 into a 4/5 identity. Nature 360, 737–741 [DOI] [PubMed] [Google Scholar]
- Miles G. B., Yohn D. C., Wichterle H., Jessell T. M., Rafuse V. F., Brownstone R. M. (2004). Functional properties of motoneurons derived from mouse embryonic stem cells. J. Neurosci. 24, 7848–7858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi N., Inoue T. (2001). Gene transfer into cultured mammalian embryos by electroporation. Methods 24, 35–42 [DOI] [PubMed] [Google Scholar]
- Peljto M., Wichterle H. (2011). Programming embryonic stem cells to neuronal subtypes. Curr. Opin. Neurobiol. 21, 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peljto M., Dasen J. S., Mazzoni E. O., Jessell T. M., Wichterle H. (2010). Functional diversity of ESC-derived motor neuron subtypes revealed through intraspinal transplantation. Cell Stem Cell 7, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin I., Schnabel C., Catron K.M., Abate C. (1994). Hox proteins have different affinities for a consensus DNA site that correlate with the positions of their genes on the hox cluster. Mol. Cell. Biol. 14, 4532–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C. Y., Yajima H., Burns C. E., Zon L. I., Sisodia S. S., Pfaff S. L., Sharma K. (2007). Notch and MAML signaling drives Scl-dependent interneuron diversity in the spinal cord. Neuron 53, 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S. F., Similowski T., Klein W., Codd J. R. (2010). The evolutionary origin of the mammalian diaphragm. Respir. Physiol. Neurobiol. 171, 1–16 [DOI] [PubMed] [Google Scholar]
- Personius K. E., Chang Q., Mentis G. Z., O’Donovan M. J., Balice-Gordon R. J. (2007). Reduced gap junctional coupling leads to uncorrelated motor neuron firing and precocious neuromuscular synapse elimination. Proc. Natl. Acad. Sci. USA 104, 11808–11813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P., Walsh C. M., Aubin J., Jeannotte L., Dasen J. S. (2012). Sustained Hox5 gene activity is required for respiratory motor neuron development. Nat. Neurosci. 15, 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price S. R., De Marco Garcia N. V., Ranscht B., Jessell T. M. (2002). Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell 109, 205–216 [DOI] [PubMed] [Google Scholar]
- Rousso D. L., Gaber Z. B., Wellik D., Morrisey E. E., Novitch B. G. (2008). Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron 59, 226–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabharwal P., Lee C., Park S., Rao M., Sockanathan S. (2011). GDE2 regulates subtype-specific motor neuron generation through inhibition of Notch signaling. Neuron 71, 1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T., Kohlhof H., Rieber N., Just U. (2003). Notch signaling induces multilineage myeloid differentiation and up-regulates PU.1 expression. J. Immunol. 170, 5538–5548 [DOI] [PubMed] [Google Scholar]
- Sharma K., Sheng H. Z., Lettieri K., Li H., Karavanov A., Potter S., Westphal H., Pfaff S. L. (1998). LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell 95, 817–828 [DOI] [PubMed] [Google Scholar]
- Sockanathan S., Jessell T. M. (1998). Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell 94, 503–514 [DOI] [PubMed] [Google Scholar]
- Soundararajan P., Miles G. B., Rubin L. L., Brownstone R. M., Rafuse V. F. (2006). Motoneurons derived from embryonic stem cells express transcription factors and develop phenotypes characteristic of medial motor column neurons. J. Neurosci. 26, 3256–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A., Wang X., Wang H., Seed B. (2010). PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38, D792–D799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sürmeli G., Akay T., Ippolito G. C., Tucker P. W., Jessell T. M. (2011). Patterns of spinal sensory-motor connectivity prescribed by a dorsoventral positional template. Cell 147, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler J., Harrison K., Sharma K., Lettieri K., Kehrl J., Pfaff S. L. (1999). Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron 23, 675–687 [DOI] [PubMed] [Google Scholar]
- Tsuchida T., Ensini M., Morton S. B., Baldassare M., Edlund T., Jessell T. M., Pfaff S. L. (1994). Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell 79, 957–970 [DOI] [PubMed] [Google Scholar]
- West B. E., Parker G. E., Savage J. J., Kiratipranon P., Toomey K. S., Beach L. R., Colvin S. C., Sloop K. W., Rhodes S. J. (2004). Regulation of the follicle-stimulating hormone beta gene by the LHX3 LIM-homeodomain transcription factor. Endocrinology 145, 4866–4879 [DOI] [PubMed] [Google Scholar]
- Wichterle H., Lieberam I., Porter J. A., Jessell T. M. (2002). Directed differentiation of embryonic stem cells into motor neurons. Cell 110, 385–397 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.