Abstract
Chromosome 1q21 aberrations have not been yet been made part of routine clinical tests and their effect in multiple myeloma is still under investigation. The prognostic value of copy number variation and percentage of plasma cells involved have remained unclear. In the present study, we analyzed the prognostic value of 1q21 in a series of 290 cases of newly diagnosed multiple myeloma treated in a prospective, non-randomized clinical trial (BDH 2008/02). We found that incidence of 1q21 aberration increased at relapse, but its copy numbers and proportion of cells involved did not change. Gains of 1q21 had no impact on survival in patients receiving thalidomide-based treatment but conferred a significantly inferior prognosis in patients under bortezomib-based chemotherapy and was an independent adverse prognostic factor for progression free survival (HR 3.831; 95%CI: 2.125–6.907; P<0.001) and overall survival (HR 3.245; 95%CI: 1.555–6.773; P=0.002). Strikingly, our results showed that the copy number variation and clone size harboring 1q21 gains carried no additional prognostic value and patients with 1q21 gains did not benefit significantly from regimens incorporating bortezomib. Our results indicate that three copies of 1q21 and 20% of plasma cells with this abnormality were enough to confer bortezomib resistance. Therefore, chromosome 1q21 gains should be considered a high-risk feature in multiple myeloma receiving bortezomib therapy.
Introduction
Extra copies of chromosome 1q21 are among the most commonly reported genetic abnormalities in multiple myeloma (MM).1 However, the prognostic value of 1q21 gains in MM remains controversial. Several studies have shown that 1q21 gain is a significant and independent poor prognostic factor, although there are also series that have failed to confirm this.2–4 Currently, testing for a gain or amplification of 1q21 locus has not been included among the high-risk genetic abnormalities listed by the International Myeloma Working Group (IMWG).5 However, heterogeneity exists among patients presenting 1q21 gains, e.g. copy number variation and the size of clones carrying 1q21 gains.
To date, the relevant genes on 1q21 remain unclear. Putative targets of this amplification include CKS1B and PMSD4 genes, mediating cell cycle progression and resistance to bortezomib, respectively.6,7 A common feature shared by CKS1B and PMSD4 is that both of their expressions are highly sensitive to copy number variation of 1q21, and higher expression levels adversely affect clinical outcome, suggesting the existence of a gene-dosage effect that is of biological and prognostic significance. However, it remains unclear whether the copy number variations of 1q21 in positive cells carry any differential risk. So far, no specific standard cut off has been applied in detection of molecular cytogenetic aberrations by fluorescence in situ hybridization (FISH). Usually, the cut-off values recommended by the European Myeloma Network (EMN) were used, i.e. for deletions and numerical aberrations the cut-off level was set at 20%.8 In one report, del(17p) is prognostic only if present in at least 60% of the plasma cells.9 However, in studies from Neben et al., del(17p) remains the most important genetic prognostic factor in MM even if the proportion of plasma cells involved is just over 10%.10,11 It is unclear whether the prognostic impact of 1q21 gains would be different if we split the patients according to the percentage of plasma cells involved.
It is also unknown whether the discrepancies in terms of prognostic value of 1q21 gains is related to the difference in proportion of patients with more than three copies of 1q21 and the percentage of involved plasma cells in each study. To address this issue, this study has analyzed the prognostic impact of copy number variation and percentage of clones carrying 1q21 gains on survival in a cohort of 290 patients with newly diagnosed MM for whom information concerning copy number of 1q21 was available. One hundred and seven patients with relapsed MM were included in order to compare the incidence of 1q21 gains between different stages of plasma cell dyscrasias.
Methods
Study design
This study is a prospective, non-randomized clinical trial. The trial was carried out in accordance with the 1996 Declaration of Helsinki and was approved by the local ethics committees of institutions. According to their request, patients were assigned to either the thalidomide-based (arm A) or bortezomib-based (arm B) treatment. Arm A consisted of 4 cycles of induction treatment with thalidomide (TAD) 200 mg/day; intravenous (i.v.) adriamycin 9 mg/m2 on Days 1–4; and oral or i.v. dexamethasone 20 mg/d on Days 1–4 and 9–12; or TCD (i.e. thalidomide 200 mg/day; i.v. cyclophosphamide 300 mg/m2 on Days 1 and 8; and oral or i.v. dexamethasone 20 mg/d on Days 1–4 and 9–12); Arm B consisted of 4 cycles of induction treatment with BCD (i.e. i.v. bortezomib 1.3 mg/m2 on Days 1, 4, 8, and 11; i.v. cyclophosphamide 300 mg/m2 on Days 1 and 8; and oral or i.v. dexamethasone 20 mg/d on Days 1, 2, 4, 5, 8, 9, 11 and 12); or PAD (i.v. bortezomib 1.3 mg/m2 on Days 1, 4, 8 and 11; i.v. adriamycin 9 mg/m2 on Days 1–4; and oral or i.v. dexamethasone, 20 mg/d on Days 1, 2, 4, 5, 8, 9, 11 and 12). After at least four cycles of treatment with partial remission or better, patients underwent consolidation therapy. This was either autologous stem cell transplant (ASCT) or chemotherapy with the patient’s original regimen according to patient request. Subsequently, patients were treated with thalidomide (100–150 mg/day) for one year to maintain response. When necessary, some of them also received supportive treatment with zoledronic acid every 1–2 months and erythropoietin or granulocyte colony-stimulating factor. All patients underwent prophylactic acyclovir treatment.
Cytogenetic analysis
All the MM samples were purified using Miltenyi technology (anti-CD138 coated magnetic beads, Paris, France) before FISH as previously reported.12 Plasma cells were then analyzed using DNA probes specific for the following chromosomal aberrations: del(13q14), del(17p), t(11;14), t(4;14), and t(14;16). Gains of 1q21 were assessed using a bacterial artificial chromosome (BAC) probe at 1q21 (RP11-307C12).3 A total of 200 interphase nuclei were analyzed and the cut-off level was set at 20%.8
The gains of 1q21 were divided into three categories based on copy numbers as follows: 1) three copies of 1q21 (the cells with at least four copies were seen in less than 20% of clonal plasma cells); 2) four copies of 1q21 (the cells with at least five copies were seen in less than 20% of clonal plasma cells); and 3) at least five copies of 1q21 (the cells with at least five copies were seen in at least 20% of clonal plasma cells).3
Statistical analysis
The primary end point was correlated with survival from the time of diagnosis. PFS was calculated from the initiation of therapy to the date of death, progression, or the last follow up. OS was measured from the initiation of treatment to the date of death or last follow up according to the international uniform response criteria.13 Two-sided Fisher’s exact tests were used to assess associations between categorical variables, with a confidence coefficient (confidence interval, CI) of 95%. The survival curves were plotted using the Kaplan-Meier method, with differences assessed by the log rank test. P≤0.05 was considered significant.
Results
Patients’ characteristics
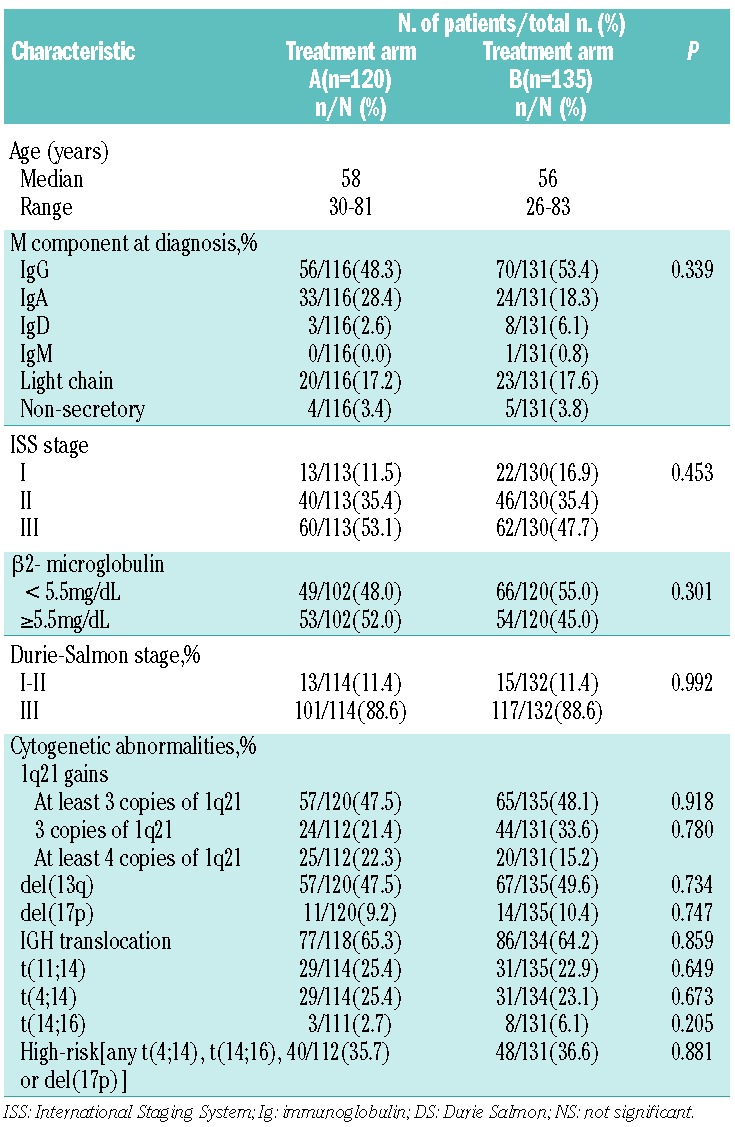
A total of 290 patients with newly diagnosed symptomatic MM were enrolled in the present study between January 2008 and December 2012, with a median follow-up time of 36 months from diagnosis. As a control, 107 MM cases in first relapse were also analyzed to compare the frequency of 1q21 gain at different stages of plasma cell dyscrasias. Paired bone marrow samples were collected at diagnosis and on relapse from 21 patients. One hundred and twenty patients were included in arm A and 3 of these underwent ASCT. One hundred and thirty-five patients were included in arm B and 22 patients received ASCT. There were no significant differences in clinical and cytogenetic characteristics between the groups (Table 1). Thirty-five patients were lost to follow up.
Table 1.
Patients’ characteristics in arm A and arm B.
Prevalence of 1q21 in patients with plasma cell dyscrasias
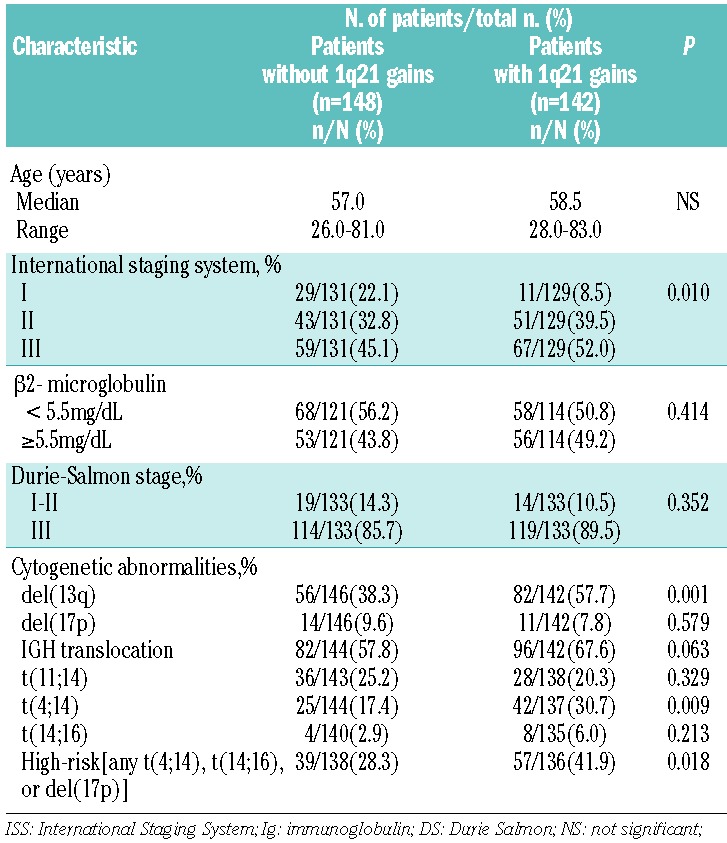
Main characteristics of patients with myeloma displaying 1q21 gains are shown in Table 2. These data suggested that cases with 1q21 gains typically had a high incidence of del(13) and t(4;14). No correlation was seen between 1q21 gains and 17p deletion. According to the Intergroup Francophone du Myelome (IFM) stratification of the myeloma model, high-risk MM was defined as the presence of any one or more of the following criteria:5,12 del(17p13) or t(4;14) or t(14;16). A high incidence of 1q21 gains was seen in patients with high-risk cytogenetic abnormalities, mainly due to the tight association between 1q21 gains and t(4;14).
Table 2.
Clinical and biological correlations of 1q21 gains in newly diagnosed multiple myeloma.
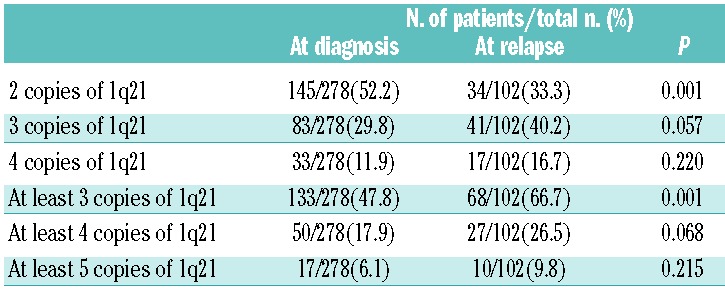
The present study investigated the frequency of 1q21 gains with different numbers of copies in newly diagnosed and relapsed MM patients. The incidence of patients carrying at least three copies of 1q21 was significantly higher in relapsed MM than in newly diagnosed MM: 73 of 107 (68.2%) versus 142 of 290 (48.9%); P=0.001. Data concerning copy number variation among the whole cohort were available for 278 newly diagnosed MM and 102 relapsed MM. No statistical difference in the frequency of different copies of 1q21 was found between relapsed and initially diagnosed cases (Table 3). The newly diagnosed MM with more than three copies of 1q21 showed a similar (P=0.216) percentage of plasma cells involved (median 76.8%, range 20.0–100.0%, n=290) when compared with relapsed MM (median 71.0%, range 21.0–100.0%, n=107).
Table 3.
Summary of interphase FISH results for 1q21 in patients with newly diagnosed and relapsed MM according to the copy number of 1q21.
Twenty-one patients had 1q21 FISH results available both at diagnosis and relapse. Among them, 4 patients evolve from two copies of 1q21 to more than three copies, 3 patients from three copies to at least four copies, and 2 patients from four copies to at least five copies. The copy numbers did not change in 7 patients with 1q21 gains, however 2 patients among them had an increased percentage of plasma cells involved. Five patients without 1q21 gains at diagnosis still did not display 1q21 gains at relapse.
Prognostic value of 1q21 gains in MM patients treated with bortezomib-based therapy
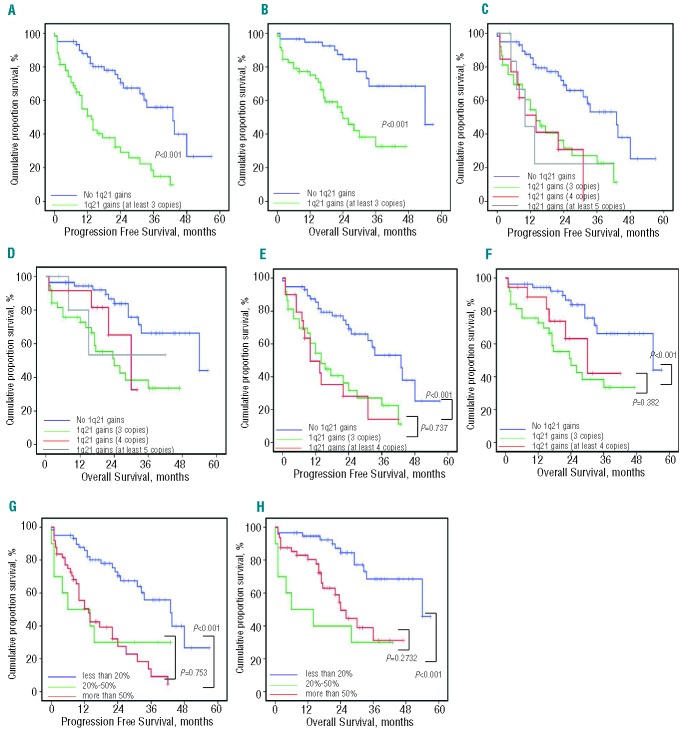
We then studied the impact of 1q21 copy number on survival in newly diagnosed MM treated with bortezomib-based therapy, and found that 1q21 gains showed a profoundly negative impact on survival. Patients with 1q21 gains had a significantly shortened PFS (13.5 vs. 43.0 months; P<0.001) and OS (24.0 vs. 54.0 months; P<0.001) than patients with two copies of 1q21. A strong correlation between 1q21 gains and t(4;14) was found in this study, and the latter was correlated with a poor prognosis, as shown in other studies.10,14 Therefore, we then analyzed the survival of patients with 1q21 gains in the absence of t(4;14), and found that the median PFS and OS among these cases were 27.5 versus 46.6 months (P=0.001) and 23.5 versus 54.0 months (P=0.002), respectively. So the poor prognostic feature of 1q21 gains was independent of t(4;14) translocation. A multivariate analysis including all the parameters shown to be associated with PFS and OS in the univariate analysis was performed. Chromosomal aberration del(17p) (HR 4.046; 95%CI: 2.022–8.096; P<0.001) and 1q21 gains (HR 3.831; 95%CI: 2.125–6.907; P<0.001) were statistically independent predictors of shortened PFS. A similar analysis for prediction of OS also identified the parameters del(17p) (HR 3.959; 95%CI: 1.890–8.292; P<0.001) and 1q21 gains (HR 3.245; 95%CI: 1.555–6.773; P=0.002).
Further investigation of the impact of copy number variation on survival indicated that the median PFS of patients who had three, four, or at least five copies of 1q21 were 14.0 months (95%CI: 8.07–19.93), 14.0 months (95%CI: 5.47–22.53), and 10.0 months (95%CI: 4.12–15.88), respectively. The median OS of patients who had three and four copies were 24.0 months (95%CI: 11.85–36.14) and 30.0 months (95%CI: 18.14–41.85), respectively, while the OS of patients with at least five copies was not obtained due to the small sample size; no statistically significant differences were found. When the patients with four and five copies were analyzed as a whole, i.e. as one group of patients with at least four copies, patients with at least four copies of 1q21 also exhibited a comparable PFS (14.0 vs. 10.0 months; P=0.737) and OS (24.0 vs. 30.0 months; P=0.382), compared with patients harboring three copies of 1q21.
Patients with 1q21 gains were divided into two groups according to the percentage of plasma cells involved (20–50% and >50%) that were seen in 28 of 290 (9.7%) and 114 of 290 (43.8%) patients with newly diagnosed MM, respectively. The median PFS was 6.0 months (95%CI: 1.00–21.5) and 13.5 months (95%CI: 8.87–18.14), respectively, and median OS was 6.0 months (95%CI: 1.00–21.49) and 24.0 months (95%CI: 18.94–29.05), respectively. There was no statistical difference in PFS and OS between the two groups (P=0.753 and 0.273, respectively) (Figure 1).
Figure 1.
PFS and OS among newly diagnosed MM patients treated with bortezomib-based chemotherapy according to copy numbers of 1q21. (A–B) Patients with 1q21 gains had a short PFS and OS. (C–D) Patients with two copies of 1q21 had a favorable survival. No difference was seen between patients with 3, 4, or at least 5 copies of 1q21. (E–F) No difference was seen between patients with 3 and at least 4 copies of 1q21. (G–H) Patients presenting 1q21 gains was artificially divided into two groups according to the percentage of plasma cells involved, but no difference was seen between both groups.
Chromosomal 1q21 gains showed no predictive value for survival of MM treated with thalidomide-based chemotherapy
Median PFS in patients with two copies of 1q21 and at least three copies of 1q21 were 22.4 and 20.0 months, respectively (P=0.625), and OS were 30.0 and 22.0 months, respectively (P=0.355). Variation in copy number did not show any influence on disease outcome. The median PFS of patients harboring three, four, and at least five copies of 1q21 were 15.0, 24.0, and 20.0 months, respectively, while OS were 17.0, 25.0, and 22.0 months, respectively. No statistically significant differences were found. We then analyzed the impact of percentage of clones carrying 1q21 gains and found that the median PFS of patients with 20–50% and over 50% cells positive for 1q21 gains were 10.0 and 21.0 months (P=0.053), respectively, and corresponding OS were 14.0 and 22.0 months (P=0.115), respectively.
Bortezomib did not significantly improve the survival of patients with 1q21 gains
Survival of patients without 1q21 gains was remarkably extended with chemotherapy incorporating the proteasome inhibitor bortezomib. The median PFS and OS of patients treated with bortezomib-based chemotherapy were 43.0 versus 22.4 months (P=0.004) and 54.0 versus 30.0 months (P=0.001), respectively. But no statistically significant differences were found in patients harboring 1q21 gains between bortezomib-based and thalidomide-based chemotherapy groups, and the median PFS and OS in patients receiving bortezomib-based regimens were 13.5 versus 20.0 months (P=0.176) and 24.0 versus 21.0 months (P=0.773), respectively (Figure 2). Therefore, bortezomib was unable to overcome the negative impact of 1q21 gains on survival, and to significantly improve the survival of these patients.
Figure 2.
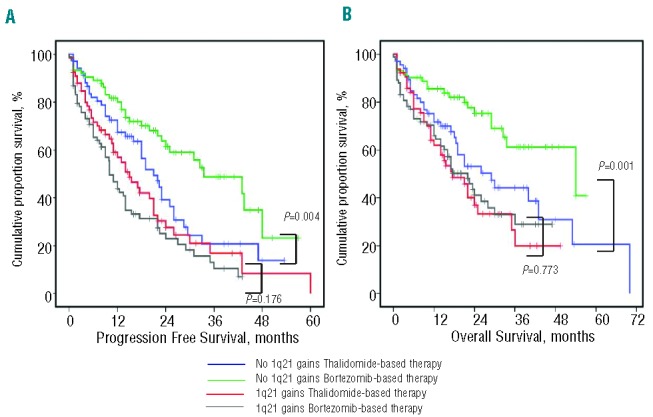
Survival of newly diagnosed MM patients with or without 1q21 gains after receiving different therapies. (A–B) PFS and OS were remarkably extended with chemotherapy incorporating the proteasome inhibitor bortezomib in patients without 1q21 gains, but not in patients carrying more than 2 copies of 1q21.
Discussion
Gains of chromosome 1q has been identified by global single-nucleotide polymorphism-based mapping.15,16 A minimally amplified region was identified between 1q21.1-q23.3 containing 679 genes, among which IL6R, KCNN3, and CKS1B had a greater than 2.5-fold increase in expression.17 There are some reports that 1q21 gains may have clinical significance as a poor-risk feature; however, 1q21 has not yet made its way into routine FISH markers to predict prognosis.18 This study has focused on the chromosome 1q21 gains in MM patients and analyzed the poor outcomes of newly diagnosed MM patients with 1q21 gains by FISH despite novel agent-based therapy.
The frequencies of chromosome 1q21 gains were consistent with those in previous studies.2,3,7 Gains of 1q21 have been investigated in many studies. However, to our knowledge, the prevalence of 1q21 gains with more than three copies and the impact of copy number variation on the survival of MM have been only mentioned in two studies, with apparently conflicting results.3,11 According to the study from Arkansas, 1q21 gains with at least four copies were detected in 88 of 479 (18%) newly diagnosed MM and 20 of 45 (44%) relapsed/refractory MM.3 However, another study from Germany showed that 1q21 gains with at least four copies were only detected in 16 of 344 (4.7%) newly diagnosed MM patients.11 Our results demonstrated that the frequency of newly diagnosed MM harboring at least four copies of 1q21 are comparable with those from Arkanas, but the frequency in relapsed MM appeared to be lower in our study. We further observed that the incidence of 1q21 gains always increases at relapse, but that there was no significant change in copy numbers or the proportion of cells with extra copies of 1q21.
The prognostic value of 1q21 gains in MM remains controversial, with some studies suggesting 1q21 gains to be a major prognostic factor for MM, while others show the opposite.18 A long-term analysis of the IFM99 trials for myeloma established that cytogenetic abnormalities 1q21 gains play a major role in defining long-term survival.19 However, the Mayo Clinic showed that this parameter was not retained in multivariate analyses and that its prognostic value disappeared when combined with other classical biological and genetic prognostic factors.2 The present study demonstrated that 1q21 gains exerted a profound effect on the outcome of patients treated with bortezomib-based therapy. In multivariate analysis, its prognostic value was almost as powerful as del(17p). However, 1q21 gains had no effect on survival treated with thalidomide-based chemotherapy. Importantly, we showed that there was no notable improvement in survival of patients with 1q21 gains when treated with chemotherapy incorporating bortezomib, implying that 1q21 gains might be linked to bortezomib resistance. Gene expression profile (GEP) found that cases with 1q21 gains showed significantly altered expression of genes involved in unfolded protein response (UPR), including upregulation of chaperone gene CLN3, UBAP2L, and UBE2Q1, proteasome degradation gene PSMD4, and CASP4 gene involved in UPR-induced apoptosis.20 Integrating GEP and DNA copy number variation data, several independent studies revealed that numerous 1q21 genes are copy number sensitive in MM.21,22 Because UPR-induced apoptosis plays an important role in the sensitivity of malignant cells to bortezomib,23 these studies strongly suggest that bortezomib resistance gene overexpression is associated with chromosome 1q21 gains. Recently, Shaughnessy et al. reported that hyperexpression of proteasome gene PSMD4 residing on chromosome 1q21 was a novel high-risk feature in myeloma treated with Total Therapy 3.6 Most laboratories using 1q probes had developed them in-house. Where laboratories had used more than one probe, the results confirmed that the majority of abnormalities are gain of the whole arm.8 Using sequential gene GEP samples, Zhan et al. defined a major functional gene group related to bortezomib resistance, and several genes, such as NEK2, FAM72A, Nuf2, CDC20, were located at chromosome 1q.24 All these findings support the important role of chromosome 1q in drug resistance to bortezomib.
The impact of copy number variation of 1q21 on the survival of MM is also rarely reported, although conflicting data are available. The impact of copy number variation on survival is still under investigation. A German group found that the clinical course of patients with more than three copies of 1q21 was characterized by a remarkably short PFS and OS, whereas exactly three copies of 1q21 were associated with only a marginal effect on outcome.11 However, the Arkansas group reported that patients with more than three copies of 1q21 at diagnosis had similar 5-year event-free survival (EFS) and OS compared with those with three copies of 1q21.3 Our data are in agreement with the conclusion of the Arkansas group that although 1q21 gains were linked to significantly inferior clinical outcomes of patients receiving bortezomib-based therapy, the copy number variation showed no additional prognostic value.
The determination of cut-off levels was the most controversial issue in detection of cytogenetic abnormalities by FISH.8 Because the prognostic value of the percentage of clones carrying cytogenetic aberration has rarely been investigated, there is still not enough evidence to determine the level that is of highest prognostic significance. In one study, del(17p) was not prognostic in all patients, and patients presenting del(17p) in less than 60% of their plasma cells did not have a specific poor outcome.9 Another study from IFM12 showed that the most powerful cut offs were 74% for del(13) and 60% for del(17p). Avet-Loiseau et al. reported that the IFM group used a 30% cut off for 1q21 gains according to their own previous results.19 As a clearly secondary cytogenetic change, 1q21 gains may only present in subpopulations. In this study, we artificially divided the patients with 1q21 gains into two groups according to the percentage of plasma cells involved and found that the percentage of cells involved in the majority of patients with 1q21 gains was over 50%. No statistical difference was found in PFS and OS between the two groups, demonstrating that the copy number variation and percentage of plasma cells involved are not of prognostic value, which may imply that three copies of 1q21 and 20% of involved plasma cells are enough to confer bortezomib resistance.
Taken together, our data demonstrated that copy numbers of 1q21 increased with progression of myeloma and predicted a poor prognosis in MM patients treated with bortezomib-based therapy. Chromosome 1q21 gains should be considered a high-risk feature in MM, and 1q21 analysis should be added to the diagnostic panel of FISH probes used in routine assessment of prognosis in patients with MM, especially those receiving bortezomib-based regimens. Our results also demonstrated that patients with three copies of 1q21 had comparable survival with patients with more than three copies, and 20% of involved plasma cells were the correct cut off to determine prognostic significance.
Supplementary Material
Footnotes
The online version of this article has a Supplementary Appendix.
Funding
The present study was supported by the Ministry of Science and Technology of China (2010DFB30270), the National Natural Science Fund (81172255, 81101794), Tianjin Science and Technology Commision (09ZCZDSF03800), Clinical Research Program from Health Ministry of China (Key project 2010 to 2012), the Scientific Research Program for Public Interests from the Health Ministry of China (Project No.201202017), Union Youth Research Fund of Peking Union Medical College (2012–2013), and the Special Key Anticancer Research and Development Program of Tianjin Municipality (12ZCDZSY17600).
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12(5):335–48 [DOI] [PubMed] [Google Scholar]
- 2.Fonseca R, Van Wier SA, Chng WJ, Ketterling R, Lacy MQ, Dispenzieri A, et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20(11):2034–40 [DOI] [PubMed] [Google Scholar]
- 3.Hanamura I, Stewart JP, Huang Y, Zhan F, Santra M, Sawyer JR, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108(5):1724–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaughnessy JD, Jr, Haessler J, van Rhee F, Anaissie E, Pineda-Roman M, Cottler-Fox M, et al. Testing standard and genetic parameters in 220 patients with multiple myeloma with complete data sets: superiority of molecular genetics. Br J Haematol. 2007;137(6):530–6 [DOI] [PubMed] [Google Scholar]
- 5.Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23 (12):2210–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaughnessy JD, Jr, Qu P, Usmani S, Heuck CJ, Zhang Q, Zhou Y, et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3. Blood. 2011;118(13):3512–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhan F, Colla S, Wu X, Chen B, Stewart JP, Kuehl WM, et al. CKS1B, overexpressed in aggressive disease, regulates multiple myeloma growth and survival through SKP2- and p27Kip1-dependent and -independent mechanisms. Blood. 2007;109(11): 4995–5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross FM, Avet-Loiseau H, Ameye G, Gutierrez NC, Liebisch P, O’Connor S, et al. Report from the European Myeloma Network on interphase FISH in multiple myeloma and related disorders. Haematologica. 2012;97(8):1272–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avet-Loiseau H, Leleu X, Roussel M, Moreau P, Guerin-Charbonnel C, Caillot D, et al. Bortezomib plus dexamethasone induction improves outcome of patients with t(4;14) myeloma but not outcome of patients with del(17p). J Clin Oncol. 2010;28(30):4630–4 [DOI] [PubMed] [Google Scholar]
- 10.Neben K, Jauch A, Bertsch U, Heiss C, Hielscher T, Seckinger A, et al. Combining information regarding chromosomal aberrations t(4;14) and del(17p13) with the International Staging System classification allows stratification of myeloma patients undergoing autologous stem cell transplantation. Haematologica. 2010;95(7):1150–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neben K, Lokhorst HM, Jauch A, Bertsch U, Hielscher T, van der Holt B, et al. Administration of bortezomib before and after autologous stem cell transplantation improves outcome in multiple myeloma patients with deletion 17p. Blood. 2012;119(4):940–8 [DOI] [PubMed] [Google Scholar]
- 12.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007;109(8):3489–95 [DOI] [PubMed] [Google Scholar]
- 13.Anderson KC, Kyle RA, Rajkumar SV, Stewart AK, Weber D, Richardson P. Clinically relevant end points and new drug approvals for myeloma. Leukemia. 2008;22(2):231–9 [DOI] [PubMed] [Google Scholar]
- 14.Jaksic W, Trudel S, Chang H, Trieu Y, Qi X, Mikhael J, et al. Clinical outcomes in t(4;14) multiple myeloma: a chemotherapy-sensitive disease characterized by rapid relapse and alkylating agent resistance. J Clin Oncol. 2005;23(28):7069–73 [DOI] [PubMed] [Google Scholar]
- 15.Walker BA, Leone PE, Jenner MW, Li C, Gonzalez D, Johnson DC, et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood. 2006;108(5):1733–43 [DOI] [PubMed] [Google Scholar]
- 16.Avet-Loiseau H, Li C, Magrangeas F, Gouraud W, Charbonnel C, Harousseau JL, et al. Prognostic significance of copy-number alterations in multiple myeloma. J Clin Oncol. 2009;27(27):4585–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116(15):e56–65 [DOI] [PubMed] [Google Scholar]
- 18.Munshi NC, Anderson KC, Bergsagel PL, Shaughnessy J, Palumbo A, Durie B, et al. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood. 2011;117(18): 4696–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avet-Loiseau H, Attal M, Campion L, Caillot D, Hulin C, Marit G, et al. Long-term analysis of the IFM 99 trials for myeloma: cytogenetic abnormalities [t(4;14), del(17p), 1q gains] play a major role in defining long-term survival. J Clin Oncol. 2012;30(16):1949–52 [DOI] [PubMed] [Google Scholar]
- 20.Fabris S, Ronchetti D, Agnelli L, Baldini L, Morabito F, Bicciato S, et al. Transcriptional features of multiple myeloma patients with chromosome 1q gain. Leukemia. 2007;21(5):1113–6 [DOI] [PubMed] [Google Scholar]
- 21.Agnelli L, Mosca L, Fabris S, Lionetti M, Andronache A, Kwee I, et al. A SNP microarray and FISH-based procedure to detect allelic imbalances in multiple myeloma: an integrated genomics approach reveals a wide gene dosage effect. Genes Chromosomes Cancer. 2009;48(7):603–14 [DOI] [PubMed] [Google Scholar]
- 22.Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, et al. High-resolution genomic profiles define distinct clinicopathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9(4):313–25 [DOI] [PubMed] [Google Scholar]
- 23.Meister S, Schubert U, Neubert K, Herrmann K, Burger R, Gramatzki M, et al. Extensive immunoglobulin production sensitizes myeloma cells for proteasome inhibition. Cancer Res. 2007;67(4):1783–92 [DOI] [PubMed] [Google Scholar]
- 24.Zhou W, Yang Y, Xia J, Wang H, Salama ME, Xiong W, et al. NEK2 Induces Drug Resistance Mainly through Activation of Efflux Drug Pumps and Is Associated with Poor Prognosis in Myeloma and Other Cancers. Cancer Cell. 2013;23(1):48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.