Abstract
Efg1 (a member of the APSES family) is an important regulator of hyphal growth and of the white-to-opaque transition in Candida albicans and very closely related species. We show that in Candida parapsilosis Efg1 is a major regulator of a different morphological switch at the colony level, from a concentric to smooth morphology. The rate of switching is at least 20-fold increased in an efg1 knockout relative to wild type. Efg1 deletion strains also have reduced biofilm formation, attenuated virulence in an insect model, and increased sensitivity to SDS and caspofungin. Biofilm reduction is more dramatic in in vitro than in in vivo models. An Efg1 paralogue (Efh1) is restricted to Candida species, and does not regulate concentric-smooth phenotype switching, biofilm formation or stress response. We used ChIP-seq to identify the Efg1 regulon. A total of 931 promoter regions bound by Efg1 are highly enriched for transcription factors and regulatory proteins. Efg1 also binds to its own promoter, and negatively regulates its expression. Efg1 targets are enriched in binding sites for 93 additional transcription factors, including Ndt80. Our analysis suggests that Efg1 has an ancient role as regulator of development in fungi, and is central to several regulatory networks.
Introduction
The transcription factor Efg1 plays a central role in regulating morphology and virulence in the major human fungal pathogen Candida albicans. It was first identified as an inducer of pseudohyphal growth in Saccharomyces cerevisiae, and subsequently shown to be required for hyphal growth in C. albicans (Stoldt et al., 1997). Strains carrying deletions of EFG1 fail to undergo a switch from yeast to hyphal growth in response to most stimuli (Lo et al., 1997; Stoldt et al., 1997; Sudbery, 2011) although filamentation is increased under hypoxic conditions, and when embedded in solid media (Doedt et al., 2004; Mulhern et al., 2006).
Efg1 has many functions in C. albicans. It is required for virulence in most infection models (Lo et al., 1997; Chamilos et al., 2006; Pukkila-Worley et al., 2009). In addition, cells lacking EFG1 are unable to invade reconstituted human epithelia, and endocytosis in epithelial and endothelial cells is reduced (Weide and Ernst, 1999; Dieterich et al., 2002).
The pleiotropic effects of Efg1 in C. albicans are related to its pivotal role in several networks controlling morphology and virulence. Efg1 is one of the core regulators of the white-opaque switch, an epigenetic switch between two genetically identical cell types. Efg1 is required for the maintenance of the default white phenotype (Sonneborn et al., 1999b; Srikantha et al., 2000). Switching to opaque cells is strongly correlated with mating efficiency (reviewed in Bennett and Johnson, 2005; Lohse and Johnson, 2009; Morschhauser, 2010). White and opaque cells differ in appearance; white cells are round or ellipsoid and opaque cells are elongated with ‘pimples’ on the surface (Anderson and Soll, 1987; Anderson et al., 1990). They also differ in behaviour; for example, white cells are preferentially phagocytosed by macrophages (Lohse and Johnson, 2008), and opaque cells do not release a chemoattractant for leukocytes (Geiger et al., 2004). The two cells types have significantly different transcriptional profiles (Tuch et al., 2010). The white-opaque switch is regulated by a network of transcription factors that includes Efg1, Wor1, Wor2 and Czf1, together with the homeodomain mating proteins a1 and alpha2 (Sonneborn et al., 1999b; Srikantha et al., 2000; Huang et al., 2006; Vinces et al., 2006; Zordan et al., 2006; 2007,).
Efg1 is also important for the growth of C. albicans cells in communities, or biofilms, on plastic or cell surfaces. Cells with an efg1 deletion form very loose biofilms on plastic consisting mostly of yeast cells (Ramage et al., 2002), and form defective biofilms on vaginal mucosa (Harriott et al., 2010). Unlike many other biofilm regulators, Efg1 is required for biofilm development under both normoxic and hypoxic (low-oxygen) conditions (Stichternoth and Ernst, 2009). Even when cells do adhere to plastic, Efg1 is required for tolerance to antifungal drugs such as azoles, amphotericin B and caspofungin (Bink et al., 2012).
A recent analysis by Nobile et al. (2012) showed that Efg1 is part of a network of six transcription factors that regulates biofilm development in C. albicans. The six regulators all control each other's expression; up to five factors bind to each promoter, and in addition each activates its own synthesis. The network regulates expression of at least 1000 genes, many of which are also expressed during biofilm growth. Nobile et al. (2012) identified several hundred intergenic regions bound by Efg1, and separately, Lassak et al. (2011) showed that Efg1 binds to the promoters of at least 53 genes in C. albicans yeast cells, including several transcription factors. Binding of Efg1 is disrupted and new binding sites appear during the yeast-to-hyphal transition. The analysis of Nobile et al. (2012) suggests that the biofilm regulatory network has evolved relatively recently, and is likely to be restricted to C. albicans.
Among the species in the Candida clade [or CTG clade, species in which CTG is translated as serine rather than leucine (Santos and Tuite, 1995)], only C. albicans and C. dubliniensis form hyphae, and white-opaque switching appears to be restricted to these species plus their close relative Candida tropicalis (Lohse and Johnson, 2009; Morschhauser, 2010; Porman et al., 2011; Xie et al., 2012). However, Efg1 is a member of the APSES family of proteins that contain a conserved basic helix–loop–helix domain (Stoldt et al., 1997). Other members of the family are involved in repression of pseudohyphal growth in S. cerevisiae (Ward et al., 1995), conidiation in Aspergillus nidulans and Penicillium marneffii (Aramayo et al., 1996; Borneman et al., 2002) and the formation of aerial hyphae and conidiation in Glomerella cingulata (Tong et al., 2007) and Wangiella dermatitidis (Wang and Szaniszlo, 2007).
We describe here the role of Efg1 in Candida parapsilosis, a major human fungal pathogen that is a member of the CTG clade and is particularly associated with infection of premature neonates (Trofa et al., 2008; van Asbeck et al., 2009). C. parapsilosis grows in yeast and pseudohyphal forms but not as true hyphae, and although variations in colony morphology have been observed (Lott et al., 1993; Enger et al., 2001; Laffey and Butler, 2005; Kim et al., 2006), no equivalent of white-opaque switching has been identified. The C. parapsilosis species group also appears to be completely asexual, and do not exhibit the parasexual cycle observed in C. albicans, C. dubliniensis and C. tropicalis (Bennett and Johnson, 2005; Butler, 2010; Porman et al., 2011; Sai et al., 2011). We show that Efg1 represses a morphological switch from concentric to smooth colony formation, and is also a repressor of filamentation in hypoxic conditions. We use a combination of RNA-seq and ChIP-seq to identify Efg1 targets. Efg1 directly regulates a significant number of transcription factors, particularly those with long promoter sequences. Targets of Efg1 are also enriched in binding sites of other transcription factors. In addition, we show that Efg1 is required for biofilm development, and for virulence.
Results
Deleting EFG1 in C. parapsilosis
The EFG1 orthologue in C. parapsilosis was identified using sequence comparison and synteny conservation (Fitzpatrick et al., 2010). Both alleles in the diploid were deleted using three different approaches. First, the alleles were replaced with a recyclable nourseothricin-resistant marker as described in Ding and Butler (2007). Second, each allele was replaced with either LEU2 (from Candida maltosa) or HIS1 (from C. dubliniensis) in an auxotrophic background strain. The auxotrophic markers were amplified using either a long oligonucleotide (approach 2) or by fusion PCR (approach 3), using methods similar to those described by Noble and Johnson (2005) (Fig. S1). EFG1 was re-introduced into one deletion strain under the control of the ACT1 promoter as shown in Fig. S1D. All phenotypes were tested using knockouts from each approach to ensure that they result from the loss of the EFG1 gene and not to a secondary effect.
The most obvious result of deleting EFG1 is that it leads to a dramatic increase in the level of morphological switching observed at the colony level (Fig. 1A). Almost all colonies of the C. parapsilosis type strain (CLIB214) are a ‘wrinkled’ or ‘concentric’ morphology (composed of a series of concentric rings). Switching to other morphologies occurs at a very low level (Lott et al., 1993; Enger et al., 2001; Kim et al., 2006). However, the efg1 deletion consists of a mixture of both concentric and smooth colonies (Fig. 1A). When grown in culture, concentric colonies give rise to > 10% of smooth colonies and smooth colonies to > 12% concentric colonies. The switching rate varies in the different knockouts (Table S1) but is always significantly higher than the wild type, which has a switching rate of approximately 0.2% (concentric to smooth) and 2% (smooth to concentric). Two of the efg1 knockouts (generated using the fusion PCR approach) switch from concentric to smooth colonies at a very high rate (65–81%). None of strains deleted for just one EFG1 allele exhibited an increased switching rate (not shown). We reintroduced EFG1 expressed from the ACT1 promoter into one of the strains with the highest switching rate (Fig. S1). EFG1 expression from this construct is approximately the same level as in a heterozygous EFG1/efg1 deletion strain (data not shown), and switching is returned to wild-type levels (Fig. 1A, Table S1).
Figure 1.
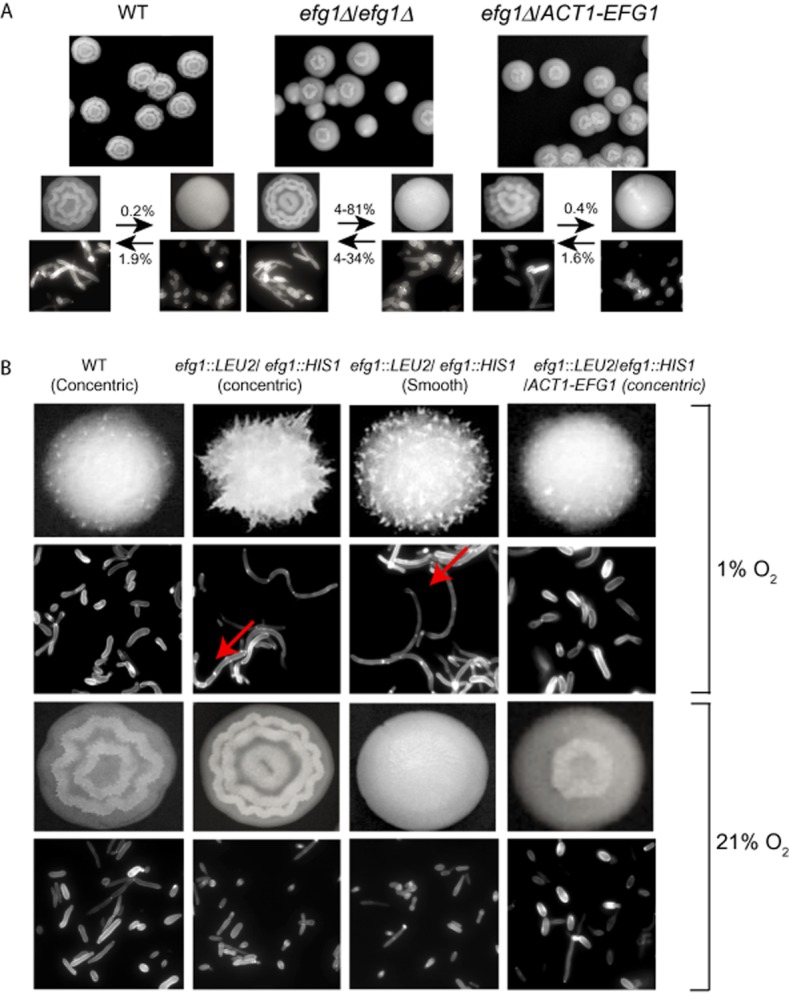
Efg1 regulates colony switching in C. parapsilosis. Both alleles of the EFG1 gene were deleted as described in Fig. S1.A. Wild-type (CLIB214) cell cultures consist of mostly concentric colonies, whereas efg1Δ/ efg1Δ (LCP7 is shown here) have a mixture of concentric and smooth colonies. The rate of switching between concentric and smooth cells varies, but can be as high as 81%, with a substantial number of sectored colonies (Table S1). Re-introducing EFG1 reduces the switching rate (LCP7_RI). Cells from concentric colonies tend to be elongated, whereas those from smooth colonies are more round and yeast-like.B. Wild-type colonies are generally smooth in hypoxic conditions (1% oxygen), and do not form the concentric phenotype obvious in normoxia (21% oxygen). Deleting both alleles of EFG1 (LCP7 is shown here) results in rough and spiked colonies in hypoxic conditions, with an increase in elongated and pseudohyphal cells (Table S2). The arrows indicate the cell wall between a mother cell and a pseudohyphal bud. The reconstituted strain is more similar to the wild-type phenotype. In normoxic conditions, few pseudohyphae are observed. The top panels show colonies and the bottom panels show corresponding calcofluor white-stained cells for growth at 1% and 21% oxygen.
Individual cells of C. parapsilosis (particularly those derived from the type strain CLIB214) are more elongated than C. albicans or S. cerevisiae yeast cells (Fig. 1A and B). However, we found that cells from the smooth colonies tend to be more round and yeast like when grown in normal atmospheric conditions (Fig. 1, Table S2). Because EFG1 is reported to be a repressor of filamentation in hypoxia in C. albicans, we also examined the morphology of the C. parapsilosis efg1 knockouts in low-oxygen conditions. As shown in Fig. 1B, growth in hypoxia induces a novel ‘spiked’ morphology in colonies that is not observed in the wild type, irrespective of the concentric/smooth morphology, independent of the level of oxygen available. However, when EFG1 is deleted, there is a significant increase in the number of pseudohyphal cells in hypoxic conditions (Fig. 1B, Table S2). Long chains of cells are very evident. The structures are reminiscent of hyphae, except that there is a cell wall between the mother (yeast) cell and the first pseudohyphal bud (indicated with arrows in Fig. 1B). Cells grown in liquid culture have the same phenotype (not shown). The phenotype of colonies of the efg1/ACT1-EFG1 complemented strain is more similar to the wild type, and the percentage of pseudohyphal cells is between that of the wild-type and the deletion strain (Table S2).
Deleting EFG1 affects biofilm development
Candida parapsilosis grows as biofilms on indwelling medical devices (Hawser and Douglas, 1994; Silva et al., 2009; Melo et al., 2011). The biofilm architecture is substantially different from that observed for C. albicans, although some of the same regulators are required. For example, the transcription factor Bcr1 is required for biofilm development in both species (Nobile et al., 2006; Ding and Butler, 2007; Finkel and Mitchell, 2011). However, the mechanism and targets are likely to be different; the CFEM family members are targets of Bcr1 in both, but have a role in biofilm development only in C. albicans (Ding and Butler, 2007; Ding et al., 2011).
Efg1 is required for biofilm formation by C. albicans (Ramage et al., 2002; Nobile et al., 2012). We therefore tested its role in C. parapsilosis. Deleting EFG1 in C. parapsilosis reduces biofilm development irrespective of colony morphology (Fig. 2). Both smooth and concentric colonies carrying an efg1 deletion have reduced biofilm formation on the surface of 24-well Nunc plates compared with wild type, with smooth cells having a slightly more pronounced defect. The phenotype is partially restored by re-introducing EFG1. The differences were observed using dry weight measurements of mature biofilms, and also crystal violet staining of adhered cells (Fig. 2A). We also determined the effect of deleting EFG1 on biofilms developed in vivo, using one deletion strain (LCP1) in the rat catheter model developed by Andes et al. (2004). As shown in Fig. 2B, in vivo biofilms generated by smooth cells carrying an efg1 deletion are mildly reduced. Concentric colonies deleted for EFG1 were not tested in vivo.
Figure 2.
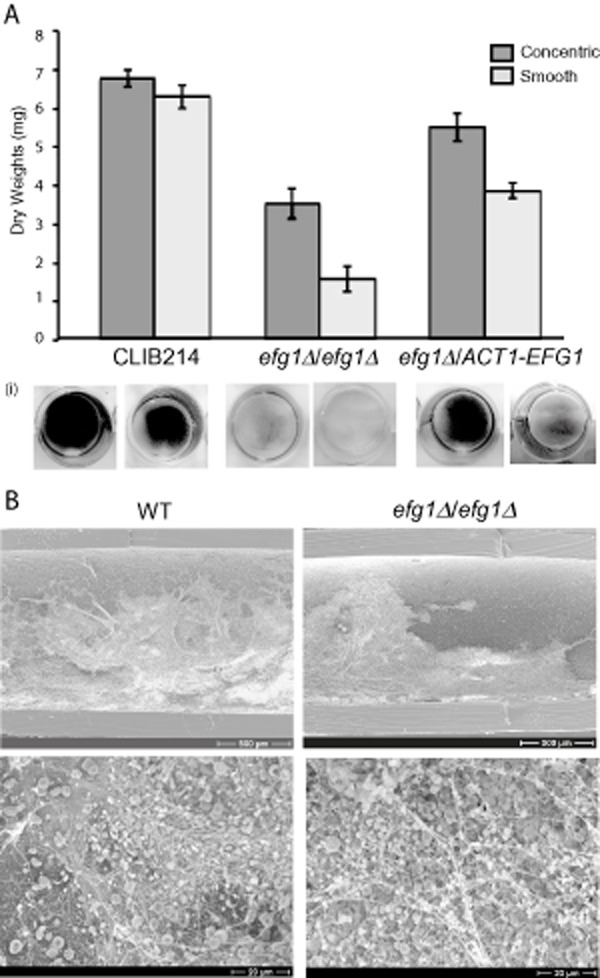
Efg1 regulates biofilm development by C. parapsilosis.A. The in vitro biofilm mass of smooth and concentric colonies from wild type (CLIB214), efg1Δ/efg1Δ deletion strains (including a mixture of strains LCP2, LCP5 and LCP8) and the reconstituted strain LCP7_RI was determined by measurement of dry weights. The average ± standard deviation from three independent measurements is shown. The panels below show biofilm growth on 24-well Nunc plates stained with crystal violet.B. In vivo biofilms were developed in the rat catheter model. Concentric wild-type (CLIB214) cells and smooth efg1/efg1 deletion (LCP1) cells were allowed to develop for 24 h and then visualized by SEM at two magnifications.
Efg1 affects cell wall function
In C. albicans, Efg1 is a major regulator of cell wall genes (Sohn et al., 2003; Zavrel et al., 2012). We therefore tested the phenotype of the efg1 knockouts under conditions of known cell wall stress. Figure 3 shows that knocking out EFG1 confers sensitivity to SDS and caspofungin, and reduces growth on calcofluor white. Both smooth and concentric cells are more susceptible to stress, but the phenotype of smooth cells is more dramatic.
Figure 3.
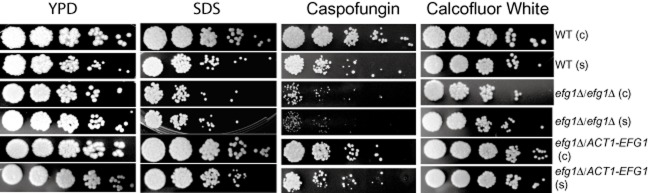
Deleting Efg1 increases sensitivity to cell wall stress. Three microlitres from serial dilutions in PBS was inoculated on YPD alone, and YPD with SDS (0.04%), caspofungin (0.1 μg ml−1) and calcofluor white (12 μM). Strains include concentric (c) and smooth (s) colonies from wild type (C. parapsilosis CLIB214), efg1Δ/efg1Δ deletion (LCP8) and the reconstituted strain (LCP7_RI).
Efg1 is required for virulence in C. parapsilsosis
In C. albicans, deleting EFG1, either alone or in combination with the transcription factor CPH1, results in greatly attenuated virulence in infection of mice (Lo et al., 1997), Caenorhabditis elegans (Pukkila-Worley et al., 2009), Drosophila (Chamilos et al., 2006) and zebrafish (Chao et al., 2010). We determined the effect of deleting EFG1 on the virulence of C. parapsilosis, using the insect Galleria mellonella, which has been shown to be a good model for candidiasis (Fallon et al., 2012).
We first confirmed that C. parapsilosis wild-type strains are capable of killing G. mellonella larvae. Following infection, survival is reduced to approximately 20% within 100 h, compared with 100% survival for uninfected larvae (Fig. 4A). Deleting EFG1 however significantly attenuates virulence (Fig. 4A – two independent deletions were tested). Interestingly, strains carrying only one intact EFG1 allele (either deleted for only one allele, or carrying only the ACT1-EFG1 allele) have virulence levels between the wild-type and the double-deletion strains (Fig. 4A). This is similar to the intermediate biofilm phenotype described in Fig. 1, and suggests that similar to C. albicans, deleting one allele of EFG1 results in a haploinsufficiency phenotype (Zavrel et al., 2012).
Figure 4.
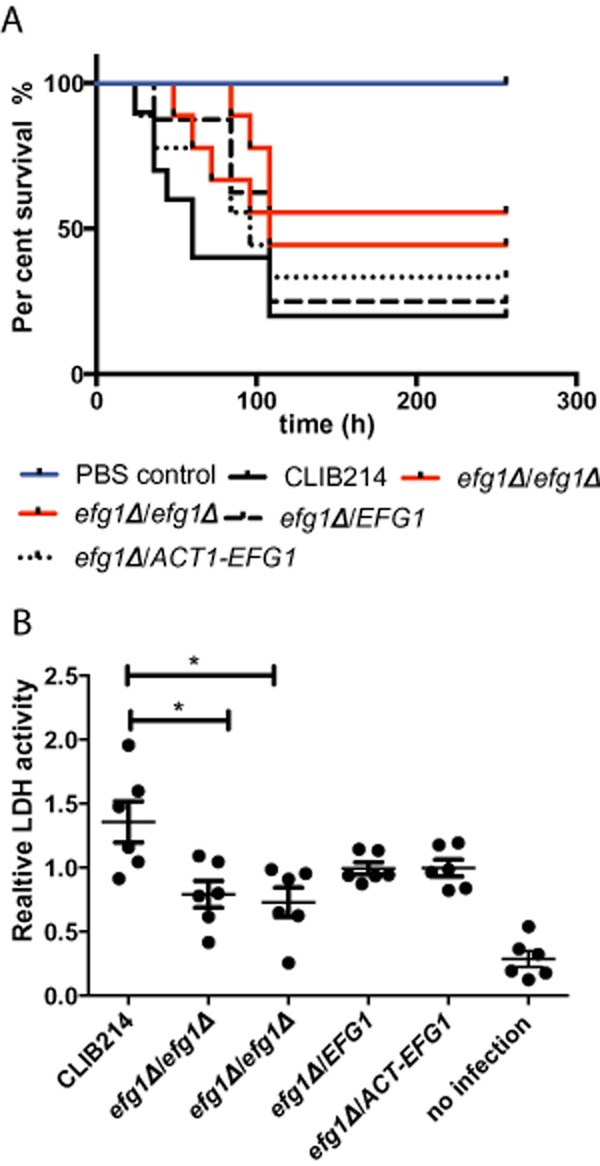
Efg1 is required for virulence.A. G. mellonella larvae were inoculated with 2 × 106 yeast cells in PBS, and survival was monitored over time. The strains tested included wild type (CLIB214), efg1Δ/efg1Δ deletion strains (LCP5 and LCP7), a heterozygote efg1Δ/EFG1 (LCPH1) and the reintegrant efg1/ACT1-EFG1 strain LCP7_RI. The survival curves for the efg1Δ/efg1Δ deletion strains are significantly different from the curves for the wild-type infections (Mantel-Cox test, P-value < 0.01).B. Human PBMC-derived macrophages from four independent donors were co-cultured with C. parapsilosis cells, and the relative levels of LDH in the media were determined after 48 h. The strains used are the same as in (A). The levels of damage induced by the efg1Δ/efg1Δ deletion strains is significantly less than that of the wild type (two-tailed Wilcoxon text, P-value < 0.05).
To determine if deleting EFG1 also affects virulence in mammalian systems, we tested the ability of C. parapsilosis to inflict damage on human PBMC-DM (peripheral blood mononuclear cell-derived macrophages) (Fig. 4B). Damage was measured by the release of Lactate Dehydrogenase (LDH). Deleting EFG1 reduces the release of LDH after 24 h (not shown) and 48 h (Fig. 4B). Once again, having only one intact EFG1 allele results in a phenotype that is intermediate between the wild type and efg1/efg1 deletions.
Identifying the targets of Efg1
We used a combination of chromatin immunoprecipitation with next-generation sequencing (ChIP-seq) to identify promoters that are directly bound by Efg1. We generated a tagged version of Efg1 in a strain carrying a deletion of one EFG1 allele. A MYC epitope tag was introduced just 5′ to the stop codon of Efg1 by homologous recombination, using tools developed for C. albicans (Lavoie et al., 2008). Two independent constructs were confirmed by PCR (Fig. S1). The strains carrying the tagged allele remain in the concentric morphology, indicating that the construct is biologically functional.
The immunoprecipitated bound targets were investigated by next-generation sequencing using Illumina technology. Reads were mapped to the current annotation of the C. parapsilosis genome (Guida et al., 2011) using BWA (Li and Durbin, 2009). Three experimental samples (from two independently tagged strains) and three control (input) samples were used. Enriched peaks were identified using MACS2 (Zhang et al., 2008; Feng et al., 2012). We used estimates of the Irreproducible Discovery Rate (IDR) to incorporate statistical measurements of reproducibility from multiple replicate samples (Landt et al., 2012). A list of bound regions [ranked by π score, fold change and log10 of the adjusted P value (Xiao et al., 2012)] is shown in Table S3.
Efg1 binds over large distances in intergenic regions, often with more than one peak per region (Fig. 5). For example, Efg1 extends over at least > 5 kb in the promoter sequence of CZF1. For ranking purposes we chose a single peak within each intergenic sequence, with the highest ranking (π score, highlighted in pink in Fig. 5). The experiments were highly reproducible, as shown by the different colour tracks in Fig. 5. We also confirmed the binding sites for six promoters using ChIP-PCR (Fig. 5).
Figure 5.
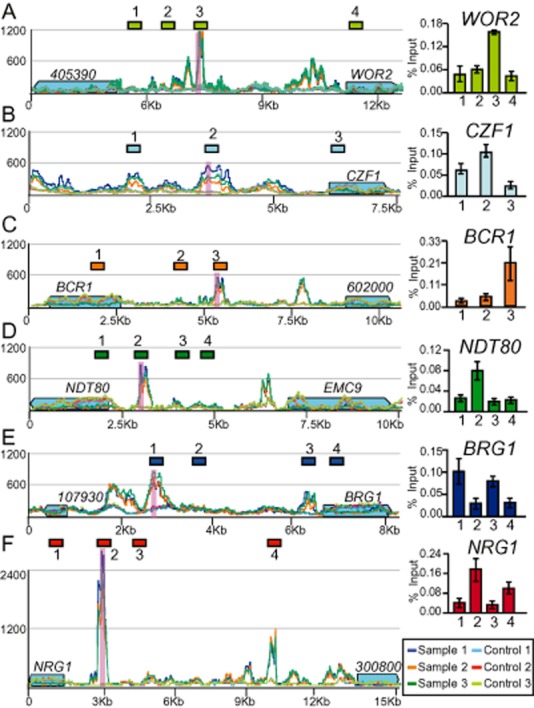
Binding of Efg1 to intergenic regions. (A)–(F): each represent a different intergenic region, with the coding sequences marked by blue boxes where the arrowhead indicates the direction of transcription. Short gene names are given where possible; long numbers (e.g. 405390) should be preceded by the identifier CPAR2. (A) WOR2/CPAR2_405390; (B) CZF1; (C) BCR1/CPAR_60200; (D) NDT80/ECM9; (E) BRG1/CPAR2_107930; (F) NRG1/CPAR2_300800. The scale shows the coverage range. Different coloured lines indicate the mapped reads from three sample (Efg1-myc) and three control (input) lanes. The peak with the highest summit fold change relative to the input in each intergenic region is indicated in pink. The images were constructed using the Integrative Genomics Viewer (IGV) (Robinson et al., 2011). Sequences within each intergenic region (indicated by numbered coloured boxes) were chosen for analysis by ChIP-PCR. The enrichment of PCR products in immunoprecipitated samples as a % of the input samples is plotted. The average ± standard deviation from three independent measurements is shown.
Of 1246 peaks identified, 53 lie within coding sequences and 38 are 3′ to two open reading frames, and were not included in subsequent analyses. The remaining 1155 peaks were assigned to 931 promoters. Because 395 of these regions are within divergent promoters, up to 1326 genes may be directly controlled by the transcription factor. Gene Ontology analysis indicates that the target genes are significantly enriched for regulatory proteins, including transcription factors (P value = 3.47e-7, Table S3C). At least 93 transcription factors are direct targets, including several previously shown to be part of networks with Efg1 that regulate white-opaque switching and biofilm development in C. albicans (Zordan et al., 2007; Nobile et al., 2012), and others associated with hyphal growth (Lassak et al., 2011) (Fig. 5).
We used DREME (Bailey, 2011) and FIMO (Grant et al., 2011) to identify conserved motifs and therefore the most likely binding site of EFG1 in C. parapsilosis, using 100 bp regions around the peak summits from the ChIP-seq experiments. Six significant motifs were identified from both analyses of all 931 promoters (Fig. 6). The most significant motifs (AYGCRC and CACAAAR) most closely resemble the binding sites for Dal82 and Ndt80 in S. cerevisiae (Dorrington and Cooper, 1993; Zhu et al., 2009) (Fig. 6A). The third most significant motif (CTGCATR) resembles the binding site for Sok2 and Phd1 from S. cerevisiae (TGCAGNNA) (Harbison et al., 2004), which are co-orthologues of Efg1. The motif is also similar to the predicted Efg1 binding sites from C. albicans recently reported by Lassak et al. (2011) (TATGCATA) and Nobile et al. (2012) (RTGCATRW). The remaining motifs are similar to binding sites for transcription factors with zinc-finger DNA-binding domains, including Zap1, which has been implicated in regulation of biofilm development in C. albicans (Nobile et al., 2009). All six motifs are present in 40% of the intergenic regions, putative Efg1 and Ndt80 binding sites are found together in 70%, and Efg1, Ndt80 and Dal82 sites co-occur in 65% (Table S4). It is therefore likely that the targets of Efg1 in C. parapsilosis are also regulated by other transcription factors.
Figure 6.
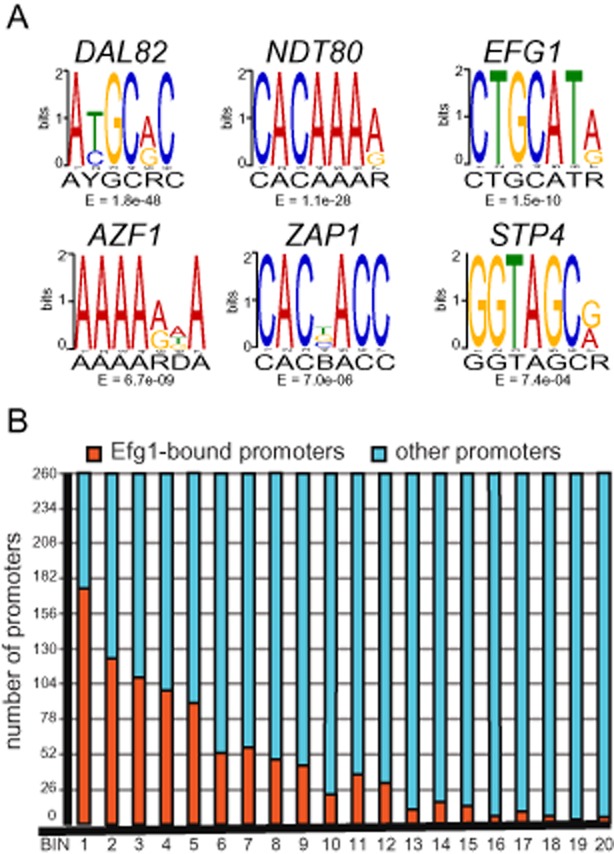
Identification of conserved motifs in Efg1-bound regions.A. Enriched motifs in Efg1-bound intergenic sequences were identified using DREME (Bailey, 2011). All significant motifs are shown, with the most likely associated regulatory protein.B. A total of 5200 intergenic regions from C. parapsilosis were divided into 20 bins depending on decreasing promoter length. Each bin contains 260 intergenic regions. Bin1 contains intergenic regions ranging from 2.7 kb to 11.5 kb, and bin 20 contains promoters ranging from 81 bp to 109 bp. The subset of intergenic regions bound by Efg1 are highlighted in orange.
The intergenic regions bound by Efg1 are significantly longer than the average intergenic region in C. parapsilosis (average of 1.7 kb compared with 0.6 kb, P value < 2.2 e-16, Welch's t-test, Fig. 6B). Nobile et al. (2012) previously found that intergenic regions bound by biofilm regulators in C. albicans are larger than average, and suggested that the combination of short regulatory motifs with larger targets for mutations may allow new genes to be incorporated in a regulatory network.
Identification of the EFG1 regulon
To evaluate the overall biological role of Efg1, we determined the transcriptional profile of efg1 knockout concentric and smooth cells and compared them to the profile of wild-type (concentric) cells. We profiled two independent deletion strains, and included two smooth and three concentric samples (Table S5). We compared the expression of concentric wild-type cells to efg1 deletions in both concentric and smooth morphologies (Table S5A). However, to ensure that the differences we observed are related to the gene deletion rather than to colony type, we predominantly analysed the comparison of concentric cells from both wild-type and efg1 deletion strains.
A total of 353 genes are differentially regulated in concentric cells from the efg1 knockout (Table S5). Efg1 acts as both an activator and a repressor of gene expression; expression of 167 genes is reduced in the efg1 deletions, and expression of 186 genes is increased. Approximately 53% of the genes that are differentially expressed (and 60% of the downregulated genes) are also bound by Efg1 (Table S6A). Similar to C. albicans (Tebarth et al., 2003), Efg1 negatively regulates its own expression. When the open reading frame is deleted, expression of the 5′ UTR region is increased in both concentric and smooth cells (Fig. S2). Upregulated genes are enriched in processes associated with ergosterol metabolism (Table S5). However, the promoter regions of most of these genes are not bound by Efg1, and so they are likely to be regulated indirectly.
There are significant differences in expression patterns between the smooth and concentric colonies from efg1 deletions; 380 genes are differentially expressed (Table S5B). Genes upregulated in concentric cells are enriched in processes associated with iron metabolism, whereas downregulated genes are enriched in metabolism of amino acid and organic acids. Less than 30% are bound by Efg1, suggesting that differential expression is likely to be related to morphology, rather than to deletion of EFG1.
Expression of some of the genes with the largest decreases in expression (which are also bound by Efg1) were confirmed using quantitative PCR. Expression is reduced in both concentric and smooth cells (Fig. S3).
Comparison of Efg1 targets in C. parapsilosis and C. albicans
We compared our ChIP-seq analysis from C. parapsilosis to the ChIP-chip analysis used to identify Efg1-bound genes in C. albicans reported by Nobile et al. (2012) and Lassak et al. (2011). A total of 218 bound genes are shared with the C. albicans data set from Nobile et al. (2012) and 29 with Lassak et al. (2011) (Table S7, Fig. 7). The shared targets are enriched for processes associated with filamentous growth, biofilm development, and cell adhesion (Table S7C, Fig. 7). Many of the shared targets are transcription factors (Fig. 7), suggesting that some regulatory networks are conserved in the two species, even if the biological processes (such as filamentation) are not.
Figure 7.
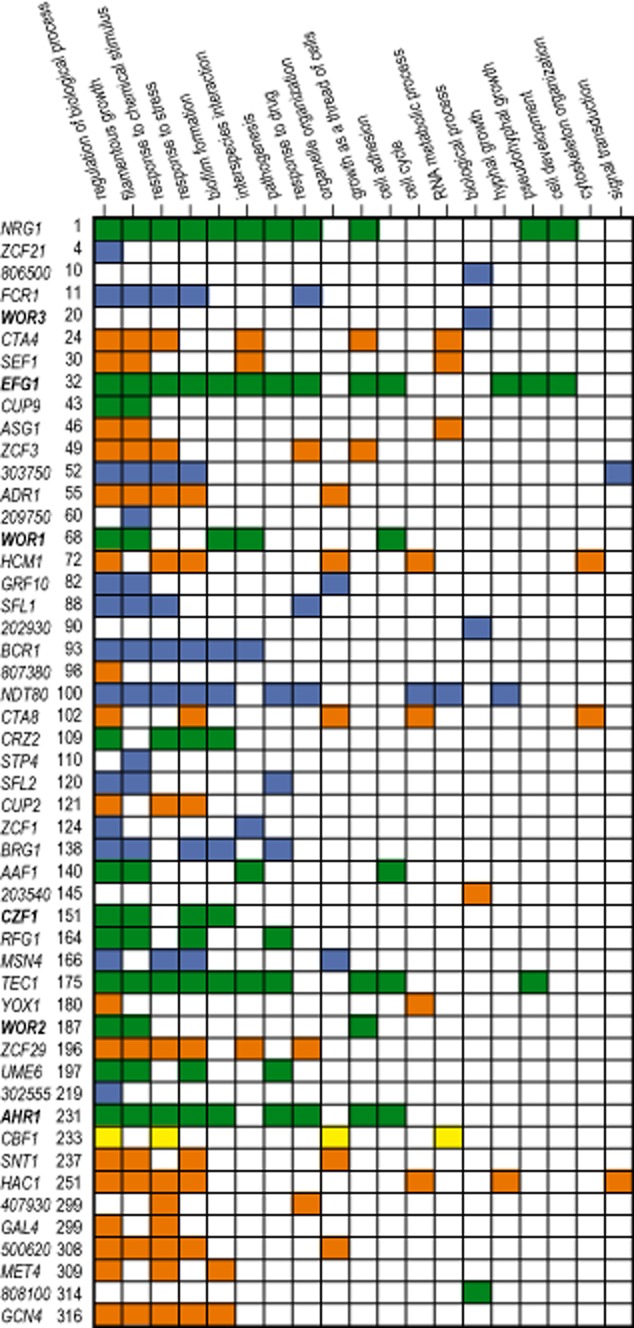
Similarities and differences between targets of Efg1 in C. parapsilosis and C. albicans. The top 50 transcription factors bound by Efg1 in C. parapsilosis are listed. The names of the C. albicans orthologues were used where possible; six-digit gene names (missing the suffix ‘CPAR2’) are used where only a C. albicans generic identifier is available (i.e. orf19). The gene name is followed by the rank from the ChIP-seq analysis (Table S2A). Processes associated with the 50 genes were determined using GO Slim Mapper at the Candida Genome Database (Inglis et al., 2012). Efg1 targets conserved in C. albicans are shown in green [identified by both Lassak et al. (2011) and Nobile et al. (2012)], yellow [identified by Lassak et al. (2011) only] and blue [identified by Nobile et al. (2012) only]. Orange boxes indicate that the relevant transcription factor is bound by Efg1 in C. parapsilosis only. Only processes including at least two genes were included. Genes highlighted in bold are part of the circuit regulating the white/opaque switch in C. albicans.
Nobile et al. (2012) suggested that the biofilm regulatory network has arisen recently in C. albicans. We confirmed that the targets of Efg1 in C. albicans are enriched for young genes, using a very restricted definition of genes that appeared on the C. albicans/C. dubliniensis/C. tropicalis branch (P value = 1.0e-4; Table S3C). In contrast, genes bound by Efg1 in C. parapsilosis are not significantly enriched for young genes (i.e. those arising in the C. parapsilosis/C. orthopsilosis/Lodderomyces elongisporus lineage). The role of Efg1 in C. parapsilosis may therefore resemble the ancestral role, rather than that observed in C. albicans.
We attempted to compare the genes differentially expressed in an efg1 deletion in C. parapsilosis with those differentially expressed in an equivalent knockout in C. albicans (Nobile et al., 2012). However, we identified only 353 differentially expressed genes from C. parapsilosis concentric cells, whereas Nobile et al. (2012) identified > 3000 in C. albicans, corresponding to half the genome. Part of the difference is probably because the profiles described by Nobile et al. (2012) are from biofilm cells. In contrast, Doedt et al. (2004) detected only 283 genes differentially expressed in a C. albicans efg1 deletion grown in similar conditions to ours. Approximately 80 are also differentially expressed in C. parapsilosis but some have different patterns of expression (e.g. downregulated in C. albicans efg1 and upregulated in C. parapsilosis efg1) (Table S5D).
The comparison of both Efg1-bound and differentially expressed genes in C. albicans and C. parapsilosis suggests that some pathways are conserved between the two species, and some are not. Many of the shared components are transcription factors, including orthologues associated with biofilm development (e.g. BCR1, NDT80, BRG1, TEC1), white-opaque switching (e.g. WOR1, WOR2, CZF1) and filamentation (e.g. NRG1, SFL1, UME6) in C. albicans (Liu, 2001; Bauer and Wendland, 2007; Zordan et al., 2007; Nobile et al., 2012; Lohse et al., 2013) (Fig. 7). However, there are also substantial differences, particularly in the RNA-seq profiles, suggesting that many of the targets are species-specific.
Efh1 shares few functions with Efg1
Candida albicans contains a paralogue of EFG1, called EFH1, that is also a member of the APSES family (Doedt et al., 2004). We identified an orthologue of EFH1 in C. parapsilosis (CPAR2_104660), and deleted both alleles in two independent strains using the fusion PCR method (Fig. S1). Deleting EFH1 does not affect biofilm development, or growth in the presence of cell wall stress (Fig. S4). The deletion does not affect switching from concentric to smooth cells, although a novel colony phenotype does arise at a low level (approximately 3%, Fig. S4, Table S1).We also found that deleting both efg1 and efh1 resulted in a phenotype that is indistinguishable from efg1 deletion alone for biofilm development and sensitivity to cell wall drugs (Fig. S4), and for cell morphology during growth in normoxic and hypoxic conditions (Table S2).
Discussion
Efg1 is a major regulator of morphogenesis in fungal species, originally characterized in C. albicans as a regulator of filamentous growth (Stoldt et al., 1997). S. cerevisiae has two Efg1 co-orthologues, called Phd1 and Sok2, and both are involved in the regulation of pseudohyphal growth (Gimeno and Fink, 1994; Ward et al., 1995; Pan and Heitman, 2000). The proteins are members of the APSES family, which contain a novel conserved DNA-binding domain (KilA-N domain) related to a basic helix-loop-helix domain that is conserved in many eukaryotic and bacterial viruses (Marchler-Bauer et al., 2011). Other members such as StuA in A. nidulans and Asm-1 in Neurospora crassa (filamentous ascomycetes, or Pezizomycotina) also control yeast-hyphal transitions (Miller et al., 1992; Aramayo et al., 1996). An orthologue from a basidiomycete, Ustilago maydis, has recently been associated with regulation of dimorphic growth in this species (Garcia-Pedrajas et al., 2010). However, in other species, the APSES proteins do not control dimorphism, but instead regulate processes such as mating and conidiation (Borneman et al., 2002). A role for APSES proteins in regulation of development has therefore been conserved across fungal species, but Borneman et al. (2002) suggest that regulation of hyphal growth is restricted to species that divide by budding. This is supported by our analysis; deleting EFG1 in C. parapsilosis increases pseudohyphal growth in hypoxic conditions (Fig. 1B, Table S2).
Both Efg1 and Efh1 in C. albicans have KilA-N/APSES domains (Doedt et al., 2004). However, unlike the two proteins in S. cerevisiae (Phd1 and Sok2), Efg1 and Efh1 fall into different clades (Fig. 8). PHD1 and SOK2 arose from an ancient duplication event in the Whole Genome Duplication clade containing S. cerevisiae (Wolfe and Shields, 1997), and both are orthologous to C. albicans EFG1 (Fig. 8). The Efg1/Efh1 proteins are poorly conserved throughout their entire lengths, and contain several polyQ regions that may be associated with transcriptional activation. However, the DNA-binding domains (KilA-N domain) are well conserved, and were used to construct the phylogenetic tree shown in Fig. 8. Whereas both Efg1 and Efh1 are present in all the Candida species, the Efg1 paralogues are similar to the APSES proteins from other fungal species, and the Efh1 proteins cluster in a separate clade that appears to be unique to Candida (Fig. 8). The Efh1 proteins are also evolving rapidly, as shown by the long branch lengths in the Efh1 cluster in Fig. 8, whereas the DNA-binding domains of Efg1 are remarkably similar (more similar than proteins within the Saccharomyces clade for example).
Figure 8.
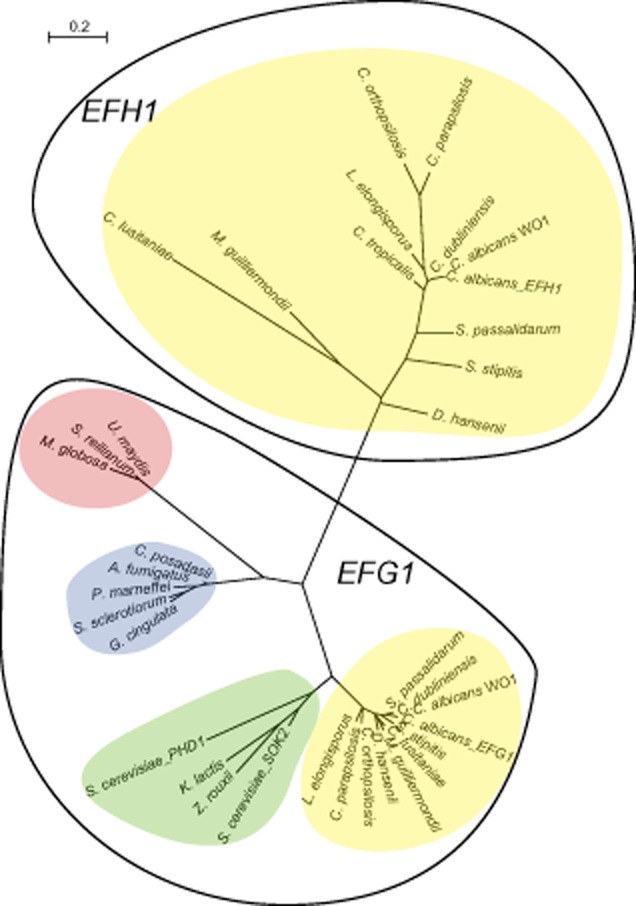
Conservation of Efg1-like proteins in fungi. Sequences were aligned using SeaView (Gouy et al., 2010), and an unrooted phylogenetic tree was constructed from the conserved bHLH domains by implementing the PhyML algorithm within SeaView. The CTG clade species are highlighted in yellow, Saccharomyces species in green, Pezizomycotina (filamentous ascomycetes) species in purple, and Basidiomycetes in peach. Gene names/accession numbers: Ustilago maydis um15042, Sporisorium reilianum CBQ71974, Malassezia globosa MGL_0313, Coccidioides posadasii XP_003066203, Aspergillus fumigatus XP_755125, Penicillium marneffei XP_002146488, Sclerotinia sclerotiourum XP_001590416, Glomerella cingulata ABQ43363, Saccharomyces cerevisiae SOK2 and PHD1, C. albicans SC5314 orf19.8243 and orf19.5498, C. albicans WO1 CAWG_02083 and CAWG_04378, C. dubliniensis CD36_33560 and CD36_20730, C. tropicalis CTRG_01780, C. parapsilosis CPAR2_701620 and CPAR2_104660, C. orthopsilosis CORT0G01760 and CORT0B05860, L. elongisporus LELG_05390 and LELG_01890, Spathaspora passalidarum SPAPADRAFT_59209 and SPAPADRAFT_66101, Scheffersomyces stipitis PICST_67427 and PICST_32309, Meyerozyma guilliermondii PGUG_03651 and PGUG_02282, Clavispora lusitaniae CLUG_02047 and CLUG_01623.
The rapid evolution of the Efh1 proteins makes it very difficult to determine their origin [due to difficulties with long-branch attraction (Fares et al., 2006)]. However, it is more likely that Efh1 arose in a common ancestor of the Candida species, rather than an earlier origin with subsequent loss in other fungal clades. The function of Efh1 remains to be determined. EFH1 has little effect on the Efg1-dependent phenotypes in C. albicans, except that a double efg1/efh1 deletion forms lateral hyphae faster and is more filamentous in hypoxic conditions, and that pseudohyphal formation caused by overproduction of EFH1 requires EFG1 (unlike filamentation caused by overproduction of EFG1, which is independent of EFH1) (Doedt et al., 2004). Efg1 and Efh1 share very few regulatory targets in C. albicans (Doedt et al., 2004), and we have shown that in C. parapsilosis, EFH1 does not regulate response to stress or biofilm development, and the phenotype of a double efg1/efh1 mutant is indistinguishable from an efg1 deletion. The switch from concentric to smooth colony morphology is also not increased by deleting EFH1, although a novel colony phenotype is observed at a very low rate (Fig. S4). EFH1 regulates colonization of the murine intestinal tract by C. albicans in a commensal model of infection (White et al., 2007); any similar role in C. parapsilosis has not been tested.
High-frequency phenotypic switching was first observed in Candida species in the 1930s (Negroni, 1935; Soll, 2002). The best-characterized system is the white-opaque switch in C. albicans; switching to opaque cells is required for efficient mating (Slutsky et al., 1987; Miller and Johnson, 2002). White and opaque cells have different transcriptional and metabolic profiles, they respond differently to phagocytosis, and they exhibit different colonization and virulence properties (reviewed in Morschhauser, 2010). Switching is a stochastic process, determined by the levels of the master regulator Wor1 (Huang et al., 2006; Srikantha et al., 2006; Zordan et al., 2007). Efg1 is a major component of the regulatory network (Srikantha et al., 2000; Doedt et al., 2004; Zordan et al., 2007). Switching is also affected by chromatin modification (Klar et al., 2001; Srikantha et al., 2001; Hnisz et al., 2009). White-opaque switching is probably restricted to C. albicans, C. dubliniensis and C. tropicalis (Pujol et al., 2004; Porman et al., 2011).
Other high-frequency switching systems have been reported in Candida species; for example in C. albicans 3135A, eight different interchangeable colony morphologies were described (Slutsky et al., 1985; Ramsey et al., 1994). Expression of some known virulence factors such as phospholipases and proteinases varies in the different phenotypes (Antony et al., 2009). Switching between unusual colony morphologies has also been reported in C. tropicalis, C. parapsilosis and C. lusitaniae (Enger et al., 2001; Miller et al., 2006; Franca et al., 2011), and outside the CTG clade, in Candida glabrata and Candida krusei (Lachke et al., 2000; Arzmi et al., 2012). However, the underlying regulatory mechanisms have not been elucidated. We showed here that Efg1, a major regulator of white-opaque switching in C. albicans, also regulates colony morphology in C. parapsilosis. The rate of switching varies between the various efg1 deletion strains (Table S1), but even the strains with high switching rates return to wild-type levels when EFG1 is re-introduced. Concentric and smooth cells deleted for EFG1 have substantially different expression profiles, enriched for genes associated with iron metabolism (Table S5). It is therefore likely that two colony morphologies differ in metabolism, similar to white/opaque cells of C. albicans.
Efg1 is required for biofilm development in C. albicans, both in in vitro and in in vivo models (Ramage et al., 2002; Ricicova et al., 2010; Nobile et al., 2012). Unlike many other biofilm regulators, Efg1 is required for biofilm development in low oxygen and high carbon dioxide, conditions that mimic systemic infection (Stichternoth and Ernst, 2009). We have shown that deleting EFG1 in C. parapsilosis also reduces biofilm formation in vitro (Fig. 2A). In hypoxic conditions wild-type strains make very little biofilm, and this is further reduced in the efg1 deletions (not shown). The defect in vivo is not as pronounced; the view of a catheter at low magnification shows that the overall mass is somewhat reduced, but the structure of the biofilm is essentially unchanged (Fig. 2B). One of the major differences between C. parapsilosis and C. albicans is that the former does not form true hyphae. Biofilms formed by C. albicans consist of a mixture of yeast, pseudohyphal and hyphal cells; the yeast cells form a basal layer, with hyphae in the upper layers of the mature biofilm (Baillie and Douglas, 1999; Blankenship and Mitchell, 2006; Ramage et al., 2009). C. parapsilosis biofilms in contrast consist of yeast and pseudohyphal cells only (Ding and Butler, 2007; Ramage et al., 2009). C. albicans strains with an EFG1 deletion are locked in yeast phase in most growth conditions, including in biofilms (Ramage et al., 2002), and other mutants with reduced hyphal development also have reduced biofilm mass (Baillie and Douglas, 1999). Although the entire biofilm network identified by Nobile et al. (2012) does not appear to be dependent on hyphal growth, it is very likely that for the EFG1 deletion some of the reduction in biofilm development is due to reduced hyphal formation. The lack of hyphae in C. parapsilosis may partly explain the difference in phenotype between the EFG1 deletions in the two species in vivo, where hyphal growth is likely to be more important for C. albicans biofilms.
We found that deleting EFG1 in C. parapsilosis results in attenuated virulence in an insect model, and reduces damage of human macrophages (Fig. 4). In C. albicans, reduction in virulence of an efg1 deletion has generally been related to a failure of the knockout to grow as hyphae (Lo et al., 1997; Chamilos et al., 2006; Pukkila-Worley et al., 2009). However, even in models where efg1 strains do form hyphae (such as gnotobiotic mice deficient in natural killer and T cells) virulence is greatly reduced (Westwater et al., 2007). The virulence phenotypes in C. albicans may therefore be linked to changes in immunogenicity of the efg1 deletion caused by alterations in the cell wall (Sohn et al., 2003; Doedt et al., 2004; Zavrel et al., 2012). Deleting EFG1 in C. parapsilosis also affects cell wall sensitivity (Fig. 3), and some of the genes differentially expressed in the efg1 deletion (such as orthologues of WSC2, WSC4, PGA26, PGA30, PGA62, ALS6, ALS7) are known cell wall proteins in C. albicans (Table S5). The role of Efg1 in virulence is therefore likely to be conserved in both species.
There is a substantial overlap between the intergenic regions bound by Efg1 in C. albicans and C. parapsilosis, even when the C. albicans experiments were carried out using biofilm cells (Table S7; Nobile et al., 2012). In both species the targets are enriched for transcription factors (Table S3) and 38 of the 93 bound by Efg1 in C. parapsilosis are also bound in C. albicans biofilms (Table S7). These include Wor1, Wor2 and Czf1, which together with Efg1 control white-opaque switching in C. albicans (Zordan et al., 2007) (Figs 5 and 7). Most of the biofilm regulatory network identified by Nobile et al. (2012) is present (Efg1, Bcr1, Ndt80, Brg1, Tec1) (Fig. 7, Table S3). Other targets of Efg1 (e.g. Nrg1) that are associated with the yeast-to-hyphal switch (Lassak et al., 2011) are also bound by Efg1 in C. parapsilosis (Fig. 7). Efg1 and Ndt80 motifs co-occur in many intergenic regions in C. parapsilosis (Table S4). This strongly suggests that the components and structure of regulatory networks are highly conserved in C. albicans and C. parapsilosis. However, the functions of the networks are not as well conserved. For example, some of the phenotypes regulated by Efg1 in C. albicans (such as true hyphal growth and white/opaque switching) do not occur in C. parapsilosis. In addition, there is relatively little overlap between genes differentially expressed in an efg1 deletion in C. parapsilosis and C. albicans (Table S5D) (Doedt et al., 2004). Even in biofilm development, where Efg1 is implicated in both species, many of the targets of the transcription factor are newly evolved in C. albicans but not in C. parapsilosis (Table S3C). It is therefore likely that the role of Efg1 in C. parapsilosis more closely resembles the ancestral state, and may have been adapted for filamentation, white-opaque switching and some aspects of biofilm regulation in C. albicans.
The intergenic regions bound by Efg1 in C. parapsilosis are significantly longer than the average intergenic region (Fig. 6B). It has also been reported that targets of Efg1 and other biofilm regulators (Nobile et al., 2012), and of Wor1 (Zordan et al., 2007), are longer in C. albicans. Nobile et al. (2012) suggest that this is partly because the biofilm targets are relatively young in evolutionary terms, and young genes tend to have long promoters. However, the association of promoter size with evolutionary age comes from an analysis of yeast species from before and after the whole-genome duplication (WGD) event. New promoters tend to be longer in post WGD species, but this is due to loss of genes within intergenic regions that occurs as part of a major rearrangement following duplication (Sugino and Innan, 2012). Candida species have not undergone genome duplication. The long promoters may be a feature of the number of regulatory proteins that bind to them; Efg1 appears to be part of many networks, and many targets are co-regulated. In C. parapsilosis, a significant number of the Efg1 targets are likely to also bind Ndt80, and possibly up to five other transcription factors. In addition, Efg1 binding, although centred on a short motif, extends over large regions of the promoters (Fig. 5). Finally, some of the transcription factors have very long 5′ untranslated regions; for EFG1 in C. albicans, the 5′UTR extend almost 1200 bp upstream of the start (Srikantha et al., 2000). Some regulation may therefore be at the post-translational level.
Efg1 has many roles in C. albicans, including controlling filamentation (Stoldt et al., 1997), hypoxic response (Stichternoth and Ernst, 2009), regulation of cell wall proteins (Sohn et al., 2003; Zavrel et al., 2012), white-opaque switch (Morschhauser, 2010), biofilm development (Ramage et al., 2002; Nobile et al., 2012), generation of chlamydospores (Sonneborn et al., 1999a), regulation of metabolism (Doedt et al., 2004) and virulence (Lo et al., 1997; Chamilos et al., 2006; Pukkila-Worley et al., 2009). Some of these phenotypes (such as biofilm development, virulence and sensitivity to cell wall-damaging agents) are conserved in C. parapsilosis (Figs 2, 3 and 5; Zavrel et al., 2012). However, there are also significant differences. The role of Efg1 in biofilm development in C. parapsilosis is not as dominant as in C. albicans. More importantly, Efg1 circuits that regulate morphological switching in C. parapsilosis may have been adapted to control white-opaque switching and filamentation in C. albicans. We look forward to identifying the other members of the network regulating concentric-smooth switching, and to explore the roles of Efg1 and its targets in multiple cellular processes in C. parapsilosis.
Experimental procedures
Strains and growth conditions
Candida parapsilosis strains were grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose) at 30°C. For colony selection 2% agar was added. To select for transformants, nourseothricin (Werner Bioagents Jena, Germany) was added to YPD agar at a final concentration of 200 μg ml−1. Transformants containing the LEU2 and HIS1 markers were selected on synthetic complete (SC; 0.67% yeast nitrogen base, 2% dextrose, 0.075% mixture of amino acids, 2% agar) media without leucine or histidine. For biofilm formation, C. parapsilosis was grown in synthetic defined (SD) medium (0.67% yeast nitrogen base) containing 50 mM glucose. Calcofluor white M2R (Sigma-Aldrich), caspofungin (from Cancidas, Merck & Co.) and Sodium n-Dodecyl Sulphate (SDS) (Biosciences) were added to the medium at the desired concentrations. For phenotype screening 500 μl of an overnight culture was washed and resuspended in phospho-buffered (PBS) at a concentration of 6.25 × 105 cells ml−1. A fivefold serial dilution was generated and 3 μl was plated onto selected agar plates, and incubated at 30°C for 3 days. The strains and oligonucleotides used are listed in Table S8 and Table S9 respectively. All hypoxia experiments were carried out at 37°C in 1% O2 in an InVivo2 400 hypoxia chamber. For microscopy, cells were taken straight from colonies on an agar plate and resuspended in 10 μl of 2 mM Calcofluor White. Images were taken using a Zeiss AxioImager M1 fluorescent microscope. All gene constructs are described in Fig. S1.
Determination of switching rate
Colonies of C. parapsilosis strains with specific morphologies were isolated and grown overnight in 5 ml YPD at 30°C, shaking at 200 r.p.m. One millilitre of overnight cultures was washed twice in PBS and diluted to ∼ 100 cells per 100 μl in PBS. One hundred microlitres of the diluted sample was spread on a YPD plate. YPD plates were incubated under various temperatures and under normoxic and hypoxic conditions. Multiple plates were used so that > 1000 colonies were counted from each phenotype under each growth condition. The plates were inspected after 2 days, 3 days and 4 days. Colonies were counted on day three to determine switching frequencies (Table S1).
Biofilm assays
Overnight cultures of C. parapsilosis strains grown in 5 ml YPD at 30°C were washed twice in 1 ml PBS buffer and diluted to an A600 of 1 in SD medium with 50 mM glucose. One millilitre of each culture was added to three wells of a 24-well Nunc Delta plate (Thermo Scientific) and incubated on a shaker (50 r.p.m.) at 37°C for 2 h (initial adhesion phase). The culture was removed and each well was washed once with 1 ml PBS buffer. One millilitre of fresh SD medium with 50 mM glucose was added to each well and the plates were shaken at 37°C for an additional 48 h. The culture was removed and wells were washed twice with 1 ml PBS buffer and allowed to dry at room temperature overnight. Biofilms were stained for 30 min with 500 μl of 0.4% (w/v) crystal violet. Crystal violet was removed and biofilms were washed twice with 1 ml PBS buffer and dried overnight at room temperature. The resulting biofilms were then photographed. For dry weight measurements of biofilms 5 ml of cultures at an A600 of 1 in SD medium with 50 mM glucose were inoculated in each well of six-well Nunc plates. Biofilms were allowed develop as above. After 48 h the wells were washed twice with 2 ml PBS buffer. One millilitre PBS buffer was added to each well and biofilm was scraped from the bottom of each well. Biofilm from two wells was combined, filtered using a 0.8 μm Millipore filter, dried for 48 h at room temperature and weighed. In vivo biofilms assays were carried out as described previously (Ding et al., 2011).
Host damage assay
Human peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats of healthy donors by Ficoll Paque PLUS (GE Healthcare) density gradient centrifugation, as described elsewhere (Schlenke et al., 1998). Experiments were performed according to the institutional regulation of the independent ethics committee of the University of Szeged. Isolated PBMCs were suspended in RPMI medium (LONZA) supplemented with 1% 100× Penicillin-Streptomycin solution (Sigma-Aldrich), and plated in 96-well cell culture plates (5 × 105 PBMC per well, in 100 μl) to isolate monocytes by plastic adherence. After 2 h of incubation (37°C, 5% CO2, 100% relative humidity), floating cells were removed, and the attached monocytes were gently washed with PBS. The isolated cells were cultured for 7 days in X-VIVO 15 serum-free medium (LONZA) supplemented with 1% 100× Penicillin-Streptomycin solution (Sigma-Aldrich), in the presence of 10 ng ml−1 human recombinant GM-CSF (Sigma-Aldrich) to enable macrophage differentiation. Host-cell damaging capacity of C. parapsilosis strains was determined by LDH (lactate dehydrogenase) activity assay. PBMC-DM (PBMC-derived macrophage) cells were co-cultured with C. parapsilosis cells at a ratio of 1:5 in X-VIVO 15 serum-free medium (LONZA) supplemented with 1% 100× Penicillin-Streptomycin solution for 24 or 48 h, or left unstimulated (negative control). Macrophage cells alone incubated under identical conditions were applied as negative controls. LDH in medium from cultures containing uninfected and infected tissues at 24 and 48 h was measured using the Cytotoxicity Detection Kit (LDH; Roche) according to the manufacturer's instructions. Relative LDH activity was calculated, as described previously (Gacser et al., 2007).
Galleria mellonella survival assay
Infection of G. mellonella larvae (Mous Livebait R.J., the Netherlands) was performed as described previously (Mylonakis, 2008; Garcia-Rodas et al., 2011). Briefly, larvae were inoculated with 10 μl of a C. parapsilosis suspension prepared in PBS containing 2 × 106 yeast cells by an injection in the last left pro-leg, using a 26-gauge needle with Hamilton syringes. After injection, caterpillars were incubated at 25°C, and the number of dead larvae was noted daily. In each experiment, a group of caterpillars were left without any manipulation (untreated control), and another group of caterpillars was inoculated with PBS only.
ChIP-seq and ChIP-PCR analysis
To introduce a tag just before the stop codon of Efg1, the MYC epitope tag and C. albicans HIS1 gene were amplified by PCR from plasmid pFA-MYC-CaHIS1 (Lavoie et al., 2008) using primers EFG1_tag_F and EFG1_tag_R (Table S9). The purified product was transformed by electroporation into C. parapsilosis LCPH3 and LCPH4 (EFG1/efg1::LEU2, his1−/his1−) generating strains LCP3M and LCP4M. Correct integration of the tag was confirmed by PCR screening (Fig. S1).
Cultures were prepared for chromatin immunoprecipitation as described by Aparicio et al. (2005), from C. parapsilosis cells grown in YPD to an A600 = 1. Samples were sonicated using a Biorupter™ (Diagenode) for 15 min at 30 s intervals at high power setting in an ice bath. The ice was then changed and the sonication was repeated. Input samples before the addition of antibody were used as control. Regions bound to Efg1 were immunoprecipitated using a mouse monoclonal antibody (Myc-tag 9B11, Cell Signaling). Three input (before immunoprecipitation) and three immunoprecipitated samples were used (Table S10).
All ChIP-seq libraries were generated according to the Illumina protocol (Illumina Guide Part # 11257047 Rev. A). A limited 18-cycle amplification of size-selected libraries was carried out. To eliminate adapter dimers libraries were further sized selected using a 2.5% TAE agarose gels. Purified libraries were quantified using a Qubit™ fluorometer (Invitrogen) and a Quant-iT™ double-stranded DNA High-Sensitivity Assay Kit (Invitrogen). Clustering and sequencing of the material was carried out as per manufacturer's instructions on the Illumina GAIIx platform in the UCD Conway Institute. On average, 22.5 million 42 bp reads were obtained from each sample (Table S10).
The reads from the three experimental samples and the three controls were mapped to the latest version C. parapsilosis genome (Guida et al., 2011) using BWA (Li and Durbin, 2009). More than 90% of reads for each library were aligned to the reference (Table S10). Approximately 6.5% of reads from the immunoprecipitated samples and 1.2% of reads from the controls mapped to the rDNA locus. The Binary Alignment/Map (BAM) files were sorted and indexed using SAMtools (Li et al., 2009). Binding sites were predicted using MACS 2.0 (Zhang et al., 2008) (Feng et al., 2012), with a q-value threshold of 0.01 and automatic estimation of reads per genomic location. Fragment size was estimated using the SPP package (Kharchenko et al., 2008). Each immunoprecipitated sample was individually compared with the three merged controls. The reproducibility of the data sets was determined by pairwise comparisons of the three samples using IDR (Irreproducible Discovery Rate method), as recommended by the Encode consortium (Landt et al., 2012) and implemented in the SPP package (Kharchenko et al., 2008). A maximum of 1246 replicable peaks were identified. The MACS2 analysis was then repeated using the merged samples as input and called peaks were ranked by P value, using a False Discovery Rate (FDR) threshold of 1%. The top 1246 peaks were selected.
A custom Python pipeline was used to associate peaks with genes in the latest C. parapsilosis annotation (Guida et al., 2011). Peaks were mapped to the gene with the nearest 5′ end. Peaks lying in a divergent promoter region were assigned to both genes. Peaks lying between two convergent genes were not assigned. Peaks completely within a coding region were also not assigned. In total, 1155 peaks were assigned to 931 intergenic regions. Overall, these map to 536 unique promoters and 395 divergent promoters. Peaks were ranked by π value (Xiao et al., 2012), i.e. a product of P-values and fold enrichment at peak summit position with respect to the control. Where there were multiple peaks in a promoter, only the peak with the highest rank was considered (Table S3).
To confirm the peaks identified by ChIP-seq analysis, quantitative real-time PCR was carried out using three independent input and three immunoprecipitated samples from strains LCP3M and LCP4M. Primers were designed to amplify regions of high and low enrichment of Efg1 identified in the ChIP-seq experiments using Primer3Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) (Table S9). For each reaction, a mastermix consisting of 5 μl 2× Brilliant III SYBR® green (Applied Biosystems), 500 nM of each primer, 0.15 μl ROX reference dye (diluted 1:500), 2.35 μl Nuclease free H2O and 1 μl of DNA (Input or Immunoprecipitated sample) was used. Samples were analysed in duplicate on a Strategene Mx3005P QPCR system (Agilent Technologies) using cycling conditions of one cycle at 95°C for 3 min followed by 40 cycles at 95°C for 10 s, 60°C for 20 s and a final cycle of 95°C for 1 min followed by melting curve analysis at 55°C to 95°C (temperature transition, 0.2°C s−1) with stepwise fluorescence detection. Enrichment in the promoter regions was calculated as the percentage of PCR product from the average of the immunoprecipitated samples normalized to the average of the input samples.
Motif prediction was carried out using the online version of DREME (Bailey, 2011) using 100 bp sequences around the summits of the best peak for each intergenic region for all samples. Shuffled input sequences were used for background comparison. Significant results were submitted to TOMTOM (Gupta et al., 2007) using default parameters. Motif occurrence was confirmed using FIMO with a significance threshold of 1e-03 (Grant et al., 2011).
RNA-seq analysis
RNA-seq library preparation for strand-specific sequencing was carried out essentially as described in Guida et al. (2011), from cultures grown in YPD. We used HTSeq and DESeq (Anders and Huber, 2010) to compare 17.7 × 106 36 bp Illumina reads from three samples of wild type (C. parapsilosis CLIB214) to 15 × 106 reads from two different efg1 deletion strains in the smooth phenotype and 16.4 × 106 reads from three deletions of the concentric phenotype (Table S10). Reads with exceptionally high coverage (rDNA) were masked. The RNA-seq data sets were submitted to the Gene Expression Omnibus (GEO) (series GSE41065). All Gene Ontology analysis was carried out at the Candida Genome Database (Inglis et al., 2012).
Gene age analysis
A total of 585 genes present in the C. albicans, C. dubliniensis and C. tropicalis and not in other CTG clade species were designated as ‘young’ on the C. albicans branch, whereas 473 genes present only in the C. parapsilosis, C. orthopsilosis and L. elongisporus branch were designated ‘young’ in C. parapsilosis. Gene orthology was defined using the Candida Gene Order Browser (version 29 August 2012) (Fitzpatrick et al., 2010; Maguire et al., 2013). Differences between observed and expected values in the list of Efg1-bound genes were determined using Fisher's exact test (Table S3C).
Acknowledgments
We are grateful to Suzanne Noble (UCSF) for providing the fusion PCR plasmids, to Sarah Maguire and Gian Luca Negri for assistance with bioinformatics, to Amanda Lohan for advice on next-generation sequencing, to Adrian Bracken (TCD) for help with ChIP-PCR, to the Imaging Facility at the Conway Institute, and to Ken Wolfe and members of the Butler group for comments. Work at UCD was supported by Science Foundation Ireland (08/N1B1865) and by the Irish Research Council. A.G. and Z.G. were supported in part by OTKA NF 84006, NN100374 (ERA-Net PathoGenomics Program) and an EMBO Installation Grant.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Mihalik R, Soll DR. Ultrastructure and antigenicity of the unique cell wall pimple of the Candida opaque phenotype. J Bacteriol. 1990;172:224–235. doi: 10.1128/jb.172.1.224-235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM, Soll DR. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J Bacteriol. 1987;169:5579–5588. doi: 10.1128/jb.169.12.5579-5588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004;72:6023–6031. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony G, Saralaya V, Gopalkrishna Bhat K, Shalini Shenoy M, Shivananda PG. Effect of phenotypic switching on expression of virulence factors by Candida albicans causing candidiasis in diabetic patients. Rev Iberoam Micol. 2009;26:202–205. doi: 10.1016/j.riam.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Aparicio O, Geisberg JV, Sekinger E, Yang A, Moqtaderi Z, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol. 2005;Chapter 21:Unit 21 23. doi: 10.1002/0471142727.mb2103s69. [DOI] [PubMed] [Google Scholar]
- Aramayo R, Peleg Y, Addison R, Metzenberg R. Asm-1+, a Neurospora crassa gene related to transcriptional regulators of fungal development. Genetics. 1996;144:991–1003. doi: 10.1093/genetics/144.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzmi MH, Abdul Razak F, Yusoff Musa M, Wan Harun WH. Effect of phenotypic switching on the biological properties and susceptibility to chlorhexidine in Candida krusei ATCC 14243. FEMS Yeast Res. 2012;12:351–358. doi: 10.1111/j.1567-1364.2011.00786.x. [DOI] [PubMed] [Google Scholar]
- Asbeck van EC, Clemons KV, Stevens DA. Candida parapsilosis: a review of its epidemiology, pathogenesis, clinical aspects, typing and antimicrobial susceptibility. Crit Rev Microbiol. 2009;35:283–309. doi: 10.3109/10408410903213393. [DOI] [PubMed] [Google Scholar]
- Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999;48:671–679. doi: 10.1099/00222615-48-7-671. [DOI] [PubMed] [Google Scholar]
- Bauer J, Wendland J. Candida albicans Sfl1 suppresses flocculation and filamentation. Eukaryot Cell. 2007;6:1736–1744. doi: 10.1128/EC.00236-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- Bink A, Govaert G, Vandenbosch D, Kucharikova S, Coenye T, Nelis H, et al. Transcription factor Efg1 contributes to the tolerance of Candida albicans biofilms against antifungal agents in vitro and in vivo. J Med Microbiol. 2012;61:813–819. doi: 10.1099/jmm.0.041020-0. [DOI] [PubMed] [Google Scholar]
- Blankenship JR, Mitchell AP. How to build a biofilm: a fungal perspective. Curr Opin Microbiol. 2006;9:588–594. doi: 10.1016/j.mib.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Hynes MJ, Andrianopoulos A. A basic helix-loop-helix protein with similarity to the fungal morphological regulators, Phd1p, Efg1p and StuA, controls conidiation but not dimorphic growth in Penicillium marneffei. Mol Microbiol. 2002;44:621–631. doi: 10.1046/j.1365-2958.2002.02906.x. [DOI] [PubMed] [Google Scholar]
- Butler G. Fungal sex and pathogenesis. Clin Microbiol Rev. 2010;23:140–159. doi: 10.1128/CMR.00053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamilos G, Lionakis MS, Lewis RE, Lopez-Ribot JL, Saville SP, Albert ND, et al. Drosophila melanogaster as a facile model for large-scale studies of virulence mechanisms and antifungal drug efficacy in Candida species. J Infect Dis. 2006;193:1014–1022. doi: 10.1086/500950. [DOI] [PubMed] [Google Scholar]
- Chao CC, Hsu PC, Jen CF, Chen IH, Wang CH, Chan HC, et al. Zebrafish as a model host for Candida albicans infection. Infect Immun. 2010;78:2512–2521. doi: 10.1128/IAI.01293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C, Schandar M, Noll M, Johannes FJ, Brunner H, Graeve T, Rupp S. In vitro reconstructed human epithelia reveal contributions of Candida albicans EFG1 and CPH1 to adhesion and invasion. Microbiology. 2002;148:497–506. doi: 10.1099/00221287-148-2-497. [DOI] [PubMed] [Google Scholar]
- Ding C, Butler G. Development of a gene knockout system in Candida parapsilosis reveals a conserved role for BCR1 in biofilm formation. Eukaryot Cell. 2007;6:1310–1319. doi: 10.1128/EC.00136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Vidanes GM, Maguire SL, Guida A, Synnott JM, Andes DR, Butler G. Conserved and divergent roles of Bcr1 and CFEM proteins in Candida parapsilosis and Candida albicans. PLoS ONE. 2011;6:e28151. doi: 10.1371/journal.pone.0028151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedt T, Krishnamurthy S, Bockmuhl DP, Tebarth B, Stempel C, Russell CL, et al. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol Biol Cell. 2004;15:3167–3180. doi: 10.1091/10.1091/mbc.E03-11-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrington RA, Cooper TG. The DAL82 protein of Saccharomyces cerevisiae binds to the DAL upstream induction sequence (UIS) Nucleic Acids Res. 1993;21:3777–3784. doi: 10.1093/nar/21.16.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enger L, Joly S, Pujol C, Simonson P, Pfaller M, Soll DR. Cloning and characterization of a complex DNA fingerprinting probe for Candida parapsilosis. J Clin Microbiol. 2001;39:658–669. doi: 10.1128/JCM.39.2.658-669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon J, Kelly J, Kavanagh K. Galleria mellonella as a model for fungal pathogenicity testing. Methods Mol Biol. 2012;845:469–485. doi: 10.1007/978-1-61779-539-8_33. [DOI] [PubMed] [Google Scholar]
- Fares MA, Byrne KP, Wolfe KH. Rate asymmetry after genome duplication causes substantial long-branch attraction artifacts in the phylogeny of Saccharomyces species. Mol Biol Evol. 2006;23:245–253. doi: 10.1093/molbev/msj027. [DOI] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA, O'Gaora P, Byrne KP, Butler G. Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC Genomics. 2010;11:290. doi: 10.1186/1471-2164-11-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca EJ, Andrade CG, Furlaneto-Maia L, Serpa R, Oliveira MT, Quesada RM, Furlaneto MC. Ultrastructural architecture of colonies of different morphologies produced by phenotypic switching of a clinical strain of Candida tropicalis and biofilm formation by variant phenotypes. Micron. 2011;42:726–732. doi: 10.1016/j.micron.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Gacser A, Trofa D, Schafer W, Nosanchuk JD. Targeted gene deletion in Candida parapsilosis demonstrates the role of secreted lipase in virulence. J Clin Invest. 2007;117:3049–3058. doi: 10.1172/JCI32294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Pedrajas MD, Baeza-Montanez L, Gold SE. Regulation of Ustilago maydis dimorphism, sporulation, and pathogenic development by a transcription factor with a highly conserved APSES domain. Mol Plant Microbe Interact. 2010;23:211–222. doi: 10.1094/MPMI-23-2-0211. [DOI] [PubMed] [Google Scholar]
- Garcia-Rodas R, Casadevall A, Rodriguez-Tudela JL, Cuenca-Estrella M, Zaragoza O. Cryptococcus neoformans capsular enlargement and cellular gigantism during Galleria mellonella infection. PLoS ONE. 2011;6:e24485. doi: 10.1371/journal.pone.0024485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger J, Wessels D, Lockhart SR, Soll DR. Release of a potent polymorphonuclear leukocyte chemoattractant is regulated by white-opaque switching in Candida albicans. Infect Immun. 2004;72:667–677. doi: 10.1128/IAI.72.2.667-677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Fink GR. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guida A, Lindstadt C, Maguire SL, Ding C, Higgins DG, Corton NJ, et al. Using RNA-seq to determine the transcriptional landscape and the hypoxic response of the pathogenic yeast Candida parapsilosis. BMC Genomics. 2011;12:628. doi: 10.1186/1471-2164-12-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott MM, Lilly EA, Rodriguez TE, Fidel PL, Jr, Noverr MC. Candida albicans forms biofilms on the vaginal mucosa. Microbiology. 2010;156:3635–3644. doi: 10.1099/mic.0.039354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawser SP, Douglas LJ. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun. 1994;62:915–921. doi: 10.1128/iai.62.3.915-921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Schwarzmuller T, Kuchler K. Transcriptional loops meet chromatin: a dual-layer network controls white-opaque switching in Candida albicans. Mol Microbiol. 2009;74:1–15. doi: 10.1111/j.1365-2958.2009.06772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci USA. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis DO, Arnaud MB, Binkley J, Shah P, Skrzypek MS, Wymore F, et al. The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res. 2012;40:D667–D674. doi: 10.1093/nar/gkr945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, El Bissati K, Ben Mamoun C. Amino acids mediate colony and cell differentiation in the fungal pathogen Candida parapsilosis. Microbiology. 2006;152:2885–2894. doi: 10.1099/mic.0.29180-0. [DOI] [PubMed] [Google Scholar]
- Klar AJ, Srikantha T, Soll DR. A histone deacetylation inhibitor and mutant promote colony-type switching of the human pathogen Candida albicans. Genetics. 2001;158:919–924. doi: 10.1093/genetics/158.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Srikantha T, Tsai LK, Daniels K, Soll DR. Phenotypic switching in Candida glabrata involves phase-specific regulation of the metallothionein gene MT-II and the newly discovered hemolysin gene HLP. Infect Immun. 2000;68:884–895. doi: 10.1128/iai.68.2.884-895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffey SF, Butler G. Phenotype switching affects biofilm formation by Candida parapsilosis. Microbiology. 2005;151:1073–1081. doi: 10.1099/mic.0.27739-0. [DOI] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassak T, Schneider E, Bussmann M, Kurtz D, Manak JR, Srikantha T, et al. Target specificity of the Candida albicans Efg1 regulator. Mol Microbiol. 2011;82:602–618. doi: 10.1111/j.1365-2958.2011.07837.x. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics. 2008;9:578. doi: 10.1186/1471-2164-9-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Transcriptional control of dimorphism in Candida albicans. Curr Opin Microbiol. 2001;4:728–735. doi: 10.1016/s1369-5274(01)00275-2. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS ONE. 2008;3:e1473. doi: 10.1371/journal.pone.0001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD. White-opaque switching in Candida albicans. Curr Opin Microbiol. 2009;12:650–654. doi: 10.1016/j.mib.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Hernday AD, Fordyce PM, Noiman L, Sorrells TR, Hanson-Smith V, et al. Identification and characterization of a previously undescribed family of sequence-specific DNA-binding domains. Proc Natl Acad Sci USA. 2013;110:7660–7665. doi: 10.1073/pnas.1221734110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott TJ, Kuykendall RJ, Welbel SF, Pramanik A, Lasker BA. Genomic heterogeneity in the yeast Candida parapsilosis. Curr Genet. 1993;23:463–467. doi: 10.1007/BF00312635. [DOI] [PubMed] [Google Scholar]
- Maguire SL, Oheigeartaigh SS, Byrne KP, Schroder MS, O'Gaora P, Wolfe KH, Butler G. Comparative genome analysis and gene finding in Candida species using CGOB. Mol Biol Evol. 2013;30:1281–1291. doi: 10.1093/molbev/mst042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo AS, Bizerra FC, Freymuller E, Arthington-Skaggs BA, Colombo AL. Biofilm production and evaluation of antifungal susceptibility amongst clinical Candida spp. isolates, including strains of the Candida parapsilosis complex. Med Mycol. 2011;49:253–262. doi: 10.3109/13693786.2010.530032. [DOI] [PubMed] [Google Scholar]
- Miller KY, Wu J, Miller BL. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 1992;6:1770–1782. doi: 10.1101/gad.6.9.1770. [DOI] [PubMed] [Google Scholar]
- Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- Miller NS, Dick JD, Merz WG. Phenotypic switching in Candida lusitaniae on copper sulfate indicator agar: association with amphotericin B resistance and filamentation. J Clin Microbiol. 2006;44:1536–1539. doi: 10.1128/JCM.44.4.1536-1539.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morschhauser J. Regulation of white-opaque switching in Candida albicans. Med Microbiol Immunol (Berl) 2010;199:165–172. doi: 10.1007/s00430-010-0147-0. [DOI] [PubMed] [Google Scholar]
- Mulhern SM, Logue ME, Butler G. The Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot Cell. 2006;5:2001–2013. [Google Scholar]
- Mylonakis E. Galleria mellonella and the study of fungal pathogenesis: making the case for another genetically tractable model host. Mycopathologia. 2008;165:1–3. doi: 10.1007/s11046-007-9082-z. [DOI] [PubMed] [Google Scholar]
- Negroni P. Variacion bacio et tipo R de Mycotorula albicans. Rev Soc Argent Biol. 1935;11:449–453. [Google Scholar]
- Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, et al. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009;7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Heitman J. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell–cell adhesion. Mol Cell Biol. 2000;20:8364–8372. doi: 10.1128/mcb.20.22.8364-8372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porman AM, Alby K, Hirakawa MP, Bennett RJ. Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc Natl Acad Sci USA. 2011;108:21158–21163. doi: 10.1073/pnas.1112076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C, Daniels KJ, Lockhart SR, Srikantha T, Radke JB, Geiger J, Soll DR. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell. 2004;3:1015–1027. doi: 10.1128/EC.3.4.1015-1027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Peleg AY, Tampakakis E, Mylonakis E. Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot Cell. 2009;8:1750–1758. doi: 10.1128/EC.00163-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, VandeWalle K, Lopez-Ribot J, Wickes B. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002;214:95. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- Ramage G, Mowat E, Jones B, Williams C, Lopez-Ribot J. Our current understanding of fungal biofilms. Crit Rev Microbiol. 2009;35:340–355. doi: 10.3109/10408410903241436. [DOI] [PubMed] [Google Scholar]
- Ramsey H, Morrow B, Soll DR. An increase in switching frequency correlates with an increase in recombination of the ribosomal chromosomes of Candida albicans strain 3153A. Microbiology. 1994;140(Part 7):1525–1531. doi: 10.1099/13500872-140-7-1525. [DOI] [PubMed] [Google Scholar]
- Ricicova M, Kucharikova S, Tournu H, Hendrix J, Bujdakova H, Van Eldere J, et al. Candida albicans biofilm formation in a new in vivo rat model. Microbiology. 2010;156:909–919. doi: 10.1099/mic.0.033530-0. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai S, Holland L, McGee CF, Lynch DB, Butler G. Evolution of mating within the Candida parapsilosis species group. Eukaryot Cell. 2011;10:578–587. doi: 10.1128/EC.00276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MA, Tuite MF. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995;23:1481–1486. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke P, Kluter H, Muller-Steinhardt M, Hammers HJ, Borchert K, Bein G. Evaluation of a novel mononuclear cell isolation procedure for serological HLA typing. Clin Diagn Lab Immunol. 1998;5:808–813. doi: 10.1128/cdli.5.6.808-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S, Henriques M, Martins A, Oliveira R, Williams D, Azeredo J. Biofilms of non-Candida albicans Candida species: quantification, structure and matrix composition. Med Mycol. 2009;47:681–689. doi: 10.3109/13693780802549594. [DOI] [PubMed] [Google Scholar]
- Slutsky B, Buffo J, Soll DR. High-frequency switching of colony morphology in Candida albicans. Science. 1985;230:666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. ‘White-opaque transition’: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K, Urban C, Brunner H, Rupp S. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol Microbiol. 2003;47:89–102. doi: 10.1046/j.1365-2958.2003.03300.x. [DOI] [PubMed] [Google Scholar]
- Soll DR. Phenotypic switching. In: Calderone RA, editor. Candida. Washington, DC: ASM; 2002. pp. 123–142. and Candidiasis. [Google Scholar]
- Sonneborn A, Bockmuhl DP, Ernst JF. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect Immun. 1999a;67:5514–5517. doi: 10.1128/iai.67.10.5514-5517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn A, Tebarth B, Ernst JF. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect Immun. 1999b;67:4655–4660. doi: 10.1128/iai.67.9.4655-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T, Tsai LK, Daniels K, Soll DR. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J Bacteriol. 2000;182:1580–1591. doi: 10.1128/jb.182.6.1580-1591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T, Tsai L, Daniels K, Klar AJ, Soll DR. The histone deacetylase genes HDA1 and RPD3 play distinct roles in regulation of high-frequency phenotypic switching in Candida albicans. J Bacteriol. 2001;183:4614–4625. doi: 10.1128/JB.183.15.4614-4625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, et al. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell. 2006;5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stichternoth C, Ernst JF. Hypoxic adaptation by Efg1 regulates biofilm formation of Candida albicans. Appl Environ Microbiol. 2009;75:3663–3672. doi: 10.1128/AEM.00098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Sugino RP, Innan H. Natural selection on gene order in the genome reorganization process after whole-genome duplication of yeast. Mol Biol Evol. 2012;29:71–79. doi: 10.1093/molbev/msr118. [DOI] [PubMed] [Google Scholar]
- Tebarth B, Doedt T, Krishnamurthy S, Weide M, Monterola F, Dominguez A, Ernst JF. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J Mol Biol. 2003;329:949–962. doi: 10.1016/s0022-2836(03)00505-9. [DOI] [PubMed] [Google Scholar]
- Tong X, Zhang X, Plummer KM, Stowell KM, Sullivan PA, Farley PC. GcSTUA, an APSES transcription factor, is required for generation of appressorial turgor pressure and full pathogenicity of Glomerella cingulata. Mol Plant Microbe Interact. 2007;20:1102–1111. doi: 10.1094/MPMI-20-9-1102. [DOI] [PubMed] [Google Scholar]
- Trofa D, Gacser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev. 2008;21:606–625. doi: 10.1128/CMR.00013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 2010;6:e1001070. doi: 10.1371/journal.pgen.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Haas C, Kumamoto CA. Expression of the Candida albicans morphogenesis regulator gene CZF1 and its regulation by Efg1p and Czf1p. Eukaryot Cell. 2006;5:825–835. doi: 10.1128/EC.5.5.825-835.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Szaniszlo PJ. WdStuAp, an APSES transcription factor, is a regulator of yeast-hyphal transitions in Wangiella (Exophiala) dermatitidis. Eukaryot Cell. 2007;6:1595–1605. doi: 10.1128/EC.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MP, Gimeno CJ, Fink GR, Garrett S. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol Cell Biol. 1995;15:6854–6863. doi: 10.1128/mcb.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide MR, Ernst JF. Caco-2 monolayer as a model for transepithelial migration of the fungal pathogen Candida albicans. Mycoses. 1999;42(Suppl. 2):61–67. [PubMed] [Google Scholar]
- Westwater C, Balish E, Warner TF, Nicholas PJ, Paulling EE, Schofield DA. Susceptibility of gnotobiotic transgenic mice (Tgepsilon26) with combined deficiencies in natural killer cells and T cells to wild-type and hyphal signalling-defective mutants of Candida albicans. J Med Microbiol. 2007;56:1138–1144. doi: 10.1099/jmm.0.47110-0. [DOI] [PubMed] [Google Scholar]
- White SJ, Rosenbach A, Lephart P, Nguyen D, Benjamin A, Tzipori S, et al. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog. 2007;3:e184. doi: 10.1371/journal.ppat.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Hsiao TH, Suresh U, Chen HI, Wu X, Wolf SE, Chen Y. A novel significance score for gene selection and ranking. Bioinformatics. 2012 doi: 10.1093/bioinformatics/btr671. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Du H, Guan G, Tong Y, Kourkoumpetis TK, Zhang L, et al. N-acetylglucosamine induces white-to-opaque switching and mating in Candida tropicalis, providing new insights into adaptation and fungal sexual evolution. Eukaryot Cell. 2012;11:773–782. doi: 10.1128/EC.00047-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavrel M, Majer O, Kuchler K, Rupp S. Transcription factor Efg1 shows a haploinsufficiency phenotype in modulating the cell wall architecture and immunogenicity of Candida albicans. Eukaryot Cell. 2012;11:129–140. doi: 10.1128/EC.05206-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci USA. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control White-Opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


