Abstract
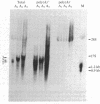
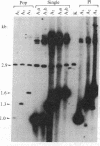
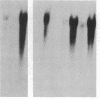
Constructs were made in which approximately equal to 500 base pairs of the 5' flanking region of the ras gene of Dictyostelium discoideum and variable amounts of the coding region were linked to a ras cDNA in a transformation vector. These constructs then were used to transform Dictyostelium cells and their regulation was examined. In Dictyostelium transformants, transcripts from the ras gene constructs were found at high levels in cells in fast-shaking cultures containing cAMP, whereas transcripts were either not detectable or present at very low levels in cultures lacking exogenous cAMP. In slow-shaking culture, a significantly lower level of ras RNA was detected. When normal developing aggregates were dissociated, RNA from the ras constructs decreased rapidly but then reaccumulated in the presence of cAMP. These results show that the sequences necessary for the response to external cAMP are present within an approximately equal to 650-base-pair region upstream from the ATG start codon and/or within portions of the protein-coding region. Moreover, the proper regulation of ras gene expression in high-copy-number transformants suggests that trans-acting factors which may control transcription are not limiting. Vector constructs were also examined in which the cDNA was present in the opposite orientation compared to the gene fragment (antisense orientation). When these were transfected into cells, no transformants were obtained, suggesting that expression of the ras gene is essential for vegetative growth.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BROOKES P., LAWLEY P. D. EVIDENCE FOR THE BINDING OF POLYNUCLEAR AROMATIC HYDROCARBONS TO THE NUCLEIC ACIDS OF MOUSE SKIN: RELATION BETWEEN CARCINOGENIC POWER OF HYDROCARBONS AND THEIR BINDING TO DEOXYRIBONUCLEIC ACID. Nature. 1964 May 23;202:781–784. doi: 10.1038/202781a0. [DOI] [PubMed] [Google Scholar]
- Barklis E., Lodish H. F. Regulation of dictyostelium discoideum mRNAs specific for prespore or prestalk cells. Cell. 1983 Apr;32(4):1139–1148. doi: 10.1016/0092-8674(83)90297-0. [DOI] [PubMed] [Google Scholar]
- Capon D. J., Chen E. Y., Levinson A. D., Seeburg P. H., Goeddel D. V. Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature. 1983 Mar 3;302(5903):33–37. doi: 10.1038/302033a0. [DOI] [PubMed] [Google Scholar]
- Charnay P., Treisman R., Mellon P., Chao M., Axel R., Maniatis T. Differences in human alpha- and beta-globin gene expression in mouse erythroleukemia cells: the role of intragenic sequences. Cell. 1984 Aug;38(1):251–263. doi: 10.1016/0092-8674(84)90547-6. [DOI] [PubMed] [Google Scholar]
- Coleman J., Green P. J., Inouye M. The use of RNAs complementary to specific mRNAs to regulate the expression of individual bacterial genes. Cell. 1984 Jun;37(2):429–436. doi: 10.1016/0092-8674(84)90373-8. [DOI] [PubMed] [Google Scholar]
- Cooper G. M. Cellular transforming genes. Science. 1982 Aug 27;217(4562):801–806. doi: 10.1126/science.6285471. [DOI] [PubMed] [Google Scholar]
- Dhar R., Ellis R. W., Shih T. Y., Oroszlan S., Shapiro B., Maizel J., Lowy D., Scolnick E. Nucleotide sequence of the p21 transforming protein of Harvey murine sarcoma virus. Science. 1982 Sep 3;217(4563):934–936. doi: 10.1126/science.6287572. [DOI] [PubMed] [Google Scholar]
- Dhar R., Nieto A., Koller R., DeFeo-Jones D., Scolnick E. M. Nucleotide sequence of two rasH related-genes isolated from the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1984 Apr 25;12(8):3611–3618. doi: 10.1093/nar/12.8.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P. Magic enhancers? DNA. 1984;3(1):1–5. doi: 10.1089/dna.1.1984.3.1. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Izant J. G., Weintraub H. Inhibition of thymidine kinase gene expression by anti-sense RNA: a molecular approach to genetic analysis. Cell. 1984 Apr;36(4):1007–1015. doi: 10.1016/0092-8674(84)90050-3. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Powers S., McGill C., Fasano O., Strathern J., Broach J., Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984 Jun;37(2):437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Manne V., Bekesi E., Kung H. F. Ha-ras proteins exhibit GTPase activity: point mutations that activate Ha-ras gene products result in decreased GTPase activity. Proc Natl Acad Sci U S A. 1985 Jan;82(2):376–380. doi: 10.1073/pnas.82.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J. P., Capon D. J., Goeddel D. V., Levinson A. D. Comparative biochemical properties of normal and activated human ras p21 protein. Nature. 1984 Aug 23;310(5979):644–649. doi: 10.1038/310644a0. [DOI] [PubMed] [Google Scholar]
- McGrath J. P., Capon D. J., Smith D. H., Chen E. Y., Seeburg P. H., Goeddel D. V., Levinson A. D. Structure and organization of the human Ki-ras proto-oncogene and a related processed pseudogene. Nature. 1983 Aug 11;304(5926):501–506. doi: 10.1038/304501a0. [DOI] [PubMed] [Google Scholar]
- Mehdy M. C., Ratner D., Firtel R. A. Induction and modulation of cell-type-specific gene expression in Dictyostelium. Cell. 1983 Mar;32(3):763–771. doi: 10.1016/0092-8674(83)90062-4. [DOI] [PubMed] [Google Scholar]
- Melton D. A. Injected anti-sense RNAs specifically block messenger RNA translation in vivo. Proc Natl Acad Sci U S A. 1985 Jan;82(1):144–148. doi: 10.1073/pnas.82.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellen W., Silan C., Firtel R. A. DNA-mediated transformation in Dictyostelium discoideum: regulated expression of an actin gene fusion. Mol Cell Biol. 1984 Dec;4(12):2890–2898. doi: 10.1128/mcb.4.12.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman-Silberberg F. S., Schejter E., Hoffmann F. M., Shilo B. Z. The Drosophila ras oncogenes: structure and nucleotide sequence. Cell. 1984 Jul;37(3):1027–1033. doi: 10.1016/0092-8674(84)90437-9. [DOI] [PubMed] [Google Scholar]
- Papageorge A., Lowy D., Scolnick E. M. Comparative biochemical properties of p21 ras molecules coded for by viral and cellular ras genes. J Virol. 1982 Nov;44(2):509–519. doi: 10.1128/jvi.44.2.509-519.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T., Amiel T., Hinze E., Auersperg N., Neave N., Sobolewski A., Weeks G. Regulation of a ras-related protein during development of Dictyostelium discoideum. Mol Cell Biol. 1985 Jan;5(1):33–39. doi: 10.1128/mcb.5.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D. H. Fifty years of benzo(a)pyrene. Nature. 1983 Jun 9;303(5917):468–472. doi: 10.1038/303468a0. [DOI] [PubMed] [Google Scholar]
- Powers S., Kataoka T., Fasano O., Goldfarb M., Strathern J., Broach J., Wigler M. Genes in S. cerevisiae encoding proteins with domains homologous to the mammalian ras proteins. Cell. 1984 Mar;36(3):607–612. doi: 10.1016/0092-8674(84)90340-4. [DOI] [PubMed] [Google Scholar]
- Reymond C. D., Gomer R. H., Mehdy M. C., Firtel R. A. Developmental regulation of a Dictyostelium gene encoding a protein homologous to mammalian ras protein. Cell. 1984 Nov;39(1):141–148. doi: 10.1016/0092-8674(84)90199-5. [DOI] [PubMed] [Google Scholar]
- Taparowsky E., Shimizu K., Goldfarb M., Wigler M. Structure and activation of the human N-ras gene. Cell. 1983 Sep;34(2):581–586. doi: 10.1016/0092-8674(83)90390-2. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Chaleff D. T., DeFeo-Jones D., Scolnick E. M. Requirement of either of a pair of ras-related genes of Saccharomyces cerevisiae for spore viability. Nature. 1984 Jun 7;309(5968):523–527. doi: 10.1038/309523a0. [DOI] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Tsuchida N., Ryder T., Ohtsubo E. Nucleotide sequence of the oncogene encoding the p21 transforming protein of Kirsten murine sarcoma virus. Science. 1982 Sep 3;217(4563):937–939. doi: 10.1126/science.6287573. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]